Abstract
This work evaluates the potential in diagnostic application of a new advanced neuroimaging method, which delineates the profile of tissue properties along the corticospinal tract (CST) in amyotrophic lateral sclerosis (ALS), by means of diffusion tensor imaging (DTI). Twenty‐four ALS patients and twenty‐four demographically matched healthy subjects were enrolled in this study. The Automated Fiber Quantification (AFQ), a tool for the automatic reconstruction of white matter tract profiles, based on a deterministic tractography algorithm to automatically identify the CST and quantify its diffusion properties, was used. At a group level, the highest non‐overlapping DTI‐related differences were detected in the cerebral peduncle, posterior limb of the internal capsule, and primary motor cortex. Fractional anisotropy (FA) decrease and mean diffusivity (MD) and radial diffusivity (RD) increases were detected when comparing ALS patients to controls. The machine learning approach used to assess the clinical utility of this DTI tool revealed that, by combining all DTI metrics measured along tract between the cerebral peduncle and the corona radiata, a mean 5‐fold cross validation accuracy of 80% was reached in discriminating ALS from controls. Our study provides a useful new neuroimaging tool to characterize ALS‐related neurodegenerative processes by means of CST profile. We demonstrated that specific microstructural changes in the upper part of the brainstem might be considered as a valid biomarker. With further validations this method has the potential to be considered a promising step toward the diagnostic utility of DTI measures in ALS. Hum Brain Mapp 38:727–739, 2017. © 2016 Wiley Periodicals, Inc.
Keywords: amyotrophic lateral sclerosis, diffusion tensor imaging, corticospinal tract, tract profile, random forest
INTRODUCTION
Amyotrophic lateral sclerosis (ALS) is a fatal neurodegenerative disease with a diagnosis based on the abnormalities in upper and lower motor neurons, which lead to a progressive atrophy and weakness of the limbs and respiratory muscles.
Neuroimaging studies have strongly contributed to our understanding of neurobiological mechanisms underlying ALS, and, in particular, diffusion tensor imaging (DTI) has proven to be the most reliable magnetic resonance imaging (MRI) technique for assessing microstructural pathological abnormalities in ALS patients [Agosta et al., 2010; Chiò et al., 2014; Turner et al., 2012]. A recent meta‐analysis pooling the results of 77 DTI studies demonstrated that the main structural abnormalities characterizing ALS patients were localized in the corticospinal tract (CST) extending to the frontal white matter as well as in a small region in the posterior limb of the internal capsule [Li et al., 2012]. The degeneration of the CST in ALS is consistent with pathologic findings [Hirano, 1991; Smith, 1960] and metabolic MRI studies [Sudharshan et al., 2011; Wang et al., 2006] and its neurodegeneration is mainly expressed by the presence of reduced values of fractional anisotropy (FA) [Ciccarelli et al., 2006, 2009; Cosottini et al., 2005; Ellis et al., 1999; Karlsborg et al., 2004; Sage et al., 2007, 2009; Sarica et al., 2014; Toosy et al., 2003; Van der Graaff et al., 2011]. Other studies have also found increased mean diffusivity (MD) values, in particular, along the internal capsule and the pons [Karlsborg et al., 2004] and the cerebral peduncle [Hong et al., 2004].
Despite the high number of DTI studies on ALS, the development of reliable diagnostic markers at an individual level remains an open question. The vast majority of ALS imaging studies, using either voxel‐based or regions of interest (ROI)‐based methods, have only reported differences between patients and controls at a group level [Bede and Hardiman, 2014; Chiò et al., 2014; Li et al., 2012]. These studies have yielded consistent neuroanatomical patterns of pathological changes in ALS patients but with overlapping MRI‐related values, not readily transferable to the clinical practice. Moreover, these neuroimaging approaches have specific shortcomings that might have increased the inconsistencies of data in the literature [Abe et al., 2010; Smith et al., 2006; Toga, 2015]. In the ROI‐based methods, the user interaction is required to specify the ROI, and the variability of ROI size, shape, number, and location could influence the significance of the group analysis [Kanaan et al., 2006]. Instead, the main drawback of the voxel‐based approach is the subjects’ registration phase in which a common coordinate system is applied and each voxel is compared. This critical phase could lead to alignment errors and make associating differences between groups to a specific tract difficult [Toga, 2015]. To overcome these limitations, the neuroimaging community has developed a new frontier of tract‐oriented methods, which quantify diffusion parameters along each fiber tract, creating a “tract profile” of diffusion parameters [Yeatman et al., 2012]. The tract‐oriented approaches provide profiles of diffusion parameters for each tract, preserving spatial information that are otherwise lost in ROI‐methods, and maintaining anatomical correspondence unlike in the voxel‐based. Thus, tract‐oriented methods provide a more detailed anatomical picture of disease progression [Yeatman et al., 2012].
The aim of this work was to evaluate the potential of a new advanced imaging method to serve as a biomarker in ALS. For this purpose, we applied, for the first time, the tract profile approach [Yeatman et al., 2012], to better define neurodegenerative processes underlying ALS. Moreover, to validate the group‐based evidence extracted from this technique in a clinically applicable diagnostic usage, we used a machine learning technique that, unlike univariate statistical methods, is able to predict the diagnosis with an estimated accuracy. Machine learning algorithms in clinical neuroimaging has been the most important computational development in the last years to satisfy the primary need of clinicians [Orrù et al., 2012]. Although several studies have assessed the diagnostic value of these techniques applied on Parkinson's disease [Cerasa, 2016] and Alzheimer's disease realms [Cuingnet et al., 2011; Salvatore et al., 2016], achieving over 90% accuracy for disease state classification, machine learning has sparsely been employed for the diagnosis of ALS. With the aim of exploring the latter, we adopted the Random Forest classifier, which has been successfully applied for reducing high dimensional dataset in many scientific realms, by providing a feature important measure [Breiman, 2001]. In the field of neuroscience, Random Forest has been used to assess the relative importance of several measures acquired with neuronal ensemble recording methods [Loh and Shih, 1997] and resulted in significantly higher accuracy compared with other classifiers in the prediction of Alzheimer's [Lebedev et al., 2014].
MATERIALS AND METHODS
Subjects and Clinical Parameters
ALS patients were enrolled from the Neurology unit of the University “Magna Graecia” of Catanzaro. An expert neurologist (P.V.), blind to the aim of this study, with 20 years of experience in motor neuron disorders performed a complete clinical examination. All patients with ALS met the revised El Escorial criteria for probable, probable laboratory supported or definite ALS [Brooks et al., 2000]. All ALS patients were right‐handed and riluzole treatment‐naïve. Motor skills were assessed according to the ALS functional rating scaled revised (ALSFRS‐R) [Cedarbaum et al., 1999]. Moreover, to define and quantify the presence of bulbar involvement, we used the score on the three bulbar items of the ALSFRS‐R (speech, sialorrhoea, and swallowing, a score of four per item representing normal function). We considered as exclusion criteria: the presence of: (a) multifocal motor neuropathy and paraneoplastic neuropathy, according to nerve conduction examinations; (b) forms of dementia, according to the structured clinical interview of the DSM‐IV [Diagnostic and Statistical Manual of Mental Disorders, 4th Edition, American Psychiatric Association, 1994]; (c) history of cerebrovascular disease, head trauma, hypertension or diabetes. Only two patients with a co‐morbid diagnosis of frontotemporal dementia according to the Neary Criteria [Neary et al., 1998] were excluded because of the confounding effects of imaging changes associated with this phenotype. Finally, 24 ALS patients were enrolled and underwent neuroimaging exam. All clinical details are reported in Table 1.
Table 1.
Patient's clinical characteristics
| Patients | Sex | Age | Onset‐symptoms |
Disease duration (months) |
ALSFRS‐R | Bulbar score |
|---|---|---|---|---|---|---|
| # 1 | F | 54 | Limb | 10 | 33 | 12 |
| # 2 | M | 69 | Limb | 36 | 18 | 8 |
| # 3 | F | 55 | Limb | 23 | 27 | 9 |
| # 4 | M | 52 | Limb | 15 | 32 | 12 |
| # 5 | F | 58 | Bulbar | 6 | 19 | 4 |
| # 6 | F | 67 | Limb | 48 | 33 | 8 |
| # 7 | M | 43 | Bulbar | 5 | 21 | 5 |
| # 8 | M | 67 | Bulbar | 3 | 38 | 8 |
| # 9 | M | 65 | Limb | 15 | 29 | 9 |
| # 10 | F | 72 | Limb | 1 | 25 | 9 |
| # 11 | M | 43 | Limb | 24 | 33 | 9 |
| # 12 | M | 60 | Limb | 24 | 30 | 9 |
| # 13 | F | 71 | Bulbar | 6 | 21 | 9 |
| # 14 | F | 53 | Bulbar | 9 | 20 | 5 |
| # 15 | M | 62 | Limb | 12 | 18 | 10 |
| # 16 | M | 58 | Limb | 6 | 37 | 12 |
| # 17 | F | 48 | Limb | 24 | 33 | 10 |
| # 18 | F | 59 | Limb | 9 | 29 | 12 |
| # 19 | F | 76 | Limb | 63 | 15 | 7 |
| # 20 | M | 67 | Bulbar | 6 | 24 | 8 |
| # 21 | F | 63 | Limb | 36 | 25 | 9 |
| # 22 | F | 72 | Bulbar | 6 | 34 | 7 |
| # 23 | F | 66 | Limb | 68 | 34 | 9 |
| # 24 | M | 55 | Limb | 5 | 35 | 12 |
| Mean ± SD | 54% Female | 60.62 ± 9.06 | 29% Bulbar | 19.16 ± 18.68 | 27.62 ± 6.81 | 8.83 ± 2.24 |
ALSFRS‐R, amyotrophic lateral sclerosis functional rating scaled revised.
Twenty‐four age‐ and sex‐matched, right‐handed, non‐demented, healthy controls [mean age ± SD 61 ± 8.3 (t‐test P‐level= 0.79); 62.5% female, (χ 2, P‐level= 0.558)] with no previous history of neurological or psychiatric diseases and with normal MRI of the brain were matched for demographical variables with patients. All participants gave written informed consent, which was approved by the Ethical Committee of the University “Magna Graecia” of Catanzaro, according to the Helsinki Declaration.
Data Acquisition
Brain MRI was performed according to our routine protocol [Sarica et al., 2014] by a 3 T scanner with an 8‐channel head coil (Discovery MR‐750, General Electric, Milwaukee, WI). The protocol included: (a) whole‐brain T1‐weighted scan MRI scanning (SPGR; TE/TR = 3.7/9.2 ms, flip angle 12°, voxel‐ size 1 × 1 × 1 mm3); (b) diffusion‐weighted volumes, acquired by using spin‐echo echo‐planar imaging (TE/TR = 87/10,000 ms, bandwidth 250 kHz, matrix size 128 × 128, 80 axial slices, voxel size 2.0 × 2.0 × 2.0 mm3) with 27 equally distributed orientations for the diffusion‐sensitizing gradients at a b‐value of 1,000 s/mm2. Particular care was taken to restrain the subject's movements with cushions and adhesive medical tape. Moreover, before further processing, we visually check the images to exclude all scans with artifacts, largely caused by subject motion.
Data Pre‐Processing
Diffusion images were first converted from DICOM to NifTI format by using the script dcm2nii from the MRIcron1 tool. The FMRIB's v5.0 (Oxford Centre for Functional MRI of the Brain2) Diffusion Toolbox (FDT) was used for correcting eddy current and motion distortion. The non‐diffusion‐weighted images, that is, the first volume image used as reference in DTI data, were skull stripped using the FMRIB's brain extraction tool (BET)3 and used to mask all diffusion‐weighted images [Smith, 2002]. A diffusion‐tensor model was fitted at each voxel using Diffusion Toolkit, generating besides fractional anisotropy (FA), mean diffusivity (MD), axial diffusivity (AD) and radial diffusivity (RD) maps, a raw T2 image with no diffusion weighting (S0). The script dtiMakeDt6FromFSL from the Mr Diffusion toolkit4 was used for aligning the T1 image—the anatomical reference—to the S0 image, obtaining a dt6 MATLAB format file for further analysis, described in the next section.
Automated Fiber Quantification
The Automated Fiber Quantification (AFQ, version 1.2 on MATLAB R2014b) tool [Yeatman et al., 2012] was used for reconstructing the CST and for evaluating diffusion metrics along the tract length. AFQ reconstructs the main white matter tracts, for both hemispheres, and measures FA, MD, AD, and RD along their trajectories. AFQ has been validated against manual tracings in healthy controls [Yeatman et al., 2012] and applied in several clinical realms, such as children born preterm [Yeatman et al., 2012], elderly individuals [Yeatman et al., 2014], autism disorders [Libero et al., 2015] and major depression [Sacchet et al., 2014].
In particular, AFQ uses a three‐step procedure to identify 24 major fiber tracts in an individual's brain. The procedure is based on a combination of the methods described by Hua et al. [2008] and Zhang et al. [2008]: (1) whole‐brain fiber tractography, (2) waypoint ROI‐based fiber tract segmentation, and (3) cleaning and refinement of fiber tracts based on a probabilistic fiber tract atlas. An example of CST reconstruction has been showed in Figure 1. For the first step, the whole‐brain fiber tractography, AFQ uses a deterministic streamline‐tracing algorithm (STT) [Mori et al., 1999] with a fourth‐order Runge–Kutta path integration method with 1‐mm step size. Voxels with FA greater than 0.3 are selected for generating a white matter mask used as a seed. Streamlines are traced bidirectionally along the principal diffusion axis, until the stopping criteria are reached (FA < 0.2 or angle between steps > 30°) [Yeatman et al., 2012]. The output of this first step is a whole‐brain tractography that is used in the next step for segmenting the fiber tracts, using a waypoint ROI‐based method [Wakana et al., 2007]. The pairs of waypoint ROIs define the fibers that pass through stereotypical locations and they are drawn on a group‐average DTI dataset in MNI space, which are then non‐linearly warped to each participant's brain [Friston and Ashburner, 2004]. In the case of the CST, the first ROI defines the entire cerebral peduncle in an axial plane at the level of the decussation of the superior cerebellar peduncle, while the second ROI is placed in the central sulcus, right after the bifurcation to the motor and sensory cortex [Wakana et al., 2007].
Figure 1.
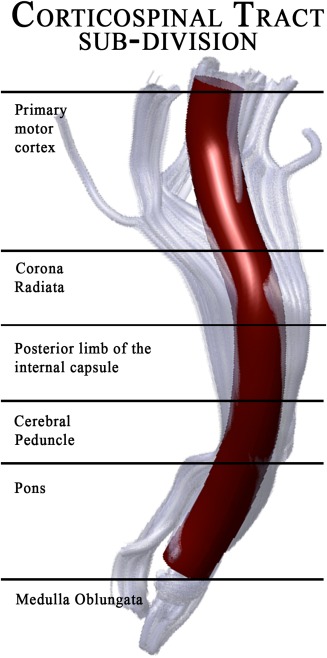
Anatomical sub‐division of the corticospinal tract as reconstructed by AFQ.
In the third step, involving the refinement of segmented fiber tracts, each candidate fiber is compared with a fiber tract probability map created by Hua et al. [2008]. This fiber tract probability map specifies the probability that a voxel is associated with a given fiber tract. Thus, estimated fibers are discarded if their probability of belonging to a fiber is low. Since the tractography process is noise prone, fiber tracts are further cleaned by iteratively discarding outlier fibers that are more than four standard deviations above the mean fiber length or that deviate more than five standard deviations from the core of the fiber tract. The Mahalanobis distance is calculated to estimate the single fiber's contribution to the core, and it corresponds to the probability that a given point belongs to the distribution. Diffusion properties are assessed at each node of each fiber, on an individual subject basis, using spline interpolation of the diffusion properties: FA, MD, AD, and RD.
Statistical and Machine Learning Analysis
To compare the diffusion properties between ALS and controls, mean values and standard deviations (±SD) were plotted [Yeatman et al., 2012] and visually analyzed and then t‐tests were conducted point‐wise along the CST for 100 voxels. A permutation‐based multiple‐comparison correction was applied to determine a statistically significant threshold for P‐values (P < 0.05) [Nichols and Holmes, 2002].
For exploring the relationship between diffusion properties in ALS patients and clinical parameters, a Pearson's correlation analysis was performed point‐wise between voxel diffusion values and ALSFRS‐R/bulbar scores and between voxel diffusion values and disease duration, with the aim of quantifying the degree of this relationship.
Although statistical separability is useful, it gives only a general evaluation of the differences between groups, and it is not able to help predict the diagnosis as well as the machine learning approach [Libero et al., 2015]. Furthermore, machine learning techniques offer the possibility to assess the clinical utility of diffusion parameters. One of the emerging methods to evaluate the prediction power of variables (i.e., how well a variable could discriminate between classes) is Random Forests [Breiman, 2001]. Random Forests is a collection or ensemble of Classification and Regression Trees (CART) [Breiman et al., 1984] trained on datasets of the same size as training set, called bootstraps, created from random resampling of the original training set. Once a tree is constructed, a set of bootstrap datasets, which does not include any particular record from the original dataset (out‐of‐bag examples), is used as test set. The error rate of the classification on test sets is the out‐of‐bag (OOB) estimate of the generalization error. Breiman [1996] showed by empirical evidence that, for the bagged classifiers, the OOB error is accurate as using a test set of the same size as the training set. To classify new input data, as depicted in Figure 2, each individual CART tree votes for one class and the forest predicts the class that has the plurality of votes.
Figure 2.
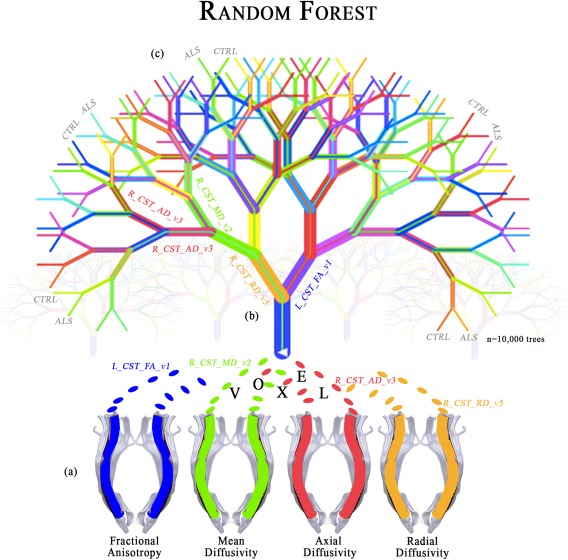
Illustration of the multivariate analysis applied in this study. (a) The dataset composed by all the corticospinal (CST) DTI voxel values of each subject, labeled by diagnosis, is used for training a forest of binary decision trees. Each voxel is labeled by side [left (L) or right (R)], followed by the related diffusion metric (FA, MD, AD, and RD) and its number (from 1 to 100). (b) Each tree is trained on a random subsample of the dataset (bootstrap). At each node a random voxel is chosen as a “splitter variable,” which attempts to separate ALS patients from healthy controls (CTRL). The more often a voxel is chosen as a splitter variable, the higher its “variable importance,” that is, the lower its Gini index. (c) Each tree of the forest gets a vote to predict new subjects. In particular, each new subject traverses each tree until it reaches a terminal node, where the class is predicted as ALS or CTRL.
Random Forest follows specific rules for tree growing, tree combination, self‐testing and post‐processing, it is robust to overfitting and it is considered more stable in the presence of outliers and in very high dimensional parameter spaces than other machine learning algorithms [Caruana and Niculescu‐Mizil, 2006; Menze et al., 2009]. The concept of variable importance is an implicit feature selection performed by Random Forest with a random subspace methodology, and it is assessed by the Gini impurity criterion index [Ceriani and Verme, 2012]. The Gini index is a measure of prediction power of variables in regression or classification, based on the principle of impurity reduction [Strobl et al., 2007]; it is non‐parametric and, therefore, does not rely on data belonging to a particular type of distribution. The Gini index is calculated as follows:
| (1) |
where D is a dataset with n classes and pj is the relative frequency of class j in D. If a dataset D, with size N and n classes, is split into two subsets D1 and D2 with sizes N1 and N2, the Gini index is defined as:
| (2) |
For splitting a node, the attribute value that provides the smallest Ginisplit(D) is chosen. A low Gini (i.e., a greater decrease in Gini) means that a particular predictor variable plays a greater role in partitioning the data into the defined classes.
In this study, the analyses were conducted by using R language (version 3.1.2) and the Random Forest package [Liaw and Wiener, 2002]. The dataset contained the CST voxel values of each subject, for each diffusion metric (FA, MD, AD, and RD). The left and right CST were kept separated in order to investigate any asymmetry and neither age nor sex were added as features owing to the almost perfect match of the three groups. In particular, this dataset has 48 rows (subjects), one diagnosis column—ALS or CTRL (healthy control)—and 800 numerical columns, that is, values of 200 voxels for each metric, 100 for each hemisphere. After a data cleaning—for removing columns with missing values (19)—the dimension of the feature space was 781. A binary classifier was then trained by Random Forest on this set, with 10,000 trees in the forest (ntree) and a recommended value for the number of variables considered at each split of [Diàz‐Uriarte & Alvarez, 2006], where number of features = 781 (after the data cleaning) and mtry = 27. Voxels were then ranked according to their Mean Decrease Gini and all voxels with a mean decrease gini ≥ 0.1 were selected and extracted for creating a new training set with the most important voxels only. This cutoff was selected so to construct a new classifier with the lowest OOB error. The accuracy of the new classifier was evaluated by the 5‐fold cross validation approach, which estimates the performance by randomly dividing for five times the set into five folds, where four folds are used for training and the remaining one for testing.
RESULTS
Univariate Tract‐Profile Analysis
Figure 3 showed comparisons of diffusion parameters between ALS patients and controls for the CST profile. We found significant differences, surviving corrections for multiple comparisons, in the FA, MD, and RD metrics. The highest differences were detected using FA and RD metrics where DTI values in some parts of CST were not overlapping between groups (Fig. 3a,d). Indeed, ALS patients showed significant bilateral FA reduction and RD increase in the cerebral peduncle, posterior limb of internal capsule, corona radiata, and the primary motor cortex with respect to controls. Otherwise, MD and AD measurements showed small regions of differences (Fig. 3b,c). Considering the former, we depicted significant differences only in the right cerebral peduncle, whereas analysis of AD revealed a very restricted—but not statistically significant—pattern of difference in the right primary motor cortex.
Figure 3.
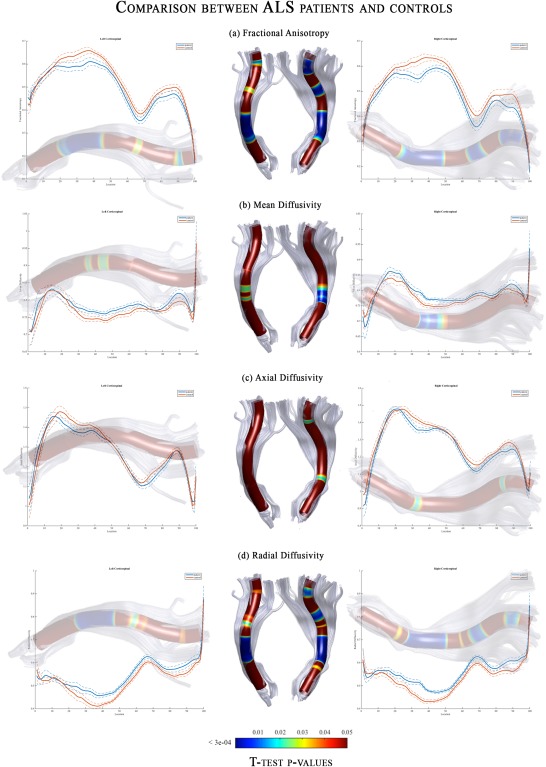
DTI‐related profiles of the CST in ALS patients compared to controls. Plots of mean values are reported voxel by voxels for each group (patients in blue and controls in orange). Dotted lines ±1 SD represent the left and right CST. The x‐axis represents the voxel location (from 1 to 100) and the y‐axis reports the subjects’ group mean values of (a) Fractional Anisotropy, (b) Mean Diffusivity, (c) Axial Diffusivity and (d) Radial Diffusivity. T‐test statistics are plotted using a 3D rendering derived from the AFQ software. The results were thresholded by a conventional criterion for correction for multiple comparisons, which implied an appropriate threshold of significance at p‐level P < 3e‐04. The 3D representation (glass effect) of the tract is added to the plot background so that each location reported on the x‐axis corresponds to the same location in the 3D view. In the middle part of the figure, the same 3D representation is reported for the left and right tract where the P‐values are associated to colors of a heat map (statistically significant differences are displayed in blue). Abbreviations: L, left; R, right; CST, corticospinal; FA, fractional anisotropy; MD, mean diffusivity; AD, axial diffusivity; RD, radial diffusivity.
Correlation Analysis
We performed correlation analyses between DTI metrics and disease severity/duration scales to evaluate the clinical relevance of the detected microstructural changes in the CST (Fig. 4). We found non‐uniform and weak relationships. Indeed, none of the correlation coefficients reached the statistically significant threshold for multiple comparisons, expect for bulbar scores. Indeed, increasing of disease severity correlated with a reduction of microstructural integrity (as measured by AD values) within the right cerebral peduncle (Fig. 4).
Figure 4.
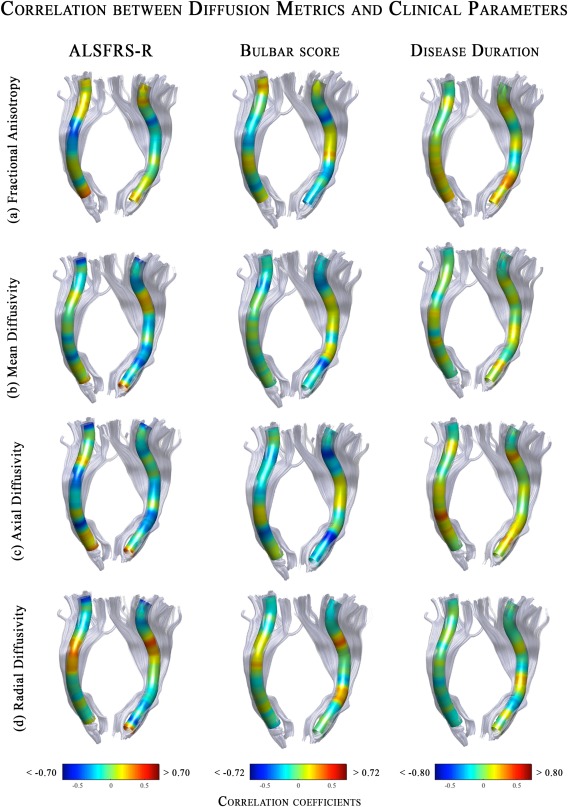
The 3D representations of correlation coefficients between DTI metrics along the CST and ALSFRS‐R scores (left side), bulbar scores (middle) and disease duration (right side) in ALS patients. The color maps report the adjusted thresholds (P < 0.05) for multiple comparisons. Significant negative correlations are colored in blue, whereas positive correlations are in red. Overall, the only significant relationship was detected between bulbar scores and Axial diffusivity values within the cerebral peduncle. Abbreviations: L, left; R, right; CST, corticospinal.
Machine Learning Applied on Tract Profile
Random Forest classifier, trained on all the features, showed an OOB error of 25% and the confusion matrix of the classification of the training set is reported in Table 2.
Table 2.
Performance of Random Forest classifiers, trained on all voxels (without feature selection) and on the most important voxels (with feature selection, mean decrease gini ≥ 0.1)
| RF classifier | OOB error |
Confusion matrix [Training set] |
|||
|---|---|---|---|---|---|
| Without feature selection | 25% | Reference | |||
| Prediction | ALS | CTRL | Class error | ||
| ALS | 19 | 5 | 0.208 | ||
| CTRL | 7 | 17 | 0.291 | ||
| With feature selection | 16.67% | Reference | |||
| Prediction | ALS | CTRL | Class error | ||
| ALS | 22 | 2 | 0.083 | ||
| CTRL | 6 | 18 | 0.25 | ||
Abbreviations: RF, random forest; OOB, out‐of‐bag.
After the feature selection, the classifier trained on the most important voxels (mean decrease gini ≥ 0.1), showed an OOB error of 16.67% (Table 2). The 5‐fold cross validation revealed a mean accuracy of 80%, with a range between 75% and 87.5%.
The analysis of the variable importance, as extracted from the classifier trained on all voxels, showed that voxels influencing the classification of ALS with respect to CTRL were localized mainly in tracts along the cerebral peduncle until the corona radiata in the FA and RD metrics (Fig. 5). The MD‐related measurements of the internal capsule in the right tract presented one significant voxel contributing to ALS classification. Furthermore, no AD voxels resulted to have a significant role in the class prediction, as it could be seen in the plot of the first fifty voxels on the left of Figure 5.
Figure 5.
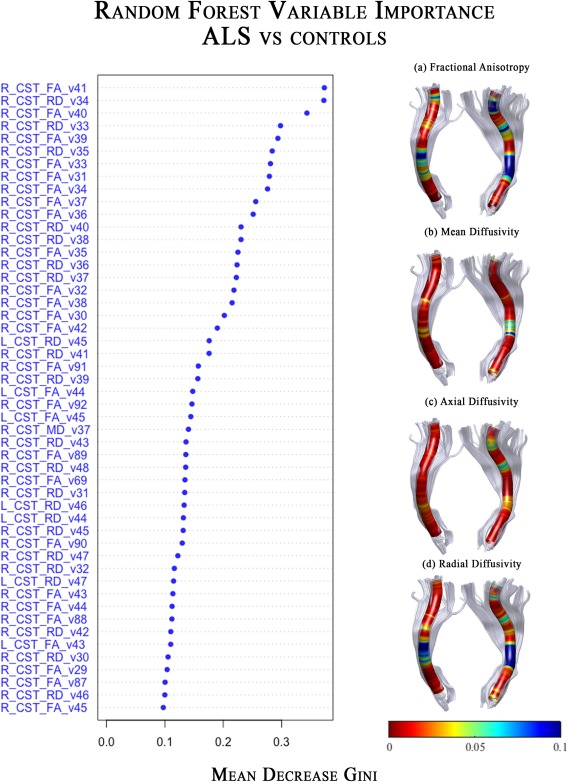
Multivariate Classification results for ALS versus controls. Random Forest classifier, trained on the most important (mean decrease gini ≥ 0.1) voxel DTI metrics of the CST, showed a good performance (mean accuracy: 80%) in discriminating ALS from controls. On the left side, we reported the variable importance plot, which presented the first 50 DTI‐related features (voxels) on the y‐axis, and their relevance for class detection on the x‐axis, ordered top‐to‐bottom as most to least important. On the right side, we plotted the same importance values, but for all voxels of each diffusion metrics, on a representative 3D map. In blue those voxels that have a mean decrease gini ≥ 0.1 on which the classifier was trained. Abbreviations: L, left; R, right; CST, corticospinal; FA, fractional anisotropy; MD, mean diffusivity; AD, axial diffusivity; RD, radial diffusivity. The number of feature corresponds to the location of the voxel along the tract.
DISCUSSION
In this work, we investigated, for the first time, the entire profile of the CST to better define the neurodegenerative processes underlying ALS. We used a double approach moving from univariate to multivariate statistical analyses. At a group level, the most relevant findings were the bilateral FA reduction and RD increase mostly localized in the cerebral peduncle, posterior limb of the internal capsule, and primary motor cortex. Taking into account all DTI measures within the CST, a machine learning algorithm (Random Forest) was able to perform an individual classification of ALS from controls with a mean accuracy of 80% (min 75%, max 87.5%), identifying as biomarkers of this optimal discrimination DTI measures extracted from the cerebral peduncle to the corona radiata.
The CST is recognized as the main pathological site of ALS in the central nervous system. In vivo evaluation of this neural sign has consistently been reported by means of DTI measures [Li et al., 2012]. Indeed, the same bilateral tendency of FA to decrease was found at the internal capsule, cerebral peduncle, pons and pyramids by Toosy et al. [2003]; from the internal capsule to the pyramids by Cosottini et al. [2005]; and in the whole tract by Ciccarelli et al. [2006], Van der Graaff et al. [2011], Wong et al. [2007], and Grapperon et al. [2014]. Our results about the tendency for MD to increase in the right cerebral peduncle matched those observed by Metwalli et al. [2010]. Again, Cirillo et al. [2012] and Grapperon et al. [2014] also consistently reported the lack of significant AD‐related pathological changes in the white matter of ALS patients. Finally, the tract profile approach proposed in this study highlighted the involvement of an additional DTI metric, which generally receives lesser consideration in neuroimaging studies: Radial Diffusivity. Overall, AD and RD are direct measures that represent, respectively, the diffusion in parallel and perpendicular directions to the tract, providing more specific neurobiological information with respect to FA or MD. A decrease in AD has been reported in both rodents and humans with axonal damage associated with axonal swelling and fragmentation [Harsan et al., 2006; Sun et al, 2006], whereas the degree of myelination as well as the changes in the axonal diameters or density are inversely correlated with RD [Alexander et al., 2007]. In our study the absence of differences between groups with AD together with the increase of RD could be explained by the so‐called pseudo‐normalization of AD that happens in acute stages when the axon and myelin debris are removed [Mac Donald et al., 2007], which is when a Wallerian degeneration is present [Pierpaoli et al., 2001]. This explanation is coherent with the clinical characteristics of our ALS cohort (high disease duration and modest motor impairment). Thus, in this scenario, a specific abnormal RD increase could represent a myelin injury with axonal loss, as proposed by Fischer et al. [2004].
Moving from a pathophysiological investigation to an individual characterization, our machine learning algorithm reached an optimal accuracy level for disease state classification. Classification analysis applied on neuroimaging data have been sparsely employed in the ALS realm. Welsh et al. [2013] reached modest results (accuracy of 71.5%) using resting‐state time‐series data for training a Support Vector Machine classifier. Otherwise, a high level of accuracy in classifying ALS patients from controls was only reached by combining DTI with other MRI metrics. Indeed, Filippini et al. [2010] and Foerster et al. [2013] obtained good results by adding whole‐brain voxel‐based volumetric analysis (accuracy 90%) or spectroscopy quantification in the motor cortex (area under the curve, AUC 75%). Taking into account this latter evidence, it should bear in mind that we reached optimal performance only by using DTI information within one specific white matter pathway.
Before drawing conclusions, some important limitations need to be discussed. First, clinically‐related parameters are not strictly connected with microstructural changes along the CST. As already discussed by Bede and Hardiman [2014] and Verstraete et al. [2015], correlation between imaging parameters (mainly DTI) and clinical metrics has far been inconsistent across studies in ALS population [Cosottini et al., 2005; Ellis et al., 1999; Graham et al., 2004; Hong et al., 2004; Metwalli et al., 2010; Sage et al., 2007; Toosy et al., 2003]. As concerns ALSFRS‐R score, one proposed explanation is that this evaluation is heavily dependent on lower motor neuron degeneration, which is not captured by the current imaging techniques. To overcome this limitation, in this study we tried to evaluate the different impact of clinical severity restricted to bulbar impairment (bulbar score from ALSFR‐R scale), reporting a slight relationship between bulbar scores and some DTI measures (i.e., AD) within the cerebral peduncle. Second, longitudinal studies need to be performed to monitor the spread of pathology along the CST in ALS patients [Van der Graaff et al., 2011]. Finally, AFQ provides a framework for combining quantitative imaging data from multiple modalities [Yeatman et al., 2012]. While diffusion imaging is quantitative, diffusion properties are not biologically specific. Future work using quantitative T1 and Proton Density (PD) in combination with DWI‐tractography based fiber tract segmentation will elucidate the precise biological underpinnings of neural injuries in ALS patients.
CONCLUSIONS
We are interested in evaluating the potential of a new advanced imaging method to serve as a biomarker in ALS. The tract profile technique described in this study allows us to both better describe pathophysiological abnormalities characterizing ALS patients and evaluate the robustness of a machine learning algorithm applied on these specific sets of data. We retain that the neuroimaging approach described in this study may open new possibilities for developing multivariate neuroimaging outcomes readily transferable to the clinical practice of ALS. Indeed, there is an urgent need to identify biomarkers, which may be used for helping and improving early diagnosis but that it is hoped will improve and create therapeutic options for ALS patients and ultimately help in the treatment of these patients. Moreover, since previous univariate neuroimaging methods have partially failed in providing definitive and clinically useful biomarkers, machine learning approaches must be employed also for improving individual differential diagnosis of ALS phenotypes with respect to other motor neuron disease disorders (i.e., PMA, PLS), as well as, ALS‐mimicking syndromes (i.e., FTD).
Supporting information
Supporting Information
ACKNOWLEDGMENTS
All authors declare no potential conflicts of interest, including any financial, personal, or other relationships with other people or organizations relevant to the subject of their manuscript. This study has been supported by Merck Serono (Drug Industry).
Footnotes
REFERENCES
- Abe O, Takao H, Gonoi W, Sasaki H, Murakami M, Kabasawa H, Kawaguchi H, Goto M, Yamada H, Yamasue H, Kasai K, Aoki S, Ohtomo K (2010): Voxel‐based analysis of the diffusion tensor. Neuroradiology 52:699–710. [DOI] [PubMed] [Google Scholar]
- Agosta F, Chio A, Cosottini M, De Stefano N, Falini A, Mascalchi M, Rocca MA, Silani V, Tedeschi G, Filippi M (2010): The present and the future of neuroimaging in amyotrophic lateral sclerosis. Am J Neuroradiol 31:1769–1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander AL, Lee JE, Lazar M, Field AS (2007): Diffusion tensor imaging of the brain. Neurotherapeutics 4:316–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bede P, Hardiman O (2014): Lessons of ALS imaging: Pitfalls and future directions—a critical review. NeuroImage: Clin 4:436–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breiman L (1996): Bagging predictors. Mach Learn 24:123–140. [Google Scholar]
- Breiman L (2001): "Random forests”. Mach Learn 45:5–32. [Google Scholar]
- Breiman L, Friedman J, Stone CJ, Olshen RA (1984): Classification and Regression Trees. Boca Raton: Crc Press. [Google Scholar]
- Brooks BR, Miller RG, Swash M, Munsat TL (2000): El Escorial revisited: Revised criteria for the diagnosis of amyotrophic lateral sclerosis. Amyotroph Lateral Scler Other Motor Neuron Disord 1:293–299. [DOI] [PubMed] [Google Scholar]
- Caruana R, Niculescu‐Mizil A (2006): An empirical comparison of supervised learning algorithms. ICML 2006 ‐ Proceedings of the 23rd International Conference on Machine Learning.
- Cedarbaum JM, Stambler N, Malta E, Fuller C, Hilt D, Thurmond B, Nakanishi (1999): The ALSFRS‐R: A revised ALS functional rating scale that incorporates assessments of respiratory function. J Neurol Sci 169:13–21. [DOI] [PubMed] [Google Scholar]
- Cerasa A (2016): Machine learning on Parkinson's disease? Let's translate into clinical practice. J Neurosci Methods 266:161–162. [DOI] [PubMed] [Google Scholar]
- Ceriani L, Verme P (2012): The origins of the gini index: Extracts from variabilità e mutabilità (1912) by corrado gini. J Econ Inequality 10:421–443. [Google Scholar]
- Chiò A, Pagani M, Agosta F, Calvo A, Cistaro A, Filippi M (2014): Neuroimaging in amyotrophic lateral sclerosis: Insights into structural and functional changes. Lancet Neurol 13:1228–1240. [DOI] [PubMed] [Google Scholar]
- Ciccarelli O, Behrens TE, Altmann DR, Orrell RW, Howard RS, Johansen‐Berg H, Miller DH, Matthews PM, Thompson AJ (2006): Probabilistic diffusion tractography: A potential tool to assess the rate of disease progression in amyotrophic lateral sclerosis. Brain 129:1859–1871. [DOI] [PubMed] [Google Scholar]
- Ciccarelli O, Behrens TE, Johansen‐Berg H, Talbot K, Orrell RW, Howard RS, Nunes RG, Miller DH, Matthews PM, Thompson AJ, Smith SM (2009): Investigation of white matter pathology in ALS and PLS using tract‐based spatial statistics. Hum Brain Mapp 30:615–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirillo M, Esposito F, Tedeschi G, Caiazzo G, Sagnelli A, Piccirillo G, Conforti R, Tortora F, Monsurrò MR, Cirillo S, Trojsi F (2012): Widespread microstructural white matter involvement in amyotrophic lateral sclerosis: A whole‐brain DTI study. Am J Neuroradiol 33:1102–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosottini M, Giannelli M, Siciliano G, Lazzarotti G, Michelassi MC, Del Corona A, Bartolozzi C, Murri L (2005): Diffusion‐tensor mr imaging of corticospinal tract in amyotrophic lateral sclerosis and progressive muscular atrophy. Radiology 237:258–264. [DOI] [PubMed] [Google Scholar]
- Cuingnet R, Gerardin E, Tessieras J, Auzias G, Lehéricy S, Habert MO, Chupin M, Benali H, Colliot O, Alzheimer's Disease Neuroimaging Initiative (2011): Automatic classification of patients with Alzheimer's disease from structural MRI: A comparison of ten methods using the ADNI database. Neuroimage 56:766–781. [DOI] [PubMed] [Google Scholar]
- Diàz‐Uriarte R, Alvarez de Andrés S (2006): Gene selection and classification of microarray data using random forest. BMC Bioinform 7:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis CM, Simmons A, Jones DK, Bland J, Dawson JM, Horsfield MA, Williams SC, Leigh PN (1999): Diffusion tensor MRI assesses corticospinal tract damage in ALS. Neurology 53:1051–1058. [DOI] [PubMed] [Google Scholar]
- Filippini N, Douaud G, Mackay CE, Knight S, Talbot K, Turner MR (2010): Corpus callosum involvement is a consistent feature of amyotrophic lateral sclerosis. Neurology 75:1645–1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer LR, Culver DG, Tennant P, Davis AA, Wang M, Castellano‐Sanchez A, Khan J, Polak MA, Glass JD (2004): Amyotrophic lateral scelrosis is a distal axonopathy: Evidence in mice and man. Exp Neurol 185:232–240. [DOI] [PubMed] [Google Scholar]
- Foerster BR, Dwamena BA, Petrou M, Carlos RC, Callaghan BC, Churchill CL, Mohamed MA, Bartels C, Benatar M, Bonzano L, Ciccarelli O, Cosottini M, Ellis CM, Ehrenreich H, Filippini N, Ito M, Kalra S, Melhem ER, Pyra T, Roccatagliata L, Senda J, Sobue G, Turner MR, Feldman EL, Pomper MG (2013): Diagnostic accuracy of diffusion tensor imaging in amyotrophic lateral sclerosis: A systematic review and individual patient data meta‐analysis. Acad Radiol 20:1099–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friston KJ, Ashburner J (2004): Generative and recognition models for neuroanatomy. Neuroimage 23:21–24. [DOI] [PubMed] [Google Scholar]
- Graham JM, Papadakis N, Evans J, Widjaja E, Romanowski CA, Paley MN, Wallis LI, Shaw PJ, Griffiths PD (2004): Diffusion tensor imaging for the assessment of upper motor neuron integrity in ALS. Neurology 63:2111–2119. [DOI] [PubMed] [Google Scholar]
- Grapperon AM, Verschueren A, Duclos Y, Confort‐Gouny S, Soulier E, Loundou AD, Guye M, Cozzone PJ, Pouget J, Ranjeva JP, Attarian S (2014): Association between structural and functional corticospinal involvement in amyotrophic lateral sclerosis assessed by diffusion tensor MRI and triple stimulation technique. Muscle Nerve 49:551–557. [DOI] [PubMed] [Google Scholar]
- Harsan LA, Poulet P, Guignard B, Steibel J, Parizel N, de Sousa PL, Boehm N, Grucker D, Ghandour MS (2006): Brain dysmyelination and recovery assessment by noninvasive in vivo diffusion tensor magnetic resonance imaging. J Neurosci Res 83:392–402. [DOI] [PubMed] [Google Scholar]
- Hirano A (1991): Cytopathology of amyotrophic lateral sclerosis. Adv Neurol 56:91–101. [PubMed] [Google Scholar]
- Hong YH, Lee KW, Sung JJ, Chang KH, Song IC (2004): Diffusion tensor MRI as a diagnostic tool of upper motor neuron involvement in amyotrophic lateral sclerosis. J Neurol Sci 227:73–78. [DOI] [PubMed] [Google Scholar]
- Hua K, Zhang J, Wakana S, Jiang H, Li X, Reich DS, Calabresi PA, Pekar JJ, van Zijl PCM, Mori S (2008): Tract probability maps in stereotaxic spaces: Analyses of white matter anatomy and tract‐specific quantification. Neuroimage 39:336–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanaan RA, Shergill SS, Barker GJ, Catani M, Ng VW, Howard R, McGuire PK, Jones DK (2006): Tract‐specific anisotropy measurements in diffusion tensor imaging. Psychiatry Res: Neuroimaging 146:73–82. [DOI] [PubMed] [Google Scholar]
- Karlsborg M, Rosenbaum S, Wiegell M, Simonsen H, Larsson H, Werdelin L, Gredal O (2004): Corticospinal tract degeneration and possible pathogenesis in ALS evaluated by MR diffusion tensor imaging. Amyotroph Lateral Scler Other Mot Neuron Disord 5:136–140. [DOI] [PubMed] [Google Scholar]
- Lebedev AV, Westman E, Van Westen GJ, Kramberger MG, Lundervold A, Aarsland D, Soininen H, Kloszewska I, Mecocci P, Tsolaki M, Vellas B, Lovestone S, Simmons A (2014): Random Forest ensembles for detection and prediction of Alzheimer's disease with a good between‐cohort robustness. Neuroimage Clin 6:115–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Pan P, Song W, Huang R, Chen K, Shang H (2012): A meta‐analysis of diffusion tensor imaging studies in amyotrophic lateral sclerosis. Neurobiol Aging 33:1833–1838. [DOI] [PubMed] [Google Scholar]
- Liaw A, Wiener M (2002): Classification and regression by Random Forest. R News 2:18–22. [Google Scholar]
- Libero LE, DeRamus TP, Lahti AC, Deshpande G, Kana RK (2015): Multimodal neuroimaging based classification of autism spectrum disorder using anatomical, neurochemical, and white matter correlates. Cortex 66:46–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh WY, Shih YS (1997): Split selection methods for classification trees. Stat Sin 7:815–840. [Google Scholar]
- Mac Donald CL, Dikranian K, Song SK, Bayly PV, Holtzman DM, Brody DL (2007): Detection of traumatic axonal injury with diffusion tensor imaging in a mouse model of traumatic brain injury. Exp Neurol 205:116–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menze BH, Kelm BM, Masuch R, Himmelreich U, Bachert P, Petrich W, Hamprecht FA (2009): A comparison of random forest and its Gini importance with standard chemometric methods for the feature selection and classification of spectral data. BMC Bioinform 10:213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metwalli NS, Benatar M, Nair G, Usher S, Hu X, Carew JD (2010): Utility of axial and radial diffusivity from diffusion tensor MRI as markers of neurodegeneration in amyotrophic lateral sclerosis. Brain Res 1348:156–164. [DOI] [PubMed] [Google Scholar]
- Mori S, Crain BJ, Chacko VP, Van Zijl P (1999): Three‐dimensional tracking of axonal projections in the brain by magnetic resonance imaging. Ann Neurol 45:265–269. [DOI] [PubMed] [Google Scholar]
- Neary D, Snowden JS, Gustafson L, Passant U, Stuss D, Black S, Freedman M, Kertesz A, Robert PH, Albert M, Boone K, Miller BL, Cummings J, Benson DF (1998): Frontotemporal lobar degeneration: A consensus on clinical diagnostic criteria. Neurology 51:1546–1554. [DOI] [PubMed] [Google Scholar]
- Nichols TE, Holmes AP (2002): Nonparametric permutation tests for functional neuroimaging: A primer with examples. Hum Brain Mapp 15:1–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orrù G, Pettersson‐Yeo W, Marquand AF, Sartori G, Mechelli A (2012): Using support vector machine to identify imaging biomarkers of neurological and psychiatric disease: A critical review. Neurosci Biobehav Rev 36:1140–1152. [DOI] [PubMed] [Google Scholar]
- Pierpaoli C, Barnett A, Pajevic S, Chen R, Penix L, Virta A, Basser P (2001): Water diffusion changes in Wallerian degeneration and their dependence on white matter architecture. Neuroimage 13:1174–1185. [DOI] [PubMed] [Google Scholar]
- Sacchet MD, Prasad G, Foland‐Ross LC, Joshi SH, Hamilton JP, Thompson PM, Gotlib IH (2014): Characterizing white matter connectivity in major depressive disorder: Automated fiber quantification and maximum density paths. In Biomedical Imaging (ISBI), 2014 IEEE 11th International Symposium on (pp. 592–595). [DOI] [PMC free article] [PubMed]
- Sage CA, Peeters RR, Gorner A, Robberecht W, Sunaert S (2007): Quantitative diffusion tensor imaging in amyotrophic lateral sclerosis. Neuroimage 34:486–499. [DOI] [PubMed] [Google Scholar]
- Sage CA, Van Hecke W, Peeters R, Sijbers J, Robberecht W, Parizel P, Marchal G, Leemans A, Sunaert S (2009): Quantitative diffusion tensor imaging in amyotrophic lateral sclerosis: Revisited. Hum Brain Mapp 30:3657–3675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvatore C, Battista P, Castiglioni I (2016): Frontiers for the early diagnosis of AD by means of MRI brain imaging and Support Vector Machines. Curr Alzheimer Res 13:509–533. [DOI] [PubMed] [Google Scholar]
- Sarica A, Cerasa A, Vasta R, Perrotta P, Valentino P, Mangone G, Guzzi PH, Rocca F, Nonnis M, Cannataro M, Quattrone A (2014): Tractography in amyotrophic lateral sclerosis using a novel probabilistic tool: A study with tract‐based reconstruction compared to voxel‐based approach. J Neurosci Methods 224:79–87. [DOI] [PubMed] [Google Scholar]
- Smith MC (1960): Nerve fibre degeneration in the brain in amyotrophic lateral sclerosis. J Neurol Neurosurg Psychiatry 23:269–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SM (2002): Fast robust automated brain extraction. Hum Brain Mapp 17:143–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SM, Jenkinson M, Johansen‐Berg H, Rueckert D, Nichols TE, Mackay CE, Watkins KE, Ciccarelli O, Cader MZ, Matthews PM, Behrens TE (2006): Tract‐based spatial statistics: Voxelwise analysis of multi‐subject diffusion data. Neuroimage 31:1487–1505. [DOI] [PubMed] [Google Scholar]
- Strobl C, Boulesteix AL, Augustin T (2007): Unbiased split selection for classification trees based on the gini index. Comput Stat Data Anal 52:483–501. [Google Scholar]
- Sudharshan N, Hanstock C, Hui B, Pyra T, Johnston W, Kalra S (2011): Degeneration of the mid‐cingulate cortex in amyotrophic lateral sclerosis detected in vivo with MR spectroscopy. AJNR Am J Neuroradiol 32:403–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun SW, Liang HF, Trinkaus K, Cross AH, Armstrong RC, Song SK (2006): Noninvasive detection of cuprizone induced axonal damage and demyelination in the mouse corpus callosum. Magn Reson Med 55:302–308. [DOI] [PubMed] [Google Scholar]
- Toga AW (2015): Brain Mapping: An Encyclopedic Reference. New York: Academic Press. [Google Scholar]
- Toosy AT, Werring DJ, Orrell RW, Howard RS, King MD, Barker GJ, Miller DH, Thompson AJ (2003): Diffusion tensor imaging detects corticospinal tract involvement at multiple levels in amyotrophic lateral sclerosis. J Neurol Neurosurg Psychiatry 74:1250–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner MR, Agosta F, Bede P, Govind V, Lulé D, Verstraete E (2012): Neuroimaging in amyotrophic lateral sclerosis. Biomark Med 6:319–337. [DOI] [PubMed] [Google Scholar]
- Van der Graaff MM, Sage CA, Caan MW, Akkerman EM, Lavini C, Majoie CB, Nederveen AJ, Zwinderman AH, Vos F, Brugman F, van den Berg LH, de Rijk MC, van Doorn PA, Van Hecke W, Peeters RR, Robberecht W, Sunaert S, de Visser M (2011): Upper and extra‐motoneuron involvement in early motoneuron disease: A diffusion tensor imaging study. Brain 134:1211–1228. [DOI] [PubMed] [Google Scholar]
- Verstraete E, Turner MR, Grosskreutz J, Filippi M, Benatar M (2015): Mind the gap: The mismatch between clinical and imaging metrics in ALS. Amyotroph Lateral Scler Frontotemporal Degener 16:524–529. [DOI] [PubMed] [Google Scholar]
- Wakana S, Caprihan A, Panzenboeck MM, Fallon JH, Perry M, Gollub RL, Hua K, Zhang J, Jiang H, Dubey P, Blitz A, van Zijl P, Mori S (2007): Reproducibility of quantitative tractography methods applied to cerebral white matter. Neuroimage 36:630–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Poptani H, Woo JH, Desiderio LM, Elman LB, McCluskey LF, Krejza J, Melhem ER (2006): Amyotrophic lateral sclerosis: Diffusion‐tensor and chemical shift MR imaging at 3.0 T. Radiology 239:831–838. [DOI] [PubMed] [Google Scholar]
- Welsh RC, Jelsone‐Swain LM, Foerster BR (2013): The utility of independent component analysis and Machine Learning in the identification of the amyotrophic lateral sclerosis diseased brain. Front Hum Neurosci 10 7:251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong JC, Concha L, Beaulieu C, Johnston W, Allen PS, Kalra S (2007): Spatial profiling of the corticospinal tract in amyotrophic lateral sclerosis using diffusion tensor imaging. J Neuroimaging 17:234–240. [DOI] [PubMed] [Google Scholar]
- Yeatman JD, Dougherty RF, Myall NJ, Wandell BA, Feldman HM (2012): Tract profiles of white matter properties: Automating fiber‐tract quantification. PLoS ONE 7:e49790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeatman JD, Wandell BA, Mezer AA (2014): Lifespan maturation and degeneration of human brain white matter. Nat Commun 5:4932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Olivi A, Hertig SJ, van Zijl P, Mori S (2008): Automated fiber tracking of human brain white matter using diffusion tensor imaging. Neuroimage 42:771–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information


