Abstract
Major depressive disorder (MDD) has been associated with disruptions in the topological organization of brain morphological networks in group‐level data. Such disruptions have not yet been identified in single‐patients, which is needed to show relations with symptom severity and to evaluate their potential as biomarkers for illness. To address this issue, we conducted a cross‐sectional structural brain network study of 33 treatment‐naive, first‐episode MDD patients and 33 age‐, gender‐, and education‐matched healthy controls (HCs). Weighted graph‐theory based network models were used to characterize the topological organization of brain networks between the two groups. Compared with HCs, MDD patients exhibited lower normalized global efficiency and higher modularity in their whole‐brain morphological networks, suggesting impaired integration and increased segregation of morphological brain networks in the patients. Locally, MDD patients exhibited lower efficiency in anatomic organization for transferring information predominantly in default‐mode regions including the hippocampus, parahippocampal gyrus, precuneus and superior parietal lobule, and higher efficiency in the insula, calcarine and posterior cingulate cortex, and in the cerebellum. Morphological connectivity comparisons revealed two subnetworks that exhibited higher connectivity strength in MDD mainly involving neocortex‐striatum‐thalamus‐cerebellum and thalamo‐hippocampal circuitry. MDD‐related alterations correlated with symptom severity and differentiated individuals with MDD from HCs with a sensitivity of 87.9% and specificity of 81.8%. Our findings indicate that single subject grey matter morphological networks are often disrupted in clinically relevant ways in treatment‐naive, first episode MDD patients. Circuit‐specific changes in brain anatomic network organization suggest alterations in the efficiency of information transfer within particular brain networks in MDD. Hum Brain Mapp 38:2482–2494, 2017. © 2017 Wiley Periodicals, Inc.
Keywords: depression, graph analysis, gray matter, structural network, single‐subject based cortical network
INTRODUCTION
Lui et al. first proposed the Psychoradiology, a new field of radiology, which seems primed to play a major clinical role in guiding diagnostic and treatment planning decisions in patients with psychiatric disorders [Lui et al., 2016]. Psychoradiological studies of depressed patients have demonstrated focal functional and structural alterations in cortical and subcortical regions, including dorsolateral prefrontal cortex [Fitzgerald et al., 2006; Groenewold et al., 2013; Hamilton et al., 2012; Wise et al., 2016], anterior cingulate cortex (ACC) [Drevets et al., 1998; Groenewold et al., 2013; Pizzagalli, 2011; Schmaal et al., 2016a], posterior cingulate cortex/precuneus (PCC/PCu) [Mah et al., 2007], amygdala [Groenewold et al., 2013; Hamilton et al., 2012; Mah et al., 2007], hippocampus [Arnone et al., 2016; Lui et al., 2009; Schmaal et al., 2016b; Wise et al., 2016] and caudate nucleus [Krishnan et al., 1992; Pizzagalli et al., 2009; Schmaal et al., 2016b]. In addition to focal abnormalities, studies of MDD patients have documented abnormal structural and functional connectivity involving the default‐mode network (DMN) [Kaiser et al., 2015; Zhu et al., 2012], prefrontal‐limbic‐thalamic [Lui et al., 2011] and ACC‐anchored networks [Connolly et al., 2013; Greicius et al., 2007]. Abnormal white‐matter structural connectivity, involving the inferior fronto‐occipital fasciculus, posterior thalamic radiation, and corpus callosum have also been reported [Bae et al., 2006; Liao et al., 2013]. More recently, graph‐based connectome studies have demonstrated MDD‐related topological anatomic disturbances of whole‐brain networks, suggesting a dysconnection syndrome in the brains of depressed individuals [Gong and He, 2015].
Whole‐brain networks can be extracted using multimodal neuroimaging techniques, among which functional MRI and diffusion MRI are most frequently used. Based on these two techniques, disrupted topological organization of large‐scale functional [Bohr et al., 2013; Cisler et al., 2013; Guo et al., 2012; Jin et al., 2011; Lord et al., 2012; Meng et al., 2014; Wang et al., 2014; Ye et al., 2015; Zhang et al., 2011] and structural brain networks [Bai et al., 2012; Korgaonkar et al., 2014; Long et al., 2015; Qin et al., 2014] have been reported in MDD.
Recently, structural MRI data has been increasingly used to delineate whole‐brain connectivity patterns by calculating inter‐regional morphological correlations [Bassett et al., 2008; He et al., 2007]. Examining morphological covariance has distinct advantages, such as its relative insensitivity to artifacts (e.g., head motion) compared with other modalities. Moreover, this method has successfully revealed MDD‐associated alterations in morphological covariance networks in groups of depressed patients [Ajilore et al., 2014; Lim et al., 2013; Singh et al., 2013]. However, this methodology to date has only been used to characterize the network alterations of a group of patients. Thus there is a need to develop and utilize approaches that characterize inter‐subject variability in specific alterations, brain‐behavior relationships and diagnostic utility of neuroanatomic network abnormalities [Alexander‐Bloch et al., 2013; Evans, 2013].
More recently, new methods have been developed to identify morphological brain networks in individual patients based on structural MRI data [Raj et al., 2010; Tijms et al., 2012], which provide an opportunity to examine associations of morphological network metrics with behavioral characteristics in depression. Specifically, Tijms et al. 2012 proposed a similarity‐based method in accordance with axon tension theory where axon‐connectivity of cortical areas has an influence on morphology [Van Essen, 1997]. This approach has been used to develop structural networks in previous studies [Evans, 2013]. This method takes not only local morphology into account (e.g., volume or thickness) but also folding structure of the cortex, which can improve characterization of morphological network topology [Alexander‐Bloch et al., 2013; Evans, 2013]. In these morphological graphs, the nodes represent small cortical areas and edges represent statistical similarities in regional grey matter volume (GMV) between these nodal regions. This method has established test–retest reliability [Tijms et al., 2012] and has been applied in studies of Alzheimer's disease [Tijms et al., 2013, 2014].
Using this similarity‐based, single‐subject morphological network method, we investigated the topological organizations of brain networks in individual patients with and, critically, whether such disruptions are associated with symptom severity and can differentiate MDD patients from healthy individuals. We conducted this study with first‐episode drug‐naive MDD patients and matched healthy controls (HCs) to reduce impact of medications and course of illness on measures. After constructing individual morphological cortical networks, graph‐based models were employed to characterize and compare their topology. We hypothesized that MDD patients would show disrupted topological architectures in single‐subject morphological networks and that these disruptions would be associated with the severity of depressive symptoms.
MATERIALS AND METHODS
Participants
A total of 66 right handed adult Chinese Han subjects were included in the present study, comprising 33 first‐episode drug‐naive MDD patients and 33 age‐, gender‐, and education‐matched HCs (see Table 1). Patients were recruited from consecutive admissions to the Department of Psychiatry of West China Hospital of Sichuan University, Chengdu, China. The diagnosis of depression was made by two psychiatrists using the Structured Clinical Interview for DSM Disorders (SCID) using DSM‐IV criteria [First et al., 1997]. The severity of depression was evaluated using the Hamilton Rating Scale for Depression (HAMD) [Williams, 1988] and the Clinical Global Impression scale [Guy, 1976]. Inclusion criteria included a first episode of MDD, having never received psychiatric treatment, a HAMD total score ≥18 on the day of MRI scanning, and a duration of depression of >2 weeks but <60 weeks. Exclusion criteria included age <18 or >60 years, history of psychosis, anxiety, or bipolar disorder, a history of major medical or neurological illness, cardiovascular disease, vasoactive medication, alcohol or drug abuse, pregnancy or nursing, and contraindications for MRI scanning. By recruiting treatment‐naive, first episode patients, we aimed to identify brain network alterations unrelated to chronic disease and treatment.
Table 1.
Demographics and clinical characteristics of study participants
| HCs (n = 33) | MDD (n = 33) | P‐value | |
|---|---|---|---|
| Age (years) | 18–59 (35.42 ± 11.64) | 18–60 (35.55 ± 12.00) | 0.967a |
| Gender (M/F) | 8/25 | 8/25 | – |
| Education (years) | 2–19 (12.03 ± 3.96) | 1–19 (11.82 ± 4.97) | 0.849a |
| Handedness (R/L) | 33/0 | 33/0 | – |
| Duration of illness (weeks) | NA | 2–60 (15.03 ± 13.87) | – |
| HAMD | NA | 18–34 (24.21 ± 4.88) | – |
| Onset age (years) | NA | 18–59 (35.21 ± 11.96) | – |
Data are presented as the range of minimum‐maximum (mean ± SD). HCs, healthy controls; MDD, major depressive disorder; M, male; F, female; R, right; L, left; HAMD, Hamilton Depression Rating Scale; NA, non‐applicable.
The P value was obtained by a two‐sample two‐tailed t test.
Healthy control subjects were recruited from the local area through poster advertisement, and screened using the SCID Non‐Patient Edition and the SCID for Personality Disorders. HCs had no current or lifetime diagnosis of any Axis I or II disorder or known family history of psychiatric illness. This study was approved by the local Medical Research Ethics Committee at West China Hospital and written informed consent was obtained from each participant.
Image Acquisition
MR scanning was performed on a 3.0 T MR scanner (EXCITE, GE Medical Systems, Milwaukee, WI, USA). Subjects were fitted with soft earplugs, positioned comfortably in the coil, and instructed to relax and remain still. Head movement was minimized with foam pads. High resolution three dimensional T1‐weighted images were acquired using a spoiled gradient recalled sequence with repetition time (TR) = 8.5 ms, echo time (TE) = 3.4 ms, flip angle (FA) = 12°, 156 axial slices with slice thickness = 1 mm, field of view (FOV) = 24 × 24 cm2, and data matrix = 256 × 256.
Data Preprocessing
Structural images were processed using the VBM8 toolbox (http://dbm.neuro.uni-jena.de/vbm8) for Statistical Parametric Mapping software (SPM8, RRID: SCR_007037; http://www.fil.ion.ucl.ac.uk/spm/software/SPM8/). Briefly, individual structural images were first segmented into grey matter (GM), white matter and cerebrospinal fluid based on an adaptive Maximum A Posteriori technique. The resultant GM maps, the focus of this research, were normalized to Montreal Neurological Institute (MNI) space using a high‐dimensional “DARTEL” approach, non‐linearly modulated to compensate for spatial normalization effects, and resampled to 2 × 2 × 2 mm3 voxels. Notably, the non‐linear modulation essentially corrected for inter‐subject variability in brain size. No spatial smoothing was performed in order to avoid inducing artifactual signal overlap among spatially adjacent regions.
Construction of Single‐Subject Morphological Networks
We used a completely automated and data driven method [Tijms et al., 2012] to construct large‐scale single‐subject morphological networks based on structural MRI images.
As shown in Figure 1, the graph's nodes were defined as small regions of interest (i.e., cubes) consisting of 3 × 3 × 3 voxels in the MNI space. Prior to this parcellation, a GM brain mask was used to exclude voxels with near zero variance in GMV across participants. A total of 2,456 cubes were then automatically extracted within this mask. The three‐dimensional structure of the cortex is kept intact within these cubes [Kiselev et al., 2003] and so both geometrical information and the GMV values could remain linked to the voxels. A standard Pearson correlation coefficient was used to measure the similarity between any pair of nodes in the graph. Considering that the cortex is a curved object, two similar cubes could be located at an angle to each other, which could decrease their similarity value. Therefore, each seed cube was rotated by an angle θ with multiples of 45° and reflection over all axes to find the maximum similarity value with the target cube [Tijms et al., 2012]. Finally, a 2,456 × 2,456 similarity matrix of correlation values was generated for each participant (Supporting Information Fig. S1). In the present study, following convention, connectivity was defined as statistical similarity of regional GMV between regions, which might exist with indirect rather than direct axonal connectivity. The definitions and mathematical methods for computing similarity measures have been described previously [Tijms et al., 2012] and are discussed in Supporting Information Methods.
Figure 1.
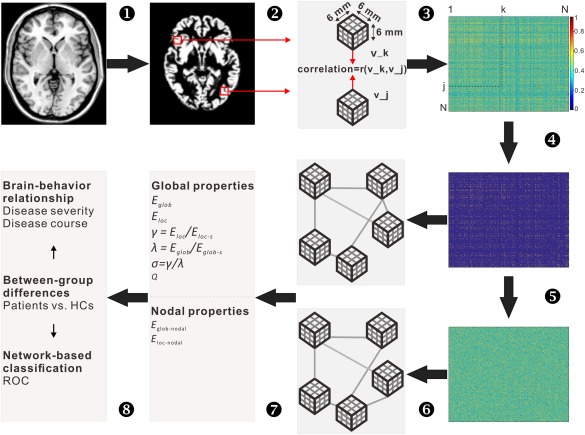
Schematic pipeline to the extraction of single subject morphological networks. T1 images (1) were first segmented into gray matter maps (2), from which a series of cubes (3 × 3 × 3 voxels) were extracted. The similarity between any pair of cubes was then calculated according to the method proposed by Tijms et al. [2012], and therefore generating a similarity matrix for each participant (3). The similarity matrices subsequently underwent a thresholding procedure to exclude non‐significant connections (P > 0.05, Bonferroni corrected) (4). For each of the resultant weighted networks, 100 random networks were generated that preserved the same number of nodes and edges and the same degree distribution to the real brain networks (5). The real and random networks can be represented as graphs (6), whose topological organization was further analyzed at both global and nodal levels (7). Finally, between‐group differences in network measures, brain‐behavior relationship and network‐based classification were performed (8). Abbreviation: mm, millimeter; E glob, global efficiency; E loc, local efficiency; E glob‐s, global efficiency of random networks; E loc‐s, local efficiency of random networks; E glob‐nodal, nodal global efficiency; E loc‐nodal, nodal local efficiency; γ, gamma; λ, Lambda; σ, small world; Q, modularity; vs., versus; HCs, healthy controls; ROC, receiver operating characteristic curves. [Color figure can be viewed at http://wileyonlinelibrary.com]
We tested the normality of GMV values within each cube for each participant with a Kolmogorov–Smirnov (Lilliefors) test. We found that among all the cubes (2,456 cubes/subject × 33 subjects = 81,048 cubes) for each group, 25% (20,636) for the healthy controls, and 26% (21,437) for the MDD patients did not follow normal distributions with respect to their GMVs. We further re‐computed individual inter‐cube similarity matrices using a non‐parametric Spearman correlation and compared them with the original Pearson‐based matrices after a Fisher's r‐to‐z transformation, and found highly similar connectivity patterns for both the MDD and HC groups as reported with the mean Pearson correlation coefficient across participants (0.96 ± 0.002 and 0.96 ± 0.002, respectively). Thus, our findings appeared to be robust to the degree of nonlinear distributions in our data.
Network Analysis
Thresholding
We employed a significance‐based procedure to convert the correlation matrices derived above to symmetrical and weighted networks. This is an effective method for excluding confounding effects of spurious correlations on network characterization [Toppi et al., 2012]. Specifically, a threshold of P < 0.05 (Bonferroni correction across connections) was used such that only correlation elements which survived this statistical threshold were retained, as in previous brain network studies [Bassett et al., 2011; Wang et al., 2013].
We used a weighted network model to construct a morphological brain network for each study participant. Weights were defined as the Pearson correlation coefficients between two cubes with respect to their regional GMV values. Compared with a binary network model whereby a connection is either present or absent, a weighted network analysis characterizes network topology more precisely to detect subtle network topological changes by taking the connectivity strength in consideration [Barrat et al., 2004; Rubinov and Sporns, 2010].
Network measures
We employed the global efficiency and local efficiency indices to characterize topological organization of the whole brain network and each network node respectively. Efficiency is a biologically relevant metric that characterizes brain networks from the view of parallel information flow. Specifically, for a given node, global efficiency (E glob) measures the capacity of information propagation of the node with all other nodes in a network, and local efficiency (E loc) measures the capacity for information exchange among the node's direct neighbors. All nodal local/global efficiency values were averaged to assess information propagation/exchange within the whole brain network [Latora and Marchiori, 2003; Newman, 2006]. To determine whether whole brain networks are non‐randomly organized into small‐worldness [Watts and Strogatz, 1998], we also calculated normalized global efficiency (λ) and normalized local efficiency (γ). This was achieved by dividing individual E glob and E loc values to their corresponding mean of 100 random networks, which preserved the same number of nodes and edges and the same degree distribution to real brain networks [Maslov and Sneppen, 2002; Sporns and Zwi, 2004]. Typically, a small‐world network shows γ > 1 and λ ≈ 1 [Humphries and Gurney, 2008; Watts and Strogatz, 1998]. In addition, we also calculated the modularity (Q), which reflects the extent to which nodes can be divided into subsets with dense connections within them but sparse connections between them [Newman, 2006]. According to Rubinov and Sporns 2010, global efficiency reflects integration and local efficiency and modularity reflects segregation of a network [Meunier et al., 2010; Rubinov and Sporns, 2010]. For detailed uses and interpretations of these network measures, see reference [Rubinov and Sporns, 2010] and Supporting Information Methods in Supporting Information.
Statistical Analysis
Between‐group differences
A non‐parametric permutation test was performed to test the statistical significance of between‐group differences in global and nodal topological properties. Briefly, the between‐group difference in the mean value was first computed for each network metric. Next we reallocated all the values randomly into two groups and recomputed the mean difference between them to test the null hypothesis that the observed group differences could occur by chance. This randomization procedure was repeated 10,000 times, and the 95th percentile points of each distribution were used as the critical values for a one‐tailed test (P < 0.05). Age, gender, and education duration were treated as covariates.
Altered inter‐cube similarity
We employed a network based statistic (NBS) approach to identify altered inter‐cube connectivity in patients compared with HCs [Zalesky et al., 2010]. The NBS is a robust approach for controlling family‐wise error rate when performing a large number of comparisons for each connection comprising a graph, similar to conventional cluster statistics. For details of the analysis, see Supporting Information Methods in Supporting Information.
Brain–behavior relationships
To examine associations with illness duration and symptom severity, we took brain network measures showing significant between‐group differences and performed multiple linear regression analyses with HAMD scores and illness duration in the MDD group, controlling for age, gender, and education.
Network‐based classification
For the network classification analysis, receiver operating characteristic curves (ROC) were plotted to determine whether graph‐based network metrics might serve as biomarkers for differentiating patients with MDD from HCs. The ROC analysis was conducted for each metric (i.e., one‐dimensional feature) showing significant between‐group differences. For each metric, a range of thresholds were used to classify each participant into the MDD or HC group. A preliminary linear discriminant analysis was performed as in previous brain network studies using these parameters to provide an overall estimate of group separation given the alterations seen in univariate studies (see Supporting Information Materials).
RESULTS
MDD‐Related Alterations in Global Topological Properties
As shown in Figure 2, both the MDD and HC groups demonstrated a small world topology in their single‐subject cortical morphological networks (i.e., γ > 1 and λ ≈ 1). However, the MDD group showed significantly lower normalized global efficiency λ (P = 0.036) and significantly higher modularity (P = 0.011), and trends toward increased normalized local efficiency in their networks. In this analysis, individual weight sums that reflect the extent of overall connectivity for a network were treated as a covariate to control for their confounding effects on between‐group comparisons of network topology. We also re‐analyzed the data without including individual weight sums as a covariate, and similar findings were observed (λ, P = 0.069; modularity, P = 0.010).
Figure 2.
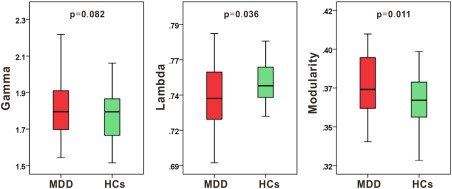
Differences in global topological properties of structural brain networks between patients with major depressive disorder (MDD) and healthy controls (HCs). MDD patients showed significantly smaller normalized global efficiency λ and significantly higher modularity, but marginally significant higher normalized local efficiency γ compared with HC group, when varied individual weight sum was controlled as a covariate. [Color figure can be viewed at http://wileyonlinelibrary.com]
Between‐Group Differences in Nodal Efficiency
Significant differences were found in nodal local efficiency between MDD and HCs. As showed in Figure 3 and Supporting Information Table S1, MDD patients had lower nodal local efficiency predominantly in default‐mode regions including left hippocampus and parahippocampal gyrus, left precuneus, right superior parietal lobule, and left middle occipital gyrus compared with HC. Additionally, higher nodal local efficiency was also observed in the left insula, bilateral calcarine and posterior cingulate cortex, right cerebellum posterior lobe, and left cerebellum anterior lobe [P < 0.05, false discovery rate (FDR) corrected]. In contrast to the nodal local efficiency analyses, there was no significant difference in nodal global efficiency between the two groups (P > 0.05) indicating that nodal efficiency alterations were restricted to specific regions.
Figure 3.
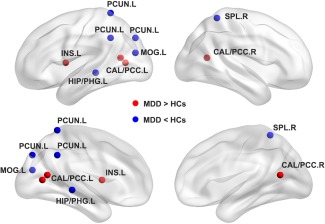
Location of significant differences in local efficiency between patients with major depressive disorder (MDD) and healthy controls (HCs). The differences are mainly in the insula and posterior default model network (DMN). R, right; L, left; CAL/PCC.L, left calcarine and posterior cingulate cortex; CAL/PCC.R, right calcarine and posterior cingulate cortex; HIP/PHG.L, left hippocampus and parahippocampal gyrus; INS.L, left insula; MOG.L, left middle occipital gyrus; PCUN.L, left precuneus; SPL.R, right superior parietal lobule. Bilateral cerebellum posterior lobes are not showed owing to the template. [Color figure can be viewed at http://wileyonlinelibrary.com]
MDD‐Related Alterations in Morphological Connectivity
Using the network based statistic (NBS) method to evaluate specific network connections impacted in MDD, we identified two brain subnetworks that exhibited higher morphological connectivity strength in patients with MDD compared with HCs (Fig. 4, Supporting Information Table S2). One subnetwork consisted of 10 nodes and 10 connections that involved cerebellum‐subcortical‐cortical structures (subnetwork 1, P = 0.012, corrected), including the left inferior temporal gyrus, left superior temporal gyrus, bilateral thalamus, right putamen, and bilateral cerebellum. The other subnetwork included 7 regions and 6 connections that were primarily related to the left hippocampus and bilateral thalamus (subnetwork 2, P = 0.045, corrected).
Figure 4.
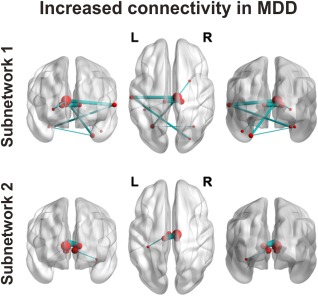
Summary of significant networks that characterize patients with major depressive disorder (MDD) using network‐based statistical analysis. Two distinct subnetworks are represented: the subnetwork depicted at the top (subnetwork 1) consists of 10 nodes and 10 edges and primarily involves neocortex, striatum, thalamus and cerebellum; the subnetwork depicted at the bottom (subnetwork 2) contains a central cluster of seven nodes involving thalamus and hippocampus. L, left; R, right. [Color figure can be viewed at http://wileyonlinelibrary.com]
Relationships Between Topological Properties and Clinical Variables
As shown in Figure 5A, normalized global efficiency (λ) was negatively correlated with HAMD scores in the depressed patients (r = −0.503, P = 0.004). There were no significant relationships between other measures examined and HAMD scores or disease duration (P > 0.05).
Figure 5.
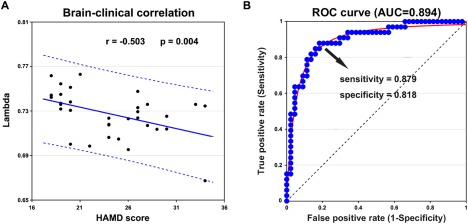
Disease state prediction and diagnostic classification of morphological cortical topology in the patients with major depressive disorder (MDD). (A) Scatter plots of rank‐transformed scores on the Hamilton Depression Scale (HAMD) with normalized global efficiency λ. A significant negative correlation was found between HAMD scores and λ for the MDD group. (B) The receiver operating characteristic curves (ROC) pattern classification analysis shows a MDD‐control discrimination sensitivity and specificity (area under the curve [AUC]) of the NBS‐based connected network. NBS, network based statistic. [Color figure can be viewed at http://wileyonlinelibrary.com]
Sensitivity and Specificity of Network Properties in Differentiating MDD Patients from HCs
Among the network measures (normalized global efficiency, normalized local efficiency, modularity, and NBS‐based subnetworks) showing MDD‐related alterations, the average connectivity strength of the NBS‐based subnetworks distinguished MDD patients from HCs (area under curve = 0.894, P < 10−3; 95% CI = [0.815 0.972]) with a sensitivity of 87.9% and a specificity of 81.8% (Fig. 5B). That is, 29 out of the 33 patients with MDD and 27 out of the 33 HCs were correctly classified. Further analysis showed that both the NBS‐based subnetworks distinguished the MDD patients from HCs with similar accuracies (subnetwork 1: area under curve = 0.825, P < 10−3; 95% CI = [0.724 0.972]; subnetwork 2: area under curve = 0.841, P < 10−3; 95% CI = [0.743 0.938]). All the other measures provided less robust group discrimination (all areas under the curve of <0.7).
DISCUSSION
Using a single‐subject, similarity‐based brain morphological network delineation method, the current study demonstrated that treatment‐naïve, first‐episode MDD patients had lower global efficiency and higher modularity of neuroanatomic brain networks. A pattern of region‐specific alterations that were higher and lower in local nodal efficiency was also seen in MDD. Two networks with higher morphological connectivity strength were identified, one widespread involving regions of neocortex, striatum, thalamus, and cerebellum, and one specifically involving thalamus and hippocampus.
The use of a single subject characterization of brain network morphology in the present study extends previous group‐based morphological brain network analyses by showing for the first time their relation to depression severity, and demonstrating that morphological alterations in brain networks differentiate individual MDD patients from healthy individuals with high sensitivity and specificity. Importantly, these findings were observed in first episode, treatment‐naïve patients free from potential confounding effects of chronic illness and pharmacological treatment.
MDD‐Associated Alterations in Global Network Properties
The human brain is organized into complex networks with a topological configuration (e.g., small‐world organization) permitting highly efficient segregated processing as well as highly integrated information processing [Bullmore and Sporns, 2012]. Despite having a common small‐world structure similar to HC subjects in general terms, MDD patients demonstrated significant alterations of global brain network organization. MDD patients showed a lower global efficiency even after controlling for individual network density, reflecting disrupted global integration of brain networks. This is consistent with previous structural brain network studies using functional [Meng et al., 2014] and diffusion MRI [Bai et al., 2012; Korgaonkar et al., 2014; Qin et al., 2014] and group‐level morphological covariance network studies [Ajilore et al., 2014] in MDD. We note however that opposite patterns have also been observed in MDD‐related network studies based on functional [Zhang et al., 2011] and diffusion MRI [Long et al., 2015] as well as EEG data [Leistedt et al., 2009; Sun et al., 2011], highlighting the need for multimodal research in future studies.
Different imaging modalities are sensitive to different mechanisms of network organization. Functional MRI‐derived networks characterize synchronized brain activity at a point in time while the structural networks measured in the present study may reflect more stable patterns of anatomical organization affected by heredity, experience‐related plasticity and mutually trophic reinforcement [Alexander‐Bloch et al., 2013; Evans, 2013]. Convergent disruptions of structural brain networks may underpin alterations seen in functional brain network studies, suggesting that functional changes may in part represent efforts to compensate for structural alterations. Alternatively, such changes might reflect the complexity of the disease and its progression, a possibility that needs to be clarified in future longitudinal studies.
We also observed higher modularity of brain networks in MDD patients compared with HCs. This is in line with a recent functional network study of medication‐free MDD patients [Ye et al., 2015], suggesting a potential structural basis for the functional alteration. Such higher modularity suggests more isolated subnetworks in MDD, with likely implications for reductions in the functional integrity of large scale neural networks and the complex behaviors they support. Previous functional network studies of MDD revealed no between‐group differences in modularity [Bohr et al., 2013] but a rearrangement of the community structure of neural network in MDD [Lord et al., 2012]. Modularity indicates an organization or natural segregation within a network [Meunier et al., 2010; Rubinov and Sporns, 2010]. Higher modularity within structural networks in MDD suggests that the illness may alter the fundamental design principle of intrinsically cohesive community network architecture by splitting larger scale modules into discrete components. This could represent a brain maturational alteration related to illness risk or a direct effect of the disease. Presumably this would indicate a breakdown in integration within specific functional networks in MDD, leading to a loss of intrinsic regulation, and coordination of brain subsystems needed for cognitive and affective processes.
MDD‐Associated Alterations in Regional Network Properties
In addition to global metrics, we also studied local and global efficiency at a nodal level to show the extent of information transmission capacity of nodes with their neighbors and all other nodes in networks. In general, both MDD and HC groups showed the highest nodal global efficiency (i.e., hubs) in similar cortical‐subcortical regions (e.g., medial prefrontal cortex, paracentral lobule, parieto‐occipital cortex, and thalamus) (see Supporting Information Results and Fig. S2 in Supporting Information), consistent with previous findings [Hagmann et al., 2008; Tomasi and Volkow, 2010].
Although no significant differences were detected in nodal global efficiency between patients and HCs, we found altered nodal local efficiency mainly in the posterior DMN, in line with previous group‐level brain structural [Korgaonkar et al., 2014] and functional [Zeng et al., 2012; Zhang et al., 2011] network studies of MDD. The posterior DMN is involved in integration between new and previously learned information, and the processing of self‐relevant information [Buckner et al., 2008; Raichle et al., 2001]. Here, using an individual‐level morphological network method, our findings provide new evidence for the view of posterior DMN abnormalities as a core brain morphometric feature of MDD [Kaiser et al., 2015; Korgaonkar et al., 2014]. Notably, opposite patterns of MDD‐related alterations were observed within the posterior DMN. Lateral DMN regions, such as the precuneus, showed lower efficiency, whereas the PCC, a core component of autobiographical associated network, showed higher efficiency in MDD. The spatial dissociation of MDD‐related abnormalities in the posterior DMN has previously been observed [Sambataro et al., 2014; Zhu et al., 2012]. Our findings are thus consistent with previous studies and may represent a structural substrate for spatially different abnormalities of the posterior DMN regions and their interactions in MDD [Li et al., 2013; Sambataro et al., 2014]. Further studies are needed in the future to help understand the differential involvements of posterior DMN in MDD.
We also found higher nodal local efficiency in left insula in MDD patients. In addition to the functions of polymodal sensory integration [Critchley et al., 2004] and monitoring of internal states [Craig, 2009], the insula also plays a critical functional role in switching between external and internal focus of cognitive control [Sridharan et al., 2008]. Abnormalities in insula structure and function have been increasingly reported in MDD [Hamilton et al., 2012]. Taken together, these morphometric alterations in posterior DMN and insula might contribute to alterations in the synchronization of information processing related to the external world and internal states in patients with MDD [Kaiser et al., 2015].
MDD patients also showed lower nodal local efficiency in occipital regions, consistent with previous reports of MDD‐associated abnormalities in visual cortex [Zeng et al., 2012]. This may be related to impairments of selective attention to emotional and non‐emotional stimuli [Desseilles et al., 2009; Furey et al., 2013]. In addition, higher nodal local efficiency was found in cerebellar regions in MDD patients. The cerebellum is involved not only in motor control but also emotional and cognitive processing through its thalamo‐cortical projections [Bostan et al., 2013; Schmahmann and Caplan, 2006]. Thus our current findings emphasize the impact of altered information transmission among the cerebellum with neighbor regions on the deficits in regulating mood and cognitive function [Bostan et al., 2013; Sweeney et al., 1998].
MDD‐Related Alterations in Morphological Connectivity
In the NBS analyses, we found higher morphological indices of connectivity strength both in neocortex‐striatum‐thalamus‐cerebellum circuitry and also in thalamo‐hippocampal circuitry. These findings are in line with previous studies showing higher white matter connection strength among cortical‐subcortical networks in MDD [Long et al., 2015] and GM alteration in related regions [Arnone et al., 2016; Wise et al., 2016]. Thalamocortical functional dysconnectivity was also observed in psychotic disorders and related to cognitive impairment [Lui et al., 2014; Woodward and Heckers, 2015]. The neocortex‐striatum‐thalamus‐cerebellum circuitry may contribute to altered regulation of emotion and cognition in mood disorders [Bostan et al., 2013; Buckner, 2013; Guo et al., 2015; Price and Drevets, 2012], while the thalamo‐hippocampal circuitry might be more specifically related to memory disturbances in depression [Price and Drevets, 2009; Stillova et al., 2015].
In contrast, a previous DTI study reported lower structural connections within the DMN and frontal‐limbic networks in a large sample of depressed patients with an average of 12 episodes [Korgaonkar et al., 2014]. Besides the use of different network node and edge definitions, varied clinical characteristics of study samples may account for the inconsistent findings as theirs was a chronically ill cohort with differences in number of prior depression episodes and medication history. Significant normalization effects of antidepressant treatment on interregional functional connectivity have been reported [Li et al., 2013; Perrin et al., 2012], but their relations to anatomic patterns remains to be clarified.
Clinical Relevance of Alterations in Single‐Subject Morphological Networks
One new finding from the present study was that lower global efficiency of single‐subject based morphological networks was associated with higher severity of depressive symptoms in MDD. To our knowledge, this is the first demonstration that global anatomic brain network features are associated with illness severity. Moreover, the higher morphological connectivity strength seen in individual patients with MDD robustly differentiated patients from controls. These findings suggest the potential of single‐subject based morphological network analysis of MDD as a promising biomarker for examining brain network alterations in MDD in relation to clinical diagnosis and illness severity.
Limitations
Several methodological issues and limitations should be noted. First, studies are needed to determine whether the novel pattern of alterations we observed can be replicated, and whether they change over the course of drug treatment, acute clinical stabilization and over the longer term course of the illness. Second, while the construction of similarity‐based individual structural networks has been successfully applied to other neuropsychiatric disorders [Tijms et al., 2013, 2014], the biological significance of these network alterations needs to be more fully understood. It has been recently documented that similarity‐based patterns of cortical morphology may arise from systematic functional co‐activation and/or mutually trophic reinforcement, genetic mediated brain maturation, as well as experience‐related plasticity [Alexander‐Bloch et al., 2013; Evans, 2013]. We need clarification as to which of these factors is related to alterations we demonstrated in MDD. Third, the sample size in the current study is not large. Although our sample has the important advantage of comprising a relatively rare group of treatment‐naive first episode patients, our study may have lacked sufficient statistical power to detect some subtle effects related to MDD. Fourth, regarding to the classification analysis, we employed the ROC method, which analyzed one‐dimensional feature independently. Multivariate based prediction analysis such as combining multidimensional features using the support vector machine method, may with larger samples provide greater clarity about how patterns of brain anatomic alteration occur in MDD. Fifth, we note that the VBM procedure we used is susceptible to false positive results [Scarpazza et al., 2015], and future studies are needed to extend our findings using other surface‐based morphological measures, such as cortical thickness or surface area to validate and clarify our findings. Finally, our MDD patients had relatively severe depressive symptoms and did not meet criteria for an anxiety disorder. Therefore, future work is need to evaluate less severe illness variants and patients with anxiety comorbidities, and other clinical factors such as responsiveness to treatments and whether these network alterations are disease specific to a degree that they can distinguish MDD from other related conditions, such as bipolar and anxiety disorders. It will also be important to examine relationships between the structural findings we have observed and similar metrics obtained in functional MRI and DTI studies.
CONCLUSIONS
Using a single‐subject grey matter morphological network approach, we demonstrated abnormal network organization in individuals with treatment‐naive, first‐episode MDD. Aspects of this alteration were associated with symptom severity and differentiated patients from healthy individuals with high sensitivity and specificity. These findings suggest promise for individual morphological network analysis in providing a new strategy for investigating brain morphometry changes in MDD and for developing reliable objective biomarkers for brain changes related to depression.
FINANCIAL DISCLOSURES
Dr. Sweeney has consulted for Takeda Pharmaceutical Co. No other disclosures were reported.
Supporting information
Supporting Information
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (Grant Nos. 81401398, 81621003, 81030027, 81227002, 81220108013, 31530032, 81301284, 81671764), Program for Changjiang Scholars and Innovative Research Team in University (PCSIRT, Grant No. IRT16R52) of China and Chinese Postdoctoral Science Foundation (Grant Nos. 2013M530401). Dr. Gong would also like to acknowledge the support from his Changjiang Scholar Professorship Award (Award No. T2014190) of China and American CMB Distinguished Professorship Award (Award No. F510000/G16916411) administered by the Institute of International Education, USA. The authors would also like to thank Dr. Betty M. Tijms for kindly providing the code to generate individual cortical structure networks.
REFERENCES
- Ajilore O, Lamar M, Leow A, Zhang A, Yang S, Kumar A (2014): Graph theory analysis of cortical‐subcortical networks in late‐life depression. Am J Geriatr Psychiatry 22:195–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander‐Bloch A, Giedd JN, Bullmore E (2013): Imaging structural co‐variance between human brain regions. Nat Rev Neurosci 14:322–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnone D, Job D, Selvaraj S, Abe O, Amico F, Cheng Y, Colloby SJ, O'Brien JT, Frodl T, Gotlib IH, Ham BJ, Kim MJ, Koolschijn PC, Perico CA, Salvadore G, Thomas AJ, Van Tol MJ, van der Wee NJ, Veltman DJ, Wagner G, McIntosh AM (2016): Computational meta‐analysis of statistical parametric maps in major depression. Hum Brain Mapp 37:1393–1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae JN, MacFall JR, Krishnan KRR, Payne ME, Steffens DC, Taylor WD (2006): Dorsolateral prefrontal cortex and anterior cingulate cortex white matter alterations in late‐life depression. Biol Psychiatry 60:1356–1363. [DOI] [PubMed] [Google Scholar]
- Bai F, Shu N, Yuan Y, Shi Y, Yu H, Wu D, Wang J, Xia M, He Y, Zhang Z (2012): Topologically convergent and divergent structural connectivity patterns between patients with remitted geriatric depression and amnestic mild cognitive impairment. J Neurosci 32:4307–4318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrat A, Barthelemy M, Pastor‐Satorras R, Vespignani A (2004): The architecture of complex weighted networks. Proc Natl Acad Sci U S A 101:3747–3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett DS, Bullmore E, Verchinski BA, Mattay VS, Weinberger DR, Meyer‐Lindenberg A (2008): Hierarchical organization of human cortical networks in health and schizophrenia. J Neurosci 28:9239–9248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett DS, Brown JA, Deshpande V, Carlson JM, Grafton ST (2011): Conserved and variable architecture of human white matter connectivity. Neuroimage 54:1262–1279. [DOI] [PubMed] [Google Scholar]
- Bohr IJ, Kenny E, Blamire A, O'Brien JT, Thomas AJ, Richardson J, Kaiser M (2013): Resting‐state functional connectivity in late‐life depression: Higher global connectivity and more long distance connections. Front Psychiatry 3:116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bostan AC, Dum RP, Strick PL (2013): Cerebellar networks with the cerebral cortex and basal ganglia. Trends Cogn Sci 17:241–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckner RL (2013): The cerebellum and cognitive function: 25 years of insight from anatomy and neuroimaging. Neuron 80:807–815. [DOI] [PubMed] [Google Scholar]
- Buckner RL, Andrews‐Hanna JR, Schacter DL (2008): The brain's default network: Anatomy, function, and relevance to disease. Ann N Y Acad Sci 1124:1–38. [DOI] [PubMed] [Google Scholar]
- Bullmore E, Sporns O (2012): The economy of brain network organization. Nat Rev Neurosci 13:336–349. [DOI] [PubMed] [Google Scholar]
- Cisler JM, James GA, Tripathi S, Mletzko T, Heim C, Hu XP, Mayberg HS, Nemeroff CB, Kilts CD (2013): Differential functional connectivity within an emotion regulation neural network among individuals resilient and susceptible to the depressogenic effects of early life stress. Psychol Med 43:507–518. [DOI] [PubMed] [Google Scholar]
- Connolly CG, Wu J, Ho TC, Hoeft F, Wolkowitz O, Eisendrath S, Frank G, Hendren R, Max JE, Paulus MP, Tapert SF, Banerjee D, Simmons AN, Yang TT (2013): Resting‐state functional connectivity of subgenual anterior cingulate cortex in depressed adolescents. Biol Psychiatry 74:898–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig AD (2009): How do you feel—now? The anterior insula and human awareness. Nat Rev Neurosci 10:59–70. [DOI] [PubMed] [Google Scholar]
- Critchley HD, Wiens S, Rotshtein P, Ohman A, Dolan RJ (2004): Neural systems supporting interoceptive awareness. Nat Neurosci 7:189–195. [DOI] [PubMed] [Google Scholar]
- Desseilles M, Balteau E, Sterpenich V, Dang‐Vu TT, Darsaud A, Vandewalle G, Albouy G, Salmon E, Peters F, Schmidt C, Schabus M, Gais S, Degueldre C, Phillips C, Luxen A, Ansseau M, Maquet P, Schwartz S (2009): Abnormal neural filtering of irrelevant visual information in depression. J Neurosci 29:1395–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drevets WC, Ongur D, Price JL (1998): Neuroimaging abnormalities in the subgenual prefrontal cortex: Implications for the pathophysiology of familial mood disorders. Mol Psychiatry 3:220–226. [DOI] [PubMed] [Google Scholar]
- Evans AC (2013): Networks of anatomical covariance. Neuroimage 80:489–504. [DOI] [PubMed] [Google Scholar]
- First M, Spitzer R, Gibbon M, Williams J (1997): Structured Clinical Interview for DSM‐IV Axis I Disorders. Washington, DC: American Psychiatric Publishing. [Google Scholar]
- Fitzgerald PB, Oxley TJ, Laird AR, Kulkarni J, Egan GF, Daskalakis ZJ (2006): An analysis of functional neuroimaging studies of dorsolateral prefrontal cortical activity in depression. Psychiat Res‐Neuroim 148:33–45. [DOI] [PubMed] [Google Scholar]
- Furey ML, Drevets WC, Hoffman EM, Frankel E, Speer AM, Zarate CA Jr (2013): Potential of pretreatment neural activity in the visual cortex during emotional processing to predict treatment response to scopolamine in major depressive disorder. JAMA Psychiatry 70:280–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Q, He Y (2015): Depression, neuroimaging and connectomics: A selective overview. Biol Psychiatry 77:223–235. [DOI] [PubMed] [Google Scholar]
- Greicius MD, Flores BH, Menon V, Glover GH, Solvason HB, Kenna H, Reiss AL, Schatzberg AF (2007): Resting‐state functional connectivity in major depression: Abnormally increased contributions from subgenual cingulate cortex and thalamus. Biol Psychiatry 62:429–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groenewold NA, Opmeer EM, de Jonge P, Aleman A, Costafreda SG (2013): Emotional valence modulates brain functional abnormalities in depression: Evidence from a meta‐analysis of fMRI studies. Neurosci Biobehav Rev 37:152–163. [DOI] [PubMed] [Google Scholar]
- Guo H, Cao X, Liu Z, Li H, Chen J, Zhang K (2012): Machine learning classifier using abnormal brain network topological metrics in major depressive disorder. Neuroreport 23:1006–1011. [DOI] [PubMed] [Google Scholar]
- Guo WB, Liu F, Zhang ZK, Liu JR, Yu MY, Zhang J, Xiao CQ, Zhao JP (2015): Unidirectionally affected causal connectivity of cortico‐limbic‐cerebellar circuit by structural deficits in drug‐naive major depressive disorder. J Affect Disord 172:410–416. [DOI] [PubMed] [Google Scholar]
- Guy W (1976): ECDEU Assessment Manual for Psychopharmacology. Rockville, MD: National Institute of Mental Health. [Google Scholar]
- Hagmann P, Cammoun L, Gigandet X, Meuli R, Honey CJ, Wedeen VJ, Sporns O (2008): Mapping the structural core of human cerebral cortex. PLoS Biol 6:e159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton JP, Etkin A, Furman DJ, Lemus MG, Johnson RF, Gotlib IH (2012): Functional neuroimaging of major depressive disorder: A meta‐analysis and new integration of base line activation and neural response data. Am J Psychiatry 169:693–703. [DOI] [PubMed] [Google Scholar]
- He Y, Chen ZJ, Evans AC (2007): Small‐world anatomical networks in the human brain revealed by cortical thickness from MRI. Cereb Cortex 17:2407–2419. [DOI] [PubMed] [Google Scholar]
- Humphries MD, Gurney K (2008): Network 'small‐world‐ness': a quantitative method for determining canonical network equivalence. PLoS ONE 3:e0002051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin C, Gao C, Chen C, Ma S, Netra R, Wang Y, Zhang M, Li D (2011): A preliminary study of the dysregulation of the resting networks in first‐episode medication‐naive adolescent depression. Neurosci Lett 503:105–109. [DOI] [PubMed] [Google Scholar]
- Kaiser RH, Andrews‐Hanna JR, Wager TD, Pizzagalli DA (2015): Large‐scale network dysfunction in major depressive disorder: A meta‐analysis of resting‐state functional connectivity. JAMA Psychiatry 72:603–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiselev VG, Hahn KR, Auer DP (2003): Is the brain cortex a fractal?. Neuroimage 20:1765–1774. [DOI] [PubMed] [Google Scholar]
- Korgaonkar MS, Fornito A, Williams LM, Grieve SM (2014): Abnormal structural networks characterize major depressive disorder: A connectome analysis. Biol Psychiatry 76:567–574. [DOI] [PubMed] [Google Scholar]
- Krishnan KR, McDonald WM, Escalona PR, Doraiswamy PM, Na C, Husain MM, Figiel GS, Boyko OB, Ellinwood EH, Nemeroff CB (1992): Magnetic resonance imaging of the caudate nuclei in depression. Preliminary observations. Arch Gen Psychiatry 49:553. [DOI] [PubMed] [Google Scholar]
- Latora V, Marchiori M (2003): Economic small‐world behavior in weighted networks. Eur Phys J B 32:249–263. [Google Scholar]
- Leistedt SJ, Coumans N, Dumont M, Lanquart JP, Stam CJ, Linkowski P (2009): Altered sleep brain functional connectivity in acutely depressed patients. Hum Brain Mapp 30:2207–2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Liu L, Friston KJ, Shen H, Wang L, Zeng LL, Hu D (2013): A treatment‐resistant default mode subnetwork in major depression. Biol Psychiatry 74:48–54. [DOI] [PubMed] [Google Scholar]
- Liao Y, Huang X, Wu Q, Yang C, Kuang W, Du M, Lui S, Yue Q, Chan RC, Kemp GJ, Gong Q (2013): Is depression a disconnection syndrome? Meta‐analysis of diffusion tensor imaging studies in patients with MDD. J Psychiatry Neurosci 38:49–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim HK, Jung WS, Aizenstein HJ (2013): Aberrant topographical organization in gray matter structural network in late life depression: A graph theoretical analysis. Int Psychogeriatr 25:1929–1940. [DOI] [PubMed] [Google Scholar]
- Long Z, Duan X, Wang Y, Liu F, Zeng L, Zhao JP, Chen H (2015): Disrupted structural connectivity network in treatment‐naive depression. Prog Neuropsychopharmacol Biol Psychiatry 56:18–26. [DOI] [PubMed] [Google Scholar]
- Lord A, Horn D, Breakspear M, Walter M (2012): Changes in community structure of resting state functional connectivity in unipolar depression. PLoS ONE 7:e41282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lui S, Parkes LM, Huang X, Zou K, Chan RC, Yang H, Zou L, Li D, Tang H, Zhang T, Li X, Wei Y, Chen L, Sun X, Kemp GJ, Gong Q (2009): Depressive disorders: Focally altered cerebral perfusion measured with arterial spin‐labeling MR imaging. Radiology 251:476–484. [DOI] [PubMed] [Google Scholar]
- Lui S, XJ Zhou, J Sweeney, Q Gong (2016): Psychoradiology: The Frontier of Neuroimaging in Psychiatry. Radiology 281:357–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lui S, Wu Q, Qiu L, Yang X, Kuang W, Chan RC, Huang X, Kemp GJ, Mechelli A, Gong Q (2011): Resting‐state functional connectivity in treatment‐resistant depression. Am J Psychiatry 168:642–648. [DOI] [PubMed] [Google Scholar]
- Lui S, Yao L, Xiao Y, Keedy SK, Reilly JL, Keefe RS, Tamminga CA, Keshavan MS, Pearlson GD, Gong Q, Sweeney JA (2014): Resting‐state brain function in schizophrenia and psychotic bipolar probands and their first‐degree relatives. Psychol Med 45:97–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mah L, Zarate CA, Jr , Singh J, Duan YF, Luckenbaugh DA, Manji HK, Drevets WC (2007): Regional cerebral glucose metabolic abnormalities in bipolar II depression. Biol Psychiatry 61:765–775. [DOI] [PubMed] [Google Scholar]
- Maslov S, Sneppen K (2002): Specificity and stability in topology of protein networks. Science 296:910–913. [DOI] [PubMed] [Google Scholar]
- Meng C, Brandl F, Tahmasian M, Shao J, Manoliu A, Scherr M, Schwerthoffer D, Bauml J, Forstl H, Zimmer C, Wohlschlager AM, Riedl V, Sorg C (2014): Aberrant topology of striatum's connectivity is associated with the number of episodes in depression. Brain 137:598–609. [DOI] [PubMed] [Google Scholar]
- Meunier D, Lambiotte R, Bullmore ET (2010): Modular and hierarchically modular organization of brain networks. Front Neurosci 4:200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman ME (2006): Finding community structure in networks using the eigenvectors of matrices. Phys Rev E Stat Nonlin Soft Matter Phys 74:036104. [DOI] [PubMed] [Google Scholar]
- Perrin JS, Merz S, Bennett DM, Currie J, Steele DJ, Reid IC, Schwarzbauer C (2012): Electroconvulsive therapy reduces frontal cortical connectivity in severe depressive disorder. Proc Natl Acad Sci U S A 109:5464–5468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pizzagalli DA (2011): Frontocingulate dysfunction in depression: Toward biomarkers of treatment response. Neuropsychopharmacology 36:183–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pizzagalli DA, Holmes AJ, Dillon DG, Goetz EL, Birk JL, Bogdan R, Dougherty DD, Iosifescu DV, Rauch SL, Fava M (2009): Reduced caudate and nucleus accumbens response to rewards in unmedicated individuals with major depressive disorder. Am J Psychiatry 166:702–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price JL, Drevets WC (2009): Neurocircuitry of mood disorders. Neuropsychopharmacology 35:192–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price JL, Drevets WC (2012): Neural circuits underlying the pathophysiology of mood disorders. Trends Cogn Sci 16:61–71. [DOI] [PubMed] [Google Scholar]
- Qin J, Wei M, Liu H, Yan R, Luo G, Yao Z, Lu Q (2014): Abnormal brain anatomical topological organization of the cognitive‐emotional and the frontoparietal circuitry in major depressive disorder. Magn Reson Med 72:1397–1407. [DOI] [PubMed] [Google Scholar]
- Raichle ME, MacLeod AM, Snyder AZ, Powers WJ, Gusnard DA, Shulman GL (2001): A default mode of brain function. Proc Natl Acad Sci U S A 98:676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj A, Mueller SG, Young K, Laxer KD, Weiner M (2010): Network‐level analysis of cortical thickness of the epileptic brain. Neuroimage 52:1302–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinov M, Sporns O (2010): Complex network measures of brain connectivity: Uses and interpretations. Neuroimage 52:1059–1069. [DOI] [PubMed] [Google Scholar]
- Sambataro F, Wolf ND, Pennuto M, Vasic N, Wolf RC (2014): Revisiting default mode network function in major depression: Evidence for disrupted subsystem connectivity. Psychol Med 44:2041–2051. [DOI] [PubMed] [Google Scholar]
- Scarpazza C, Tognin S, Frisciata S, Sartori G, Mechelli A (2015): False positive rates in Voxel‐based Morphometry studies of the human brain: Should we be worried?. Neurosci Biobehav Rev 52:49–55. [DOI] [PubMed] [Google Scholar]
- Schmaal L, Hibar DP, Samann PG, Hall GB, Baune BT, Jahanshad N, Cheung JW, van Erp TG, Bos D, Ikram MA, Vernooij MW, Niessen WJ, Tiemeier H, Hofman A, Wittfeld K, Grabe HJ, Janowitz D, Bulow R, Selonke M, Volzke H, Grotegerd D, Dannlowski U, Arolt V, Opel N, Heindel W, Kugel H, Hoehn D, Czisch M, Couvy‐Duchesne B, Renteria ME, Strike LT, Wright MJ, Mills NT, de Zubicaray GI, McMahon KL, Medland SE, Martin NG, Gillespie NA, Goya‐Maldonado R, Gruber O, Kramer B, Hatton SN, Lagopoulos J, Hickie IB, Frodl T, Carballedo A, Frey EM, van Velzen LS, Penninx BW, van Tol MJ, van der Wee NJ, Davey CG, Harrison BJ, Mwangi B, Cao B, Soares JC, Veer IM, Walter H, Schoepf D, Zurowski B, Konrad C, Schramm E, Normann C, Schnell K, Sacchet MD, Gotlib IH, MacQueen GM, Godlewska BR, Nickson T, McIntosh AM, Papmeyer M, Whalley HC, Hall J, Sussmann JE, Li M, Walter M, Aftanas L, Brack I, Bokhan NA, Thompson PM, Veltman DJ (2016a. ): Cortical abnormalities in adults and adolescents with major depression based on brain scans from 20 cohorts worldwide in the ENIGMA Major Depressive Disorder Working Group. Mol Psychiatry. http://www.nature.com/mp/journal/vaop/ncurrent/full/mp201660a.html. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmaal L, Veltman DJ, van Erp TG, Samann PG, Frodl T, Jahanshad N, Loehrer E, Tiemeier H, Hofman A, Niessen WJ, Vernooij MW, Ikram MA, Wittfeld K, Grabe HJ, Block A, Hegenscheid K, Volzke H, Hoehn D, Czisch M, Lagopoulos J, Hatton SN, Hickie IB, Goya‐Maldonado R, Kramer B, Gruber O, Couvy‐Duchesne B, Renteria ME, Strike LT, Mills NT, de Zubicaray GI, McMahon KL, Medland SE, Martin NG, Gillespie NA, Wright MJ, Hall GB, MacQueen GM, Frey EM, Carballedo A, van Velzen LS, van Tol MJ, van der Wee NJ, Veer IM, Walter H, Schnell K, Schramm E, Normann C, Schoepf D, Konrad C, Zurowski B, Nickson T, McIntosh AM, Papmeyer M, Whalley HC, Sussmann JE, Godlewska BR, Cowen PJ, Fischer FH, Rose M, Penninx BW, Thompson PM, Hibar DP (2016b): Subcortical brain alterations in major depressive disorder: Findings from the ENIGMA Major Depressive Disorder working group. Mol Psychiatry 21:806–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmahmann JD, Caplan D (2006): Cognition, emotion and the cerebellum. Brain 129:290–292. [DOI] [PubMed] [Google Scholar]
- Singh MK, Kesler SR, Hadi Hosseini SM, Kelley RG, Amatya D, Hamilton JP, Chen MC, Gotlib IH (2013): Anomalous gray matter structural networks in major depressive disorder. Biol Psychiatry 74:777–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sporns O, Zwi JD (2004): The small world of the cerebral cortex. Neuroinformatics 2:145–162. [DOI] [PubMed] [Google Scholar]
- Sridharan D, Levitin DJ, Menon V (2008): A critical role for the right fronto‐insular cortex in switching between central‐executive and default‐mode networks. Proc Natl Acad Sci U S A 105:12569–12574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stillova K, Jurak P, Chladek J, Chrastina J, Halamek J, Bockova M, Goldemundova S, Riha I, Rektor I (2015): The role of anterior nuclei of the thalamus: A subcortical gate in memory processing: An intracerebral recording study. PLoS ONE 10:e0140778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Hu S, Chambers J, Zhu Y, Tong S (2011): Graphic patterns of cortical functional connectivity of depressed patients on the basis of EEG measurements. Conf Proc IEEE Eng Med Biol Soc 2011:1419–1422. [DOI] [PubMed] [Google Scholar]
- Sweeney JA, Strojwas MH, Mann JJ, Thase ME (1998): Prefrontal and cerebellar abnormalities in major depression: Evidence from oculomotor studies. Biol Psychiatry 43:584–594. [DOI] [PubMed] [Google Scholar]
- Tijms BM, Series P, Willshaw DJ, Lawrie SM (2012): Similarity‐based extraction of individual networks from gray matter MRI scans. Cereb Cortex 22:1530–1541. [DOI] [PubMed] [Google Scholar]
- Tijms BM, Moller C, Vrenken H, Wink AM, de Haan W, van der Flier WM, Stam CJ, Scheltens P, Barkhof F (2013): Single‐subject grey matter graphs in Alzheimer's disease. PLoS ONE 8:e58921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tijms BM, Yeung HM, Sikkes SA, Moller C, Smits LL, Stam C, Scheltens P, van der Flier WM, Barkhof F (2014): Single‐subject grey matter graph properties and their relationship with cognitive impairment in early‐ and late onset Alzheimer's disease. Brain Connect 4:337–346. [DOI] [PubMed] [Google Scholar]
- Tomasi D, Volkow ND (2010): Functional connectivity density mapping. Proc Natl Acad Sci U S A 107:9885–9890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toppi J, De Vico Fallani F, Vecchiato G, Maglione AG, Cincotti F, Mattia D, Salinari S, Babiloni F, Astolfi L (2012): How the statistical validation of functional connectivity patterns can prevent erroneous definition of small‐world properties of a brain connectivity network. Comput Math Methods Med 2012:130985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Essen DC (1997): A tension‐based theory of morphogenesis and compact wiring in the central nervous system. Nature 385:313–318. [DOI] [PubMed] [Google Scholar]
- Wang J, Zuo X, Dai Z, Xia M, Zhao Z, Zhao X, Jia J, Han Y, He Y (2013): Disrupted functional brain connectome in individuals at risk for Alzheimer's disease. Biol Psychiatry 73:472–481. [DOI] [PubMed] [Google Scholar]
- Wang L, Dai Z, Peng H, Tan L, Ding Y, He Z, Zhang Y, Xia M, Li Z, Li W, Cai Y, Lu S, Liao M, Zhang L, Wu W, He Y, Li L (2014): Overlapping and segregated resting‐state functional connectivity in patients with major depressive disorder with and without childhood neglect. Hum Brain Mapp 35:1154–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts DJ, Strogatz SH (1998): Collective dynamics of 'small‐world' networks. Nature 393:440–442. [DOI] [PubMed] [Google Scholar]
- Williams JB (1988): A structured interview guide for the Hamilton Depression Rating Scale. Arch Gen Psychiatry 45:742–747. [DOI] [PubMed] [Google Scholar]
- Wise T, Radua J, Via E, Cardoner N, Abe O, Adams TM, Amico F, Cheng Y, Cole JH, de Azevedo Marques Perico C, Dickstein DP, Farrow TF, Frodl T, Wagner G, Gotlib IH, Gruber O, Ham BJ, Job DE, Kempton MJ, Kim MJ, Koolschijn PC, Malhi GS, Mataix‐Cols D, McIntosh AM, Nugent AC, O'Brien JT, Pezzoli S, Phillips ML, Sachdev PS, Salvadore G, Selvaraj S, Stanfield AC, Thomas AJ, van Tol MJ, van der Wee NJ, Veltman DJ, Young AH, Fu CH, Cleare AJ, Arnone D (2016): Common and distinct patterns of grey‐matter volume alteration in major depression and bipolar disorder: Evidence from voxel‐based meta‐analysis. Mol Psychiatry. http://www.nature.com/mp/journal/vaop/ncurrent/full/mp201672a.html. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward ND, Heckers S (2015): Mapping thalamocortical functional connectivity in chronic and early stages of psychotic disorders. Biol Psychiatry 79:1016–. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye M, Yang T, Qing P, Lei X, Qiu J, Liu G (2015): Changes of functional brain networks in major depressive disorder: A graph theoretical analysis of resting‐state fMRI. PLoS ONE 10:e0133775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalesky A, Fornito A, Bullmore ET (2010): Network‐based statistic: Identifying differences in brain networks. Neuroimage 53:1197–1207. [DOI] [PubMed] [Google Scholar]
- Zeng LL, Shen H, Liu L, Wang L, Li B, Fang P, Zhou Z, Li Y, Hu D (2012): Identifying major depression using whole‐brain functional connectivity: A multivariate pattern analysis. Brain 135:1498–1507. [DOI] [PubMed] [Google Scholar]
- Zhang JR, Wang J, Wu Q, Kuang W, Huang X, He Y, Gong Q (2011): Disrupted brain connectivity networks in drug‐naive, first‐episode major depressive disorder. Biol Psychiatry 70:334–342. [DOI] [PubMed] [Google Scholar]
- Zhu X, Wang X, Xiao J, Liao J, Zhong M, Wang W, Yao S (2012): Evidence of a dissociation pattern in resting‐state default mode network connectivity in first‐episode, treatment‐naive major depression patients. Biol Psychiatry 71:611. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information


