Abstract
Background
Several studies have shown that the in utero environment, which can be indexed by birth weight (BW), is associated with cortical morphology in adolescence and adulthood. Work in monozygotic (MZ) twins suggests that this association is driven by non‐shared environmental factors. This correlation could be the result of in utero impacts on DNA methylation. The aim of the present study with MZ twins is to replicate the association between discordance in BW and brain morphology and test whether discordance in DNA methylation mediates this relationship.
Methods
One hundred and four adolescent MZ twins (52 pairs, of which 42% were male pairs) who have been followed regularly since birth underwent T1 weighted structural MRI, and epigenome‐wide assessment of DNA methylation from saliva at age 15.
Results
Co‐twins had very similar measures of DNA methylation and cortical morphology. Higher BW members of a twin pair had increased total cortical surface area, and decreased cortical thickness compared to their lower BW sibling. BW Discordance was positively associated with both cortical surface area and cortical volume discordance. Genes involved in neurodevelopment were tentatively identified as mediators of both the BW ‐ cortical volume, and BW‐ cortical surface area relationships.
Conclusions
The association between BW and cortical morphology in adolescence appears to be attributable to in utero environmental effects, and DNA methylation may play a role in mediating this relationship. Hum Brain Mapp 38:2037–2050, 2017. © 2017 Wiley Periodicals, Inc.
Keywords: epigenetics, child development, brain development, MRI, twin designs, genome‐wide, brain morphometry, neurodevelopment, birth weight
INTRODUCTION
There is substantial research interest in the effects of the in utero environment on brain development. Birth weight (BW) is among the best‐studied indices of the quality of the in utero environment [Allin et al., 2004; Dunkel Schetter, 2011; Himpel et al., 2006]. Low BW is predictive of a range of adverse outcomes throughout maturation and into adulthood [Abernethy et al., 2002; Bjuland et al., 2014; Breslau, 1994; Løhaugen et al., 2013; Nosarti et al., 2008; Taylor et al., 2011], including behavioral issues [McCormick et al., 1990; Wiener et al., 1968] and health problems [Barker et al., 1989].
Recent work suggests that BW is strongly associated with cortical morphology. Specifically, individuals born with very low BW (<1,500 g) have smaller brains and altered cortical morphology as adolescents [Martinussen et al., 2005; Nagy et al., 2009]. Low BW is associated with reduced cortical thickness [Bjuland et al., 2013], surface area [Ajayi‐Obe et al., 2000; Skranes et al., 2013], and volume [Ball et al., 2012] in infants and adolescents, and these reductions appear to be related to cognitive deficits [Bjuland et al., 2013; Schlotz et al., 2014; Skranes et al., 2013; Walhovd et al., 2004]. Preterm children with very low BW show significant delays in the development of cortical thickness, relative to normal weight children [Mürner‐Lavanchy et al., 2014].
The biology underpinning the association between BW and cortical morphology is unclear. One possibility is that adversity experienced during gestation induces epigenetic changes, which alter the development of the cortex. Alterations in DNA methylation are well positioned to play such a role. They are capable of altering gene expression [Comb and Goodman, 1990; Nan et al., 1998], and can be induced in utero [Waterland and Jirtle, 2003]. A difficulty in interpreting these associations is the possibility of passive gene‐environment correlations where heritable factors influence both the quality of intrauterine environment and brain development.
Both DNA methylation [Kaminsky et al., 2009; Kerkel et al., 2008] and cortical morphology [Joyner et al., 2009; Panizzon et al., 2009; Pezawas et al., 2004; Winkler et al., 2010] are sensitive to differences in genetic sequence, which could lead to conflation of genetic and environmental effects. Studies of monozygotic (MZ) twins avoid this confound. Numerous recent studies have examined DNA methylation in twins [Baranzini et al., 2010; Dempster et al., 2011; Gervin et al., 2012; Gordon et al., 2012; Marsit et al., 2013; Souren et al., 2013; Sugawara et al., 2011; van Dongen et al., 2014; Yu et al., 2012], and have generally reported highly similar methylation patterns between co‐twins, with MZ twins being more similar than dizygotic (DZ) twins. We previously published data from a subsample of the twins studied here, showing that DNA methylation patterns in adolescent MZ twins are highly similar, but that there is substantial, and potentially biologically relevant variability at a fraction of cytosine‐guanine dinucleotides (CpGs) [Lévesque et al., 2014].
The MZ twin approach has also been applied to the study of brain structure, the results suggest that both genetics and environmental influences make substantial contributions [Baaré et al., 2001; Geschwind et al., 2002; Lenroot et al., 2009; Pennington et al., 2000; Wright et al., 2002; Yoon et al., 2012]. To our knowledge, the only study to have assessed the association between BW and cortical morphology in a MZ twin sample is by Raznahan et al. [2012], and found that discordance in BW is associated with discordance in cortical surface area and intelligence. Here, we extend the work of Raznahan et al. by examining possible BW‐associated epigenetic changes that could contribute to alterations in brain development.
Recent evidence suggests that normal variation in BW is associated with differences in DNA methylation [Engel et al., 2014; Simpkin et al., 2015]. A few groups have looked for evidence of methylation differences associated with BW discordance in MZ twins [Gordon et al., 2012; Souren et al., 2013; Tan et al., 2014; see also Chiarella et al., 2015], but have not reported robust effects. These studies, and the present work, which search for epigenome‐wide differences between MZ twins are complicated by the small effect sizes expected and the massive multiple comparisons problem inherent in using microarrays with hundreds of thousands of probes. Here we have taken steps to mitigate these issues by repeating DNA methylation samples to identify CpGs where methylation is stable over time. We also assessed the technical test‐retest stability of each probe, and investigated the subset of probes where inter‐twin variability could be distinguished from technical noise. We used this approach to increase our power of identifying methylation differences that could plausibly contribute to ongoing alterations in brain development.
The aim of the present study of adolescent MZ twins is to test the hypothesis that discordance in BW is associated with discordance in cortical morphology, and that discordance in DNA methylation is a potential mediator of this association.
MATERIALS AND METHODS
Participants
Participants were 104 fifteen‐year old twins (52 pairs of MZ twins: 22 male and 30 female, mean age ± SD: 15.7 ± 0.3 year, range 15.3–16.7 years) recruited from the Quebec Newborn Twin Study (QNTS) [Boivin et al., 2005; Brendgen et al., 2005]. The QNTS used the Quebec Ministry of Health and Social Services registry of new births occurring in the Province of Quebec, between April 1, 1995 and December 31, 1998 to recruit participants and followed them longitudinally [Boivin et al., 2013]. All twins who underwent scanning reported good current health, and denied any history of medical or neurological illness or use of psychotropic medications. They were determined to be free of any psychiatric disorders (verified by the Dominic [Scott et al., 2006] and the Kiddie‐Schedule for Affective Disorders and Schizophrenia (K‐SADS [Endicott and Spitzer, 1978]). Written informed consent and assent was obtained from the parents and twins, respectively. The study protocol was approved by the appropriate ethics committees (Montreal Neurological Institute [MNI] and Sainte‐Justine Hospital research center) and was in compliance with the Code of Ethics of the World Medical Association (Declaration of Helsinki).
MRI
Scans were acquired at the MNI Brain Imaging Centre, with a Siemens Magnetom 3T Tim Trio scanner (http://www.medical.siemens.com) using a magnetization‐prepared rapid acquisition gradient‐echo (MPRAGE) 9 min sequence (176 slices; 1‐mm thickness, TR = 2,300 ms, TE = 2.98 ms, TI = 900 ms, flip‐angle = 9°, FOV = 240 × 256 mm).
MRI Analysis
Native T1‐weighted MRIs were processed through the CIVET automated pipeline (version 1.1.11) [Ad‐Dab'bagh et al., 2006]. This pipeline includes the CLASP algorithm for generating cortical thickness measurements at 40,962 vertices per hemisphere [Ad‐Dab'bagh et al., 2006; Collins et al., 1994; Kim et al., 2005; Lyttelton et al., 2007; MacDonald et al., 2000]. Cortical thickness was calculated as the distance between the outer CSF–gray matter and gray matter–white matter interfaces [Kim et al., 2005; Lyttelton et al., 2007; MacDonald et al., 2000]. Cortical surface area was measured at the middle cortical surface, which minimized bias toward sulcal or gyral regions [Im et al., 2008; Van Essen et al., 2006]. Statistical analyses were implemented using SurfStat [Worsley et al., 2009] (http://www.math.mcgill.ca/keith/surfstat/), and Matlab statistics toolbox. The primary outcomes measures were the discordance in each twin pair's absolute native‐space cortical thickness image (blurred to 20 mm), as well as surface area and volume images (blurred to 40 mm). These data were analyzed in two ways; once at every vertex on the cortical surface, and once using the sum of the cortical morphology measure across the whole cortical surface.
DNA Methylation
Saliva samples
Whole saliva was collected using the OrageneTM DNA self‐collection kit following the manufacturer's instructions (DNA Genotek). Participants were asked not to eat or drink anything but water or chew gum for 30 minutes before the samples were taken. Each participant was asked to provide 2 ml of saliva, which was mixed with 2 ml of the Oragene solution, beginning the initial stage of DNA isolation and stabilizing the sample until extraction could be performed. Extraction was accomplished using the Promega Genomic DNA Purification kit, and sent to Genome Quebec for whole‐genome analysis using Illumina. In a sub‐sample of eight twin pairs, we took a second saliva sample 3–6 months following the first to perform a test‐retest‐analysis (see [Lévesque et al., 2014]) to identify temporally stable probes that might contain a record of long lasting epigenetic differences between the twins. We also performed technical replications on three sample three times each to estimate the technical stability of each probe on the chip. This approach allowed us to study a subset of the data that was both technically and temporally stable. It also allowed us to significantly reduce the multiple comparisons problem.
Illumina
We made use of the available Illumina Infinium HumanMethylation450 BeadChip Kit, which covers more than 480,000 methylation sites per sample, including 96% of CpG islands as well as additional coverage in island shores and surrounding regions, at single‐nucleotide resolution. Analysis was conducted at the Genome Quebec Innovation Centre. The manual protocol supplied by Illumina was followed for all steps except for Single Base Extension and Staining, which were conducted using the automated protocol. Briefly, the isolated DNA was first checked for quality with picogreen, and bisulfite converted using the Zymo EZ‐96 DNA Methylation‐Gold Kit. Samples were transferred to BCD and then MSA4 plates, and neutralized before overnight amplification. MSA4 plates were fragmented, precipitated, and re‐suspended before hybridization and transfer to Multi BeadChips. The Multi BeadChips then underwent washing, single‐base extension, and staining, before imaging using the HiScan array scanner. Sample preparation prior to genome wide analyses was performed in the McGill University Department of Pharmacology and Therapeutics CFI‐ Imaging and Molecular Biology Platform.
DNA Methylation Analysis
Data analysis
The raw Illumina output was processed using the R package Minfi, a part of the Bioconductor project (http://bioconductor.org). Stratified quantile normalization preprocessing was implemented using preprocessQuantile within Minfi. The main outcome measures were beta‐values at each probe, numbers ranging from zero to one, which represent the proportion of methylated cells detected. Each probe was annotated with a particular genomic location and gene based on the manifest files provided by Illumina (http://support.illumina.com/downloads/humanmethylation450_15017482_v1-2_product_files.ilmn). The beta values and their positional information were then exported to Matlab for analysis (http://mathworks.com, version 15a).
Cellular composition of saliva
In this protocol, DNA samples were collected from saliva. This has the advantage of being non‐invasive, particularly in an adolescent population. However, saliva contains a heterogeneous mixture of cell types that differ in proportion in each sample. The different cell types have differentially methylated subsets of probes which can confound inference about differences in methylation between the populations being studied. Correction for cell type variability can be accomplished by reference to an external validation set of purified cell types (e.g., [Houseman et al., 2012]), which is becoming standard practice in samples of whole blood. However, reference samples for saliva have not been published. Here, we have implemented reference free correction using RefFreeCellMixArray [Houseman et al., 2014, 2016], and estimated proportions of six cell types in each sample. We regressed these values against the beta values at each probe, and removed their estimated contribution from the beta values to produce a dataset that is linearly independent of these cell type effects.
Assessment of test‐retest variability
The values from the replicated samples were isolated and test–retest differences were calculated for each pair of samples for a given individual (sample A–sample B, sample A–sample C, sample B–sample C). This allowed us to calculate both the maximum observed pairwise difference, and a standard deviation for this difference distribution. These numbers were used in the data filtering steps below.
Data filtering
After removing the replicates and technical control samples from our dataset we had a matrix of 482,421 probes by 102 twins (52 twin pairs). We next removed data from probes where the technical variation at a probe exceeded two standard deviations of the observed values at that probe (264,454 probes). We removed the 11,135 probes annotated to the X chromosome, and the 416 probes annotated to the Y chromosome. Because our aim was to examine biologically relevant genes and conduct pathway analysis, we analyzed probes associated with known genes according to the Illumina manifest separately from unannotated probes (117,778 unannotated probes). Because methylation is not necessarily stable over time, we took advantage of test‐retest in 16 samples (8 twin pairs). A second saliva sample was collected 3–6 months following the first. After correction for differences in the buccal epithelial cell content of the sample, we compared the temporal stability of each probe. We were interested in examining probes that were stable over time, as these might contribute to more trait‐like phenotypes [Lévesque et al., 2014]. In the present analysis, probes showing the trait‐like pattern were examined. The distinction between state‐like and trait‐like was made using a median split of the mean test‐retest difference; the choice of this threshold was arbitrary. After removing probes that met any of these exclusion criteria, we were left with datasets of 70,502 annotated probes, and 24,642 unannotated probes. We were also interested in assessing altered methylation within the CpG islands and shores associated with known genes from the illumina manifest. A third dataset consisted of mean methylation within the CpG islands and shores of annotated genes (5,544 genes).
Statistics
Linear mixed models
All statistics were calculated using linear mixed models. Paired t‐test comparisons between high and low BW members of twin pairs were calculated using model 1.
| (1) |
where Y = 104 brain morphology or epigenetic measures, HiLo is a grouping variable with two levels Hi is the higher BW member of the twin pair, and Lo is the lower BW member, Sex is male or female, and Family are 104 family names of which 52 are unique. High versus Low BW twins were contrasted in this model.
Twin discordance measures were calculated by subtracting the lower BW twin's value from the higher BW twin's value on all measures. Thus, discordance in BW is always greater than zero, but discordance in other measures could take on any value. Linear effects of BW discordance on brain morphology, and epigenetics were calculated using model 2.
| (2) |
where Y was 52 measures of discordance in morphology at both vertex‐wise and whole cortex values, and epigenetics was the discordance in beta values at each probe. The contrast tested in this model was BW discordance.
Mediation
Mediation analysis was also implemented using linear mixed models. Here, mediation of the relationship between BW discordance and whole cortex brain morphometry discordance by DNA methylation discordance was considered. In all models, the independent variable (X) was BW discordance, and discordance in beta values at each probe was considered as a possible mediator (M), these models were fit separately for three dependent variables (1) discordance in cortical thickness, (2) discordance in cortical volume, and (3) discordance in cortical surface area (Y). Not all possible analyses were conducted, establishing mediation in models where there was not a significant X–Y relationship (path C) was not considered to be of research interest, and mediation can only be established in the presence of a significant X–M relationship (path A). If these conditions were not met no further analysis was conducted. Mediation was established by comparing the effect size of the indirect pathway (β indirect) calculated using the Sobel product of coefficients approach [Sobel, 1982] to a permutation‐based distribution under the null hypothesis.
Adjustment for multiple comparisons
All statistical inference was made using permutation‐based testing using the Max T method [Nichols and Holmes, 2002]. Briefly, linear mixed models where used in all analysis describing (1) cortical morphometry; both voxel‐wise and at the whole cortex level, (2) DNA methylation at the per‐probe level, and (3) mediation of the relationship between BW discordance and brain morphometry by DNA methylation. Each model was permuted 10,000 times; the variable being tested in the contrast was randomly reordered at each permutation. This procedure was used to generate (1) a null distribution across all tests and all permutations which was used to calculate unadjusted P‐value, and (2) a distribution of maximum absolute t‐values for each permutation which was used to calculate false discovery rate adjusted Q values. For example, in the case of voxel‐wise morphometry measures a matrix of 81,924 vertices × 10,000 permutations was calculated, and the raw P value for each vertex in the original image was calculated as that vertex's position in the resulting sorted null distribution, divided by the size of the distribution (size 1 by 819,240,000). At each iteration of the permutation, the maximum value of the absolute t‐statistic from the 81,924 vertices was also saved, and used to build an array of 10,000 maximum t‐statistics (Max T method). The absolute t value of each vertex in the original image was then compared to this distribution, and its position within the distribution of maximum t values divided by 10,000 resulted in the Q value for that vertex. The same procedure was used for all linear models under consideration where the T statistic was being evaluated. In the case of mediation, statistical inference was based on the magnitude of the β indirect term as compared to the permutation‐based distribution of β indirect under the null hypothesis.
RESULTS
Birth Weight
Twins weighed 2.54 ± 0.51 kg (mean ± SD) at birth (Table 1). On average the twins' discordance in BW was 0.31 ± 0.24 kg. This represented an average discordance of 15.36 ± 15.53% (mean ± SD), calculated as a percentage of the larger twins BW. The range in BW was between 1.00 and 3.73 kg. Discordance ranged between 0.01 and 1.07 kg, and between 0.36% and 78.41%. Thirty nine of the 104 individuals had a BW below 2,500 g. The twins were born at 37 ± 2.35 weeks of gestation (range 30–40 weeks), with 17 of the twin pairs being born before 37 weeks gestation. Gestational age was highly correlated with BW (r = 0.672, P = 2.20 e − 013), but was unrelated to the discordance in BW between the twins (r = −0.155, P = 0.302). As within‐pair comparisons and measures of discordance were the primary measures of interest, gestational age was not considered in any further analysis.
Table 1.
Sample means and discordance values (higher birth weight twin – lower birth weight twin)
| Sample mean ± SD | Discordance ± SD | |
|---|---|---|
| Birth weight | 2.54 ± 0.51 kg | 0.31 ± 0.24 kg |
|
Cortical thickness Cortical surface area Cortical volume |
3.38 ± 0.12 mm 1949.36 ± 169.68 cm2 5820.48 ± 489.23 cm3 |
−0.03 ± 0.09 mm 24 ± 62.38 cm2 27.94 ± 227.95 cm3 |
| DNA methylation (β) Annotated CpGs | 0.5364 ± 0.302 | 0 ± 0.0037 |
| DNA methylation (β) unnnotated CpGs | 0.6519 ± 0.2434 | 0 ± 0.0037 |
| DNA methylation (β) collapsed CpG islands | 0.6458 ± 0.1509 | 0 ± 0.0019 |
Cortical Morphometry
When the twins were separated into high and low BW members of each twin pair, and this grouping was used to compare measures of cortical morphometry in adolescence, we found that higher BW twins had increased cortical surface area across the whole cortex (t (101) = 2.77, P = 0.01), but decreased average cortical thickness (t (101) = −2.08, P = 0.04), relative to their lower BW siblings. As cortical volume is a convolution of these two measures, it unsurprisingly was not different (t (101) = 0.88, P = 0.38, Fig. 1). When BW discordance was regressed against discordance in morphology, significant positive associations were observed for both cortical surface area (t (49) = 4.07, P = 8.40 e − 005) and cortical volume (t (49) = 4.45, P = 2.40 e − 005, Fig. 2). Twin pairs with larger differences in BW had larger differences in brain morphometry. This relationship was not observed for cortical thickness (t (49) = 1.23, P = 0.230). When this same regression model was fitted vertex‐wise, a significant cluster of association was observed between discordance in BW and discordance in cortical volume in the right operculum extending into the lateral orbitofrontal cortex and inferior frontal gyrus (maximum t (49) = 4.70, P = 2.76 e − 005, Q = 0.034; world coordinates x = 41.0 y = 23.9 z = −11.86; 44 vertices Q < 0.05, Fig. 3).
Figure 1.
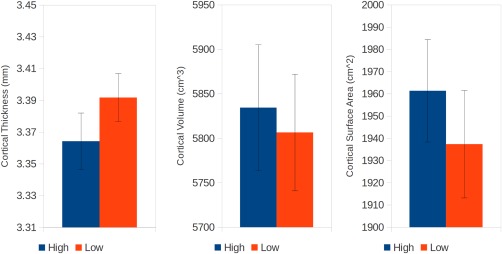
Plots of average cortical thickness (left), cortical volume (middle), and cortical surface area (right) in the High (blue) versus Low (red) birth weight members of each twin pair (mean ± SEM). [Color figure can be viewed at http://wileyonlinelibrary.com]
Figure 2.
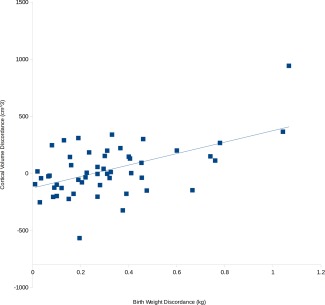
Discordance in birth weight is correlated with discordance in the total volume of the cortex. [Color figure can be viewed at http://wileyonlinelibrary.com]
Figure 3.
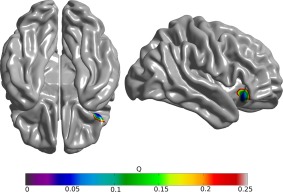
Areas of significant association between discordance in birth weight and cortical volume, false discovery rate corrected Q values. [Color figure can be viewed at http://wileyonlinelibrary.com]
DNA Methylation
When discordance measures of BW, cortical thickness, cortical surface area, and cortical volume were regressed against discordance in DNA methylation (separately for annotated CpGs, unannotated CpGs, and collapsed CpG islands) no significant associations were observed after correction for multiple comparisons. Candidate probes and genes with unadjusted P‐values below 1 e − 004 are presented in Table 2.
Table 2.
Probes DNA methylation discordance may be associated with discordance in birth weight, or discordance in cortical morphometry
| Measure | Probe | Chromosome | Symbol | G island_position | t | P | Q |
|---|---|---|---|---|---|---|---|
| Annotated probes | |||||||
| Birth weight discordance | cg06313433 | 3 | FLNB | Island | −5.414 | 2.38 E − 006 | 0.15285 |
| cg11967457 | 1 | LEFTY2 | S_Shore | −4.6472 | 2.89 E − 005 | 0.82917 | |
| cg18755581 | 1 | TATDN3 | S_Shore | −4.3127 | 8.95 E − 005 | 0.997 | |
| cg09608383 | 15 | FAM189A1 | S_Shelf | 4.2842 | 9.89 E − 005 | 0.999 | |
| Cortical thickness discordance | cg14845385 | 1 | MRPL55 | Island | 5.185 | 5.04 E − 006 | 0.31768 |
| cg01113680 | 13 | ENOX1 | 4.455 | 4.87 E − 005 | 0.97103 | ||
| cg22656126 | 17 | WDR81 | Island | −4.3535 | 6.67 E − 005 | 0.99101 | |
| Cortical volume discordance | cg14063817 | 6 | EXOC2 | Island | −5.4769 | 4.20 E − 006 | 0.25175 |
| cg19708984 | 12 | CACNA2D4 | N_Shore | −5.262 | 8.30 E − 006 | 0.44555 | |
| cg14217589 | 4 | TBC1D1 | S_Shore | −5.2111 | 9.50 E − 006 | 0.48851 | |
| cg14364926 | 9 | ZER1 | −4.7562 | 3.77 E − 005 | 0.92408 | ||
| cg23036385 | 19 | EFNA2 | Island | 4.5392 | 7.19 E − 005 | 0.99301 | |
| cg05520031 | 9 | FAM125B | Island | −4.4462 | 9.42 E − 005 | 0.999 | |
| cg16668397 | 16 | JPH3 | S_Shore | −4.4436 | 9.48 E − 005 | 0.999 | |
| Cortical surface area discordance | cg14217589 | 4 | TBC1D1 | S_Shore | −4.6157 | 2.14 E − 005 | 0.76124 |
| cg05801648 | 9 | SYK | Island | 4.5732 | 2.49 E − 005 | 0.82018 | |
| cg19978105 | 11 | DLG2 | 4.4606 | 3.70 E − 005 | 0.91508 | ||
| cg26138500 | 7 | PTPRN2 | S_Shore | −4.4326 | 4.10 E − 005 | 0.93606 | |
| cg07950855 | 6 | PPP1R2P1 | N_Shore | −4.4204 | 4.27 E − 005 | 0.94306 | |
| cg13636907 | 3 | MITF | 4.3372 | 5.74 E − 005 | 0.97502 | ||
| cg14479344 | 3 | MYH15 | −4.3301 | 5.88 E − 005 | 0.97602 | ||
| cg18049045 | 5 | FCHO2 | N_Shore | −4.3296 | 5.88 E − 005 | 0.97602 | |
| cg12848118 | 14 | FOXN3 | N_Shelf | −4.3306 | 5.85 E − 005 | 0.97602 | |
| cg23537932 | 10 | GRK5 | 4.2984 | 6.52 E − 005 | 0.98601 | ||
| cg25972205 | 7 | VIPR2 | N_Shelf | 4.1966 | 9.16 E − 005 | 0.996 | |
| Unannotated probes | |||||||
| Birth weight discordance | cg18790856 | 2 | −5.014 | 1.14 E − 005 | 0.23077 | ||
| cg17316316 | 5 | Island | −4.978 | 1.27 E − 005 | 0.25574 | ||
| Cortical thickness discordance | cg05191217 | 5 | Island | −4.4848 | 3.70 E − 005 | 0.58442 | |
| cg06915545 | 12 | 4.3398 | 5.68 E − 005 | 0.73926 | |||
| cg08236766 | 11 | Island | 4.268 | 7.15 E − 005 | 0.80919 | ||
| Cortical volume discordance | cg23762657 | 6 | 5.0141 | 3.06 E − 005 | 0.54246 | ||
| cg20301308 | 1 | S_Shore | 4.8211 | 5.28 E − 005 | 0.74026 | ||
| cg05963100 | 7 | 4.791 | 5.69 E − 005 | 0.75924 | |||
| cg14740251 | 19 | −4.6474 | 8.39 E − 005 | 0.87512 | |||
| Cortical surface area discordance | cg23506042 | 5 | N_Shelf | −4.5681 | 4.34 E − 005 | 0.63936 | |
| cg26600461 | 16 | −4.5557 | 4.49 E − 005 | 0.65235 | |||
| Collapsed CpG islands | |||||||
| Birth weight discordance | No association | ||||||
| Cortical thickness discordance | No association | ||||||
| Cortical volume discordance | 4 | C4orf47 | −4.6659 | 3.57 E − 005 | 0.26773 | ||
| 5 | PCDHB10 | 4.4777 | 6.68 E − 005 | 0.45355 | |||
| 20 | NKX2‐4 | 4.442 | 7.41 E − 005 | 0.48951 | |||
| Cortical surface area discordance | No association | ||||||
All probes with an unadjusted P‐value < 1 e − 004 are presented.
Mediation
DNA methylation was considered as a potential mediator of the relationship between BW and cortical morphometry. Discordance in BW correlated with discordance in DNA methylation at an unadjusted P < 0.005 in 306 annotated CpGs, 120 unannotated CpGs, and 46 collapsed CpG islands; only these probes were considered as mediation candidates. As mentioned above, BW discordance was significantly associated with discordance in cortical volume and cortical surface area, but not cortical thickness, so cortical thickness was not analyzed further. Mediation candidates with unadjusted P‐values below 1 e − 004 are presented in Table 3. This threshold yielded a list of 13 annotated CpGs that were potential mediators of the relationship between BW discordance and discordance in cortical volume. Nine annotated CpGs were potential mediators of the relationship between BW discordance and discordance in cortical surface area.
Table 3.
Probes where discordance in DNA methylation may mediates the relationship between discordance in birth weight and discordance in cortical morphometry
| Measure | Probe | Chromosome | Symbol | G island_position | P | Q |
|---|---|---|---|---|---|---|
| Annotated probes | ||||||
| Cortical thickness discordance | No association | |||||
| Cortical volume discordance | cg15384717 | 1 | PRPF3 | Island | 9.80 E−007 | 0.00029997 |
| cg07670722 | 7 | RELN | 4.90 E−006 | 0.0014999 | ||
| cg14063817 | 6 | EXOC2 | Island | 1.18 E−005 | 0.0035996 | |
| cg11967457 | 1 | LEFTY2 | S_Shore | 1.86 E−005 | 0.0056994 | |
| cg07690455 | 5 | CAST | N_Shore | 1.96 E−005 | 0.0059994 | |
| cg12208381 | 19 | CSNK1G2 | Island | 1.99 E−005 | 0.0060994 | |
| cg23064481 | 6 | TRIM15 | 3.14 E−005 | 0.009599 | ||
| cg14458783 | 17 | ABR | Island | 3.30 E−005 | 0.010099 | |
| cg19124225 | 6 | TNF | N_Shelf | 7.65 E−005 | 0.022898 | |
| cg21798061 | 6 | C6orf10 | 9.22 E−005 | 0.027597 | ||
| cg11155374 | 13 | FREM2 | 0.00012124 | 0.036196 | ||
| cg17612569 | 21 | GABPA | Island | 0.00013072 | 0.038896 | |
| cg18361975 | 5 | EFNA5 | 0.00014673 | 0.043596 | ||
| Cortical surface area discordance | cg11312353 | 13 | TMCO3 | Island | 6.21 E−006 | 0.0018998 |
| cg23064481 | 6 | TRIM15 | 1.05 E−005 | 0.0031997 | ||
| cg25597833 | 11 | KDM2A | S_Shore | 1.70 E−005 | 0.0051995 | |
| cg27641018 | 7 | EPDR1 | N_Shore | 5.62 E−005 | 0.017198 | |
| cg09690321 | 3 | PARP14 | S_Shelf | 6.37 E−005 | 0.019398 | |
| cg26097210 | 10 | PBLD | S_Shore | 6.54 E−005 | 0.019898 | |
| cg07670722 | 7 | RELN | 8.01 E−005 | 0.023998 | ||
| cg12184450 | 17 | ACACA | 0.0001232 | 0.036196 | ||
| cg12760995 | 1 | CASZ1 | 0.00013529 | 0.039596 | ||
| Unannotated probes | ||||||
| Cortical thickness discordance | No association | |||||
| Cortical volume discordance | cg11412793 | 2 | 9.17 E−006 | 0.0010999 | ||
| cg23939182 | 4 | 5.75 E−005 | 0.0068993 | |||
| cg12757676 | 2 | 6.50 E−005 | 0.0077992 | |||
| cg11182874 | 8 | 9.58 E−005 | 0.011499 | |||
| cg03520683 | 13 | N_Shore | 9.75 E−005 | 0.011699 | ||
| cg02897008 | 1 | N_Shelf | 0.0001225 | 0.014599 | ||
| cg12995004 | 5 | 0.0002575 | 0.029997 | |||
| cg03408785 | 3 | S_Shelf | 0.00033833 | 0.039296 | ||
| cg09514588 | 10 | 0.00036583 | 0.042196 | |||
| cg09355008 | 1 | 0.000385 | 0.044196 | |||
| Cortical surface area discordance | cg17915189 | 7 | 6.67 E−006 | 0.00079992 | ||
| cg14073706 | 1 | N_Shelf | 2.50 E−005 | 0.0029997 | ||
| cg09163412 | 10 | Island | 4.50 E−005 | 0.0053995 | ||
| cg03408785 | 3 | S_Shelf | 4.92 E−005 | 0.0058994 | ||
| cg16197202 | 10 | N_Shore | 0.0002275 | 0.026497 | ||
| cg11412793 | 2 | 0.00037583 | 0.042696 | |||
| Collapsed CpG islands | ||||||
| Cortical thickness discordance | No association | |||||
| Cortical volume discordance | 20 | GGT7 | 0.00034783 | 0.015598 | ||
| 7 | PGAM2 | 0.00067826 | 0.030297 | |||
| Cortical surface area discordance | 12 | DBX2 | 0.00040217 | 0.018398 | ||
| 7 | PGAM2 | 0.00098913 | 0.043996 | |||
All probes with an unadjusted P‐value less than 1 e − 004 are presented.
Pathway Analysis
The lists of gene names of the annotated CpGs identified as candidate mediators in Table 3 were used as inputs to Ingenuity Pathway Analysis. This suggested that 12 of the 13 genes that might mediate the relationship between BW discordance and discordance in cortical volume were part of a network involved in “Cell morphology, cellular assembly and organization, cellular development” (network score is 36, Fig. 4). All nine candidate mediators of the BW discordance, cortical surface area discordance relationship were included in a network involved in “Cell cycle, cell death and survival, liver necrosis/cell death” (network score 27, Fig. 5). The unannotated probes were not suitable for pathway analysis, and the list of genes identified by collapsing across CpG islands was not sufficiently different from the individual CpG analysis to warrant a separate pathway analysis.
Figure 4.
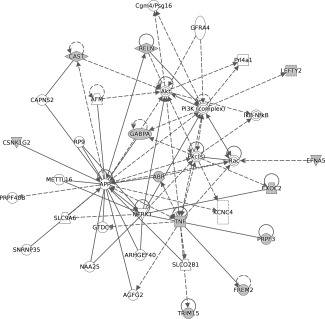
Top network identified through Ingenuity Pathway Analysis of the 13 genes that might mediate the relationship between birth weight discordance and discordance in cortical volume (threshold P < 1 e − 004). Twelve of these genes were part of a network involved in “Cell morphology, cellular assembly and organization, cellular development” (network score is 36).
Figure 5.
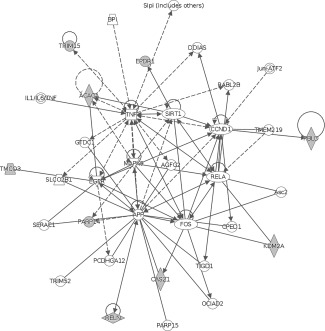
Second network identified through Ingenuity Pathway Analysis of the nine genes that might mediate the relationship between birth weight discordance and discordance in cortical volume (threshold P < 1 e − 004). All nine of these genes were part of a network involved in “Cell cycle, cell death and survival, liver necrosis/cell death” (network score 27).
DISCUSSION
The present study indicates that discordance in BW in MZ twins is associated with differences in brain development when tested during adolescence. We found that compared to their lower BW siblings, the higher BW member of a twin pair had on average increased cortical surface area, and decreased cortical thickness. The magnitude of the BW discordance was correlated with the degree of discordance in both total cortical surface area and total cortical volume. This association was most pronounced in the right operculum and orbitofrontal cortex. We found evidence suggesting that DNA methylation at certain probes may mediate the relationship between BW discordance and cortical morphometry.
A series of papers has established compelling evidence of an association between BW and cortical morphometry [Ajayi‐Obe et al., 2000; Ball et al., 2012; Bjuland et al., 2013; Martinussen et al., 2005; Mürner‐Lavanchy et al., 2014; Schlotz et al., 2014; Skranes et al., 2013]. All of these studies investigated single births, and all but one [Schlotz et al., 2014], studied preterm or very‐low‐BW births. While studying preterm or very low BW individuals makes detecting large effects likely, the design of single births makes interpretation more complicated, as these designs cannot control for genotypic differences between participants, as well as for many other sources of environmental variation. We studied healthy adolescent MZ twins, which allowed us to investigate a range of BWs among twins, and examine their relationships to cortical morphology.
To our knowledge, only one other group has performed comparable work. Raznahan et al. [Raznahan et al., 2012] studied normal variation in BW in 85 pairs of MZ twins with a wide age range (5 years to 25.6 years; mean 13.7) many of whom were studied longitudinally (one to more than 3 MRI scans) compared to the present cross sectional study with tight constraints on the subjects' ages (15.3 to 16.7 years: mean 15.7). The ranges in BW and BW discordances studied were roughly similar. Both studies examined the same cortical morphometry variables and found comparable results; Raznahan et al. also found substantial increases in cortical surface area among high BW members of a twin pair. However, they observed higher cortical thickness and volume among high BW members of a twin pair relative to the lower BW members of a twin pair, while we observed lower cortical thickness and no change in volume. Cortical thickness in healthy populations is thought to peak at age 7 and then decline, while measures of cortical surface area are believed to peak at age 13 [Shaw et al., 2012]. The wider age range and longitudinal scanning approach used by Raznahan et al. allowed them to report curvilinear increases in cortical thickness modeled over time, while we found evidence of decreases in cortical thickness at one particular time point. This discrepancy might be explained by the important age differences and approaches used.
The present study innovated by examining the putative mediating role of DNA methylation on the BW–cortical morphometry relationship. To our knowledge, only a few other studies examining BW related variations in epigenome‐wide DNA methylation in MZ twins have been published [Chen et al., 2016; Gordon et al., 2012; Souren et al., 2013; Tan et al., 2014]. The most directly comparable work is by Souren et al. [Souren et al., 2013], who studied 17 adult twin pairs with methylation data from saliva samples. They did not find any associations between BW discordance and methylation that survived false discovery rate correction, and were not able to replicate any of their top candidates using deep bisulfite sequencing. Our study had two significant advantages that allowed us to focus on a smaller list of target probes. One complication they noted was the proportion of probes that show statistically significant effects, but with very limited variability in ß values that are likely within the range of technical variation. We used a set of technical replicates to establish expected variability at each probe, and used this expected variability to identify probes that distinguish a signal from technical noise. We took this idea further by retesting a subset of the twins 3–6 months later to look for probes that were stable over time, expecting that methylation changes established at birth and influencing the course of development would be stable over a period of 3–6 months. This approach allowed us to study a subset of the data that was both technically and temporally stable. It also allowed us to significantly reduce the multiple comparisons problem. However, even with these advantages we could not establish a statistically robust association between BW discordance and DNA methylation. Another study of blood samples from 150 adult MZ twin pairs of various ages (30–74 years) found no association between BW discordance and differences in methylation [Tan et al., 2014], although using differentially methylated region‐based association analysis, one significantly altered region covering two genes, CRYZ and TYW3 on chromosome 1 was identified [Chen et al., 2016]. Another study examined human umbilical vein endothelial cells from 22 newborn MZ twin pairs and 12 DZ twin pairs [Gordon et al., 2012] and found evidence for an association at only one probe at an FDR threshold of Q = 0.07. Taken together these studies suggest that the variability in DNA methylation and BW within MZ twin pairs may not be sufficient to consistently establish robust and strong associations in epigenome‐wide analyses.
However, our study design allowed us to use DNA methylation information in a more targeted fashion. When considering methylation as a mediator of BW discordance–cortical morphometry relationships, we were able to select a subset of probes that were associated with BW discordance at an unadjusted statistical threshold, and to search for evidence of mediation only in these probes. It should be noted that none of the probes entered into mediation analysis survived adjustment for multiple comparison at the genome wide level, they were entered into the mediation analysis using a much lower statistical threshold. We consider this to be an informative approach, but replication of these results, and follow up studies of gene expression and protein synthesis in brain tissue will be required. This approach suggested mediation candidates for discordance in both cortical surface area, and cortical volume.
Pathway analysis suggested that mediators of discordance in cortical volume were primarily involved in networks regulating “Cell morphology, cellular assembly and organization, cellular development.” Ingenuity analysis suggests that several of these genes were implicated in biological functions relevant to the central nervous system and neurodevelopment including reelin (RELN), active BCR‐related (ABR), calpastatin (CAST), Ephrin A5 (EFNA5), GA binding protein transcription factor alpha subunit (GABPA), and tumor necrosis factor (TNF). These genes are involved in the abnormal morphology of the nervous system (ABR, CAST, EFNA5, TNF, P = 2.92 e − 03), and the development of neurons (CAST, EFNA5, GABPA, RELN, TNF, P = 2.92 e − 03).
The network identified in the mediation analysis of cortical surface area discordance is implicated in “Cell cycle, cell death and survival, liver necrosis/cell death.” Notably reelin (RELN) was identified in both the analysis of cortical volume, and cortical surface area. Ingenuity analysis suggests that three of the nine genes identified, castor zinc finger 1 (CASZ1), lysine demethylase 2A (KDM2A), and reelin are involved in the differentiation of the nervous system (P = 2.02 e − 03). When the methylation data was collapsed across CPG islands and shores, two of the eight identified genes which could mediate the BW discordance cortical volume relationship are thought to be involved in nervous system development and function (neurofibromin 2 (NF2) and Kruppel like factor 9 (KLF9), P = 2.84 e − 04). While these associations should be considered tentative at this point, the identified genes appear to have reasonable face‐validity for their proposed role in regulating the development of the cortex.
The following limitations should be considered when interpreting these findings. First, The MZ twins were highly similar in terms of all of the variables under investigation, including BW and cortical morphology. This is not surprising as we carefully selected MZ twins for the present study who were mentally and physically healthy. Nevertheless, it would be of interest to study to what extent the results can be generalized to more heterogeneous populations, particularly those at high risk of psychiatric illness. Methodologically, the high similarity of the twins means that the effect sizes expected in this study are relatively small. Given the massive multiple comparisons problems inherent in both neuroimaging and DNA microarray studies, it is difficult to distinguish small real variations against a background of substantial noise. We have taken precautions in this regard by carefully screening our data, and using targeted subset or summary measures whenever possible. A related issue is that less than 20% of probes on the Illumina chip were included in our analysis, while we believe aggressively filtering the data is a strength of our work, it inherently increases the possibility of type II error. Second, we measured DNA methylation during adolescence, at the same time as we measured brain structure. The strongest case for mediation can be made when the independent variable is measured first, followed by the mediator, and then the dependent variable. That was not possible in our study, as the Illumina 450K microarrays have only relatively recently been developed. To mitigate this concern, we measured the temporal stability of our probes, and selected those that were stable over a period of 6 months, which we expected, would enrich our sample for longer lasting epigenetic modifications that are expected to be associated with trait differences. However, our sample is undoubtedly a mixture of probes that differed at birth, and probes that have been differentially regulated over time scales of months to years. Ongoing work with newborn twins will allow us to validate these findings, and discriminate these populations, using multiple blood samples collected over a period of years. A third related issue is the possibility of non‐shared environmental influences that occurred after birth influencing cortical development of the twins. Because the population being studied has been carefully followed since birth, examining the influence of additional intervening variables and gene by environment interactions is possible, and is the subject of ongoing study. Fourth, our work should be interpreted in the larger context of studies using peripheral methylation markers in a particular tissue type and the degree to which these results apply to other cell types which may undergo differential methylation. There is reason to believe that careful analysis and interpretation of these types of studies (see [Liang and Cookson, 2014] for review) can produce valid discoveries, which are best used to guide follow‐up investigation using more targeted measures.
CONCLUSION
Together these findings add to the growing body of literature suggesting an association between BW and cortical morphology, irrespective of DNA sequence effects. We further extend these findings by presenting initial evidence of epigenetic mechanisms through which the in utero environment could influence adolescent cortical development.
ACKNOWLEDGMENTS
We are grateful to the children and parents of the Quebec Newborn Twin Study (QNTS), and the participating teachers and schools. We thank Jocelyn Malo and Marie‐Élyse Bertrand for coordinating the data collection, as well as Hélène Paradis and Nadine Forget‐ Dubois for managing the data bank of QNTS. We thank Victoria Ly, B.Sc. and Dr. Farida Vaisheva for technical assistance.
Conflict of interest: The authors declare no conflict of interest.
REFERENCES
- Abernethy LJ, Palaniappan M, Cooke RWI (2002): Quantitative magnetic resonance imaging of the brain in survivors of very low birth weight. Arch Dis Child 87:279–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ad‐Dab'bagh Y, Einarson D, Lyttelton O, Muehlboeck J‐S, Mok K, Ivanov O, Vincent R, Lepage C, Lerch JP, Fombonne E, Evans AC (2006): The CIVET Image‐Processing Environment: A Fully Automated Comprehensive Pipeline for Anatomical Neuroimaging Research. In Proceedings of 12th Annual Meeting of the Human Brain Mapping Organization. Florence, Italy. Available at: http://www.doctoraddabbagh.ca/Publications.html, Accessed August 7, 2012.
- Ajayi‐Obe M, Saeed N, Cowan F, Rutherford M, Edwards A (2000): Reduced development of cerebral cortex in extremely preterm infants. The Lancet 356:1162–1163. [DOI] [PubMed] [Google Scholar]
- Allin M, Henderson M, Suckling J, Nosarti C, Rushe T, Fearon P, Stewart AL, Bullmore ET, Rifkin L, Murray R (2004): Effects of very low birthweight on brain structure in adulthood. Dev Med Child Neurol 46:46–53. [DOI] [PubMed] [Google Scholar]
- Baaré WF, van Oel CJ, Hulshoff Pol HE, Schnack HG, Durston S, Sitskoorn MM, Kahn RS (2001): Volumes of brain structures in twins discordant for schizophrenia. Arch Gen Psychiatry 58:33–40. [DOI] [PubMed] [Google Scholar]
- Ball G, Boardman JP, Rueckert D, Aljabar P, Arichi T, Merchant N, Gousias IS, Edwards AD, Counsell SJ (2012): The effect of preterm birth on thalamic and cortical development. Cereb Cortex 22:1016–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baranzini SE, Mudge J, van Velkinburgh JC, Khankhanian P, Khrebtukova I, Miller NA, Zhang L, Farmer AD, Bell CJ, Kim RW, May GD, Woodward JE, Caillier SJ, McElroy JP, Gomez R, Pando MJ, Clendenen LE, Ganusova EE, Schilkey FD, Ramaraj T, Khan OA, Huntley JJ, Luo S, Kwok P‐Y, Wu TD, Schroth GP, Oksenberg JR, Hauser SL, Kingsmore SF (2010): Genome, epigenome and RNA sequences of monozygotic twins discordant for multiple sclerosis. Nature 464:1351–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker DJ, Winter PD, Osmond C, Margetts B, Simmonds SJ (1989): Weight in infancy and death from ischaemic heart disease. Lancet Lond Engl 2:577–580. [DOI] [PubMed] [Google Scholar]
- Bjuland KJ, Løhaugen GC, Martinussen M, Skranes J (2013): Cortical thickness and cognition in very‐low‐birth‐weight late teenagers. Early Hum Dev 89:371–380. [DOI] [PubMed] [Google Scholar]
- Bjuland KJ, Rimol LM, Løhaugen GCC, Skranes J (2014): Brain volumes and cognitive function in very‐low‐birth‐weight (VLBW) young adults. Eur J Paediatr Neurol 18:578–590. [DOI] [PubMed] [Google Scholar]
- Boivin M, Pérusse D, Dionne G, Saysset V, Zoccolillo M, Tarabulsy GM, Tremblay N, Tremblay RE (2005): The genetic‐environmental etiology of parents' perceptions and self‐assessed behaviours toward their 5‐month‐old infants in a large twin and singleton sample. J Child Psychol Psychiatry 46:612–630. [DOI] [PubMed] [Google Scholar]
- Boivin M, Brendgen M, Dionne G, Dubois L, Pérusse D, Robaey P, Tremblay RE, Vitaro F (2013): The Quebec Newborn Twin Study into adolescence: 15 years later. Twin Res Hum Genet 16:64–69. [DOI] [PubMed] [Google Scholar]
- Brendgen M, Dionne G, Girard A, Boivin M, Vitaro F, Pérusse D (2005): Examining genetic and environmental effects on social aggression: A study of 6‐year‐old twins. Child Dev 76:930–946. [DOI] [PubMed] [Google Scholar]
- Breslau N (1994): A gradient relationship between low birth weight and IQ at age 6 years. Arch Pediatr Adolesc Med 148:377. [DOI] [PubMed] [Google Scholar]
- Chen M, Baumbach J, Vandin F, Röttger R, Barbosa E, Dong M, Frost M, Christiansen L, Tan Q (2016): Differentially methylated genomic regions in birth‐weight discordant twin pairs. Ann Hum Genet 80:81–87. [DOI] [PubMed] [Google Scholar]
- Chiarella J, Tremblay RE, Szyf M, Provencal N, Booij L (2015): Impact of early environment on children's mental health: Lessons from DNA methylation studies with monozygotic twins. Twin Res Hum Genet 18:623–634. [DOI] [PubMed] [Google Scholar]
- Collins DL, Neelin P, Peters TM, Evans AC (1994): Automatic 3D intersubject registration of MR volumetric data in standardized Talairach space. J Comput Assist Tomogr 18:192–205. [PubMed] [Google Scholar]
- Comb M, Goodman HM (1990): CpG methylation inhibits proenkephalin gene expression and binding of the transcription factor AP‐2. Nucleic Acids Res 18:3975–3982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dempster EL, Pidsley R, Schalkwyk LC, Owens S, Georgiades A, Kane F, Kalidindi S, Picchioni M, Kravariti E, Toulopoulou T, Murray RM, Mill J (2011): Disease‐associated epigenetic changes in monozygotic twins discordant for schizophrenia and bipolar disorder. Hum Mol Genet 20:4786–4796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunkel Schetter C (2011): Psychological science on pregnancy: Stress processes, biopsychosocial models, and emerging research issues. Annu Rev Psychol 62:531–558. [DOI] [PubMed] [Google Scholar]
- Endicott J, Spitzer RL (1978): A diagnostic interview: The schedule for affective disorders and schizophrenia. Arch Gen Psychiatry 35:837–844. [DOI] [PubMed] [Google Scholar]
- Engel SM, Joubert BR, Wu MC, Olshan AF, Håberg SE, Ueland PM, Nystad W, Nilsen RM, Vollset SE, Peddada SD, London SJ (2014): Neonatal genome‐wide methylation patterns in relation to birth weight in the Norwegian Mother and Child Cohort. Am J Epidemiol 179:834–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gervin K, Vigeland MD, Mattingsdal M, Hammerø M, Nygård H, Olsen AO, Brandt I, Harris JR, Undlien DE, Lyle R (2012): DNA methylation and gene expression changes in monozygotic twins discordant for psoriasis: Identification of epigenetically dysregulated genes. PLoS Genet 8:e1002454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geschwind DH, Miller BL, DeCarli C, Carmelli D (2002): Heritability of lobar brain volumes in twins supports genetic models of cerebral laterality and handedness. Proc Natl Acad Sci USA 99:3176–3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon L, Joo JE, Powell JE, Ollikainen M, Novakovic B, Li X, Andronikos R, Cruickshank MN, Conneely KN, Smith AK, Alisch RS, Morley R, Visscher PM, Craig JM, Saffery R (2012): Neonatal DNA methylation profile in human twins is specified by a complex interplay between intrauterine environmental and genetic factors, subject to tissue‐specific influence. Genome Res 22:1395–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himpel S, Bartels J, Zimdars K, Huether G, Adler L, Dawirs RR, Moll GH (2006): Association between body weight of newborn rats and density of serotonin transporters in the frontal cortex at adulthood. J Neural Transm (Vienna) 113:295–302. [DOI] [PubMed] [Google Scholar]
- Houseman EA, Accomando WP, Koestler DC, Christensen BC, Marsit CJ, Nelson HH, Wiencke JK, Kelsey KT (2012): DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics 13:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseman EA, Molitor J, Marsit CJ (2014): Reference‐free cell mixture adjustments in analysis of DNA methylation data. Bioinformatics 30:1431–1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseman EA, Kile ML, Christiani DC, Ince TA, Kelsey KT, Marsit CJ (2016): Reference‐free deconvolution of DNA methylation data and mediation by cell composition effects. BMC Bioinformatics 17:259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im K, Lee J‐M, Lyttelton O, Kim SH, Evans AC, Kim SI (2008): Brain size and cortical structure in the adult human brain. Cereb Cortex N Y N 1991 18:2181–2191. [DOI] [PubMed] [Google Scholar]
- Joyner AH, J CR, Bloss CS, Bakken TE, Rimol LM, Melle I, Agartz I, Djurovic S, Topol EJ, Schork NJ, Andreassen OA, Dale AM (2009): A common MECP2 haplotype associates with reduced cortical surface area in humans in two independent populations. Proc Natl Acad Sci USA 106:15483–15488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminsky ZA, Tang T, Wang S‐C, Ptak C, Oh GHT, Wong AHC, Feldcamp LA, Virtanen C, Halfvarson J, Tysk C, McRae AF, Visscher PM, Montgomery GW, Gottesman II, Martin NG, Petronis A (2009): DNA methylation profiles in monozygotic and dizygotic twins. Nat Genet 41:240–245. [DOI] [PubMed] [Google Scholar]
- Kerkel K, Spadola A, Yuan E, Kosek J, Jiang L, Hod E, Li K, Murty VV, Schupf N, Vilain E, Morris M, Haghighi F, Tycko B (2008): Genomic surveys by methylation‐sensitive SNP analysis identify sequence‐dependent allele‐specific DNA methylation. Nat Genet 40:904–908. [DOI] [PubMed] [Google Scholar]
- Kim JS, Singh V, Lee JK, Lerch J, Ad‐Dab'bagh Y, MacDonald D, Lee JM, Kim SI, Evans AC (2005): Automated 3‐D extraction and evaluation of the inner and outer cortical surfaces using a Laplacian map and partial volume effect classification. NeuroImage 27:210–221. [DOI] [PubMed] [Google Scholar]
- Lenroot RK, Schmitt JE, Ordaz SJ, Wallace GL, Neale MC, Lerch JP, Kendler KS, Evans AC, Giedd JN (2009): Differences in genetic and environmental influences on the human cerebral cortex associated with development during childhood and adolescence. Hum Brain Mapp 30:163–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lévesque ML, Casey KF, Szyf M, Ismaylova E, Ly V, Verner M‐P, Suderman M, Brendgen M, Vitaro F, Dionne G, Boivin M, Tremblay RE, Booij L (2014): Genome‐wide DNA methylation variability in adolescent monozygotic twins followed since birth. Epigenetics 9:1410–1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang L, Cookson WOC (2014): Grasping nettles: Cellular heterogeneity and other confounders in epigenome‐wide association studies. Hum Mol Genet 23:R83–R88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Løhaugen GCC, Østgård HF, Andreassen S, Jacobsen GW, Vik T, Brubakk A‐M, Skranes J, Martinussen M (2013): Small for gestational age and intrauterine growth restriction decreases cognitive function in young adults. J Pediatr 163:447–453. [DOI] [PubMed] [Google Scholar]
- Lyttelton O, Boucher M, Robbins S, Evans A (2007): An unbiased iterative group registration template for cortical surface analysis. NeuroImage 34:1535–1544. [DOI] [PubMed] [Google Scholar]
- MacDonald D, Kabani N, Avis D, Evans AC (2000): Automated 3‐D extraction of inner and outer surfaces of cerebral cortex from MRI. NeuroImage 12:340–356. [DOI] [PubMed] [Google Scholar]
- Marsit CJ, Koestler DC, Watson‐Smith D, Boney CM, Padbury JF, Luks F (2013): Developmental genes targeted for epigenetic variation between twin‐twin transfusion syndrome children. Clin Epigenetics 5:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinussen M, Fischl B, Larsson HB, Skranes J, Kulseng S, Vangberg TR, Vik T, Brubakk A‐M, Haraldseth O, Dale AM (2005): Cerebral cortex thickness in 15‐year‐old adolescents with low birth weight measured by an automated MRI‐based method. Brain J Neurol 128:2588–2596. [DOI] [PubMed] [Google Scholar]
- McCormick MC, Gortmaker SL, Sobol AM (1990): Very low birth weight children: Behavior problems and school difficulty in a national sample. J Pediatr 117:687–693. [DOI] [PubMed] [Google Scholar]
- Mürner‐Lavanchy I, Steinlin M, Nelle M, Rummel C, Perrig WJ, Schroth G, Everts R (2014): Delay of cortical thinning in very preterm born children. Early Hum Dev 90:443–450. [DOI] [PubMed] [Google Scholar]
- Nagy Z, Ashburner J, Andersson J, Jbabdi S, Draganski B, Skare S, Böhm B, Smedler A‐C, Forssberg H, Lagercrantz H (2009): Structural correlates of preterm birth in the adolescent brain. Pediatrics 124:e964–e972. [DOI] [PubMed] [Google Scholar]
- Nan X, Ng HH, Johnson CA, Laherty CD, Turner BM, Eisenman RN, Bird A (1998): Transcriptional repression by the methyl‐CpG‐binding protein MeCP2 involves a histone deacetylase complex. Nature 393:386–389. [DOI] [PubMed] [Google Scholar]
- Nichols TE, Holmes AP (2002): Nonparametric permutation tests for functional neuroimaging: A primer with examples. Hum Brain Mapp 15:1–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nosarti C, Giouroukou E, Healy E, Rifkin L, Walshe M, Reichenberg A, Chitnis X, Williams SCR, Murray RM (2008): Grey and white matter distribution in very preterm adolescents mediates neurodevelopmental outcome. Brain J Neurol 131:205–217. [DOI] [PubMed] [Google Scholar]
- Panizzon MS, Fennema‐Notestine C, Eyler LT, Jernigan TL, Prom‐Wormley E, Neale M, Jacobson K, Lyons MJ, Grant MD, Franz CE, Xian H, Tsuang M, Fischl B, Seidman L, Dale A, Kremen WS (2009): Distinct genetic influences on cortical surface area and cortical thickness. Cereb Cortex 19:2728–2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennington BF, Filipek PA, Lefly D, Chhabildas N, Kennedy DN, Simon JH, Filley CM, Galaburda A, DeFries JC (2000): A twin MRI study of size variations in human brain. J Cogn Neurosci 12:223–232. [DOI] [PubMed] [Google Scholar]
- Pezawas L, Verchinski BA, Mattay VS, Callicott JH, Kolachana BS, Straub RE, Egan MF, Meyer‐Lindenberg A, Weinberger DR (2004): The brain‐derived neurotrophic factor val66met polymorphism and variation in human cortical morphology. J Neurosci 24:10099–10102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raznahan A, Greenstein D, Lee NR, Clasen LS, Giedd JN (2012): Prenatal growth in humans and postnatal brain maturation into late adolescence. Proc Natl Acad Sci USA 109:11366–11371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlotz W, Godfrey KM, Phillips DI (2014): Prenatal origins of temperament: Fetal growth, brain structure, and inhibitory control in adolescence. PLoS One 9:e96715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott TJL, Short EJ, Singer LT, Russ SW, Minnes S (2006): Psychometric properties of the Dominic interactive assessment: A computerized self‐report for children. Assessment 13:16–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw P, Malek M, Watson B, Sharp W, Evans A, Greenstein D (2012): Development of cortical surface area and gyrification in attention‐deficit/hyperactivity disorder. Biol Psychiatry 72:191–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpkin AJ, Suderman M, Gaunt TR, Lyttleton O, McArdle WL, Ring SM, Tilling K, Davey Smith G, Relton CL (2015): Longitudinal analysis of DNA methylation associated with birth weight and gestational age. Hum Mol Genet 24:3752–3763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skranes J, Lø haugen GCC, Martinussen M, H\aa berg A, Brubakk A‐M, Dale AM (2013): Cortical surface area and IQ in very‐low‐birth‐weight (VLBW) young adults. Cortex 49:2264–2271. [DOI] [PubMed] [Google Scholar]
- Sobel ME (1982): Asymptotic confidence intervals for indirect effects in structural equation models In: Leinhardt S, editor. Sociological Methodology. Washington, DC: American Sociological Association; pp 290–312. [Google Scholar]
- Souren NY, Lutsik P, Gasparoni G, Tierling S, Gries J, Riemenschneider M, Fryns J‐P, Derom C, Zeegers MP, Walter J (2013): Adult monozygotic twins discordant for intra‐uterine growth have indistinguishable genome‐wide DNA methylation profiles. Genome Biol 14:R44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawara H, Iwamoto K, Bundo M, Ueda J, Miyauchi T, Komori A, Kazuno A, Adati N, Kusumi I, Okazaki Y, Ishigooka J, Kojima T, Kato T (2011): Hypermethylation of serotonin transporter gene in bipolar disorder detected by epigenome analysis of discordant monozygotic twins. Transl Psychiatry 1:e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Q, Frost M, Heijmans BT, von Bornemann Hjelmborg J, Tobi EW, Christensen K, Christiansen L (2014): Epigenetic signature of birth weight discordance in adult twins. BMC Genomics 15:1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor HG, Klein N, Anselmo MG, Minich N, Espy KA, Hack M (2011): Learning problems in kindergarten students with extremely preterm birth. Arch Pediatr Adolesc Med 165:819–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dongen J, Ehli EA, Slieker RC, Bartels M, Weber ZM, Davies GE, Slagboom PE, Heijmans BT, Boomsma DI (2014): Epigenetic variation in monozygotic twins: A genome‐wide analysis of DNA methylation in buccal cells. Genes 5:347–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Essen DC, Dierker D, Snyder AZ, Raichle ME, Reiss AL, Korenberg J (2006): Symmetry of cortical folding abnormalities in Williams syndrome revealed by surface‐based analyses. J Neurosci 26:5470–5483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walhovd KB, Psychol C, Fjell AM, Reinvang I, Lundervold A, Fischl B, Quinn BT, Dale AM (2004): Size does matter in the long run Hippocampal and cortical volume predict recall across weeks. Neurology 63:1193–1197. [DOI] [PubMed] [Google Scholar]
- Waterland RA, Jirtle RL (2003): Transposable elements: Targets for early nutritional effects on epigenetic gene regulation. Mol Cell Biol 23:5293–5300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiener G, Rider RV, Oppel WC, Harper PA (1968): Correlates of low birth weight. Psychological status at eight to ten years of age. Pediatr Res 2:110–118. [DOI] [PubMed] [Google Scholar]
- Winkler AM, Kochunov P, Blangero J, Almasy L, Zilles K, Fox PT, Duggirala R, Glahn DC (2010): Cortical thickness or grey matter volume? The importance of selecting the phenotype for imaging genetics studies. NeuroImage 53:1135–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worsley KJ, Taylor JE, Carbonell F, Chung MK, Duerden E, Bernhardt B, Lyttelton O, Boucher M, Evans AC (2009): SurfStat: A Matlab toolbox for the statistical analysis of univariate and multivariate surface and volumetric data using linear mixed effects models and random field theory. NeuroImage 47:S102. [Google Scholar]
- Wright IC, Sham P, Murray RM, Weinberger DR, Bullmore ET (2002): Genetic contributions to regional variability in human brain structure: Methods and preliminary results. NeuroImage 17:256–271. [DOI] [PubMed] [Google Scholar]
- Yoon U, Perusse D, Evans AC (2012): Mapping genetic and environmental influences on cortical surface area of pediatric twins. Neuroscience 220:169–178. [DOI] [PubMed] [Google Scholar]
- Yu C‐C, Furukawa M, Kobayashi K, Shikishima C, Cha P‐C, Sese J, Sugawara H, Iwamoto K, Kato T, Ando J, Toda T (2012): Genome‐wide DNA methylation and gene expression analyses of monozygotic twins discordant for intelligence levels. PLoS One 7:e47081. [DOI] [PMC free article] [PubMed] [Google Scholar]


