Abstract
Obesity is related to altered functional connectivity of resting state brain networks that are involved in reward and motivation. It is unknown to what extent these associations reflect genetic confounding and whether the obesity‐related connectivity changes are associated with differences in dietary intake. In this study, resting state functional MRI was performed after an overnight fast in 16 female monozygotic twin pairs (aged 48.8 ± 9.8 years) with a mean BMI discordance of 3.96 ± 2.1 kg/m2 (range 0.7–8.2). Functional connectivity of the salience, basal ganglia, default mode and anterior cingulate–orbitofrontal cortex networks was examined by independent component analysis. Dietary intake was assessed using 3‐day 24‐hour recalls. Results revealed that within the basal ganglia network, heavier versus leaner co‐twins have decreased functional connectivity strength in bilateral putamen (P < 0.05, FWE‐corrected). There were no differences in connectivity in the other networks examined. In the overall group, lower functional connectivity strength in the left putamen was correlated with higher intake of total fat (P < 0.01). It was concluded that, after eliminating genetic effects, overweight is associated with lower resting state functional connectivity in bilateral putamen in the basal ganglia network. The association between lower putamen connectivity and higher fat intake suggests an important role of the putamen in appetitive mechanisms. The cross‐sectional nature of our study cannot discriminate cause and consequence, but the findings are compatible with an effect of lower putamen connectivity on increased BMI and associated higher fat intake. Hum Brain Mapp 38:5069–5081, 2017. © 2017 Wiley Periodicals, Inc.
Keywords: obesity, discordant, monozygotic, resting state networks, functional connectivity, putamen
INTRODUCTION
Obesity is a major public health problem due to its pandemic occurrence and its association with the prevalence of diabetes, cardiovascular disease and musculoskeletal disorders [Haslam and James, 2005]. Causes of obesity are a mixture of genetic and environmental factors and its interactions [Barsh et al., 2000]. Heritability estimates have indicated that 40%–70% of inter‐individual variation in BMI is explained by genetic factors [Stunkard et al., 1986], and recent meta‐analyses of genome‐wide association studies have identified 97 genetic loci associated with obesity [Locke et al., 2015; Speliotes et al., 2010].
The brain has shown to be an important regulator of food intake and it is hypothesized that excessive eating in obese individuals is due to dysfunction of the reward system in the brain [Kenny, 2011; Morton et al., 2006]. We and other investigators using task‐based functional magnetic resonance imaging (fMRI) demonstrated that obese compared with lean individuals have altered brain reward region responses to food stimuli [Killgore et al., 2003; Stice et al., 2008a; Stoeckel et al., 2008; Van Bloemendaal et al., 2014, 2015].
Previous studies also reported obesity‐related alterations in spontaneous brain activity when no task was performed and participants were at rest [Garcia‐Garcia et al., 2013; Hogenkamp et al., 2016; Kullmann et al., 2014; Lips et al., 2014]. This so‐called resting state fMRI allows for the investigation of brain networks that comprise spatially distinct but functionally connected brain areas [Lee et al., 2013]. The degree of functional connectivity provides information on the integrity of these resting state networks [Fox and Raichle, 2007]. These resting state networks are commonly identified using methods such as Independent Component Analysis (ICA) [Beckmann et al., 2005]. Such methods determine connectivity between two or more groups, within the spatially distinct networks. It does not indicate connectivity between location A and B, for which a seed‐based analysis is more appropriate. Advantages of ICA over seed‐based analyses included the data‐driven approach instead of predefined seeds, networks are less sensitive to noise and motion, and the ICA networks converge largely with results from task‐related activation studies.
Most previous resting state studies on obesity focussed on networks implicated in reward and attention, that is, the salience, basal ganglia and default mode network (DMN). The salience network comprises the insula and anterior cingulate cortex (ACC) and integrates sensory, emotional and cognitive information which contributes to decision making [Seeley et al., 2007]. The basal ganglia network comprises subcortical structures such as the caudate nucleus and putamen and is involved in movement, cognition and reward‐related motivation [Robinson et al., 2009]. The DMN comprises the precuneus and posterior cingulate cortex and is active during resting, awake conditions but deactivates during the performance of a task [Raichle et al., 2001].
Comparable to BMI variation, the inter‐individual variation in functional connectivity of resting state networks has shown to be in part under genetic control, as recently demonstrated by a classical twin study [Fu et al., 2015]. More evidence for a genetic contribution to network connectivity comes from studies in individuals with rare genetic mutations, such as Prader–Willi syndrome [Zhang et al., 2013], and individuals carrying risk alleles of the FTO‐gene [Olivo et al., 2016], the obesity‐associated gene with the largest effect on BMI in common obesity.
Together, these studies suggest an overlap between genetic factors that influence BMI and genetic factors that influence functional connectivity of resting state networks. Such an overlap could reflect causal effects of connectivity on body weight, but is also compatible with the situation in which genetic factors exert their effects independently on body weight and on resting state networks, that is, through pleiotropy. In that case, the association between obesity and altered resting state connectivity in previous observational studies may have resulted not from a true causal relationship, but rather, from genetic confounding. Consequently, this would indicate that interventions aimed at improving the integrity of the brain reward system would not contribute to a reduced risk of obesity.
Discordant monozygotic twins provide a unique opportunity to investigate the association between BMI and resting state functional connectivity independent of genetic effects, since these twins are genetically identical [Kaprio and Koskenvuo, 1989; Nordstrom et al., 2016; Van Dongen et al., 2012]. If, within discordant twin pairs, differences in resting state functional connectivity exist, this is supportive for a causal relationship between resting state connectivity and obesity. Accordingly, we studied BMI‐discordant monozygotic twins on resting state functional connectivity of the salience network, basal ganglia network and DMN. In addition, we studied a network comprising the ACC and orbitofrontal cortex (OFC), as this network has shown involvement in gustation, reward and response inhibition [Laird et al., 2011]. A secondary aim was to investigate whether alterations in connectivity within these networks are related to differences in dietary intake, which has not been investigated previously.
MATERIALS AND METHODS
Participants
In the present study we included 16 female monozygotic twin pairs with a previously measured BMI discordance. Subjects were selected from the Netherlands Twins Registry, as described in detail previously [Doornweerd et al., 2016]. In short, out of 2,775 monozygotic twin pairs 54 pairs were selected based on relatively large intra‐pair differences (≥2 kg/m2) in previously measured BMI [Van Dongen et al., 2015]. We selected only females to achieve a study population that is homogeneous with respect to gender, considering earlier reported gender‐related differences in brain responses to food cues, with females showing higher activations than men [Pursey et al., 2014]. Sixteen pairs were willing to participate and fulfilled our inclusion criteria, that is, no history of metabolic, neurological or psychiatric disease including eating disorders and depression (as assessed with the Centre for Epidemiologic Studies Depression Scale [Schroevers et al., 2000]), no drug dependence, no MRI contra‐indications (metal implants or claustrophobia), no pregnancy or recent weight change (>5% self‐reported weight change in the previous 3 months).
The study was approved by the ethics committee of the VU University Medical Centre and was performed in accordance with the Helsinki Declaration. All subjects provided written informed consent.
Clinical Assessments
Both co‐twins of a single pair arrived at the test visit between 8:00 and 10:00 AM after an overnight fast. Weight, height, waist‐to‐hip ratio and body composition were measured in a standardized manner as described in detail previously [Doornweerd et al., 2016]. Handedness was assessed using a validated questionnaire [Van Strien, 1992]. Venous blood samples were drawn for the assessment of fasting glucose and lipid spectrum. Before the scanning session feelings of appetite and hunger were scored on a visual analogue scale (VAS) that ranged from 0 (“not at all”) to 10 (“extremely”). Participants were asked the questions: (1) How hungry are you now?; (2) How full are you now?; (3) How much could you eat right now?; (4) How much is your desire right now to eat something sweet/savoury/fat?
Dietary Intake
During the weeks following the test visit dietary intake was examined using the validated Unites States Department of Agriculture (USDA) five‐step multiple‐pass 24‐hour recalls method by unannounced telephone calls on 2 weekdays and one Sunday, as described previously [Doornweerd et al., 2016; Moshfegh et al., 2008]. To reduce misreporting, we additionally used a food portion size photo book, a table scale and extensive tableware as an aid in portion‐size estimation. Food items were coded and analysed using the Dutch Food Composition Table (NEVO) [RIVM, 2013]. We calculated daily intake of total energy (kcal), percentages of kcals (en%) derived from protein, carbohydrates and total fat, and percentages of total fat (%) derived from saturated, mono‐ and polyunsaturated fatty acids.
Data Acquisition
Magnetic resonance imaging data were obtained using a 3.0 Tesla GE Signa HDxt scanner (General Electric, Milwaukee, WI). For structural imaging, T1 weighted scans were acquired using a 3D fast spoiled gradient‐echo sequence. For the resting state scan an echo‐planar imaging sequence (repetition time/echo time = 1,800/35 ms, flip angle 80°, slice thickness 3 mm, matrix size 64 × 64, voxel size 3 × 3 × 3 mm, 34 slices) was used to acquire 202 images. During the resting state scan participants were instructed to keep their eyes closed and not fall asleep.
Data Analysis
Clinical data
Clinical data were analysed using IBM SPSS Statistics (version 20, IBM Corp., 2011, Armonk, NY). Results are expressed as mean ± SD, unless otherwise stated. Differences between the leaner and heavier co‐twins were tested with paired t‐tests for continuous variables [Altman, 1991], McNemar tests for dichotomous variables and Wilcoxon signed‐ranks tests for ordinal data.
Imaging data
Imaging data were preprocessed using SPM8 software (Wellcome Trust Centre for Neuroimaging, London) run within Matlab R2012a (Mathworks, Inc.). A first inspection of the data revealed that images of one participant had artefacts due to a metal implant in the spinal cord. The data of this participant (and her twin sister in case of paired analyses) was excluded from further imaging analyses. Of the remaining subjects, the origin of each imaging volume was aligned to the anterior commissure. The first two images of functional time series were discarded for steady‐state magnetization. Functional images of each subject were then slice time corrected and realigned to the mean image to correct for head motion. No subject had more than the maximum allowed displacement of 3 mm in translation and 2.5° in rotation. Following this step, all data were co‐registered with the structural scan and segmented to be spatially normalized to the standard Montreal Neurological institute (MNI) template. Finally, images were smoothed using an 8 mm full‐width at half maximum Gaussian kernel.
We used the Group ICA of fMRI Toolbox (GIFT) (http://icatb.sourceforge.net) [Calhoun et al., 2001] on preprocessed images to identify 25 spatially independent resting state components. The minimum description length criterion [Li et al., 2007], implemented in GIFT, indicated that the optimal number of components in our data set was 25. Data from all subjects were concatenated and the aggregated data set reduced using principal component analysis. Independent group components were estimated using the Infomax algorithm [Bell and Sejnowski, 1995]. Consistency of the derived networks were analysed using the ICASSO software, implemented in GIFT.
Identification of the salience network, basal ganglia network, DMN and ACC‐OFC network was performed in two steps. First, networks were visually identified through comparison with previous literature based on their spatial configuration [Beckmann et al., 2005; Damoiseaux et al., 2006; Laird et al., 2011]. Next, the 25 independent components were spatially correlated with masks of the intrinsic connectivity networks (ICN) previously described by Laird et al. (http://www.brainmap.org/icns) [Laird et al., 2011]. Components with the highest correlation with Laird's ICN 4, ICN 3, ICN 13 or ICN 2 were identified as the salience network, basal ganglia network, DMN and ACC‐OFC network, respectively.
Since ICA identifies aggregated component spatial maps of the whole group, it is not capable of capturing connectivity strength at a subject‐level. Therefore, we used GICA‐based back‐reconstruction, as implemented in the GIFT software, to regress back the time‐course and spatial distribution of our networks of interest to the subjects' own fMRI scan. With back‐reconstructing, every voxel of each spatial component is given a value that quantifies the relationship between that voxel and the time‐course of the component. Finally, each subject component image and time course was converted to Z‐values, to obtain voxel values that are comparable across subjects.
For group comparisons Z‐value images for each component were entered into a second level analysis model in SPM8. One sample t‐tests, with a threshold of P < 0.05 family‐wise error (FWE) whole brain corrected, were performed on each network to visualize the network and to create a mask of each network containing all brain regions that contributed to the network, which served as a region of interest in further analyses. Group differences between leaner and heavier co‐twins were investigated using paired t‐tests [Altman, 1991]. An explicit mask of the network‐specific group map was used to investigate results within brain regions that contributed to the network only. Results were considered statistically significant when P < 0.05 FWE‐corrected at the cluster level.
To correlate resting state functional connectivity with dietary intake, Z values of individual component maps were extracted using MarsBaR (MRC Cognition and Brain Sciences Unit, Cambridge) for each anatomical brain region (WFU Pickatlas) that showed significant group differences between leaner and heavier co‐twins. Linear regression analyses in the total group of twins were performed in Stata13, using family ID as a cluster variable to correct for non‐independence of family members.
RESULTS
Clinical Characteristics
Clinical characteristics of the 16 included twin pairs are presented in Table 1. Twin pairs had a mean age of 48.8 ± 9.8 years, ranging from 37 to 70 years. The selection of discordant twin pairs resulted in expected significant differences between co‐twins in weight, BMI, waist‐hip ratio and body fat percentage. The mean BMI discordance was 3.96 kg/m2 and ranged from 0.7 to 8.2 kg/m2. After excluding the twin pair comprising the participant with imaging artefacts, the mean BMI discordance was 4.2 ± 1.9 kg/m2 (range 1.0–8.2). There were no significant differences in biochemical assessments except for HDL‐cholesterol and total/HDL‐cholesterol ratio, which were less favourable in the heavier versus the leaner co‐twins. VAS‐scores on hunger and appetite showed that the heavier co‐twins had stronger feelings of hunger (P < 0.05) and a stronger desire to eat something sweet (P < 0.05) as compared with the leaner co‐twins prior to the scanning session. Leaner and heavier co‐twins were comparable for self‐reported daily smoking (P = 0.5), handedness (P = 1.0) and menopausal status (P = 0.7). Of the included women, 6 were daily smokers: in 2 pairs both co‐twins smoked and in 2 pairs only the leaner co‐twin smoked. Two women were left handed (1 leaner and 1 heavier co‐twin in different pairs). Thirteen women were premenopausal (defined as having a regular menstrual cycle): 7 leaner and 6 heavier co‐twins in 7 pairs. In premenopausal women we initially aimed to perform all scans during the follicular phase, defined as on day 1–12 counting forward from the start of the menstruation. However, since both co‐twins of a pair were scanned on the same day, this was not always feasible. Nevertheless, no significant group differences were present in menstrual cycle phase, with 3 women being scanned during the follicular phase in each group (P = 0.3).
Table 1.
Clinical characteristics
|
Leaner co‐twins (n = 16) |
Heavier co‐twins (n = 16) |
P‐value | |
|---|---|---|---|
| Age (y) | 49.8 ± 9.8 | 49.8 ± 9.8 | – |
| Weight (kg) | 68.9 ± 9.2 | 80.5 ± 11.0 | <0.001 |
| BMI (kg/m2) | 24.4 ± 3.1 | 28.4 ± 3.5 | <0.001 |
| Waist‐to‐hip ratio | 0.80 ± 0.1 | 0.84 ± 0.1 | <0.05 |
| Body fat (%) | 32.0 ± 6.1 | 37.8 ± 6.1 | <0.001 |
| Glucose (mmol/L) | 4.7 ± 0.3 | 4.8 ± 0.3 | 0.5 |
| Total cholesterol (mmol/L) | 5.2 ± 1.1 | 5.3 ± 1.2 | 0.8 |
| HDL cholesterol (mmol/L) | 2.0 ± 0.4 | 1.7 ± 0.4 | 0.05 |
| LDL cholesterol (mmol/L) | 2.9 ± 1.0 | 3.2 ± 1.2 | 0.3 |
| Ratio total/HDL cholesterol | 2.7 ± 0.6 | 3.2 ± 1.0 | 0.01 |
| Triglycerides (mmol/L) | 0.8 ± 0.2 | 0.9 ± 0.3 | 0.1 |
| VAS‐scores | |||
| Hunger | 3.3 ± 2.1 | 5.0 ± 2.2 | <0.05 |
| Fullness | 3.5 ± 2.1 | 3.4 ± 2.0 | 0.8 |
| Prospective food consumption | 5.3 ± 1.4 | 5.6 ± 0.9 | 0.5 |
| Desire for sweet food | 2.8 ± 2.6 | 4.5 ± 3.1 | <0.05 |
| Desire for savoury food | 4.3 ± 2.9 | 5.2 ± 3.0 | 0.08 |
| Desire for high fat food | 0.6 ± 1.4 | 1.6 ± 2.0 | 0.06 |
Mean ± SD, all biochemical assessments are done in the fasted state. HDL, high‐density lipoprotein; LDL, low‐density lipoprotein; VAS, visual analogue scale.
Resting State Networks Identification
Components with the highest correlation with intrinsic connectivity networks previously described [Laird et al., 2011] were identified as the salience network (correlation coefficient, r = 0.41), basal ganglia network (r = 0.58), DMN (r = 0.54) and ACC‐OFC network (r = 0.57). Selected components based on spatial correlation matched our initially selected components based on visual inspection. The identified independent components are visualized with threshold P < 0.05 whole brain FWE corrected and presented in Figure 1. Brain regions that contributed to the networks are listed in detail in Table 2.
Figure 1.
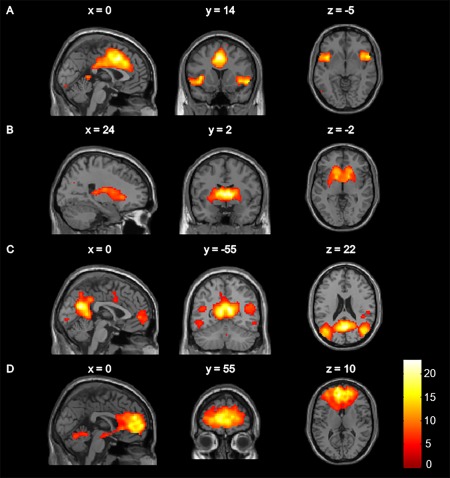
Spatial maps of the resting state networks of interest: (A) salience network, (B) basal ganglia network, (C) default mode network, (D) anterior cingulate cortex–orbitofrontal cortex network. Colour bar represents t statistics. Maps were visualized using a threshold of P < 0.05 whole‐brain family‐wise error corrected.
Table 2.
Brain regions contributing to resting state networks of interest
| MNI coordinates (mm) | ||||||
|---|---|---|---|---|---|---|
| Side | K | T | x | y | z | |
| Salience network | ||||||
| Anterior and middle cingulate cortex | L/R | 2366 | 23.02 | −3 | 11 | 40 |
| Insula | R | 299 | 16.02 | 54 | 14 | −5 |
| Insula | L | 288 | 15.79 | −42 | 11 | −5 |
| Middle frontal gyrus | R | 192 | 13.43 | 33 | 44 | 22 |
| Inferior parietal lobe | R | 468 | 13.09 | 66 | −31 | 31 |
| (Pre)cuneus | R | 163 | 12.19 | 15 | −70 | 34 |
| Cuneus | L | 161 | 10.91 | −12 | −70 | 22 |
| Posterior cingulate cortex | L | 38 | 9.64 | −3 | −46 | 4 |
| Inferior frontal gyrus | L | 48 | 9.50 | −51 | 5 | 28 |
| Inferior frontal gyrus | R | 77 | 9.27 | 51 | 8 | 31 |
| Middle occipital gyrus | L | 37 | 8.69 | −45 | −70 | −14 |
| Lingual gyrus | R | 12 | 7.50 | 18 | −28 | −11 |
| Cerebellum | L | 34 | 7.31 | −36 | −61 | −26 |
| Superior frontal gyrus | L | 26 | 7.11 | −21 | 2 | 61 |
| Inferior parietal lobe | L | 11 | 6.97 | −48 | −31 | 25 |
| Middle occipital gyrus | L | 9 | 6.69 | −45 | −79 | 19 |
| Inferior temporal gyrus | R | 3 | 6.57 | 54 | −55 | −11 |
| Middle frontal gyrus | L | 7 | 6.43 | −36 | −4 | 49 |
| Middle occipital gyrus | R | 1 | 6.02 | 42 | −76 | 22 |
| Superior parietal gyrus | R | 2 | 6.00 | 42 | −46 | 61 |
| Inferior parietal lobe | L | 1 | 5.99 | −51 | −46 | 25 |
| Thalamus | R | 1 | 5.92 | 6 | −13 | 10 |
| Basal ganglia network | ||||||
| Caudate, putamen, thalamus | L/R | 2442 | 26.22 | 9 | 2 | 7 |
| Cerebellum | R | 40 | 9.48 | 0 | −58 | −29 |
| Precuneus | L | 24 | 8.77 | −21 | −76 | 22 |
| Middle cingulate cortex | R | 16 | 7.46 | 3 | −37 | 49 |
| Cerebellum | R | 22 | 7.28 | 12 | −46 | −26 |
| Superior occipital gyrus | R | 4 | 6.82 | 27 | −70 | 28 |
| Superior parietal lobe | L | 6 | 6.76 | −24 | −70 | 43 |
| Insula | L | 5 | 6.44 | −39 | −4 | −11 |
| Inferior parietal lobe | L | 3 | 6.37 | −39 | −58 | 40 |
| Calcarine gyrus | L | 1 | 6.22 | 0 | −79 | 10 |
| Lingual gyrus | R | 1 | 6.07 | 15 | −73 | −11 |
| Middle temporal gyrus | R | 1 | 5.94 | 39 | −64 | 28 |
| Default mode network | ||||||
| Precuneus, posterior cingulate cortex | L/R | 4030 | 27.26 | 9 | −55 | 16 |
| Middle and superior frontal gyrus | R | 832 | 15.01 | 27 | 29 | 43 |
| Middle and superior frontal gyrus | L | 833 | 14.49 | −24 | 29 | 43 |
| Orbitofrontal cortex | L | 254 | 10.92 | −6 | 62 | −5 |
| Middle temporal gyrus | R | 58 | 9.59 | 57 | −10 | −20 |
| Middle occipital and fusiform gyrus | L | 83 | 9.06 | −6 | −91 | −2 |
| Calcarine gyrus | R | 61 | 8.43 | 15 | −94 | −2 |
| Postcentral gyrus | R | 79 | 8.09 | 54 | −25 | 25 |
| Insula | L | 13 | 7.68 | −42 | −10 | −2 |
| Inferior parietal lobe | R | 11 | 7.53 | 45 | −37 | 22 |
| Insula | R | 33 | 7.31 | 45 | −1 | −2 |
| Caudate | R | 9 | 6.92 | 6 | 5 | 10 |
| Cerebellum | R | 6 | 6.70 | 27 | −61 | −32 |
| Inferior temporal gyrus | R | 14 | 6.70 | 54 | −55 | −8 |
| Cerebellum | R | 1 | 6.16 | 3 | −55 | −32 |
| Precentral gyrus | L | 4 | 6.14 | −48 | 2 | 31 |
| Inferior parietal lobe | R | 1 | 5.92 | 36 | −40 | 43 |
| ACC–OFC network | ||||||
| Anterior cingulate cortex, orbitofrontal cortex, medial frontal gyrus | L/R | 4952 | 27.92 | −6 | 47 | 10 |
| Cerebellum | R | 222 | 10.51 | −9 | −67 | −14 |
| Precuneus | L | 33 | 10.38 | −9 | −55 | 19 |
| Pre− and postcentral gyrus | R | 159 | 9.54 | 45 | −31 | 55 |
| Precuneus | R | 3 | 7.03 | 12 | −58 | 58 |
| Insula | R | 16 | 6.95 | 36 | −22 | 13 |
| Rolandic operculum | L | 8 | 6.59 | −39 | −31 | 16 |
| Cerebellum | R | 1 | 6.13 | 12 | −52 | −35 |
| Precuneus | L | 1 | 6.10 | −15 | −46 | 4 |
Brain regions as identified by the Wake Forest University (WFU) Pickatlas. Montreal Neurological Institute (MNI) coordinates and T‐values are presented of the peak voxel within each cluster. Results are P < 0.05 whole‐brain family‐wise error corrected. K, cluster size; ACC, anterior cingulate cortex; OFC, orbitofrontal cortex.
Group Differences
In the basal ganglia network, we observed significantly lower functional connectivity strength in the heavier as compared with the leaner co‐twins in two different independent clusters. The first cluster was found in the left putamen (MNI coordinates peak voxel (mm): x = −27, y = −4, z = 7, cluster size 23 voxels, T‐value 4.69, P = 0.017 FWE‐corrected on cluster level). The second cluster was located in the right putamen (MNI coordinates peal voxel (mm): x = 24, y = −4, z = 13, cluster size 21 voxels, T‐value 5.61, P = 0.022 FWE‐corrected on cluster level) (Fig. 2). In the other networks no statistically significant differences were observed between the groups.
Figure 2.
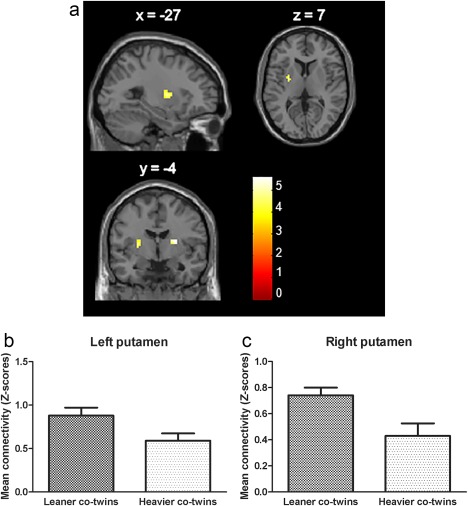
Decreased functional connectivity strength in heavier versus leaner co‐twins in left and right putamen of the basal ganglia network. Left putamen: MNI −27 −4 7, cluster size 23 voxels, T‐value 4.69, P = 0.017 FWE‐corrected on cluster level. Right putamen: MNI 24 −4 13, cluster size 21 voxels, T‐value 5.61, P = 0.022 FWE‐corrected on cluster level. Colour bar represents t statistics. (A) difference in connectivity strength of heavier < leaner co‐twins presented in sagittal, coronal and axial plane, (B) means and SEM of functional connectivity strength (in Z‐scores) in anatomically located left putamen, and (C) right putamen (after extracting data using MarsBaR).
We added an analysis to investigate whether the observed difference in functional connectivity in the bilateral putamen was explained by the difference in individual hunger and appetite scores by performing an additional regression analysis between VAS‐scores of hunger and appetite and mean connectivity strength in all individuals (n = 31). No significant associations were observed.
Relation Between Functional Connectivity and Dietary Intake
As we have previously published [Doornweerd et al., 2016], dietary intake assessments showed that heavier compared with leaner co‐twins had a higher intake of total fat (35.6 ± 6.7 vs. 31.2 ± 4.1 en%, P < 0.05). Intake of total energy (kcal), and relative intake of other macronutrients (en%) and types of fatty acids (% of total fat) did not differ between groups. Linear regression analysis in the total group of twins revealed that lower functional connectivity strength in the left putamen within the basal ganglia network was associated with higher intake of total fat (r = −0.37 P < 0.01), see Figure 3. After adjustment for BMI this association remained significant (r = −0.32 P = 0.02). We performed an additional regression analysis to investigate whether the variability of left putamen connectivity within pairs correlated with variability of intra‐pair differences in fat intake. No such intra‐pair association was observed.
Figure 3.
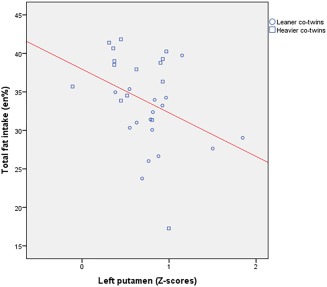
Relationship between functional connectivity strength in the left putamen within the basal ganglia network (in Z‐scores) and intake of total fat (in percentages of total energy intake). [Color figure can be viewed at http://wileyonlinelibrary.com]
DISCUSSION
Using a unique design of monozygotic twin pairs highly discordant for BMI, we investigated whether the association between altered functional connectivity of reward‐related resting state networks and obesity could reflect a causal relationship or simply resulted from genetic confounding. We observed that, in the basal ganglia network, heavier co‐twins had lower functional connectivity in bilateral putamen as compared with leaner co‐twins. The fact that these differences were observed in monozygotic twins with identical genetic backgrounds implies that the relation between altered functional connectivity and obesity is not influenced by genetic confounding [Kaprio and Koskenvuo, 1989; Nordstrom et al., 2016]. Inherent to our twin design is that the observed lower putamen connectivity in heavier females resulted from environmental factors, which have the benefit of being potentially modifiable and, thereby, offering opportunities for future research on obesity prevention and treatment strategies. We, furthermore, observed that lower functional connectivity of the putamen was associated with higher intake of total fat, a finding that was not done before.
Our findings are in line with some but not all previous studies investigating obesity‐related alterations in resting state connectivity in singletons [Garcia‐Garcia et al., 2013; Hogenkamp et al., 2016; Kullmann et al., 2014; Lips et al., 2014; Wijngaarden et al., 2015; Zhang et al., 2015]. In correspondence with our results are two studies that also reported lower functional connectivity strength in obese versus lean individuals in brain regions implicated in food reward and taste processing, in specific the ACC and insula [Kullmann et al., 2014], and the inter‐regional connectivity between the amygdala and the ventromedial prefrontal cortex (PFC) [Wijngaarden et al., 2015]. However, despite this similarity in direction of the observed effects, there are also differences between our study and previous reports. In the particular study that used independent component analysis (ICA) similar to our approach [Kullmann et al., 2014], the observed differences were found in the DMN and temporal lobe network, rather than in the basal ganglia network. In fact, this study observed no BMI‐related differences in the basal ganglia network, similar to a ICA‐based resting state study performed a year later [Garcia‐Garcia et al., 2013]. Interestingly, the latter study did observe differences in connectivity in the putamen, although this was done when the salience network was investigated. Specifically, in this study the obese individuals showed increased connectivity in the putamen relative to lean individuals [Garcia‐Garcia et al., 2013], which contrasts our observations of decreased putamen connectivity in the heavier co‐twins. Two other following studies reported BMI‐related alterations in putamen connectivity in singletons [Hogenkamp et al., 2016; Zhang et al., 2015]. In both studies obese individuals had higher putamen connectivity compared with lean individuals.
Several explanations could be put forward for the discrepancy in findings between our study and previous studies in singletons. First, since monozygotic twins are identical in their genome, the lack of differences in our monozygotic twin design in functional connectivity of, for instance, the DMN and salience network, suggests that positive findings in previous studies in unrelated individuals may be explained by genetic factors. Further, as monozygotic twins are also identical in age, gender and shared environment, the use of our monozygotic twin model allows for the elimination of confounding by all such factors. Thus, whereas previous studies may have been influenced by factors that exert their effect independently on both resting state network connectivity and on body weight, the observations in our current study are independent of such confounding [Kaprio and Koskenvuo, 1989; Nordstrom et al., 2016]. Second, given the different connectivity patterns in the putamen when studied in either the basal ganglia network or the salience network [Garcia‐Garcia et al., 2013], it might be speculated that the degree of connectivity in the putamen dependents on the specific network involved. The salience network typically comprises the insula and ACC [Laird et al., 2011; Seeley et al., 2007], brain regions that showed to be highly activated in response to watching palatable food pictures or cues that predicted the delivery of a palatable food stimulus [Pursey et al., 2014] in task‐based fMRI studies comparing obese and lean individuals. In contrast, the basal ganglia network comprises the dorsal striatum, which has repeatedly been related to decreased brain activations in response to palatable food receipt in previous task‐based fMRI studies [Stice et al., 2008a; Van Bloemendaal et al., 2015]. Although some might say that resting state fMRI and task‐based fMRI evaluate different aspects of brain functioning, numerous studies have demonstrated that brain regions that are co‐activated during a task tend to be positively correlated during rest [Cole et al., 2014; Fair et al., 2007]. Thus, it is tempting to speculate that, when implicated in the salience network, the putamen in obese versus lean individuals shows higher connectivity [Garcia‐Garcia et al., 2013], whereas, when implicated in the basal ganglia network, the putamen in obese individuals shows lower connectivity. However, this explanation is speculative and it would be interesting to investigate this theory in ICA‐based studies further. Furthermore, differences in results between our study and previous studies could be ascribed to differences in participant characteristics and study designs which have shown to influence the regulation of brain reward, for example, gender [Pursey et al., 2014], scheduled time of the scanning session [De Graaf et al., 1993] and fasting duration [Pursey et al., 2014]. Specifically, whereas previous studies investigated males [Zhang et al., 2015] or both males and females [Garcia‐Garcia et al., 2013; Kullmann et al., 2014; Wijngaarden et al., 2015], performed scans throughout the day [Garcia‐Garcia et al., 2013; Zhang et al., 2015] after only short fasts [Garcia‐Garcia et al., 2013; Zhang et al., 2015], in our study we investigated only females, performed all scans in the early morning after an overnight fast. Finally, the discrepant study outcomes may have resulted from the use of different analytical approaches [Lee et al., 2013]. In contrast to some previous studies using seed‐based analyses [Hogenkamp et al., 2016; Lips et al., 2014; Wijngaarden et al., 2015; Zhang et al., 2015], analyses in the current study were done using ICA, which has the benefit of not having a priori assumptions about regions of interest, thereby reducing the influence of selection bias.
The lower functional connectivity in the putamen in heavier versus lean individuals was related to a higher intake of total fat. This observation extends findings of a recent resting state study that found a link between dorsal striatum functional connectivity and cravings for palatable food, as assessed with ratings of pictures of appetizing foods [Contreras‐Rodriguez et al., 2015]. Our finding of higher fat intake is consistent with the proposed hypothesis of an obesity‐related hypo‐functioning reward system [Burger and Stice, 2011; Stice et al., 2008b], which postulates that obese individuals have reduced reward system activation during actual food consumption, which induces compensatory overeating of highly rewarding foods. Evidence for this theory comprises studies that observed reduced striatal response to the receipt of a palatable milkshake in obese versus lean individuals, as observed with task‐based fMRI [Stice et al., 2008a; Van Bloemendaal et al., 2015], and reduced striatal dopamine receptor availability in morbid obesity, as assessed with positron emission tomography [Wang et al., 2001]. Thus, in line with this reasoning, the lower functional connectivity we observed in bilateral putamen might reflect a dysfunction of the reward system, which may result in increased intake of palatable high‐fat foods to compensate for a lack of reward.
Although we have successfully eliminated genetic confounding, the cross sectional nature of our study does not allow us to determine whether lower functional connectivity of the putamen, and its associated increased fat intake, is causal to obesity development or, rather, results as a consequence of it. Support for a causal effect of altered connectivity on obesity comes from a previous resting state study which showed that altered functional connectivity between the dorsal striatum and the somatosensory cortex predicted future weight gain [Contreras‐Rodriguez et al., 2015]. In the context of these findings, the altered functional connectivity of the putamen observed in our study may be explained as a vulnerability factor that might lead to weight gain, possibly through mediating effects of an acquired increased preference for high fat foods. However, our findings are also compatible with the hypothesis that impaired connectivity in the brain results as a consequence of high‐fat diet consumption, for instance through inflammatory processes in neurons and glia, as observed previously in a rodent model [Dorfman and Thaler, 2015]. In line with this finding, an observational MRI study using diffusion tensor imaging reported an inverse correlation between systemic inflammation and integrity of brain structures involved in food reward [Cazettes et al., 2011]. Future studies on resting state, ideally using longitudinal data in genetically informative designs, should elucidate whether altered network connectivity is mainly a cause or consequence of obesity and high fat intake.
A further limitation of our study was a relatively small sample size, although similar and even smaller sample sizes were used in previous studies [Garcia‐Garcia et al., 2013; Hogenkamp et al., 2016; Kullmann et al., 2014; Wijngaarden et al., 2015].The power of our study, however, should be evaluated within the context of the study design, that is, monozygotic twins being highly matched for possible confounding factors such as age, gender and genetic background, but, in the same time, ultimately discordant for BMI, which together enhance the power of this study to a great extent [Van Dongen et al., 2012; Zwijnenburg et al., 2010].
In conclusion, our study extends evidence for altered functional connectivity of brain regions involved in food reward in overweight and obesity. We demonstrated that, when genetic factors are eliminated, overweight is associated with lower functional connectivity of bilateral putamen within the basal ganglia network. Furthermore, the lower connectivity in the left putamen showed a link with subsequent higher intake of total fat, suggesting that the putamen is involved in the regulation of appetite and food preferences. Since monozygotic twins have identical genomes, the observed alterations in functional connectivity of the putamen in females with higher BMI should be ascribed to unique environmental factors that exert their influence on the brain either direct or indirect, for example, through epigenetic mechanisms [Vucetic et al., 2012]. Identification of the environmental factors that are responsible for the observed alterations in brain network connectivity may contribute to developing new strategies in the management of obesity, such as pharmacotherapies or neuromodulation techniques.
CONFLICTS OF INTEREST
None of the authors declared a conflict of interest.
ACKNOWLEDGMENTS
We thank all twin pairs who participated in this study.
REFERENCES
- Altman D (1991): Comparing Groups ‐ Continuous Data. Practical Statistics for Medical Research. London: Chapman and Hall; pp 179–228. [Google Scholar]
- Barsh GS, Farooqi IS, O'Rahilly S (2000): Genetics of body‐weight regulation. Nature 404:644–651. [DOI] [PubMed] [Google Scholar]
- Beckmann CF, DeLuca M, Devlin JT, Smith SM (2005): Investigations into resting‐state connectivity using independent component analysis. Philos Trans R Soc Lond B Biol Sci 360:1001–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell AJ, Sejnowski TJ (1995): An information‐maximization approach to blind separation and blind deconvolution. Neural Comput 7:1129–1159. [DOI] [PubMed] [Google Scholar]
- Burger KS, Stice E (2011): Variability in reward responsivity and obesity: Evidence from brain imaging studies. Curr Drug Abuse Rev 4:182–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calhoun VD, Adali T, Pearlson GD, Pekar JJ (2001): A method for making group inferences from functional MRI data using independent component analysis. Hum Brain Mapp 14:140–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazettes F, Cohen JI, Yau PL, Talbot H, Convit A (2011): Obesity‐mediated inflammation may damage the brain circuit that regulates food intake. Brain Res 1373:101–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole MW, Bassett DS, Power JD, Braver TS, Petersen SE (2014): Intrinsic and task‐evoked network architectures of the human brain. Neuron 83:238–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contreras‐Rodriguez O, Martin‐Perez C, Vilar‐Lopez R, Verdejo‐Garcia A (2015): Ventral and dorsal striatum networks in obesity: Link to food craving and weight gain. Biol Psychiatry 81:789–796. [DOI] [PubMed] [Google Scholar]
- Damoiseaux JS, Rombouts SA, Barkhof F, Scheltens P, Stam CJ, Smith SM, Beckmann CF (2006): Consistent resting‐state networks across healthy subjects. Proc Natl Acad Sci U S A 103:13848–13853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Graaf C, Jas P, van der Kooy K, Leenen R (1993): Circadian rhythms of appetite at different stages of a weight loss programme. Int J Obes Relat Metab Disord 17:521–526. [PubMed] [Google Scholar]
- Doornweerd S, van der Eijk L, Neter JE, van Dongen J, van der Ploeg HP, de Geus EJ (2016): Physical activity and dietary intake in BMI discordant identical twins. Obesity (Silver Spring) 24:1349–1355. [DOI] [PubMed] [Google Scholar]
- Dorfman MD, Thaler JP (2015): Hypothalamic inflammation and gliosis in obesity. Curr Opin Endocrinol Diabetes Obes 22:325–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fair DA, Schlaggar BL, Cohen AL, Miezin FM, Dosenbach NU, Wenger KK, Fox MD, Snyder AZ, Raichle ME, Petersen SE (2007): A method for using blocked and event‐related fMRI data to study “resting state” functional connectivity. Neuroimage 35:396–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox MD, Raichle ME (2007): Spontaneous fluctuations in brain activity observed with functional magnetic resonance imaging. Nat Rev Neurosci 8:700–711. [DOI] [PubMed] [Google Scholar]
- Fu Y, Ma Z, Hamilton C, Liang Z, Hou X, Ma X, Hu X, He Q, Deng W, Wang Y, Zhao L, Meng H, Li T, Zhang N (2015): Genetic influences on resting‐state functional networks: A twin study. Hum Brain Mapp 36:3959–3972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia‐Garcia I, Jurado MA, Garolera M, Segura B, Sala‐Llonch R, Marques‐Iturria I, Pueyo R, Sender‐Palacios MJ, Vernet‐Vernet M, Narberhaus A, Ariza M, Junque C (2013): Alterations of the salience network in obesity: A resting‐state fMRI study. Hum Brain Mapp 34:2786–2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haslam DW, James WP (2005): Obesity. Lancet 366:1197–1209. [DOI] [PubMed] [Google Scholar]
- Hogenkamp PS, Zhou W, Dahlberg LS, Stark J, Larsen AL, Olivo G, Wiemerslage L, Larsson EM, Sundbom M, Benedict C, Schioth HB (2016): Higher resting‐state activity in reward‐related brain circuits in obese versus normal‐weight females independent of food intake. Int J Obes (Lond) 40:1687–1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaprio J, Koskenvuo M (1989): Twins, smoking and mortality: A 12‐year prospective study of smoking‐discordant twin pairs. Soc Sci Med 29:1083–1089. [DOI] [PubMed] [Google Scholar]
- Kenny PJ (2011): Reward mechanisms in obesity: New insights and future directions. Neuron 69:664–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killgore WD, Young AD, Femia LA, Bogorodzki P, Rogowska J, Yurgelun‐Todd DA (2003): Cortical and limbic activation during viewing of high‐ versus low‐calorie foods. Neuroimage 19:1381–1394. [DOI] [PubMed] [Google Scholar]
- Kullmann S, Heni M, Linder K, Zipfel S, Haring HU, Veit R, Fritsche A, Preissl H (2014): Resting‐state functional connectivity of the human hypothalamus. Hum Brain Mapp 35:6088–6096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laird AR, Fox PM, Eickhoff SB, Turner JA, Ray KL, McKay DR, Glahn DC, Beckmann CF, Smith SM, Fox PT (2011): Behavioral interpretations of intrinsic connectivity networks. J Cogn Neurosci 23:4022–4037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MH, Smyser CD, Shimony JS (2013): Resting‐state fMRI: A review of methods and clinical applications. AJNR Am J Neuroradiol 34:1866–1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YO, Adali T, Calhoun VD (2007): Estimating the number of independent components for functional magnetic resonance imaging data. Hum Brain Mapp 28:1251–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lips MA, Wijngaarden MA, van der Grond J, van Buchem MA, de Groot GH, Rombouts SA, Pijl H, Veer IM (2014): Resting‐state functional connectivity of brain regions involved in cognitive control, motivation, and reward is enhanced in obese females. Am J Clin Nutr 100:524–531. [DOI] [PubMed] [Google Scholar]
- Locke AE, Kahali B, Berndt SI, Justice AE, Pers TH, Day FR, Powell C, Vedantam S, Buchkovich ML, Yang J, Croteau‐Chonka DC, Esko T, Fall T, Ferreira T, Gustafsson S, Kutalik Z, Luan J, Magi R, Randall JC, Winkler TW, Wood AR, Workalemahu T, Faul JD, Smith JA, Hua ZJ, Zhao W, Chen J, Fehrmann R, Hedman AK, Karjalainen J, Schmidt EM, Absher D, Amin N, Anderson D, Beekman M, Bolton JL, Bragg‐Gresham JL, Buyske S, Demirkan A, Deng G, Ehret GB, Feenstra B, Feitosa MF, Fischer K, Goel A, Gong J, Jackson AU, Kanoni S, Kleber ME, Kristiansson K, Lim U, Lotay V, Mangino M, Mateo L, I, Medina‐Gomez C, Medland SE, Nalls MA, Palmer CD, Pasko D, Pechlivanis S, Peters MJ, Prokopenko I, Shungin D, Stancakova A, Strawbridge RJ, Ju SY, Tanaka T, Teumer A, Trompet S, van der Laan SW, van SJ, Van Vliet‐Ostaptchouk JV, Wang Z, Yengo L, Zhang W, Isaacs A, Albrecht E, Arnlov J, Arscott GM, Attwood AP, Bandinelli S, Barrett A, Bas IN, Bellis C, Bennett AJ, Berne C, Blagieva R, Bluher M, Bohringer S, Bonnycastle LL, Bottcher Y, Boyd HA, Bruinenberg M, Caspersen IH, Ida Chen YD, Clarke R, Daw EW, de Craen AJ, Delgado G, Dimitriou M, Doney AS, Eklund N, Estrada K, Eury E, Folkersen L, Fraser RM, Garcia ME, Geller F, Giedraitis V, Gigante B, Go AS, Golay A, Goodall AH, Gordon SD, Gorski M, Grabe HJ, Grallert H, Grammer TB, Grassler J, Gronberg H, Groves CJ, Gusto G, Haessler J, Hall P, Haller T, Hallmans G, Hartman CA, Hassinen M, Hayward C, Heard‐Costa NL, Helmer Q, Hengstenberg C, Holmen O, Hottenga JJ, James AL, Jeff JM, Johansson A, Jolley J, Juliusdottir T, Kinnunen L, Koenig W, Koskenvuo M, Kratzer W, Laitinen J, Lamina C, Leander K, Lee NR, Lichtner P, Lind L, Lindstrom J, Sin LK, Lobbens S, Lorbeer R, Lu Y, Mach F, Magnusson PK, Mahajan A, McArdle WL, McLachlan S, Menni C, Merger S, Mihailov E, Milani L, Moayyeri A, Monda KL, Morken MA, Mulas A, Muller G, Muller‐Nurasyid M, Musk AW, Nagaraja R, Nothen MM, Nolte IM, Pilz S, Rayner NW, Renstrom F, Rettig R, Ried JS, Ripke S, Robertson NR, Rose LM, Sanna S, Scharnagl H, Scholtens S, Schumacher FR, Scott WR, Seufferlein T, Shi J, Vernon SA, Smolonska J, Stanton AV, Steinthorsdottir V, Stirrups K, Stringham HM, Sundstrom J, Swertz MA, Swift AJ, Syvanen AC, Tan ST, Tayo BO, Thorand B, Thorleifsson G, Tyrer JP, Uh HW, Vandenput L, Verhulst FC, Vermeulen SH, Verweij N, Vonk JM, Waite LL, Warren HR, Waterworth D, Weedon MN, Wilkens LR, Willenborg C, Wilsgaard T, Wojczynski MK, Wong A, Wright AF, Zhang Q, Brennan EP, Choi M, Dastani Z, Drong AW, Eriksson P, Franco‐Cereceda A, Gadin JR, Gharavi AG, Goddard ME, Handsaker RE, Huang J, Karpe F, Kathiresan S, Keildson S, Kiryluk K, Kubo M, Lee JY, Liang L, Lifton RP, Ma B, McCarroll SA, McKnight AJ, Min JL, Moffatt MF, Montgomery GW, Murabito JM, Nicholson G, Nyholt DR, Okada Y, Perry JR, Dorajoo R, Reinmaa E, Salem RM, Sandholm N, Scott RA (2015): Genetic studies of body mass index yield new insights for obesity biology. Nature 518:197–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton GJ, Cummings DE, Baskin DG, Barsh GS, Schwartz MW (2006): Central nervous system control of food intake and body weight. Nature 443:289–295. [DOI] [PubMed] [Google Scholar]
- Moshfegh AJ, Rhodes DG, Baer DJ, Murayi T, Clemens JC, Rumpler WV, Paul DR, Sebastian RS, Kuczynski KJ, Ingwersen LA, Staples RC, Cleveland LE (2008): The US Department of Agriculture Automated Multiple‐Pass Method reduces bias in the collection of energy intakes. Am J Clin Nutr 88:324–332. [DOI] [PubMed] [Google Scholar]
- Nordstrom P, Pedersen NL, Gustafson Y, Michaelsson K, Nordstrom A (2016): Risks of myocardial infarction, death, and diabetes in identical twin pairs with different body mass indexes. JAMA Intern Med 176:1522–1529. [DOI] [PubMed] [Google Scholar]
- Olivo G, Wiemerslage L, Nilsson EK, Solstrand DL, Larsen AL, Olaya BM, Gustafsson VP, Titova OE, Bandstein M, Larsson EM, Benedict C, Brooks SJ, Schioth HB (2016): Resting‐state brain and the fto obesity risk allele: Default mode, sensorimotor, and salience network connectivity underlying different somatosensory integration and reward processing between genotypes. Front Hum Neurosci 10:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pursey KM, Stanwell P, Callister RJ, Brain K, Collins CE, Burrows TL (2014): Neural responses to visual food cues according to weight status: A systematic review of functional magnetic resonance imaging studies. Front Nutr 1:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raichle ME, MacLeod AM, Snyder AZ, Powers WJ, Gusnard DA, Shulman GL (2001): A default mode of brain function. Proc Natl Acad Sci U S A 98:676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RIVM (2013): NEVO‐online version 2013/4.0. Rijksinstituut voor volksgezondheid en milieu.
- Robinson S, Basso G, Soldati N, Sailer U, Jovicich J, Bruzzone L, Kryspin‐Exner I, Bauer H, Moser E (2009): A resting state network in the motor control circuit of the basal ganglia. BMC Neurosci 10:137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroevers MJ, Sanderman R, van SE, Ranchor AV (2000): The evaluation of the center for epidemiologic studies depression (ces‐d) scale: Depressed and positive affect in cancer patients and healthy reference subjects. Qual Life Res 9:1015–1029. [DOI] [PubMed] [Google Scholar]
- Seeley WW, Menon V, Schatzberg AF, Keller J, Glover GH, Kenna H, Reiss AL, Greicius MD (2007): Dissociable intrinsic connectivity networks for salience processing and executive control. J Neurosci 27:2349–2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speliotes EK, Willer CJ, Berndt SI, Monda KL, Thorleifsson G, Jackson AU, Lango AH, Lindgren CM, Luan J, Magi R, Randall JC, Vedantam S, Winkler TW, Qi L, Workalemahu T, Heid IM, Steinthorsdottir V, Stringham HM, Weedon MN, Wheeler E, Wood AR, Ferreira T, Weyant RJ, Segre AV, Estrada K, Liang L, Nemesh J, Park JH, Gustafsson S, Kilpelainen TO, Yang J, Bouatia‐Naji N, Esko T, Feitosa MF, Kutalik Z, Mangino M, Raychaudhuri S, Scherag A, Smith AV, Welch R, Zhao JH, Aben KK, Absher DM, Amin N, Dixon AL, Fisher E, Glazer NL, Goddard ME, Heard‐Costa NL, Hoesel V, Hottenga JJ, Johansson A, Johnson T, Ketkar S, Lamina C, Li S, Moffatt MF, Myers RH, Narisu N, Perry JR, Peters MJ, Preuss M, Ripatti S, Rivadeneira F, Sandholt C, Scott LJ, Timpson NJ, Tyrer JP, van WS, Watanabe RM, White CC, Wiklund F, Barlassina C, Chasman DI, Cooper MN, Jansson JO, Lawrence RW, Pellikka N, Prokopenko I, Shi J, Thiering E, Alavere H, Alibrandi MT, Almgren P, Arnold AM, Aspelund T, Atwood LD, Balkau B, Balmforth AJ, Bennett AJ, Ben‐Shlomo Y, Bergman RN, Bergmann S, Biebermann H, Blakemore AI, Boes T, Bonnycastle LL, Bornstein SR, Brown MJ, Buchanan TA, Busonero F, Campbell H, Cappuccio FP, Cavalcanti‐Proenca C, Chen YD, Chen CM, Chines PS, Clarke R, Coin L, Connell J, Day IN, den HM, Duan J, Ebrahim S, Elliott P, Elosua R, Eiriksdottir G, Erdos MR, Eriksson JG, Facheris MF, Felix SB, Fischer‐Posovszky P, Folsom AR, Friedrich N, Freimer NB, Fu M, Gaget S, Gejman PV, Geus EJ, Gieger C, Gjesing AP, Goel A, Goyette P, Grallert H, Grassler J, Greenawalt DM, Groves CJ, Gudnason V, Guiducci C, Hartikainen AL, Hassanali N, Hall AS, Havulinna AS, Hayward C, Heath AC, Hengstenberg C, Hicks AA, Hinney A, Hofman A, Homuth G, Hui J, Igl W, Iribarren C, Isomaa B, Jacobs KB, Jarick I, Jewell E, John U, Jorgensen T, Jousilahti P, Jula A, Kaakinen M, Kajantie E, Kaplan LM, Kathiresan S, Kettunen J, Kinnunen L, Knowles JW, Kolcic I, Konig IR, Koskinen S, Kovacs P, Kuusisto J, Kraft P, Kvaloy K, Laitinen J, Lantieri O, Lanzani C, Launer LJ, Lecoeur C, Lehtimaki T, Lettre G, Liu J, Lokki ML, Lorentzon M, Luben RN, Ludwig B, Manunta P, Marek D, Marre M, Martin NG, McArdle WL, McCarthy A, McKnight B, Meitinger T, Melander O, Meyre D, Midthjell K, Montgomery GW, Morken MA, Morris AP, Mulic R, Ngwa JS, Nelis M, Neville MJ, Nyholt DR, O'Donnell CJ, O'Rahilly S, Ong KK, Oostra B, Pare G, Parker AN, Perola M, Pichler I, Pietilainen KH, Platou CG, Polasek O, Pouta A, Rafelt S, Raitakari O, Rayner NW, Ridderstrale M, Rief W, Ruokonen A, Robertson NR, Rzehak P, Salomaa V, Sanders AR, Sandhu MS, Sanna S, Saramies J, Savolainen MJ, Scherag S, Schipf S, Schreiber S, Schunkert H, Silander K, Sinisalo J, Siscovick DS, Smit JH, Soranzo N, Sovio U, Stephens J, Surakka I, Swift AJ, Tammesoo ML, Tardif JC, Teder‐Laving M, Teslovich TM, Thompson JR, Thomson B, Tonjes A, Tuomi T, van Meurs JB, van Ommen GJ (2010): Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet 42:937–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stice E, Spoor S, Bohon C, Small DM (2008a): Relation between obesity and blunted striatal response to food is moderated by TaqIA A1 allele. Science 322:449–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stice E, Spoor S, Bohon C, Veldhuizen MG, Small DM (2008b): Relation of reward from food intake and anticipated food intake to obesity: A functional magnetic resonance imaging study. J Abnorm Psychol 117:924–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoeckel LE, Weller RE, Cook EW, III , Twieg DB, Knowlton RC, Cox JE (2008): Widespread reward‐system activation in obese women in response to pictures of high‐calorie foods. Neuroimage 41:636–647. [DOI] [PubMed] [Google Scholar]
- Stunkard AJ, Foch TT, Hrubec Z (1986): A twin study of human obesity. Jama 256:51–54. [PubMed] [Google Scholar]
- Van Bloemendaal L, Ijzerman RG, Ten Kulve JS, Barkhof F, Konrad RJ, Drent ML, Veltman DJ, Diamant M (2014): GLP‐1 receptor activation modulates appetite‐ and reward‐related brain areas in humans. Diabetes 63:4186–4196. [DOI] [PubMed] [Google Scholar]
- Van Bloemendaal L, Veltman DJ, Ten Kulve JS, Groot PF, Ruhe HG, Barkhof F, Sloan JH, Diamant M, Ijzerman RG (2015): Brain reward‐system activation in response to anticipation and consumption of palatable food is altered by glucagon‐like peptide‐1 receptor activation in humans. Diabetes Obes Metab 17:878–886. [DOI] [PubMed] [Google Scholar]
- Van Dongen J, Slagboom PE, Draisma HH, Martin NG, Boomsma DI (2012): The continuing value of twin studies in the omics era. Nat Rev Genet 13:640–653. [DOI] [PubMed] [Google Scholar]
- Van Dongen J, Willemsen G, Heijmans BT, Neuteboom J, Kluft C, Jansen R, Penninx BW, Slagboom PE, de Geus EJ, Boomsma DI (2015): Longitudinal weight differences, gene expression and blood biomarkers in BMI‐discordant identical twins. Int J Obes (Lond) 39:899–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Strien JW (1992): Classificatie van links‐ en rechtshandige proefpersonen. Ned Tijdschr Psychol 47:88–92. [Google Scholar]
- Vucetic Z, Carlin JL, Totoki K, Reyes TM (2012): Epigenetic dysregulation of the dopamine system in diet‐induced obesity. J Neurochem 120:891–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GJ, Volkow ND, Logan J, Pappas NR, Wong CT, Zhu W, Netusil N, Fowler JS (2001): Brain dopamine and obesity. Lancet 357:354–357. [DOI] [PubMed] [Google Scholar]
- Wijngaarden MA, Veer IM, Rombouts SA, van Buchem MA, Willems van DK, Pijl H, van der Grond J (2015): Obesity is marked by distinct functional connectivity in brain networks involved in food reward and salience. Behav Brain Res 287:127–134. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Zhao H, Qiu S, Tian J, Wen X, Miller JL, von Deneen KM, Zhou Z, Gold MS, Liu Y (2013): Altered functional brain networks in Prader‐Willi syndrome. NMR Biomed 26:622–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Tian D, Yu C, Zhang J, Tian X, von Deneen KM, Zang Y, Walter M, Liu Y (2015): Altered baseline brain activities before food intake in obese men: A resting state fMRI study. Neurosci Lett 584:156–161. [DOI] [PubMed] [Google Scholar]
- Zwijnenburg PJ, Meijers‐Heijboer H, Boomsma DI (2010): Identical but not the same: The value of discordant monozygotic twins in genetic research. Am J Med Genet B Neuropsychiatr Genet 153:1134–1149. [DOI] [PubMed] [Google Scholar]


