Abstract
Objectives
Positron emission tomography (PET) and volume of interest (VOI) analysis have recently shown in amyotrophic lateral sclerosis (ALS) an accuracy of 93% in differentiating patients from controls. The aim of this study was to disclose by spatial independent component analysis (ICA) the brain networks involved in ALS pathological processes and evaluate their discriminative value in separating patients from controls. Experimental design: Two hundred fifty‐nine ALS patients and 40 age‐ and sex‐matched control subjects underwent brain 18F‐2‐fluoro‐2‐deoxy‐D‐glucose PET (FDG‐PET). Spatial ICA of the preprocessed FDG‐PET images was performed. Intensity values were converted to z‐scores and binary masks were used as data‐driven VOIs. The accuracy of this classifier was tested versus a validated system processing intensity signals in 27 brain meta‐VOIs. A support vector machine was independently applied to both datasets and the ‘leave‐one‐out’ technique verified the general validity of results. Principal observations: The 8 components selected as pathophysiologically meaningful discriminated patients from controls with 99.0% accuracy, the discriminating value of bilateral cerebellum/midbrain alone representing 96.3%. Among the meta‐VOIs, right temporal lobe alone reached an accuracy of 93.7%. Conclusions: Spatial ICA identified in a very large cohort of ALS patients distinct spatial networks showing a high discriminatory value, improving substantially on the previously obtained accuracy. The cerebellar/midbrain component accounted for the highest accuracy in separating ALS patients from controls. Spatial ICA and multivariate analysis perform better than univariate semi‐quantification methods in identifying the neurodegenerative features of ALS and pave the way for inclusion of PET in clinical trials and early diagnosis. Hum Brain Mapp 37:942–953, 2016. © 2015 Wiley Periodicals, Inc.
Keywords: amyotrophic lateral sclerosis, positron emission tomography, independent component analysis, meta‐volumes of interest, discriminant analysis
INTRODUCTION
Amyotrophic lateral sclerosis (ALS) is a neurodegenerative disease characterized by a progressive muscle paralysis associated with degeneration of motor neurons in the primary motor cortex, brainstem, and spinal cord [Chio et al., 2014], with an incidence of 2‐3/100,000 new cases per year [Chio et al., 2013]. Two‐thirds of patients present with spinal onset (hypotrophy and weakness of upper or lower limb muscles), one‐third with bulbar onset (dysphagia and dysarthria). In the latter group, a moderate‐to‐severe impairment of frontal lobe functions occurs in a variable percentage of cases [Abrahams et al., 2005; Quartuccio et al., 2014].
The diagnosis of ALS remains largely based on the demonstration of the involvement of upper and lower motor neurons, associated with a relentlessly progressive course. In the diagnostic work‐up and to differentiate ALS from mimicking diseases, brain positron emission tomography (PET) could represent a valuable tool and various radiopharmaceuticals have been used to assess changes in regions affected by ALS [Corcia et al., 2012; Johansson et al., 2007; Lloyd et al., 2000; Turner et al., 2004].
Most of the PET studies in ALS have been performed using 18F‐2‐fluoro‐2‐deoxy‐D‐glucose (FDG‐PET), recruiting normal subjects as a control group and looking for ALS‐related metabolic patterns. Patients with ALS usually demonstrate a decreased [18F]FDG uptake in primary and supplementary motor, premotor and frontal cortices, as well as clusters of relative hypermetabolism in the corticospinal white matter tracts, mesiotemporal regions, cerebellum, and upper brain stem, reflecting a complex pathophysiology involving degeneration of gray matter and areas of reactive glial activation [Braak et al., 2013; Chio et al., 2014; Cistaro et al., 2012].
The potential of FDG‐PET as an important biomarker for ALS diagnosis has been recently confirmed by two investigations in rather large cohorts [Pagani et al., 2014; Van Laere et al., 2014]. Both studies showed FDG‐PET in ALS to have a diagnostic accuracy of over 90% in separating patients from controls and confirmed a mixed hypometabolic‐hypermetabolic pattern.
The majority of the above‐mentioned studies were largely based on univariate linear models in which voxels or regions of interest were analyzed independently of each other, but the high accuracy obtained by discriminant analysis, also involving nonlinear techniques, highlighted the potential of a multivariate approach [Pagani et al., 2014; Van Laere et al., 2014].
A recent study reported both structural MRI and FDG‐PET voxel‐based analyses in a limited number of patients with ALS and fronto‐temporal dementia. Gray matter volume reduction was mainly due to cortical thinning rather than to reduced brain volume and this was paralleled by concomitant glucose hypometabolism. However, isolated changes at FDG PET were detected in motor and extramotor regions suggesting that functional abnormalities of gray matter may precede cortical atrophy [Rajagopalan and Pioro, 2015].
In brain disorders, analyses of cross‐sectional data of multiple images aim at disclosing the characteristic patterns of pathophysiological changes. FDG‐PET reflects changes in synaptic activity in the brain: correlated metabolic changes in specific brain regions might provide an index of neuropathological processes.
Spatial functional networks delineating coherent interregional spontaneous fluctuations in different brain areas have been investigated mainly by functional magnetic resonance imaging (fMRI). In this respect, coactivation of spatially disparate brain regions is well captured by independent component analysis (ICA), a data‐driven technique blindly separating mixed signals into independent sources. ICA does not require any initial topographic assumption and intrinsically determines distinct components by dividing the image data into spatially independent activation maps without the need of seed regions [Margulies et al., 2010].
Connectivity analyses have been performed in ALS by diffusion tensor imaging (DTI) and resting state fMRI [Douaud et al., 2011; Verstraete et al., 2010] with findings of white matter tract changes associated to cortical ones [Douaud et al., 2011]. ICA also showed, by fMRI, changes in the default mode network (DMN) [Mohammadi et al., 2009]. More recently, Welsh et al. [2013] utilized a support vector machine applied to fMRI data to discriminate ALS patients and controls by ICA‐derived networks, achieving an accuracy of 71% and revealing the DMN and motor network as the main discriminant components.
However, to date no connectivity analysis has been performed in ALS patients by PET, although some studies have suggested that ICA could be applied to a series of PET images, one image per subject or recording condition, in order to disclose brain networks involved in physiological and pathological processes [Gray et al., 2012; Park et al., 2003]. The aim of this study was to identify covariant spatial patterns by ICA in resting state FDG‐PET derived from a large sample of ALS patients and to test their accuracy in discriminating patients from healthy controls. The results were compared in the same groups of subjects to those obtained with a more conventional volume‐of‐interest (VOI)‐based discriminant analysis.
MATERIALS AND METHODS
We performed FDG‐PET in 259 ALS patients (mean age 63.3 ± 11.8 years, range 21–86; females 107/259) referred to the Turin ALS center and prospectively recruited between June 2011 and February 2014. Diagnoses of laboratory‐supported probable, probable, or definite ALS were made according to El Escorial ALS revised criteria [Brooks et al., 2000]. Most patients underwent FDG‐PET within 5 months of diagnosis. Median time between disease onset and PET examination was 15.3 months.
According to the clinical presentation at the time of the PET scan, 165 patients were diagnosed as spinal (mean age 61.9 ± 12.4 years, range 21–86; females 52/165) and 94 as bulbar (mean age 65.6 ± 10.2 years, range 40–82; females 55/94). To be classified in the bulbar group, patients with bulbar symptoms had to have less than 4 points loss on the spinal domain of the ALSFRS‐R scale (items 4 to 12). The mean disease duration was 15 ± 12.4 months, range 2–76; mean ALS‐FRS score was 39.3 ± 6.4, range 10–48. Seventeen patients were carrying the C9ORF72 mutation.
The control group was composed of 40 subjects (mean age 62 ± 14.4 years, range 27–84; females 11/40) undergoing FDG‐PET due to suspected diagnosis of lung cancer, but in which no abnormal metabolism was found in any body or brain region. All subjects had normal neurological assessment and the exclusion criteria were the same as applied in studies of other neurodegenerative disorders: presence of major systemic illness, major vision disturbances, psychiatric illnesses, and diseases impacting on brain functioning and metabolism [Nobili et al., 2010; Pagani et al., 2010].
The study design was approved by the institutional ethics committee in compliance with the Code of Ethics of the World Medical Association (Declaration of Helsinki). Patients and controls signed written informed consent.
Image Acquisition and Preprocessing
FDG‐PET examinations were performed following current guidelines [Varrone et al., 2009]. PET/computerized tomography (CT) brain scans were acquired by a Discovery ST‐E System (General Electric) in two sequential scans: PET scan (1 FOV of 30 transaxial cm) was initiated immediately after CT (thickness of 3.75 mm, 140 kVolt, 60–80 mA/s) used for attenuation correction of PET data. Data were collected in 128 × 128 matrices with a reconstructed voxel of 2.34 x 2.34 x 2.00 mm.
PET scans were normalized using a customized brain FDG‐PET template, obtained using FDG‐PET scans from a group of healthy subjects performed at the same center. Template editing was performed according to published procedures [Morbelli et al., 2012]. The spatially normalized PET images were further normalized to the average of the brain voxels with the highest 20% of intensity and smoothed with an 8 mm isotropic Gaussian filter, an ideal parameter for favoring in PET pooled analyses the optimal detectability of separated spatial signals [Woods, 1996].
Statistical Analyses
Independent component analysis (ICA)
ICA models observe data as a linear combination of a set of statistically independent components having non‐Gaussian (mainly sparse) distribution [Brown et al., 2001]. In the analysis of neuroimages, ICA results in spatial components representing metabolic networks varying coherently across subjects, the distribution of voxels in each component being statistically independent of the distribution in other components [McKeown et al., 1998]. This approach enforces the delineation of borders between possibly overlying networks [Kalcher et al., 2012].
Spatial ICA of the preprocessed FDG‐PET images was performed using the GIFT toolbox (http://mialab.mrn.org/software/). Image data from each subject were arranged in a sequence and were whitened and reduced by principal component analysis (PCA). Then an Infomax ICA algorithm, minimizing the mutual information, was used to estimate the independent spatial components (ICs) [Bell and Sejnowski, 1995; Brown et al., 2001]. The basic assumption of the infomax algorithm is that the underlying sources are sparsely distributed, i.e. the distribution of each source is super‐Gaussian [Park et al., 2003]. The optimal number of components was determined from the data using the GIFT dimensionality estimation tool, this number representing a good tradeoff between the preservation of the information intrinsically contained in the data and the reduction of its size. To display the voxels that most contributed to the resulting spatial maps, intensity values in each of them were converted to z‐scores and image values were visualized using a threshold of z = 1.96 creating binary masks. From the resulting components, those with either pathophysiological or anatomo‐functional meaning were selected and each component was named according to its anatomy.
The masks were then used as data‐driven VOIs. ICA was applied to the series of ALS patients in order to disclose brain networks affected by pathological processes as well as components which are supposedly shared with healthy subjects. The resulting transformation matrix was then applied to compute these components for each subject in the whole group in order to compare patients and controls by ICA‐based discriminant analysis. This approach is in agreement with the one‐class‐classification model, constructed to describe a reference group and where differences, or ‘novelities’, are detected as deviations from this model [Tax and Duin, 1999]. One‐class‐classification is particularly suitable for datasets in which a very well‐sampled reference group with a large number of samples is compared to an undersampled alternative group. In the present study, the number of ALS patients far exceeded that of normal controls and we could consider ALS as the reference group in order to explore independent components as one‐class descriptors and to verify how well these descriptors could perform in two‐class discriminant analysis.
Region of interest identification and data preprocessing
In order to test the ICA‐based classifier versus an already validated system, we analyzed the FDG‐PET data of both ALS patients and controls by an in‐house created Matlab‐based script [Pagani et al., 2015] that automatically extracted and processed mean relative FDG uptake signals intensity in 45 anatomical volumes of interest (VOIs) in each hemisphere (90 VOIs) as identified by the Automated Anatomical Labelling (AAL) Atlas [Tzourio‐Mazoyer et al., 2002]. In order to reduce the number of variables and to verify to what extent the analysis of specific and functionally meaningful regions could provide information about pathological processes, based on a priori criteria we merged regions with similar anatomo‐functional characteristics into meta‐VOIs [Pagani et al., 2015].
Twelve meta‐VOIs were constructed in each hemisphere:
Occipital cortex (calcarine/lingual/inferior occipital/middle occipital/superior occipital gyri);
Putamen/pallidum/caudate;
Parahippocampal gyrus/amygdala/hippocampus/insula;
Orbito‐frontal cortex (inferior frontal/medial frontal/superior‐orbital frontal gyri);
Frontal cortex (middle frontal/superior frontal/superior‐medial frontal gyri);
Cuneus/fusiform gyrus/precuneus;
Postcentral/precentral gyri/supplementary motor area;
Parietal lobe (inferior parietal/superior parietal gyri);
Anterior cingulate gyrus;
Posterior cingulate gyrus;
Temporal lobe (inferior temporal/middle temporal/superior temporal gyri), and
Temporal pole (middle temporal pole/superior temporal pole gyri).
Since in previous investigations on a sub‐group of the present ALS patient sample an involvement of the cerebellum was found [Cistaro et al., 2014; Pagani et al., 2014], we added into the dataset three further VOIs, the two cerebellar hemispheres, separately, and the cerebellar vermis, and they were included in the discriminant analysis.
Discriminant analysis (DA)
DA was performed by a non‐linear classifier based on the support vector machine (SVM) method with radial basis functions [Cortes and Vapnik, 1995] in order to discriminate ALS patients and controls based on FDG‐PET. PET data were preprocessed by ICA and by meta‐VOI, producing two different datasets. DA was independently applied to the two datasets (ICA‐based and meta‐VOI‐based) drawn from the same groups of PET images in order to evaluate the influence of the feature extraction method and to identify the regions or networks with the highest discriminatory value.
The discrimination based on each dataset was followed by the application of a stepwise backward selection procedure in which the least influential variable was removed and replaced by the best variable (if any) at each step. The effectiveness of the discrimination was evaluated by ‘leave‐one‐out’ cross‐validation by which each subject was classified using a model fitted to all the remaining ones. Cross‐validation was adopted in order to verify the general validity of results and to prevent accuracy overrating due to overfitting, i.e. an excessive adaptation to the characteristics of the study sample.
Receiver operating characteristic (ROC) curves and area under the curve (AUC) were computed at each classification step and relevant confidence intervals were estimated by the bootstrap method described by Qin and Hotilovac [2008]. For a comparison between the results obtained with two different procedures, the ROC‐AUCs of the two analyses were compared by evaluating the z‐value for paired scores according to Hanley and McNeil's formula [Hanley and McNeil, 1982, 1983] in which the standard error of the difference between the AUCs is evaluated as function of their own values and of the correlation between scores.
DA by SVM and the following ROC curve analysis and accuracy measurements were performed using the Statistics Toolbox of Matlab R2014a (MathWorks, Natick, MA).
The effectiveness of the discrimination in the first stage of the disease was further explored by excluding the patients with a disease duration < 6 months from the training set: the model was then fitted using the sample made up of the patients with longer disease duration along with controls, while the test was performed on the patients with short disease duration.
RESULTS
ICA
The optimal number of components was set by GIFT to 18 and all were visually inspected. Ten components appeared to be noise or scattered small clusters without any pathophysiological meaning and were discarded. The remaining 8 components were classified as follows:
Bilateral cerebellum/midbrain;
Bilateral basal ganglia/thalami;
Bilateral temporal cortex;
Bilateral frontal/prefrontal cortex;
Bilateral parietal/posterior cingulate cortices (posterior portion of DMN);
Bilateral medial temporal/sub‐genual anterior cingulate cortex;
Bilateral occipital cortex;
Bilateral motor cortex.
When all 8 components were included in the DA the accuracy in discriminating ALS patients from controls was 99.0% [confidence interval, CI 0.979–1.000] with an AUC of 0.996 [0.965–1.000] and these values remained more or less constant until the 4 most significant components were selected by the backward stepwise procedure (bilateral cerebellum/midbrain; bilateral medial temporal/sub‐genual anterior cingulate cortex; bilateral parietal/posterior cingulate cortices; bilateral motor cortex) (Figs. 1 and 2). When only the component corresponding to bilateral cerebellum/midbrain was left, the accuracy was still 96.3% [94.2–98.5] with an AUC of 0.992 [0.973–0.997] (Fig. 2). The difference between the AUC associated with this best component and the AUC associated with the whole set of 8 components was not significant.
Figure 1.
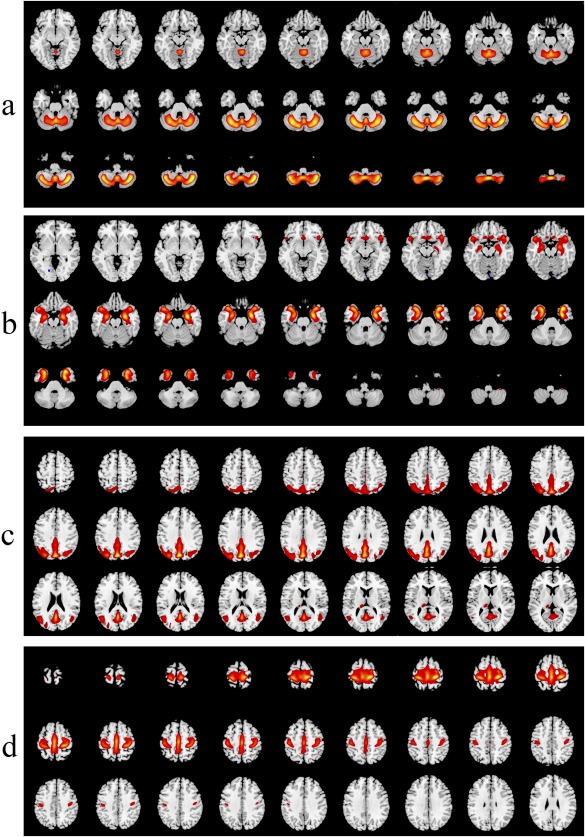
Component maps showing the voxels which contributed most to each component as evaluated by z‐scores. The figure shows the sequence of slides with the spatial maps for the following components: a) Bilateral cerebellum/midbrain; b) Bilateral medial temporal/sub‐genual anterior cingulate cortex; c) Bilateral parietal/posterior cingulate cortices (posterior portion of DMN); d) Bilateral motor cortex.
Figure 2.
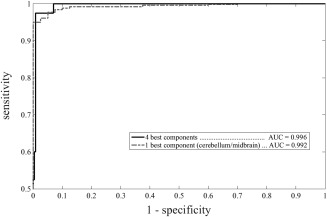
Receiver operating characteristic curves expressing classifier performance. Independent‐component based classification: optimal performance is obtained with the four best components and does not decrease significantly when the single best component is used.
Meta‐VOIs
The 27 meta‐VOIs showed an accuracy of 99.8% [99.0–100.0] with an AUC of 1.00 [0.998–1.000] until the backward stepwise procedure identified the 4 most influential regions (postcentral/precentral gyri/supplementary motor area; left anterior cingulate cortex; left temporal lobe; and right cerebellum) with an accuracy of 99.3% and AUC 0.999 (Fig. 3). Once the backward stepwise procedure selected only one meta‐VOI (right temporal cortex), the accuracy in discriminating ALS patients from controls was 93.65% [88.5–94.8] and the AUC was 0.969 [0.946–0.982] (Fig. 3). The difference between the AUC associated with this best discriminant meta‐VOI and the AUC associated with the whole set of 27 regions was significant (z = 3.43, P < 0.001).
Figure 3.
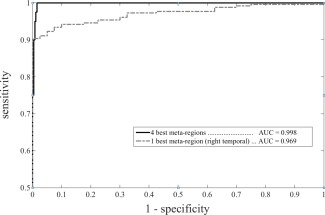
Receiver operating characteristic curves expressing classifier performance. Meta‐region based classification: optimal performance is obtained with at least four meta‐regions while the performance slightly but significantly decreases using only the best meta‐region. [Color figure can be viewed in the online issue, which is available at http://wileyonlinelibrary.com.]
Comparison Between ICA and meta‐VOI Classification Performance
Considering the very high accuracy consistently achieved by both the ICA and meta‐VOI models when all or many components or regions were included, the first comparison made was between the 4‐component and the 4‐region models obtained by backward stepwise selection. The difference in performance between the ICA‐based model (AUC = 0.996) and the meta‐VOI‐based model (AUC=0.993) was not significant (z = 1.022, N.S.). A second comparison was performed in the extreme case of models including one single feature: in this case the difference between the ICA‐based model (AUC=0.992) and the meta‐VOI‐based one (AUC = 0.969) was significant (z = 2.31, P < 0.02).
Discrimination in the Early Stage of the Disease
The previously selected best component was analyzed in the sample of controls and long disease‐duration patients in order to set the discriminant threshold that was then applied to the sample of patients in the early stage of the disease. Out of these 28 early‐ALS patients, only one was undetected (versus 11/299 misclassifications in the whole sample as tested by ‘leave‐one‐out’ cross‐validation). When the same procedure was applied using the previously selected best meta‐VOI region, 4 patients were undetected (versus 25/299 misclassifications).
DISCUSSION
In the present study, we implemented ICA in resting state FDG‐PET to disclose the networks and meta‐region activities characterizing ALS and to assess their possible diagnostic value. ICA uncovered several spatially distinct networks that hierarchically showed an extremely high discriminative value, the cerebellar/midbrain network being the most accurate (AUC >0.992) in separating ALS patients from controls. This value was obtained including in the analysis 259 ALS patients and it exceeded the already highly significant one found in a previous investigation in which 195 ALS patients were investigated by FDG‐PET showing an AUC of 0.945 [Pagani et al., 2014]. The latter study was performed using 51 VOIs derived from 65 Brodmann functional areas and all variables were taken into account in the analysis. In the present investigation, in order to compare and validate the ICA‐based analysis against an established method, we performed discriminant analysis on 27 meta‐VOIs derived from an anatomic atlas, AAL, utilizing the same methodology proven able to discriminate mild cognitive‐impaired patients from healthy controls [Pagani et al., 2015]. The slightly lower accuracy of the previous ALS study [Pagani et al., 2014] might be partially due to overfitting (cross‐validation was applied to a smaller sample with a larger number of VOIs) and partially to the difference in meta‐region definition. In fact, the present choice was consistent with the nature of the ICs extracted by FDG‐PET analysis, in which disease‐specific spatial patterns were evaluated in resting state condition possibly not superimposing the functional mapping derived from the segmentation of brain by means of Brodmann areas.
Resting state interregional correlations reflect neural interactions occurring when brain activity is not externally induced or voluntarily generated, yet the subject is cognitively active, mind wandering, meditating or ruminating during tracer infusion and uptake. Since the brain at rest contains several functionally discrete networks, intrinsically organized into covariance patterns, ICA is an ideal technique since it identifies the underlying activity unique to each network using data alone without the constraint of spatial bounds or a priori knowledge [Storti et al., 2013].
In neurodegenerative diseases the interregional correlations of metabolic rates among cross‐sectional data of subjects sharing common independent metabolic patterns indicate the association of spatially connected synaptic activities affected by common processes and appearing in two or more regions. This might be of particular relevance in disorders of the ALS spectrum in which concrete connections between brain regions are certainly affected by the degeneration of neurons and white matter fibers.
Neurodegenerative processes integrate activity in networks rather than in isolated regions, causing brain areas to reorganize the reciprocal connections. In this respect, in FDG‐PET investigations spatial connectivity representing the metabolic “signature” of the disease is better captured by multivariate methods revealing distributed regional patterns of metabolic integration not shown by the univariate subtraction methods, the latter ignoring inter‐regional metabolic coherence [Toussaint et al., 2012].
Discriminant analysis of the AAL‐based meta‐VOIs showed a very high accuracy as well, being still very high when the most statistically meaningful 4 meta‐VOIs were included in the analysis (AUC = 0.999) and slightly lower when only one meta‐VOI (right temporal) was selected by the backward stepwise procedure (AUC = 0.969).
Our analysis shows that the main metabolic changes can also be detected in the early stages of the disease. Considering the very high accuracy of the method (i.e., few misclassifications) the simplest models (using only one component or one meta‐VOI) were fitted to patients with long disease duration and then applied to classify the patients with a disease duration less than 6 months. The simplest models, in fact, might have given rise to more errors, but the number of undetected patients was still low.
Besides the outcome of the discriminant analyses, we were able to reveal by ICA highly concomitant metabolic activity in sparse networks resulting from the high degree of interconnectivity across the entire brain. Despite the different origin of the independent spatial components with respect to the meta‐VOIs (data‐driven vs. a priori segmentation, respectively) the most discriminant regions selected by the stepwise backward selection procedure were in both analyses the cerebellum and the temporal and sensorimotor cortices. These regions, showing the most metabolic coherence, were among the ones previously described to account for the significant differences between ALS patients and control subjects [Pagani et al., 2014; Van Laere et al., 2014] and represent the site where the neuropathological processes peculiar to the disease take place.
The component including cerebellum and midbrain encompasses two regions found in ALS to show changes in gray [Ellis et al., 2001; Mioshi et al., 2013] and white [Kassubek et al., 2005] matter volume, fractional anisotropy [Keil et al., 2012; Prell et al., 2013; Prudlo et al., 2012] and metabolism [Cistaro et al., 2012; Hoffman et al., 1992; Pagani et al., 2014; Van Laere et al., 2014]. In a previous FDG‐PET study performed on a subset of the present dataset of ALS patients, these two structures were found to be relatively hypermetabolic in comparison to controls [Pagani et al., 2014]. The relative hypermetabolism was ascribed to a possible excessive presence of activated astrocytes and microglia, as also described in structural [Kushner et al., 1991; Philips and Robberecht 2011; Yamanaka et al., 2008] and functional [Corcia et al., 2012; Johansson et al., 2007; Turner et al., 2004] studies, resulting in higher relative FDG uptake than in controls. This has been considered to be the characteristic metabolic signature of ALS, and both ICA and meta‐VOI analyses confirmed the preeminent role of these regions in the pathophysiology of the disease. Indeed, the cerebellum, along with the sensorimotor network and DMN, represented one of the 10 resting state networks described by Welsh et al. [2013] in the only previous fMRI study implementing ICA and SVM, to discriminate 32 ALS patients from 31 controls. It showed the possibility of reproducible and consistent results in ALS research also when different methods are implemented.
On the other hand, a model of how ALS spreads pathologically has recently identified the initial lesions to appear in the frontal agranular neocortex, somatomotor neurons of the spinal cord and brainstem diffusing to precerebellar nuclei of lower brainstem, reticular formation, red nucleus and eventually to the prefrontal cortex and sensory area to finally develop in the anterior temporal and hippocampal cortex [Braak et al., 2013]. This hypothesis of anterograde propagation from pyramidal cells would support our findings of changes in the corticospinal tracts and the primary role of somatosensory cortex, midbrain and medial temporal cortex in ALS pathology.
The sensorimotor cortex has been extensively found by MRI to be affected in ALS [Chang et al., 2005; Thivard et al., 2007; Turner et al., 2007] and functional studies have reported connectivity to be both increased [Agosta et al., 2013] and decreased [Zhou et al., 2013] in this region. In keeping with previous findings [Douaud et al., 2011] and a recent FDG‐PET investigation in Alzheimer's disease (AD) [Ashraf et al., 2015], in the present study the normalized signal intensity of the bilateral motor cortex component was found to be hypermetabolic compared to the other ICs, and higher in ALS patients than in controls, suggesting the relative hypermetabolism to be a compensatory mechanism to neuronal damage.
A neurophysiological correlate of these findings is the hyperexcitability of the motor cortex in ALS patients identified in the early phases of the disease [Geevasinga et al., 2015] and even preclinically in subjects carrying SOD1 mutations [Vucic et al., 2008]. Interestingly, the hyperexcitability of the motor cortex is specific to ALS when compared to ALS‐mimic syndromes [Vucic et al., 2008]. It has been proposed that this early event is related to a loss of inhibitory cortical interneurons in motor neuron disease patients [Nihei et al., 1993] or to a glutamate‐mediated down‐regulation of short‐interval intracortical inhibition [Stefan et al., 2001].
The other important component found to play a significant role in both ALS‐ICA studies was the posterior part of the default mode network (described as temporoparietal lobe/posterior cingulate cortex in our study). This region was found to significantly discriminate mild cognitive‐impaired (MCI) patients converting vs. non converting to AD both in PET‐ICA [Shaffer et al., 2013], PET‐PCA [Nobili et al., 2008] and PET‐metaVOIs [Pagani et al., 2015] investigations. Furthermore, it was one of the ICs that best predicted conversion from stable to progressing‐to‐AD MCI patients [Willette et al., 2014]. In ALS fMRI studies, enhanced connectivity [Agosta et al., 2013] and decreased activity [Mohammadi et al., 2009] were reported in this region with respect to healthy subjects, underscoring its role in neurodegenerative pathologies, with and without cognitive impairment.
This posterior portion of the DMN is confirmed to be a “hub” of functional connectivity coming from distant frontal and temporal regions and thus largely suffering from synaptic disconnection, consistently shown by means of FDG‐PET, similarly as happens in AD [Drzezga et al., 2011].
The right temporal cortex in meta‐VOI analysis and the bilateral medial temporal/sub‐genual anterior cingulate cortex along with bilateral cerebellum/midbrain in the ICA were the regions showing the highest discriminative value in differentiating ALS patients from controls. In the present study, we found a relatively higher intensity signal in these regions compared to the average of the whole brain. The medial temporal and hippocampal region showed a significant relative metabolic increase in ALS patients, as compared to controls [Pagani et al., 2014], and a significant increase in activated glial cells [Corcia et al., 2012]. These regions are immediately adjacent to the corticospinal tracts running into the corona radiata and the smearing of the hypermetabolic PET signal present in white matter tracts, specific of ALS, might enclose the most medial part of the temporal and parahippocampal cortex and be the cause for the voxels composing the ICA to share similar metabolic coherence.
As compared to previous ICA investigations in ALS and other neurodegenerative disorders [Toussaint et al., 2012], the estimates of inter‐subject random variability in the present study were more robust based on the greater number of subjects, increasing the power of the analysis since the sampling error decreases with the increase of sample size. Moreover, the exploratory analysis of a relatively large number of regions was supported by the large sample size, reducing sampling error and overfitting, while the generalization of findings and effectiveness of classification were supported by the “leave‐one‐out” cross validation. Further, use of the stepwise backward selection procedure identified the minimal number of components (or meta‐VOIs) necessary for a suitable group discrimination focusing on the most distinctive and robust pathophysiological changes caused by ALS. Cross‐correlation analysis allowed for the discrimination of the pathology at individual level, paving the way to a clinical application of the method based on large datasets of patients and healthy controls in which subjects can be assigned to either group.
Although an expert system based on predetermined regions of interest provided a discrimination accuracy between 97 and 99% using 1 or 4 meta‐VOIs, respectively, the neurophysiological value of the ICA findings cannot be overlooked. A single component is more representative than a single meta‐VOI since it results from a data‐driven connectivity heavily influenced by the pathophysiology of the disease. The spatial correlations expressed by the ICs found in the present study are informative beyond the pure diagnostic utility of the study. ICA can help to detect in vivo different patterns of involvement of functional networks beyond the “classical” motor system (i.e., primary motor cortex and pyramidal tracts) and the modality of spreading of the functional alterations over time. ICs can also be used to determine the different endophenotypes of ALS, improving our ability to classify patients for clinical and trial purposes. In this perspectives, when implemented in neurodegenerative disorders, ICA‐derived (or more generally: multivariately derived) profiles can satisfy both needs of (i) high diagnostic utility and (ii) mechanistic insight into the etiology of the various diseases.
Pattern‐based classification, providing insight into the nature of neural representation, has a strong potential to become the standard approach in functional neuroimaging overcoming the limitations of distinct cortical disease localization treating voxels as independent units and discarding most of the information potentially gathered by neuroimaging [O'Toole et al., 2007].
The components defined by the present analysis are somewhat similar to the ones reported by investigations on large datasets of resting state studies performed on healthy controls by fMRI [Kalcher et al., 2012], PET [Martinez‐Murcia et al., 2013] or both [Di and Biswal, 2012]. As compared to those ICs, the ones identified by our analysis were larger and more symmetric in accordance with the high correlations found in metabolic studies of homotropic homologous regions [Horwitz et al., 1984] and encompassed regions found to be implicated in ALS pathology. In fact, among the physiologically meaningful, even if less discriminant, we identified the bilateral frontal/prefrontal cortex and occipital cortex, described by previous studies to be implicated in significant metabolic differences between ALS patients and controls [Pagani et al., 2014; Van Laere et al., 2014].
The implementation of the one‐class‐discrimination approach, in which the very well‐sampled group of patients served as a training set, isolated the ICs characteristic of the pathology, connoting the analysis as ALS‐driven and underscoring the differences with respect to the less homogeneous group of control subjects.
Limitations
Even though the regions included in the same component are spatially correlated we cannot exclude that the extraction of too many or too few components caused regions not present in the same component to be arbitrarily separated by the algorithm [Di and Biswal, 2012]. However low model orders of up to 100 components seem to be robust and repeatable [Abou‐Elseoud et al., 2010].
The analyses did not take into account in this cohort of ALS patients the possible gray and white matter changes of volume. Anyhow, the volumetric correction of metabolic data was found to have no impact on the PET data, the two parameters being dissociated [Di and Biswal, 2012].
One possible drawback of exploratory analyses is that data were analyzed first and labeled post‐hoc subjectively by visual assessment of the experimenters according to their pathophysiological interpretability, potentially biasing the results. However, the interpretation of the findings and the choice of the meaningful components fits with the known neuropathological features of ALS, confirming, in a large cohort of patients and by an objective data‐driven method, the involvement in ALS of the brain regions described by several functional studies [Cistaro et al., 2012; Lloyd et al., 2000; Pagani et al., 2014; Turner et al., 2005; Van Laere et al., 2014].
This study investigated only ALS patients without taking into account other forms of motor neuron disease. Future studies comparing PET patterns in ALS patients and in subjects with ALS‐mimic syndromes will help us to better understand whether PET can be of support in the differential diagnosis of ALS. This will be feasible when there will be a sufficient number of the latter patients to guarantee reliable results.
CONCLUSION
This work shows that FDG‐PET brain patterns in ALS are consistently identified by means of ICA, which is not possible with conventional voxel‐based univariate analysis. The data confirm, in a very large ALS cohort, the capability of FDG‐PET and ICA to identify the neurodegenerative features typical of patients, paving the way to a correct inclusion of ALS patients in clinical trials and early diagnosis, as in the past occurred for AD. We have shown that ICA, networking and multivariate analysis perform better in comparison to other univariate semi‐quantification methods. This might be of particular relevance in disorders of the ALS spectrum in which concrete connections between brain regions are certainly affected by the degeneration of neurons and white matter fibers.
In addition, the metabolic network patterns identified in this study indicate the functional involvement in ALS of several motor and non‐motor areas, including “atypical” areas such as the cerebellum/midbrain, as well as the presence of hypermetabolism in the classically involved area, i.e., the bilateral motor cortex. These latter findings, which parallel previous neurophysiological findings, could represent a relevant target for future therapies aimed at modulating the pathological hyperactivity/hypermetabolism of the motor cortex.
ACKNOWLEDGMENTS
The authors thank the patients and the control subjects for participating in the study as well as the technician of IRMET S.p.A for providing assistance and making the investigations possible.
The authors wish also to thank Dr. Alessandro Giuliani for his wise and valuable statistical advice and Fondazione Vialli e Mauro e Fondazione Magnetto for the valuable support.
REFERENCES
- Abou‐Elseoud A, Starck T, Remes J, Nikkinen J, Tervonen O, Kiviniemi V (2010): The effect of model order selection in group PICA. Hum Brain Mapp 31:1207–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abrahams S, Leigh PN, Goldstein LH (2005): Cognitive change in ALS: A prospective study. Neurology 64:1222–1226. [DOI] [PubMed] [Google Scholar]
- Agosta F, Canu E, Valsasina P, Riva N, Prelle A, Comi G, Filippi M (2013): Divergent brain network connectivity in amyotrophic lateral sclerosis. Neurobiol Aging 34:419–427. [DOI] [PubMed] [Google Scholar]
- Ashraf A, Fan Z, Brooks DJ, Edison P (2015): Cortical hypermetabolism in MCI subjects: A compensatory mechanism? Eur J Nucl Med Mol Imaging 42:447–458. [DOI] [PubMed] [Google Scholar]
- AJ Bell, TJ Sejnowski (1995): An information‐maximazation approach to blind separation and blind deconvolution. Neural Comput 7:1129–1159. [DOI] [PubMed] [Google Scholar]
- Braak H, Brettschneider J, Ludolph AC, Lee VM, Trojanowski JQ, Del Tredici K (2013): Amyotrophic lateral sclerosis—A model of corticofugal axonal spread. Nat Rev Neurol 9:708–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks BR, Miller RG, Swash M, Munsat TL (2000): El Escorial revisited: Revised criteria for the diagnosis of amyotrophic lateral sclerosis. Amyotroph Lateral Scler Other Motor Neuron Disord 1:293–299. [DOI] [PubMed] [Google Scholar]
- Brown GD, Yamada S, Sejnowski TJ (2001): Independent component analysis at the neural cocktail party. Trends Neurosci 24:54–63. [DOI] [PubMed] [Google Scholar]
- Chang JL, Lomen‐Hoerth C, Murphy J, Henry RG, Kramer JH, Miller BL, Gorno‐Tempini ML (2005): A voxel‐based morphometry study of patterns of brain atrophy in ALS and ALS/FTLD. Neurology 65:75–80. [DOI] [PubMed] [Google Scholar]
- Chio A, Logroscino G, Traynor BJ, Collins J, Simeone JC, Goldstein LA, White LA (2013): Global epidemiology of amyotrophic lateral sclerosis: A systematic review of the published literature. Neuroepidemiology 41:118–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chio A, Pagani M, Agosta F, Calvo A, Cistaro A, Filippi M (2014): Neuroimaging in amyotrophic lateral sclerosis: insights into structural and functional changes. Lancet Neurol 13:1228–1240. [DOI] [PubMed] [Google Scholar]
- Cistaro A, Pagani M, Montuschi A, Calvo A, Moglia C, Canosa A, Restagno G, Brunetti M, Traynor BJ, Nobili F, G Carrara, P Fania, L Lopiano, MC Valentini, A Chiò (2014): The metabolic signature of C9ORF72‐related ALS: FDG PET comparison with nonmutated patients. Eur J Nucl Med Mol Imaging 41:844–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cistaro A, Valentini MC, Chio A, Nobili F, Calvo A, Moglia C, Montuschi A, Morbelli S, Salmaso D, Fania P, G Carrara, M Pagani (2012): Brain hypermetabolism in amyotrophic lateral sclerosis: A FDG PET study in ALS of spinal and bulbar onset. Eur J Nucl Med Mol Imaging 39:251–259. [DOI] [PubMed] [Google Scholar]
- Corcia P, Tauber C, Vercoullie J, Arlicot N, Prunier C, Praline J, Nicolas G, Venel Y, Hommet C, Baulieu JL, JP Cottier, C Roussel, M Kassiou, D Guilloteau, MJ Ribeiro (2012): Molecular imaging of microglial activation in amyotrophic lateral sclerosis. PLoS One 7:e52941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortes C, Vapnik V (1995): Support‐Vector Networks. Machine Learn 20:273–297. [Google Scholar]
- Di X, Biswal BB (2012): Metabolic brain covariant networks as revealed by FDG‐PET with reference to resting‐state fMRI networks. Brain Connect 2:275–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douaud G, Filippini N, Knight S, Talbot K, Turner MR (2011): Integration of structural and functional magnetic resonance imaging in amyotrophic lateral sclerosis. Brain 134:3470–3479. [DOI] [PubMed] [Google Scholar]
- Drzezga A, Becker JA, Van Dijk KR, Sreenivasan A, Talukdar T, Sullivan C, Schultz AP, Sepulcre J, Putcha D, Greve D, KA Johnson, RA Sperling (2011): Neuronal dysfunction and disconnection of cortical hubs in non‐demented subjects with elevated amyloid burden. Brain 134:1635–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis CM, Suckling J, Amaro E,J, Bullmore ET, Simmons A, Williams SC, Leigh PN (2001): Volumetric analysis reveals corticospinal tract degeneration and extramotor involvement in ALS. Neurology 57:1571–1578. [DOI] [PubMed] [Google Scholar]
- Geevasinga N, Menon P, Sue CM, Kumar KR, Ng K, Yiannikas C, Kiernan MC, Vucic S (2015): Cortical excitability changes distinguish the motor neuron disease phenotypes from hereditary spastic paraplegia. Eur J Neurol 22:826–831. [DOI] [PubMed] [Google Scholar]
- Gray KR, Wolz R, Heckemann RA, Aljabar P, Hammers A, Rueckert D, Neuroimaging AD (2012): Multi‐region analysis of longitudinal FDG‐PET for the classification of Alzheimer's disease. Neuroimage 60:221–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanley JA, McNeil BJ (1982): The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology 143:29–36. [DOI] [PubMed] [Google Scholar]
- Hanley JA, McNeil BJ (1983): A method of comparing the areas under receiver operating characteristic curves derived from the same cases. Radiology 148:839–843. [DOI] [PubMed] [Google Scholar]
- Hoffman JM, Mazziotta JC, Hawk TC, Sumida R (1992): Cerebral glucose utilization in motor neuron disease. Arch Neurol 49:849–854. [DOI] [PubMed] [Google Scholar]
- Horwitz B, Duara R, Rapoport SI (1984): Intercorrelations of glucose metabolic rates between brain regions: Application to healthy males in a state of reduced sensory input. J Cereb Blood Flow Metab 4:484–499. [DOI] [PubMed] [Google Scholar]
- Johansson A, Engler H, Blomquist G, Scott B, Wall A, Aquilonius SM, Langstrom B, Askmark H (2007): Evidence for astrocytosis in ALS demonstrated by [11C](L)‐deprenyl‐D2 PET. J Neurol Sci 255:17–22. [DOI] [PubMed] [Google Scholar]
- Kalcher K, Huf W, Boubela RN, Filzmoser P, Pezawas L, Biswal B, Kasper S, Moser E, Windischberger C (2012): Fully exploratory network independent component analysis of the 1000 functional connectomes database. Front Hum Neurosci 6:301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassubek J, Unrath A, Huppertz HJ, Lule D, Ethofer T, Sperfeld AD, Ludolph AC (2005): Global brain atrophy and corticospinal tract alterations in ALS, as investigated by voxel‐based morphometry of 3‐D MRI. Amyotroph Lateral Scler Other Motor Neuron Disord 6:213–220. [DOI] [PubMed] [Google Scholar]
- Keil C, Prell T, Peschel T, Hartung V, Dengler R, Grosskreutz J (2012): Longitudinal diffusion tensor imaging in amyotrophic lateral sclerosis. BMC Neurosci 13:141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushner PD, Stephenson DT, Wright S (1991): Reactive astrogliosis is widespread in the subcortical white matter of amyotrophic lateral sclerosis brain. J Neuropathol Exp Neurol 50:263–277. [DOI] [PubMed] [Google Scholar]
- Lloyd CM, Richardson MP, Brooks DJ, Al‐Chalabi A, Leigh PN (2000): Extramotor involvement in ALS: PET studies with the GABA(A) ligand [(11)C]flumazenil. Brain 123:2289–2296. [DOI] [PubMed] [Google Scholar]
- Margulies DS, Bottger J, Long X, Lv Y, Kelly C, Schafer A, Goldhahn D, Abbushi A, Milham MP, Lohmann G, A Villringer (2010): Resting developments: a review of fMRI post‐processing methodologies for spontaneous brain activity. Magma 23:289–307. [DOI] [PubMed] [Google Scholar]
- Martinez‐Murcia FJ, Gorriz JM, Ramirez J, Puntonet CG, Illan IA (2013): Functional activity maps based on significance measures and Independent Component Analysis. Comput Methods Programs Biomed 111:255–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKeown MJ, Makeig S, Brown GG, Jung TP, Kindermann SS, Bell AJ, Sejnowski TJ (1998): Analysis of fMRI data by blind separation into independent spatial components. Hum Brain Mapp 6:160–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mioshi E, Lillo P, Yew B, Hsieh S, Savage S, Hodges JR, Kiernan MC, Hornberger M (2013): Cortical atrophy in ALS is critically associated with neuropsychiatric and cognitive changes. Neurology 80:1117–1123. [DOI] [PubMed] [Google Scholar]
- Mohammadi B, Kollewe K, Samii A, Krampfl K, Dengler R, Munte TF (2009): Changes of resting state brain networks in amyotrophic lateral sclerosis. Exp Neurol 217:147–153. [DOI] [PubMed] [Google Scholar]
- Morbelli S, Drzezga A, Perneczky R, Frisoni GB, Caroli A, van Berckel BN, Ossenkoppele R, Guedj E, Didic M, Brugnolo A, G Sambuceti, M Pagani, E Salmon, F Nobili (2012): Resting metabolic connectivity in prodromal Alzheimer's disease. A European Alzheimer Disease Consortium (EADC) project. Neurobiol Aging 33:2533–2550. [DOI] [PubMed] [Google Scholar]
- Nihei K, McKee AC, Kowall NW (1993): Patterns of neuronal degeneration in the motor cortex of amyotrophic lateral sclerosis patients. Acta Neuropathol 86:55–64. [DOI] [PubMed] [Google Scholar]
- Nobili F, Mazzei D, Dessi B, Morbelli S, Brugnolo A, Barbieri P, Girtler N, Sambuceti G, Rodriguez G, Pagani M (2010): Unawareness of memory deficit in amnestic MCI: FDG‐PET findings. J Alzheimer's Dis JAD 22:993–1003. [DOI] [PubMed] [Google Scholar]
- Nobili F, Salmaso D, Morbelli S, Girtler N, Piccardo A, Brugnolo A, Dessi B, Larsson SA, Rodriguez G, Pagani M (2008): Principal component analysis of FDG PET in amnestic MCI. Eur J Nucl Med Mol Imaging 35:2191–2202. [DOI] [PubMed] [Google Scholar]
- O'Toole AJ, Jiang F, Abdi H, Penard N, Dunlop JP, Parent MA (2007): Theoretical, statistical, and practical perspectives on pattern‐based classification approaches to the analysis of functional neuroimaging data. J Cogn Neurosci 19:1735–1752. [DOI] [PubMed] [Google Scholar]
- Pagani M, Chio A, Valentini MC, Oberg J, Nobili F, Calvo A, Moglia C, Bertuzzo D, Morbelli S, De Carli F, P Fania, A Cistaro (2014): Functional pattern of brain FDG‐PET in amyotrophic lateral sclerosis. Neurology 83:1067–1074. [DOI] [PubMed] [Google Scholar]
- Pagani MD, Carli F, Morbelli S, Oberg J, Chincarini A, Frisoni GB, Galluzzi S, Perneczky R, Drzezga A, van Berckel BN, R Ossenkoppele, E Guedj, M Didic, A Brugnolo, A Picco, D Arnaldi, M Ferrara, A Buschiazzo, G Sambuceti, F Nobili (2015): Volume of interest‐based [(18)F]fluorodeoxyglucose PET discriminates MCI converting to Alzheimer's disease from healthy controls. A European Alzheimer's Disease Consortium (EADC) study. Neuroimage Clin 7:34–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagani M, Dessi B, Morbelli S, Brugnolo A, Salmaso D, Piccini A, Mazzei D, Villavecchia G, Larsson SA, Rodriguez G, F Nobili (2010): MCI patients declining and not‐declining at mid‐term follow‐up: FDG‐PET findings. Curr Alzheimer Res 7:287–294. [DOI] [PubMed] [Google Scholar]
- Park HJ, Kim JJ, Youn T, Lee DS, Lee MC, Kwon JS (2003): Independent component model for cognitive functions of multiple subjects using [15O]H2O PET images. Hum Brain Mapp 18:284–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philips T, Robberecht W (2011): Neuroinflammation in amyotrophic lateral sclerosis: Role of glial activation in motor neuron disease. Lancet Neurol 10:253–263. [DOI] [PubMed] [Google Scholar]
- Prell T, Peschel T, Hartung V, Kaufmann J, Klauschies R, Bodammer N, Kollewe K, Dengler R, Grosskreutz J (2013): Diffusion tensor imaging patterns differ in bulbar and limb onset amyotrophic lateral sclerosis. Clin Neurol Neurosurg 115:1281–1287. [DOI] [PubMed] [Google Scholar]
- Prudlo J, Bissbort C, Glass A, Grossmann A, Hauenstein K, Benecke R, Teipel SJ (2012): White matter pathology in ALS and lower motor neuron ALS variants: A diffusion tensor imaging study using tract‐based spatial statistics. J Neurol 259:1848–1859. [DOI] [PubMed] [Google Scholar]
- Qin GS, Hotilovac L (2008): Comparison of non‐parametric confidence intervals for the area under the ROC curve of a continuous‐scale diagnostic test. Stat Methods Med Res 17:207–221. [DOI] [PubMed] [Google Scholar]
- Quartuccio N, Van Weehaeghe D, Cistaro A, Jonsson C, Van Laere K, Pagani M (2014): Positron emission tomography neuroimaging in amyotrophic lateral sclerosis: What is new? Q J Nucl Med Mol Imaging 58:344–354. [PubMed] [Google Scholar]
- Rajagopalan V, Pioro EP (2015): Comparing brain structural MRI and metabolic FDG‐PET changes in patients with ALS‐FTD: 'the chicken or the egg?' question. J Neurol Neurosurg Psychiatry 86:952–958. [DOI] [PubMed] [Google Scholar]
- Shaffer JL, Petrella JR, Sheldon FC, Choudhury KR, Calhoun VD, Coleman RE, Doraiswamy PM (2013): Predicting cognitive decline in subjects at risk for Alzheimer disease by using combined cerebrospinal fluid, MR imaging, and PET biomarkers. Radiology 266:583–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefan K, Kunesch E, Benecke R, Classen J (2001): Effects of riluzole on cortical excitability in patients with amyotrophic lateral sclerosis. Ann Neurol 49:536–539. [PubMed] [Google Scholar]
- Storti SF, Formaggio E, Nordio R, Manganotti P, Fiaschi A, Bertoldo A, Toffolo GM (2013): Automatic selection of resting‐state networks with functional magnetic resonance imaging. Front Neurosci 7:72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tax DMJ, Duin RPW (1999): Support vector domain description. Pattern Recogn Lett 20:1191–1199. [Google Scholar]
- Thivard L, Pradat PF, Lehericy S, Lacomblez L, Dormont D, Chiras J, Benali H, Meininger V (2007): Diffusion tensor imaging and voxel based morphometry study in amyotrophic lateral sclerosis: Relationships with motor disability. J Neurol Neurosurg Psychiatry 78:889–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toussaint PJ, Perlbarg V, Bellec P, Desarnaud S, Lacomblez L, Doyon J, Habert MO, Benali H (2012): Resting state FDG‐PET functional connectivity as an early biomarker of Alzheimer's disease using conjoint univariate and independent component analyses. Neuroimage 63:936–946. [DOI] [PubMed] [Google Scholar]
- Turner MR, Cagnin A, Turkheimer FE, Miller CC, Shaw CE, Brooks DJ, Leigh PN, Banati RB (2004): Evidence of widespread cerebral microglial activation in amyotrophic lateral sclerosis: An [11C](R)‐PK11195 positron emission tomography study. Neurobiol Dis 15:601–609. [DOI] [PubMed] [Google Scholar]
- Turner MR, Hammers A, Allsop J, Al‐Chalabi A, Shaw CE, Brooks DJ, Leigh PN, Andersen PM (2007): Volumetric cortical loss in sporadic and familial amyotrophic lateral sclerosis. Amyotroph Lateral Scler 8:343–347. [DOI] [PubMed] [Google Scholar]
- Turner MR, Rabiner EA, Hammers A, Al‐Chalabi A, Grasby PM, Shaw CE, Brooks DJ, Leigh PN (2005): [11C]‐WAY100635 PET demonstrates marked 5‐HT1A receptor changes in sporadic ALS. Brain 128:896–905. [DOI] [PubMed] [Google Scholar]
- Tzourio‐Mazoyer N, Landeau B, Papathanassiou D, Crivello F, Etard O, Delcroix N, Mazoyer B, Joliot M (2002): Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single‐subject brain. Neuroimage 15:273–289. [DOI] [PubMed] [Google Scholar]
- Van Laere K, Vanhee A, Verschueren J, De Coster L, Driesen A, Dupont P, Robberecht W, Van Damme P (2014): Value of 18fluorodeoxyglucose‐positron‐emission tomography in amyotrophic lateral sclerosis: A prospective study. JAMA Neurol 71:553–561. [DOI] [PubMed] [Google Scholar]
- Varrone A, Asenbaum S, Vander Borght T, Booij J, Nobili F, Nagren K, Darcourt J, Kapucu OL, Tatsch K, Bartenstein P, Van Laere K (2009): EANM procedure guidelines for PET brain imaging using [18F]FDG, version 2. Eur J Nucl Med Mol Imaging 36:2103–2110. [DOI] [PubMed] [Google Scholar]
- Verstraete E, van den Heuvel MP, Veldink JH, Blanken N, Mandl RC, Hulshoff Pol HE, van den Berg LH (2010): Motor network degeneration in amyotrophic lateral sclerosis: A structural and functional connectivity study. PLoS One 5:e13664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vucic S, Nicholson GA, Kiernan MC (2008): Cortical hyperexcitability may precede the onset of familial amyotrophic lateral sclerosis. Brain 131:1540–1550. [DOI] [PubMed] [Google Scholar]
- Welsh RC, Jelsone‐Swain LM, Foerster BR (2013): The utility of independent component analysis and machine learning in the identification of the amyotrophic lateral sclerosis diseased brain. Front Hum Neurosci 7:251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willette AA, Calhoun VD, Egan JM, Kapogiannis D (2014): Prognostic classification of mild cognitive impairment and Alzheimer's disease: MRI independent component analysis. Psychiatry Res 224:81–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods RP (1996): Modeling for intergroup comparisons of imaging data. Neuroimage 4:S84–S94. [DOI] [PubMed] [Google Scholar]
- Yamanaka K, Chun SJ, Boillee S, Fujimori‐Tonou N, Yamashita H, Gutmann DH, Takahashi R, Misawa H, Cleveland DW (2008): Astrocytes as determinants of disease progression in inherited amyotrophic lateral sclerosis. Nat Neurosci 11:251–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F, Gong H, Li F, Zhuang Y, Zang Y, Xu R, Wang Z (2013): Altered motor network functional connectivity in amyotrophic lateral sclerosis: A resting‐state functional magnetic resonance imaging study. Neuroreport 24:657–662. [DOI] [PubMed] [Google Scholar]


