Abstract
The hippocampus is composed of distinct subfields linked to diverse functions and disorders. The subfields can be mapped using high‐resolution magnetic resonance images, and their volumes can potentially be used as quantitative phenotypes for genetic investigation of hippocampal function. We estimated the heritability of hippocampus subfield volumes of 465 subjects from the Human Connectome Project (twins and non‐twin siblings) using two methods. The first used a univariate model to estimate heritability with and without adjustment for total brain volume (TBV) and ipsilateral hippocampal volume to determine if heritability was uniquely attributable to subfield volume rather than confounds that attributed to global volumes. We observed the right: subiculum, cornu ammonis 2/3, and cornu ammonis 4/dentate gyrus subfields had the highest significant heritability estimates after adjusting for ipsilateral hippocampal volume. In the second analysis, we used a bivariate model to investigate the shared heritability and genetic correlation of the subfield volumes with TBV and ipsilateral hippocampal volume. Genetic correlation demonstrates shared genetic architecture between phenotypes and shared heritability is what proportion of the genetic architecture of one trait is shared by the other. Highest genetic correlations were between subfield volumes and ipsilateral hippocampal volume than with TBV. The pattern was opposite for shared heritability suggesting that subfields share greater proportion of the genetic architecture with TBV than with ipsilateral hippocampal volume. The relationship between the genetic architecture of TBV, hippocampal volume, and of individual subfields should be accounted for when using hippocampal subfield volumes as quantitative phenotypes for imaging genetics studies. Hum Brain Mapp 38:4337–4352, 2017. © 2017 Wiley Periodicals, Inc.
Keywords: genetic variation, genetic correlation, quantitative phenotypes, extended twin design, hippocampal subfield segmentation, univariate and bivariate model
INTRODUCTION
The hippocampus plays a key role in cognitive functioning, influencing processes such as learning, episodic and working memory [Braskie et al., 2013]. Structural variation within the hippocampal structure and function has been implicated in neurodegenerative and neuropsychiatric disorders such as Alzheimer's disease [Braak and Braak, 1991; Mouiha and Duchesne, 2011], depression [Bremner et al., 2000; Campbell et al., 2004; Treadway et al., 2015] and schizophrenia [Altshuler et al., 1998; Bogerts et al., 1993; Haukvik et al., 2015; Tamminga et al., 2010]. Although the hippocampus is often referred to in neuroimaging studies as a single unitary structure, it is composed of several different subfields. While there are varying definitions for the delineation and the nomenclature of these subfields, there is a general consensus in the neuroimaging research discipline that subfields can be resolved using magnetic resonance imaging (MRI) techniques to include some combination of cornu ammonis (CA) 1–4, dentate gyrus (DG), subiculum and the molecular layers composed of the stratum: radiatum, lacunosum and moleculare (SRLM) [Duvernoy, 2005]. Subfields within the hippocampus differ from each other in terms of connection to other regions of the brain, their cytoarchitectonic structure, and their role in memory formation and cognitive function [Amunts et al., 2005; Duvernoy, 2005; Fatterpekar et al., 2002; La Joie et al., 2010; Mai et al., 2008; Mueller et al., 2007; Mueller and Weiner, 2009; Voineskos et al., 2015; Yang et al., 2013].
As improved techniques for automated mapping of the hippocampal subfields emerge [Iglesias et al., 2015; Pipitone et al., 2014; Van Leemput et al., 2008; Yushkevich et al., 2010] in concert with large‐scale consortia dedicated to genome‐wide association analysis [Thompson et al., 2014], it is critical to determine whether the individual subfields can be used as quantitative traits in such studies. Prior to using volumetric estimates of the hippocampal subfields as quantitative phenotypes, we must first investigate whether these volumes are indeed heritable. Heritability estimates are defined by the proportion of observed variation in a trait that can be attributed to inherited genetic factors [Jacquard, 1983] and can be estimated using twin studies, family studies and genome wide association data from large unrelated populations based on three factors: genetics (A), common (C), and unique environment (E). Typically, structural equation modeling (SEM) is used in twin studies to estimate variation of the phenotype of interest [Neale and Cardon, 1992].
While several other studies on the heritability of brain structures exist [Baare et al., 2001; Pennington et al., 2000; Sullivan et al., 2001], there is only one study that examines the heritability of the hippocampal subfields [Whelan et al., 2015]. Whelan et al., [2015] used standard T1‐weighted MRI data from the Queen Twins Imaging (QTIM) and also investigated the reliability of automatically segmented human hippocampus with a newly developed FreeSurfer subfield segmentation tool. Whelan et al. [2015] demonstrated moderate 4T QTIM test‐retest reliability scores of intraclass correlation coefficient (ICC) in the range of 0.50–0.86, where the CA1 region had the highest ICC of 0.86 and hippocampal‐amygdaloid transition (HATA) area had the lowest ICC of 0.5. In addition Whelan et al. [2015] found high heritability estimates ranging from 0.67 (HATA region) to 0.85 (molecular layer of DG) of hippocampal subregion volumes. Our study differs from the study of Whelan et al. based on the previously validated segmentation technique [Pipitone et al., 2014] and the use of high‐resolution and contrast of MRI scans, from the Human Connectome Project (HCP) [Van Essen et al., 2013]. Furthermore, when exploring heritability it is important to consider the relationship of the hippocampal subfields, with overall volume of the brain and or the entire hippocampus; both of which vary between individuals [Blatter et al., 1995]. Recent studies that have examined the heritability of neuroanatomical volume measurements have not adjusted for total brain volume (TBV) or volumes of larger structures encompassing the region of interest. Baare et al. demonstrated that the whole brain is highly heritable, estimated to be 90% [Baare et al., 2001], therefore not accounting for TBV on regions of interest can influence the heritability estimates of the target regions resulting in inaccurate heritability scores.
In our study, we explored how heritability of individual hippocampal subfields is influenced by both TBV and ipsilateral (left or right) hippocampus volume to the subfield being examined using two different methods. Our first approach was using a univariate model examining the heritability of a single phenotype at a time; in our case, we examined the heritability of hippocampal subfields. First, we adjusted hippocampal subfield volume for TBV, then we adjusted for only ipsilateral hippocampal volume and finally the last step was adjusting for both TBV and ipsilateral hippocampal volumes. In our second approach, we used a bivariate model where we examined the heritability of two structures together instead of one structure alone. We went on to quantify the relationship of the shared genetic variation between hippocampal subfield volumes with TBV and ipsilateral hippocampal volume by examining the genetic correlation and shared heritability in a bivariate model. Both models give us heritability estimates which can reveal the genetic architecture of a phenotype by itself but also in relation to other phenotypes (see Box 1 for definitions of heritability terms). In the univariate model, the heritability estimates are based on the genetic variation of one phenotype. Heritability estimates in our univariate model are based on genetic variation of subfield volumes alone. We removed the genetic influence of TBV and hippocampal volume which provided us with unique heritability estimates for subfield volumes. Conversely, the bivariate model allows us to account for the genetic influence of TBV and hippocampal volume by estimating both the genetic correlation and shared heritability between the two phenotypes. Genetic correlation measures the genetic relationship in terms of how similar the genetic architecture is between the two phenotypes. Shared heritability measures the shared genetic variation between the two phenotypes as a proportion of the heritability of one phenotype. Using both the univariate and bivariate model to investigate the genetic variation within the hippocampal subfields allows us to identify potential quantitative phenotypes to be used in imaging genetic studies.
Box 1. Key Heritability Concepts.
Heritability estimates the amount of genetic variation that is seen in a phenotypic trait within a population. Total variation includes genetics [A], common environment [C] and unique environment [E]. Heritability is the proportion of the variation that can be attributed to genetics.
Unique heritability accounts for the genetic variation out of total variation (genetic and common and unique environment) of one trait only. This can be calculated in a univariate model where only one phenotype is being analyzed. In a univariate model, factors that may confound heritability estimates such as total brain volume can be controlled for within the model.
In a bivariate model, the genetic variation is accounted for between two phenotypes instead of focusing on only one trait. In these models we can calculate the genetic correlation between the two phenotypes and the shared heritability. Genetic correlation is the genetic relationship between the two traits. It indicates how similar the genetic variation (genetic architecture) is between both traits. Shared heritability measures the amount of variation from the genetic correlation that is common from trait one and present in trait two. Both genetic correlation and shared heritability estimates should be considered when examining heritability estimates in bivariate model.
METHODS
Human Connectome Project Dataset
Data from the HCP was used for this study. The aim of the HCP is to investigate properties of human brain connectivity and function. To better understand the interaction of brain circuits and human behavior, structural and functional properties of neuroanatomical structures as well as genetic factors can be studied. The HCP investigators are recruiting 1,200 healthy twin and non‐twin sibling adults. The HCP consortium aims to have a healthy population that represents the ethnic and racial composition of United States and diversity in terms of behavioral, ethnic, and socioeconomic status. Sibling relationships were removed if the individuals within the relationship had neuropsychiatric, neurodevelopmental, or neurological disorders. Individuals having other illnesses such as diabetes and high blood pressure were excluded. Premature twins (born before 34 weeks gestation) and non‐twins (born before 37 weeks gestation) were excluded. Individuals who smoked or who were overweight were included in the study. Also individuals who have not experienced severe symptoms but have a history of heavy drinking or recreational drug use were included. Reason for including individuals who smoked, who are overweight or use of recreational substances can be used for future psychiatric studies [Van Essen et al., 2013]. For more information on the inclusion and exclusion criteria, see Supporting Information Table S1 of Van Essen DC et al. [2013].
The sample used for this study contained 542 individuals of which imaging data was available on 511 individuals (data release June 2014). Imaging data were collected using a Siemens 3 Tesla (T) Skyra scanner which has been modified with a Siemens SC72 gradient coil to increase the maximum gradient strength from 40 to 100 mT/m [Van Essen et al., 2012, 2013]. For our study, we used the 3T, high‐resolution T1 weighted MRI data (0.7mm isotropic voxel dimensions). Acquisition parameters are: inversion time = 1,000 ms, echo time = 2.14 ms, repetition time = 2,400 ms, acquisition time = 7 min 40 sec, flip angle = 8 degrees and field of view = 224 mm × 224 mm [Van Essen et al., 2012].
After quality control (QC) of the segmented imaging data and removal of families with one individual and no siblings (described below), the final sample consisted of 100 monozygotic (MZ) twins, 94 dizygotic (DZ) twins and 271 non‐twin siblings (277 women and 188 men with age range of 22‐ to 36‐year old and the average age was 29.24‐ (± 3.49 SD) year old). Average handedness for our sample was 65.39 (± 46.42 SD). The scale for handedness ranges from −100 (left‐hand dominant) to 100 (right‐hand dominant). The measure of fluid intelligence was represented using Raven's Progressive Matrices test where the final score represents the number of correct responses out of 24 questions. Demographic information is summarized in Table 1. Handedness and fluid intelligence measures are not used in subsequent heritability calculations, but simply to demonstrate that MZ, DZ, and non‐twin sibling subgroups are well matched to one another and represent a healthy population.
Table 1.
Demographic breakdown of monozygotic twins (MZ), dizygotic twins (DZ), and non‐twin siblings from the subset data of the HCP, including averages and standard deviation (± SD)
| N | Average age (year ± SD) | Age range | Gender female: male | Average handedness (± SD) | Average fluid intelligence (± SD) | |
|---|---|---|---|---|---|---|
| MZ | 100 | 29.97 (3.11) | 22–36 | 74:26 | 72.4 (41.76) | 16.26 (4.64) |
| DZ | 94 | 29.86 (2.93) | 22–35 | 67:27 | 63.19 (46.06) | 16.71 (4.81) |
| Non‐twin siblings | 271 | 28.75 (3.72) | 22–36 | 136:135 | 63.56 (48.06) | 16.12 (5.11) |
| Total | 465 | 29.24 (3.49) | 22–36 | 277:188 | 65.39 (46.42) | 16.27 (4.95) |
Average fluid intelligence is a measure of number of correct responses out of 24 questions.
Image Processing
Hippocampal subfield segmentation was estimated using Multiple Automatically Generated Templates (MAGeT Brain) [Chakravarty et al., 2013; Pipitone et al., 2014]. The segmentation procedure consists of three steps. First, five independent high‐resolution MRI atlases (0.3mm isotropic voxels) of the hippocampus and hippocampal subfields containing detailed segmentations of the left and right CA1, CA2/CA3, CA4/DG, subiculum, SRLM are used as inputs [Winterburn et al., 2013]. Then, a “template library” is generated where a subset of individuals from the HCP dataset is first selected (n = 21; a number demonstrated as being optimal in previous work [Pipitone et al., 2014]). The 21 templates were selected to represent the HCP dataset (12 females, 9 males, aged range: 22 to 36 years). Each template undergoes a model‐based segmentation procedure with each atlas yielding five candidate labels for each template. The next step is similar to a regular multi‐atlas segmentation strategy, where each subject is nonlinearly matched to template, thereby growing the number of candidate segmentations to 105 (5 atlases × 21 templates) for each subject. The 105 labels per subject are then fused using a majority vote [Collins and Pruessner, 2010], a method which was previously demonstrated to be accurate relative to weighted voting. Pipitone et al. [2014] showed that using weighted voting methods did not significantly improve MAGeT Brain segmentation when compared to using majority vote labeling [Bhagwat et al., 2016; Pipitone et al., 2014]. Recent work by Bhagwat et al. [2016] has also demonstrated accuracies to be homologous to Joint Label Fusion [Wang et al., 2013] when used within the MAGeT Brain framework [Bhagwat et al., 2016]. In Winterburn et al. [2013] protocol for human hippocampal subfields segmentation, intra reliability was measured via test‐retest and results ranged from 0.64 in the CA2/CA3 region to 0.83 mean Kappa in the CA4/DG region. The second lowest test‐retest was the SRLM region with mean Kappa of 0.71. The subiculum and the CA1 region had a Kappa mean of 0.75 and 0.78, respectively.
QC of hippocampal segmentation was completed for 511 subjects with imaging data and 502 passed QC. Images were rated on a three point scale of 0, 0.5, and 1, where 1 was a pass with very negligible errors, 0.5 was satisfactory with few errors but still considered pass and images scored 0 is a fail and the subject was removed for the final analysis. Subjects scored as 1 had less than 5 slices with minor errors whereas subjects that contained minor errors found in 5 to 10 slices were assigned 0.5. Subjects that contain errors in more than 10 slices unilaterality were scored 0. Minor errors included small deviation from the correct segmentation within subjects which are scored 1 and 0.5. Major deviation from the correct segmentation where large portions of the hippocampus are missing or segmented in the wrong location on the slice was scored as 0. In the Supporting Information section Figure S1, A–C show examples of subjects scored 0, 0.5, and 1.
MAGeT Brain was implemented using the publicly available pipeline (https://github.com/CobraLab/MAGeTbrain) and atlases (https://github.com/CobraLab/atlases/tree/master/hippocampus-subfields). In addition, TBV was extracted using the automated BEaST pipeline [Eskildsen et al., 2012] along with minc‐bpipe‐library and within BEaST output all subjects passed QC. Nonlinear registration operations were performed using Automatic Normalization Tools (ANTs) [Avants et al., 2008]. All images were converted to the MINC format and MAGeT Brain was implemented using tools included in MINC (http://www.bic.mni.mcgill.ca/ServicesSoftware/ServicesSoftwareMincToolKit), including a version of ANTs adapted to work with MINC‐tools.
Method 1: Heritability of Subfields After Adjusting for Ipsilateral Hippocampal Volume and TBV Using a Univariate Model
Before adjusting for TBV and ipsilateral hippocampal volume on hippocampal subfield volumes, we examined the correlation between hippocampal subfields volumes with ipsilateral hippocampal volume and TBV; we also investigated correlation between left and right hippocampal subfield volumes. Pearson's correlation along with a P‐value was calculated using R 3.2.1 statistical computing software [R Core Development Team, 2013]. Hippocampal subfield volumes were adjusted for: (1) sex and age alone, hereafter called “unadjusted,” (2) TBV, (3) ipsilateral hippocampal volume, (4) ipsilateral hippocampal volume and TBV together. A residual approach in R was used for each adjustment on the hippocampal subfields volumes and then heritability was calculated for each type of adjustment in a univariate ACE model (calculating the: genetics [A], common environment [C], and unique environment [E] variation). The resulting analysis examines the influence of overall brain and hippocampus volume on subfield heritability measures.
Broad sense heritability (H 2) of hippocampal subfield volumes was estimated using SEM implemented with the OpenMx 2.3.1 [Neale et al., 2016] package within R. Following the imaging QC described above, the number of subjects was reduced to 465 healthy subjects from 502, based on removing subjects in which the family only had one individual. The sample included: 100 MZ twins, 94 DZ twins, and 271 siblings (from 177 families, 96 families had siblings added to the twin pair and 81 families consisted of non‐twin siblings only).
Heritability is the ratio of variance of a phenotypic measurement in which the numerator is the variation attributable to genetics [A] and the denominator is the total observed variation (genetics [A], common environment [C], and unique environment [E]). In classical twin studies, the model assumes that twins (both MZ and DZ twins) are raised together; therefore, the common environment is based on twin status and is the same for each individual within the twin pair. When siblings are introduced the environment is also assumed to be identical but this may be a less valid assumption. However, heritability estimates were similar whether or not we included siblings lending weight to the robustness of the test. Therefore, common environment [C] was assumed to be identical within a family. MZ twins share 100% of their genes whereas DZ twins share 50% of segregating genes, similar to DZ twins, non‐twin siblings share on average 50% of their genes. To account for the genetic [A] component on the twins and non‐twin siblings in the model, MZ twins had a coefficient of 1 and DZ twins and non‐twin siblings a coefficient of 0.5. In the dataset, some families had data for only non‐twin siblings; in these cases, a coefficient of 0.5 was used for both family members. Heritability was calculated for volume of both ipsilateral hippocampal subfields (CA1, CA2/CA3, CA4/DG, subiculum, and SRLM) along with TBV and ipsilateral hippocampal volumes.
A full univariate ACE model was applied to each hippocampal subfield and compared against a CE model to examine the significance of the genetic variation when “A” is removed from the full model using a likelihood ratio test. A significant P‐value (P < 0.05) for the difference in fit of the two models indicates that the “A” component plays an important role in explaining the variance of the trait thereby demonstrating the significance of the heritability. This was applied to all four models (1) unadjusted subfield volumes, (2) adjusted for TBV, (3) adjusted for ipsilateral hippocampal volume, (4) adjusted for TBV and ipsilateral hippocampal volume together. Confidence intervals of 95% on the heritability estimates were calculated for each hippocampal subfield in all four models within the univariate model.
For both univariate model and bivariate models (shown below), we simply observed the heritability estimate of the subfields to see which regions are heritable within the hippocampus instead of statistically comparing the heritability estimates between regions. As the subfield volumes are highly correlated with TBV and ipsilateral hippocampal volume, the tests would not be independent and applying Bonferroni correction would be too stringent. Therefore, the P values reported when comparing the full ACE model against the CE model are uncorrected values.
Method 2: Heritability Using a Bivariate Model
A bivariate Cholesky decomposition ACE model was used to examine the shared genetic variation (H 2) and the genetic correlation (r g) between the hippocampal subfield volumes with TBV and ipsilateral hippocampal volume. Genetic correlation measures the shared genetics effects (genetic architecture) between the two volumes. Shared heritability estimates the proportion of genetic variation of one trait that is present in the other. Therefore, two structures can have a high genetic correlation such as 0.8 but a low shared heritability of 0.55. This means that there is a large overlap in genes that may influence the variation on both structures but the proportion of shared genetic variation between the structures is low. For the calculation of shared heritability, the order of the phenotypic variables under study is important within the model. We selected the larger global structure as the reference trait while controlling for sex and age. Volumes of the hippocampus subfields and TBV were normalized using z‐scores. To examine the significance of heritability scores, the bivariate ACE model was compared to a bivariate CE model and confidence intervals (95%) on the heritability scores were calculated.
RESULTS
Segmentation and Volumes of Hippocampal Subfields
From the 511 images segmented using MAGeT Brain, 502 passed QC (example of a segmented hippocampus in Fig. 1). Table 2 shows the average volumes and standard deviation of each hippocampal subfield including left and right hippocampal volume and TBV. From the five subfield regions and as expected, the CA1 region showed the largest average volume (left CA1 = 771.51 mm3, ± 92.15 SD, right CA1 = 836.47 mm3, ± 98.14 SD) and the CA2/CA3 subfield was the smallest (left CA2/CA3 = 147.91 mm3, ± 25.11 SD, right CA2/CA3 = 179.22 mm3, ± 25.18 SD). Table 2 also includes average volumes and standard deviation for MZ, DZ and non‐twin siblings. Higher TBV found in non‐twin siblings compared to both twin groups is accounted by having a higher ratio of females whereas the ratio between males and females are the same in the non‐twin sibling group. It has been shown that females have smaller TBV compared to males [Kretschmann et al., 1979; Swaab and Hofman, 1984], which explains the differences in average volumes between both the twins groups and non‐twin siblings.
Figure 1.
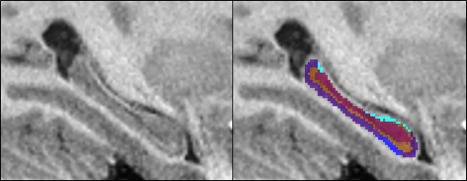
Hippocampal subfield segmentation, T1 sagittal scan. Left image is non‐segmented and right image is segmented. Hippocampal subfields: CA1, CA2/CA3, CA4 and DG, SRLM, Subiculum.
Table 2.
Average volume and standard deviation (±SD) of unadjusted left and right hippocampal subfields in monozygotic twins (MZ), dizygotic twins (DZ), and non‐twin siblings
| Region | MZ average volume (mm3 ± SD) N = 100 | DZ average volume (mm3 ± SD) N = 94 | Non‐twin sibling average volume (mm3 ± SD) N = 271 | Total Sample average volume mm3 (± SD) N = 465 |
|---|---|---|---|---|
| Left CA1 | 755.71 (89.43) | 761.50 (74.00) | 780.82 (97.77) | 771.51 (92.15) |
| Right CA1 | 821.41 (90.97) | 826.97 (87.04) | 845.33 (103.51) | 836.47 (98.14) |
| Left CA2CA3 | 145.9705 (22.55) | 146.69 (21.56) | 149.04 (27.10) | 147.91 (25.12) |
| Right CA2CA3 | 174.82 (22.61) | 174.13 (24.04) | 182.61 (25.98) | 179.22 (25.18) |
| Left CA4DG | 623.61 (70.02) | 621.75 (55.12) | 649.06 (73.27) | 638.07 (70.34) |
| Right CA4GD | 615.00 (65.91) | 616.62 (55.99) | 649.88 (72.54) | 635.67 (70.00) |
| Left SRLM | 577.99 (67.90) | 581.60 (61.98) | 589.51 (73.93) | 585.44 (70.42) |
| Right SRLM | 543.56 (64.33) | 546.66 (63.48) | 558.37 (68.70) | 552.81 (66.94) |
| Left subiculum | 331.43 (46.22) | 327.84 (43.27) | 335.89 (50.83) | 333.31 (48.43) |
| Right subiculum | 329.45 (43.63) | 327.48 (38.08) | 333.92 (45.87) | 331.66 (43.92) |
| Left hippocampus | 2434.72 (253.57) | 2439.39 (216.75) | 2504.33 (277.15) | 2476.23 (262.56) |
| Right hippocampus | 2484.25 (249.13) | 2491.87 (233.02) | 2570.11 (277.67) | 2535.82 (265.78) |
| Total brain | 1344941.77 (138663.72) | 1351379.79 (117637.94) | 1393633.69 (150431.05) | 1374620.66 (143368.64) |
Hippocampal Subfield Correlation With TBV and Ipsilateral Hippocampal Volume
Both the left and right hippocampal subfield volumes were highly positively correlated with left and right hippocampal volume, respectively (Table 3). The highest correlation coefficient between hippocampus subfields and ipsilateral hippocampal volume was the right and left CA1 and SRLM region (r = 0.96, P < 0.001) and the lowest correlation coefficients were left CA2/CA3 (r = 0.56, P < 0.001) and right subiculum (r = 0.60, P < 0.001). In contrast, the correlation coefficient was lower between hippocampal subfields and TBV but still positive. The highest coefficient between hippocampus subfield and TBV was left and right CA1 (r = 0.69, r = 0.66, respectively) and lowest correlation coefficient was the left and right CA2/CA3 region (r = 0.32, r = 0.47, respectively). In addition, the left and right hippocampal volume was highly correlated with TBV (r = 0.72, r = 0.70, respectively).
Table 3.
Pearson correlations (r) between subfield with ipsilateral hippocampal volume and total brain volume (TBV)
| Left hippocampal volume | TBV | |
|---|---|---|
| Left CA1 | 0.96 | 0.69 |
| Left CA2CA3 | 0.56 | 0.32 |
| Left CA4DG | 0.91 | 0.64 |
| Left SRLM | 0.96 | 0.60 |
| Left subiculum | 0.58 | 0.63 |
| Left hippocampus | 1.00 | 0.72 |
| Right hippocampal volume | TBV | |
| Right CA1 | 0.96 | 0.66 |
| Right CA2CA3 | 0.74 | 0.47 |
| Right CA4DG | 0.89 | 0.63 |
| Right SRLM | 0.96 | 0.60 |
| Right subiculum | 0.60 | 0.61 |
| Right hippocampus | 1.00 | 0.71 |
All subfield volumes showed significant correlation between ipsilateral hippocampal volume and TBV (P < 0.001).
Univariate Model: Heritability Estimates of Hippocampal Subfields Volumes
Unadjusted hippocampal subfield volumes demonstrated moderate to high heritability estimates. The highest heritability estimate within hippocampal subfield volumes was the right subiculum at 85% and the lowest estimate was the left CA2/CA3 at 38% (Table 4, Fig. 2). Observed heritability estimates of left and right hippocampal volume were 80% and 81%, respectively, and the heritability of TBV was 92%. When adjusted for TBV and ipsilateral hippocampal volume the heritability of subfield volumes ranged from 38% (left CA1) to 79% (right subiculum) (Table 4, Fig. 2) and the heritability of left and right hippocampal volume was lower with estimates of 65% and 63%, respectively, when adjusting for TBV. When both TBV and ipsilateral hippocampal volume were adjusted for, the observed heritability estimate of each subfield volume was similar to only adjusting for ipsilateral hippocampal volume (Table 4, Fig. 2).
Table 4.
Heritability estimates of left and right hippocampal subfield volumes with 95% confidence intervals (sample size N = 465)
| No volume adjustment (only adjusted for sex and age) | Total brain volume adjustment | ipsilateral hippocampal volume adjustment | ipsilateral hippocampal and total brain volume adjustment | |||||
|---|---|---|---|---|---|---|---|---|
| Region | H 2 (%) | 95% confidence interval | H 2 (%) | 95% confidence interval | H 2 (%) | 95% confidence interval | H 2 (%) | 95% confidence interval |
| Left CA1 | 73** | 55–83 | 56** | 33–72 | 39** | 21–55 | 39 | 21–55 |
| Right CA1 | 70** | 50–83 | 54** | 29–71 | 59* | 42–72 | 59* | 42–72 |
| Left CA2CA3 | 38* | 18–58 | 41** | 21–59 | 53* | 35–67 | 52* | 34–67 |
| Right CA2CA3 | 70** | 51–82 | 68** | 51–80 | 67* | 52–78 | 66* | 50–78 |
| Left CA4DG | 77** | 58–87 | 65** | 41–81 | 57** | 38–73 | 57** | 37–73 |
| Right CA4DG | 80** | 65–88 | 72** | 54–83 | 65** | 50–76 | 65** | 50–76 |
| Left SRLM | 64** | 42–78 | 53** | 30–70 | 47* | 30–62 | 40* | 22–56 |
| Right SRLM | 74** | 56–84 | 64** | 43–78 | 59** | 42–72 | 55** | 38–69 |
| Left subiculum | 50* | 16–77 | 63** | 26–75 | 60* | 20–72 | 57** | 22–71 |
| Right subiculum | 85** | 68–90 | 76** | 62–84 | 82** | 71–88 | 78** | 65–86 |
| Left hippocampus | 80** | 66–88 | 65** | 43–80 | ||||
| Right hippocampus | 81** | 66–89 | 63** | 41–78 | ||||
| Total brain | 92** | 75–95 | ||||||
Significant heritability P values are represented by * indicates P < 0.05, and ** indicates P < 0.001 when ACE model is compared to CE model. Heritability estimates highlighted in yellow are not significant, but show a trend toward significance.
Figure 2.
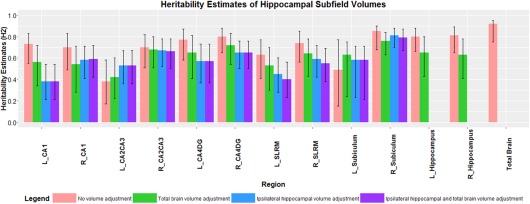
Heritability scores (H 2) of left (L) and right (R) hippocampal subfields volumes, L and R hippocampal volumes and TBV with 95% confidence intervals (sample size N = 465). Pink bars indicate unadjusted subfield volume, whereas green bars indicate TBV adjustment, blue bars indicates ipsilateral (left or right) hippocampal volume adjustment and purple bars indicates ipsilateral hippocampal and TBV adjustment on subfield volumes.
Overall, a trend of lower heritability scores of the hippocampal subfields was seen when adjusting for ipsilateral hippocampal volume compared to adjusting for only TBV. For example, unadjusted left CA2/CA3 volumes (Table 4, Fig. 2) had a heritability estimate of 38% but after adjusting for TBV and left hippocampal volume separately, the estimate was higher (41% and 53%, respectively). When the left CA2/CA3 volume was adjusted for both left hippocampal volume and TBV together, the heritability estimate was similar to when adjusting for left hippocampal volume only (52%). Similarly, heritability of the left subiculum unadjusted volume was 50%, but when adjusted for TBV the estimate was higher (63%). However, adjusting for the left hippocampal volume along with TBV, the heritability estimate of left subiculum volume was also higher than unadjusted left subiculum volume but lower than accounting for only TBV. When the ACE model is compared to the CE model, the heritability was significant for all regions whether or not the subfield volume was adjusted for TBV (P < 0.05). When subfield volumes were adjusted for ipsilateral hippocampal volume all but one subfield had a significant heritability estimate (the left CA1 region P = 0.09) (Table 4).
Bivariate Model: Shared Heritability Estimates of Hippocampal Subfields Volumes With TBV and Ipsilateral Hippocampal Volume
In the bivariate model, both the genetic correlation and heritability was measured between hippocampal subfield volumes with TBV and ipsilateral hippocampal volume. The shared and unique heritability estimates were significant (P < 0.05) when the full ACE bivariate model was compared to the CE model across all subfields with both the TBV and ipsilateral hippocampal volume.
The genetic correlation (r g) between TBV and hippocampal subfield volumes (Table 5) was positive and slightly higher than the Pearson correlation performed in the univariate model (Table 3). The shared heritability between TBV and subfield was high ranging from 86% in left subiculum to 99% heritability in right subiculum and left CA2/CA3 region (Table 6, Fig. 3). Overall the genetic correlation between ipsilateral hippocampal volume and hippocampal subfield volumes showed a higher positive genetic correlation trend compared to TBV and hippocampal subfield. The genetic correlation of left hippocampal volume and left hippocampal subfield volumes ranged from 0.75 in left CA2/CA3 to 0.98 in left CA1 and left SRLM. The range in the right was from 0.66 in the right subiculum to 0.97 in the right CA1 and SRLM region (Table 5). The shared heritability between left hippocampal and left hippocampal subfield ranged from 77% in the CA2/CA3 region to 93% in the left subiculum region (Table 7, Fig. 4). For the right, the range was 83% in the CA1 region to 94% in the right subiculum region (Table 7, Fig. 4).
Table 5.
Bivariate genetic correlation (r g) between hippocampal subfield volumes with ipsilateral hippocampal volume and total brain volume (TBV)
| Left hippocampal volume | TBV | |
|---|---|---|
| Left CA1 | 0.98 | 0.77 |
| Left CA2CA3 | 0.75 | 0.49 |
| Left CA4DG | 0.94 | 0.69 |
| Left SRLM | 0.98 | 0.70 |
| Left subiculum | 0.76 | 0.71 |
| Left hippocampus | 0.77 | |
| Right hippocampal volume | TBV | |
| Right CA1 | 0.97 | 0.77 |
| Right CA2CA3 | 0.79 | 0.55 |
| Right CA4DG | 0.93 | 0.69 |
| Right SRLM | 0.97 | 0.68 |
| Right subiculum | 0.66 | 0.67 |
| Right hippocampus | 0.78 |
All subfield volumes showed significant correlation between ipsilateral hippocampal volume and TBV (P < 0.001).
Table 6.
Bivariate heritability estimates between TBV and subregions (left and right hippocampal subfield volumes and whole hippocampal volumes) with 95% confidence intervals (sample size N = 465)
| TBV | Subregion | Shared (TBV and subregion) | ||||
|---|---|---|---|---|---|---|
| Region | H 2 (%) | 95% confidence interval | H 2 (%) | 95% confidence interval | H 2 (%) | 95% confidence interval |
| Left CA1 | 95 | 87–97 | 77 | 62–86 | 96 | 84–100 |
| Right CA1 | 96 | 86–97 | 76 | 62–86 | 97 | 87–100 |
| Left CA2CA3 | 95 | 84–97 | 52 | 24–68 | 99 | 80–100 |
| Right CA2CA3 | 95 | 85–97 | 75 | 56–84 | 95 | 82–100 |
| Left CA4DG | 95 | 87–97 | 79 | 63–88 | 95 | 83–100 |
| Right CA4DG | 96 | 86–97 | 85 | 73–91 | 97 | 87–100 |
| Left SRLM | 95 | 86–97 | 70 | 53–81 | 95 | 81–100 |
| Right SRLM | 96 | 86–97 | 79 | 65–87 | 98 | 87–100 |
| Left subiculum | 93 | 82–97 | 62 | 29–82 | 86 | 63–100 |
| Right subiculum | 95 | 85–97 | 88 | 75–92 | 99 | 86–100 |
| Left hippocampus | 95 | 87–97 | 83 | 70–90 | 96 | 85–99 |
| Right hippocampus | 96 | 86–97 | 84 | 73–91 | 98 | 89–100 |
All values were significant at P < 0.001, when ACE model is compared to CE model.
Figure 3.
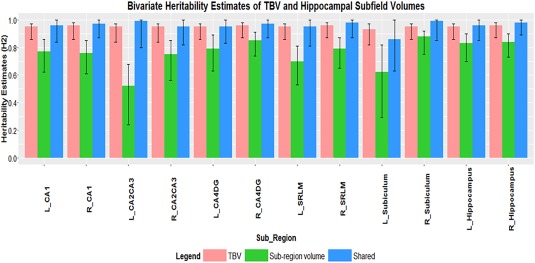
Bivariate heritability scores (H 2) between TBV and left (L) and right (R) hippocampal subfields volumes, with 95% confidence intervals (sample size N = 465). Pink bars indicate heritability estimates of TBV, whereas green bars indicate heritability estimates of subregions (hippocampal subfields and whole hippocampal volumes) and blue bars indicates shared heritability estimates between TBV and subregions.
Table 7.
Bivariate heritability estimates between ipsilateral hippocampal volume and subregions (left and right hippocampal subfield volumes) with 95% confidence intervals (sample size N = 465)
| Ipsilateral hippocampal volume | Subregion | Shared (ipsilateral hippocampal volume and subregion) | ||||
|---|---|---|---|---|---|---|
| Region | H 2 (%) | 95% confidence interval | H 2 (%) | 95% confidence interval | H 2 (%) | 95% confidence interval |
| Left CA1 | 86 | 76–92 | 82 | 69–89 | 86 | 75–92 |
| Right CA1 | 85 | 73–91 | 78 | 62–87 | 83 | 69–90 |
| Left CA2CA3 | 80 | 64–89 | 43 | 9–67 | 77 | 45–98 |
| Right CA2CA3 | 86 | 74–92 | 76 | 53–85 | 84 | 67–93 |
| Left CA4DG | 87 | 76–92 | 83 | 70–90 | 87 | 76–93 |
| Right CA4DG | 87 | 77–92 | 87 | 75–92 | 89 | 79–94 |
| Left SRLM | 85 | 75–91 | 75 | 62–84 | 82 | 70–89 |
| Right SRLM | 87 | 76–92 | 81 | 69–88 | 85 | 74–91 |
| Left subiculum | 81 | 67–89 | 63 | 32–82 | 93 | 67–100 |
| Right subiculum | 84 | 72–91 | 88 | 75–92 | 94 | 80–100 |
All values were significant at P < 0.001, when ACE model is compared to CE model.
Figure 4.
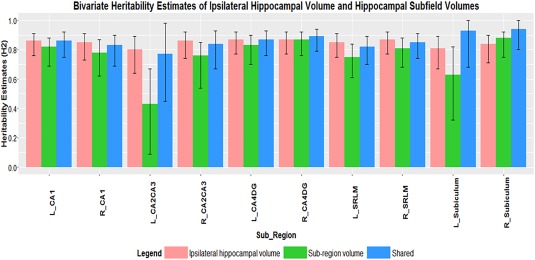
Bivariate heritability scores (H 2) between ipsilateral hippocampal volume and left (L) and right (R) hippocampal subfields volumes, with 95% confidence intervals (sample size N = 465). Pink bars indicate heritability estimates of ipsilateral hippocampal volume, whereas green bars indicate heritability estimates of subregions (hippocampal subfield volumes) and blue bars indicates shared heritability estimates between ipsilateral hippocampal volume and subregions.
DISCUSSION
In this manuscript, we used 465 healthy twin and non‐twin siblings from the HCP dataset to estimate the heritability of hippocampal subfield volumes. Overall in the univariate ACE model, unadjusted hippocampal subfield volumes showed moderate to high heritability estimates ranging from 38% (left CA2/CA3) to 85% (right subiculum). Furthermore, within each subfield the right subfields showed higher heritability estimates compared to left subfields when adjusting for both TBV and ipsilateral hippocampal volume. However, the confidence intervals between left and right corresponding subfields overlap suggesting that the amount of genetic variation is similar. A trend of lower heritability estimates was seen when adjusting for both TBV and ipsilateral hippocampal volume. The TBV and ipsilateral hippocampal volume was also highly heritable. Interestingly, hippocampal subfield heritability was often lower when adjusting for ipsilateral hippocampal volume compared to adjusting for only TBV alone. This trend suggests that once you have accounted for the hippocampal volume, the overall brain volume minimally influences the outputs of hippocampal subfield heritability measures. These results suggest when investigating heritability estimate of small brain structures such as hippocampal subfields it is important to account for the influence of larger structures surrounding the region of interest. In addition, these results also provide indirect evidence of the shared heritability between each of the subfields and TBV and ipsilateral hippocampal volume.
To further investigate how TBV and ipsilateral hippocampal volume influence heritability estimates of hippocampal subfield volumes, we used a bivariate model to examine the shared genetics variance within the two structures. Shared heritability between ipsilateral hippocampal volume and hippocampal subfields was quite high (77%–94%) (Table 7), albeit lower than shared heritability between TBV and hippocampal subfields volume (86%–99%) (Table 6). However, overall the genetic correlation between ipsilateral hippocampal volume and hippocampal subfields volume was higher (0.66–0.98) than TBV and subfield volumes (0.49–0.77) (Table 5), which is similar to the general correlation without accounting for the genetic effects (Table 3). The highest phenotypic and genetic correlation between TBV and hippocampal subfield region was the CA1 region (0.69 and 0.77, respectively). Similarly, Harding et al. [1998] also reported high correlation between CA1 and cerebral volume of 0.68 when exploring the variation in the number of hippocampal neurons with age and brain volume [Harding et al., 1998]. These correlations suggest a possible similarity in genetic etiologies governing the size between CA1 region and total brain which can be seen in the high genetic correlation observed in our study. Overall the genetic correlation trend suggests that there is less overlap in genetic effects (genetic variation) that influence TBV and hippocampal subfield volume than those that influence ipsilateral hippocampal volume and subfields based on genetic correlation estimates. However, in terms of the proportion of the genetic correlation estimates, the shared heritability between subfield volume and TBV demonstrates a greater proportion of shared genetic variation from the genetics correlation than ipsilateral hippocampal volume. This could be due to the fact that the genetic architecture related to TBV is simpler than that of the ipsilateral hippocampal volume and warrants more study. For example, there can be different genetic signaling mechanisms for both TBV and ipsilateral hippocampal development, where the types of genes and gene expression can differ [Cipriani et al., 2016; Roy et al., 2000; Stein et al., 2012]. In addition, the role of genes that influence brain development processes [Miller et al., 2014] is another area of research that can be used to understand the genetic architecture. To tease apart these differences, more studies are required comparing the development of the brain as a whole and the subfields of the hippocampus at pre‐natal stages to adult hood.
The recent study by Whelan et al. [2015] also analyzed heritability of hippocampal subfields using the QTIM dataset (T1‐weighted MRI from 132 MZ twins pairs and 232 DZ twin pairs [Whelan et al., 2015]). The differences between our study and the Whelan study relate to the quality of the dataset used and the study design. A significant advantage in the use of the HCP dataset is that it not only includes twins but also their non‐twin siblings. Families within our data consisted of twin pairs, non‐twin siblings and twin pairs with their non‐twin sibling. Posthuma and Boomsma [2000] have shown that within a family, adding one or two non‐twin siblings along with their twin siblings is an advantage in heritability analysis by reducing the sample size needed and increase the statistical power to detect heritability [Posthuma and Boomsma, 2000]. Also the HCP dataset has high‐resolution MRI T1‐weighted scans with voxel dimensions of 0.7 × 0.7 × 0.7 mm, (whereas QTIM data has a voxel dimension of 0.94 × 0.98 × 0.98mm). Implicitly, the specific subfields can actually be resolved using HCP data whereas this has yet to be proven to be the case in more standard T1‐weighted acquisition [Pipitone et al., 2014]. Furthermore, in our study the heritability estimates are presented bilaterally and we explore the effects of ipsilateral hippocampal volume and TBV on subfield volume heritability, whereas only the overall heritability of each subfield volumes was described by Whelan et al. [2015]. This is a critical distinction as we clearly show a significant impact of ipsilateral hippocampal volume and a moderate impact of TBV on subfield heritability. We note here that our findings report similar results for the heritability of the hippocampus, where Whelan et al. [2015] reported an estimate of 88% heritability in total hippocampal volume and our study estimates slightly lower heritability of 80% and 81% for left and right hippocampal volume, respectively. Similarly across subfields the heritability estimates were lower in our study compared to Whelan et al. [2015]. Heritability estimates for unadjusted left (77%) and right (80%) CA4/DG were similar to estimates reported by Whelan et al., for the CA4 region (79%) and the granule cells of the DG region (82%). Our article also extends the analysis through the use of the bivariate model. This approach has previously only been used to examine TBV with total hippocampal volumes [DeStefano et al., 2009]. Lastly, the segmentation process differs between both studies. We used MAGeT Brain, the Whelan study used FreeSurfer (which was previously validated against our own Winterburn protocol [Iglesias et al., 2015]), which results in differences between the hippocampal subfield definitions and the methodology used to define these subfields. In our study, we define five subfields which included the CA1, CA2/CA3, CA4/DG, SRLM, and subiculum [Winterburn et al., 2013]. Whereas, subfield delineation in the Whelan et al. study did not include the CA2 region in their heritability estimates. However, the Whelan paper examined other regions which we did not, such as the fimbra (white matter tracts) and HATA area. The framework provided in our study allows us to examine the role of TBV and overall hippocampal volume to the heritability of individual subfields, which will allow for further investigation with respect to newer subfield definitions that are the product of the ongoing subfield segmentation harmonization effort [Yushkevich et al., 2015a].
The use of twin/non‐twin sibling HCP design and dataset presents limitations due to variation in intrauterine environment and pre/postnatal complications. Unfortunately, to the best of our knowledge, this information is not provided for HCP subjects. As part of the exclusion process, individuals that were premature (see method section) was excluded but other prenatal or postnatal information was not collected which may affect hippocampal volume and in turn affect heritability estimates. For example, the CA1 region is hypoxia‐sensitive and prenatal hypoxia may be related to greater neuronal loss in the CA1 region compared to other regions [Kuchna, 1994].
The HCP scans used in this study are of high‐resolution and ‐contrast with 0.7 mm isotropic voxels. Figure 1 and Figure S2 in the Supporting Information section demonstrate that the internal structure of the hippocampus can be distinguished in the sagittal and coronal view. While work from our group has assessed the feasibility of subfield segmentation using standard T1‐weighted images [Amaral et al., in press; Pipitone et al., 2014], as has work from the FreeSurfer group [Iglesias et al., 2015, 2016] (although see refs for criticism with respect to FreeSurfer subfield segmentations [de Flores et al., 2015; Wisse et al., 2014]), it is important to note that there are other options that are often used for subfield level segmentations. Other prevalent techniques use T2‐weighted images that are high‐resolution in the coronal plane and low‐resolution outside of this plane [Goubran et al., 2016; Yushkevich et al., 2015b]. However, it is likely that many of the well‐documented limitations inherent to other subfield segmentation techniques [Yushkevich et al., 2015a], are also relevant here. These limitations include ambiguity in the identification of the CA2/3 definition and the CA1/subiculum border.
A limitation found in MRI‐based neuroanatomical studies is that it cannot account for all cytoarchitectonic features within the subfields of the hippocampus. In our segmentation process, a combination of intensity, contrast, and geometric rules based on known cytoarchitectonic definitions were used to define the hippocampal subfields [Winterburn et al., 2013]; however, there are specific boundaries where there is less certainty across subfield protocols. For example, defining the boundary between the CA1 region and subiculum is difficult and is one of the most variable delineations in hippocampal subfield definitions [Yushkevich et al., 2015a]. This is not only true at the level and resolution offered by MRI data, but even when attempting to identify this boundary at the microscopic‐level using histological data [Adler et al., 2014]. This heterogeneity in the ability to identify the subiculum will logically contribute to possible measurement errors and higher uncertainty in structure‐specific heritability estimates. The ambiguity in this boundary at the level of subiculum/CA1 delineation may have also contributed to the lack of significance in our left CA1 heritability estimates after adjusting for TBV and left hippocampal volume together in the ACE univariate model. However, when we did not adjust for TBV or left hippocampal volume our heritable estimates for the CA1 region are significant (comparing univariate ACE against CE model) for both right and left CA1 volume, 73% and 72%, respectively. Similarly, Whelan et al. [2015] reported a slightly higher and significant heritability estimate of 84% for CA1 volume. The CA2/CA3 and SRLM subfields are also regions that are difficult to define based on the thin and complex structure on T1 weighted images. Winterburn et al. [2013] showed low test‐retest results of the manual segmentation protocol for the CA2/CA3 and the SRLM region. Also Pipitone et al. [2014] showed that the CA2/CA3 and SRLM region are less reliably reproduced segmentation on standard 1 × 1 × 1 mm3 T1‐weighted images out of the five subfields. In addition, Wisse et al. [2014] demonstrated lower accuracy through automated segmentation of hippocampal subfields on 7T images on small subfield such as the CA2/CA3 regions which are undersegmented. In our study, the heritability was higher in the left CA2/CA3 region when adjusting for total hippocampal volume compared to no adjustment which is the opposite trend compared to other subfields regions in the ACE univariate model. These limitations in defining the CA2/CA3 and SRLM region can potentially influence the accuracy of the heritability scores. Therefore, higher resolution images, the use of T2 weighted images, and methodological improvements to segmentation techniques may further help with defining these regions. As such, direct comparison of results between studies need to be made with caution and a detailed understanding of these different protocols to define the subfields.
There are different types of segmentation tools available and used to segment the hippocampus such as FreeSurfer [Fischl et al., 2002] and Automatic Segmentation of Hippocampal Subfields (ASHS [Yushkevich et al., 2015b]). Comparisons have been done between FreeSurfer and MAGeT Brain by Pipitone et al. [2014] demonstrating the robustness of MAGeT brain on whole hippocampal segmentations. A recent comparison of automated segmentation approaches, including MAGeT Brain, ASHS, FreeSurfer, and a Bayesian inference model [Van Leemput et al., 2009] has not been done. Therefore, in future research a comparison study on automatic segmentations protocols will be of great value. Furthermore, it will be interesting to compare the automated segmentation methods on the HCP dataset, along with calculating heritability estimates of hippocampal subfield volumes across each segmentation protocol to evaluate the reliability in the heritability scores.
In conclusion, we have demonstrated the heritability of hippocampal subfield volumes using the HCP data in a twin and non‐twin sibling design using a univariate and bivariate model. The univariate model allowed us to examine the heritability of each subfield itself adjusting for TBV and ipsilateral hippocampal volume, whereas in the bivariate model we were able to examine the shared heritability and genetic correlation between two traits. The univariate model demonstrated the heritability of the subfield volumes was lower but significant compared to the heritability of the left or right hippocampal volume. From the bivariate model, shared heritability between many subfields and the hippocampal volume was high and significant. Identifying subfields that have significant and high heritability estimates such as the right subiculum, right CA2/CA3, and right CA4/DG demonstrates their utility as quantitative phenotypes in neurological and psychiatric illnesses.
The use of both univariate and bivariate models in the future allows for the examination of different aspects of the genetic architecture on the target traits. The univariate model demonstrates the heritability of hippocampal subfield volumes in isolation after removing the influence of TBV and ipsilateral hippocampal volume. However, the bivariate model allows us to capture the influence one trait has on another by examining the shared heritability between the two traits. Our univariate model has shown that volumes of smaller target structures are influenced by larger structures that contain the smaller targeted structure and this was quantified by shared heritability estimates in the bivariate model. Therefore, it is important to look at the heritability of a structure in isolation but also in relation to other neuroanatomical structures to get a full understanding of the genetic architecture and genetic interaction found within and between brain structures. As segmentation and image acquisition techniques improve, sample sizes available will also further improve heritability and genome‐wide association analysis. Our work provides a basis for similar ongoing studies, such as those pursued through the ENIGMA consortium [Stein et al., 2012; Thompson et al., 2014]. Therefore, the data presented in this manuscript further motivate the association between the genetic basis of the structure and function of hippocampal subfields in cases of normal brain function and dysfunction.
Supporting information
Supporting Information Figure 1
Supporting Information Figure 2
ACKNOWLEDGMENTS
Data were provided [in part] by the Human Connectome Project, WU‐Minn Consortium (Principal Investigators: David Van Essen and Kamil Ugurbil; 1U54MH091657). Computations were performed on the gpc supercomputer at the SciNet HPC Consortium [Loken et al., 2010]. JK holds the Joanne Murphy Professorship in Behavioral Science. The authors report no conflict of interest.
Contributor Information
Sejal Patel, Email: Sejal.Patel@camh.ca.
M. Mallar Chakravarty, Email: mallar@cobralab.ca.
REFERENCES
- Adler DH, Pluta J, Kadivar S, Craige C, Gee JC, Avants BB, Yushkevich PA (2014): Histology‐derived volumetric annotation of the human hippocampal subfields in postmortem MRI. Neuroimage 84:505–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altshuler LL, Bartzokis G, Grieder T, Curran J, Mintz J (1998): Amygdala enlargement in bipolar disorder and hippocampal reduction in schizophrenia: An MRI study demonstrating neuroanatomic specificity. Arch Gen Psychiatry 55:663–664. [DOI] [PubMed] [Google Scholar]
- Amaral RS, Park MT, Devenyi GA, Lynn V, Pipitone J, Winterburn J, Chavez S, Schira M, Lobaugh NJ, Voineskos AN, Pruessner JC, Chakravarty MM; Alzheimer's Disease Neuroimaging Initiative (2016): Manual segmentation of the fornix, fimbria, and alveus on high‐resolution 3T MRI: Application via fully‐automated mapping of the human memory circuit white and grey matter in healthy and pathological aging. Neuroimage. [DOI] [PubMed] [Google Scholar]
- Amunts K, Kedo O, Kindler M, Pieperhoff P, Mohlberg H, Shah NJ, Habel U, Schneider F, Zilles K (2005): Cytoarchitectonic mapping of the human amygdala, hippocampal region and entorhinal cortex: Intersubject variability and probability maps. Anat Embryol (Berl) 210:343–352. [DOI] [PubMed] [Google Scholar]
- Avants BB, Epstein CL, Grossman M, Gee JC (2008): Symmetric diffeomorphic image registration with cross‐correlation: Evaluating automated labeling of elderly and neurodegenerative brain. Med Image Anal 12:26–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baare WF, Hulshoff Pol HE, Boomsma DI, Posthuma D, de Geus EJ, Schnack HG, van Haren NE, van Oel CJ, Kahn RS (2001): Quantitative genetic modeling of variation in human brain morphology. Cereb Cortex 11:816–824. [DOI] [PubMed] [Google Scholar]
- Bhagwat N, Pipitone J, Winterburn JL, Guo T, Duerden EG, Voineskos AN, Lepage M, Miller SP, Pruessner JC, Chakravarty MM (2016): Manual‐protocol inspired technique for improving automated MR image segmentation during label fusion. Front Neurosci 10:325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blatter DD, Bigler ED, Gale SD, Johnson SC, Anderson CV, Burnett BM, Parker N, Kurth S, Horn SD (1995): Quantitative volumetric analysis of brain MR: Normative database spanning 5 decades of life. AJNR Am J Neuroradiol 16:241–251. [PMC free article] [PubMed] [Google Scholar]
- Bogerts B, Lieberman JA, Ashtari M, Bilder RM, Degreef G, Lerner G, Johns C, Masiar S (1993): Hippocampus‐amygdala volumes and psychopathology in chronic schizophrenia. Biol Psychiatry 33:236–246. [DOI] [PubMed] [Google Scholar]
- Braak H, Braak E (1991): Neuropathological stageing of Alzheimer‐related changes. Acta Neuropathol 82:239–259. [DOI] [PubMed] [Google Scholar]
- Braskie MN, Toga AW, Thompson PM (2013): Recent advances in imaging Alzheimer's disease. J Alzheimers Dis 33(Suppl 1):S313–S327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bremner JD, Narayan M, Anderson ER, Staib LH, Miller HL, Charney DS (2000): Hippocampal volume reduction in major depression. Am J Psychiatry 157:115–118. [DOI] [PubMed] [Google Scholar]
- Campbell S, Marriott M, Nahmias C, MacQueen GM (2004): Lower hippocampal volume in patients suffering from depression: A meta‐analysis. Am J Psychiatry 161:598–607. [DOI] [PubMed] [Google Scholar]
- Chakravarty MM, Steadman P, van Eede MC, Calcott RD, Gu V, Shaw P, Raznahan A, Collins DL, Lerch JP (2013): Performing label‐fusion‐based segmentation using multiple automatically generated templates. Hum Brain Mapp 34:2635–2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cipriani S, Nardelli J, Verney C, Delezoide AL, Guimiot F, Gressens P, Adle‐Biassette H (2016): Dynamic expression patterns of progenitor and pyramidal neuron layer markers in the developing human hippocampus. Cereb Cortex 26:1255–1271. [DOI] [PubMed] [Google Scholar]
- Collins DL, Pruessner JC (2010): Towards accurate, automatic segmentation of the hippocampus and amygdala from MRI by augmenting ANIMAL with a template library and label fusion. Neuroimage 52:1355–1366. [DOI] [PubMed] [Google Scholar]
- de Flores R, La Joie R, Landeau B, Perrotin A, Mezenge F, de La Sayette V, Eustache F, Desgranges B, Chetelat G (2015): Effects of age and Alzheimer's disease on hippocampal subfields: Comparison between manual and FreeSurfer volumetry. Hum Brain Mapp 36:463–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeStefano AL, Seshadri S, Beiser A, Atwood LD, Massaro JM, Au R, Wolf PA, DeCarli C (2009): Bivariate heritability of total and regional brain volumes: The Framingham Study. Alzheimer Dis Assoc Disord 23:218–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duvernoy HM. (2005) The Human Hippocampus: Functional Anatomy, Vascularization and Serial Sections with MRI. Berlin Heidelberg New York: Springer Science & Business Media. [Google Scholar]
- Eskildsen SF, Coupe P, Fonov V, Manjon JV, Leung KK, Guizard N, Wassef SN, Ostergaard LR, Collins DL (2012): BEaST: Brain extraction based on nonlocal segmentation technique. Neuroimage 59:2362–2373. [DOI] [PubMed] [Google Scholar]
- Fatterpekar GM, Naidich TP, Delman BN, Aguinaldo JG, Gultekin SH, Sherwood CC, Hof PR, Drayer BP, Fayad ZA (2002): Cytoarchitecture of the human cerebral cortex: MR microscopy of excised specimens at 9.4 Tesla. AJNR Am J Neuroradiol 23:1313–1321. [PMC free article] [PubMed] [Google Scholar]
- Fischl B, Salat DH, Busa E, Albert M, Dieterich M, Haselgrove C, van der Kouwe A, Killiany R, Kennedy D, Klaveness S, Montillo A, Makris N, Rosen B, Dale AM (2002): Whole brain segmentation: Automated labeling of neuroanatomical structures in the human brain. Neuron 33:341–355. [DOI] [PubMed] [Google Scholar]
- Goubran M, Bernhardt BC, Cantor‐Rivera D, Lau JC, Blinston C, Hammond RR, de Ribaupierre S, Burneo JG, Mirsattari SM, Steven DA, Parrent AG, Bernasconi A, Bernasconi N, Peters TM, Khan AR (2016): In vivo MRI signatures of hippocampal subfield pathology in intractable epilepsy. Hum Brain Mapp 37:1103–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding AJ, Halliday GM, Kril JJ (1998): Variation in hippocampal neuron number with age and brain volume. Cereb Cortex 8:710–718. [DOI] [PubMed] [Google Scholar]
- Haukvik UK, Westlye LT, Morch‐Johnsen L, Jorgensen KN, Lange EH, Dale AM, Melle I, Andreassen OA, Agartz I (2015): In vivo hippocampal subfield volumes in schizophrenia and bipolar disorder. Biol Psychiatry 77:581–588. [DOI] [PubMed] [Google Scholar]
- Iglesias JE, Augustinack JC, Nguyen K, Player CM, Player A, Wright M, Roy N, Frosch MP, McKee AC, Wald LL, Fischl B, Van Leemput K; Alzheimer's Disease Neuroimaging Initiative (2015): A computational atlas of the hippocampal formation using ex vivo, ultra‐high resolution MRI: Application to adaptive segmentation of in vivo MRI. Neuroimage 115:117–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iglesias JE, Van Leemput K, Augustinack J, Insausti R, Fischl B, Reuter M (2016): Bayesian longitudinal segmentation of hippocampal substructures in brain MRI using subject‐specific atlases. Neuroimage 141:542–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquard A (1983): Heritability: One word, three concepts. Biometrics 39:465–477. [PubMed] [Google Scholar]
- Kretschmann HJ, Schleicher A, Wingert F, Zilles K, Loblich HJ (1979): Human brain growth in the 19th and 20th century. J Neurol Sci 40:169–188. [DOI] [PubMed] [Google Scholar]
- Kuchna I (1994): Quantitative studies of human newborns' hippocampal pyramidal cells after perinatal hypoxia. Folia Neuropathol 32:9–16. [PubMed] [Google Scholar]
- La Joie R, Fouquet M, Mezenge F, Landeau B, Villain N, Mevel K, Pelerin A, Eustache F, Desgranges B, Chetelat G (2010): Differential effect of age on hippocampal subfields assessed using a new high‐resolution 3T MR sequence. Neuroimage 53:506–514. [DOI] [PubMed] [Google Scholar]
- Loken C, Gruner D, Groer L, Peltier R, Bunn N, Craig M, Henriques T, Dempsey J, Yu C‐H, Chen J (2010): SciNet: Lessons learned from building a power‐efficient top‐20 system and data centre. J Phys Conf Ser 2010;256. [Google Scholar]
- Mai J, Paxinos G, Voss T (2008) Atlas of the Human Brain, 3rd ed London, UK: Elsevier/Academic Press. [Google Scholar]
- Miller JA, Ding SL, Sunkin SM, Smith KA, Ng L, Szafer A, Ebbert A, Riley ZL, Royall JJ, Aiona K, Arnold JM, Bennet C, Bertagnolli D, Brouner K, Butler S, Caldejon S, Carey A, Cuhaciyan C, Dalley RA, Dee N, Dolbeare TA, Facer BA, Feng D, Fliss TP, Gee G, Goldy J, Gourley L, Gregor BW, Gu G, Howard RE, Jochim JM, Kuan CL, Lau C, Lee CK, Lee F, Lemon TA, Lesnar P, McMurray B, Mastan N, Mosqueda N, Naluai‐Cecchini T, Ngo NK, Nyhus J, Oldre A, Olson E, Parente J, Parker PD, Parry SE, Stevens A, Pletikos M, Reding M, Roll K, Sandman D, Sarreal M, Shapouri S, Shapovalova NV, Shen EH, Sjoquist N, Slaughterbeck CR, Smith M, Sodt AJ, Williams D, Zöllei L, Fischl B, Gerstein MB, Geschwind DH, Glass IA, Hawrylycz MJ, Hevner RF, Huang H, Jones AR, Knowles JA, Levitt P, Phillips JW, Sestan N, Wohnoutka P, Dang C, Bernard A, Hohmann JG, Lein ES (2014): Transcriptional landscape of the prenatal human brain. Nature 508:199–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouiha A, Duchesne S (2011): Hippocampal atrophy rates in Alzheimer's disease: Automated segmentation variability analysis. Neurosci Lett 495:6–10. [DOI] [PubMed] [Google Scholar]
- Mueller SG, Weiner MW (2009): Selective effect of age, Apo e4, and Alzheimer's disease on hippocampal subfields. Hippocampus 19:558–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller SG, Stables L, Du AT, Schuff N, Truran D, Cashdollar N, Weiner MW (2007): Measurement of hippocampal subfields and age‐related changes with high resolution MRI at 4T. Neurobiol Aging 28:719–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale M, Cardon L. (1992) Methodology for Genetic Studies of Twins and Families. Springer Science & Business Media. [Google Scholar]
- Neale MC, Hunter MD, Pritikin JN, Zahery M, Brick TR, Kirkpatrick RM, Estabrook R, Bates TC, Maes HH, Boker SM (2016): OpenMx 2.0: Extended structural equation and statistical modeling. Psychometrika 81:535–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennington BF, Filipek PA, Lefly D, Chhabildas N, Kennedy DN, Simon JH, Filley CM, Galaburda A, DeFries JC (2000): A twin MRI study of size variations in human brain. J Cogn Neurosci 12:223–232. [DOI] [PubMed] [Google Scholar]
- Pipitone J, Park MT, Winterburn J, Lett TA, Lerch JP, Pruessner JC, Lepage M, Voineskos AN, Chakravarty MM (2014): Multi‐atlas segmentation of the whole hippocampus and subfields using multiple automatically generated templates. Neuroimage 101:494–512. [DOI] [PubMed] [Google Scholar]
- Posthuma D, Boomsma DI (2000): A note on the statistical power in extended twin designs. Behav Genet 30:147–158. [DOI] [PubMed] [Google Scholar]
- R Core Development Team (2013) A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- Roy NS, Wang S, Jiang L, Kang J, Benraiss A, Harrison‐Restelli C, Fraser RA, Couldwell WT, Kawaguchi A, Okano H, Nedergaard M, Goldman SA (2000): In vitro neurogenesis by progenitor cells isolated from the adult human hippocampus. Nat Med 6:271–277. [DOI] [PubMed] [Google Scholar]
- Stein JL, Medland SE, Vasquez AA, Hibar DP, Senstad RE, Winkler AM, Toro R, Appel K, Bartecek R, Bergmann O, Bernard M, Brown AA, Cannon DM, Chakravarty MM, Christoforou A, Domin M, Grimm O, Hollinshead M, Holmes AJ, Homuth G, Hottenga JJ, Langan C, Lopez LM, Hansell NK, Hwang KS, Kim S, Laje G, Lee PH, Liu X, Loth E, Lourdusamy A, Mattingsdal M, Mohnke S, Maniega SM, Nho K, Nugent AC, O'Brien C, Papmeyer M, Pütz B, Ramasamy A, Rasmussen J, Rijpkema M, Risacher SL, Roddey JC, Rose EJ, Ryten M, Shen L, Sprooten E, Strengman E, Teumer A, Trabzuni D, Turner J, van Eijk K, van Erp TG, van Tol MJ, Wittfeld K, Wolf C, Woudstra S, Aleman A, Alhusaini S, Almasy L, Binder EB, Brohawn DG, Cantor RM, Carless MA, Corvin A, Czisch M, Curran JE, Davies G, de Almeida MA, Delanty N, Depondt C, Duggirala R, Dyer TD, Erk S, Fagerness J, Fox PT, Freimer NB, Gill M, Göring HH, Hagler DJ, Hoehn D, Holsboer F, Hoogman M, Hosten N, Jahanshad N, Johnson MP, Kasperaviciute D, Kent JW Jr, Kochunov P, Lancaster JL, Lawrie SM, Liewald DC, Mandl R, Matarin M, Mattheisen M, Meisenzahl E, Melle I, Moses EK, Mühleisen TW, Nauck M, Nöthen MM, Olvera RL, Pandolfo M, Pike GB, Puls R, Reinvang I, Rentería ME, Rietschel M, Roffman JL, Royle NA, Rujescu D, Savitz J, Schnack HG, Schnell K, Seiferth N, Smith C, Steen VM, Valdés Hernández MC, Van den Heuvel M, van der Wee NJ, Van Haren NE, Veltman JA, Völzke H, Walker R, Westlye LT, Whelan CD, Agartz I, Boomsma DI, Cavalleri GL, Dale AM, Djurovic S, Drevets WC, Hagoort P, Hall J, Heinz A, Jack CR Jr, Foroud TM, Le Hellard S, Macciardi F, Montgomery GW, Poline JB, Porteous DJ, Sisodiya SM, Starr JM, Sussmann J, Toga AW, Veltman DJ, Walter H, Weiner MW; Alzheimer's Disease Neuroimaging Initiative; EPIGEN Consortium; IMAGEN Consortium; Saguenay Youth Study Group, Bis JC, Ikram MA, Smith AV, Gudnason V, Tzourio C, Vernooij MW, Launer LJ, DeCarli C, Seshadri S; Cohorts for Heart and Aging Research in Genomic Epidemiology Consortium, Andreassen OA, Apostolova LG, Bastin ME, Blangero J, Brunner HG, Buckner RL, Cichon S, Coppola G, de Zubicaray GI, Deary IJ, Donohoe G, de Geus EJ, Espeseth T, Fernández G, Glahn DC, Grabe HJ, Hardy J, Hulshoff Pol HE, Jenkinson M, Kahn RS, McDonald C, McIntosh AM, McMahon FJ, McMahon KL, Meyer‐Lindenberg A, Morris DW, Müller‐Myhsok B, Nichols TE, Ophoff RA, Paus T, Pausova Z, Penninx BW, Potkin SG, Sämann PG, Saykin AJ, Schumann G, Smoller JW, Wardlaw JM, Weale ME, Martin NG, Franke B, Wright MJ, Thompson PM; Enhancing Neuro Imaging Genetics through Meta‐Analysis Consortium (2012): Identification of common variants associated with human hippocampal and intracranial volumes. Nat Genet 44:552–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan EV, Pfefferbaum A, Swan GE, Carmelli D (2001): Heritability of hippocampal size in elderly twin men: Equivalent influence from genes and environment. Hippocampus 11:754–762. [DOI] [PubMed] [Google Scholar]
- Swaab DF, Hofman MA (1984): Sexual differentiation of the human brain. A historical perspective. Prog Brain Res 61:361–374. [DOI] [PubMed] [Google Scholar]
- Tamminga CA, Stan AD, Wagner AD (2010): The hippocampal formation in schizophrenia. Am J Psychiatry 167:1178–1193. [DOI] [PubMed] [Google Scholar]
- Thompson PM, Stein JL, Medland SE, Hibar DP, Vasquez AA, Renteria ME, Toro R, Jahanshad N, Schumann G, Franke B, Wright MJ, Martin NG, Agartz I, Alda M, Alhusaini S, Almasy L, Almeida J, Alpert K, Andreasen NC, Andreassen OA, Apostolova LG, Appel K, Armstrong NJ, Aribisala B, Bastin ME, Bauer M, Bearden CE, Bergmann O, Binder EB, Blangero J, Bockholt HJ, Bøen E, Bois C, Boomsma DI, Booth T, Bowman IJ, Bralten J, Brouwer RM, Brunner HG, Brohawn DG, Buckner RL, Buitelaar J, Bulayeva K, Bustillo JR, Calhoun VD, Cannon DM, Cantor RM, Carless MA, Caseras X, Cavalleri GL, Chakravarty MM, Chang KD, Ching CR, Christoforou A, Cichon S, Clark VP, Conrod P, Coppola G, Crespo‐Facorro B, Curran JE, Czisch M, Deary IJ, de Geus EJ, den Braber A, Delvecchio G, Depondt C, de Haan L, de Zubicaray GI, Dima D, Dimitrova R, Djurovic S, Dong H, Donohoe G, Duggirala R, Dyer TD, Ehrlich S, Ekman CJ, Elvsåshagen T, Emsell L, Erk S, Espeseth T, Fagerness J, Fears S, Fedko I, Fernández G, Fisher SE, Foroud T, Fox PT, Francks C, Frangou S, Frey EM, Frodl T, Frouin V, Garavan H, Giddaluru S, Glahn DC, Godlewska B, Goldstein RZ, Gollub RL, Grabe HJ, Grimm O, Gruber O, Guadalupe T, Gur RE, Gur RC, Göring HH, Hagenaars S, Hajek T, Hall GB, Hall J, Hardy J, Hartman CA, Hass J, Hatton SN, Haukvik UK, Hegenscheid K, Heinz A, Hickie IB, Ho BC, Hoehn D, Hoekstra PJ, Hollinshead M, Holmes AJ, Homuth G, Hoogman M, Hong LE, Hosten N, Hottenga JJ, Hulshoff Pol HE, Hwang KS, Jack CR Jr, Jenkinson M, Johnston C, Jönsson EG, Kahn RS, Kasperaviciute D, Kelly S, Kim S, Kochunov P, Koenders L, Krämer B, Kwok JB, Lagopoulos J, Laje G, Landen M, Landman BA, Lauriello J, Lawrie SM, Lee PH, Le Hellard S, Lemaître H, Leonardo CD, Li CS, Liberg B, Liewald DC, Liu X, Lopez LM, Loth E, Lourdusamy A, Luciano M, Macciardi F, Machielsen MW, Macqueen GM, Malt UF, Mandl R, Manoach DS, Martinot JL, Matarin M, Mather KA, Mattheisen M, Mattingsdal M, Meyer‐Lindenberg A, McDonald C, McIntosh AM, McMahon FJ, McMahon KL, Meisenzahl E, Melle I, Milaneschi Y, Mohnke S, Montgomery GW, Morris DW, Moses EK, Mueller BA, Muñoz Maniega S, Mühleisen TW, Müller‐Myhsok B, Mwangi B, Nauck M, Nho K, Nichols TE, Nilsson LG, Nugent AC, Nyberg L, Olvera RL, Oosterlaan J, Ophoff RA, Pandolfo M, Papalampropoulou‐Tsiridou M, Papmeyer M, Paus T, Pausova Z, Pearlson GD, Penninx BW, Peterson CP, Pfennig A, Phillips M, Pike GB, Poline JB, Potkin SG, Pütz B, Ramasamy A, Rasmussen J, Rietschel M, Rijpkema M, Risacher SL, Roffman JL, Roiz‐Santiañez R, Romanczuk‐Seiferth N, Rose EJ, Royle NA, Rujescu D, Ryten M, Sachdev PS, Salami A, Satterthwaite TD, Savitz J, Saykin AJ, Scanlon C, Schmaal L, Schnack HG, Schork AJ, Schulz SC, Schür R, Seidman L, Shen L, Shoemaker JM, Simmons A, Sisodiya SM, Smith C, Smoller JW, Soares JC, Sponheim SR, Sprooten E, Starr JM, Steen VM, Strakowski S, Strike L, Sussmann J, Sämann PG, Teumer A, Toga AW, Tordesillas‐Gutierrez D, Trabzuni D, Trost S, Turner J, Van den Heuvel M, van der Wee NJ, van Eijk K, van Erp TG, van Haren NE, van 't Ent D, van Tol MJ, Valdés Hernández MC, Veltman DJ, Versace A, Völzke H, Walker R, Walter H, Wang L, Wardlaw JM, Weale ME, Weiner MW, Wen W, Westlye LT, Whalley HC, Whelan CD, White T, Winkler AM, Wittfeld K, Woldehawariat G, Wolf C, Zilles D, Zwiers MP, Thalamuthu A, Schofield PR, Freimer NB, Lawrence NS, Drevets W; Alzheimer's Disease Neuroimaging Initiative, EPIGEN Consortium, IMAGEN Consortium, Saguenay Youth Study (SYS) Group (2014): The ENIGMA Consortium: Large‐scale collaborative analyses of neuroimaging and genetic data. Brain Imaging Behav 8:153–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treadway MT, Waskom ML, Dillon DG, Holmes AJ, Park MT, Chakravarty MM, Dutra SJ, Polli FE, Iosifescu DV, Fava M, Gabrieli JD, Pizzagalli DA (2015): Illness progression, recent stress, and morphometry of hippocampal subfields and medial prefrontal cortex in major depression. Biol Psychiatry 77:285–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Essen DC, Ugurbil K, Auerbach E, Barch D, Behrens TE, Bucholz R, Chang A, Chen L, Corbetta M, Curtiss SW, Della Penna S, Feinberg D, Glasser MF, Harel N, Heath AC, Larson‐Prior L, Marcus D, Michalareas G, Moeller S, Oostenveld R, Petersen SE, Prior F, Schlaggar BL, Smith SM, Snyder AZ, Xu J, Yacoub E; WU‐Minn HCP Consortium (2012): The Human Connectome Project: A data acquisition perspective. Neuroimage 62:2222–2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Essen DC, Smith SM, Barch DM, Behrens TE, Yacoub E, Ugurbil K (2013): The WU‐Minn Human Connectome Project: An overview. Neuroimage 80:62–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Leemput K, Bakkour A, Benner T, Wiggins G, Wald LL, Augustinack J, Dickerson BC, Golland P, Fischl B (2008): Model‐based segmentation of hippocampal subfields in ultra‐high resolution in vivo MRI. Med Image Comput Comput Assist Interv 11:235–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Leemput K, Bakkour A, Benner T, Wiggins G, Wald LL, Augustinack J, Dickerson BC, Golland P, Fischl B (2009): Automated segmentation of hippocampal subfields from ultra‐high resolution in vivo MRI. Hippocampus 19:549–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voineskos AN, Winterburn JL, Felsky D, Pipitone J, Rajji TK, Mulsant BH, Chakravarty MM (2015): Hippocampal (subfield) volume and shape in relation to cognitive performance across the adult lifespan. Hum Brain Mapp 36:3020–3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Suh JW, Das SR, Pluta JB, Craige C, Yushkevich PA (2013): Multi‐Atlas Segmentation with Joint Label Fusion. IEEE Trans Pattern Anal Mach Intell 35:611–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whelan CD, Hibar DP, van Velzen LS, Zannas AS, Carrillo‐Roa T, McMahon K, Prasad G, Kelly S, Faskowitz J, deZubiracay G, Iglesias JE, van Erp TG, Frodl T, Martin NG, Wright MJ, Jahanshad N, Schmaal L, Sämann PG, Thompson PM; Alzheimer's Disease Neuroimaging Initiative (2015): Heritability and reliability of automatically segmented human hippocampal formation subregions. Neuroimage 128:125–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winterburn JL, Pruessner JC, Chavez S, Schira MM, Lobaugh NJ, Voineskos AN, Chakravarty MM (2013): A novel in vivo atlas of human hippocampal subfields using high‐resolution 3 T magnetic resonance imaging. Neuroimage 74:254–265. [DOI] [PubMed] [Google Scholar]
- Wisse LE, Biessels GJ, Geerlings MI (2014): A Critical Appraisal of the Hippocampal Subfield Segmentation Package in FreeSurfer. Front Aging Neurosci 6:261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Goh A, Chen SH, Qiu A (2013): Evolution of hippocampal shapes across the human lifespan. Hum Brain Mapp 34:3075–3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yushkevich PA, Wang H, Pluta J, Das SR, Craige C, Avants BB, Weiner MW, Mueller S (2010): Nearly automatic segmentation of hippocampal subfields in in vivo focal T2‐weighted MRI. Neuroimage 53:1208–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yushkevich PA, Amaral RS, Augustinack JC, Bender AR, Bernstein JD, Boccardi M, Bocchetta M, Burggren AC, Carr VA, Chakravarty MM, Chételat G, Daugherty AM, Davachi L, Ding SL, Ekstrom A, Geerlings MI, Hassan A, Huang Y, Iglesias JE, La Joie R, Kerchner GA, LaRocque KF, Libby LA, Malykhin N, Mueller SG, Olsen RK, Palombo DJ, Parekh MB, Pluta JB, Preston AR, Pruessner JC, Ranganath C, Raz N, Schlichting ML, Schoemaker D, Singh S, Stark CE, Suthana N, Tompary A, Turowski MM, Van Leemput K, Wagner AD, Wang L, Winterburn JL, Wisse LE, Yassa MA, Zeineh MM; Hippocampal Subfields Group (HSG) (2015a): Quantitative comparison of 21 protocols for labeling hippocampal subfields and parahippocampal subregions in in vivo MRI: Towards a harmonized segmentation protocol. Neuroimage 111:526–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yushkevich PA, Pluta JB, Wang H, Xie L, Ding SL, Gertje EC, Mancuso L, Kliot D, Das SR, Wolk DA (2015b): Automated volumetry and regional thickness analysis of hippocampal subfields and medial temporal cortical structures in mild cognitive impairment. Hum Brain Mapp 36:258–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information Figure 1
Supporting Information Figure 2


