ABSTRACT
The Endoplasmic Reticulum (ER)-Golgi apparatus of plants is the site of synthesis of non-cellulosic polysaccharides that then traffic to the cell wall. A two-step protocol of flotation centrifugation followed by free-flow electrophoresis (FFE) resolved ER and Golgi proteins into three profiles: an ER-rich fraction, two Golgi-rich fractions, and an intermediate fraction enriched in cellulose synthases. Nearly three dozen Rab-like proteins of eight different subgroups were distributed differentially in ER- vs. Golgi-rich fractions, whereas seven 14-3-3 proteins co-fractionated with cellulose synthases in the intermediate fraction. FFE offers a powerful means to classify resident and transient proteins in cell-free assays of cellular location.
KEYWORDS: Maize (Zea mays), Endoplasmic reticulum (ER), Golgi, Small GTPase Rab proteins, 14-3-3 proteins, Calcium-dependent proteins, Cellulose synthase (CesA)
The Endoplasmic Reticulum (ER)-Golgi apparatus of all eukaryotic cells is the site of synthesis and packaging of glycoproteins and proteoglycans to be trafficked to the cell surface. In plants, the Golgi is also the site of synthesis of non-cellulosic polysaccharides of the cell wall. We combined flotation centrifugation with subsequent free-flow electrophoresis (FFE) to provide highly enriched ER-Golgi membranes from etiolated maize coleoptiles for glycome and proteome analysis.1 Of more than 2,000 proteins identified in the Golgi membranes, over 200 were associated with nucleotide-sugar synthesis and transport, polysaccharide synthesis, and a host of enzymes associated with downstream cell wall metabolic processes. FFE resolved the ER-Golgi membranes by flotation centrifugation into four fractions that comprise two fractions of Golgi membranes enriched in glycosyl transferases and non-cellulosic polysaccharide synthases, an ER fraction, and an intervening fraction uniquely rich in cellulose synthase (CesA) proteins.1 Consistent with role of the Golgi apparatus as the secretory organelle of the cell, we report here that, in addition to the protein machinery for cell-wall polysaccharide synthesis, numerous proteins that function in trafficking of vesicles to the cell surface and in cell signaling were identified. Using the MapMan functional annotations in the Plant Proteome Database,2 62 Golgi-associated proteins were classified as trafficking and signaling-related, including small GTPases, including several homologs of Rat Sarcoma (Ras) and Ras-binding (Rab) proteins, 14-3-3-like proteins, calcium-binding and calcium-dependent proteins, and several other proteins associated with vesicle fusion and trafficking (Table 1). FFE separation of ER and Golgi membranes revealed that many proteins of same class were distributed differentially across the four fractions of ER-Golgi based on the electrophoretic mobility of their membranes of residence or transport.
Table 1.
Maize signaling and trafficking proteins identified in ER- and Golgi membranes from etiolated maize coleoptiles.
| Maize Accession number1 | Description | Mascot Spectral Counts | MaxQuant MS/MS Counts | Arabidopsis Putative Orthologs2 |
|---|---|---|---|---|
| Small GTPases3 | ||||
| GRMZM2G071071 | Rho GTPase1 | nd | 11 | At5g27540 (MIRO1) |
| GRMZM2G079817 | Ran GTPase-activating protein | 5 | nd | At3g63130 (RANGAP1) |
| GRMZM2G157334 | Ran GTP binding protein | 6 | nd | At5g55190 (RAN3) |
| GRMZM2G127648 | Rab RIC2-like (RabA1b) | 46 | 5 | At1g16920 (RabA1b) |
| GRMZM2G101938 | Rab 11B-like (RabA1d-1) | 64 | 48 | At4g18800 (RabA1d) |
| GRMZM2G018619 | Rab RIC2-like (RabA1d-2) | 62 | 5 | At4g18800 (RabA1d) |
| GRMZM2G020661 | Rab 11B-like (RabA1f) | 45 | nd | At5g60860 (RabA1f) |
| GRMZM2G029486 | Rab 11A-like (RabA2a-1) | 30 | nd | At1g09630 (RabA2a) |
| GRMZM2G020544 | Rab11C-like (RabA2a-2) | 19 | 6 | At1g09630 (RabA2a) |
| GRMZM2G144008 | Rab11A-like (RabA2b-1) | 39 | 5 | At1g07410 (RabA2b) |
| GRMZM2G154960 | Rab11C-like (RabA2b-2) | 42 | nd | At1g07410 (RabA2b) |
| GRMZM2G061912 | Rab RGP2-like (RabA2b-3) | 25 | nd | At1g07410 (RabA2b) |
| GRMZM2G093186 | Rab RGP2 11A-like (RabA2b-4) | 27 | 9 | At1g07410 (RabA2b) |
| GRMZM2G164527 | Rab A4A-like (RabA4a-1) | nd | 19 | At5g65270 (RabA4a) |
| GRMZM2G061280 | Rab11D-like (RabA4a-2) | 47 | nd | At5g65270 (RabA4a) |
| GRMZM2G122805 | Rab11D-like(RabA4a-3) | 43 | nd | At5g65270 (RabA4a) |
| At4g39990 (RabA4b) | ||||
| GRMZM2G335738 | Rab ARA4-like (RabA5a) | 14 | 2 | At5g47520 (RabA5a) |
| GRMZM2G173878 | Rab 2B-like (RabB1c-1) | 9 | nd | At4g17170 (RabB1c) |
| GRMZM2G330430 | Rab B1C-like (RabB1c-2) | nd | 6 | At4g17170 (RabB1c) |
| GRMZM5G836471 | Rab 18-like (RabC1) | nd | 4 | At1g43890 (RabC1) |
| GRMZM2G097728 | GTP-binding protein YPTM2 (RabD2a-1) | 36 | nd | At1g02130 (RabD2a) |
| GRMZM2G097746 | Rab 1A-like (RabD2a-2) | nd | 12 | At1g02130 (RabD2a) |
| GRMZM2G106960 | Rab 1B-like (RabD2c) | 40 | 19 | At4g17530 (RabD2c) |
| GRMZM2G061900 | Rab ARA-3-like (RabE1c) | 48 | nd | At3g46060 (RabE1c) |
| GRMZM2G362088 | Rab RHN1(RABF1-like) (RabF1) | 17 | 2 | At3g54840 (RabF1) |
| GRMZM2G158887 | Rab G3F (RAB7B-like) (RabG3f) | nd | 49 | At3g18820 (RabG3f) |
| GRMZM2G169694 | Rab 6A-like (RabH1b) | 24 | 35 | At2g44610 (RabH1b) |
| AC166636.1_FGP008 | Rab-6A-like (RabH1a) | 17 | nd | At5g64990 (RabH1a) |
| At4g39890 (RabH1c) | ||||
| At2g22290 (RabH1d) | ||||
| At5g10260 (RabH1e) | ||||
| AC197246.3_FGP001 | Rab ARA4-like | 12 | nd | |
| GRMZM2G357399 | ADP-ribosylation factor (ARFA1e) | nd | 4 | At3g62290 (ARFA1e) |
| GRMZM2G081622 | ADP-ribosylation factor (ARLA1b-1) | nd | 6 | At3g49860 (ARLA1b) |
| At3g49870 (ARLA1c) | ||||
| At5g67560 (ARLA1d) | ||||
| GRMZM2G007188 | ADP-ribosylation factor 8B (ARLA1b-2) | 9 | nd | At3g49860 (ARLA1b) |
| At3g49870 (ARLA1c) | ||||
| At5g67560 (ARLA1d) | ||||
| Rab protein regulators4 | ||||
| GRMZM2G050890 | Prenylated Rab acceptorF2 | 49 | 33 | At1g55190 (PRA1F2) |
| GRMZM2G432662 | Prenylated Rab receptorB4-1 | 14 | 12 | At2g38360 (PRA1B4) |
| GRMZM5G831519 | Prenylated Rab receptorB4-2 | 24 | nd | |
| GRMZM2G089783 | Prenylated Rab receptorB4-3 | 3 | 3 | At2g38360 (PRA1B4) |
| ER-Golgi Protein Trafficking5 | ||||
| GRMZM2G178618 | Coatomer γ-subunit2 | nd | 3 | At2g16200 |
| GRMZM2G042089 | Coatomer δ-subunit delta | 8 | 2 | At5g05010 |
| GRMZM2G036034 | Coatomer γ-subunit gamma | 5 | nd | At2g16200 |
| GRMZM2G141587 | Coatomer β-subunit-2C | nd | 2 | |
| GRMZM2G115775 | SNARE domain Syntaxin51(SYP51) | 28 | 13 | At1g16225, At1g16230 |
| At1g16240, At1g79590 | ||||
| GRMZM5G838961 | Golgi SNAP receptor complex1-2 | nd | 3 | At2g45200 |
| GRMZM2G063420 | SNARE coiled-coil Bet1-like | nd | 2 | At1g29060 At4g14600 |
| GRMZM2G064268 | Protein YIF1A-1(HRF1-like) | 6 | 4 | At1g30890, At3g59500 |
| GRMZM2G135599 | Protein YIF1A-2 (HRF1-like) | 3 | nd | At1g30890, At3g59500 |
| Molecular Chaperones | ||||
| AC217050.4_FGP006 | 14-3-3c | 16 | nd | |
| GRMZM2G091155 | 14-3-3m | 8 | 2 | At3g02520 (GF14η) |
| GRMZM2G106424 | 14-3-3q | 8 | nd | |
| GRMZM2G140545 | 14-3-3p | 7 | nd | At1g78300 (GF14ω) |
| GRMZM2G078641 | 14-3-3z | 15 | nd | At3g02520 (GF14η) |
| GRMZM2G102499 | 14-3-3f | 51 | 19 | At3g02520 (GF14η) |
| GRMZM5G866082 | 14-3-3o | 8 | nd | At3g02520 (GF14η) |
| Calcium Binding | ||||
| GRMZM2G010093 | Calcineurin B-like 3 | 3 | nd | At4g26570 (CBL3) |
| GRMZM2G025387 | Calcium-dependent protein kinase3 | 8 | 6 | At4g23650 (CPK3) |
| GRMZM2G430600 | Calmodulin5 | nd | 4 | At2g27030 (CAM5) |
| GRMZM2G391364 | Calmodulin13a | 5 | nd | At1g12310 (CML13) |
| GRMZM2G324643 | Calmodulin13b | 2 | 3 | At1g12310 (CML13) |
| At1g62820 (CML13) | ||||
| GRMZM2G115628 | Calmodulin7 | 2 | nd | |
| GRMZM2G004703 | Calmodulin7 | 2 | nd | |
| GRMZM2G134668 | Calnexin | 156 | 19 | |
| GRMZM2G358059 | Calreticulin1 | 47 | 32 | At1g56340 (CRT1) |
| GRMZM2G028516 | Calreticulin3 | 2 | nd | At1g08450 (CRT3) |
| GRMZM2G058305 | Calcium-dependent protein kinase3ʹ | 7 | nd | At4g23650 (CPK3) |
| GRMZM2G058870 | Guanine-nucleotide-exchange factor (GEF) | 8 | 5 | At3g60860 (BIG2) |
| Signal Cleavage | ||||
| GRMZM2G112366 | Signal peptidase complex1 (SPC1) | 7 | nd | At2g22425 (SPS1) |
| GRMZM2G131321 | Signal peptidase complex2(SPC2) | nd | 3 | |
| GRMZM2G077463 | Signal peptidase complex3(SPC3) | 3 | 3 | |
| Protein Sorting | ||||
| GRMZM2G068489 | Vacuolar protein sorting29 (VPS29) | 3 | nd | At3g47810 (VPS29) |
| GRMZM5G825524 | Vacuolar protein sorting35 (VPS35) | 2 | nd | At1g75850 (VPS35B) |
| GRMZM2G389362 | Vacuolar sorting receptor1 (VSR1) | nd | 6 | At3g52850 (VSR1) |
aMaize accession numbers as in MaizeGDB v.2/v.3 (https://www.maizegdb.org/)
bArabidopsis nomenclature as in UniProtKB (https://www.uniprot.org/)
cNomenclature defined in Vernoud et al.3 following Periera-Leal and Seabra.4
dNomenclature as in Kamei et al.5
eNomenclature as in by Brandizzi and Barlowe,6 and Barozzi et al.7
fMaize nomenclature defined by Kumar et al.8
Small GTPases of the maize ER-Golgi
Small GTPases related to heterotrimeric G proteins are established components of signal transduction in eukaryotes and comprise a superfamily of proteins of diverse and non-redundant function.9 Small GTPases form five subfamilies in human and yeast genomes, including the Ras- and Rab- proteins that serve as regulators of vesicle budding, motility and fusion, the ADP-ribosylation factor (ARF) GTPases that function as regulators in vesicle trafficking, the Rho GTPases involved in actin dynamics, and Ran GTPases involved in nuclear trafficking.3 The Arabidopsis genome contains 93 of these proteins in four of the subgroups, with true Ras proteins absent from plant genomes.3 We identified 32 small GTPases in the ER-Golgi of elongating maize coleoptiles, including a Rho1, a Ran and Ran-activating protein (RANGAP1), three ARF and ARF-like (ARL) proteins (Table 1). Rab proteins form eight subgroups based on homology,4 and at least one member of all eight was identified. Arabidopsis RabA1, RabA2 and RabA4 are thought to be involved in the regulation of cargo content in Golgi vesicles trafficked to the wall,10 with RabA1 specifically regulating TGN-plasma membrane transport.11
Bidirectional transport between ER and Golgi is mediated by COPI and COPII carrier protein complexes.6 COPI retrograde transport involves ARF/ARL ribosylation factors and several different coatomer proteins, for which we identified a β-, a δ-, and two γ-subunits (Table 1). RabA6 has been shown to function as a regulator of COPI-independent retrograde transport from the Golgi to the ER.12
We identified four Prenylated Rab Acceptor (PRA) proteins in maize Golgi membranes (Table 1). PRAs are membrane-associated proteins that modulate vesicle trafficking as receptors of Rab GTPases and SNARE family Vesicle-Associated Membrane Protein2 (VAMP2), where they are thought to assist in release of Rab GTPases from the Rab-GDI protein.5 Arabidopsis PRA1 is implicated in both secretory and endocytic intracellular trafficking in the endosomal/prevacuolar compartments of ER and Golgi apparatus, with PRA1 F2 and PRA1 B4 localized in vesicular structures with networks of ER strands.5 PRA1 is implicated in the promotion of small GTPases trafficking through the endomembrane system by binding to the hydrophobic isoprenoid moieties of the small GTPases.13
Flotation density-gradient centrifugation alone provided a rich source of ER and Golgi membranes, and FFE resolved these proteins into four major fractions (Figure 1(a)). Established marker proteins indicated that the most electrophoretically mobile ‘Fraction 28 (F28)’ was enriched with ER-proteins, whereas F34 and F37, representing the bulk of the protein recovered, contained known Golgi markers and glycosyl transferases and non-cellulosic polysaccharide synthases.1 A concentration of cellulose synthases (CesAs) was found in F31. We report here that signaling- and trafficking-related proteins were differentially distributed across the four fractions containing ER-Golgi membranes. Thirteen Rab proteins, representing the A, D, E and H classes were more highly enriched in the ER fraction F28 than in those of the Golgi (Figure 1(b)). By contrast, six other Rabs, three RabAs, a RabB1, and a RabF1, were more highly enriched in the Golgi fractions (Figure 1(c)). These findings underscore that substantial sub- and neofunctionalization might have occurred within in each subgroup, and, thus, membrane locations are not subgroup-specific.
Figure 1.
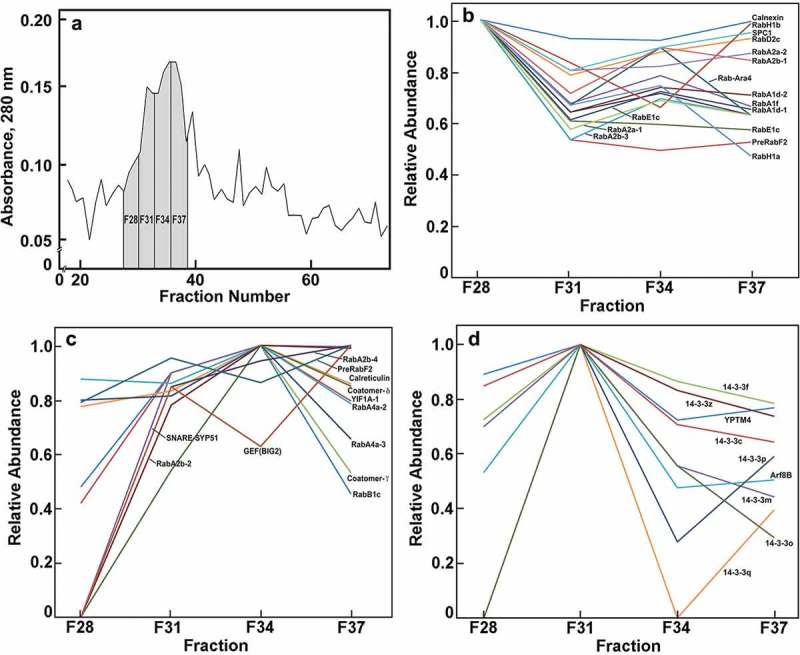
Relative distribution of ER-Golgi-associated proteins across four major fractions recovered after FFE. Three fractions from FFE were pooled; fractions 27–29 were pooled to give F28, and so forth. (a). Relative abundance of proteins estimated by Absorbance at 280 nm. (b). Relative abundance of proteins most abundant in F28. (c). Relative abundance of proteins most abundant in F34 or F37. (d). Relative abundance of proteins most abundant in F31. Nomenclature as in Table 1. Methods used for flotation centrifugation, FFE, and proteomics analysis are described in Reference 1. The MS proteomics data have been deposited to the ProteomeXchange Consortium via the partner repository,20 with the data set identifier PXD007612 and null.
Other diverse trafficking proteins enriched in the Golgi fractions included YIF1-like transport proteins, SNARE Syntaxin51 (SYP51) proteins, a PRA1, a guanine-nucleotide exchange protein, and two coatomer proteins (Figure 1(c)). Members of the SNARE family of proteins are typically associated with the ER or terminal locations of tonoplast and plasma membrane, and with retrograde Golgi to ER transport as components of COPI complexes.6,7 We show here that they partition primarily with the bulk Golgi membranes rather than with the ER (Figure 1(c)). Conversely, many known ER markers that concentrate in F28 also show substantial abundance in the Golgi fraction, indicating the possibility that the ER specifically associates with the cis-Golgi face and remains tightly associated during FFE. This finding is consistent with laser-trapping studies that showed strong interactions of Golgi with ER tubules.15 Also consistent with strong ER-Golgi interactions is the presence in the Golgi-rich fractions of the signal peptidase complex subunit1, YIF1, several Rab proteins, and two coatomer proteins associated with retrograde transport from Golgi to ER (Figure 1(b,c)).
Molecular chaperones in Golgi trafficking
We identified seven 14-3-3-like proteins in maize Golgi (Table 1). The 14-3-3 proteins are a family of cellular scaffolds with diverse regulatory functions in eukaryotes.16 They are a major class of molecular chaperones linked to signal transduction pathways regulating cell cycle checkpoints, MAP kinase activation, apoptosis and programs of gene expression.17 The 30 kDa proteins form homo- and heterodimers, which can aggregate further into homo- and heterotetramers.18 All seven maize 14-3-3 proteins showed a distinct peak of proteins in F31 between the major ER and Golgi fractions (Figure 1(d)). The only two other proteins enriched in F31were an ARF8B and a YPT-type Rab protein. Fraction 31 is specifically associated with an enrichment of CesA proteins.1 As the Golgi is the site of the formation of large multi-membered ‘particle rosette’ cellulose-synthase complexes, an intriguing possibility emerges that 14-3-3 proteins are not associated with non-cellulosic pectin- and hemicellulosic polysaccharide synthases of the Golgi, but uniquely involved in the assembly of rosette complexes for export to the PM. The 14-3-3 proteins function by binding to sites of phosphorylation to induce conformational changes and protein-protein interactions,8,19 and it is known that phosphorylation of CesA1 at specific sites is required for normal cellulose synthesis and microtubule-dependent mobility of the rosette.20
Outlook
Flotation centrifugation in sucrose density gradient steps results in rapid enrichment of ER and Golgi membranes that are resolved electrophoretically into sub-fractions by FFE.1 In reports to date, proteomic analyses of these fractions have given snapshots of resident and transiting components of the Golgi. The unexpected finding of an enrichment of Ras-like and Rab proteins in the ER might reflect the sites of insertion for subsequent transport to sites of residence. We have inferred that cohorts of associated proteins are co-fractionating by FFE, but this remains to be tested by flux measurements in pulse-chase experiments. FFE is also expected to be helpful in defining the full complements of proteins in post-Golgi processing compartments, such as the trans-Golgi network, endosomes, and other compartments involved in exo- and endocytosis.
Funding Statement
This work was supported by the Center for Direct Catalytic Conversion of Biomass to Biofuels (C3Bio), an Energy Frontier Research Center funded by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, Award Number DE-SC0000997, the U.S. Department of Energy, Office of Science, Office of Biological and Environmental Research for the Golgi isolation, linkage analyses, proteomics analyses and RNA-seq experiments. The Lawrence Berkeley National Laboratory and the U.S. Department of Energy (DE-AC02-05CH11231) supported the FFE experiments.
Acknowledgments
We thank Vicki Hedrick (Proteomics Facility, Purdue) for her technical expertise in proteome analysis.
Author Contributions
I.O.O., J.L.H., and N.C.C. designed research; I.O.O., S.M.G.F-N., U.K.A., B.W.P., J.L., and N.C.C. performed experiments; I.O.O., U.K.A., B.W.P., J.L.H, M.C.M., and N.C.C. analyzed data; I.O.O., U.K.A., B.W.P., J.L.H., M.C.M., and N.C.C. wrote the article.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed
References
- 1.Okekeogbu IO, Pattathil S, Gonzalez SM, Aryal UK, Penning BW, Lao J, Heazlewood JL, Hahn MG, McCann MC, Carpita NC.. Glycome and proteomic components of Golgi membranes are common between two angiosperms with distinct cell wall structures. Plant Cell. 2019;31(5):1–5. doi: 10.1105/tpc.18.00755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sun Q, Zybailov B, Majeran W, Friso G, Olinares PD, van Wijk KJ.. PPDB, the plant proteomics database at Cornell. Nucleic Acids Res. 2009;37:D969–D974. doi: 10.1093/nar/gkn654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vernoud V, Horton AC, Yang Z, Nielsen E. Analysis of the small GTPase gene superfamily of Arabidopsis. Plant Physiol. 2003;131(3):1191–1208. doi: 10.1104/pp.013052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pereira-Leal JB, Seabra MC. Evolution of the Rab family of small GTP-binding proteins. J Mol Biol. 2001;313:889–901. doi: 10.1006/jmbi.2001.5060. [DOI] [PubMed] [Google Scholar]
- 5.Kamei CLA, Boruc J, Vandepoele K, Den Daele HV, Maes S, Russinova E, Inzé D, de Veylder L. The PRA1 gene family in Arabidopsis. Plant Physiol. 2008;147(4):1735–1749. doi: 10.1104/pp.107.115535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brandizzi F, Barlowe C. Organization of the ER-Golgi interface for membrane traffic control. Nat Rev Mol Cell Biol. 2013;14(6):382–392. doi: 10.1038/nrm3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barozzi F, Papadia P, Stefano G, Renna L, Brandizzi F, Migoni D, Fanizzi FP, Piro G, Di Sansebastiano G-P. Variation in membrane trafficking linked to SNARE AtSYP51 interaction with Aquaporin NIP1;1. Frontiers Plant Sci. 2019;9:1949. doi: 10.3389/fpls.2018.01949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kumar K, Muthamilarasan M, Bonthala VS, Roy R, Prasad M. Unraveling 14-3-3 proteins in C4 panicoids with emphasis on model plant Setaria italica reveals phosphorylation-dependent subcellular localization of RS splicing factor. PLoS One. 2015;10(4):e0123236. doi: 10.1371/journal.pone.0123236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Small YZ, Page MD, Alder NP, Eriksson M, Quinn J, Soto F, Theg SM, Hippler M, Merchant S. GTPases: versatile signaling switches in plants. Plant Cell. 2002;14:S375–S388. doi: 10.1105/tpc.010420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Asaoka R, Uemura T, Ito J, Fujimoto M, Ito E, Ueda T, Nakano A. Arabidopsis RABA1 GTPases are involved in transport between the trans-Golgi network and the plasma membrane, and are required for salinity stress tolerance. Plant J. 2013;73(2):240–249. doi: 10.1111/tpj.12023. [DOI] [PubMed] [Google Scholar]
- 11.Lunn D, Gaddipati SR, Tucker GA, Lycett GW. Null mutants of individual raba genes impact the proportion of different cell wall components in stem tissue of Arabidopsis thaliana. PLoS One. 2013;8(10):e75724. doi: 10.1371/journal.pone.0075724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matsuto M, Kano F, Murata M. Reconstitution of the targeting of Rab6A to the Golgi apparatus in semi-intact HeLa cells: a role of BICD2 in stabilizing Rab6A on Golgi membranes and a concerted role of Rab6A/BICD2 interactions in Golgi-to-ER retrograde transport. Biochim Biophys Acta. 2015;1853(10):2592–2609. doi: 10.1016/j.bbamcr.2015.05.005. [DOI] [PubMed] [Google Scholar]
- 13.Figueroa C, Taylor J, Vojtek AB. Prenylated Rab acceptor protein is a receptor for prenylated small GTPases. J Biol Chem. 2001;276(30):28219–28225. doi: 10.1074/jbc.M101763200. [DOI] [PubMed] [Google Scholar]
- 14.Perez-Riverol Y, Csorda A, Bai J, Bernal-Llinares M, Hewapathirana S, Kundu DJ, Inuganti A, Griss J, Mayer G, Eisenacher M, et al. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 2019;47(D1):D442–D450. doi: 10.1093/nar/gky1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sparkes IA, Ketelaar T, deRuiter NCA, Hawes C. Grab a Golgi: laser trapping of Golgi bodies reveals in vivo interactions with the endoplasmic reticulum. Traffic. 2009;10:567–571. doi: 10.1111/j.1600-0854.2009.00891.x. [DOI] [PubMed] [Google Scholar]
- 16.Wang W, Shakes DC. Molecular evolution of the 14-3-3 protein family. J Mol Evol. 1996;43(4):384–398. doi: 10.1007/bf02339012. [DOI] [PubMed] [Google Scholar]
- 17.Hondermarck H. 14-3-3 proteins In: handbook of cell signaling. 2nd ed. editors, Bradshaw RA, Dennis EA. Acad Press: 2009. [Google Scholar]
- 18.Aryal UK, McBride Z, Chen D, Xie J, Szymanski DB. Analysis of protein complexes in Arabidopsis leaves using size exclusion chromatography and label-free protein correlation profiling. J Proteom. 2017;166:8–18. doi: 10.1016/j.jprot.2017.06.004. [DOI] [PubMed] [Google Scholar]
- 19.MacIntosh C. Dynamic interactions between 14-3-3 proteins and phosphoproteins regulate diverse cellular processes. Biochem J. 2004;381:329–342. doi: 10.1042/BJ20031332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen S, Ehrhardt DW, Somerville CR. Mutations of cellulose synthase (CESA1) phosphorylation sites modulate anisotropic cell expansion and bidirectional mobility of cellulose synthase. Proc Natl Acad Sci USA. 2010;107(40):17188–17193. doi: 10.1073/pnas.1012348107. [DOI] [PMC free article] [PubMed] [Google Scholar]


