Abstract
The structural organization of the brain can be characterized as a hierarchical ensemble of segregated modules linked by densely interconnected hub regions that facilitate distributed functional interactions. Disturbances to this network may be an important marker of abnormal development. Recently, several neurodevelopmental disorders, including autism spectrum disorder (ASD), have been framed as disorders of connectivity but the full nature and timing of these disturbances remain unclear. In this study, we use non‐negative matrix factorization, a data‐driven, multivariate approach, to model the structural network architecture of the brain as a set of superposed subnetworks, or network components. In an openly available dataset of 196 subjects scanned between 5 and 85 years we identify a set of robust and reliable subnetworks that develop in tandem with age and reflect both anatomically local and long‐range, network hub connections. In a second experiment, we compare network components in a cohort of 51 high‐functioning ASD adolescents to a group of age‐matched controls. We identify a specific subnetwork representing an increase in local connection strength in the cingulate cortex in ASD (t = 3.44, P < 0.001). This work highlights possible long‐term implications of alterations to the developmental trajectories of specific cortical subnetworks. Hum Brain Mapp 38:4169–4184, 2017. © 2017 Wiley Periodicals, Inc.
Keywords: connectivity, brain development, networks, non‐negative matrix factorization
INTRODUCTION
Diffusion MRI allows the non‐invasive inference of white matter pathways in the human brain. At a millimetre‐scale, the structural connections between brain regions can be conceptualized as a complex network and interrogated with graph theoretical approaches [Bullmore and Sporns, 2009]. Graph theory provides a mathematical framework for the analysis of brain networks, with brain regions defined as a set of nodes, connected by a set of edges. From this perspective, a number of metrics exist to define both global and local properties of the brain network: from simple measures of connectivity (or degree), to more complex measures of regional integration, segregation, and influence.
This has led to the broad characterization of the macroscale organization of the mammalian brain as a near‐decomposable system built on multiple, parallel and partially segregated modules [Meunier et al., 2010; Simon, 1962]. Brain networks are organized hierarchically [Bassett et al., 2011] and linked by a set of overarching, densely interconnected hub regions that facilitate distributed interactions across the network [Bullmore and Sporns, 2012; Sporns et al., 2005; van den Heuvel and Sporns, 2011].
This view is supported by evidence that cerebral regions can be clustered together based on the extent of their shared connections into communities [Bullmore and Sporns, 2012; Hilgetag et al., 2000]. Anatomical connectivity between regions reflects a shared functional specialization [Hilgetag et al., 2000; Honey et al., 2009], and connected regions tend to have similar metabolic demands, and gene expression profiles [Collin et al., 2013; French and Pavlidis, 2011; Fulcher and Fornito, 2016; Vaishnavi et al., 2010]. Furthermore, anatomically connected regions tend to mature in tandem across development [Raznahan et al., 2011], resulting in common patterns of cortical growth and functional coordination over the lifespan [Alexander‐Bloch et al., 2013; Hagmann et al., 2010; Zielinski et al., 2010]. Taken together, this evidence suggests that connections within complex brain networks can be decomposed, or clustered, into subnetworks that link modules with distinct roles and developmental trajectories.
Long‐distance cortico‐cortical connections are established during gestation, and complex network architecture is evident at birth [Ball et al., 2014]. The effects of cerebral maturation on increasingly distributed connectivity is marked in the first years of life [Yap et al., 2011], after which the large‐scale topological organization of the structural connectome remains relatively stable [Baker et al., 2015; Dennis et al., 2013]. Over the full lifespan, measures of network efficiency and modularity follow a distinct inverted U trajectory, peaking in the third decade and mirrored by microstructural markers of the underlying white matter [Imperati et al., 2011; Kochunov et al., 2012; Zhao et al., 2015]. In elderly individuals, although network topology remains relatively consistent with younger adults, simulations suggest a preference for local communication compared to long‐range hub‐to‐hub connectivity, correspondent to evidence from functional analyses [Cao et al., 2014; Perry et al., 2015].
The early establishment of structural connectivity and long‐term stability of structural networks suggests that disturbances to network organization may be an important marker of abnormal cerebral development. A number of neurodevelopmental disorders, including autism spectrum disorder (ASD) and attention deficit hyperactivity disorder, have been linked to alterations in the development of structural and functional brain connectivity [Ecker et al., 2015; Konrad and Eickhoff, 2010; Tomasi and Volkow, 2012]. ASD is a complex, multifactorial disorder characterized by social, behavioral, and language impairments evident from an early age. Although the aetiology of ASD remains unknown, neuropathological studies have identified cortical alterations including laminar and columnar disorganization and increased neuronal density in frontal, temporal and cingulate cortices [Bailey et al., 1998; Casanova et al., 2002; Stoner et al., 2014; Uppal et al., 2014]. Early evidence from MRI studies suggested that head growth is accelerated in ASD during infancy but differences appear to dissipate with age [Courchesne et al., 2001; Ecker et al., 2015]. More recently, ASD has been framed as a disorder of connectivity [Belmonte et al., 2004; Vissers et al., 2012], based on accumulating evidence of disruptions to both functional and structural networks in autistic populations [Di Martino et al., 2014; Mueller et al., 2013; Rudie et al., 2012; Supekar et al., 2013]. Although the nature and extent of these alterations remain unclear with a number of conflicting observations, previous studies have described complex patterns of disrupted white matter organization in ASD that appear to be dependent on age and mirrored by evidence of both hypo‐ and hyper‐connectivity between functional networks and differences in electrophysiological recordings [for review, see Vissers et al., 2012].
In this study, we apply an unsupervised and data‐driven approach to model complex networks derived from whole‐brain tractography as a set of components, or subnetworks, that vary together across the population. We first demonstrate that network components can be robustly and reliably identified in a large cohort. We then model component strength across the human lifespan, defining specific developmental trajectories for subnetwork connectivity. Finally, we explore how disruptions to typical development may impact structural connectivity by testing the hypothesis that ASD is associated with alterations in subnetwork component strength. By identifying a set of network components in a group of adolescents with high‐functioning ASD and age‐matched, typically‐developing controls, we find a specific cortical subnetwork with significantly increased connection strength in the autistic population.
METHODS
Data
Preprocessed connectivity data were downloaded from the USC Multimodal Connectivity database (http://umcd.humanconnectomeproject.org) [Brown et al., 2012]. Full MRI acquisition and image processing details are given elsewhere [Brown et al., 2012; Rudie et al., 2012] but are reported in brief below.
NKI‐Rockland lifespan sample
In total, connectivity matrices from 196 healthy participants (114 male; age range: 4–85 years) were available, collected as part of the NKI/Rockland lifespan study [Nooner et al., 2012]. Diffusion MRI was acquired at 3 T with 64 gradient directions and the following parameters: TR, 10,000 ms; TE, 91 ms; voxel size, 2 mm3; b‐value, 1,000 s/mm2. After correction for motion and eddy current distortions using linear registration, diffusion tensors were modeled and tractography performed using fiber assignment by continuous tracking with an angular threshold of 45° [Mori et al., 1999].
To construct each connectivity matrix, 188 regions‐of‐interest (ROI) were defined using a group‐based functional MRI (fMRI) parcelation [Craddock et al., 2012], and structural connectivity was calculated as the total number of streamlines connecting any two ROI [Brown et al., 2012]. Prior to analysis, ROI in the brainstem were removed and fiber counts were log‐transformed resulting in a 182 × 182 connectivity matrix for each participant.
UCLA autism sample
Connectivity data from a total of 43 typically developing children and adolescents (36 male; age range: 8.9–17.9 years) and 51 with high‐functioning ASD (45 male; age range: 8.4–18.2 years) were available for analysis [Rudie et al., 2012]. DTI data of 3 T were acquired with: 32 gradient directions; TR, 9,500 ms; TE, 87 ms; voxel size, 2 mm3; b‐value, 1,000 s/mm2. Motion and eddy current correction, diffusion tensor modeling, and tractography were performed as above but with an angular threshold of 50°.
ROI were defined as 10 mm radius spheres placed at 264 coordinates in MNI space and transformed to individual diffusion data [Power et al., 2011] and connectivity was defined as streamline count between connected ROI. As above, fiber counts were log‐transformed before analysis, resulting in a 264 × 264 connectivity matrix for each participant.
Non‐Negative Matrix Factorization
Non‐negative matrix factorization (NMF) is an unsupervised, multivariate decomposition technique that models an data matrix, , as the product of two non‐negative matrices: and :
where is the number of features and is the number of samples and and have the dimensions and , respectively, where is the number of network components or basis images (Fig. 1). The optimal solution is sought by iteratively updating and to minimize the (Euclidean) distance between the original and reconstructed matrices, subject to non‐negativity constraints:
Figure 1.
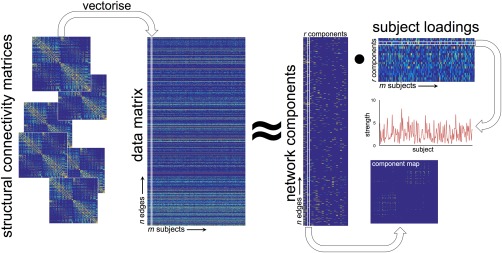
Projective NMF pipeline. Individual connectivity matrices are concatenated into a large data matrix. Projective NMF is used to decompose the data into a set of network components. A map of connections shows the topological organization of each component, or subnetwork, and a subject‐specific weighting estimates the component's contribution each individual's full network. [Color figure can be viewed at http://wileyonlinelibrary.com]
Generally, , thus represents a low‐rank approximation of the original data in [Lee and Seung, 1999]. NMF offers a natural setting for exploration of data that is inherently non‐negative and, as such, is particularly well‐suited to neuroimage analysis allowing an intuitive understanding of image‐derived, non‐negative features including, for example: tissue volume, image intensity, cortical thickness, fractional anisotropy, and structural connectivity [Sotiras et al., 2015].
In a recently introduced variant, projective NMF (PNMF), the subject loading matrix, is replaced with such that:
PNMF confers a number of benefits over standard NMF including fewer learned parameters, and increased sparsity and orthogonality of the resulting component matrix [Yang and Oja, 2010].
In terms of a network analysis, PNMF results in a set of highly orthogonal network components, each comprising a sparse set of (topologically) localized connections (i.e., the edge structure of different components does not overlap) and a corresponding subject‐specific weighting (Fig. 1). Together, these elements can then be combined to approximately reconstruct the full connectivity network of any given subject.
In addition, Yang et al. introduced a method to estimate the rank of the factorizing matrix, , using automatic relevance determination (ARD‐PNMF) [Yang et al., 2010]. Here, we use ARD‐PNMF to perform an exploratory analysis of network structure and extract a set of effective network components for further analysis.
Network Decomposition
The analysis pipeline is shown in Figure 1. For each study, the lower triangles of each symmetric structural connectivity network were vectorized and collated into an matrix before normalizing to . To reduce computation time and remove noisy connections, edges that were present in less than 10% of the study population were removed. ARD‐PNMF was initialized with non‐negative double singular value decomposition (NNDSVD), a procedure that speeds up NMF convergence compared to a random initialization and ensures consistent results across runs [Boutsidis and Gallopoulos, 2008]. We chose an initial rank estimate of 50 and performed a maximum of 20,000 PNMF iterations, or until the algorithm converged.
Network decomposition was performed in Matlab R2015b using PNMF code available at: http://sites.google.com/site/zhirongyangcs/pnmf and NNDSVD code available at: http://www.boutsidis.org/software.html).
Simulations
To demonstrate the application of PNMF to structural connectivity data, we performed a set of simulation experiments. We created a set of 150 “networks,” each comprising a weighted combination of six network components (Fig. 2). Each network component was constructed by adding binary edges between 10 and 20 randomly selected nodes in a 100 100 empty network. The weighted contribution of each component to an individual network was varied according to a set of predefined patterns that varied across the population.
Figure 2.
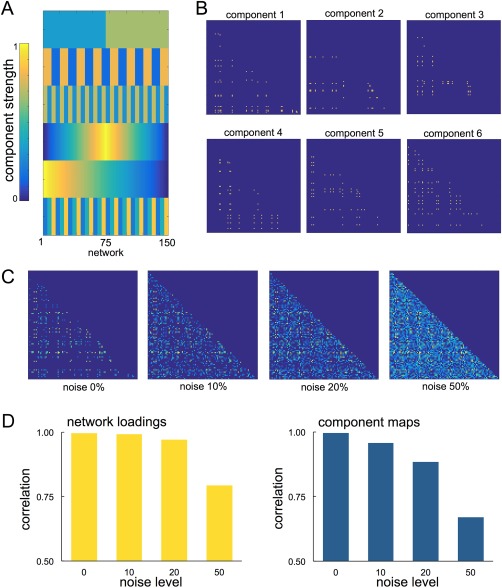
Simulating networks for PNMF decomposition. Component weights (A) and spatial maps (B) for simulating connectivity networks. Each component was weighted according to the corresponding component strength and summed to form a network. Noise was added at four levels to the final network (C). The mean correlation between recovered component loadings and the original network weights is shown in D, alongside the spatial correlation between recovered maps and the original component maps at each noise level. [Color figure can be viewed at http://wileyonlinelibrary.com]
For a given network, each of the six components were multiplied by the corresponding component weight and linearly summed to create the final network. Additionally, noise was added to each network by constructing a symmetric noise matrix with edge density set at 0, 10, 20, or 50%, and edge strength drawn from a normal distribution with mean and variance defined by existing network edge strengths.
The simulated networks (with or without additional noise) were concatenated into data matrix , removing any edges that were empty across all networks. ARD‐PNMF was then initialized using NNSVD with rank 6 and 2,500 iterations.
We found that PNMF was able to recover both the spatial maps and pattern of population variation (network loadings) of all components even under noisy conditions, achieving an average correlation between the original and recovered component maps of 0.996, 0.957, 0.884, and 0.669 (for 0, 10, 20, and 50% noise, respectively) and a correlation between original and recovered component loadings of 0.997, 0.992, 0.971, and 0.794 (Fig. 2).
Split‐Half Reliability
To investigate if the network components can be robustly and reliability identified across population subsamples, we performed a split‐half reliability assessment [Groppe et al., 2009; Groves et al., 2012]. The NKI‐Rockland dataset was split into two, randomly selected and equal size samples and PNMF performed independently on each. The resulting split‐half components were then greedily paired with components obtained from the full dataset. Components were matched based on the correlation between the component loadings of overlapping subject populations in the half and full datasets to produce a triplet, with each original component paired with a single component from each half sample. Reliability of the original components was evaluated by measuring the edgewise (cosine) similarity between correspondent split‐half component maps. Component reliability was compared to a null distribution built by randomly permuting edges in corresponding split‐half pairs before calculating spatial similarity, 1,000 permutations were performed for each pair.
Rich Club Analysis
To further investigate the topological organization of extracted network components, we performed a rich club analysis [van den Heuvel and Sporns, 2011]. Using the group mean structural network, nodes were sorted by degree and low degree nodes incrementally removed in steps. At each step, the density of the remaining network connections was compared to a set of 100 randomized networks of the same size to give a normalized rich club coefficient, . The rich club was defined as nodes with degree > , where . Following Collin et al., after identifying network nodes comprising the rich club, edges in each network component were defined as either “rich” if they connected two rich club nodes to each other, “local” if they connected two non‐rich club nodes, of “feeder” connections if they connected rich club nodes to non‐rich club nodes [Collin et al., 2013]. The overall “richness” or “locality” of each component was determined by comparing the number of rich or local edges to a set of 1,000 equivalent random networks. Network analysis was performed with the Brain Connectivity Toolbox [Rubinov and Sporns, 2010].
Statistical Analysis
For extracted components in the NKI‐Rockland sample, component strength over the lifespan was modeled using polynomial regression (up to power 3) with age as a covariate and sex as an additional factor. The Akaike Information Criterion (AIC) was used to select the best model for each component (i.e., linear, quadratic, or cubic; with or without sex). Statistical analysis was performed with the package in R 3.31.
In the UCLA autism cohort, component strength was compared between groups using an independent samples t‐test. Analysis was performed in JASP 0.7.5.6.
Data Visualization
The edge structure of network components were visualized with Circos [Krzywinski et al., 2009], graph nodes were ordered according to cerebral lobe and the x, y, z coordinates of the central voxel of each cerebral ROI supplied with the connectivity matrices.
In addition, to visualize the anatomical location of connected ROI in each network component, we calculated node degree (i.e., the total number of connections of each ROI) and projected the values onto standard space masks of the cortical gray matter, subcortical structures, and cerebellum. Mask voxels were assigned an ROI membership based on Euclidean distance to the nearest ROI center and assigned the corresponding nodal degree value. Node degree images were then smoothed with a Gaussian kernel of FWHM 5mm and projected onto a 3D representation of the smoothed cortical surface using Surf Ice (http://www.nitrc.org/projects/surfice).
RESULTS
Network Components across the Lifespan
Reliability assessment
In total, 22 network components were identified in the NKI‐Rockland dataset (Supporting Information Figure S1). Reliability scores were calculated for each component using a split‐half framework [Groppe et al., 2009]. Network components identified in two independent group samples were matched to the original set of components and the spatial similarity between the respective edge structures compared as a marker of reliability. Both groups were matched in age and sex (group one: mean age = 35.4 years; 55/98 male; group two: 34.5 years; 59/98 male). In total, 19 components were identified in group one and 21 in group two. Of the 19 original components matched with corresponding pairs in the split‐half sample, 15 demonstrated a significantly higher spatial correlation than would be expected between two random matrices with the same number of edges (all P < 0.001, 1,000 permutations; Supporting Information, Figure S2; Table S1). The mean spatial similarity between matched components was 0.49 (range: 0.14–0.96).
Developmental trajectories
The subject loading of each network component (i.e., the contribution of a given component to the individual's full network) was modeled as a function of age using polynomial regression. Of 22 components, 16 demonstrated significant age‐related variation (all P < 0.01; Table 1). Of these, nine components followed nonlinear trajectories over the lifespan best described by quadratic models; six increased linearly with age, and one decreased. The addition of sex as a factor improved the model fit in 4 of the 16 significant components. Table 1 shows the best model selected for each component and Figure 3 highlights some of these trends; networks and modeled trajectories for all components are shown in Supporting Information Figure S1 in order of reliability.
Table 1.
Modeling development trajectories of network components
| Regression coefficients | ||||||||
|---|---|---|---|---|---|---|---|---|
| Component | Best model | AIC | Adjusted R 2 | P | –log10(p) | β 1 | β 2 | β 3 |
| Aa | age + age2 + sex | 544.28 | 0.09 | <0.01 | 3.89 | 2.13 | −3.13 | −0.15 |
| Ba | age + age2 + sex | 539.04 | 0.11 | <0.01 | 4.96 | −2.28 | −2.63 | 0.29 |
| Ca | age + age2 | 552.15 | 0.05 | <0.01 | 2.37 | 2.71 | −1.83 | — |
| Da | age + age2 | 534.21 | 0.13 | <0.01 | 6.21 | −4.81 | −1.93 | — |
| Ea | age + age2 | 533.10 | 0.13 | <0.01 | 6.45 | 4.18 | −3.21 | — |
| Fa | age + age2 | 544.10 | 0.08 | <0.01 | 4.09 | 0.47 | −4.23 | — |
| Ga | age | 520.00 | 0.19 | <0.01 | 9.75 | 6.08 | — | — |
| Ha | age | 508.38 | 0.23 | <0.01 | 12.29 | 6.79 | — | — |
| Ia | age + age2 | 534.87 | 0.13 | <0.01 | 6.07 | 4.61 | −2.24 | — |
| Ja | age + age2 + sex | 526.97 | 0.16 | <0.01 | 7.46 | 5.01 | −1.90 | −0.17 |
| K | sex | 558.34 | 0.01 | 0.09 | 1.04 | — | — | −0.12 |
| La | age | 545.73 | 0.07 | <0.01 | 4.04 | 3.85 | — | — |
| M | age + sex | 556.79 | 0.02 | 0.04 | 1.37 | 1.41 | — | −0.15 |
| Na | age | 545.37 | 0.07 | <0.01 | 4.12 | −3.89 | — | — |
| Oa | age + sex | 515.58 | 0.21 | <0.01 | 10.19 | 6.10 | — | −0.17 |
| Pa | age | 551.21 | 0.04 | <0.01 | 2.78 | 3.12 | — | — |
| Qa | age | 538.22 | 0.11 | <0.01 | 5.73 | 4.65 | — | — |
| R | age + age2 | 557.08 | 0.02 | 0.05 | 1.31 | 1.97 | −1.46 | — |
| Sa | age + age2 | 543.80 | 0.09 | <0.01 | 4.16 | 4.00 | −1.56 | — |
| T | age | 555.30 | 0.02 | 0.02 | 1.81 | −2.41 | — | — |
| U | intercept only | 559.23 | 0.00 | — | — | — | — | — |
| V | sex | 557.16 | 0.02 | 0.05 | 1.34 | — | — | −0.14 |
Model significant at P < 0.01.
Figure 3.
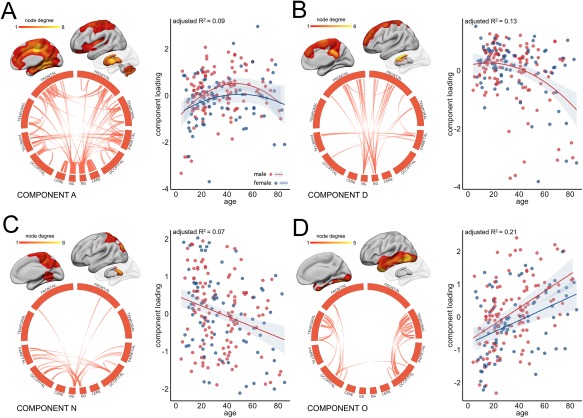
Age‐related variation in component strength. Four network components are highlighted as examples of age‐related variation in subnetwork connectivity (see Supporting Information Figure S1 for all components). For visualization, components maps were thresholded at the 95th percentile and connections are shown in circular format. To show the anatomical location of connected regions, node degree was calculated for each cerebral ROI as the sum of its connections in the thresholded component and projected onto cortical/subcortical surfaces. The relationship between (normalized) component strength and age was modeled using polynomial regression. The best model fit is shown for each component. Separate model fits indicate when sex was included as an additional factor. Red indicates male; blue, female. Cere = cerebellum, BG = basal ganglia and thalamus. [Color figure can be viewed at http://wileyonlinelibrary.com]
In Figure 3, component A comprises a robust (split‐half similarity: 0.96) and relatively dense pattern of connectivity including both local, within‐lobe, and longer, between‐lobe, connections. Inter‐hemispheric connections are apparent between frontal and parietal lobes. Highly connected cerebral regions in this component include the cingulate and paracingulate, insular, medial temporal, and superior parietal cortices, with dense connectivity also evident within the subcortical structures, and between these structures and higher cerebral cortex. Over the lifespan, connectivity between these regions increases rapidly over childhood and adolescence, peaking between 40 and 50 and declining into older age. Component loading is slightly higher in males than in females across the lifespan although follows a steeper decline with old age. Similar trends in connectivity are seen in components E, F and J (Supporting Information Figure S1). Components D and N (Fig. 3B,C) capture bilateral patterns of connectivity between subcortical gray matter structures and frontal and parietal cortices, respectively. Edges in component D predominantly connect the caudate nucleus and superior, medial frontal regions. The strength of this component remains relatively stable until middle age before declining. The strength of component N (Fig. 3C) monotonically decreases with age with edges connecting the thalamus to post‐central cortex and superior parietal regions. In contrast, component O follows a linearly increasing trend with age (Fig. 3D) Connections in this component are primarily local, connecting anatomically adjacent cortical regions within the temporal lobe and temporo‐occipital junction in both hemispheres. A similar pattern can be seen in component L (Supporting Information Figure S1).
Component topology
Qualitative assessment of the patterns of connectivity within components reveals a generally bilateral and symmetric organization across hemispheres. Unilateral components appear to reflect the topology of corresponding components in the opposite hemisphere (e.g., I and S; O and L).
To quantify whether inter‐regional connections within individual network components revealed a preferential support for connectivity between network hubs over topologically local nodes, we performed a rich club analysis. Sixty‐five nodes with degree greater than 100 were defined as the rich club (maximum = 1.09) and included nodes bilaterally in: frontal pole; cingulate and paracingulate cortex; insula; hippocampus; precuneus and superior parietal lobule; lateral occipital cortex, and subcortical structures (caudate, pallidum, and thalamus).
Component edges were defined as rich, feeder or local connections, and the “richness” or “locality” of each component defined as the number of rich or local connections compared to a set of 1,000 random networks (Table 2). Four components were found to contain significantly more rich‐club connections than expected by chance (A, E, F, and I; all P = 0.001). These components are shown in Figure 4; richness and locality indices for all components are shown in Table 2. All rich components show a similar inverted “U” trajectory with age (Table 1, Supporting Information Figure S1). Edge strength distributions in Figure 4 show that component richness is associated with both an increased number and strength of rich compared to local edges. In contrast, four components were to found to have significantly more local connections than expected by chance (Fig. 5; M, O, L, and J; P = 0.001). These components all had increased number and strength of local compared to rich connections and increased in strength across the lifespan (Table 1, Supporting Information Figure S1).
Table 2.
Rich club analysis of network components
| Component | Number of edges | Rich edges | Richness a | P | Local edges | Localitya | P |
|---|---|---|---|---|---|---|---|
| A | 2549 | 780 | 1.34 | 0.001a | 658 | 1.00 | 0.418 |
| B | 788 | 115 | 0.64 | 1.000 | 223 | 1.10 | 0.035 |
| C | 530 | 61 | 0.50 | 1.000 | 152 | 1.12 | 0.059 |
| D | 605 | 155 | 1.12 | 0.049 | 134 | 0.87 | 0.987 |
| E | 1119 | 434 | 1.69 | 0.001a | 158 | 0.55 | 1.000 |
| F | 835 | 238 | 1.25 | 0.001a | 169 | 0.79 | 1.000 |
| G | 1583 | 396 | 1.09 | 0.015 | 333 | 0.82 | 1.000 |
| H | 757 | 160 | 0.92 | 0.882 | 211 | 1.08 | 0.071 |
| I | 520 | 155 | 1.30 | 0.001a | 119 | 0.89 | 0.917 |
| J | 623 | 73 | 0.51 | 1.000 | 193 | 1.21 | 0.001a |
| K | 1400 | 293 | 0.91 | 0.967 | 309 | 0.86 | 1.000 |
| L | 628 | 86 | 0.60 | 1.000 | 210 | 1.30 | 0.001a |
| M | 576 | 44 | 0.33 | 1.000 | 254 | 1.71 | 0.001a |
| N | 535 | 119 | 0.97 | 0.612 | 103 | 0.75 | 1.000 |
| O | 761 | 93 | 0.53 | 1.000 | 316 | 1.62 | 0.001a |
| P | 710 | 161 | 0.99 | 0.540 | 184 | 1.01 | 0.446 |
| Q | 498 | 111 | 0.98 | 0.599 | 153 | 1.19 | 0.007 |
| R | 342 | 70 | 0.89 | 0.830 | 94 | 1.07 | 0.236 |
| S | 572 | 132 | 1.01 | 0.425 | 120 | 0.82 | 0.999 |
| T | 599 | 157 | 1.15 | 0.019 | 132 | 0.86 | 0.987 |
| U | 481 | 41 | 0.37 | 1.000 | 120 | 0.97 | 0.648 |
| V | 345 | 55 | 0.70 | 0.999 | 100 | 1.13 | 0.073 |
aNumber of edges compared to 1,000 random networks of the same size.
Significant at P = 0.001.
Figure 4.
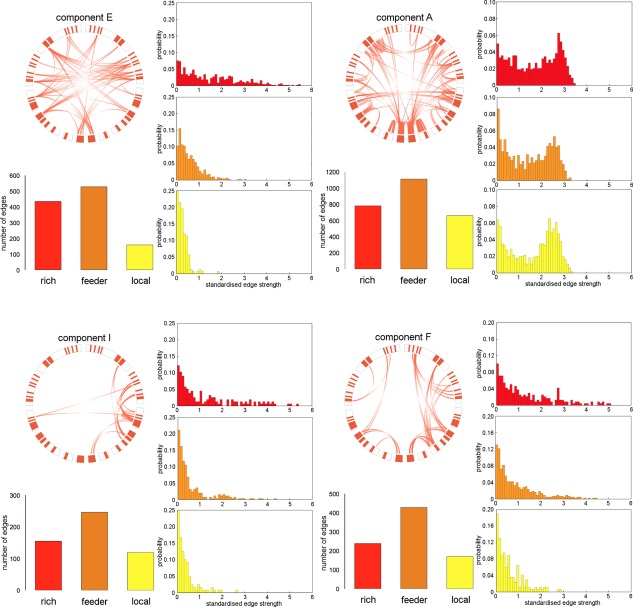
Rich network components. Network components with significantly more rich‐club edges than in a set of 1,000 equivalent random networks are shown. Thresholded connectivity maps are displayed in circular diagrams as in Figure 3, with rich club nodes highlighted in red. The number and probability distribution of rich, feeder, and local edges are displayed for each component. [Color figure can be viewed at http://wileyonlinelibrary.com]
Figure 5.

Local network components. Network components with significantly more local edges than in a set of 1,000 equivalent random networks are shown. Connectivity maps and edge distributions are shown as in Figure 4. [Color figure can be viewed at http://wileyonlinelibrary.com]
Network Components in Autism Spectrum Disorder
In our second experiment, we applied PNMF to a dataset comprising high‐functioning individuals with ASD and a set of age‐ and sex‐matched controls. In total, 24 components were identified (Table 3). One component was found to have a significantly higher loading in the ASD group compared to typically developing individuals (t (1,92)=3.161; P = 0.002; P < 0.05 after Bonferroni correction for multiple comparisons). Additionally controlling for age did not alter this relationship (ANCOVA: F (1,91) = 11.05, P < 0.001). To control for possible gender effects, we repeated the statistical analysis after excluding female participants (n = 7 TD; n = 6 ASD). The difference in component strength remained significant (t (1,79)=3.44; P < 0.001).
Table 3.
Between‐group comparison of component loadings
| Component | t (1,92) | P | Cohen's d |
|---|---|---|---|
| 1 | 1.769 | 0.080 | 0.366 |
| 2 | 1.576 | 0.119 | 0.326 |
| 3 | 0.809 | 0.421 | 0.168 |
| 4 | 0.308 | 0.759 | 0.064 |
| 5 | 1.388 | 0.168 | 0.287 |
| 6 | −0.735 | 0.464 | −0.152 |
| 7 | −0.342 | 0.733 | −0.071 |
| 8 | −1.180 | 0.241 | −0.244 |
| 9 | 0.698 | 0.487 | 0.145 |
| 10 | 3.161 | 0.002a | 0.654 |
| 11 | 0.730 | 0.467 | 0.151 |
| 12 | 1.421 | 0.159 | 0.294 |
| 13 | −0.095 | 0.925 | −0.020 |
| 14 | −0.019 | 0.985 | −0.004 |
| 15 | 1.670 | 0.098 | 0.346 |
| 16 | −0.376 | 0.708 | −0.078 |
| 17 | 0.146 | 0.884 | 0.030 |
| 18 | 0.328 | 0.744 | 0.068 |
| 19 | 0.283 | 0.778 | 0.059 |
| 20 | 0.162 | 0.872 | 0.033 |
| 21 | 1.459 | 0.148 | 0.302 |
| 22 | 1.162 | 0.248 | 0.241 |
| 23 | 0.410 | 0.683 | 0.085 |
| 24 | −0.528 | 0.599 | −0.109 |
Significant at P < 0.05 after Bonferroni correction for multiple comparisons.
This component is shown in detail in Figure 6 and comprised a bilateral and symmetric pattern of connectivity with edges predominantly linking nodes in the anterior and posterior cingulate cortex, paracingulate cortex, supplemental motor areas, and parietal cortex. Both intra‐ and inter‐hemispheric connections are visible with additional connections between the putamen and parietal cortex in both hemispheres. The mean (±S.D.) component loadings were 2.9 ± 0.69 in the ASD group and 2.5 ± 0.57 in the typically‐developing group (Fig. 6B). Rich club analysis revealed that this component had significantly more local connections than expected by chance (locality: 1.32, P = 0.001).
Figure 6.
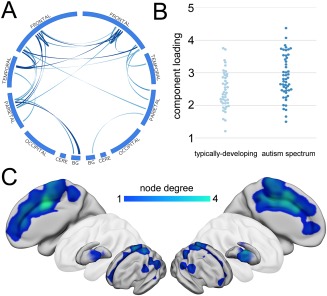
Structural connectivity is significantly increased in ASD. A single subnetwork was found to be significantly stronger in the ASD cohort. The component map is shown in A (thresholded at 95th percentile), and component loadings for both groups compared in B. The anatomical locations of connected regions are visualized as in Figure 3 and shown in C. [Color figure can be viewed at http://wileyonlinelibrary.com]
As noted above, a threshold was applied to the network data before performing PNMF, limiting the analysis to edges shared by at least 10% of participants. We additionally performed PNMF after applying two alternate thresholds: 5% and 20%. At both thresholds, a similar pattern was observed and found to be significantly different between groups (at 5%: t = 2.732, P = 0.008; excluding females: t = 3.16, P = 0.002; at 20%: t = 2.40, P = 0.018; excluding females: t = 2.77, P = 0.007). The spatial patterns associated with this component at each threshold match closely to that described above.
DISCUSSION
In this study, we modeled structural connectivity networks as the combination of separable subnetworks, or network components, using a data‐driven and multivariate approach. We first showed that complex networks could be reliably decomposed into network components using simulated data. We then applied this method to two independent cohorts, demonstrating that network components follow specific developmental trajectories with age and can capture disruptions to neurodevelopment in autism spectrum disorder.
NMF provides a natural setting for the analysis of neuroimaging data due to the inherent non‐negativity common to many imaging‐derived metrics (e.g., tissue volume, fiber count) [Sotiras et al., 2015]. In image analysis, NMF leads to a parts‐based representation of the data, extracting sparse image components with localized spatial support [Lee and Seung, 1999]. In this way, NMF confers a relatively simple interpretation of the data, namely that the complex whole can be approximated by the summation of the localized parts. We show that decomposing structural connectivity networks with NMF results in a soft clustering of connections that co‐vary together across the population forming relatively sparse subnetworks. We also find that these components are biologically relevant, capturing known subsystems (e.g., subcortical‐cortical projections; components C, D, N, and P), local patterns of connectivity between anatomically or functionally homologous regions (e.g., components L, O, and T), and varying with age across the lifespan.
MR studies have found that both white and gray matter tissue volumes follow an inverted U trajectory over the human lifespan, rising rapidly in development, peaking in the second or third decade and declining into older age [Giedd et al., 1999; Westlye et al., 2010; Ziegler et al., 2012]. Markers of microstructural maturation in cerebral white matter also follow similar trends [Lebel et al., 2012; Westlye et al., 2010]. In the NKI‐Rockland cohort, a recent graph theoretical analysis of structural connectivity found several network properties, including efficiency, varied along the similar polynomial trajectories [Zhao et al., 2015]. In contrast, the modularity—or extent to which a network could be segregated into regional communities—remained stable over time.
In this study, using a subsample of the NKI‐Rockland dataset, we found that several network components followed an inverted U‐shaped trajectory. Significant variation in component strength with age was found in 16 of 22 components, of which 9 followed a nonlinear trajectory. When ordered by reliability across subsamples, age variation in 8 of the top 10 most robust components was best described by a polynomial relationship. Of the remaining components, 6 increased linearly with age, and 1 decreased. We found that components linking densely‐connected network hub regions tended to follow a nonlinear trajectory with age, decreasing in strength in later life. In contrast, components that increased in strength with age reflected local connectivity patterns between neighboring regions, or between corresponding regions in the opposite hemisphere (e.g., components H, L, M, O, and Q), compared to more global connectivity patterns (A, E, F, J). This increase in component strength may represent a relative sparing of local connections. It may also reflect age‐related alterations to the underlying microstructural white matter organization. For example, a consequence of regional decreases in the number of crossing fibers in older subjects could be the improved resolution of the predominant fiber direction, resulting in an apparent increase in fiber number in short, local connections.
These observations support evidence of an increasing dependence on local connectivity in the elderly connectome with corresponding decreases in long‐range connection strength between cortical hubs [Betzel et al., 2014; Cao et al., 2014; Perry et al., 2015; Zhao et al., 2015]. Analyses of functional connectivity networks have shown that hub connectivity follows a U shaped trajectory over the human lifespan alongside a decrease in network modularity, suggesting a less segregated network topology in old age [Cao et al., 2014; Chan et al., 2014]. Performing a comparative analysis of both functional and structural networks, Betzel et al. showed structural connectivity of hub regions decreases dramatically with age, while local connectivity is relatively spared. In addition, functional connectivity within resting state networks (RSN) decreased with age whereas connectivity between RSN increased. This increase in between‐network connectivity was subserved by multi‐step structural connections. Similarly, Perry et al. showed that alterations to edge strength in the aging structural connectome likely lead to an increased preference for network communication via multiple, non‐hub, local pathways [Perry et al., 2015]. Taken together, this evidence suggests that the decline of long‐distance, hub‐to‐hub connections with a relative sparing of topologically, local connections results in less efficient network communication in older age, a process that a may underlie progressive cognitive decline [Andrews‐Hanna et al., 2007; O'Sullivan et al., 2001].
Structural connectivity is established early in development [Ball et al., 2014; Kostovic and Jovanov‐Milosevic, 2006], and early disturbances to network organization may be a marker of abnormal neurodevelopment [Ecker et al., 2015]. In our second experiment, we performed a network decomposition in an adolescent cohort and found a single subnetwork with significantly greater connectivity in ASD. This network was composed primarily of multiple, local connections between neighboring regions in the cingulate and paracingulate cortices, both within and between hemispheres, along with connections between supplementary motor areas, parietal cortex and the putamen. The cingulum, and in particular the anterior cingulate cortex, has been linked to ASD due mainly to its role in social interaction and attention [Mundy, 2003]. Indeed, fMRI studies have found that altered anterior cingulate activation in ASD is associated with errors in response inhibition and repetitive behavior [Agam et al., 2010; Kana et al., 2007; Thakkar et al., 2008] and performance during social tasks [Di Martino et al., 2009; Dichter et al., 2009]. Similarly, metabolic [Tebartz van Elst et al., 2014], neuropathological [Simms et al., 2009] and neuroanatomical disturbances [Schumann et al., 2010] have also been reported in the cingulate in ASD. In a comprehensive diffusion tractography study of the white matter tracts of the limbic system, Pugliese et al. found significantly increased tract volume bilaterally in the cingulum bundle (determined by streamline count) of adults with Asperger's syndrome [Pugliese et al., 2009]. This observation is convergent with post‐mortem findings in autistic cases of excessive axonal connectivity between neighboring cortical regions in the cingulate [Zikopoulos and Barbas, 2010]. This is of particular interest, given the putative neurodevelopmental origins of ASD, as the cingulum bundle forms early in gestation, followed by short range cortico‐cortical connections in the third trimester [Takahashi et al., 2012; Vasung et al., 2010] suggesting that any early disruptions to white matter development in utero could have significant long‐term implications on neurodevelopment.
Previous structural network analyses in autistic populations have found significantly increased structural connectivity, inferred from tractography streamline count, among cortical regions including anterior and posterior cingulate cortex, superior frontal, superior parietal, and insula cortex [Ray et al., 2014]. In a subset of the UCLA cohort, Watanabe and Rees found evidence for delayed, or immature, hub connectivity in ASD [Watanabe and Rees, 2015] whereas Ghanbari et al. used a supervised variant of NMF coupled with a graph embedding approach to define discriminative network components and found decreased interhemispheric connection strength in subcortical subnetworks in ASD [Ghanbari et al., 2014]. Similarly, other studies have reported decreases in structural connectivity, dependent on measures of white matter microstructure [Lo et al., 2011; Noriuchi et al., 2010; Thakkar et al., 2008]. These discrepancies may relate, in part, to the uncertain correspondence between different measures of structural connectivity. In the original study of this cohort, Rudie et al. found that streamline count was significantly increased in four times as many connections in ASD subjects compared to controls [Rudie et al., 2012]. However, a concomitant decrease in fractional anisotropy and increase in mean diffusivity was also noted in white matter connections on average in ASD. These differences resulted in an atypical age‐related development of network efficiency in ASD, a factor that related to symptom severity [Rudie et al., 2012].
NMF belongs to a class of multivariate matrix decomposition and dimension reduction techniques that include principal component analysis and independent component analysis (ICA). Recently, exploratory multivariate analysis methods have proven well‐suited to the discovery of complex organizational relationships in the brain [Beckmann and Smith, 2005; Calhoun et al., 2009; McIntosh and Mišić, 2013]. Previous studies have shown the potential of matrix factorization techniques to isolate topological subnetworks from functional and structural connectivity matrices on an individual or group level [Clayden et al., 2013; Ghanbari et al., 2014; Park et al., 2014]. We performed simulations to demonstrate that PNMF is particularly able to retrieve superposed spatial patterns and the corresponding, population‐varying component weights from a set of noisy connectivity networks. In addition, although we were unable to compare network components identified in the UCLA and NKI‐Rockland samples directly, due to the different parcelation schemes used in each study, [Meskaldji et al., 2013;Wijk et al., 2010; Zalesky et al., 2010], a split‐half reliability analysis showed that the majority of structural subnetworks in the NKI cohort could be consistently identified across population subsamples with shared network structure.
Additionally, an important aspect of any dimension reduction task is determining the optimal dimensionality of the solution. In this article, we chose to use an automatic model selection process that iteratively updates model rank, removing components with low spatial variance that do not contribute significantly to the final reconstruction [Yang et al., 2010]. This resulted in around 20 effective components selected in both experimental cohorts. Previous studies have shown that, for ICA‐based decomposition of fMRI data, higher‐dimensional decompositions can reveal nested subsystems within functional networks [Abou Elseoud et al., 2011; Dipasquale et al., 2015; Kiviniemi et al., 2009]. Indeed, in the present study we observed apparently correspondent components that were split between opposite hemispheres (e.g., components I and S), whereas others formed bilateral symmetric patterns (components A, U, and H) suggesting some hierarchical structure within components. We also note that several lateralized components contributed to rich club connectivity. Although there is some evidence for laterality in the human rich club when hemispheric connectivity is analyzed separately, with a stronger rich club present in the right hemisphere [Goulas et al., 2014], it is generally accepted that the rich club is symmetric in organization [van den Heuvel and Sporns, 2011]. Although these components contain rich club edges, they appear to represent a mixture of rich, local and feeder connections that vary together with age. Future exploration of PNMF network decomposition and the relationship between network components derived from PNMF and modules/communities and hierarchies derived from graph theoretical approaches are warranted. Likewise, PNMF network decomposition at multiple dimensionalities would provide a framework to investigate the nested, or hierarchical nature of structural connectivity subnetworks [Betzel et al., 2013; Betzel and Bassett, 2016; Meunier et al., 2010]. Future analyses may also consider the impact of group structure on PNMF decomposition. In this study, we applied PNMF jointly across both typically developing and ASD networks to examine components that were present in all participants. An alternative strategy may be to produce separate decompositions for each group to uncover group‐specific network structures.
STUDY LIMITATIONS
The use of (log‐transformed) streamline counts to estimate structural connectivity could be considered a limitation of this study. Is it important to note that fiber counts do not necessarily reflect true anatomical connectivity, and tractography is prone to mapping false positive connections due to local accumulation of modeling errors [Jones, 2010; Jones et al., 2013]. However, using available open access datasets, we have shown that PNMF is able to extract subnetworks that can provide insight into biological variability, with developmental trajectories that suggest streamline count, in part, can reflect maturational processes in the brain. Importantly, PNMF is generalisable to any inherently non‐negative data. This opens future avenues to explore network components derived from modern, probabilistic tractographic algorithms that better reflect true anatomical connectivity [Pestilli et al., 2014; Smith et al., 2015].
MRI studies in ASD may be at risk of selection bias due to the additional difficulties in scanning non‐compliant, or low‐functioning individuals, who may require sedation [Erbetta et al., 2014]. In this study, only data from high‐functioning ASD participants were available. As such we may underestimate any morphological traits of ASD through the exclusion of low‐functioning participants. Conversely, given the phenotypic heterogeneity of ASD, it may be that specific alterations in brain morphology are not present in all ASD subtypes [Vissers et al., 2012], and as such studies will benefit from identifying endophenotypic variations in specific sub‐populations of the ASD population. We believe this is an exciting avenue for further research.
CONCLUSIONS
In conclusion, we present a multivariate analysis of structural connectivity in two cohorts. We demonstrate that complex networks can be decomposed into robust and reliable subnetworks that vary in strength with age. Furthermore, we identify a specific subnetwork with increased connection strength in autism spectrum disorder. We propose that this form of network component analysis shows good potential for further exploration of the human structural connectome.
Supporting information
Supporting Information
ACKNOWLEDGMENTS
The authors would like to thank Dr. Jesse Brown and all involved in the collection and aggregation of data for the USC Multimodal Connectivity Database. This research was conducted within the Developmental Imaging research group, Murdoch Childrens Research Institute and the Children's MRI Centre, Royal Children's Hospital, Melbourne, Victoria. It was supported by the Murdoch Childrens Research Institute, the Royal Children's Hospital, Department of Paediatrics, The University of Melbourne and the Victorian Government's Operational Infrastructure Support Program.
REFERENCES
- Abou Elseoud A, Littow H, Remes J, Starck T, Nikkinen J, Nissilä J, Timonen M, Tervonen O, Kiviniemi V (2011): Group‐ICA model order highlights patterns of functional brain connectivity. Front Syst Neurosci 5:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agam Y, Joseph RM, Barton JJS, Manoach DS (2010): Reduced cognitive control of response inhibition by the anterior cingulate cortex in autism spectrum disorders. NeuroImage 52:336–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander‐Bloch A, Raznahan A, Bullmore E, Giedd J (2013): The convergence of maturational change and structural covariance in human cortical networks. J Neurosci 33:2889–2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews‐Hanna JR, Snyder AZ, Vincent JL, Lustig C, Head D, Raichle ME, Buckner RL (2007): Disruption of large‐scale brain systems in advanced aging. Neuron 56:924–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey A, Luthert P, Dean A, Harding B, Janota I, Montgomery M, Rutter M, Lantos P (1998): A clinicopathological study of autism. Brain J Neurol 121 (Pt 5):889–905. [DOI] [PubMed] [Google Scholar]
- Baker STE, Lubman DI, Yücel M, Allen NB, Whittle S, Fulcher BD, Zalesky A, Fornito A (2015): Developmental changes in brain network hub connectivity in late adolescence. J Neurosci off J Soc Neurosci 35:9078–9087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball G, Aljabar P, Zebari S, Tusor N, Arichi T, Merchant N, Robinson EC, Ogundipe E, Rueckert D, Edwards AD, Counsell SJ (2014): Rich‐club organization of the newborn human brain. Proc Natl Acad Sci USA 111:7456–7461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett DS, Brown JA, Deshpande V, Carlson JM, Grafton ST (2011): Conserved and variable architecture of human white matter connectivity. NeuroImage 54:1262–1279. [DOI] [PubMed] [Google Scholar]
- Beckmann CF, Smith SM (2005): Tensorial extensions of independent component analysis for multisubject FMRI analysis. NeuroImage 25:294–311. [DOI] [PubMed] [Google Scholar]
- Belmonte MK, Allen G, Beckel‐Mitchener A, Boulanger LM, Carper RA, Webb SJ (2004): Autism and abnormal development of brain connectivity. J Neurosci 24:9228–9231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betzel RF, Bassett DS (2016): Multi‐scale brain networks. Neuroimage 2016. doi: 10.1016/j.neuroimage.2016.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betzel RF, Griffa A, Avena‐Koenigsberger A, Goñi J, Thiran J‐P, Hagmann P, Sporns O (2013): Multi‐scale community organization of the human structural connectome and its relationship with resting‐state functional connectivity. Netw Sci 1:353–373. [Google Scholar]
- Betzel RF, Byrge L, He Y, Goñi J, Zuo X‐N, Sporns O (2014): Changes in structural and functional connectivity among resting‐state networks across the human lifespan. NeuroImage 102:345–357. [DOI] [PubMed] [Google Scholar]
- Boutsidis C, Gallopoulos E (2008): SVD based initialization: A head start for nonnegative matrix factorization. Pattern Recognit 41:1350–1362. [Google Scholar]
- Brown JA, Rudie JD, Bandrowski A, Van Horn JD, Bookheimer SY (2012): The UCLA multimodal connectivity database: A web‐based platform for brain connectivity matrix sharing and analysis. Front Neuroinformatics 6:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullmore E, Sporns O (2009): Complex brain networks: Graph theoretical analysis of structural and functional systems. Nat Rev 10:186–198. [DOI] [PubMed] [Google Scholar]
- Bullmore E, Sporns O (2012): The economy of brain network organization. Nat Rev Neurosci 13:336–349. [DOI] [PubMed] [Google Scholar]
- Calhoun VD, Liu J, Adalı T (2009): A review of group ICA for fMRI data and ICA for joint inference of imaging, genetic, and ERP data. NeuroImage 45:S163–S172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao M, Wang J‐H, Dai Z‐J, Cao X‐Y, Jiang L‐L, Fan F‐M, Song X‐W, Xia M‐R, Shu N, Dong Q, Milham MP, Castellanos FX, Zuo X‐N, He Y (2014): Topological organization of the human brain functional connectome across the lifespan. Dev Cogn Neurosci 7:76–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanova MF, Buxhoeveden DP, Switala AE, Roy E (2002): Minicolumnar pathology in autism. Neurology 58:428–432. [DOI] [PubMed] [Google Scholar]
- Chan MY, Park DC, Savalia NK, Petersen SE, Wig GS (2014): Decreased segregation of brain systems across the healthy adult lifespan. Proc Natl Acad Sci USA 111:E4997–E5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayden JD, Dayan M, Clark CA (2013): Principal networks. PLoS One 8:e60997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collin G, Sporns O, Mandl RCW, van den Heuvel MP (2013): Structural and functional aspects relating to cost and benefit of rich club organization in the human cerebral cortex. Cereb Cortex 24:2258–2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courchesne E, Karns CM, Davis HR, Ziccardi R, Carper RA, Tigue ZD, Chisum HJ, Moses P, Pierce K, Lord C, Lincoln AJ, Pizzo S, Schreibman L, Haas RH, Akshoomoff NA, Courchesne RY (2001): Unusual brain growth patterns in early life in patients with autistic disorder: An MRI study. Neurology 57:245–254. [DOI] [PubMed] [Google Scholar]
- Craddock RC, James GA, Holtzheimer PE, Hu XP, Mayberg HS (2012): A whole brain fMRI atlas generated via spatially constrained spectral clustering. Hum Brain Mapp 33:1914–1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis EL, Jahanshad N, McMahon KL, de Zubicaray GI, Martin NG, Hickie IB, Toga AW, Wright MJ, Thompson PM (2013): Development of brain structural connectivity between ages 12 and 30: A 4‐Tesla diffusion imaging study in 439 adolescents and adults. NeuroImage 64:671–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Martino A, Ross K, Uddin LQ, Sklar AB, Castellanos FX, Milham MP (2009): Functional brain correlates of social and nonsocial processes in autism spectrum disorders: An activation likelihood estimation meta‐analysis. Biol Psychiatry 65:63–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Martino A, Fair DA, Kelly C, Satterthwaite TD, Castellanos FX, Thomason ME, Craddock RC, Luna B, Leventhal BL, Zuo X‐N, Milham MP (2014): Unraveling the miswired connectome: A developmental perspective. Neuron 83:1335–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dichter GS, Felder JN, Bodfish JW (2009): Autism is characterized by dorsal anterior cingulate hyperactivation during social target detection. Soc Cogn Affect Neurosci 4:215–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dipasquale O, Griffanti L, Clerici M, Nemni R, Baselli G, Baglio F (2015): High‐dimensional ICA analysis detects within‐network functional connectivity damage of default‐mode and sensory‐motor networks in Alzheimer's disease. Front Hum Neurosci 9:43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ecker C, Bookheimer SY, Murphy DGM (2015): Neuroimaging in autism spectrum disorder: Brain structure and function across the lifespan. Lancet Neurol 14:1121–1134. [DOI] [PubMed] [Google Scholar]
- Erbetta A, Bulgheroni S, Contarino V, Chiapparini L, Esposito S, Vago C, Riva D (2014): Neuroimaging findings in 41 low‐functioning children with autism spectrum disorder: A single‐center experience. J Child Neurol 29:1626–1631. [DOI] [PubMed] [Google Scholar]
- French L, Pavlidis P (2011): Relationships between Gene Expression and Brain Wiring in the Adult Rodent Brain. PLOS Comput Biol 7:e1001049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulcher BD, Fornito A (2016): A transcriptional signature of hub connectivity in the mouse connectome. Proc Natl Acad Sci USA 113:1435–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghanbari Y, Smith AR, Schultz RT, Verma R (2014): Identifying group discriminative and age regressive sub‐networks from DTI‐based connectivity via a unified framework of non‐negative matrix factorization and graph embedding. Med Image Anal 18:1337–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giedd JN, Blumenthal J, Jeffries NO, Castellanos FX, Liu H, Zijdenbos A, Paus T, Evans AC, Rapoport JL (1999): Brain development during childhood and adolescence: A longitudinal MRI study. Nat Neurosci 2:861–863. [DOI] [PubMed] [Google Scholar]
- Goulas A, Bastiani M, Bezgin G, Uylings HBM, Roebroeck A, Stiers P (2014): Comparative analysis of the macroscale structural connectivity in the macaque and human brain. PLOS Comput Biol 10:e1003529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groppe DM, Makeig S, Kutas M (2009): Identifying reliable independent components via split‐half comparisons. NeuroImage 45:1199–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groves AR, Smith SM, Fjell AM, Tamnes CK, Walhovd KB, Douaud G, Woolrich MW, Westlye LT (2012): Benefits of multi‐modal fusion analysis on a large‐scale dataset: Life‐span patterns of inter‐subject variability in cortical morphometry and white matter microstructure. NeuroImage 63:365–380. [DOI] [PubMed] [Google Scholar]
- Hagmann P, Sporns O, Madan N, Cammoun L, Pienaar R, Wedeen VJ, Meuli R, Thiran J‐P, Grant PE (2010): White matter maturation reshapes structural connectivity in the late developing human brain. Proc Natl Acad Sci USA 107:19067–19072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilgetag C‐C, Burns GAPC, O'Neill MA, Scannell JW, Young MP (2000): Anatomical connectivity defines the organization of clusters of cortical areas in the macaque and the cat. Philos Trans R Soc Lond B Biol Sci 355:91–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honey CJ, Sporns O, Cammoun L, Gigandet X, Thiran JP, Meuli R, Hagmann P (2009): Predicting human resting‐state functional connectivity from structural connectivity. Proc Natl Acad Sci USA 106:2035–2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imperati D, Colcombe S, Kelly C, Martino AD, Zhou J, Castellanos FX, Milham MP (2011): Differential development of human brain white matter tracts. PLoS One 6:e23437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DK (2010): Challenges and limitations of quantifying brain connectivity in vivo with diffusion MRI. Imaging Med 2:341–355. [Google Scholar]
- Jones DK, Knösche TR, Turner R (2013): White matter integrity, fiber count, and other fallacies: The do's and don'ts of diffusion MRI. NeuroImage 73:239–254. [DOI] [PubMed] [Google Scholar]
- Kana RK, Keller TA, Minshew NJ, Just MA (2007): Inhibitory control in high‐functioning autism: Decreased activation and underconnectivity in inhibition networks. Biol Psychiatry 62:198–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiviniemi V, Starck T, Remes J, Long X, Nikkinen J, Haapea M, Veijola J, Moilanen I, Isohanni M, Zang Y‐F, Tervonen O (2009): Functional segmentation of the brain cortex using high model order group PICA. Hum Brain Mapp 30:3865–3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochunov P, Williamson DE, Lancaster J, Fox P, Cornell J, Blangero J, Glahn DC (2012): Fractional anisotropy of water diffusion in cerebral white matter across the lifespan. Neurobiol Aging 33:9–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konrad K, Eickhoff SB (2010): Is the ADHD brain wired differently? A review on structural and functional connectivity in attention deficit hyperactivity disorder. Hum Brain Mapp 31:904–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostovic I, Jovanov‐Milosevic N (2006): The development of cerebral connections during the first 20–45 weeks' gestation. Semin Fetal Neonatal Med 11:415–422. [DOI] [PubMed] [Google Scholar]
- Krzywinski MI, Schein JE, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA (2009): Circos: An information aesthetic for comparative genomics. Genome Res 19:1639–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebel C, Gee M, Camicioli R, Wieler M, Martin W, Beaulieu C (2012): Diffusion tensor imaging of white matter tract evolution over the lifespan. NeuroImage 60:340–352. [DOI] [PubMed] [Google Scholar]
- Lee DD, Seung HS (1999): Learning the parts of objects by non‐negative matrix factorization. Nature 401:788–791. [DOI] [PubMed] [Google Scholar]
- Lo Y‐C, Soong W‐T, Gau SS‐F, Wu Y‐Y, Lai M‐C, Yeh F‐C, Chiang W‐Y, Kuo L‐W, Jaw F‐S, Tseng W‐YI (2011): The loss of asymmetry and reduced interhemispheric connectivity in adolescents with autism: A study using diffusion spectrum imaging tractography. Psychiatry Res 192:60–66. [DOI] [PubMed] [Google Scholar]
- McIntosh AR, Mišić B (2013): Multivariate statistical analyses for neuroimaging data. Annu Rev Psychol 64:499–525. [DOI] [PubMed] [Google Scholar]
- Meskaldji DE, Fischi‐Gomez E, Griffa A, Hagmann P, Morgenthaler S, Thiran J‐P (2013): Comparing connectomes across subjects and populations at different scales. NeuroImage 80:416–425. [DOI] [PubMed] [Google Scholar]
- Meunier D, Lambiotte R, Bullmore ET (2010): Modular and hierarchically modular organization of brain networks. Front Neurosci 4:200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori S, Crain BJ, Chacko VP, van Zijl PC (1999): Three‐dimensional tracking of axonal projections in the brain by magnetic resonance imaging. Ann Neurol 45:265–269. [DOI] [PubMed] [Google Scholar]
- Mueller S, Keeser D, Samson AC, Kirsch V, Blautzik J, Grothe M, Erat O, Hegenloh M, Coates U, Reiser MF, Hennig‐Fast K, Meindl T (2013): Convergent findings of altered functional and structural brain connectivity in individuals with high functioning autism: A multimodal MRI study. PLoS One 8:e67329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mundy P (2003): Annotation: The neural basis of social impairments in autism: The role of the dorsal medial‐frontal cortex and anterior cingulate system. J Child Psychol Psychiatry 44:793–809. [DOI] [PubMed] [Google Scholar]
- Nooner KB, Colcombe SJ, Tobe RH, Mennes M, Benedict MM, Moreno AL, Panek LJ, Brown S, Zavitz ST, Li Q, Sikka S, Gutman D, Bangaru S, Schlachter RT, Kamiel SM, Anwar AR, Hinz CM, Kaplan MS, Rachlin AB, Adelsberg S, Cheung B, Khanuja R, Yan C, Craddock CC, Calhoun V, Courtney W, King M, Wood D, Cox CL, Kelly AMC, Di Martino A, Petkova E, Reiss PT, Duan N, Thomsen D, Biswal B, Coffey B, Hoptman MJ, Javitt DC, Pomara N, Sidtis JJ, Koplewicz HS, Castellanos FX, Leventhal BL, Milham MP (2012): The NKI‐Rockland sample: A model for accelerating the pace of discovery science in psychiatry. Front Neurosci 6:152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noriuchi M, Kikuchi Y, Yoshiura T, Kira R, Shigeto H, Hara T, Tobimatsu S, Kamio Y (2010): Altered white matter fractional anisotropy and social impairment in children with autism spectrum disorder. Brain Res 1362:141–149. [DOI] [PubMed] [Google Scholar]
- O'Sullivan M, Jones DK, Summers PE, Morris RG, Williams SCR, Markus HS (2001): Evidence for cortical “disconnection” as a mechanism of age‐related cognitive decline. Neurology 57:632–638. [DOI] [PubMed] [Google Scholar]
- Park B, Kim D‐S, Park H‐J (2014): Graph independent component analysis reveals repertoires of intrinsic network components in the human brain. PLoS One 9:e82873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry A, Wen W, Lord A, Thalamuthu A, Roberts G, Mitchell PB, Sachdev PS, Breakspear M (2015): The organisation of the elderly connectome. NeuroImage 114:414–426. [DOI] [PubMed] [Google Scholar]
- Pestilli F, Yeatman JD, Rokem A, Kay KN, Wandell BA (2014): Evaluation and statistical inference for human connectomes. Nat Methods 11:1058–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power JD, Cohen AL, Nelson SM, Wig GS, Barnes KA, Church JA, Vogel AC, Laumann TO, Miezin FM, Schlaggar BL, Petersen SE (2011): Functional network organization of the human brain. Neuron 72:665–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugliese L, Catani M, Ameis S, Dell'Acqua F, de Schotten MT, Murphy C, Robertson D, Deeley Q, Daly E, Murphy DGM (2009): The anatomy of extended limbic pathways in Asperger syndrome: A preliminary diffusion tensor imaging tractography study. NeuroImage 47:427–434. [DOI] [PubMed] [Google Scholar]
- Ray S, Miller M, Karalunas S, Robertson C, Grayson DS, Cary RP, Hawkey E, Painter JG, Kriz D, Fombonne E, Nigg JT, Fair DA (2014): Structural and functional connectivity of the human brain in autism spectrum disorders and attention‐deficit/hyperactivity disorder: A rich club‐organization study. Hum Brain Mapp 35:6032–6048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raznahan A, Lerch JP, Lee N, Greenstein D, Wallace GL, Stockman M, Clasen L, Shaw PW, Giedd JN (2011): Patterns of coordinated anatomical change in human cortical development: A longitudinal neuroimaging study of maturational coupling. Neuron 72:873–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinov M, Sporns O (2010): Complex network measures of brain connectivity: Uses and interpretations. NeuroImage 52:1059–1069. [DOI] [PubMed] [Google Scholar]
- Rudie JD, Brown JA, Beck‐Pancer D, Hernandez LM, Dennis EL, Thompson PM, Bookheimer SY, Dapretto M (2012): Altered functional and structural brain network organization in autism. NeuroImage Clin 2:79–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumann CM, Bloss CS, Barnes CC, Wideman GM, Carper RA, Akshoomoff N, Pierce K, Hagler D, Schork N, Lord C, Courchesne E (2010): Longitudinal MRI study of cortical development through early childhood in autism. J Neurosci off J Soc Neurosci 30:4419–4427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simms ML, Kemper TL, Timbie CM, Bauman ML, Blatt GJ (2009): The anterior cingulate cortex in autism: Heterogeneity of qualitative and quantitative cytoarchitectonic features suggests possible subgroups. Acta Neuropathol (Berl) 118:673–684. [DOI] [PubMed] [Google Scholar]
- Simon HA (1962): The architecture of complexity. Proc Am Philos Soc 106:467–482. [Google Scholar]
- Smith RE, Tournier J‐D, Calamante F, Connelly A (2015): SIFT2: Enabling dense quantitative assessment of brain white matter connectivity using streamlines tractography. NeuroImage 119:338–351. [DOI] [PubMed] [Google Scholar]
- Sotiras A, Resnick SM, Davatzikos C (2015): Finding imaging patterns of structural covariance via Non‐Negative Matrix Factorization. NeuroImage 108:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sporns O, Tononi G, Kotter R (2005): The human connectome: A structural description of the human brain. PLoS Comput Biol 1:e42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoner R, Chow ML, Boyle MP, Sunkin SM, Mouton PR, Roy S, Wynshaw‐Boris A, Colamarino SA, Lein ES, Courchesne E (2014): Patches of disorganization in the neocortex of children with autism. N Engl J Med 370:1209–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supekar K, Uddin LQ, Khouzam A, Phillips J, Gaillard WD, Kenworthy LE, Yerys BE, Vaidya CJ, Menon V (2013): Brain hyperconnectivity in children with autism and its links to social deficits. Cell Rep 5:738–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi E, Folkerth RD, Galaburda AM, Grant PE (2012): Emerging cerebral connectivity in the human fetal brain: An MR tractography study. Cereb Cortex 22:455–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tebartz van Elst L, Maier S, Fangmeier T, Endres D, Mueller GT, Nickel K, Ebert D, Lange T, Hennig J, Biscaldi M, Riedel A, Perlov E (2014): Disturbed cingulate glutamate metabolism in adults with high‐functioning autism spectrum disorder: Evidence in support of the excitatory/inhibitory imbalance hypothesis. Mol Psychiatry 19:1314–1325. [DOI] [PubMed] [Google Scholar]
- Thakkar KN, Polli FE, Joseph RM, Tuch DS, Hadjikhani N, Barton JJS, Manoach DS (2008): Response monitoring, repetitive behaviour and anterior cingulate abnormalities in autism spectrum disorders (ASD). Brain J Neurol 131:2464–2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasi D, Volkow ND (2012): Abnormal functional connectivity in children with attention‐deficit/hyperactivity disorder. Biol Psychiatry 71:443–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uppal N, Wicinski B, Buxbaum JD, Heinsen H, Schmitz C, Hof PR (2014): Neuropathology of the anterior midcingulate cortex in young children with autism. J Neuropathol Exp Neurol 73:891–902. [DOI] [PubMed] [Google Scholar]
- Vaishnavi SN, Vlassenko AG, Rundle MM, Snyder AZ, Mintun MA, Raichle ME (2010): Regional aerobic glycolysis in the human brain. Proc Natl Acad Sci USA 107:17757–17762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Heuvel MP, Sporns O (2011): Rich‐club organization of the human connectome. J Neurosci off J Soc Neurosci 31:15775–15786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasung L, Huang H, Jovanov‐Milošević N, Pletikos M, Mori S, Kostović I (2010): Development of axonal pathways in the human fetal fronto‐limbic brain: Histochemical characterization and diffusion tensor imaging. J Anat 217:400–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vissers ME, Cohen MX, Geurts HM (2012): Brain connectivity and high functioning autism: A promising path of research that needs refined models, methodological convergence, and stronger behavioral links. Neurosci Biobehav Rev 36:604–625. [DOI] [PubMed] [Google Scholar]
- Watanabe T, Rees G (2015): Age‐associated changes in rich‐club organisation in autistic and neurotypical human brains. Sci Rep 5:16152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westlye LT, Walhovd KB, Dale AM, Bjørnerud A, Due‐Tønnessen P, Engvig A, Grydeland H, Tamnes CK, Ostby Y, Fjell AM (2010): Life‐span changes of the human brain white matter: Diffusion tensor imaging (DTI) and volumetry. Cereb Cortex N Y N 1991 20:2055–2068. [DOI] [PubMed] [Google Scholar]
- Wijk BCM. v, Stam CJ, Daffertshofer A (2010): Comparing brain networks of different size and connectivity density using graph theory. PLoS One 5:e13701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Oja E (2010): Linear and nonlinear projective nonnegative matrix factorization. IEEE Trans Neural Netw 21:734–749. [DOI] [PubMed] [Google Scholar]
- Yang Z, Zhu Z, Oja E (2010): Automatic rank determination in projective nonnegative matrix factorization In: Vigneron V, Zarzoso V, Moreau E, Gribonval R, Vincent E, editors. Latent Variable Analysis and Signal Separation. Lecture Notes in Computer Science. Berlin, Heidelberg: Springer; pp 514–521. [Google Scholar]
- Yap P‐T, Fan Y, Chen Y, Gilmore JH, Lin W, Shen D (2011): Development trends of white matter connectivity in the first years of life. PLoS One 6:e24678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalesky A, Fornito A, Harding IH, Cocchi L, Yücel M, Pantelis C, Bullmore ET (2010): Whole‐brain anatomical networks: Does the choice of nodes matter? NeuroImage 50:970–983. [DOI] [PubMed] [Google Scholar]
- Zhao T, Cao M, Niu H, Zuo X‐N, Evans A, He Y, Dong Q, Shu N (2015): Age‐related changes in the topological organization of the white matter structural connectome across the human lifespan. Hum Brain Mapp 36:3777–3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler G, Dahnke R, Jäncke L, Yotter RA, May A, Gaser C (2012): Brain structural trajectories over the adult lifespan. Hum Brain Mapp 33:2377–2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielinski BA, Gennatas ED, Zhou J, Seeley WW (2010): Network‐level structural covariance in the developing brain. Proc Natl Acad Sci USA 107:18191–18196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zikopoulos B, Barbas H (2010): Changes in prefrontal axons may disrupt the network in autism. J Neurosci 30:14595–14609. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information


