Abstract
Use of functional magnetic resonance imaging (fMRI) in studies of aging is often hampered by uncertainty about age‐related differences in the amplitude and timing of the blood oxygenation level dependent (BOLD) response (i.e., hemodynamic impulse response function (HRF)). Such uncertainty introduces a significant challenge in the interpretation of the fMRI results. Even though this issue has been extensively investigated in the field of neuroimaging, there is currently no consensus about the existence and potential sources of age‐related hemodynamic alterations. Using an event‐related fMRI experiment with two robust and well‐studied stimuli (visual and auditory), we detected a significant age‐related difference in the amplitude of response to auditory stimulus. Accounting for brain atrophy by circumventing spatial normalization and processing the data in subjects' native space eliminated these observed differences. In addition, we simulated fMRI data using age differences in brain morphology while controlling HRF shape. Analyzing these simulated fMRI data using standard image processing resulted in differences in HRF amplitude, which were eliminated when the data were analyzed in subjects' native space. Our results indicate that age‐related atrophy introduces inaccuracy in co‐registration to standard space, which subsequently appears as attenuation in BOLD response amplitude. Our finding could explain some of the existing contradictory reports regarding age‐related differences in the fMRI BOLD responses. Hum Brain Mapp 38:3402–3414, 2017. © 2017 Wiley Periodicals, Inc.
Keywords: hemodynamic impulse response function, fMRI, aging, native space, spatial normalization, registration
INTRODUCTION
Functional magnetic resonance imaging (fMRI) is a popular noninvasive tool for in vivo imaging of brain function through the blood oxygenation level dependent (BOLD) signal. Controlling for refractory and adaptation effects via effective experimental design, there is evidence for rough linearity in the BOLD response to neuronal stimulation [Huettel et al., 2004]. General linear models (GLM) of fMRI data evolved from this linearity assumption, and became the most widely used models for analyzing fMRI data. The most basic element of a GLM is the hemodynamic impulse response function (HRF), which is defined as the BOLD response to an extremely brief stimulus. Under the assumption of linearity, one can predict the BOLD response to any combination of stimuli with varying amplitude, duration, or delay. Importantly, most standard GLM analyses also assume that the shape of the HRF is the same across brain regions and subjects, utilizing a “canonical” double‐gamma function. However, the actual shape of the HRF arises through a complex interaction of multiple physiological processes including blood flow, blood volume, and cerebral metabolic rate of oxygen [Buxton et al., 1998; Donahue et al., 2009]. The involvement of multiple physiological processes in shaping the final HRF curve warrants the possibility of its inter‐regional and intersubject variability [Buckner et al., 2000]. However, the majority of fMRI analyses are currently performed under the assumption of HRF invariance. Results of any group comparison without this assumption can be challenging to interpret, as it makes it impossible to distinguish whether or not the observed differences in activation are due to underlying neuronal activity or differences in the amplitude and timing of the HRF that can result from other physiological variations.
There is evidence that the human cerebrovascular system goes through multiple age‐related changes and deterioration including but not limited to increased tortuosity, atherosclerosis, and reduced reactivity [Desjardins, 2015; D'Esposito et al., 2003]. These changes pose problems for fMRI as well as other hemodynamic methods such as near‐infrared spectroscopy [Bonnéry et al., 2012]. While these factors represent substantial evidence that the shape of HRF is subject to change with age [Desjardins, 2015; D'Esposito et al., 2003; Handwerker et al., 2012], the existing results in the literature about age‐related alterations in the HRF are often mixed or even contradictory. For instance, age‐related reductions in the amplitude of the HRF to visual stimuli have been reported in many studies [Ances et al., 2009; Handwerker et al., 2007; Raemaekers et al., 2006; Ross et al., 1997; Tekes et al., 2005]. Reductions have also been reported for motor stimuli [Buckner et al., 2000; Handwerker et al., 2007; Hesselmann et al., 2001; Raemaekers et al., 2006; Riecker et al., 2003]. In contrast, some studies report increases in the amplitude of the HRF [Mattay et al., 2002; Mohtasib et al., 2012, Restom et al., 2007], while others have found no age‐related differences [Aizenstein et al., 2004; D'Esposito et al., 2003, 1999; Huettel et al., 2001; Stefanova et al., 2013; Yan et al., 2011]. There are also reports claiming that age‐related differences are most pronounced in the variance of the HRF amplitude rather than the mean [Ances et al., 2009; D'Esposito et al., 2003]. In addition to the amplitude of the HRF, its temporal properties have also been investigated. For instance, Huettel et al. [2001] detected a faster baseline to peak in the HRF of older subjects and another study demonstrated systematic increases in the duration of the postpeak return to baseline [Richter and Richter, 2003].
Cortical regions involved in different tasks are composed of distinct types of neurons and probably different vasculature structure [Brodmann, 2007; von Economo et al., 2008]. Therefore, it seems logical to assume that different brain regions exhibit distinct age‐related alterations in HRF shape. In the investigation of regional variability in age‐related alterations of the HRF, one study suggested that age is associated with decreases in the amplitude of the HRF in visual cortex but not in motor cortex [Ross et al., 1997], whereas another study suggested the opposite [Morsheddost et al., 2015].
Even though it is challenging to draw conclusions from across these studies, it seems that most studies processing their fMRI data by transforming them to standard space show some significant age‐related difference in the amplitude of the HRF [Buckner et al., 2000; Hu et al., 2013; Mattay et al., 2002; Mohtasib et al., 2012; Raemaekers et al., 2006; Riecker et al., 2003; Zysset et al., 2007], whereas most studies processing their fMRI data in subjects’ native space show no significant difference [Aizenstein et al., 2004; D'Esposito et al., 1999; Huettel et al., 2001; Restom et al., 2007; Yan et al., 2011]. There are also reports that transformation into standard space significantly affects the results of structural and functional analyses [Hutchison et al., 2014; Krishnan et al., 2006; Razlighi et al., 2014]. In addition, there is a strong regional dependency in age‐related brain atrophy measured by cortical thickness, wherein the primary visual cortex exhibits minimal or no age‐related cortical thinning and the primary motor and auditory cortices show high levels of age‐related atrophy [Fjell et al., 2009; Salat et al., 2004]. This evidence raises the possibility that differential age‐related morphological changes in brain structure (brain atrophy) might be responsible for some of the observed age‐related alterations of the BOLD response reported in the literature, and that this relationship may be mediated by the misregistration of the older participants’ functional data to a standard anatomical template.
In this study, we used a jittered event‐related fMRI experiment with two robust and well‐studied sensory stimuli (visual and auditory) to investigate the effect of brain atrophy in the observed age‐related alterations in the amplitude and timing of HRF. We hypothesize that extensive brain atrophy introduces inaccuracy in the co‐registration of the older participants’ brain to standard space and that this inaccuracy subsequently manifests as attenuation of the amplitude of the HRF. We aimed to investigate this hypothesis using both real and simulated fMRI data from stimuli specifically selected to evoke activation in primary visual and auditory cortices. These two sensory cortices are well suited for this study, as little or no age‐related atrophy has been demonstrated in the visual cortex, whereas the auditory cortex is among the regions that show the most age‐related atrophy. Furthermore, we investigated whether or not slice‐timing differences could influence the observed delay in the BOLD response reported in some of the literature and also found in our experiments.
METHODS
Participants and Data Acquisition
Thirty‐six right‐handed healthy subjects (18/18 younger/older; percentage female: 0.56/0.61, age mean + std: 26.1/64.9 + 2.8/2.2 years) were presented with visual (6 Hz flashing checker boards) and auditory (6 Hz alternating tones with 600 and 1000 Hz frequency) stimuli in a jittered event‐related design while undergoing functional magnetic resonance imaging. The event duration was sampled from a uniform distribution (range: 0.4/0.6–4.0/4.0 s for visual/auditory) and the onset of the stimuli were jittered by applying a uniformly distributed interstimulus interval (range: 2.5 ∼ 9.0 s). To ensure attention to the stimuli, subjects were asked to respond with a button press at the conclusion of each visual stimulus.
Functional and structural images were acquired using the same 3.0 T Philips Achieva magnetic resonance scanner. A T1‐weighted magnetization‐prepared rapid gradient‐echo sequence [TE/TR = 3/6.5 ms, flip angle = 8°, 256 × 256 matrix size, in‐plane resolution = 1 × 1 mm, and 180 slices in axial plane with slice‐thickness/gap = 1/0 mm] was used to acquire structural images for the localization of fMRI data. Functional images were acquired with a fast field echo echo‐planar imaging (FFE‐EPI) sequence [TE/TR = 20/2000 ms, flip angle = 72°, 112 × 112 matrix size, in‐plane resolution = 2 × 2 mm, slice thickness = 3.0 mm (no gap), 41 axial slices per volume, 6:1 Philips interleaved, in ascending order]. Participants were scanned for 5.5 min, with at least 37 events for each of the visual and auditory stimuli. All images were reviewed by a radiologist, and any participant with vascular or neurological pathology was excluded from the study. Two participants were excluded by this process.
Preprocessing
Structural images were reconstructed using Freesurfer (v 5.1.0) and visually inspected for any inaccuracy in the cortical parcellation and subcortical segmentation according to the provided guidelines (http://surfer.nmr.mgh.harvard.edu/fswiki/Edits). The gray matter thickness of the cerebral cortex was obtained at each vertex on the white matter surface by computing its shortest distance from the pia mater surface. The cortical thickness of each parcellated region was obtained by averaging the vertices within that region. Functional images were preprocessed using FMRIB software library (FSL). FMRI volumes were aligned using rigid body registration to the first volume to correct for subject head motion, slice‐time corrected using Sinc interpolation, and smoothed using a 5 mm3 nonlinear kernel. Intensity normalization was carried out such that the 50th percentile of all volume intensities after removing the background noise was 104 for every subject. The time‐series were then high‐pass filtered using a Gaussian filter with cutoff frequency of 0.008 Hz. Spatial normalization was performed by initial rigid‐body registration of the subject's functional mean image to the structural image of the same subject and subsequent linear (affine) registration of the structural image to MNI152 standard space. We also performed nonlinear registration between structural images and the MNI152 template to test the effects of different registration methods. Both registrations were performed with default parameters of the FSL (v 5.0.7) software package.
Statistical Analysis of fMRI Data
We used multiple regression analysis to obtain the age‐related differences in mean cortical thickness of the primary visual (lateral‐occipital) and primary auditory (transverse‐temporal) cortices, controlling for gender and intracranial volume effects.
The standard two‐level GLM technique, also implemented in FSL, was used for the statistical analysis of the preprocessed fMRI data. The first level linear model fits individual voxel time‐series to multiple regressors of interest (visual, auditory, and motor response stimuli, convolved with the canonical double‐gamma HRF) and nuisance regressors (6 sets of motion parameters). The first‐level activation maps were extracted for every subject after prewhitening using an autocorrelation matrix given by a first‐order autoregressive (AR(1)) model of residuals from a preliminary GLM.
The second‐level linear analysis was performed on the obtained activation maps for all participants and each age group separately to compute a contrast map for between‐group differences in activation. Obtained maps were corrected for multiple comparisons using cluster analysis (Z threshold = 2.3/5.0 for real/simulated data, with the clusterwise P < 0.01). Binarizing the group‐level activation maps gives the region of interest masks used to extract the fMRI time‐series in standard space for deconvolving the HRF. These masks were visually inspected to ensure their locations matched the primary visual and auditory cortices in standard space. Because this procedure is biased to select voxels with HRF shapes similar to the canonical double gamma, we also repeated the computation of these ROI masks using two separate approaches: (1) with a Gaussian HRF and (2) without any a priori assumption on the shape of the HRF using independent component analysis (ICA).
Time‐Series From Subject's Native Space
To analyze fMRI data in subjects’ native space, the first‐level statistical analyses were repeated for fMRI data without spatial normalization to standard template space. To obtain the regional activation masks in the subjects’ native space localized to the primary visual and auditory cortices, the subject‐wise Freesurfer extracted ROI masks were used (superior‐temporal and transverse gyri for auditory cortex and lateral occipital, fusiform gyrus, pericalcarine, and inferior parietal for visual cortex). Only significantly activated (P < 0.01 uncorrected) voxels within the primary visual and auditory cortices were selected to extract the time‐series for the native space analysis. Using manually drawn ROI masks to select the activated voxels within the primary visual and auditory cortices is often used in the literature analyzing their fMRI data in subjects’ native space [Ances et al., 2009; D'Esposito et al., 1999; Huettel et al., 2001; Mehagnoul‐Schipper et al., 2002; Ross et al., 1997]. However, we feel that using subject‐wise Freesurfer ROI masks is more consistent and less susceptible to human error.
HRF Deconvolution
Deconvolution can extract subject‐ and region‐specific HRFs from a voxel's time‐series given the time‐course of the associated stimulus. Under the assumption of linearity, the fMRI signal at location i can be formulated as
| (1) |
where is the binary sequence that represents the time‐course of the stimuli of interest, is the HRF, is the residual time course, and is the preprocessed fMRI signal after regressing out the six standard motion parameters as nuisance variables. Deconvolution, in general, is an ill‐posed problem that can have multiple solutions. There are different deconvolution techniques that have been utilized in fMRI data [Glover, 1999; Goutte et al., 2000; Josephs et al., 1997; Woolrich et al., 2004] and each of them is based on an underlying assumption. For instance, given a fixed shape of the HRF (e.g., double gamma) with amplitude being the only varying parameter, Eq. (1) can be reformatted as a simple regression model; . In this case, the solution for is estimated by ordinary least squares (OLS) under the assumption of independent noise, or generalized least squares assuming autocorrelated errors, as in the subject‐level GLM described above. This is the simplest solution for Eq. (1), and is thus commonly used in the fMRI data analysis. However, it assumes a rigid shape for the HRF across regions or subjects and would be biased in cases where there is systematic deviation from this assumed shape, making interpretation of the obtained age‐related differences in the BOLD response problematic.
For the sake of our experiment, in this study, we chose a highly constrained model, FMRIB's linear optimal basis sets (FLOBS) introduced in Woolrich et al. [2004], and a moderately relaxed model, finite impulse response (FIR) filtering introduced in Goutte et al. [2000] and Josephs et al. [1997], to make sure that our results are not confounded by specific deconvolution methods. These two techniques have been commonly used in the literature to extract the HRF for visual, auditory, and motor cortex. We used these techniques to estimate the HRF in young and old subjects using both native space and standard space extracted time‐series.
FMRIB's Linear Optimal Basis Sets
FLOBS are basis sets derived such that their linear combinations explain a large proportion of the variance for the most physiologically plausible shapes of the HRF [Woolrich et al., 2004]. FLOBS were introduced to relax the HRF rigidity assumption in GLM analysis. They allow differences in delay and amplitude for four parts of the HRF curve. In general, they can be formulated as
| (2) |
where are the different basis functions generated to fit a range of the most physiologically plausible HRFs. Six standard motion nuisance regressors were also included in the analysis. FLOBS weightings, , can also be estimated by OLS which is an advantage for this method. Once are estimated, they can be used to construct the final shape of the HRF for each subject and brain region [Grinband et al., 2008].
Finite Impulse Response Filtering
Using an FIR filter does not impose any particular constraint to the shape of the HRF, instead it assumes that the HRF is a special case of an autoregressive model with an exogenous input [Goutte et al., 2000]. It is thus more flexible but has a higher chance of producing a physiologically meaningless shape. Equation (1) can be simply reformulated as an FIR filter:
| (3) |
It is clear that can be considered as the weighting of an FIR filter which is applied to . In this case, is the order of the filter, which is determined by the length of the HRF and the repetition time (TR) of the EPI sequence. To estimate FIR filter weightings, , Eq. (3) can be reformulated as a product of a vector of weighting factors and a Toeplitz matrix , where , and . Thus,
| (4) |
where , N is the total number of sample points in the fMRI signal (Goutte et al., 2000), and follows a similar notation as . Under this matrix formulation and normally distributed noise, maximum likelihood estimation of the weighting factors is obtained as
| (5) |
Six standard motion nuisance regressors were also included in the analysis. Once the FIR filter weightings, , are computed, the shape of the HRF is given using those weighting factors.
Simulated fMRI Data
Real fMRI data are problematic for evaluating the quality of one method over another because the true underlying BOLD signal is not known. Because of this, simulated data were used so that processed images could be compared to a known underlying BOLD signal. The morphology of each simulated fMRI scan comes from a real subject's data by temporally averaging all volumes. Reconstructing the same subject's structural image with FreeSurfer [Fischl et al., 2002, 2004a] and intermodal rigid‐body registration with FSL [Greve and Fischl, 2009] gives us the ROI masks in the fMRI space. We used the lateral occipital region as primary visual cortex and the third quarter of superior temporal plus transverse temporal gyri in the anterior to posterior direction as primary auditory cortex. Then neuronal activity stimuli consisting of sequences of 20 boxcar pulses with jittered onsets (at least 10 s apart) and randomly generated durations (uniformly distributed with a range of 0.5–3.5 s) were created for each ROI. The neuronal stimulus for each voxel was convolved with the canonical HRF (e.g., double gamma) to generate the hemodynamic response. Identical HRFs were used for both age groups, ensuring no age‐related alteration in the neurovascular structure. In this case, any age‐related difference obtained from the simulated data must be caused by data processing. Both HRF and neuronal stimuli were sampled at 20 Hz to simulate interleaved slice acquisition timing for a given TR and number of slices. Null data (random noise) were assigned to voxels outside ROIs. To simulate cardiac and respiratory variations in the fMRI signal, a single sinusoid at f c = 1.23 Hz for cardiac and another sinusoid at f r = 0.25 Hz for respiratory noise have been added to the data. The amplitude of the cardiac signal is modulated with the inverse of the distance of the voxel from the nearest artery. Additionally, 5% Gaussian thermal noise was added to the signal. The temporally averaged volume of the real data was also used to obtain the mean value at each voxel, which was used to shift the mean of the hemodynamic signal and to scale the standard deviation of the signal to 1% of the mean value, comparable to a robust signal in the visual cortex.
We simultaneously simulated motion, interleaved slice acquisition, and their interaction. Motion parameters were extracted from real subjects. We then upsampled these motion parameters (6 parameters) using spline interpolation to the same sampling frequency as the simulated BOLD signals (20 Hz). By applying the upsampled parameters to the volume at the initial point, we can specify the exact position of the volume at any fractional time (20× TR) between the two original sampling points to realistically simulate the interaction between slice‐timing and motion. More detail on the fMRI simulator used in this project can be found in Parker et al. [2017].
Slice Acquisition Timing and HRF Delay
Because some studies have reported differences in HRF delay rather than amplitude across different age groups, we also explored the effects of data processing on HRF timing. In addition to the effects of spatial normalization on HRF delay, we also specifically examined the effects of slice acquisition timing. In fMRI data acquired with interleaved slice acquisition, adjacent slices are acquired with delay up to half of the repetition time (TR = 2 s). If the slices are acquired in transverse plane, as is usually done, then one can expect the primary visual cortex to be spatially extended into multiple slices, whereas the primary auditory cortex often extends to fewer slices, as the transverse gyri only spatially extend through coronal or sagittal planes. Therefore, any systematic age‐related differences in brain morphology could potentially generate differences in the timing of the extracted HRF. While there are reports suggesting that slice‐timing correction should be included in the fMRI preprocessing pipeline [Parker et al., 2017; Sladky et al., 2011], the exact timings of the slice acquisition are often missing from image headers [Biswal et al., 2010] and may be difficult to recover retrospectively [Parker et al., 2014]. In addition, many researchers prefer to eliminate the suboptimal slice‐timing correction step and instead include slice‐dependent predictors in their GLM design to account for this delay. To investigate whether or not such timing differences in the acquisition of the adjacent slices can induce delay in the extracted HRF, we extracted the HRF from different slices in each subject's primary auditory cortex and tested whether or not the delay in slice acquisition relates to the timing of the extracted HRFs. We further examined whether any induced delay could be removed with the proper application of slice‐timing correction.
RESULTS
Brain Atrophy in Primary Visual and Auditory Cortices
Our results revealed a significant age‐related decrease in the mean cortical thickness of the primary auditory cortex (transverse‐temporal) (t = −2.5, P <0.01), whereas primary visual cortex (lateral occipital) did not present any significant age‐related alteration (t = −1.64, P = 0.11).
Group Level Activation Maps
The two sensory stimuli (visual and auditory) induced significant activation in the primary visual and primary auditory cortices. Figure 1a shows the group level activation area with significant z‐statistics for visual and auditory stimuli of all subjects in the standard space. Figure 1b,c shows the same statistical activation maps for younger and older groups separately, and Figure 1d illustrates significant activation differences for primary visual and auditory cortices. There were no significant differences between the activation maps of the younger and older group for visual stimuli that would survive clusterwise multiple comparison correction, whereas significant differences were present within the auditory cortex in response to auditory stimuli. The age‐related differences in the activation maps of the primary auditory cortex can be interpreted as differences in neural activity if we assume older and younger participants have identical HRFs. However, if age‐related changes in other physiological processes result in HRF differences, interpretation of these results becomes challenging.
Figure 1.
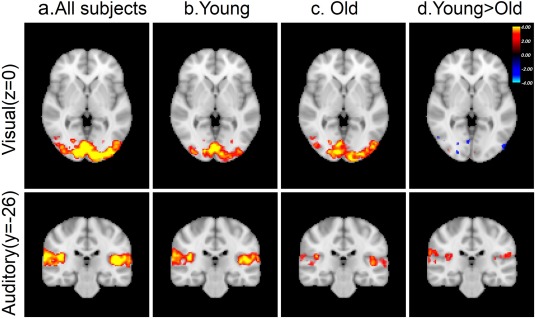
Brain activation induced by visual (top row) and auditory (bottom row) stimuli illustrated by color‐coded z‐statistics overlaid on the MNI template for (a) all subject combined, (b) only young subjects, (c) only old subjects, and (d) the contrast between young and old subjects. [Color figure can be viewed at http://wileyonlinelibrary.com]
Age‐Related Differences in HRF Amplitude and Timing
To investigate the age‐related differences in HRF amplitude, we used both FLOBS and FIR to extract the HRF from both age groups. Figure 2a illustrates the differences between the extracted HRF from younger and older groups. Compared to the younger group, there was a significant reduction in the amplitude of the auditory HRF in older participants (Δ = 0.36, P < 0.05), and a marginal increase in amplitude of the visual HRF (Δ = 0.29, P = 0.08). Figure 2a also shows a nonsignificant delay in the BOLD response (from peak to baseline) for both visual (Δt = 2.1 s, P = 0.93) and auditory (Δt = 0.90 s, P = 0.26) stimuli using FLOBS. These delays in BOLD responses to both stimuli are eliminated or reduced when the FIR technique is used to extract the HRF. While not passing the threshold for statistical significance, any differences in the return to baseline could pose problems for interpreting activation differences between groups using GLM models that assume HRF invariance.
Figure 2.
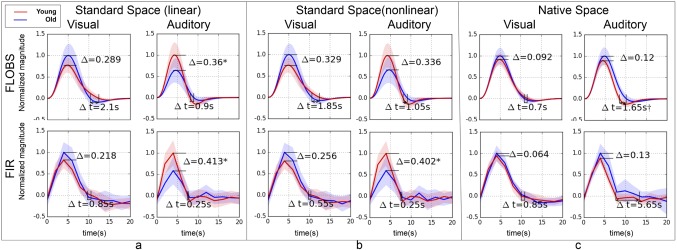
Differences between extracted HRFs from young (red curves) and old (blue curves) subjects using visual and auditory stimulation in real fMRI data. For comparison, two separate deconvolution techniques—FLOBS (top row) and FIR (bottom row)—are used for HRF extraction. Three different fMRI spatial preprocessing pipelines were used: (a) standard space with linear (affine) registration, (b) standard space with nonlinear (warped) registration, and (c) native space analysis without registration. The significant difference in the magnitude is marked with * and in the delay (from peak to baseline) is marked by (P < 0.05, outliers excluded). [Color figure can be viewed at http://wileyonlinelibrary.com]
Figure 2b shows the analogous results using nonlinear registration for spatial normalization. Nonlinear registration resulted in a slight attenuation in the magnitude difference observed in the auditory HRF using linear registration, which made the difference nonsignificant (Δ = 0.34, P = 0.051). For the visual HRF, the nonlinear registration increased the HRF amplitude difference, yet remained non‐significant (Δ = 0.33, P = 0.051). This serves as a preliminary indication that the obtained difference in age groups may be at least partially due to the inaccuracy in the registration of old subjects to standard space, as nonlinear registration has been shown to be more accurate than registration using affine transformation [Andersson et al., 2007].
Figure 2c shows the results without spatial normalization, using time‐series extracted from the subjects’ native space. The observed age‐related differences in the amplitude of the extracted HRF were virtually eliminated by circumventing the spatial normalization step, suggesting that these differences may be the byproduct of misregistration of older participants to standard space. Supporting Information, Figure S1 shows the comparative results obtained using a Gaussian HRF suggesting a minimal effect of HRF shape selection in the first‐level analysis in our final results.
Using the native space time‐series did not eliminate the observed nonsignificant delay of HRF for auditory stimuli in the standard space. In fact, it slightly increased this delay to the point of significance (Δt = 1.65 s, P < 0.05). FIR analysis on the native space time‐series also resulted in a large but nonsignificant delay (Δt = 5.65 s, P = 0.26) in the return to baseline. Because it was relatively unaffected by registration, this delay can be interpreted as an underlying neurovascular process shaping the HRF curve obtained from the auditory cortex, as has been reported elsewhere [Taoka et al., 1998]. Next, we used simulated fMRI data to provide evidence that age‐related differences in the brain morphology could generate an age‐related difference in the HRF amplitude extracted from spatially normalized fMRI data.
HRF Amplitude and Timing Differences in Simulated Data
The above results indicate that age‐related differences in the BOLD response can be accounted for by analyzing the fMRI data in subject's native space. To further test our hypothesis that differences in age‐related brain atrophy could lead to HRF differences through the co‐registration step in spatial normalization, we used simulated fMRI data. Though identical HRFs were used to generate the underlying BOLD signal for both young and old subjects in the simulated data, Figure 3a shows that the previously observed age differences in the amplitude and delay of the HRF were present for both visual (FLOBS: Δ = 0.12, P = 0.12; Δt = 1.95 s, p = 0.80) and auditory (FLOBS: Δ = 0.18, P < 0.05; Δt = 1.75 s, P = 0.37) stimuli using each deconvolution technique. Furthermore, Figure 3b illustrates that the difference between the magnitude of the young and old HRF was reduced by performing non‐linear registration (visual: Δ = 0.01, P = 0.45; audio: Δ = 0.12, P = 0.10). Finally, the native space analysis (Fig, 3c) virtually eliminated the observed differences in the amplitude (visual: Δ = 0.02, P = 0.40; audio: Δ = 0.00, P = 0.49) and the delay (visual: Δt = 0.00 s, P = 0.53, audio: Δt = 0.05 s, P = 0.79) of the HRF. Supporting Information, Figure S2 shows the comparative results obtained using a Gaussian HRF as a convolution kernel for our regressor in the first‐level analysis of the simulated fMRI data. It is noteworthy to mention that the BOLD response in the simulated data was generated with a double‐gamma HRF. The similarity of the results in Figure 3 and Supporting Information, Figure S1 suggest a minimal effect of HRF shape selection on our final results.
Figure 3.
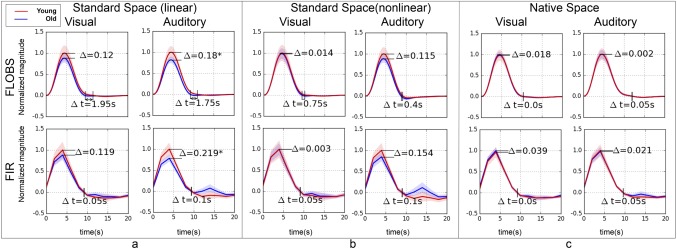
Recapitulating the results in Figure 2 with simulated data. Extracted HRFs from young (red curves) and old (blue curves) subjects using visual and auditory stimulation in simulated fMRI data. For comparison, two separate deconvolution techniques—FLOBS (top row) and FIR (bottom row)—are used for extracting the HRF. Three different fMRI spatial preprocessing pipelines were used: (a) standard space with linear (affine) registration, (b) standard space with nonlinear (warped) registration, and (c) native space analysis without registration. The significant difference in the magnitude is marked with *. None of the observed difference in the delay (from peak to baseline) was significant. [Color figure can be viewed at http://wileyonlinelibrary.com]
The results of the simulated fMRI data indicated that age‐related brain atrophy can introduce marginal delay in the extracted HRF (visual: Δt = 1.95 s, P = 0.80, audio: Δt = 1.75 s, P = 0.37) using standard space time‐series; however, this is only observed with the FLOBS deconvolution technique (Fig. 3a). The observed delay in simulated data was completely removed by extracting the HRF from subject's native space time‐series (visual: Δt = 0.00 s, P = 0.53, audio: Δt = 0.05 s, P = 0.79). These findings may be used as evidence that any atrophy‐related difference observed in the amplitude and timing of the HRF extracted from standard space time‐series will be eliminated by using the native space time‐series. Next, due to the previously mentioned differences in slice acquisition timing between visual and auditory cortices, we sought to investigate whether or not the slice acquisition timing could cause the observed delay of the HRF extracted from native space time‐series in real data.
HRF Delay Due to Slice Acquisition Timing
Figure 4a,b shows the HRF extracted from significant voxels in two adjacent slices within the auditory cortex of a typical subject, with and without slice‐timing correction. As shown in Figure 4a, the extracted HRFs from the uncorrected slices exhibit a delay equal to the delay in their slice acquisition time. Figure 4b shows that the obtained delay in the extracted slicewise HRF was completely eliminated when slice‐timing correction is properly applied in the preprocessing pipeline. Figure 4c,d plots the time to peak of every subjects’ auditory HRF from each of the same two slices with and without slice timing correction. Slice‐timing correction eliminated any slice‐dependent differences in the extracted HRF delay, suggesting that the observed significant age‐related delay in the HFR from native space time‐series of the real fMRI data (Δt = 1.65 s, P < 0.05 Figure 2c), cannot be explained by the slice acquisition delay if proper slice timing was applied.
Figure 4.
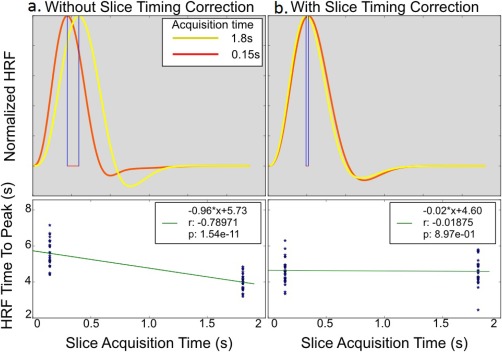
Demonstrating slice‐dependent delay in the extracted HRF from two adjacent slice (slices 17 and 18) in the significantly activated brain area due to auditory stimulus of a typical subject's (top row) unsmoothed data (a) with and (b) without proper slice timing correction. Without slice timing correction, FLOBS accurately captures the same amount of delay in the extracted HRF that are imposed during their acquisition. Bottom row illustrates a linear regression of slice‐dependent HRF time to peak vs the acquisition delay of that slice with and without STC for 35 subjects. Without STC, there is a one‐to‐one (slope is equal to one, β = −0.96*) relationship between the slice‐dependent HRF time to peak and the slice acquisition delay. With STC, this relationship is completely removed. [Color figure can be viewed at http://wileyonlinelibrary.com]
DISCUSSION
Using standard fMRI processing in a small number of participants, we detected significant age‐related differences in the BOLD response to auditory stimuli, with older subjects’ BOLD response being significantly weaker than younger subjects. The simplest interpretation of this finding is that the older subjects present weaker neuronal activity than younger subjects in response to the same auditory stimulus. While we cannot completely rule out this possibility, many researchers have argued that for a brief sensory stimulus with no high‐level information processing, the observed differences might be due to age‐related changes in the neurovascular structure. Numerous pieces of evidence for age‐related changes in the neurovascular structure support this hypothesis [Abernethy et al., 1993; D'Esposito et al., 2003; Farkas and Luiten, 2001; Kalaria, 1996; Knox et al., 1980; Masawa et al., 1994; Moody et al., 1997; S et al., 1979; Sonntag et al., 1997]. A direct consequence of this hypothesis is that if the age‐related neurovascular changes in the brain cause alterations in the BOLD response and HRF amplitude and timing, then the interpretation of the fMRI results in studies of aging becomes challenging. This is because it is hard to distinguish whether any age‐related findings in higher level association cortices are due to changes in the neuronal activity or due to changes in the HRF caused by underlying neurovascular structures.
In this study, we illustrated that age‐related changes in the neuroanatomical structure of the brain (atrophy) represents a confounding factor that can manifest as attenuation in the BOLD response. We showed that the significant age‐related differences in the amplitude of the HRF extracted from subjects’ primary auditory cortex could be eliminated by analyzing the data in subjects’ native space. Our analysis of simulated fMRI data confirmed these findings and further demonstrated that differential brain atrophy alone can cause attenuation in the amplitude of the HRF extracted from older participants. By processing the simulated fMRI data in native space, which used identical HRFs to generate the underlying BOLD signal for both young and old participants, we eliminated the observed attenuation.
The fact that no significant age‐related differences were observed in the amplitude of the HRF extracted from primary visual cortex is in agreement with age‐related brain atrophy literature showing that the lateral occipital region (visual cortex) remains intact with age, whereas the superior temporal and temporal transverse gyri (auditory cortex) have the maximum level of age‐related cortical thickness loss [Fischl et al., 2004b; Fjell et al., 2009]. Our results suggest that greater atrophy in auditory cortex introduces more inaccuracy in their registration to standard template space. The excessive misregistration in older participants causes more time‐series to be extracted from voxels outside the primary auditory cortex that do not present any level of BOLD activation. In more severe misregistration cases, parts of the primary auditory cortex that do in fact exhibit strong BOLD activation can also fall outside of the group‐level ROI mask. We have already demonstrated both these effects in a previous publication [Razlighi et al., 2014]. Incorporation of the signals from areas outside the activated regions and partially neglecting signals from activated areas could attenuate the mean BOLD signal, which would manifest as a decrease in the amplitude of the extracted HRF. Another possibility is that excessive brain deformation in older participants enforces much stronger warping to transfer their morphology to the standard space. Such differential warping may result in differential partial voluming effects that can be the cause of the observed attenuation. However, the latter mechanism assumes that registration has optimally transformed the older participants’ brains to standard space. We have shown in recent work that the correspondence of the cortical regions after spatial normalization is 38%/55% using affine/nonlinear registration [Razlighi, 2016]. Therefore, we think that the actual misregistration rather than the partial voluming during the transformation is the primary cause of the effect detected in our data.
Our finding that brain atrophy induces differences in the amplitude of the HRF between the two age groups could explain some of the divergent findings between studies using co‐registration to standard space, which tend to show significant age‐related amplitude differences in sensory cortices [Buckner et al., 2000; Hu et al., 2013; Mattay et al., 2002; Mohtasib et al., 2012; Raemaekers et al., 2006; Riecker et al., 2003; Zysset et al., 2007], and research using native space analysis, which tend to show no significant differences [Aizenstein et al., 2004; D'Esposito et al., 1999; Huettel et al., 2001; Restom et al., 2007; Yan et al., 2011]. However, it should be noted that not all existing studies can be explained by this categorization [Ances et al., 2009; Handwerker et al., 2007; Hesselmann et al., 2001; Hutchison et al., 2013; Ross et al., 1997; Stefanova et al., 2013; Tekes et al., 2005].
Owing to the limited number of participants in our study, we cannot currently rule out the possibility that age‐related changes in the neurovascular structure either attenuate or magnify the BOLD response. However, it is likely that the effect of age‐related changes in neuroanatomical structure has a stronger effect than age‐related changes in neurovascular structure, as we were able not only to detect an age difference but also to eliminate it with a native space approach even with a small number of participants. However, these effects should be examined in larger samples to rule out the possibility of small vascular effects on the HRF. Therefore, we refrain from making any conclusions about actual age‐related differences in the shape of the HRF. Our goal is to show the effect of brain atrophy as confounding factor for studies investigating age‐related changes in the amplitude of the HRF.
We used two different deconvolution techniques to extract the HRF from fMRI data, selected from the most restricted (FLOBS) and the most relaxed (FIR) methods to examine whether or not the method of deconvolution would have any effect on the observed atrophy‐related changes on HRF amplitude and timing. As seen in Figures 2 and 3, both FLOBS and FIR techniques produced comparable results for the amplitude of the extracted HRF (with one exception in Fig. 2b). However, the timing of the extracted HRF is somewhat different between the two techniques. The main difference is in the temporal resolution of the extracted HRF: while the FIR filter samples the HRF curve with the same sampling rate as the fMRI data, FLOBS can produce results with much higher temporal resolution. While some researchers may consider this to be very beneficial when the timing of the HRF is being investigated, others might raise the possibility that the observed delay is not real but rather enforced by the prior shape constraints applied onto the basis functions in FLOBS. For instance, Figure 2a,b illustrates that using FLOBS detects a rather large but not significant delay in the return to baseline of the BOLD response for older participants. However, all the detected delays in Figure 2a,b were suppressed to smaller or negligible levels using the FIR technique. On the other hand, with such a low temporal resolution (TR = 2 s), the FIR technique also generates some results that do not seem physiologically plausible. For instance, the extremely large, but not significant, delay detected in the older subjects’ BOLD response to auditory stimuli in native space (Fig. 2c) seems unlikely, and could be a direct result of the coarse sampling of the fMRI data. Using simulated fMRI data, we have demonstrated the same level of inconsistency in the timing of the extracted HRF (Fig. 3a,b). However, all the observed (nonsignificant) differences in the timing of the HRF were eliminated by processing the simulated fMRI data in subjects’ native space (Fig. 3c). This suggests that processing data in subjects’ native space can eliminate any atrophy‐related delay in HRF timing. Given these simulated results, it is possible that the significant delay detected in the HRF obtained from real data in subjects’ native space (Fig. 2c) is not due to the age‐related brain atrophy, but is caused by age‐related changes in the neurovascular system. Similar findings have been reported earlier [Taoka et al., 1998]. While we cannot rule out such a possibility at this time, we should emphasize again that with the moderate sampling rate used in our fMRI acquisition (TR = 2 s), it is not possible for us to investigate whether or not the observed delay is real or an artifact of the deconvolution techniques. In the future, fMRI data with higher sampling rate could be used for such an investigation.
Finally, we investigated whether or not the slice‐dependent delay during fMRI data acquisition could account for the observed significant age‐related delay of the HRF extracted from subject's native space auditory cortex. Our results suggest that the delay in the acquisition of each slice is reflected in the extracted HRF from that slice, if slice‐timing correction has not been applied. However, the appropriate application of slice‐timing correction removed the slice‐dependent delay. This finding highlights the fact that neglecting slice‐timing correction from the preprocessing pipeline, or applying it incorrectly, has the potential to introduce delay in the HRF extracted from different groups. Therefore, a proper slice‐timing correction should be considered an essential preprocessing step when investigating any alterations in the timing of the HRF. Among the studies investigating age‐related differences in the HRF, only a few specifically indicated an application of slice‐timing correction during preprocessing [D'Esposito et al., 1999; Hu et al., 2013; Huettel et al., 2001; Riecker et al., 2003; Zysset et al., 2007]. In addition, our results indicate that if slice‐timing correction has been substituted by slice‐dependent predictors in the first‐level GLM analysis, then the HRF extracted from each slice still needs to be corrected for slice acquisition delay.
The process of obtaining group‐level ROI masks when the double‐gamma HRF is used in the first‐level analysis may present a potential bias toward selecting voxels with the same HRF shape. To address this issue, we recomputed the group‐level ROI mask with two different methods; first using a Gaussian HRF and then using ICA, which does not require any prior knowledge about the HRF shape. Repeating the first‐level and second‐level analyses with a Gaussian kernel as the new HRF gave a near‐perfectly overlapping ROI mask (Dice = 95%), which generated similar results for both real and simulated data (see Supporting Information, Figs. S1 and S2). Furthermore, we used group ICA (temporal concatenation) to extract the activated areas for both visual and auditory stimulation. ICA is a multivariate technique that does not require any prior assumption about the shape of the HRF, therefore eliminating any bias toward the selected shape in GLM analysis. The ICA‐generated ROI mask was larger than the one generated by Gaussian or double‐gamma HRFs yet it contained 100% of both ROI masks. Therefore, repeating the standard space analysis using the ICA‐derived ROI mask should generate similar results. Both these experiments provide evidence that while the selected ROI could generate bias toward the double‐gamma canonical HRF, our conclusions are not affected by such a bias.
Any de/convolution method is based on an underlying assumption of linearity. Therefore, our experiments in this work and all existing studies investigating age‐related differences in the HRF are based on the linearity of the BOLD response in old participants. The linearity of the BOLD response to stimuli of a specific duration has been demonstrated for young participants [Boynton et al., 1996; Dale and Buckner, 1997; Robson et al., 1998]. However, to our knowledge, the appropriate linearity has not been examined in old participants. Future studies should consider examination of the linearity assumption in older participants to clarify this issue.
CONCLUSION
Using both simulated and real data, we have shown that age‐related changes in the neuroanatomical structure can introduce changes in the amplitude and timing of the extracted HRF. We also showed that analyzing fMRI data in subjects’ native space eliminates these effects. This finding addresses a number of discrepancies in the literature regarding age‐related alterations in the amplitude and timing of the BOLD response. We also highlighted the importance of proper slice‐timing correction in studies investigating alterations in the timing of the HRF. Our findings strongly suggest that fMRI data for studies of aging should be analyzed in subjects’ native space, as even advanced nonlinear co‐registration could not remove the effect of brain atrophy on the BOLD response.
Supporting information
Supporting Information Figure 1.
Supporting Information Figure 2.
REFERENCES
- Abernethy WB, Bell MA, Morris M, Moody DM (1993): Microvascular density of the human paraventricular nucleus decreases with aging but not hypertension. Exp Neurol 121:270–274. [DOI] [PubMed] [Google Scholar]
- Aizenstein HJ, Clark KA, Butters MA, Cochran J, Stenger VA, Meltzer CC, Reynolds CF, Carter CS (2004): The BOLD hemodynamic response in healthy aging. J Cogn Neurosci 16:786–793. [DOI] [PubMed] [Google Scholar]
- Ances BM, Liang CL, Leontiev O, Perthen JE, Fleisher AS, Lansing AE, Buxton RB (2009): Effects of aging on cerebral blood flow, oxygen metabolism, and blood oxygenation level dependent responses to visual stimulation. Hum Brain Mapp 30:1120–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson JL, Jenkinson M, Smith S (2007): Non‐linear registration, aka Spatial normalisation FMRIB technical report TR07JA2. FMRIB Anal. Group Univ. Oxf. 2.
- Biswal BB, Mennes M, Zuo X‐N, Gohel S, Kelly C, Smith SM, Beckmann CF, Adelstein JS, Buckner RL, Colcombe S, Dogonowski A‐M, Ernst M, Fair D, Hampson M, Hoptman MJ, Hyde JS, Kiviniemi VJ, Kötter R, Li S‐J, Lin C‐P, Lowe MJ, Mackay C, Madden DJ, Madsen KH, Margulies DS, Mayberg HS, McMahon K, Monk CS, Mostofsky SH, Nagel BJ, Pekar JJ, Peltier SJ, Petersen SE, Riedl V, Rombouts SARB, Rypma B, Schlaggar BL, Schmidt S, Seidler RD, Siegle GJ, Sorg C, Teng G‐J, Veijola J, Villringer A, Walter M, Wang L, Weng X‐C, Whitfield‐Gabrieli S, Williamson P, Windischberger C, Zang Y‐F, Zhang H‐Y, Castellanos FX, Milham MP (2010): Toward discovery science of human brain function. Proc Natl Acad Sci USA 107:4734–4739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnéry C, Leclerc P‐O, Desjardins M, Hoge R, Bherer L, Pouliot P, Lesage F (2012): Changes in diffusion path length with old age in diffuse optical tomography. J Biomed Opt 17:0560021–0560028. [DOI] [PubMed] [Google Scholar]
- Boynton GM, Engel SA, Glover GH, Heeger, DJ (1996): Linear Systems Analysis of Functional Magnetic Resonance Imaging in Human V1. J Neurosci 16:4207–4221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodmann K (2007): Brodmann's: Localisation in the Cerebral Cortex. Springer Science & Business Media. [Google Scholar]
- Buckner RL, Snyder AZ, Sanders AL, Raichle ME, Morris JC (2000): Functional brain imaging of young, nondemented, and demented older adults. J Cogn Neurosci 12:24–34. [DOI] [PubMed] [Google Scholar]
- Buxton RB, Wong EC, Frank LR (1998): Dynamics of blood flow and oxygenation changes during brain activation: The balloon model. Magn Reson Med 39:855–864. [DOI] [PubMed] [Google Scholar]
- Dale AM, Buckner RL (1997): Selective averaging of rapidly presented individual trials using fMRI. Hum Brain Mapp 5:329–340. [DOI] [PubMed] [Google Scholar]
- Desjardins M (2015): Vascular correlates of aging in the brain: Evidence from imaging data. IRBM 36:158–165. [Google Scholar]
- D'Esposito M, Deouell LY, Gazzaley A (2003): Alterations in the BOLD fMRI signal with ageing and disease: A challenge for neuroimaging. Nat Rev Neurosci 4:863–872. [DOI] [PubMed] [Google Scholar]
- D'Esposito M, Zarahn E, Aguirre GK, Rypma B (1999): The effect of normal aging on the coupling of neural activity to the bold hemodynamic response. NeuroImage 10:6–14. [DOI] [PubMed] [Google Scholar]
- Donahue MJ, Blicher JU, Østergaard L, Feinberg DA, MacIntosh BJ, Miller KL, Günther M, Jezzard P (2009): Cerebral blood flow, blood volume, and oxygen metabolism dynamics in human visual and motor cortex as measured by whole‐brain multi‐modal magnetic resonance imaging. J Cereb Blood Flow Metab 29:1856–1866. [DOI] [PubMed] [Google Scholar]
- Farkas E, Luiten PGM (2001): Cerebral microvascular pathology in aging and Alzheimer's disease. Prog Neurobiol 64:575–611. [DOI] [PubMed] [Google Scholar]
- Fischl B, Salat DH, Busa E, Albert M, Dieterich M, Haselgrove C, van der Kouwe A, Killiany R, Kennedy D, Klaveness S, Montillo A, Makris N, Rosen B, Dale AM (2002): Whole brain segmentation: Automated labeling of neuroanatomical structures in the human brain. Neuron 33:341–355. [DOI] [PubMed] [Google Scholar]
- Fischl B, Salat DH, van der Kouwe AJW, Makris N, Ségonne F, Quinn BT, Dale AM (2004): Sequence‐independent segmentation of magnetic resonance images. NeuroImage 23:S69–S84. [DOI] [PubMed] [Google Scholar]
- Fjell AM, Westlye LT, Amlien I, Espeseth T, Reinvang I, Raz N, Agartz I, Salat DH, Greve DN, Fischl B, Dale AM, Walhovd KB (2009): High consistency of regional cortical thinning in aging across multiple samples. Cereb Cortex 19:2001–2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover GH (1999): Deconvolution of impulse response in event‐related BOLD fMRI1. NeuroImage 9:416–429. [DOI] [PubMed] [Google Scholar]
- Goutte C, Nielsen FA, Hansen KH (2000): Modeling the hemodynamic response in fMRI using smooth FIR filters. IEEE Trans Med Imaging 19:1188–1201. [DOI] [PubMed] [Google Scholar]
- Greve DN, Fischl B (2009): Accurate and robust brain image alignment using boundary‐based registration. NeuroImage 48:63–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grinband J, Wager TD, Lindquist M, Ferrera VP, Hirsch J (2008): Detection of time‐varying signals in event‐related fMRI designs. NeuroImage 43:509–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handwerker DA, Gazzaley A, Inglis BA, D'Esposito M (2007): Reducing vascular variability of fMRI data across aging populations using a breathholding task. Hum Brain Mapp 28:846–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handwerker DA, Gonzalez‐Castillo J, D'Esposito M, Bandettini PA (2012): The continuing challenge of understanding and modeling hemodynamic variation in fMRI. NeuroImage 62:1017–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesselmann V, Zaro Weber O, Wedekind C, Krings T, Schulte O, Kugel H, Krug B, Klug N, Lackner KJ (2001): Age related signal decrease in functional magnetic resonance imaging during motor stimulation in humans. Neurosci Lett 308:141–144. [DOI] [PubMed] [Google Scholar]
- Hu S, Chao HH‐A, Zhang S, Ide JS, Li C‐SR (2013): Changes in cerebral morphometry and amplitude of low‐frequency fluctuations of BOLD signals during healthy aging: Correlation with inhibitory control. Brain Struct Funct 219:983–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huettel SA, Obembe OO, Song AW, Woldorff MG (2004): The BOLD fMRI refractory effect is specific to stimulus attributes: Evidence from a visual motion paradigm. Neuroimage 23:402–408. [DOI] [PubMed] [Google Scholar]
- Huettel SA, Singerman JD, McCarthy G (2001): The effects of aging upon the hemodynamic response measured by functional MRI. NeuroImage 13:161–175. [DOI] [PubMed] [Google Scholar]
- Hutchison JL, Hubbard NA, Brigante RM, Turner M, Sandoval TI, Hillis GAJ, Weaver T, Rypma B (2014): The efficiency of fMRI region of interest analysis methods for detecting group differences. J Neurosci Methods 226:57–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison JL, Lu H, Rypma B (2013): Neural mechanisms of age‐related slowing: The ΔCBF/ΔCMRO2 ratio mediates age‐differences in BOLD signal and human performance. Cereb Cortex. 23:2337–2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josephs O, Turner R, Friston K (1997): Event‐related fMRI. Hum Brain Mapp 5:243–248. [DOI] [PubMed] [Google Scholar]
- Kalaria RN (1996): Cerebral vessels in ageing and Alzheimer's disease. Pharmacol Ther 72:193–214. [DOI] [PubMed] [Google Scholar]
- Knox CA, Yates RD, Chen I.‐l, Klara PM (1980): Effects of aging on the structural and permeability characteristics of cerebrovasculature in normotensive and hypertensive strains of rats. Acta Neuropathol (Berl) 51:1–13. [DOI] [PubMed] [Google Scholar]
- Krishnan S, Slavin MJ, Tran T‐TT, Doraiswamy PM, Petrella JR (2006): Accuracy of spatial normalization of the hippocampus: Implications for fMRI research in memory disorders. NeuroImage 31:560–571. [DOI] [PubMed] [Google Scholar]
- Masawa N, Yoshida Y, Yamada T, Joshita T, Sato S, Mihara B (1994): Morphometry of structural preservation of tunica media in aged and hypertensive human intracerebral arteries. Stroke 25:122–127. [DOI] [PubMed] [Google Scholar]
- Mattay VS, Fera F, Tessitore A, Hariri AR, Das S, Callicott JH, Weinberger DR (2002): Neurophysiological correlates of age‐related changes in human motor function. Neurology 58:630–635. [DOI] [PubMed] [Google Scholar]
- Mehagnoul‐Schipper DJ, van der Kallen BFW, Colier WNJM, van der Sluijs MC, van Erning LJTO, Thijssen HOM, Oeseburg B, Hoefnagels WHL, Jansen RWMM (2002): Simultaneous measurements of cerebral oxygenation changes during brain activation by near‐infrared spectroscopy and functional magnetic resonance imaging in healthy young and elderly subjects. Hum Brain Mapp 16:14–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohtasib RS, Lumley G, Goodwin JA, Emsley HCA, Sluming V, Parkes LM (2012): Calibrated fMRI during a cognitive Stroop task reveals reduced metabolic response with increasing age. NeuroImage 59:1143–1151. [DOI] [PubMed] [Google Scholar]
- Moody DM, Brown WR, Challa VR, Ghazi‐Birry HS, Reboussin DM (1997): Cerebral microvascular alterations in aging, leukoaraiosis, and Alzheimer's diseases. Ann N Y Acad Sci 826:103–116. [DOI] [PubMed] [Google Scholar]
- Morsheddost H, Asemani D, Shalchy MA (2015): Evaluation of hemodynamic response function in vision and motor brain regions for the young and elderly adults. Basic Clin Neurosci 6:58–68. [PMC free article] [PubMed] [Google Scholar]
- Parker D, Liu X, Razlighi QR (2017): Optimal slice timing correction and its interaction with fMRI parameters and artifacts. Med Image Anal 35:434–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker D, Rotival G, Laine A, Razlighi QR (2014). Retrospective detection of interleaved slice acquisition parameters from fMRI data. Proc IEEE Int Symp Biomed Imaging Nano Macro IEEE Int Symp Biomed Imaging 2014:37–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raemaekers M, Vink M, van den Heuvel MP, Kahn RS, Ramsey NF (2006): Effects of aging on BOLD fMRI during Prosaccades and Antisaccades. J Cogn Neurosci 18:594–603. [DOI] [PubMed] [Google Scholar]
- Razlighi QR, Habeck C, Steffener J, Gazes Y, Zahodne LB, MacKay‐Brandt A, Stern Y (2014): Unilateral disruptions in the default network with aging in native space. Brain Behav 4:143–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razlighi QR (2016): Region‐Based Spatial Normalization for fMRI Research in Brain Aging. Organ Hum Brain Mapp 4061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Restom K, Bangen KJ, Bondi MW, Perthen JE, Liu TT (2007): Cerebral blood flow and BOLD responses to a memory encoding task: A comparison between healthy young and elderly adults. NeuroImage 37:430–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter W, Richter M (2003): The shape of the fMRI BOLD response in children and adults changes systematically with age. NeuroImage 20:1122–1131. [DOI] [PubMed] [Google Scholar]
- Riecker A, Grodd W, Klose U, Schulz JB, Gröschel K, Erb M, Ackermann H, Kastrup A (2003): Relation between regional functional MRI activation and vascular reactivity to carbon dioxide during normal aging. J Cereb Blood Flow Metab 23:565–573. [DOI] [PubMed] [Google Scholar]
- Robson MD, Dorosz JL, Gore JC (1998): Measurements of the temporal fMRI response of the human auditory cortex to trains of tones. NeuroImage 7:185–198. [DOI] [PubMed] [Google Scholar]
- Ross MH, Yurgelun‐Todd DA, Renshaw PF, Maas LC, Mendelson JH, Mello NK, Cohen BM, Levin JM (1997): Age‐related reduction in functional MRI response to photic stimulation. Neurology 48:173–176. [DOI] [PubMed] [Google Scholar]
- S N, H H, A O, Y N, K M, K H (1979): Mechanical properties of human cerebral arteries. Part 1: Effects of age and vascular smooth muscle activation. Surg Neurol 12:297–304. [PubMed] [Google Scholar]
- Salat DH, Buckner RL, Snyder AZ, Greve DN, Desikan RSR, Busa E, Morris JC, Dale AM, Fischl B (2004): Thinning of the cerebral cortex in aging. Cereb Cortex 14:721–730. [DOI] [PubMed] [Google Scholar]
- Sladky R, Friston KJ, Tröstl J, Cunnington R, Moser E, Windischberger C (2011): Slice‐timing effects and their correction in functional MRI. NeuroImage 58:588–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonntag WE, Lynch CD, Cooney PT, Hutchins PM (1997): Decreases in cerebral microvasculature with age are associated with the decline in growth hormone and insulin‐like growth factor 1*. Endocrinology 138:3515–3520. [DOI] [PubMed] [Google Scholar]
- Stefanova I, Stephan T, Becker‐Bense S, Dera T, Brandt T, Dieterich M (2013): Age‐related changes of blood‐oxygen‐level–dependent signal dynamics during optokinetic stimulation. Neurobiol Aging 34:2277–2286. [DOI] [PubMed] [Google Scholar]
- Taoka T, Iwasaki S, Uchida H, Fukusumi A, Nakagawa H, Kichikawa K, Takayama K, Yoshioka T, Takewa M, Ohishi H (1998): Age correlation of the time lag in signal change on EPI‐fMRI. J Comput Assist Tomogr 22:514–517. [DOI] [PubMed] [Google Scholar]
- Tekes A, Mohamed MA, Browner NM, Calhoun VD, Yousem DM (2005): Effect of age on visuomotor functional MR imaging1. Acad Radiol 12:739–745. [DOI] [PubMed] [Google Scholar]
- von Economo CF, Koskinas GN, Triarhou LC (2008): Atlas of cytoarchitectonics of the adult human cerebral cortex. Karger Basel [Google Scholar]
- Woolrich MW, Behrens TE, Smith SM (2004): Constrained linear basis sets for HRF modelling using Variational Bayes. NeuroImage 21:1748–1761. [DOI] [PubMed] [Google Scholar]
- Yan L, Zhuo Y, Wang B, Wang DJ (2011): Loss of coherence of low frequency fluctuations of BOLD FMRI in visual cortex of healthy aged subjects. Open Neuroimaging J 5:105–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zysset S, Schroeter ML, Neumann J, Yves von Cramon D (2007): Stroop interference, hemodynamic response and aging: An event‐related fMRI study. Neurobiol Aging 28:937–946. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information Figure 1.
Supporting Information Figure 2.


