Abstract
Vascular endothelial growth factor (VEGF) plays a critical role in the angiogenesis and proliferation of various types of cells such as neurons, astroglia, and endothelial cells in the brain. A common polymorphism in the VEGF gene (−2578 C/A) is associated with circulating VEGF levels, cancers and Alzheimer's disease. Nonetheless, the effects of this polymorphism on normal human brain volume, arterial blood volume, and blood supply remain unclear. In this study, the effects of this polymorphism on the total gray matter volume (TGMV) and total white matter volume (TWMV) using T1‐weighted structural images and the total arterial blood volume (TABV) and mean cerebral blood flow (mCBF) during rest using arterial spin labeling (ASL) in 765 young adult humans were investigated. Voxel‐by‐voxel whole‐brain analyses of these measures were also performed. Multiple regression analyses with age and sex as covariates revealed that the VEGF genotype (number of C alleles) was significantly and positively correlated with TGMV, TWMV, and TABV as well as with regional gray and white matter volumes in widespread areas and regional arterial blood volume in some areas with high arterial blood volume. However, these regional associations were not seen when the corresponding global signal was included as a covariate in the multiple regression analyses, indicating that we failed to obtain evidence of region‐specific associations between these brain measures and the genotype. The results suggest that the VEGF‐2578C allele, is associated with changes in the vascular system that lead to increased blood volume and larger brain volume. Hum Brain Mapp 38:3516–3526, 2017. © 2017 Wiley Periodicals, Inc.
Keywords: VEGF, brain, polymorphism, blood volume, brain volume
INTRODUCTION
Vascular endothelial growth factor (VEGF) plays a critical role in the angiogenesis [Ferrara et al., 2003]. Moreover, independent of vessel‐mediated effects, VEGF has direct effects on neural tissues [Rosenstein et al., 2010]. VEGF plays a key role in the proliferation of various types of cells such as neurons, astroglia, and endothelial cells in the brain [Jin et al., 2002] and promotes a wide range of neuronal functions, both in vitro and in vivo, including neurogenesis, neuronal migration, neuronal survival, and axon guidance [Mackenzie and Ruhrberg, 2012]. The role of VEGF is not limited to these during development but extends into the plasticity of vessels for the increased demand for the supply of blood, both in neural systems and in pathological states (tumors) requiring nutritional supply [Ferrara et al., 2003; Hillman et al., 2008]. A common polymorphism in the VEGF gene (−2578 C/A) has been shown to robustly affect circulating VEGF levels [Steffensen et al., 2010], VEGF gene expression, and VO2 max [Prior et al., 2006]. Circulating VEGF is known to be associated with not only cancer and tumor size [Broll et al., 2001], but also several brain diseases [Greenberg and Jin, 2004; Mateo et al., 2007]. Consistently, this VEGF polymorphism has been shown to be associated with cancer [for a meta analysis, see Zhao et al., 2012] and several brain diseases such as Alzheimer's disease [for meta analyses, see Del Bo et al., 2009; Liu et al., 2013]. Multiple VEGF polymorphisms are reported to be associated with specific phenotypes or endophenotypes; for example, −460 T/C and +405 G/C are both associated with circulating VEGF levels [Steffensen et al., 2010]. However, we focused on −2578 C/A because associations between this polymorphism and various phenotypes and endophenotypes have been confirmed (often by meta‐analyses).
However, despite the importance of VEGF and its polymorphism in the formation of blood vessels, effects on cell proliferations in the brain, and pathological states (including those in neural systems), the effects of this VEGF gene polymorphism (−2578 C/A) on normal human structural brain volume, arterial blood volume in the default state (which could partly be associated with vessel volume), and blood supply remain unclear. The present study aimed to investigate these issues. This polymorphism, the 2578C allele, is known to lead to increased VEGF circulating levels [Steffensen et al., 2010], and subsequent associated phenotypes can be understood from the consequence of the increased capacity for blood supply through the increased angiogenesis. In addition, as described, VEGF is known to facilitate the proliferation of various types of cells such as neurons, astroglia, and endothelial cells in the brain [Jin et al., 2002]. Thus, we hypothesized the 2578C allele to be associated with increased blood vessel volume (which in turn leads to increased arterial blood volume), increased blood supply, and larger brain volume.
For this purpose, we recruited a large sample of several hundred normal young adult subjects and scanned their brain through magnetic resonance imaging (MRI). Brain structural volume of each individual was assessed by the total gray matter volume (TGMV) and total white matter volume (TWMV). In addition, to assess the arterial blood volume and blood supply, we measured the total arterial blood volume (TABV) and mean cerebral blood flow (mCBF) during rest using arterial spin labeling (ASL; see Methods). We focused on these total or mean measures of the brain based on the assumed global effects of VEGF on brain and vascular mechanisms. However, there is an important body of evidence supporting the local effects of VEGF. In animal models, VEGF has been shown to play a role in both paracrine and autocrine signaling in the maintenance of neurons and endothelia in the central nervous system [Ogunshola et al., 2002]. It also has both indirect and direct vessel‐mediated effects on neural tissues [Rosenstein et al., 2010]. Also, following cerebral ischemia, VEGF enhanced the delayed survival of newborn neurons in the dentate gyrus and subventricular zone, and stimulated angiogenesis in the striatal ischemic penumbra, but not in the dentate gyrus, suggesting the existence of region‐specific effects [Sun et al., 2003]. VEGF promotes hippocampal cell proliferation [Oosthuyse et al., 2001]. In addition, VEGF increases long‐term potentiation (LTP) responses in the dentate gyrus and improves memory primarily a result of increasing plasticity of mature neurons rather than through the contribution of newly added hippocampal neurons [Licht et al., 2011]. In addition, insufficient VEGF‐dependent neuroprotection specifically leads to motor neuron degeneration [Ogunshola et al., 2002] and insufficient VEGF levels plays an important role in the progression of amyotrophic lateral sclerosis [Cleveland and Rothstein, 2001]. Therefore, we also performed supplementary voxel‐by‐voxel whole‐brain analyses.
Based on these investigations, we were able to demonstrate the role of VEGF and its gene polymorphism on normal human structures and the vascular system. Furthermore, these investigations will provide new insights into the mechanism underlying the protective and facilitative effects of this polymorphism on pathological states.
MATERIAL AND METHODS
Subjects
Data from 765 healthy, right‐handed individuals (425 men and 340 women; 20.7 ± 1.9 years of age) were used in this study as a part of an ongoing project, consisting of various types of MRI scanning and psychological test batteries besides the ones analyzed in this manuscript, to investigate associations among brain imaging, cognitive functions, aging, genetics, and daily habits [Takeuchi et al., 2013a, 2013c, 2014]. Data derived from the subjects in this study are also going to be used in other studies that are irrelevant to this study. Some of the subjects who participated in this study also became subjects of intervention studies (psychological and imaging data recorded before the intervention were used in this study) [Takeuchi et al., 2013b]. All subjects were university, college, or postgraduate students or subjects who had graduated from these institutions within 1 year before the experiment and had normal vision. None had a history of neurological or psychiatric illness. Handedness was evaluated using the Edinburgh Handedness Inventory [Oldfield, 1971]. Written informed consent was obtained from each subject in accordance with the Declaration of Helsinki (1991). This study was approved by the Ethics Committee of Tohoku University.
Genotyping of Subjects
Saliva samples from the participants were collected in Oragene containers (DNA Genotek Inc., Ottawa, Canada), and high‐molecular‐weight DNA was isolated using Oragene saliva DNA extraction kit (DNA Genotek Inc., Ottawa, Canada) according to the manufacturer's protocol. Genotype of the common VEGF promoter SNP rs699947 (22578C/A) was determined using the assay‐on‐demand (Applied Biosystems, Foster City, CA)‐based allelic (VIC‐ and FAM‐labeled) discrimination method. The reaction was performed in 10 ml volume, containing genomic DNA samples (20 ng/2.5 mL), 0.25 µL of the above assay‐on‐demand assay mixture, and 5 µM of TaqMan Universal PCR Master Mix (Applied Biosystems, Foster City, CA), on 96‐well plates using the CFX96 real‐time PCR detection system, according to manufacturer's instructions (Bio‐Rad Laboratories, Inc., Hercules, CA). Reaction conditions used with the thermal cycler were as follows: an initial incubation at 95°C for 10 min; 50 cycles of 95°C for 15 s, and 60°C for 60 s. As a negative control, PCR mixtures without DNA sample were run in several wells of every 96‐well plates to ensure that the mixture was free from any contamination. The genotyping was replicated for all DNA samples to assess reproducibility of the genotyping procedure, and the concordance was 100%. The PCR products of representative subjects were verified by DNA sequencing.
Among the 777 participants in this study, data for the polymorphism was successfully obtained from 765 subjects (425 men and 340 women; 20.7 ± 1.9 years of age); genotyping data of 12 subjects were not available, due to failures either in proper extraction of DNA sample from saliva (10 subjects), or in amplification in the PCR procedure (2 subjects), probably due to the quality of saliva sample. The genotypic distributions of the 765 subjects were as follows: VEGF(−2578)A/A (n = 90, 11.8%), VEGF(−2578)A/C (n = 292, 38.2%), and VEGF(−2578)C/C (n = 383, 50.1%). Allele frequencies of A and C alleles were 31% and 69%, respectively, which were concordant with previous findings. Tests for the Hardy–Weinberg equilibrium exhibited no deviations from the expected genotype distribution (P > 0.05).
Assessment of Psychometric Measures of General Intelligence
Raven's Advanced Progressive Matrix [Raven, 1998], which is often shown to be the measure most correlated with general intelligence and thus the best measure of general intelligence [Raven, 1998], was used to assess intelligence and provide subjects' basic characteristics. We did not have a particular hypothesis regarding the psychological measures in this study. For additional details on administration of Raven's Advanced Progressive Matrix, refer to Takeuchi et al. [2010a, 2010b].
Image acquisition
All MRI data acquisition was performed using a 3‐T Philips Achieva scanner. Using a magnetization‐prepared rapid gradient echo sequence, high‐resolution T1‐weighted structural images (240 × 240 matrix, TR = 6.5 ms, TE = 3 ms, FOV = 24 cm, 162 slices, slice thickness = 1 mm) were acquired. ASL was performed to measure resting state CBF and ABV. As described elsewhere [Takeuchi et al., 2011a], it was performed with quantitative signal‐intensity targeting by alternating the radio‐frequency pulse labeling of arterial regions (QUASAR), a pulsed ASL method [Petersen et al., 2006a]. Details of the sequence and the method for calculating perfusion parameters have been outlined elsewhere [Petersen et al., 2006a, 2006b, 2010]. The actual imaging parameters were as follows: 64 × 64 matrix, TR = 300 ms, TE = 22 ms, FOV = 24 cm, 7 slices, slice thickness = 7 mm (2.0 mm gap), SENSE = 2.5, 84 averages, scan duration = 5 min 52 s. We determined the position of the slice by putting the fourth of seven slices on the body of the corpus callosum in the coronal scout view [Taki et al., 2011]. During ASL scan, the subjects were instructed to remain still with their eyes closed, as motionless as possible, and not to sleep or think about anything in particular. Maps of raw resting‐rCBF, ABV and the longitudinal relaxivity [R1(=1/T1)] of each subject were obtained using dedicated software running on IDL (Research Systems, Boulder, Colorado) [Petersen et al., 2006a; National Neuroscience Institute, Singapore]. The R1 map was obtained as part of the ASL scan. The technical details of this scan were described in the previous study [Petersen et al., 2006a]1. The following constants were used in CBF calculation: T1 of arterial blood, 1.65 s; inversion efficiency, 95%; blood–brain partition coefficients for GM and WM (0.98 and 0.82, respectively) [Petersen et al., 2006a].
Preprocessing of Imaging Data
Preprocessing of structural and ASL data was performed using SPM8 implemented in Matlab.
Preprocessing for T1‐weighted structural images for VBM analyses was performed as has been fully described in our previous studies [Takeuchi et al., 2015]. In brief, using a new segmentation algorithm [Ashburner and Friston, 2005] implemented in SPM8, T1‐weighted structural images (Fig. 1a) obtained for each subject were segmented into six tissues [Fig. 1b: gray matter, white matter, cerebrospinal fluid (CSF), skull, soft tissue outside the brain, and air and other matter outside the head]. We then proceeded to the Diffeomorphic Anatomical Registration Through Exponentiated Lie Algebra (DARTEL) registration process implemented in SPM8, and each subject's image was registered using this procedure. The resulting images were spatially normalized to the Montreal Neurological Institute (MNI) space to images with 1.5 × 1.5 × 1.5 mm3 voxels (Fig. 1c). In addition, we performed a volume change correction (modulation) by modulating each voxel with the Jacobian determinants derived from spatial normalization, which allowed us to determine regional differences in the absolute amount of brain tissue [Ashburner and Friston, 2000]. Next, TGMV and TWMV were calculated from these normalized rGMV and rWMV maps and used in analyses. On the other hand, normalized rGMV and rWMV maps were smoothed (8 mm full‐width half‐maximum) and subjected to the whole brain group‐level analyses of the genotype. For additional details of these preprocessing procedures and rationales, please see our previous study [Takeuchi et al., 2015].
Figure 1.

The images during preprocessing. (a) A raw T1‐weighted structural image. (b) Segmented tissues. (c) Normalized segmented tissues. (d) An artificial image created from the function of “1 × GM map + 2 × WM map.” (e) A raw R1 map. (f) A raw CBF map. (g) A raw aBV map. (h) Normalized rCBF and aBV maps. (i) A mask image of the areas where subjects were commonly scanned in ASL scans. (j) Normalized and masked rCBF and aBV maps. (k) Normalized and masked segmented tissues.
For analyses of ASL measures using each individual's unnormalized segmented GM map and WM map, which were produced from the abovementioned segmentation process, we created an artificial image created from the function of “1 × GM map + 2 × WM map” for each individual (Fig. 1d, we call this artificial “1 × GM + 2 × WM map” in this subsection). The contrast of this artificial “1 × GM + 2 × WM map” was similar to that of the R1 map of ASL (as confirmed by visual inspection) and was used in the following preprocessing of ASL images. In addition, the R1 map of ASL that is preprocessed by the abovementioned software lacked the skull and skin sections (Fig. 1e).
Next, using the within‐subject registration method, each individual's R1 map of ASL [which is in alignment with each individual's rCBF map (Fig. 1f) and ABV map (Fig. 1g)] was coregistered to the individual's artificial “1 × GM + 2 × WM map” together with the rCBF map and ABV map (this means that each individual's ASL images were coregistered to each individual's T1‐weighted structural image). The R1 map, rCBF map, and ABV map from each subject were then resliced to 3 × 3 × 3 mm3 voxels. Next, these coregistered ASL‐related images were normalized using the normalization parameter of the T1‐weighted structural image created in the abovementioned preprocessing procedure of the T1‐weighted structural image (Fig. 1h). For the volume measure of ASL (ABV), we performed a volume change correction (modulation) by modulating each voxel using the Jacobian determinants derived from spatial normalization, allowing for the determination of regional differences in the absolute amount of ABV [Ashburner and Friston, 2000].
We resliced the R1, rCBF, and ABV maps to 3 × 3 × 3 mm3 voxels before normalization, since without this procedure, normalization of these ASL maps to standard space of 3 × 3 × 3 mm3 voxels resulted in image distortions.
Following this, using the normalized R1 map, a mask image of the areas where subjects were commonly scanned in ASL scans was created (Fig. 1i). The areas of this mask were defined as the areas where all the 63 subjects of this project (from which the template of this experiment was created [see Takeuchi et al., 2015 for details]) had signals larger than 0 in the normalized R1 maps. We applied this common mask to the normalized rCBF map and ABV map of each participant (Fig. 1j). Next, the mean rCBF value and TABV within this mask were calculated for each subject. This masking procedure was performed because the ASL scan cannot cover the entire brain and we had to avoid the possibility that the differences of the areas scanned affect individual differences in values of ASL measures.
In addition, we applied this mask to the normalized gray matter‐ and white matter‐segmented images (from the preprocessing procedure of T1‐weighted structural images; Fig. 1k) and extracted the mean gray matter signal and mean white matter signal in the masked normalized gray and white matter segmented images. We then calculated the ratio of the total signal of gray matter and total signal of white matter in the masked area. This value was added as a covariate in the analysis of the associations between genotype and TABV or mean rCBF value. This procedure was performed to rule out the possibility that the difference of extent (density) of gray matter and white matter, (there are substantial differences in rCBF values in gray matter and white matter areas) impacts mCBF and TABV,
Voxel‐by‐voxel analyses of normalized and masked rCBF maps and the regional ABV map were smoothed (8 mm full‐width half‐maximum) and subjected to whole‐brain group‐level analyses of the genotype.
Statistical Group‐Level Analysis of Genetic Data
Genetic data were analyzed using SPSS 16.0 (SPSS Inc., Chicago, IL). Multiple regression analyses were employed to investigate the association between the VEGF gene polymorphism [VEGF(−−2578)A/A = 0, VEGF(−2578)A/C = 1, and VEGF(−2578)C/C = 2] and imaging values (TGMV, TWMV, mCBF, and TABV). We assumed this type of quantitative model (treating three types of genotypes as continuous variables), based on the results of the meta‐analysis of this polymorphism using the largest sample [Cao et al., 2011], which suggested the order of effects of VEGF(−−2578)A/A, VEGF(−2578)A/C, and VEGF(−2578)C/C. However, even when analyses of covariance (ANCOVAs) models that treated the three genotypes as a fixed group factor with other variables that are described below are added as covariates, were employed, all the significant associations between the VEGF gene polymorphism and imaging values in this study remain significant (P values of the effects of VEGF polymorphism on TGMV, TWMV, and TABV were 0.004, 0.004, and 0.044, respectively). We nonetheless chose quantitative model to increase the sensitivity of the analyses because increasing the sensitivity of analyses is critical for studies of polymorphisms, which usually have to find tiny effects using a large sample.
Analyses of T1W1‐related measures were performed with covariates of age and sex, while analyses of mCBF measures were performed with covariates of age, sex, and the ratio between the mean gray matter density and mean white matter density of the analyzed area. Results with a threshold of P < 0.05, were considered statistically significant.
Whole‐brain imaging data analysis
Whole‐brain statistical analyses of the brain imaging data were performed using SPM8. Whole‐brain multiple regression analyses were performed to investigate the associations between the VEGF genotype (number of C alleles) and rGMV, rWMV, rCBF, and ABV.
For each imaging measure, we performed two multiple regression analyses. In one type of multiple regression analysis, the covariates were age and sex. In another type of multiple regression analysis, the covariates were age, sex, and global signal (the mean imaging signal in the analyzed area). We used the former procedure because we were interested in the absolute differences (compared with the relative difference between imaging measures such as rGMV of one area and those of other brain regions) with regard to the VEGF genotype [Mechelli et al., 2005]. Removing the global differences would correspond to assuming that the total GMV (etc.) would be the same in all participants, and in this case, rGMV represents the relative difference compared with other areas.
In the analysis of rGMV (or rWMV), we only included voxels that showed rGMV (or rWMV) values of greater than 0.05 in all subjects. The analyses of rCBF and ABV were limited to the mask of the scanned area that was created above. A multiple comparison correction of the cross‐sectional analyses was performed using threshold‐free cluster enhancement (TFCE) [Smith and Nichols, 2009], with randomized (5,000 permutations) nonparametric permutation testing in the TFCE toolbox (http://dbm.neuro.uni-jena.de/tfce/). We applied the threshold of an FWE corrected P < 0.05.
RESULTS
Genetic Analysis
The basic demographic variables were presented in Table 1.
Table 1.
Subject's basic demographics as a function of the VEGF genotype
| VEGF(−2578)A/A | VEGF(−2578)A/C | VEGF(−2578)C/C | P value[Link], uncorrected | |
|---|---|---|---|---|
| Male/Female | 42/48 | 164/128 | 219/164 | 0.132 |
|
Age (mean ± SD) (range) |
20.3 ± 1.7 18‐26 |
20.8 ± 1.9 18‐27 |
20.7 ± 1.8 18‐27 |
0.228 |
|
Raven's Advanced Progressive Matrix (mean ± SD) (range) |
28.6 ± 3.8 20‐36 |
28.7 ± 3.8 15‐36 |
28.6 ± 3.6 17‐36 |
0.884 |
VEGF, vascular endothelial growth factor; SD, standard deviation.
Simple regression analysis between these variables and the VEGF genotype.
We employed multiple regression analyses that coded genotype as a continuous variable [VEGF(−2578)A/A = 0, VEGF(−2578)A/C = 1, VEGF(−2578)C/C = 2]. Multiple regression analysis revealed that after controlling for confounding variables, the VEGF genotype (number of C alleles) was significantly and positively correlated with TGMV, TWMV, and TABV but not with mCBF (Fig. 2a–c, for average and statistical values, see Table 2).
Figure 2.

Associations between VGEF genotype (number of C alleles) and brain measures. Partial regression plots with trend lines depicting the correlations between residuals in the multiple regression analyses with (a) total gray matter volume, (b) total white matter volume, and (c) mean arterial blood volume in the analyzed area as the dependent variable and number of C alleles and other confounding variables as independent variables.
Table 2.
Subject characteristics as a function of VEGF genotype and statistical values from multiple regression analyses
| VEGF(−2578)A/A | VEGF(−2578)A/C | VEGF(−2578)C/C |
P value (uncorrected) t value, β |
|
|---|---|---|---|---|
| Total gray matter volume | 690.1 ± 56.1 | 696.2 ± 57.9 | 707.7 ± 56.8 |
1.83 × 10−3 2.915, 0.081 |
| Total white matter volume | 492.5 ± 41.4 | 499.1 ± 43.4 | 507.7 ± 43.2 |
9.50 × 10−4 3.116, 0.091 |
| Mean cerebral blood flow | 29.1 ± 4.3 | 29.7 ± 4.5 | 29.7 ± 4.8 |
0.122 1.163, 0.041 |
| Total arterial blood volume (a.u.) | 151.6 ± 49.1 | 162.6 ± 54.4 | 168.4 ± 53.1 |
6.83 × 10−3 2.472, 0.089 |
VEGF, vascular endothelial growth factor; SD, standard deviation.
Whole‐Brain Analysis of the Correlation Between Regional Gray Matter Volume and VEGF Genotype
A whole‐brain multiple regression analysis corrected for age and sex revealed that the VEGF genotype (number of C alleles) was significantly and positively correlated with rGMV in extensive gray matter areas across the whole brain, particularly in subcortical areas (Fig. 3a, b). For statistical values and anatomical areas where significant correlations were found, see Supporting Information Table 1.
Figure 3.
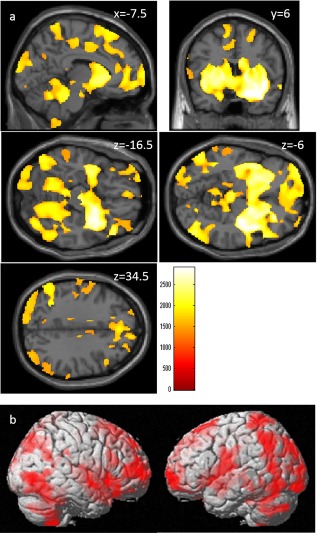
Positive correlations between VGEF genotype (number of C alleles) and rGMV. The results shown were obtained using a threshold of threshold‐free cluster enhancement (TFCE) of P < 0.05, based on 5,000 permutations. The results were corrected at the whole‐brain level. (a) Regions with significant correlations are overlaid on a “single subject” T1 image from SPM8. The color represents the strength of the TFCE value. Significant correlations were found in the extensive gray matter areas across the whole brain, particularly in subcortical areas. (b) Regions with significant correlations are projected onto the rendered brain from SPM8. Significant correlations were found in the extensive gray matter areas across the whole brain.
In a whole‐brain multiple regression analysis corrected for age, sex, and global rGMV signal, the VEGF genotype (number of C alleles) was not significantly correlated with any of the regions.
Whole‐Brain Analysis of the Correlation Between Regional White Matter Volume and VEGF Genotype
A whole‐brain multiple regression analysis corrected for age and sex revealed that the VEGF genotype (number of C alleles) was significantly and positively correlated with rWMV in the extensive white matter areas across the whole brain (Fig. 4). For statistical values and anatomical areas where significant correlations were found, see Supporting Information Table 2.
Figure 4.

Positive correlations between VGEF genotype (number of C alleles) and rWMV. The results shown were obtained using a threshold of threshold‐free cluster enhancement (TFCE) of P < 0.05, based on 5,000 permutations. The results were corrected at the whole‐brain level. Regions with significant correlations are overlaid on a “single subject” T1 image from SPM8. The color represents the strength of the TFCE value. Significant correlations were found in the extensive white matter areas across the whole brain.
In a whole‐brain multiple regression analysis corrected for age, sex, and global rWMV signal, the VEGF genotype (number of C alleles) was not significantly correlated with WMV in any of the regions.
Whole‐Brain Analysis of the Correlation between Regional Arterial Blood Volume and VEGF Genotype
A whole‐brain multiple regression analysis corrected for age and sex revealed that the VEGF genotype (number of C alleles) was significantly and positively correlated with ABV in the anterior midline and bilateral temporal areas (Fig. 5). Areas of significant correlations were concentrated in the areas with greater signals (see right panel of the Fig. 5), possibly because of a greater signal to noise ratio in these areas. For statistical values and anatomical areas where significant correlations were found, see Supporting Information Table 3.
Figure 5.
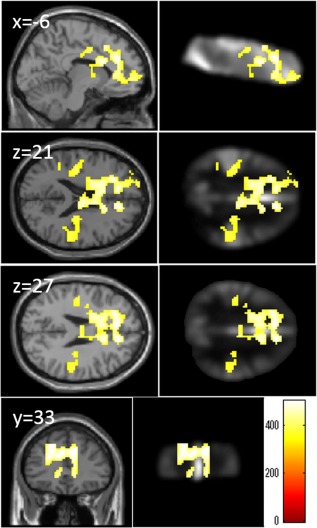
Positive correlations between VGEF genotype (number of C alleles) and regional ABV. The results shown were obtained using a threshold of threshold‐free cluster enhancement (TFCE) of P < 0.05, based on 5,000 permutations. The color represents the strength of the TFCE value. The results were corrected at the whole‐brain level. In the left panels, regions with significant correlations are overlaid on a “single subject” T1 image from SPM8. In the right panels, regions with significant correlations are overlaid on the mean of preprocessed images acquired from those subjects used to create the template. Significant correlations were found in the anterior midline areas and bilateral temporal areas. Note that only part of the whole brain was scanned and analyzed, as can be seen in the right panel.
In a whole‐brain multiple regression analysis corrected for age, sex, and global ABV signal, the VEGF genotype (number of C alleles) was not significantly correlated with ABV in any of the regions.
DISCUSSION
In this study, we demonstrated that the VEGF −2578C allele was associated with significantly larger TGMV, TWMV, and TABV, which was consistent with our hypothesis. Voxel‐by‐voxel analyses in which the global signals were not regressed out revealed widespread associations between the VEGF −2578C allele and greater rWMV and rWMV and the associations between the VEGF −2578C allele and greater regional ABV in some of the areas where ABV is larger. However, in any of these cases, there were no significant associations with the VEGF −2578C allele after regressing out the corresponding global signal, indicating that we failed to obtain the evidence for region‐specific associations between these brain measures and the genotype. However, the VEGF −2578C allele was not associated with mCBF, though a statistical result close to a tendency toward the expected direction (P = 0.122) was observed. The results suggest that the VEGF −2578C allele, which is robustly known to increase the risk of cancer and decrease the risk of Alzheimer's disease, is associated with changes in the vascular system that lead to increased blood volume and larger brain volume. There were no significant differences in the performance of general intelligence measures between genotypes, suggesting lack of evidence that the genotypic neural difference is substantially affecting general cognitive function in the present sample.
As reasoned in the Introduction, the VEGF −2578C allele is known to lead to increased VEGF levels [Steffensen et al., 2010]. VEGF is critical for the formation of blood vessels, and this may lead to increased cerebral ABV. However, other possibilities such as increased vasodilation in the default state cannot be excluded from this study. In addition, VEGF is known to facilitate the proliferation of various types of cells such as neurons, astroglia, and endothelial cells in the brain [Jin et al., 2002]. Increased capacity for blood supply through the increased blood vessels throughout development may enable efficient formation of brain volume, as observed in this study, while increasing the risk of cancer in later life [Broll et al., 2001].
One interesting speculation is that greater cortical and vessel structures caused by the VEGF −2578C allele may work as an extra reserve to reduce the risk of Alzheimer's disease. Meta‐analyses have shown the association between the VEGF −2578A allele and the increased risk of Alzheimer's disease [Del Bo et al., 2009; Liu et al., 2013] as well as other diseases [Borroni et al., 2010; Borroni et al., 2008]. It is known that Alzheimer's disease is associated with cortical atrophy [Frisoni et al., 2005] and reduced blood supply [Yoshiura et al., 2009]. The declines in these measures in old age are essentially associated with functional losses [Head et al., 2008]. Moreover, Katzman et al. [1988] showed that individuals with larger brains were more likely to remain unaffected by dementia despite having similar numbers of Alzheimer lesions to those who were suffering dementia. Perhaps, greater brain volume (structures) and changes in the vascular system that lead to increased arterial blood volume caused by the VEGF −2578C allele may work as an extra reserve against the development of Alzheimer's disease because subjects with the allele have a greater preservation of those tissues and need to undergo greater tissue loss before showing symptoms of dementia.
The reason why we could not obtain a significant association between VEGF gene polymorphism and mCBF remains unclear. Because a P value of 0.128 in the expected direction was obtained and given that single common polymorphisms usually have small effect sizes at least on phenotypes [Terracciano et al., 2008], the lack of significance may just reflect a lack of statistical power. Second, CBF is affected by many transient factors such as mood of the day [Schneider et al., 1994], and the test–retest reliability of CBF is not as high as that of brain structure measures [Petersen et al., 2010]. These factors may partly explain the lack of a significant association in this study.
This study had at least one limitation, which was common to our previous studies and other studies that used college cohorts [Jung et al., 2010; Song et al., 2008; Takeuchi et al., 2010a, b, 2011b], that is, limited sampling among young, healthy subjects with a high educational background. As described previously, limited sampling from the full range of intellectual abilities is a common hazard when sampling from college cohorts [Jung et al., 2010]. Nonetheless, it should be considered that limited sampling may be an important step to rule out the effects of age or education level that could strongly influence the brain structures and increase the sensitivity of the analyses. Also, this is a cross‐sectional genetic study, and although the polymorphism is associated with phenotype, phenotype cannot affect polymorphism (also, false correlation due to mediation by a third factor is usually unlikely in studies of polymorphisms). However, how the change in gene sequence leads to the observed changes in brain structure cannot be revealed in this kind of study. This needs to be revealed in future studies.
Supporting information
Supporting Information Figure 1.
Supporting Information Tables.
ACKNOWLEDGMENTS
We thank Yuki Yamada for operating the MRI scanner, Mutsumi Oohashi for performing the genotyping, all other assistants for helping with the experiments and the study, and the study participants and all our other colleagues at IDAC, Tohoku University for their support.
Footnotes
In this previous study [Petersen ET, Lim T, Golay X (2006a): Model‐free arterial spin labeling quantification approach for perfusion MRI. Magn Reson Med 55(2):219–232], the R1,app,eff map is referred. However, it is proper to call the map we used in this study as R1 map. The R1 map is calculated from the Look‐Locker readout. In order to get R1 and not R1,app,eff the control images are used and scans are acquired at nominal flip angle as well as 1/3 of nominal as part of the sequence. This allow to estimate the actual flip angle in the Look‐Locker readout on a voxel by voxel basis and thereby calculate R1 from the acquired R1,eff (Personal communication with the Dr. Esben Thade Petersen).
REFERENCES
- Ashburner J, Friston KJ (2000): Voxel‐based morphometry‐the methods. Neuroimage 11:805–821. [DOI] [PubMed] [Google Scholar]
- Ashburner J, Friston KJ (2005): Unified segmentation. Neuroimage 26:839–851. [DOI] [PubMed] [Google Scholar]
- Borroni B, Ghezzi S, Agosti C, Archetti S, Fenoglio C, Galimberti D, Scarpini E, Di Luca M, Bresolin N, Comi G (2008): Preliminary evidence that VEGF genetic variability confers susceptibility to frontotemporal lobar degeneration. Rejuven Res 11:773–780. [DOI] [PubMed] [Google Scholar]
- Borroni B, Del Bo R, Goldwurm S, Archetti S, Bonvicini C, Agosti C, Bigni B, Papetti A, Ghezzi S, Sacilotto G (2010): VEGF haplotypes are associated with increased risk to progressive supranuclear palsy and corticobasal syndrome. J Alzheim Dis 21:87–94. [DOI] [PubMed] [Google Scholar]
- Broll R, Erdmann H, Duchrow M, Oevermann E, Schwandner O, Markert U, Bruch H, Windhövel U (2001): Vascular endothelial growth factor (VEGF)–a valuable serum tumour marker in patients with colorectal cancer?. Eur J Surg Oncol (EJSO) 27:37–42. [DOI] [PubMed] [Google Scholar]
- Cao C, Ying T, Fang J‐J, Sun S‐F, Lv D, Chen Z‐B, Ma H‐Y, Yu Y‐M, Ding Q‐L, Shu L‐H (2011): Polymorphism of vascular endothelial growth factor–2578C/A with cancer risk: Evidence from 11263 subjects. Med Oncol 28:1169–1175. [DOI] [PubMed] [Google Scholar]
- Cleveland DW, Rothstein JD (2001): From Charcot to Lou Gehrig: Deciphering selective motor neuron death in ALS. Nat Rev Neurosci 2:806–819. [DOI] [PubMed] [Google Scholar]
- Del Bo R, Ghezzi S, Scarpini E, Bresolin N, Comi GP (2009): VEGF genetic variability is associated with increased risk of developing Alzheimer's disease. J Neurol Sci 283:66–68. [DOI] [PubMed] [Google Scholar]
- Ferrara N, Gerber H‐P, LeCouter J (2003): The biology of VEGF and its receptors. Nat Med 9:669–676. [DOI] [PubMed] [Google Scholar]
- Frisoni G, Testa C, Sabattoli F, Beltramello A, Soininen H, Laakso M (2005): Structural correlates of early and late onset Alzheimer's disease: Voxel based morphometric study. J Neurol Neurosurg Psychiatry 76:112–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg DA, Jin K (2004): VEGF and ALS: The luckiest growth factor?. Trends Mol Med 10:1–3. [DOI] [PubMed] [Google Scholar]
- Head D, Rodrigue KM, Kennedy KM, Raz N (2008): Neuroanatomical and cognitive mediators of age‐related differences in episodic memory. Neuropsychology 22:491–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillman CH, Erickson KI, Kramer AF (2008): Be smart, exercise your heart: Exercise effects on brain and cognition. Nat Rev: Neurosci 9:58–65. [DOI] [PubMed] [Google Scholar]
- Jin K, Zhu Y, Sun Y, Mao XO, Xie L, Greenberg DA (2002): Vascular endothelial growth factor (VEGF) stimulates neurogenesis in vitro and in vivo. Proc Natl Acad Sci U S A 99:11946–11950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung RE, Segall JM, Bockholt HJ, Flores RA, Smith SM, Chavez RS, Haier RJ (2010): Neuroanatomy of creativity. Hum Brain Mapp 31:398–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzman R, Terry R, DeTeresa R, Brown T, Davies P, Fuld P, Renbing X, Peck A (1988): Clinical, pathological, and neurochemical changes in dementia: A subgroup with preserved mental status and numerous neocortical plaques. Ann Neurol 23:138–144. [DOI] [PubMed] [Google Scholar]
- Licht T, Goshen I, Avital A, Kreisel T, Zubedat S, Eavri R, Segal M, Yirmiya R, Keshet E (2011): Reversible modulations of neuronal plasticity by VEGF. Proc Natl Acad Sci 108:5081–5086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu SY, Zeng FF, Chen ZW, Wang CY, Zhao B, Li KS (2013): Vascular endothelial growth factor gene promoter polymorphisms and alzheimer's disease risk: A meta‐analysis. CNS Neurosci Therap 19:469–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie F, Ruhrberg C (2012): Diverse roles for VEGF‐A in the nervous system. Development 139:1371–1380. [DOI] [PubMed] [Google Scholar]
- Mateo I, Llorca J, Infante J, Rodríguez‐Rodríguez E, Fernández‐Viadero C, Pena N, Berciano J, Combarros O (2007): Low serum VEGF levels are associated with Alzheimer's disease. Acta Neurol Scand 116:56–58. [DOI] [PubMed] [Google Scholar]
- Mechelli A, Price CJ, Friston KJ, Ashburner J (2005): Voxel‐based morphometry of the human brain: Methods and applications. Curr Med Imaging Rev 1:105–113. [Google Scholar]
- Ogunshola OO, Antic A, Donoghue MJ, Fan S‐Y, Kim H, Stewart WB, Madri JA, Ment LR (2002): Paracrine and autocrine functions of neuronal vascular endothelial growth factor (VEGF) in the central nervous system. J Biol Chem 277:11410–11415. [DOI] [PubMed] [Google Scholar]
- Oldfield RC (1971): The assessment and analysis of handedness: The Edinburgh inventory. Neuropsychologia 9:97–113. [DOI] [PubMed] [Google Scholar]
- Oosthuyse B, Moons L, Storkebaum E, Beck H, Nuyens D, Brusselmans K, Van Dorpe J, Hellings P, Gorselink M, Heymans S (2001): Deletion of the hypoxia‐response element in the vascular endothelial growth factor promoter causes motor neuron degeneration. Nat Genet 28:131–138. [DOI] [PubMed] [Google Scholar]
- Petersen ET, Lim T, Golay X (2006a): Model‐free arterial spin labeling quantification approach for perfusion MRI. Magn Reson Med 55:219–232. [DOI] [PubMed] [Google Scholar]
- Petersen ET, Zimine I, Ho YC, Golay X (2006b): Non‐invasive measurement of perfusion: A critical review of arterial spin labelling techniques. Br J Radiol 79:688–701. [DOI] [PubMed] [Google Scholar]
- Petersen ET, Mouridsen K, Golay X (2010): The QUASAR reproducibility study, Part II: Results from a multi‐center Arterial Spin Labeling test‐retest study. Neuroimage 49:104–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prior SJ, Hagberg JM, Paton CM, Douglass LW, Brown MD, McLenithan JC, Roth SM (2006): DNA sequence variation in the promoter region of the VEGF gene impacts VEGF gene expression and maximal oxygen consumption. Am J Physiol‐Heart Circ Physiol 290:H1848–H1855. [DOI] [PubMed] [Google Scholar]
- Raven J ( 1998): Manual for Raven's Progressive Matrices and Vocabulary Scales. Oxford: Oxford Psychologists Press. [Google Scholar]
- Rosenstein JM, Krum JM, Ruhrberg C (2010): VEGF in the nervous system. Organogenesis 6:107–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider F, Gur RC, Jaggi JL, Gur RE (1994): Differential effects of mood on cortical cerebral blood flow: A 133 xenon clearance study. Psychiatry Res 52:215–236. [DOI] [PubMed] [Google Scholar]
- Smith SM, Nichols TE (2009): Threshold‐free cluster enhancement: Addressing problems of smoothing, threshold dependence and localisation in cluster inference. Neuroimage 44:83–98. [DOI] [PubMed] [Google Scholar]
- Song M, Zhou Y, Li J, Liu Y, Tian L, Yu C, Jiang T (2008): Brain spontaneous functional connectivity and intelligence. Neuroimage 41:1168–1176. [DOI] [PubMed] [Google Scholar]
- Steffensen K, Waldstrøm M, Brandslund I, Jakobsen A (2010): The relationship of VEGF polymorphisms with serum VEGF levels and progression‐free survival in patients with epithelial ovarian cancer. Gynecol Oncol 117:109–116. [DOI] [PubMed] [Google Scholar]
- Sun Y, Jin K, Xie L, Childs J, Mao XO, Logvinova A, Greenberg DA (2003): VEGF‐induced neuroprotection, neurogenesis, and angiogenesis after focal cerebral ischemia. J Clin Investig 111:1843–1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi H, Taki Y, Sassa Y, Hashizume H, Sekiguchi A, Fukushima A, Kawashima R (2010a): Regional gray matter volume of dopaminergic system associate with creativity: Evidence from voxel‐based morphometry. Neuroimage 51:578–585. [DOI] [PubMed] [Google Scholar]
- Takeuchi H, Taki Y, Sassa Y, Hashizume H, Sekiguchi A, Fukushima A, Kawashima R (2010b): White matter structures associated with creativity: Evidence from diffusion tensor imaging. Neuroimage 51:11–18. [DOI] [PubMed] [Google Scholar]
- Takeuchi H, Taki Y, Hashizume H, Sassa Y, Nagase T, Nouchi R, Kawashima R. (2011a): Cerebral blood flow during rest associates with general intelligence and creativity. PLoS ONE 6(9):e25532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi H, Taki Y, Hashizume H, Sassa Y, Nagase T, Nouchi R, Kawashima R (2011b): Failing to deactivate: The association between brain activity during a working memory task and creativity. Neuroimage 55:681–687. [DOI] [PubMed] [Google Scholar]
- Takeuchi H, Taki Y, Nouchi R, Hashizume H, Sekiguchi A, Kotozaki Y, Nakagawa S, Miyauchi CM, Sassa Y, Kawashima R (2013a): Anatomical correlates of self‐handicapping tendency. Cortex 49:1148–1154. [DOI] [PubMed] [Google Scholar]
- Takeuchi H, Taki Y, Nouchi R, Hashizume H, Sekiguchi A, Kotozaki Y, Nakagawa S, Miyauchi CM, Sassa Y, Kawashima R (2013b): Effects of working memory‐training on functional connectivity and cerebral blood flow during rest. Cortex 49:2106–2125. [DOI] [PubMed] [Google Scholar]
- Takeuchi H, Taki Y, Sassa Y, Hashizume H, Sekiguchi A, Fukushima A, Kawashima R (2013c): Brain structures associated with executive functions during everyday events in a non‐clinical sample. Brain Struct Funct 218:1017–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi H, Taki Y, Nouchi R, Sekiguchi A, Kotozaki Y, Miyauchi C, Yokoyama R, Iizuka K, Hashizume H, Nakagawa S (2014): Regional gray matter density is associated with achievement motivation: Evidence from voxel‐based morphometry. Brain Struct Funct 219:71–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi H, Taki Y, Sekiguchi A, Nouchi R, Kotozaki Y, Nakagawa S, Miyauchi CM, Iizuka K, Yokoyama R, Shinada T, Yamamoto Y, Hanawa S, Araki T, Hashizume H, Kunitoki K, Sassa Y, Kawashima R (2015): Amygdala and cingulate structure is associated with stereotype on sex‐role. Sci Rep 5:14220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taki Y, Hashizume H, Sassa Y, Takeuchi H, Wu K, Asano M, Asano K, Fukuda H, Kawashima R. (2011): Correlation between gray matter density‐adjusted brain perfusion and age using brain MR images of 202 healthy children. Hum Brain Mapp 32:1973–1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terracciano A, Sanna S, Uda M, Deiana B, Usala G, Busonero F, Maschio A, Scally M, Patriciu N, Chen W‐M (2008): Genome‐wide association scan for five major dimensions of personality. Mol Psychiatry 15:647–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshiura T, Hiwatashi A, Yamashita K, Ohyagi Y, Monji A, Takayama Y, Nagao E, Kamano H, Noguchi T, Honda H (2009): Simultaneous measurement of arterial transit time, arterial blood volume, and cerebral blood flow using arterial spin‐labeling in patients with Alzheimer disease. Am J Neuroradiol 30:1388–1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, Ba C, Wang W, Wang X, Xue R, Wu X (2012): Vascular endothelial growth factor (VEGF) gene polymorphisms and colorectal cancer: A meta‐analysis of epidemiologic studies. Genet Test Mol Biomark 16:1390–1394. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information Figure 1.
Supporting Information Tables.


