Abstract
Introduction
Friedreich's ataxia (FRDA) is the most common autosomal‐recessive ataxia worldwide. It is characterized by early onset, sensory abnormalities, and slowly progressive ataxia. However, some individuals manifest the disease after the age of 25 years and are classified as late‐onset FRDA (LOFA). Therefore, we propose a transversal multimodal MRI‐based study to investigate which anatomical substrates are involved in classical (cFRDA) and LOFA.
Methods
We enrolled 36 patients (13 with LOFA) and 29 healthy controls. All subjects underwent magnetic resonance imaging in a 3 T device; three‐dimensional high‐resolution T1‐weighted images and diffusion tensor images were used to assess gray and white matter, respectively. We used T1 multiatlas approach to assess deep gray matter and cortical thickness measures to evaluate cerebral cortex and DTI multiatlas approach to assess white matter. All analyses were corrected for multiple comparisons.
Results
Group comparison showed that both groups presented gray matter atrophy mostly in the motor cortex. Regarding white matter, we found abnormalities in the cerebellar peduncles, pyramidal tracts, midbrain, pons, and medulla oblongata for both groups, but the microstructural abnormalities in the cFRDA group were more widespread. In addition, we found that the corticospinal tract presented more severe microstructural damage in the LOFA group. Finally, the midbrain volume of the cFRDA, but not of the LOFA group, correlated with disease duration (R = −0.552, P = 0.012) and severity (R = −0.783, P < 0.001).
Conclusion
The cFRDA and LOFA groups have similar, but not identical neuroimaging damage pattern. These structural differences might help to explain the phenotypic variability observed in FRDA. Hum Brain Mapp 38:4157–4168, 2017. © 2017 Wiley Periodicals, Inc.
Keywords: Friedreich's ataxia, LOFA, MRI, cortical thickness, multiatlas approach
INTRODUCTION
Friedreich's ataxia (FRDA) is the most common autosomal‐recessive ataxia worldwide, with prevalence ranging from 1 to 3 for each 100,000 people (Abrahão et al., 2015; Bhidayasiri et al., 2005; Campuzano et al., 1996). It is characterized by early onset, slowly progressive ataxia, and deep sensory abnormalities. In 1996, the genetic basis of FRDA was described: homozygous GAA expansions in the first intron of the FXN gene (Campuzano et al., 1996). This mutation reduces the expression of the protein Frataxin, leading to mitochondrial dysfunction and neurodegeneration (Pandolfo, 2008). The disease typically begins in late childhood or adolescence (classical FRDA, cFRDA), but there is a subgroup of patients with FRDA that manifest the disease symptoms after the age of 25 years. The latter group is known as late‐onset Friedreich's Ataxia (LOFA) (de Michele et al., 1994). LOFA patients are characterized by slower disease progression, milder nonneurological symptoms, smaller GAA expansions, spasticity, and sustained reflexes (Bhidayasiri et al., 2005).
Neurodegeneration in FRDA extends to the central and peripheral nervous system. Previous MRI‐based studies showed gray matter (GM) and white matter (WM) atrophy in portions of the cerebellum, brainstem, and cerebellar peduncles (Della Nave et al., 2011; Pagani et al., 2010). Gray matter supratentorial damage has been described in some studies as well, mainly in the precentral gyri (França et al., 2009; Rezende et al., 2016). Furthermore, Bonilha da Silva et al. (2014) described progressive alterations in the T2 relaxometry of dentate nuclei and Rezende et al. (2016) showed progressive abnormalities at the corpus callosum, pyramidal tracts, and superior cerebellar peduncles in a short follow‐up of 1 year. Despite that, none of these studies compared specifically classical FRDA (cFRDA) with LOFA patients or, in general, compared LOFA patients with healthy controls. Therefore, it is not clear whether these conditions have different patterns of structural damage, or how the CNS damage correlates with clinical parameters in LOFA.
The characterization of different phenotypes of a given disease (FRDA) is potentially helpful on revealing underpinning pathology, and, in the long term, may be of practical importance for prognosis prediction and design of treatments. In this study, we investigated which anatomical substrates are selectively involved in cFRDA and LOFA, and possible associations of these features with clinical parameters. By using multimodal MRI, whole‐brain segmentation, and techniques able to evaluate both GM and WM, we performed a comprehensive, holistic, and unbiased study of the brain anatomy in FRDA using atlas‐based analysis (ABA) (Faria et al., 2010). This is a technique used to automatically segment the entire brain into anatomical structures, allowing multimodal analyses within the same anatomical framework. For instance, for a given anatomical structure, it gives measures from T1‐ and T2‐weighted images, Diffusion Tensor Imaging, and Susceptibility Weighted Images (Djamanakova et al., 2014; Faria et al., 2015; Lim et al., 2013; Miller et al., 2013). Still, anatomical heterogeneity can insert errors into the registration between each patient brain and the atlas. These errors may be ameliorated by using highly accurate image mapping, such as Large Deformation Diffeomorphic Metric Mapping (LDDMM), and multiple atlases with heterogeneous features, so some of the atlases are anatomically closer to the patient (Tang et al., 2014), providing accurate and reliable measures.
MATERIALS
Study Design
Figure 1 is the flowchart that describes the sequential experimental steps performed in this study.
Figure 1.
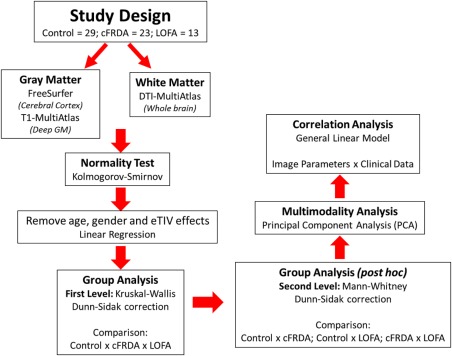
Study design. [Color figure can be viewed at http://wileyonlinelibrary.com]
Participants
This study was approved by our institution research ethics committee and a written informed consent was obtained from all participants. Thirty‐two patients with molecular confirmation of FRDA (23 cFRDA and 13 LOFAs) and 29 healthy controls (mean age = 27.1 ± 10.3 years; 10 males) were enrolled to this study. All patients are regularly followed at the neurogenetics outpatient clinics at the University of Campinas (UNICAMP) and the Federal University of São Paulo (UNIFESP) hospitals between 2009 and 2016. Clinical (age at disease onset, duration of disease, and clinical subtype) and genetic data (length of expanded repeat in both shorter, GAA1, and longer, GAA2 alleles) were recorded (Table 1). Severity of ataxia was quantified using the Friedreich's Ataxia Rating Scale (FARS) (Subramony et al., 2005) (Table 1). Patients with concomitant neurological disorders, unable to perform MRI scans and those with significant motion artifacts on images were excluded.
Table 1.
Clinical, demographic, and genetic data of patients
| cFRDA (n = 23) | LOFA (n = 13) | P‐value | |
|---|---|---|---|
| Age (median ± IQR, years) | 23.0 ± 9.0 | 40.0 ± 10.0 | <0.001b |
| Gender (M/F) | 8/15 | 8/5 | 0.169a |
| Onset (median ± IQR, years) | 12.0 ± 8.0 | 27.0 ± 6.0 | <0.001b |
| Duration (median ± IQR, years) | 10.0 ± 5.0 | 14.0 ± 15.5 | 0.626b |
| GAA1 | 1063 ± 287 | 750 ± 310 | 0.471b |
| GAA2 | 899 ± 194 | 593 ± 161 | 0.013b |
| FARS III subscore (median ± IQR) | 59.0 ± 30.0 | 40.0 ± 33.5 | 0.021b |
Chi‐square test with Yates correction.
Mann–Whitney Test.
Image Acquisition
All subjects were submitted to a high‐resolution MRI on a 3 T Phillips Achieva Scanner. Routine T2‐weighted sequences were performed for all subjects to exclude unrelated abnormalities (e.g., white matter disease, minor stroke). A board‐certified neuroradiologist (FC) carefully reviewed the images that were all acquired using a standard eight‐channel head coil.
For FreeSurfer and T1 Multi‐Atlas analyses, we used high‐resolution T1 volumetric images of the brain with sagittal orientation, voxel matrix 240 × 240 × 180, voxel size 1 × 1 × 1 mm3, TR/TE 7/3.201 ms, and flip angle of 8°.
For DTI multiatlas analyses, we used a spin echo DTI sequence: 2 × 2 × 2 mm3 acquiring voxel size, interpolated to 1 × 1 × 2 mm3; reconstructed matrix 256 × 256; 70 slices; TE/TR 61/8500 ms; flip angle 90°; 32 gradient directions; no averages; max b‐factor = 1000 s/mm2; 6 min scan.
Image Processing
Gray matter analysis
T1 multiatlas
The images were processed using “MRICloud” (http://MRICloud.org), a public web‐based service for multicontrast imaging segmentation and quantification. The image processing involves orientation correction (sagittal to axial), to match the atlas orientation, homogeneity correction by the N4 algorithm (Tustison et al., 2010), and two‐level brain segmentation: first, skull‐stripping (Tang et al., 2015), then whole brain segmentation. In addition to linear and nonlinear algorithms for brain co‐registration, the LDDMM algorithm (Miller et al., 2005) is also used. To identify the brain regions, a multiatlas labeling fusion (MALF) algorithm was employed (Tang et al., 2015), followed by a last step of labeling adjusting with PICSL (https://masi.vuse.vanderbilt.edu/workshop2013/images/1/1b/SATA_2013_Proceedings.pdf). Forty‐five atlases (JHU adult atlas version 7A) were used to generate 289 structural definitions in a five‐level ontological hierarchical relationship (Djamanakova et al., 2014; Wu et al., 2016). We chose to use the third level segmentation, which basically defines the lobes, because it showed the highest correlation with human evaluation in previous studies (Faria et al., 2015). All analyses were performed in native space. The computations were done on the Gordon cluster of XSEDE (Towns et al., 2014).
Cortical thickness
Cortical thickness was determined using FreeSurfer software v.5.3. The choice of this measure for assessing cortical damage was due to the fact that this parameter is more sensitive to cortical variations than area and volume (Hutton et al., 2009). Measurements were performed according to the protocol suggested by Fischl and Dale (2000).
Images were corrected for magnetic field inhomogeneity, aligned to the Talairach and Tournoux atlas (Talairach and Tournoux, 1988), and skull‐stripped. Next, voxels were labeled as gray matter (GM), white matter (WM), or cerebral spinal fluid (CSF). From these, using triangle meshes, two surfaces were created: the white surface, which is the interface between GM and WM, and the pial surface (Fischl and Dale, 2000). Cortical thickness was calculated as the shortest distance between the pial and white surface at each vertex across the cortical mantle. For all analyses, a Gaussian filter with 10 mm FWHM was used for smoothing the surface. Furthermore, estimated total intracranial volume (eTIV) (Buckner et al., 2004) and the volume for subcortical regions (Fischl et al., 2002) were calculated.
White matter analysis
DTI multiatlas
Raw DTI‐weighted images were co‐registered and corrected for eddy currents (Zhuang et al., 2006) and subject motion using a 12‐parameter affine transform (Woods et al., 1998). The DTI‐parameters were calculated using a multivariate linear fitting and skull‐stripped using the b = 0 image, by intensity threshold, a tool of RoiEditor software (Li, X.; Jiang, H.; Yue, Li.; and Mori, S.; Johns Hopkins University, http://www.MriStudio.org or http://www.kennedykrieger.org). This preprocessing was performed using DTIStudio software (H. Jiang and S. Mori, Johns Hopkins University, Kennedy Krieger Institute) (Jiang et al., 2006). After that, a nonlinear registration using a multicontrast LDDMM (Tang et al., 2014) was performed and, then, the parcellation, which employs a DLFA algorithm (Tang et al., 2014). Eight atlases (JHU adult atlas version 1) were used to generate 168 structures. All analyses are performed in native space. The computations were processed on the Gordon cluster of XSEDE (Towns et al., 2014).
Statistical analysis
We removed age, gender, and eTIV effects using a linear regression and, then, the Kruskal–Wallis test was initially employed to assess group differences. This test was chosen because most MRI‐derived parameters of the cFRDA and LOFA groups did not follow a normal distribution according to the Kolmogorov–Smirnov test. In order to correct for multiple comparisons, we employed the Dunn–Sidak test with level of significance adjusted to 0.05. We then used a post‐hoc Mann–Whitney test for each pairwise group combination (control vs FRDA, control vs LOFA, FRDA vs LOFA) to identify where these group differences are. For these post‐hoc analyses, we also employed the Dunn–Sidak correction for multiple comparisons. After we identified these differences, we performed the principal component analysis (PCA) to evaluate qualitatively the separation between patients and controls. To accomplish that, we employed a z‐score transformation to put all measures into the same scale and evaluated all parameters together (just those with group differences) (Figure 1).
PCA is an exploratory method to analyze, mainly, a large pool of data, reducing its dimensionality (Jolliffe, 1986). It means that there is some level of redundancy among the variables. In this sense, some variables might be correlated with one another, possibly because they are measuring the same thing. Then, PCA reduces the number of observed variables into a smaller number of uncorrelated principal components that will account for most of the variance in the observed variables. However, one of the most important uses of PCA is to find relationships between objects, that is, finding classes or, more specifically, clinical subtypes of a disease (Faria et al., 2015).
The general linear model was performed to assess possible correlations between imaging measures and clinical data (disease duration and disease severity). In all regressions, we used age and gender as covariates. To adjust for multiple comparisons, we employed the FDR correction (level of significance, α = 0.05).
RESULTS
Gray Matter Analysis
T1 multiatlas
T1 multiatlas showed volumetric reduction at the midbrain (including the red nuclei and substantia nigra), medulla oblongata (Mann–Whitney, P < 0.001), pons (P < 0.001), and thalami (right: P < 0.001; left: P < 0.001) in patients with cFRDA when compared to controls (Figure 2). The LOFA patients, in comparison with healthy controls, showed volumetric reductions on the same structures found in the cFRDA group, except by the pons that did not show significant difference when compared to controls (Figure 2). Finally, LOFA patients did not present any significant difference when compared to cFRDA patients.
Figure 2.
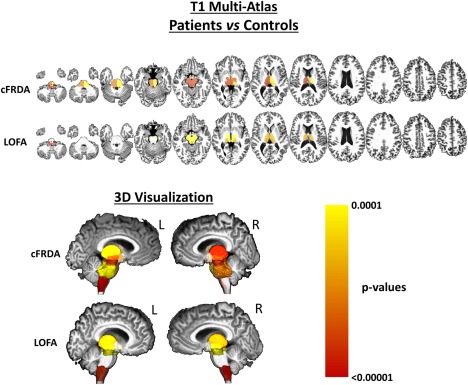
Results of ROI‐based analyses using T1 multiatlas approach to assess deep GM. The cFRDA and LOFA patients were compared to healthy controls. All results were corrected for multiple comparisons, using Dunn–Sidak test, and the measures were linearly regressed to age, gender, and total intracranial volume. [Color figure can be viewed at http://wileyonlinelibrary.com]
Cortical thickness
We observed cortical thinning in the motor cortex, particularly in the left precentral gyrus and left subcentral gyrus and sulcus, for cFRDA and LOFA groups when compared to controls (Table 2). Furthermore, we did not find any significant difference between cFRDA and LOFA groups (Table 2).
Table 2.
Results of FreeSurfer analyses showing significant cortical thinning in patients with cFRDA and LOFA when compared to healthy controls
| Cortical thickness analysis | |||
|---|---|---|---|
| Structure | Controls | cFRDA | LOFA |
| Left subcentral gyrus and sulcus | 2.67 ± 0.20 | 2.53 ± 0.15 | 2.44 ± 0.13 |
| Left precentral gyrus | 2.69 ± 0.13 | 2.48 ± 0.21 | 2.49 ± 0.20 |
| P‐values from group analyses | |||
| Structure | Controls vs cFRDA | Controls vs LOFA | cFRDA vs LOFA |
| Left subcentral gyrus and sulcus | <0.001 | <0.001 | 0.542 |
| Left precentral gyrus | <0.001 | 0.010 | 0.084 |
Results corrected for multiple comparison (Dunn–Sidak) and linearly regressed to age and gender.
White Matter Analysis
DTI multiatlas
Axial diffusivity
We only found WM abnormalities in the insular cortex (P < 0.001) to the cFRDA group when compared to control group. However, to make clear the interpretation of this result, we are just considering the WM beneath the GM in the insular cortex.
Fractional anisotropy
In the cFRDA group, when compared to the control group, we identified widespread FA decrease in several brain regions, particularly those related to motor control. These abnormalities include damage in afferent areas of cerebellum, mostly at superior and inferior cerebellar peduncles in both hemispheres (both P < 0.001), medulla (P < 0.001), midbrain (P < 0.001), and pyramidal tracts (P < 0.001). We also found decreased FA in bilateral splenium and body of corpus callosum (both P < 0.001) and the ramifications of the thalamus (P < 0.001) (Figure 3).
Figure 3.
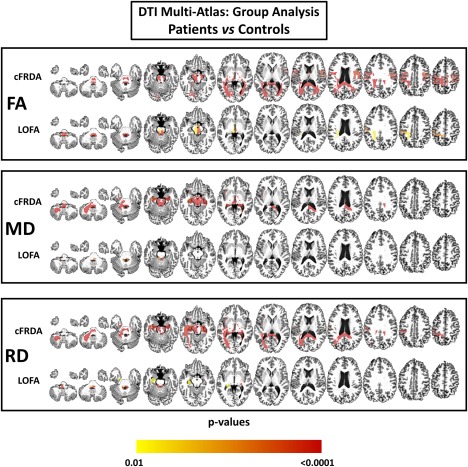
Results of DTI multiatlas approach showing areas of reduced fractional anisotropy (FA), increased mean diffusivity (MD), and incresead mean diffusivity (RD) in patients with cFRDA and LOFA after comparison with controls. Effects of age and gender were removed and all results were corrected for multiple comparisons using Dunn–Sidak test. [Color figure can be viewed at http://wileyonlinelibrary.com]
In the LOFA group, we found FA decrease, when compared with control group, at similar structures to those found altered in cFRDA group. However, we did not observe WM abnormalities in bilateral splenium and body of corpus callosum, cerebral peduncles, and corticospinal tract (Figure 3). In addition, we found significantly decreased FA in the cFRDA group when compared to the LOFA group at the left corticospinal tract (P = 0.004), bilateral splenium of corpus callosum (right: P = 0.002; left: P = 0.007), cerebral peduncles (right: P = 0.004, left: P = 0.004), right inferior cerebellar peduncle (P = 0.011), left posterior thalamic radiation (P = 0.015), and the WM beneath postcentral (P = 0.002) and precentral gyri (P < 0.001) (Figure 4).
Figure 4.
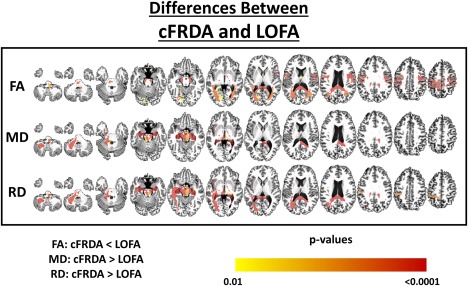
Structural differences between cFRDA and LOFA patients. The yellow–red scale shows structures more severely altered in the cFRDA group. All results were corrected for multiple comparisons using the Dunn–Sidak test. The effects of age and gender were regressed in the model. [Color figure can be viewed at http://wileyonlinelibrary.com]
Mean diffusivity
In the cFRDA group, when compared to the control group, the mean diffusivity (MD) was increased at superior (right: P < 0.001; left: P < 0.001) and inferior (right: P < 0.001; left: P < 0.001) cerebellar peduncles, left middle cerebellar peduncle (P <0.001), cerebral peduncles (right: P < 0.001; left: P < 0.001), medial lemnisci (right: P < 0.001; left: P < 0.001), left corticospinal tract (P < 0.001), midbrain (P < 0.001) and medulla (P < 0.001). The cFRDA patients also presented with increased MD in the right splenium of corpus callosum (P < 0.001) and bilateral hippocampi (right: P < 0.001; left: P = 0.001) (Figure 3). In contrast, the LOFA group showed increased MD only at superior cerebellar peduncles (right: P < 0.001; left: P < 0.001), left inferior cerebellar peduncle (P = 0.001) and right medial lemniscus when compared to healthy controls (Figure 3).
Now, comparing cFRDA and LOFA groups, we found significant between‐group MD differences at midbrain (P = 0.005), hippocampi (right: P < 0.001; left: P < 0.001), medulla oblongata (P = 0.001), left medial lemniscus (P = 0.004), left superior (P = 0.008) and middle (P < 0.001) cerebellar peduncle, cerebral peduncles (right: P < 0.001; left: P < 0.001), right splenium of corpus callosum (P < 0.001), and left corticospinal tract (P = 0.001) (Figure 4). In addition, evaluating the median MD for each one of these structures, we found that values were all higher in the cFRDA group. The only remarkable exception was the left corticospinal tract that presented higher MD value in the LOFA group (Figure 5).
Figure 5.
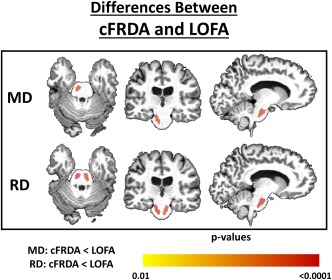
Structural differences between cFRDA and LOFA patients. The yellow–red scale shows structures more severely altered in the LOFA group. All results were corrected for multiple comparisons using the Dunn–Sidak test. The effects of age and gender were regressed in the model. [Color figure can be viewed at http://wileyonlinelibrary.com]
Radial diffusivity
By comparing cFRDA group with control group, we found widespread increased RD in regions associated with motor control, such as pyramidal tracts (right: P < 0.001; left: P < 0.001), inferior (P < 0.001) and superior (right: P < 0.001; left: P < 0.001) cerebellar peduncle, medial lemnisci (right: P < 0.001; left: P < 0.001) and ramifications, medulla (P < 0.001) and midbrain (P < 0.001) (Figure 3). We also found increased RD in hippocampi (P < 0.001), splenium of corpus callosum (right: P < 0.001; left: P < 0.001), and cerebellum (P < 0.001). The LOFA group showed increased RD at left inferior cerebellar peduncle (P < 0.001), the superior cerebellar peduncles (right: P < 0.001; left: P < 0.001), left hippocampus (P = 0.012), left medial lemniscus (P = 0.007), medulla oblongata (P = 0.001), and right fornix (P = 0.001) in contrast to matched controls (Figure 3).
We also found increased RD, in cFRDA group, at the left inferior cerebellar peduncle (P = 0.017), left middle cerebellar peduncle (P < 0.001), cerebral peduncles (right: P < 0.001; left: P < 0.001), both hippocampi (right: P < 0.001; left: P < 0.001), medulla oblongata (P = 0.002), bilateral fornix (right: P = 0.012; left: P < 0.001), and bilateral splenium of corpus callosum (right: P < 0.001; left: P < 0.001) when compared to LOFA group (Figure 4). In contrast, the LOFA group showed higher RD values at the bilateral corticospinal tract (right: P = 0.001; left: P < 0.001) compared to the cFRDA group (Figure 5).
Multimodality Analysis
Merging information from all approaches employed in this study (T1 multiatlas, DTI multiatlas, and cortical thickness), we found a clear tendency of segregation between cFRDA and healthy controls (Figure 6). In contrast, the LOFA group is placed in a region between cFRDA patients and the control group with a smooth tendency to remain close (Figure 6).
Figure 6.
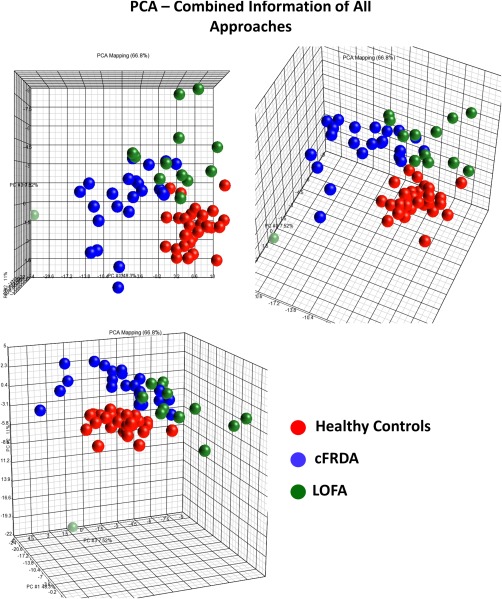
PCA plot with features selected by the group comparisons, colored by diagnosis. [Color figure can be viewed at http://wileyonlinelibrary.com]
Correlation Analyses
In the cFRDA group, volumetric reduction at midbrain and pons correlated with disease duration (midbrain: R = −0.553, P = 0.016; pons: R = −0.569, P = 0.016) and disease severity (midbrain: R = −0.784, P < 0.001; pons: R = −0.683, P < 0.001), respectively. In addition, disease duration also correlated with volumetric reduction at thalami (left: R = −0.603, P = 0.003; right: R = −0.548, P = 0.007) and medulla oblongata (R = −0.693, P < 0.001). In contrast, we did not find any significant correlation for the LOFA group. We did not find significant correlation between clinical and DTI parameters. All results were corrected for multiple comparisons using FDR test.
DISCUSSION
This study is devoted to investigate cerebral damage in patients with cFRDA and LOFA using a multimodal MRI‐based analysis. We found GM atrophy in both groups at the left precentral gyrus, left subcentral gyrus, and sulcus and the thalami. White matter damage in cFRDA group was much more widespread than in the LOFA group, but the superior and inferior cerebellar peduncles, pyramidal tracts, midbrain, pons, and medulla oblongata were compromised in both groups. These results are in line with previous pathological reports that showed the reduction of giant Betz cells in the layer V of primary motor cortex, degeneration of cerebellar peduncles, and the reduction of spinal cord area in FRDA (Klawiter et al., 2011; Koeppen et al., 2011).
Our results for cFRDA corroborate previous MRI reports (Akhlaghi et al., 2014; Bonilha da Silva et al., 2014; Della Nave et al., 2011; França et al., 2009; Pagani et al., 2010; Rezende et al., 2016; Silva et al., 2012; Zalesky et al., 2014). Major targets of neurodegeneration were the corticospinal tracts and cerebellar connections, structures that help to explain the cardinal motor features of the disease (ataxia + pyramidal signs). In addition, the white matter abnormalities in the corpus callosum, hippocampi, fornix, pathways linking the cerebellum to thalamus and the prefrontal cortices may also explain the subtle, but increasingly recognized cognitive deficits in the disease, such as impairment of visuospatial and executive domains (Nieto et al., 2013). We were also able to show that limbic connections are involved as well, areas that are tightly related to mood disorders.
Overall, the pattern of damage in the LOFA group was similar to the cFRDA group. The key targets of neurodegeneration were the cerebellar peduncles, fornix, thalami, pyramidal tracts, and motor cortex in LOFA. Interestingly, these results might help to explain the mild cognitive impairment (executive dysfunction) described by Nieto et al. (2013) in these patients. We have indeed found abnormal imaging results in regions known to be associated to specific cognitive functions, such as the posterior part of the corpus callosum and parieto‐occipital association areas, which is in line with the results described by Nieto et al. (2013). The milder signs of neurological damage in LOFA patients are also in line with their genetic background, as shorter GAA repeats result in less severe under expression of functional frataxin (Koeppen et al., 2011).
Corticospinal tracts were found to be altered in both groups. However, diffusivity parameters (MD and RD) pointed to more severe microstructural damage in the LOFA group. A clinical correlate of this MRI feature are the pyramidal signs (spasticity, hyperreflexia, and Babinski sign), which were much more frequent and severe in our LOFA group as described by Martinez et al. (2017). In terms of disease pathophysiology, this result looks counterintuitive at first, but we believe that it sheds some light in the basis of Frataxin‐related neuronal loss. It seems that low frataxin levels determine a selective neuronal damage depending on both the magnitude of deficiency and the duration of such deficiency. Patients with FRDA are under the effects of low frataxin expression since their conception. So, the exposure to the low levels of frataxin is longer in the LOFA group as they are older than the cFRDA group. In this sense, we hypothesize that corticospinal tracts are more resistant to the deleterious effects of frataxin loss in the short term, and therefore damage would only appear late in the disease course, what characterize a survivor effect as the LOFA patients experiment, on average, longer life span than cFRDA patients (Koeppen et al, 2011; Martinez et al., 2017). An alternative explanation would be that small and large (GAA) expansions lead to neurodegeneration through distinct biochemical mechanisms. Some specific tracts, such as the corticospinal tracts might be preferentially affected in a scenario of small (GAA) expansions, which are typical for LOFA. Previous reports indeed indicate that neuropathological signature at the cellular level in cFRDA and LOFA is slightly different (Koeppen et al, 2011). Further experimental studies are needed to address this question.
We did not find any correlation between clinical data and DTI parameters or cortical thickness data. This lack of correlation with cortical thickness measures was also observed in a previous study from our group (Rezende et al., 2016). At least two possibilities might help to explain this finding. There was reduced cortical thickness in FRDA only around primary motor cortices, but the clinical severity scale employed (FARS) is actually a measure of ataxia and some pyramidal signs, but not spasticity that is an important pyramidal sign found in our group of LOFA patients. Perhaps, if we look at clinical scales that truly quantify upper motor neuron damage or specifically spasticity per se, then such correlations might appear. Another hypothesis is a possible floor effect, because mean disease duration was around 13 years for each group. Regarding DTI data, we believe that ROI‐based analysis might have been an important factor for not finding clinical correlations. The computed DTI measures were indeed a mean value for each label (structures), and this might have diluted effects that were found only in part of the label (Rezende et al, 2016).
There was no significant correlation between clinical and neuroimaging parameters (of those regions found atrophic) for the LOFA group. This is in striking contrast to the cFRDA group, where we found significant correlations, such as the midbrain volume with both disease duration and severity. One must take into account, however, that we enrolled a small cohort of LOFA subjects and therefore, the lack of correlation might be simply due to reduced statistical power. Another possible reason is that clinical‐neuroimaging correlation truly behaves in different ways for LOFA and cFRDA. This is an important finding if we consider that neuroimaging markers are emerging as potential surrogate outcome measures for drug tests. For heredodegenerative diseases, our results indicate that any potential MRI‐based metric must be validated across the whole phenotypic spectrum, before attempting to use it in clinical trials. Again, longitudinal studies with long follow‐up periods would be extremely helpful.
In conclusion, cFRDA and LOFA have similar, but not identical neuroimaging signatures. Although subtle, the structural differences might help to explain the phenotypic variability seen in both conditions. The corticospinal tracts are damaged in both conditions, but more severely in the LOFA group. These results provide meaningful insights into disease biology and also add relevant information on the use of neuroimaging metrics as biomarkers for FRDA. The multiatlas approach proved to be a useful tool to identify biomarkers in FRDA, which might help upcoming clinical trials.
FUNDING
Fundação de Amparo à Pesquisa do Estado de São Paulo – FAPESP, São Paulo, Brazil. Drs MCFJ, JLP, IL‐C, FC, JLP, OGB, and ILC are supported by FAPESP and CNPq (Conselho Nacional de Pesquisa‐BRAZIL). Dr AVF is supported by NIH (National Institute of Health). TJRB and ARMM receive a PhD scholarship from FAPESP (Grant #2014/19786–7 and 2013/26410–0), IF receives a PhD scholarship from CAPES (Coordenação de Aperfeiçoamento de pessoal de nível superior‐BRAZIL). The funding agencies did not interfere with the design of the study, collection of data, or drafting of the manuscript.
DISCLOSURES
Dr Rezende receives a PhD scholarship from FAPESP.
Dr Martinez receives a PhD scholarship from FAPESP.
Dr Faber receives a PhD scholarship from CAPES.
Dr Girotto reports no disclosures.
Dr Pedroso received research support from CNPq and FAPESP.
Dr Barsottini received research support from CNPq and FAPESP.
Dr Lopes‐Cendes received research support from CNPq and FAPESP.
Dr Cendes serves as an editorial board member of Neurology and received research support from CNPq and FAPESP.
Dr Faria received research support from NIH.
Dr França Jr received research support from CNPq and FAPESP.
CONTRIBUTION OF THE AUTHORS
Research project: A. Conception, B. Organization, C. Execution
Statistical Analysis: A. Design, B. Execution, C. Review and Critique
Manuscript: A. Writing of the first draft, B. Review and Critique
Thiago Junqueira R. Rezende: 1, 2A, 2B, 3A
Alberto R. Muro Martinez: 1C, 2B, 3A
Ingrid Faber: 1C, 2B, 3B
Karen Girotto: 1C, 2B, 3B
José Luiz Pedroso: 1A, 1B, 2C, 3B
Orlando G. Barsottini: 1A, 1B, 2C, 3B
Iscia Lopes‐Cendes: 1A, 1B, 2C, 3B
Fernando Cendes: 1A, 1B, 2C, 3B
Andreia V. Faria: 1A, 1B, 2C, 3B
Marcondes C. França Jr: 1, 2A, 2B, 3B
FULL FINANCIAL DISCLOSURES OF ALL AUTHORS FOR THE PAST YEAR
Thiago Junqueira R. Rezende: none
Alberto R. Muro Martinez: none
Ingrid Faber: none
Karen Girotto: none
José Luiz Pedroso: none
Orlando G. Barsottini: none
Iscia Lopes‐Cendes: none
Fernando Cendes: none
Andreia V. Faria: none
Marcondes C. França Jr: none
The authors report no conflict of interest regarding this research.
REFERENCES
- Abrahão A, Pedroso JL, Braga‐Neto P, Bor‐Seng‐Shu E, de Carvalho Aguiar P, Barsottini OG (2015): Milestones in Friedreich ataxia: More than a century and still learning. Neurogenetics 16:151–160. [DOI] [PubMed] [Google Scholar]
- Akhlaghi H, Yu J, Corben L, Georgiou‐Karistianis N, Bradshaw JL, Storey E, Delatycki MB, Egan GF (2014): Cognitive deficits in Friedreich ataxia correlate with microstructural changes in dentatorubral tract. Cerebellum 13:187–198. [DOI] [PubMed] [Google Scholar]
- Bhidayasiri R, Perlman SL, Pulst SM, Geschwind DH (2005): Late‐onset Friedreich ataxia: Phenotypic analysis, magnetic resonance imaging findings, and review of the literature. Arch Neurol 629:1865–1869. [DOI] [PubMed] [Google Scholar]
- Bonilha da Silva C, Bergo FP, D'Abreu A, Cendes F, Lopes‐Cendes I, França MC, Jr. (2014): Dentate nuclei T2 relaxometry is a reliable neuroimaging marker in Friedreich's ataxia. Euro J Neurol 21:1131–1136. [DOI] [PubMed] [Google Scholar]
- Buckner RL, Head D, Parker J, Fotenos AF, Marcus D, Morris JC, Snyder AZ (2004): A unified approach for morphometric and functional data analysis in young, old, and demented adults using automated atlas‐based head size normalization: Reliability and validation against manual measurement of total intracranial volume. Neuroimage 23:724–738. [DOI] [PubMed] [Google Scholar]
- Campuzano V, Montermini L, Moltò MD, Pianese L, Cossée M, Cavalcanti F, Monros E, Rodius F, Duclos F, Monticelli A, Zara F, Cañizares J, Koutnikova H, Bidichandani SI, Gellera C, Brice A, Trouillas P, De Michele G, Filla A, De Frutos R, Palau F, Patel PI, Di Donato S, Mandel JL, Cocozza S, Koenig M, Pandolfo M (1996): Friedreich's ataxia: Autosomal recessive disease caused by anintronic GAA triplet repeat expansion. Science 271:1423–1427. [DOI] [PubMed] [Google Scholar]
- De Michele G, Filla A, Cavalcanti F, Di Maio L, Pianese L, Castaldo I, Calabrese 0, Monticelli A, Varrone S, Campanella G, Leone M, Pandolfo M, Cocozza S (1994): Late onset Friedreich's disease: Clinical features and mapping of mutation to the FRDA locus. J Neurol Neurosurg Psychiatry 57:977–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Della Nave R, Ginestroni A, Diciotti S, Salvatore E, Soricelli A, Mascalchi M (2011): Axial diffusivity in increased in the degenerating superior cerebellar peduncles of Friedreich's ataxia. Neuroradiology 53:367–372. [DOI] [PubMed] [Google Scholar]
- Djamanakova A, Tang X, Li X, Faria AV, Ceritoglu C, Oishi K, Hillis AE, Albert M, Lyketsos C, Miller MI, Mori S (2014): Tools for multiple granularity analysis of brain MRI data for individualized image analysis. Neuroimage 101:168–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faria AV, Oishi K, Yoshida S, Hillis A, Miller MI, Mori S (2015): Content‐based image retrieval for brain MRI: An image‐searching engine and population‐based analysis to utilize past clinical data for future diagnosis. Neuroimage Clin 15:367–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faria AV, Zhang J, Oishi K, Li X, Jiang H, Akhter K, Hermoye L, Lee SK, Hoon A, Stashinko E, Miller MI, van Zijl PC, Mori S (2010): Atlas‐based analysis of neurodevelopment from infancy to adulthood using diffusion tensor imaging and applications for automated abnormality detection. Neuroimage 15:415–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl B, Dale AM (2000): Measuring the thickness of the human cerebral cortex from magnetic resonance images. Proc Natl Acad Sci USA 97:11050–11055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl B, Salat DH, Busa E, Albert M, Dieterich M, Haselgrove C, van der Kouwe A, Killiany R, Kennedy D, Klaveness S, Montillo A, Makris N, Rosen B, Dale AM (2002): Whole brain segmentation: Automated labeling of neuroanatomical structures in the human brain. Neuron 33:341–355. [DOI] [PubMed] [Google Scholar]
- França MC, Jr. , D'Abreu A, Yasuda CL, Bonadia LC, Santos da Silva M, Nucci A, Lopes‐Cendes I, Cendes F (2009): A combined voxel‐based morphometry and 1H MRS study in patients with Friedreich's ataxia. J Neurol 256:1114–1120. [DOI] [PubMed] [Google Scholar]
- Hutton C, Draganski B, Ashburner J, Weiskopf N (2009): A comparison between voxel‐based cortical thickness and voxel‐based morphometry in normal aging. Neuroimage 48:371–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, van Zijl PC, Kim J, Pearlson GD, Mori S (2006): DtiStudio: Resource program for diffusion tensor computation and fiber bundle tracking. Comput. Methods Prog. Biomed 81:106–116. [DOI] [PubMed] [Google Scholar]
- Jolliffe IT (1986): Principal Component Analysis. New York: Springer‐Verlag. [Google Scholar]
- Klawiter EC, Schmidt RE, Trinkaus K, Liang HF, Budde MD, Naismith RT, Song SK, Cross AH, Benzinger TL (2011): Radial diffusivity predicts demyelination in ex vivo multiple sclerosis spinal cords. Neuroimage 15:1454–1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koeppen AH, Morral JA, McComb RD, Feustel PJ (2011): The neuropathology of late‐onset Friedreich's ataxia. Cerebellum 10:96–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim IA, Faria AV, Li X, Hsu JT, Airan RD, Mori S, van Zijl PC (2013): Human brain atlas for automated region of interest selection in quantitative susceptibility mapping: Application to determine iron content in deep gray matter structures. Neuroimage 15:449–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez AR, Moro A, Abrahao A, Faber I, Borges CR, Rezende TJ, Martins CR, Jr. , Moscovich M, Munhoz RP, Segal SL, Arruda WO, Saraiva‐Pereira ML, Karuta S, Pedroso JL, D'Abreu A, Jardim LB, Lopes‐Cendes Í, Barsottini OG, Teive HA, França MC, Jr. (2017): Nonneurological involvement in late‐onset Friedreich ataxia (LOFA): Exploring the phenotypes. Cerebellum 16:253–256. [DOI] [PubMed] [Google Scholar]
- Miller MI, Beg MF, Ceritoglu C, Stark C (2005): Increasing the power of functional maps of the medial temporal lobe by using large deformation diffeomorphic metric mapping. Proc. Natl. Acad. Sci. USA 102:9685–9690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MI, Faria AV, Oishi K, Mori S (2013): High‐throughput neuroimaging informatics. Front Neuroinform 17:7–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto A, Correia R, de Nóbrega E, Montón F, Barroso J (2013): Cognition in late‐onset Friedreich ataxia. Cerebellum 12:504–512. [DOI] [PubMed] [Google Scholar]
- Pagani E, Ginestroni A, Della Nave R, Agosta F, Salvi F, De Michele G, Piacentini S, Filippi M, Mascalchi M (2010): Assessment of brain white matter bundle atrophy in patients with Friedreich ataxia. Radiology 255:882–887. [DOI] [PubMed] [Google Scholar]
- Pandolfo M (2008): Friedreich ataxia. Arch Neurol 65:1296–1303. [DOI] [PubMed] [Google Scholar]
- Rezende TJ, Silva CB, Yassuda CL, Campos BM, D'Abreu A, Cendes F, Lopes‐Cendes I, França MC, Jr. (2016): Longitudinal magnetic resonance imaging study shows progressive pyramidal and callosal damage in Friedreich's ataxia. Mov Disord 31:70–78. [DOI] [PubMed] [Google Scholar]
- Silva CB, Yasuda CL, D'Abreu A, Cendes F, Lopes‐Cendes I, França MC, Jr. (2012): Neuroanatomical correlates of depression in Friedreich's ataxia: A voxel‐based morphometry study. Cerebellum 12:429–436. [DOI] [PubMed] [Google Scholar]
- Subramony SH, May W, Lynch D, Gomez C, Fischbeck K, Hallett M, Taylor P, Wilson R, Ashizawa T; Cooperative Ataxia Group (2005): Measuring Friedreich ataxia: Interrater reliability of a neurologic rating scale. Neurology 64:1261–1262. [DOI] [PubMed] [Google Scholar]
- Talairach J, Tournoux P. (1988): Co‐planar Stereotaxic Atlas of the Human Brain. NY: Thieme. [Google Scholar]
- Tang X, Crocetti D, Kutten K, Ceritoglu C, Albert MS, Mori S, Mostofsky SH, Miller MI (2015): Segmentation of brain magnetic resonance images based on multi‐atlas likelihood fusion: Testing using data with a broad range of anatomical and photometric profiles. Front Neurosci 3:61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X, Yoshida S, Hsu J, Huisman TA, Faria AV, Oishi K, Kutten K, Poretti A, Li Y, Miller MI, Mori S (2014): Multi‐contrast multi‐atlas parcellation of diffusion tensor imaging of the human brain. Plos One 9:e96985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towns J, Cockerill T, Dahan M, Foster I, Gaither K, Grimshaw A, Hazlewood V, Lathrop S, Lifka D, Peterson GD, Roskies R, Scott JR, Wilkins‐Diehr N (2014): XSEDE: Accelerating scientific discovery. Comput Sci Eng 16:62–74. [Google Scholar]
- Tustison NJ, Avants BB, Cook PA, Zheng Y, Egan A, Yushkevich PA, Gee JC (2010): N4ITK: Improved N3 bias correction. IEEE Trans Med Imaging 29:1310–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods RP, Grafton ST, Holmes CJ, Cherry SR, Mazziotta JC (1998): Automated image registration: I. General methods and intrasubject, intramodality validation. J Comput Assist Tomogr 22:139–152. [DOI] [PubMed] [Google Scholar]
- Wu D, Ma T, Ceritoglu C, Li Y, Chotiyanonta J, Hou Z, Hsu J, Xu X, Brown T, Miller MI, Mori S (2016): Resource atlases for multi‐atlas brain segmentations with multiple ontology levels based on T1‐weighted MRI. Neuroimage 125:120–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalesky A, Akhlaghi H, Corben LA, Bradshaw JL, Delatycki MB, Storey E, Georgiou‐Karistianis N, Egan GF (2014): Cerebello‐cerebral connectivity deficits in Friedreich ataxia. Brain Struct Funct 219:969–981. [DOI] [PubMed] [Google Scholar]
- Zhuang J, Hrabe J, Kangarlu A, Xu D, Bansal R, Branch CA, Peterson BS (2006): Correction of eddy–current distortions in diffusion tensor images using the knowndirections and strengths of diffusion gradients. J. Magn. Reson. Imaging 24:1188–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]


