Abstract
Juvenile myoclonic epilepsy (JME) is a common subtype of idiopathic generalized epilepsies (IGEs) and is characterized by myoclonic jerks, tonic‐clonic seizures and infrequent absence seizures. The network notion has been proposed to better characterize epilepsy. However, many issues remain not fully understood in JME, such as the associations between discharge‐affecting networks and the relationships among resting‐state networks. In this project, eigenspace maximal information canonical correlation analysis (emiCCA) and functional network connectivity (FNC) analysis were applied to simultaneous EEG‐fMRI data from JME patients. The main findings of our study are as follows: discharge‐affecting networks comprising the default model (DMN), self‐reference (SRN), basal ganglia (BGN) and frontal networks have linear and nonlinear relationships with epileptic discharge information in JME patients; the DMN, SRN and BGN have dense/specific associations with discharge‐affecting networks as well as resting‐state networks; and compared with controls, significantly increased FNCs between the salience network (SN) and resting‐state networks are found in JME patients. These findings suggest that the BGN, DMN and SRN may play intermediary roles in the modulation and propagation of epileptic discharges. These roles further tend to disturb the switching function of the SN in JME patients. We also postulate that emiCCA and FNC analysis may provide a potential analysis platform to provide insights into our understanding of the pathophysiological mechanism of epilepsy subtypes such as JME. Hum Brain Mapp 37:3515–3529, 2016. © 2016 Wiley Periodicals, Inc.
Keywords: Juvenile myoclonic epilepsy, EEG‐fMRI, nonlinearity, complex discharge‐affecting networks, functional network connectivity
INTRODUCTION
Juvenile myoclonic epilepsy (JME), characterized by myoclonic jerks occurring at full consciousness within hours after awakening or sleep deprivation [Koepp et al., 2014], tonic‐clonic seizures and infrequent absence seizures, is a common subtype of idiopathic generalized epilepsies (IGE) associated with an age‐related onset of seizures [1989; Genton et al., 2013; Janz, 1985]. The interictal and ictal electrophysiological features of JME involve rapid, often irregular 3–6 Hz generalized spike‐wave discharges (GSWDs) or polyspike‐wave discharges, with a fronto‐central predominance [Janz, 1985; Koepp et al., 2014]. Because of no underlying structural brain lesion and/or other neurologic signs, the mechanism of JME is not fully understood till now.
Recently, the notion of network abnormality has been suggested to further understand the mechanism of epilepsy without lesion such as JME [Centeno and Carmichael, 2014; Spencer, 2002]. Using network concept, generalized epileptic seizures are suggested as occurring in and rapidly engaging bilaterally distributed networks [Berg et al., 2010]. The thalamocortical network has been found to be characteristically involved in the epileptic activity in a number of studies in IGE [Aghakhani et al., 2004; Gotman et al., 2005; Hamandi et al., 2006; Li et al., 2009; Xue et al., 2014], and the thalamofrontal circuit, which is a frontal lobe variant of thalamocortical network, is further suggested to be involved in JME [Koepp, 2005; Koepp et al., 2014; Pulsipher et al., 2009]. Using the network framework, there are also various possible ways to interpret the generation and propagation of epileptic discharges. For example, discharges generated from a focal epileptic zone may entrain a large neural network [Spencer, 2002]; additionally, the discharges may be driven by the pattern of connections in brain networks [Terry et al., 2012]. Therefore, we presume that abnormality of networks in JME may be affected by epileptic discharges.
With the development of functional magnetic resonance imaging (fMRI), resting‐state fMRI has been widely used as a powerful tool for studying mechanisms of functional alterations in epilepsy patients. Abnormalities in several resting‐state networks, including the default modal, basal ganglia, sensorimotor networks, etc., have been found in epilepsy patients [Luo et al., 2012, 2015; Zhang et al., 2014, 2015]. Furthermore, using functional network connectivity (FNC) [Jafri et al., 2008], effects of epileptic discharges on interactions between resting‐state networks has been investigated to gain a deeper understanding of epilepsy [Centeno and Carmichael, 2014; Li et al., 2015; Luo et al., 2011a; Zhang et al., 2014]. These findings may imply potential impacts of epileptic discharges on resting‐state networks in epilepsy patients. However, because of nonrecording of electroencephalography (EEG) during fMRI scanning, these effects are less directly investigated in JME. Although EEG data are simultaneously recorded, the EEG data are only simply utilized for analysis such as identification of discharge subsessions, group classification for comparative investigation or correlation analysis in epilepsy [Luo et al., 2012; Zhang et al., 2014].
Due to the noninvasiveness and complementarity of the spatio‐temporal resolution, simultaneous EEG and fMRI are quickly becoming a popular method to study human brain function. The traditional approach of combining simultaneous EEG and fMRI in epilepsy such as general linear model (GLM) assumes that the relationships between EEG and fMRI are linear [Gotman and Pittau, 2011; Huster et al., 2012]. However, potential nonlinearity in EEG‐fMRI integration should be acknowledged, and the nonlinearity between epileptic discharges and blood oxygen level‐dependent (BOLD) signals may be an important objective of JME studies. Because nonlinearity may be caused by neurovascular coupling [Liu et al., 2010], HRF variety in epilepsy patient and spike location [Beers et al., 2015] and even by the variety of methods available to quantify the multimodal signals [He et al., 2011]. Currently, a new method, named eigenspace maximal information canonical correlation analysis (emiCCA) [Dong et al., 2015], was developed to uncover the underlying linear and nonlinear relationships between two datasets. As an unsupervised multivariate method, the advantages of emiCCA are: (1) it unbiasedly uncovers the relationships between EEG discharges and fMRI networks in JME, while univariate method such as GLM cannot tackle the multivariate problem of comparing two datasets; (2) it further captures the nonlinear relationships between datasets to cope with possible nonlinear effects; (3) it is a data‐driven method without the practical choices of the key parameter settings, thus, it may be suitable for studying of a disease without being fully understood such as JME; (4) it identifies associated variables of datasets in the original data space, thus, it may have potential to quantify the effects of epileptic discharges on networks; and (5) it has the potential for applications in other types of epilepsy involving multimodal fusion.
The main aims of the current study are (1) to detect linear and nonlinear relationships between datasets from EEG and fMRI in JME; some networks, termed discharge‐affecting networks, refer to the brain regions/resting‐state networks affected by the generation and propagation of epileptic discharges in JME; and (2) to investigate the associations between identified discharge‐affecting networks as well as the relationships among the resting‐state networks which may be influenced by aspects of epilepsy in JME. In this study, group level independent component analysis (ICA) [Calhoun et al., 2001] was used to extract spatiotemporal features (i.e., spatial components and the corresponding time courses); then, emiCCA was utilized to investigate the potential discharge‐affecting networks in spatially independent components. Further, associations between discharge‐affecting networks as well as the relationships among meaningful resting‐state networks were investigated using functional network connectivity analysis.
MATERIALS AND METHODS
Subjects
Eighteen JME patients (12 females/6 males; mean age = 21 years; standard deviation = 7 years; age range = 15–34 years) were recruited for this study. All patients were diagnosed by neurologists (P.W. and Y.Z.) based on the clinical information consistent with the International League Against Epilepsy (ILAE) guidelines [Engel and International League Against, 2001]. Routine examinations of CT and MRI scanning showed no structural abnormalities, and 24‐h scalp EEG recordings demonstrated 3–6 Hz generalized spike‐wave or polyspike‐wave discharges. The detailed demographic information and the clinical characteristics of JME patients are summarized in Table 1. Age‐ and gender‐matched, healthy subjects were also recruited as controls (12 females/6 males; mean age = 23 years; standard deviation = 5.5 years; age range = 17–35 years). A written consent form was obtained from each of the patients and controls. The study was approved by the Ethics Committee of the University of Electronic Science and Technology of China (UESTC).
Table 1.
Detailed demographic information and clinical characteristics of JME patients
| Patient # |
Male/Female |
Age | Age of onset (years) | Family history | Antiepileptic drugs | Frequency of GSWDs | No. of volumes with GSWDs in selected run |
|---|---|---|---|---|---|---|---|
| 1 | F | 17 | 10 | — | VPA | 2 Hz | 1 |
| 2 | F | 17 | 14 | — | LTG | 3 Hz | 20 |
| 3 | F | 33 | 20 | — | VPM | 6 Hz | 6 |
| 4 | M | 22 | 8 | — | VPM | 4 Hz | 3 |
| 5 | F | 19 | 12 | Uncle with GTCS | MgV | 3 Hz | 6 |
| 6 | F | 20 | 6 | — | VPM/LTG | 2 Hz | 111 |
| 7 | M | 15 | 5 | — | — | 3∼3.5 Hz | 12 |
| 8 | F | 22 | 14 | — | VPA | 3 Hz | 2 |
| 9 | F | 17 | 3 | Sister with JME | VPA | 3∼3.5 Hz | 2 |
| 10 | F | 17 | 13 | Sister with JME | VPA | 2 Hz | 8 |
| 11 | F | 29 | 10 | — | TCM/VPM | 3∼3.5 Hz | 2 |
| 12 | M | 18 | 14 | — | VPM | 5 Hz | 2 |
| 13 | F | 27 | 16 | Daughter with GTCS | — | 4 Hz | 3 |
| 14 | F | 21 | 11 | — | VPM | 2 Hz | 117 |
| 15 | M | 10 | 5 | Brother with JME | VPM | 4 Hz | 2 |
| 16 | M | 13 | 9 | — | VPA | 3∼3.5 Hz | 4 |
| 17 | M | 34 | 14 | — | TCM/VPM | 3.5∼4 Hz | 4 |
| 18 | F | 34 | 18 | — | — | 3 Hz | 6 |
GSWDs: generalized spike‐wave discharges; GTCS: generalized tonic‐clonic seizures; VPA: valproic acid; LTG: lamotrigine; VPM: valpromide; MgV: magnesium valproate; TCM, traditional Chinese medicine.
EEG and fMRI Recording
In this study, simultaneous EEG data of JME patients were recorded using a 64‐channel MR compatible EEG system (Neuroscan, Charlotte, NC). Sixty‐two EEG electrodes were distributed according to 10–20 cap system, and two additional electrodes, one placed below the left eye and another attached approximately 4 cm below the clavicle, were used for electrocardiogram (ECG) recording. The sampling rate was set at 5,000 Hz.
Simultaneous fMRI data of JME patients were collected using a 3‐T MRI scanner (Discovery MR750, GE) in the Center for Information in Medicine of UESTC. T1‐weighted images were acquired using a 3‐dimensional fast spoiled gradient echo (3D‐FSPGR) sequence, and the scan parameters were as follows: 152 axial slices, TR/TE= 5.936 ms/1.956 ms, flip angle = 9°, field of view = 25.6 × 25.6 cm2, voxel size = 1 × 1 × 1 mm3 and slice thickness (no gap) = 1 mm. Functional images were collected using a gradient‐echo echo‐planar imaging (EPI) sequence, and the scan parameters were as follows: 35 slices per volume, TR/TE = 2,000 ms/30 ms, flip angle = 90°, matrix size = 64 × 64, field of view = 24 × 24 cm2 and slice thickness = 4 mm. A total of 255 volumes were obtained over each run period (510 s), and 5 repeated runs (a total of 40.25 mins) were conducted for each JME patient. During scanning, all JME patients were instructed to close their eyes and relax without falling asleep. In addition, for controls, resting‐state fMRI data were separately collected over a run period using identical scan parameters, as described above, and EEG data were not recorded.
Data Preprocessing
All EEG data were primarily analyzed using Curry 7 (Neuroscan software). Briefly, gradient artifacts were removed using a local average artifact template procedure [Allen et al., 2000]; then, the EEG data were pass‐band filtered (1–30 Hz) and down‐sampled to 250 Hz and the ballistocardiogram (BCG) artifacts were removed by using the OBS‐based BCG correction [Niazy et al., 2005]. Finally, all EEG data were re‐referenced to the Cz reference, and onsets of generalized spike‐wave discharges (GSWDs) were independently identified by two experienced neurologists (both in agreement).
FMRI data of both the patients and controls were analyzed in an identical fashion. The fMRI data preprocessing comprised slice time correction, 3D motion detection and correction, spatial normalization (3 × 3 × 3 mm3) and spatial smoothing (8‐mm full‐width at half maximum (FWHM) of an isotropic Gaussian filter). The analysis was conducted using SPM8 software (http://www.fil.ion.ucl.ac.uk/spm/software/spm8/).
In JME patients, runs with small head motion (translation < 1 mm and rotation < 1°), high cooperation of patients (e.g., seldom complained of discomfort, no eye blinks, no falling asleep, etc.) and high‐quality of both EEG and fMRI data were first identified. Then, one run with maximum amount of GSWDs was selected for further analysis. In addition, two sample t‐test yielded no significant differences between head motions of JME patients and controls (P > 0.05).
Discharge‐Affecting Network Analysis
To ascertain discharge‐affecting networks in JME patients, a specific approach was conducted in this work (Fig. 1). To remove potential artifacts on individual‐subject data, nuisance parameters such as six head motion parameters, linear trend, individual white matter (WM) and cerebrospinal fluid (CSF) mean signals were firstly regressed out from fMRI data. Then, because epileptic discharges only occurred in JME patients, group spatial independent component analysis (ICA) [Calhoun et al., 2001] was separately applied in JME patients and controls to extract the spatiotemporal features (i.e., spatial components and the corresponding time courses). The optimal number of independent components (ICs) was estimated at 42 based on the minimum description length criteria [Li et al., 2007], and reliability of the ICs was achieved by using ICASSO (30 times) [Himberg et al., 2004]. Then, IC time courses were concatenated across JME patients to increase the statistical power and were defined as dataset Y. Three components were first visually discarded due to possible residual artifacts (e.g., located in the cerebrospinal fluid, Supporting Information Fig. S4) for further investigation. For EEG data, the onset times of GSWDs were convolved with 4 SPM canonical HRFs peaking at 3, 5, 7 and 9 s [Bagshaw et al., 2004], 1 Glover HRF [Glover, 1999] and 1 single Gamma HRF. And the dataset X was defined by a design matrix containing the convolved onsets of GSWDs, which were also concatenated across JME patients. Then, emiCCA [Dong et al., 2015] was applied (http://www.neuro.uestc.edu.cn/emiCCA.html). Briefly, the crucial point of emiCCA is that the eigenvectors and eigenvalues from the eigenspaces of the maximal information coefficient (MIC) [Reshef et al., 2011] matrix of two datasets are utilized as a new measure for assessing the relationships between datasets. Using emiCCA, weight values of each IC time courses in dataset Y, which represented its weightiness to the relationships between two datasets, were estimated. And, ICs with weights exceeding the 1.5 standard deviations of weights values corresponding to significant (P < 0.001, Bonferroni‐corrected) maximal information eigen coefficients (MIECs, which quantify the relationships between the two datasets) were identified as the discharge‐affecting networks. More details of emiCCA can be seen in Supporting Information A. In addition, to assess the performance of the aforementioned approach, traditional EEG‐informed fMRI analysis in epilepsy was also conducted (discharge‐affecting brain regions were studied using GLM, which implemented 4 canonical HRFs peaking at 3, 5, 7 and 9 s).
Figure 1.
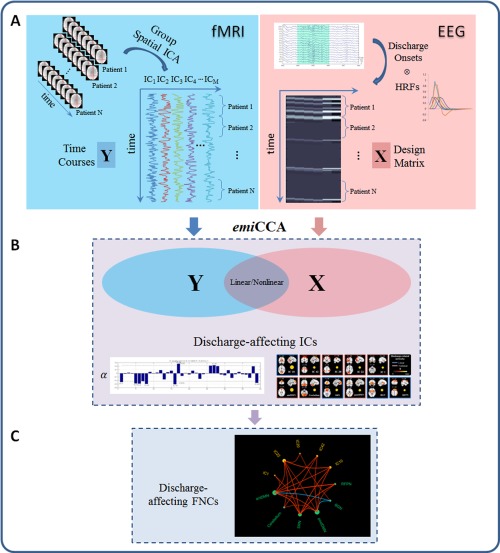
The framework of discharge‐affecting network analysis using emiCCA. A: For fMRI data, group ICA was first applied to extract the spatiotemporal features of the fMRI data; then, the IC time courses were concatenated across JME patients and defined as dataset Y. For EEG data, the onsets of GSWDs were first identified by neurologists. Then, the dataset X was defined by a design matrix containing the onsets of GSWDs, which were convolved with 4 SPM canonical HRFs (peaking at 3, 5, 7 and 9 s), 1 Glover HRF and 1 single Gamma HRF. B: emiCCA was applied to identify the linear and nonlinear discharge‐affecting components with weights (α) exceeding the 1.5 standard deviations of weight values corresponding to the significant maximal information eigen coefficients (MIECs). C: The maximal time‐lagged correlation method was used to examine the possible functional network connectivity between those discharge‐affecting networks identified by emiCCA. [Color figure can be viewed in the online issue, which is available at http://wileyonlinelibrary.com.]
Functional Network Connectivity Analysis
Spatial ICA assumes that the time courses of brain regions within one component are synchronous [Calhoun et al., 2004]. However, the components are spatially independent, and significant temporal dependency may exist between the time courses of components. To examine the possible associations between those discharge‐affecting networks identified by emiCCA, a constrained maximal time‐lagged correlation method, known as functional network connectivity analysis [Jafri et al., 2008], was performed. Briefly, for each subject, time courses of the identified ICs were first filtered (0.01–0.08 Hz), interpolated and circularly shifted from −5 to +5 s. The absolute maximal lagged Pearson's correlation was then calculated and Fisher z‐shifted. Lastly, temporal associations between any 2 discharge‐affecting networks were examined by the one‐sample t‐test [P < 0.05, false discovery rate (FDR) corrected]. To further assess the impacts of JME on associations between resting‐state networks, FNC analysis was also performed between meaningful resting‐state networks in JME patients and controls. Two‐sample t‐tests were performed to study the difference of FNCs between JME patients and controls (P < 0.05, FDR‐corrected). The lag values of FNCs in each group were examined by the two‐sided sign test (P < 0.05). Further, a two‐sided Wilcoxon rank sum test was used to study the difference of lags between JME patients and controls (P < 0.05). In addition, resting‐state networks were first visually inspected and matched (independently by L.D. and C.L.) according to the networks described in earlier studies [Biswal et al., 2010; D'Argembeau et al., 2005; Damoiseaux et al., 2006, 2008; Luo et al., 2011a, 2012; Smith et al., 2009]. Next, for the disagreement of spatially matched resting‐state networks, the eta2 coefficient [Cohen et al., 2008] between networks of each group were further calculated. And the networks were finally matched corresponding to the maximal eta2. The eta2 is equal to the fraction of the variance in one image accounted for by variance in a second image where comparisons are done on a point by point basis. In this work, 12 components were considered as meaningful resting‐state networks, which included the posterior part of the default mode network (postDMN), anterior part of the DMN (antDMN), self‐referential network (SRN), salience network (SN), left and right lateral frontoparietal networks (LFPN/RFPN), basal ganglia network (BGN), sensorimotor network (SMN), primary and extra‐striate visual networks (primVN/extraVN), auditory network (AN) and Cerebellum. Here, six resting‐state networks in JME patients were labeled as follows (other six resting‐state networks can be seen in the section of results):
LFPN: the left lateral frontoparietal network showed spatial patterns consisting of the left superior frontal gyrus [Brodmann area (BA) 6/8], angular gyrus (BA39) and precuneus (BA7). The LFPN and RFPN were the only maps to be strongly lateralized and were largely left–right mirrors of each other.
SN: the salience network [Luo et al., 2014; Manoliu et al., 2013; Seeley et al., 2007; Uddin, 2015], showed spatial patterns mainly consisting of the bilateral insula (BA47) and supplementary motor area (BA6).
AN: the auditory network primarily encompassed the bilateral superior temporal gyrus (BA22/41).
SMN: the sensorimotor network, corresponding to sensory‐motor function [Biswal et al., 1995; Fox et al., 2006], included the bilateral medial frontal gyrus (BA6) and paracentral lobule (BA6).
primVN: the primary visual network showed spatial patterns consisting of the bilateral lingual gyrus (BA18) and cuneus (BA18).extraVN: the extra‐striate visual network included the bilateral fusiform gyrus (BA19) and inferior occipital gyrus (BA19).
Correlation between Clinical Features and FNCs
To detect the underlying relationships between discharge‐affecting FNCs (Fisher's z‐score) and clinical features (including the age of epilepsy onset and the duration of epilepsy), partial correlation analysis was performed in JME patients, while controlling for the gender (P < 0.05).
RESULTS
Discharge‐Affecting Networks
Using emiCCA, significant relationships (P < 0.001, Bonferroni‐corrected for all MIECs) existed between the EEG epileptic discharges (dataset X) and the fMRI time courses (dataset Y). The EEG figure of discharge waves of a typical JME patient can be seen in Supporting Information B (Supporting Information Fig. S6). Eleven discharge‐affecting networks that corresponded to larger weights (> 1.5 standard deviations of weights) were also identified and presented as follows [P < 0.05, family wise error (FWE) corrected, Fig. 2 and Table 2]:
Figure 2.
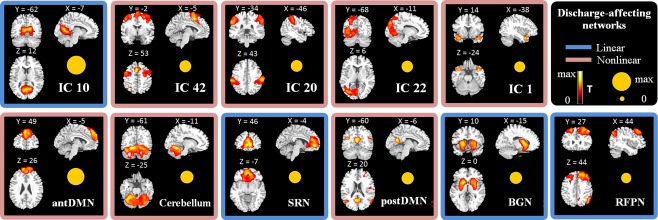
Discharge‐affecting networks identified by emiCCA in JME patients. The T‐maps of these spatial components (P < 0.05, FWE‐corrected) are shown. The size of the yellow circle presents the weight of each time course in emiCCA, which represent the weightiness of each piece of dataset Y. Linear (blue border) or nonlinear (red border) relationships between the fMRI time courses of networks and the EEG discharges are also shown. [Color figure can be viewed in the online issue, which is available at http://wileyonlinelibrary.com.]
Table 2.
Spatial distributions of discharge‐affecting networks identified by emiCCA in JME patients (one‐sample t‐test, P < 0.05, FWE‐corrected)
| MNI coordinates | L/R | Lobe |
Brodmannarea |
T | Voxels | |||
|---|---|---|---|---|---|---|---|---|
| x | y | z | ||||||
| IC 10 | 12 | −51 | −3 | R | Lingual Gyrus | BA18/19 | 30.16 | 2,807 |
| −21 | −54 | 29 | L | Lingual Gyrus | BA19 | 26.97 | ||
| IC 42 | −3 | 12 | 54 | L | Supplementary Motor Area | BA6 | 24.19 | 3,033 |
| −27 | −72 | 54 | L | Superior Parietal Lobule | BA7 | 11.07 | 56 | |
| −30 | 54 | 21 | L | Middle Frontal Gyrus | BA10 | 10.58 | 36 | |
| 57 | −39 | 21 | R | Superior Temporal Gyrus | BA22 | 9.48 | 42 | |
| 51 | −63 | −15 | R | Middle Occipital Gyrus | BA37 | 8.97 | 27 | |
| −42 | −66 | −27 | L | Declive | 8.87 | 28 | ||
| IC 20 | −60 | −27 | 30 | L | Inferior Parietal Lobule | BA40 | 26.78 | 1,037 |
| 66 | −18 | 24 | R | Inferior Parietal Lobule | BA40 | 22.42 | 1,228 | |
| 57 | 12 | 15 | R | Inferior Frontal Gyrus | BA44 | 14.89 | 226 | |
| IC 22 | −24 | −81 | 27 | L | Cuneus | BA18/19 | 24.93 | 3,534 |
| −24 | −81 | 39 | L | Middle Occipital Gyrus | BA19 | 21.62 | ||
| 39 | 18 | −15 | R | Inferior Frontal Gyrus | BA47 | 14.01 | 26 | |
| −45 | −51 | 60 | L | Inferior Parietal Lobule | BA40 | 10.72 | 27 | |
| IC 1 | 39 | 6 | −27 | R | Superior Temporal Gyrus | BA38 | 17.37 | 261 |
| −39 | 9 | −30 | L | Superior Temporal Gyrus | BA38 | 14.82 | 294 | |
| antDMN | −12 | 60 | 21 | L | Superior Frontal Gyrus | BA9/10 | 24.82 | 2,033 |
| 0 | −9 | 42 | Cingulate Gyrus | BA24 | 11.29 | 63 | ||
| 51 | 36 | 9 | R | Inferior Frontal Gyrus | BA46 | 11.07 | 25 | |
| Cerebellum | −18 | −66 | −15 | L | Cerebellum Posterior Lobe | 22.33 | 4,520 | |
| 30 | −54 | −18 | R | Declive | 22.19 | |||
| SRN | 9 | 42 | 0 | R | Anterior Cingulate | BA32 | 27.06 | 2,739 |
| −3 | 48 | −6 | L | Medial Frontal Gyrus | BA10 | 25.42 | ||
| postDMN | 42 | −75 | 30 | R | Angular Gyrus | BA39 | 20.21 | 300 |
| −6 | −60 | 15 | L | Precuneus | BA31 | 19.49 | 439 | |
| −48 | −66 | 21 | L | Middle Temporal Gyrus | BA39 | 15.36 | 408 | |
| 36 | 48 | 24 | R | Middle Frontal Gyrus | BA10 | 10.74 | 114 | |
| 3 | −9 | 78 | R | Superior Frontal Gyrus | BA6 | 9.57 | 23 | |
| 48 | −9 | 15 | R | Sub‐Gyral | BA43 | 9.46 | 62 | |
| BGN | −21 | 0 | 9 | L | Putamen | 28.08 | 1,549 | |
| 27 | 3 | −12 | R | Putamen | 21.87 | 1,401 | ||
| RFPN | 48 | 24 | 51 | R | Middle Frontal Gyrus | BA8/9 | 24.8 | 1,793 |
| 51 | −60 | 36 | R | Angular Gyrus | BA39 | 19.89 | 512 | |
| −24 | 30 | 48 | L | Middle Frontal Gyrus | BA8 | 17.3 | 823 | |
| −51 | −12 | 3 | L | Superior Temporal Gyrus | BA22 | 10.12 | 69 | |
| 9 | −66 | 45 | R | Precuneus | BA7 | 9.73 | 42 | |
IC10: the spatial distributions consisting of the bilateral lingual gyrus (BA18/19). IC42: the spatial patterns consisting of the supplementary motor area (BA6), superior parietal lobule (BA7), middle frontal gyrus (BA10) and superior temporal gyrus (BA22). IC20: the spatial distributions consisting of the bilateral inferior parietal lobule (BA40) and right inferior frontal gyrus (BA44). IC22: the spatial distributions mainly consisting of the left cuneus (BA18/19), middle occipital gyrus (BA19) and inferior parietal lobule (BA40). IC1: the spatial patterns primarily encompassing the bilateral superior temporal gyrus (temporal polar, BA38).
antDMN: the anterior part of the DMN; the spatial patterns mainly consisting of the bilateral superior frontal gyrus and middle frontal gyrus (BA9/10).
Cerebellum: the spatial patterns primarily encompassing the cerebellum posterior lobe and declive.
SRN: the self‐referential network putatively related to self‐referential mental activity [D'Argembeau et al., 2005], mainly including the bilateral medial‐ventral prefrontal cortex (BA10) and anterior cingulate (BA32).
postDMN: the spatial patterns primarily encompassing the precuneus (BA31) and bilateral angular gyrus (BA39), as well as the middle frontal gyrus (BA10) and sub‐gyral (BA43). Interestingly, the DMN was split into two components, the anterior areas (antDMN) and the posterior areas (postDMN), in the current study. A similar decomposition of the DMN has been observed previously [Damoiseaux et al., 2006, 2008; Luo et al., 2011a; Rombouts et al., 2009].
BGN: the basal ganglia network; including the putamen, caudate nucleus, pallidum and parahippocampus, which are the main nuclei of the basal ganglia [Luo et al., 2012; Robinson et al., 2009].
RFPN: the right lateral frontoparietal network; the spatial patterns consisting of the bilateral middle frontal gyrus (BA8/9), right angular gyrus (BA39) and precuneus (BA7). These clusters lateralized to the right hemisphere putatively associated with the dorsal attention network [Corbetta and Shulman, 2002].
In addition, the discharge‐affecting brain regions in JME patients revealed by traditional EEG‐informed fMRI analysis were mainly located in the left superior frontal gyrus (BA6), bilateral anterior cingulate (BA32), supplementary motor area (BA32), insula (BA47), temporal polar (BA28), thalamus, pallidum, putamen and cerebellum (Supporting Information Fig. S1 and Table S1). Furthermore, to illustrate that linearity and nonlinearity may co‐exist in neural imaging, trend lines (referenced by polynomial curve fitting, order = 1, 2, 3 or 4) between the fMRI IC time courses of discharge‐affecting networks and the assumed discharge‐affecting BOLD responses (onset times of GSWDs convolved with the optimal HRF) were also analyzed; these trend lines are shown in Supporting Information Fig. S2. Briefly, the IC10, SRN, BGN and RFPN were linearly related to the assumed discharge‐affecting BOLD responses, while the relationships of the IC42, IC20, IC22, IC1, antDMN, cerebellum and postDMN were parabolic, cubic or biquadrate.
Results of FNC Analysis
To understand the associations among the discharge‐affecting networks, FNC analysis was performed. Using one‐sample t‐test, 24 out of the 55 possible combinations were significant (P < 0.05, FDR‐corrected) in the discharge‐affecting FNC diagram for JME patients (Fig. 3). A large number of positive significant associations were detected among discharge‐affecting networks, while only two negative connectivities between BGN and antDMN/postDMN were found. Figure 3 also shows the distribution of degrees across these networks and the lags. The direction of each arrow represents the lag between the two components. For example, in Figure 3, an arrow connects IC10 and antDMN (points at antDMN), representing that antDMN lags IC10 by certain time units. Notably, the antDMN, postDMN and SRN show more connections with networks, while the remaining networks show fewer connections.
Figure 3.
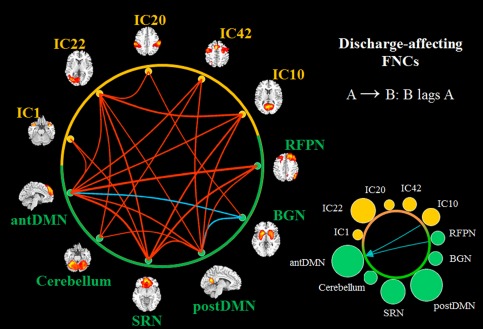
Discharge‐affecting FNCs in JME patients (P < 0.05, FDR‐corrected). The lines show the significant positive (red) and negative (blue) correlation connections in JME patients from the 55 possible correlation combinations. The size of circles in the bottom right represents the degree of the node in functional network connectivity (containing positive and negative), and the green color represents the networks, which are also the resting‐state networks. The lags, which are the amount of delay between component time courses, are also shown on the right (P < 0.05). A → B: component B lags component A by some calculated seconds. [Color figure can be viewed in the online issue, which is available at http://wileyonlinelibrary.com.]
To further understand the impact of JME on the relationships among resting‐state networks, six other resting‐state networks, which were not discharge‐affecting networks, were identified from networks described in earlier studies [Biswal et al., 2010; Damoiseaux et al., 2008; Smith et al., 2009] (Fig. 4 and Table 3).
Figure 4.
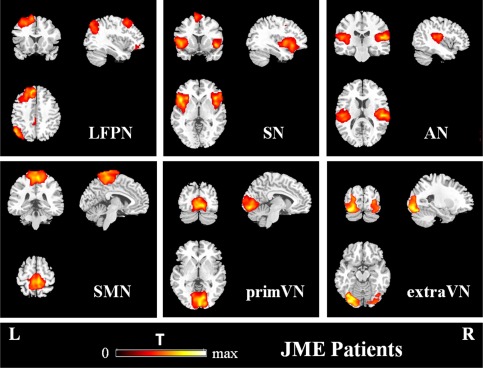
Other 6 meaningful resting‐state networks in JME patients (P < 0.05, FWE‐corrected). L: left; R: right. [Color figure can be viewed in the online issue, which is available at http://wileyonlinelibrary.com.]
Table 3.
Other meaningful resting‐state networks in JME patients (one‐sample t‐test, P < 0.05, FWE‐corrected)
| MNI coordinates | L/R | Lobe |
Brodmannarea |
T | Voxels | |||
|---|---|---|---|---|---|---|---|---|
| x | y | z | ||||||
| LPFN | −12 | 24 | 60 | L | Superior Frontal Gyrus | BA6/8 | 26.49 | 2,068 |
| −51 | −66 | 27 | L | Angular Gyrus | BA39 | 22.86 | 715 | |
| −6 | −63 | 30 | L | Precuneus | BA31 | 13.66 | 63 | |
| 60 | −3 | 6 | R | Superior Temporal Gyrus | BA22 | 9.98 | 36 | |
| −3 | −45 | 42 | L | Precuneus | BA31 | 9.24 | 52 | |
| SN | −45 | 18 | −9 | L | Insula | BA47 | 27.41 | 1,603 |
| 42 | 21 | −12 | R | Insula | BA47 | 28.62 | 1,081 | |
| −9 | 6 | 60 | L | Supplementary Motor Area | BA6 | 16.56 | 373 | |
| −57 | −45 | 27 | L | Supramarginal Gyrus | BA40 | 11.3 | 41 | |
| −42 | 3 | 42 | L | Precentral Gyrus | BA6 | 10.54 | 86 | |
| −27 | 54 | 24 | L | Middle Frontal Gyrus | BA10 | 8.69 | 39 | |
| AN | 51 | −12 | 3 | R | Superior Temporal Gyrus | BA22 | 28.17 | 1,067 |
| −51 | −24 | 12 | L | Superior Temporal Gyrus | BA41 | 17.78 | 1,011 | |
| SMN | 0 | −12 | 81 | Medial Frontal Gyrus | BA6 | 25.59 | 2,186 | |
| 6 | −27 | 66 | R | Paracentral Lobule | BA6 | 22.44 | ||
| primVN | 3 | −96 | 3 | R | Cuneus | BA18 | 28.3 | 1,921 |
| −9 | −87 | −12 | L | Lingual Gyrus | BA18 | 21.94 | ||
| extraVN | −36 | −72 | −18 | L | Fusiform Gyrus | BA19 | 19.77 | 1,871 |
| 33 | −78 | −6 | R | Inferior Occipital Gyrus | BA19 | 10.84 | ||
Figure 5 depicts the FNC diagram for both JME patients and controls (spatial distributions of resting‐state networks in controls can be observed in Supporting Information Fig. S5 and Table S2). Significantly correlated resting‐state networks are represented by red (positive) or blue (negative) lines. Both controls and JME patients showed significant positive network connectivities between resting‐state networks; however, some of the connections between networks in JME patients and controls were different. Briefly, (1) negative connectivities between the BGN and LFPN/antDMN/postDMN/extraVN and positive connectivity of the BGN and AN were only found in JME patients; (2) negative connectivity of the antDMN and AN was only found in controls, while positive connectivity of the SRN and SMN was only found in JME patients; (3) negative connectivities between the RFPN and SN/SMN/extraVN were only found in controls; (4) negative connectivities between the SN and LFPN/antDMN/postDMN, were only found in controls, while positive connectivities between the SN and SMN/primVN were only found in JME patients. To understand the difference of FNC between groups, two‐sample t‐tests were further performed (P < 0.05, FDR‐corrected). Figure 5 shows that (1) JME patients had higher mean correlations than controls for connections between SRN and antDMN/RFPN/primVN, (2) JME patients had greater correlations than controls for connections between the SN and postDMN/antDMN/RFPN/LFPN/SMN/primVN, (3) controls had higher correlations than JME patients for connections between the postDMN and BGN/RFPN/LFPN/antDMN and (4) controls had a lower correlation than JME patients for the connection between the postDMN and extraVN. Figure 5 also shows the significant lags between the two networks in each group. An arrow connects components A and B (points at component B), representing that component B lags component A by certain time units. The significant difference of lags between JME patients and controls are also shown. An arrow (from component A to B) represents that the delay (component B lags component A) in JME patients is larger than controls. In addition, the FNCs of all 17 meaningful ICs in JME patients can be seen in Fig. S3 (Supporting Information B). It showed that wide associations between discharge‐affecting networks and resting‐state networks existed in JME patients (P < 0.05, FDR‐corrected).
Figure 5.
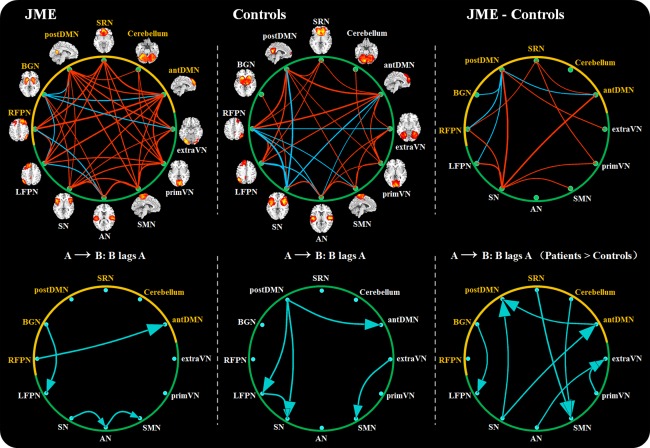
The FNCs of resting‐state networks in JME patients and controls (P < 0.05, FDR‐corrected) and the differences of FNCs between them. For FNCs in each group, the red lines represent positive connections, and blue lines represent negative connections. The lag (P < 0.05), A →; B, represents that component B lags component A by some calculated seconds. For the differences between two groups, red lines represent that JME patients show greater correlations than controls, while blue lines representing that JME patients show lower connections. The yellow color represents the discharge‐affecting resting‐state networks. The delay (P < 0.05), A → B (Patients > Controls), represents that the delay (component B lags component A) in JME patients is larger than controls. [Color figure can be viewed in the online issue, which is available at http://wileyonlinelibrary.com.]
Relationship between FNCs and Clinical Features
The z‐scores of discharge‐affecting FNCs were partially correlated with the epilepsy duration and the age of epilepsy onset, while controlling for the gender. Significant partial correlations were found between the age of epilepsy onset and FNC between the IC10 and SRN (R = −0.54, P = 0.013) as well as the onset age and FNC between IC20 and BGN (R = −0.15, P = 0.007) (Fig. 6). The partial correlations between epilepsy duration and the z‐scores of FNC were not significant.
Figure 6.
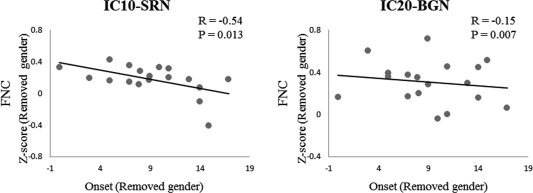
Partial correlations between discharge‐affecting FNCs (Fisher's z‐score) and the age of epilepsy onset in JME patients, while controlling for the gender. R: partial correlation coefficient; P: P‐value.
DISCUSSION
In this study, we investigated the complex discharge‐affecting networks and FNCs in patients suffering from JME using simultaneous EEG‐fMRI. Briefly, the main findings of the present study were as follows: (1) several discharge‐affecting networks (the BGN, DMN and SRN, among others) in JME were identified using emiCCA, in which linear and nonlinear relationships between EEG and fMRI features existed; (2) discharge‐affecting FNCs in JME patients perhaps revealed the crucial roles of specific networks, including the DMN, SRN and BGN; (3) significant alterations of the FNCs of resting‐state networks (e.g., the SN) in JME patients were discovered; and (4) relationships between FNCs and the age of epilepsy onset were also found.
Identified Discharge‐Affecting Networks
The concept of network abnormality refers to the brain regions/resting‐state networks affected by the generation and propagation of epileptic activity; such notion has been used to further understand epilepsy [Centeno and Carmichael, 2014; Spencer, 2002]. Using emiCCA, several linear or nonlinear discharge‐affecting networks in JME were identified in the current study (Fig. 2). Frontal lobe structural and functional changes in JME patients have been widely reported in various studies [Koepp et al., 2014; Wolf et al., 2015], and these changes may indicate the distinct cognitive impairment pattern in JME. The identified component IC42, which mainly contained the supplementary motor area and middle frontal gyrus, perhaps was a part of the thalamofrontal network involved in JME, as suggested in the previous studies [Koepp, 2005; Koepp et al., 2014; Pulsipher et al., 2009], and reflected the effects of JME on the cognitive functions of the frontal lobe. The BGN, which contained the main nuclei of the basal ganglia, was found to be associated with the epileptic discharges in JME. It has been suggested that the basal ganglia play important roles in the regulation of epileptic discharges [Norden and Blumenfeld, 2002]. Using resting‐state fMRI, Luo et al., found that IGE patients show significantly enhanced integration within the BGN during periods with and without discharges and suggested that BGN perhaps plays a role as an important modulator in IGE [Luo et al., 2012]. Further evidence for the involvement of the basal ganglia in the modulation of epileptic discharges has been suggested in the studies of diffusion tensor imaging (DTI) in absence epilepsy [Luo et al., 2011b], neuropharmacology [Danober et al., 1998; Deransart et al., 1998], deep brain stimulation in epilepsy [Loddenkemper et al., 2001] and computational evidence of absence seizures [Chen et al., 2014]. In this study, our findings indicated that the BGN may also be associated with the modulation and propagation of epileptic discharges in JME. The widespread deactivation of regions such as the bilateral precuneus/posterior cingulate cortex, angular gyrus and middle frontal gyrus, so called “default mode” networks [Raichle et al., 2001], has been commonly found in a number of studies of epilepsy [Aghakhani et al., 2004; Gotman et al., 2005; Hamandi et al., 2006; Li et al., 2009]. These previous findings perhaps represent downstream consequences of GSWDs. Here, our results indicate that DMN may also be influenced by epileptic discharges of JME. Furthermore, various components such as the SRN, which is putatively related to self‐referential processing [D'Argembeau et al., 2005], the RFPN, which is associated with dorsal attention network [Corbetta and Shulman, 2002], the cerebellum, visual components (IC10 and IC22), the IC20 and the IC1 were found to be influenced by epileptic discharges in JME; these findings may indicate that epilepsy has a widespread impact on brain function in terms of networks. In addition, in JME patients, the aforementioned results are similar to the discharge‐affecting regions revealed by traditional EEG‐informed fMRI analysis, such as the basal ganglia, superior frontal gyrus, supplementary motor area, temporal polar and cerebellum (Supporting Information Fig. S1 and Table S1).
FNCs Between Discharge‐Affecting Networks
As an extension of functional connectivity, FNC analysis has been widely used as an effective method to evaluate the temporal associations between resting‐state networks in a healthy population [Liao et al., 2010], patients with schizophrenia [Jafri et al., 2008; Sakoglu et al., 2010] and epilepsy patients [Li et al., 2015; Luo et al., 2011a] using fMRI. To understand the associations between the identified discharge‐affecting networks, functional network connectivity analysis was performed in the current study. Dense FNCs were observed among the discharge‐affecting networks constructed in JME patients (Fig. 3); this finding may indicate strong communication of information between these discharge‐affecting networks in JME. Because discharges generated from a focal epileptic zone may entrain a large neural network [Spencer, 2002] and the epileptic discharges perhaps are driven by the pattern of connections in brain networks [Terry et al., 2012], these widespread associations between discharge‐affecting networks may to some extent contribute to the generation and propagation of epileptic activity in JME. Furthermore, it is worth noting that the antDMN, postDMN and SRN have larger degrees of connectivity in the discharge‐affecting networks. Regions from the DMN have been suggested to serve as cortical hubs for information integration, especially in posterior cingulate cortex [Buckner et al., 2009]. In previous resting‐state fMRI studies [Li et al., 2011; Liao et al., 2010], the DMN and SRN were also found to be involved in the high flow of information among the meaningful resting‐state networks. The DMN and SRN may play important roles in the process of resting‐state information transfer and integration, respectively. Therefore, our results indicated that the DMN and SRN might play crucial roles in the propagation of epileptic activity between discharge‐affecting networks. Another interesting finding of our results was the negative connectivity between the BGN and DMN. This finding further supported that the BGN might play an important role in the regulation [Norden and Blumenfeld, 2002] and modulation [Luo et al., 2011b, 2012] of epileptic discharges in JME. Moreover, interesting links between FNCs (SRN‐IC10 and BGN‐IC20) and clinical feature have been provided by significant negative correlations with the age of epilepsy onset. This finding further supported the crucial roles of the SRN and BGN in the propagation of epileptic activity in JME.
FNCs Between Resting‐State Networks
To further understand the potential impacts of JME on the relationships between resting‐state networks, the maximal time‐lagged correlation method was performed in JME patients and controls and alterations of FNC in JME patients were further investigated. As shown in Figure 5, overall, JME patients preferentially displayed impaired connections between the systems of lower‐level sensory (e.g., AN, SMN, primVN and extraVN) and higher‐order cognitive networks (e.g., DMN, RPFN, LPFN, SRN and SN), while the intra‐system connections were preserved to some extent. This hierarchical impairment in epilepsy was in accordance with a previous study, in which the loss of interactions among the intersystem were found in patients with partial epilepsy [Luo et al., 2011a]. This phenomenon might indicate that the FNC impairment in JME patients also had a hierarchical selectivity and that the selective impairment had an important functional and theoretical implication; therefore suitable to understand JME from the perspective of a complex network. Another interesting finding was that negative connectivities of the BGN and LFPN/antDMN/postDMN/extraVN and the positive connectivity of the BGN and AN were only found in JME patients. Because of the important roles of the BGN in the regulation [Norden and Blumenfeld, 2002] and modulation [Luo et al., 2011b, 2012] of epileptic discharges, the results further indicated that the BGN may related to the modulation and propagation of epileptic discharges in JME.
Furthermore, compared with controls, meaningful findings indicated that JME patients had higher correlations for connections between the SRN and antDMN/RFPN/primVN, and lower mean correlations for connections between the postDMN and antDMN/RPFN/LPFN/BGN. The SRN has been suggested to have an intermediary role in information exchange between the systems of lower‐level sensory (primVN, etc.) and higher‐order cognitive (DMN and RFPN, etc.) networks [Li et al., 2011; Liao et al., 2010], and the DMN is pivotal in the process of information transfer and integration [Buckner et al., 2009; Li et al., 2011; Liao et al., 2010]. Therefore, our findings suggest that the DMN and SRN may contribute to the propagation of epileptic discharges between resting‐state networks in JME. Moreover, the findings of increased correlations for connections between the SN and DMN/RFPN/LFPN/SMN/primVN in JME patients were also found in the current study. The salience network, with key nodes in the insular cortices, has a crucial role in salience processing across multiple sensory and cognitive domains [Seeley et al., 2007; Uddin, 2015] and in switching between the DMN and central executive network [Sridharan et al., 2008]. Disturbance of the SN system, which has been posited to contribute to the aberrant salience processing, has been observed in patients with certain disorders such as schizophrenia [Manoliu et al., 2013] and childhood absence epilepsy [Luo et al., 2014]. In addition, the differences of lags between JME patients and controls were also found between resting‐state networks. It perhaps demonstrated the impact of JME on the delay relationships between two resting‐state networks. Combining these findings, we presume that in JME patients, the DMN and SRN may play important roles to extend the propagation of epileptic discharges into resting‐state networks, especially into the SN system, thus affecting the switching function of the SN.
Methodological Considerations and Limitations
Several methodological issues and limitations should be considered in this study. First, many potential factors may lead to nonlinearity in neural imaging, for example, the nonlinearity may be caused by neurovascular coupling [Liu et al., 2010] or even the variety of methods used to quantify the multimodal signals [Dong et al., 2014; He et al., 2011]. Moreover, HRFs vary in epilepsy patients and spike locations [Beers et al., 2015; Masterton et al., 2010] and across brain regions and subjects [Handwerker et al., 2004], and this variation may also potentially lead to nonlinearity. Therefore, the nonlinearity may be acknowledged in the EEG‐fMRI study of epilepsy. Because emiCCA can tolerate several variably shaped HRFs and has the ability to perform nonlinear processing [Dong et al., 2015], this method may extend our perspectives from linearity to nonlinearity and help us to further understand epilepsy. Second, notably, open questions about ICA remain, such as the number of independent components and the reliability of IC maps. In this work, the minimum description length criterion was used to determine the number of ICs [Li et al., 2007], and ICASSO was used to achieve reliable ICs [Himberg et al., 2004]. In addition, considering group ICA itself has capability for separating noises, we also directly conducted group ICA on fMRI data without regressing out nuisance signals, and then calculated the results using the same procedure. Together with the results of unregressed (Supporting Information C) and regressed fMRI data, it perhaps also supported that the important roles of BGN, SRN and DMN in JME and the potentials of emiCCA and FNC analysis to provide important insights into understanding JME. Third, due to the limited sample size (only 18 JME patients), future studies should include larger sample sizes to determine these mechanisms underlying the connections found in the current study. In addition, because neuropsychological evaluations in JME patients were not conducted in this study, relationships between SN system and cognitive impairment in JME patients would be investigated in the future. Lastly, antiepileptic drugs may influence the functional and discharge‐affecting networks despite the fact that the JME patients had discontinued medication for approximately 24 h in the current study. Therefore, this issue may confound the findings in this study due to the potential effects of antiepileptic drugs on brain cognitive function.
CONCLUSION
In conclusion, the present study provided evidence of complex discharge‐affecting networks (including the BGN, DMN, SRN and frontal networks, among others) in JME patients, in which linear and nonlinear relationships between EEG and fMRI features existed. Furthermore, our results suggested that BGN may be associated with the modulation and propagation of epileptic discharges in JME. Moreover, the DMN and SRN may play intermediary roles in the propagation of epileptic discharges between discharge‐affecting networks, as well as propagation between resting‐state networks. These roles may contribute to the increased FNCs between the SN and resting‐state networks in JME patients. The switching function of SN may be disturbed by these roles, which account for the cognitive impairment of JME patients. We presumed that emiCCA and FNC analysis may be potential tools to investigate the epilepsy using simultaneous EEG‐fMRI and provided important insights into understanding the pathophysiological mechanism of epilepsy such as JME.
Supporting information
Supporting Information
ACKNOWLEDGMENT
All of the authors have no conflict of interest. We are also grateful to Benjamin Klugah‐Brown for English language help.
REFERENCES
- (1989): Proposal for revised classification of epilepsies and epileptic syndromes. Commission on classification and terminology of the International League against Epilepsy. Epilepsia 30:389–399. [DOI] [PubMed] [Google Scholar]
- Aghakhani Y, Bagshaw AP, Benar CG, Hawco C, Andermann F, Dubeau F, Gotman J (2004): fMRI activation during spike and wave discharges in idiopathic generalized epilepsy. Brain 127:1127–1144. [DOI] [PubMed] [Google Scholar]
- Allen PJ, Josephs O, Turner R (2000): A method for removing imaging artifact from continuous EEG recorded during functional MRI. Neuroimage 12:230–239. [DOI] [PubMed] [Google Scholar]
- Bagshaw AP, Aghakhani Y, Benar CG, Kobayashi E, Hawco C, Dubeau F, Pike GB, Gotman J (2004): EEG‐fMRI of focal epileptic spikes: Analysis with multiple haemodynamic functions and comparison with gadolinium‐enhanced MR angiograms. Hum Brain Mapp 22:179–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beers CA, Williams RJ, Gaxiola‐Valdez I, Pittman DJ, Kang AT, Aghakhani Y, Pike GB, Goodyear BG, Federico P (2015): Patient specific hemodynamic response functions associated with interictal discharges recorded via simultaneous intracranial EEG‐fMRI. Hum Brain Mapp 36(12): 5252–5264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg AT, Berkovic SF, Brodie MJ, Buchhalter J, Cross JH, van Emde Boas W, Engel J, French J, Glauser TA, Mathern GW, Moshe SL, Nordli D, Plouin P, Scheffer IE (2010): Revised terminology and concepts for organization of seizures and epilepsies: Report of the ILAE Commission on Classification and Terminology, 2005‐2009. Epilepsia 51:676–685. [DOI] [PubMed] [Google Scholar]
- Biswal B, Yetkin FZ, Haughton VM, Hyde JS (1995): Functional connectivity in the motor cortex of resting human brain using echo‐planar MRI. Magn Reson Med 34:537–541. [DOI] [PubMed] [Google Scholar]
- Biswal BB, Mennes M, Zuo XN, Gohel S, Kelly C, Smith SM, Beckmann CF, Adelstein JS, Buckner RL, Colcombe S, Dogonowski AM, Ernst M, Fair D, Hampson M, Hoptman MJ, Hyde JS, Kiviniemi VJ, Kotter R, Li SJ, Lin CP, Lowe MJ, Mackay C, Madden DJ, Madsen KH, Margulies DS, Mayberg HS, McMahon K, Monk CS, Mostofsky SH, Nagel BJ, Pekar JJ, Peltier SJ, Petersen SE, Riedl V, Rombouts SA, Rypma B, Schlaggar BL, Schmidt S, Seidler RD, Siegle GJ, Sorg C, Teng GJ, Veijola J, Villringer A, Walter M, Wang L, Weng XC, Whitfield‐Gabrieli S, Williamson P, Windischberger C, Zang YF, Zhang HY, Castellanos FX, Milham MP (2010): Toward discovery science of human brain function. Proc Natl Acad Sci USA 107:4734–4739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckner RL, Sepulcre J, Talukdar T, Krienen FM, Liu H, Hedden T, Andrews‐Hanna JR, Sperling RA, Johnson KA (2009): Cortical hubs revealed by intrinsic functional connectivity: Mapping, assessment of stability, and relation to Alzheimer's disease. J Neurosci 29:1860–1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calhoun VD, Adali T, Pearlson GD, Pekar JJ (2001): A method for making group inferences from functional MRI data using independent component analysis. Human Brain Mapp 14:140–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calhoun VD, Kiehl KA, Liddle PF, Pearlson GD (2004): Aberrant localization of synchronous hemodynamic activity in auditory cortex reliably characterizes schizophrenia. Biol Psychiatry 55:842–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centeno M, Carmichael DW (2014): Network connectivity in epilepsy: Resting state fMRI and EEG‐fMRI contributions. Front Neurol 5:93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Guo D, Wang T, Jing W, Xia Y, Xu P, Luo C, Valdes‐Sosa PA, Yao D (2014): Bidirectional control of absence seizures by the basal ganglia: A computational evidence. PLoS Comput Biol 10:e1003495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen AL, Fair DA, Dosenbach NU, Miezin FM, Dierker D, Van Essen DC, Schlaggar BL, Petersen SE (2008): Defining functional areas in individual human brains using resting functional connectivity MRI. Neuroimage 41:45–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbetta M, Shulman GL (2002): Control of goal‐directed and stimulus‐driven attention in the brain. Nat Rev Neurosci 3:201–215. [DOI] [PubMed] [Google Scholar]
- D'Argembeau A, Collette F, Van der Linden M, Laureys S, Del Fiore G, Degueldre C, Luxen A, Salmon E (2005): Self‐referential reflective activity and its relationship with rest: A PET study. Neuroimage 25:616–624. [DOI] [PubMed] [Google Scholar]
- Damoiseaux JS, Rombouts SA, Barkhof F, Scheltens P, Stam CJ, Smith SM, Beckmann CF (2006): Consistent resting‐state networks across healthy subjects. Proc Natl Acad Sci USA 103:13848–13853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damoiseaux JS, Beckmann CF, Arigita EJ, Barkhof F, Scheltens P, Stam CJ, Smith SM, Rombouts SA (2008): Reduced resting‐state brain activity in the “default network” in normal aging. Cereb Cortex 18:1856–1864. [DOI] [PubMed] [Google Scholar]
- Danober L, Deransart C, Depaulis A, Vergnes M, Marescaux C (1998): Pathophysiological mechanisms of genetic absence epilepsy in the rat. Prog Neurobiol 55:27–57. [DOI] [PubMed] [Google Scholar]
- Deransart C, Vercueil L, Marescaux C, Depaulis A (1998): The role of basal ganglia in the control of generalized absence seizures. Epilepsy Res 32:213–223. [DOI] [PubMed] [Google Scholar]
- Dong L, Gong D, Valdes‐Sosa PA, Xia Y, Luo C, Xu P, Yao D (2014): Simultaneous EEG‐fMRI: Trial level spatio‐temporal fusion for hierarchically reliable information discovery. Neuroimage 99:28–41. [DOI] [PubMed] [Google Scholar]
- Dong L, Zhang Y, Zhang R, Zhang X, Gong D, Valdes‐Sosa PA, Xu P, Luo C, Yao D (2015): Characterizing nonlinear relationships in functional imaging data using eigenspace maximal information canonical correlation analysis (emiCCA). Neuroimage 109:388–401. [DOI] [PubMed] [Google Scholar]
- Engel J Jr, International League Against E (2001): A proposed diagnostic scheme for people with epileptic seizures and with epilepsy: Report of the ILAE Task Force on Classification and Terminology. Epilepsia 42:796–803. [DOI] [PubMed] [Google Scholar]
- Fox MD, Snyder AZ, Zacks JM, Raichle ME (2006): Coherent spontaneous activity accounts for trial‐to‐trial variability in human evoked brain responses. Nat Neurosci 9:23–25. [DOI] [PubMed] [Google Scholar]
- Genton P, Thomas P, Kasteleijn‐Nolst Trenite DG, Medina MT, Salas‐Puig J (2013): Clinical aspects of juvenile myoclonic epilepsy. Epilepsy Behav 28:S8–14. [DOI] [PubMed] [Google Scholar]
- Glover GH (1999): Deconvolution of impulse response in event‐related BOLD fMRI. Neuroimage 9:416–429. [DOI] [PubMed] [Google Scholar]
- Gotman J, Grova C, Bagshaw A, Kobayashi E, Aghakhani Y, Dubeau F (2005): Generalized epileptic discharges show thalamocortical activation and suspension of the default state of the brain. Proc Natl Acad Sci USA 102:15236–15240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotman J, Pittau F (2011): Combining EEG and fMRI in the study of epileptic discharges. Epilepsia 52:38–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamandi K, Salek‐Haddadi A, Laufs H, Liston A, Friston K, Fish DR, Duncan JS, Lemieux L (2006): EEG‐fMRI of idiopathic and secondarily generalized epilepsies. Neuroimage 31:1700–1710. [DOI] [PubMed] [Google Scholar]
- Handwerker DA, Ollinger JM, D'Esposito M (2004): Variation of BOLD hemodynamic responses across subjects and brain regions and their effects on statistical analyses. Neuroimage 21:1639–1651. [DOI] [PubMed] [Google Scholar]
- He B, Yang L, Wilke C, Yuan H (2011): Electrophysiological imaging of brain activity and connectivity‐challenges and opportunities. IEEE Trans Biomed Eng 58:1918–1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himberg J, Hyvarinen A, Esposito F (2004): Validating the independent components of neuroimaging time series via clustering and visualization. Neuroimage 22:1214–1222. [DOI] [PubMed] [Google Scholar]
- Huster RJ, Debener S, Eichele T, Herrmann CS (2012): Methods for Simultaneous EEG‐fMRI: An introductory review. J Neurosci 32:6053–6060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jafri MJ, Pearlson GD, Stevens M, Calhoun VD (2008): A method for functional network connectivity among spatially independent resting‐state components in schizophrenia. Neuroimage 39:1666–1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janz D (1985): Epilepsy with impulsive petit mal (juvenile myoclonic epilepsy). Acta Neurol Scand 72:449–459. [DOI] [PubMed] [Google Scholar]
- Koepp MJ (2005): Juvenile myoclonic epilepsy–a generalized epilepsy syndrome? Acta Neurol Scand Suppl 181:57–62. [DOI] [PubMed] [Google Scholar]
- Koepp MJ, Thomas RH, Wandschneider B, Berkovic SF, Schmidt D (2014): Concepts and controversies of juvenile myoclonic epilepsy: Still an enigmatic epilepsy. Expert Rev Neurother 14:819–831. [DOI] [PubMed] [Google Scholar]
- Li Q, Luo C, Yang T, Yao Z, He L, Liu L, Xu H, Gong Q, Yao D, Zhou D (2009): EEG‐fMRI study on the interictal and ictal generalized spike‐wave discharges in patients with childhood absence epilepsy. Epilepsy Res 87:160–168. [DOI] [PubMed] [Google Scholar]
- Li Q, Cao W, Liao X, Chen Z, Yang T, Gong Q, Zhou D, Luo C, Yao D (2015): Altered resting state functional network connectivity in children absence epilepsy. J Neurol Sci 354:79–85. [DOI] [PubMed] [Google Scholar]
- Li R, Chen K, Fleisher AS, Reiman EM, Yao L, Wu X (2011): Large‐scale directional connections among multi resting‐state neural networks in human brain: A functional MRI and Bayesian network modeling study. Neuroimage 56:1035–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YO, Adali T, Calhoun VD (2007): Estimating the number of independent components for functional magnetic resonance imaging data. Hum Brain Mapp 28:1251–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao W, Mantini D, Zhang Z, Pan Z, Ding J, Gong Q, Yang Y, Chen H (2010): Evaluating the effective connectivity of resting state networks using conditional Granger causality. Biol Cybern 102:57–69. [DOI] [PubMed] [Google Scholar]
- Liu Z, Rios C, Zhang N, Yang L, Chen W, He B (2010): Linear and nonlinear relationships between visual stimuli, EEG and BOLD fMRI signals. Neuroimage 50:1054–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loddenkemper T, Pan A, Neme S, Baker KB, Rezai AR, Dinner DS, Montgomery EB Jr, Luders HO (2001): Deep brain stimulation in epilepsy. J Clin Neurophysiol 18:514–532. [DOI] [PubMed] [Google Scholar]
- Luo C, Qiu C, Guo Z, Fang J, Li Q, Lei X, Xia Y, Lai Y, Gong Q, Zhou D, Yao D (2011a): Disrupted functional brain connectivity in partial epilepsy: A resting‐state fMRI study. PLoS One 7:e28196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo C, Xia Y, Li Q, Xue K, Lai Y, Gong Q, Zhou D, Yao D (2011b): Diffusion and volumetry abnormalities in subcortical nuclei of patients with absence seizures. Epilepsia 52:1092–1099. [DOI] [PubMed] [Google Scholar]
- Luo C, Li Q, Xia Y, Lei X, Xue K, Yao Z, Lai Y, Martinez‐Montes E, Liao W, Zhou D, Valdes‐Sosa PA, Gong Q, Yao D (2012): Resting state basal ganglia network in idiopathic generalized epilepsy. Hum Brain Mapp 33:1279–1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo C, Yang T, Tu S, Deng J, Liu D, Li Q, Dong L, Goldberg I, Gong Q, Zhang D, An D, Zhou D, Yao D (2014): Altered intrinsic functional connectivity of the salience network in childhood absence epilepsy. J Neurol Sci 339:189–195. [DOI] [PubMed] [Google Scholar]
- Luo C, Zhang Y, Cao W, Huang Y, Yang F, Wang J, Tu S, Wang X, Yao D (2015): Altered Structural and Functional Feature of Striato‐Cortical Circuit in Benign Epilepsy with Centrotemporal Spikes. Int J Neural Syst 25:1550027. [DOI] [PubMed] [Google Scholar]
- Manoliu A, Riedl V, Doll A, Bauml JG, Muhlau M, Schwerthoffer D, Scherr M, Zimmer C, Forstl H, Bauml J, Wohlschlager AM, Koch K, Sorg C (2013): Insular dysfunction reflects altered between‐network connectivity and severity of negative symptoms in schizophrenia during psychotic remission. Front Hum Neurosci 7:216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masterton RA, Harvey AS, Archer JS, Lillywhite LM, Abbott DF, Scheffer IE, Jackson GD (2010): Focal epileptiform spikes do not show a canonical BOLD response in patients with benign rolandic epilepsy (BECTS). Neuroimage 51:252–260. [DOI] [PubMed] [Google Scholar]
- Niazy RK, Beckmann CF, Iannetti GD, Brady JM, Smith SM (2005): Removal of FMRI environment artifacts from EEG data using optimal basis sets. Neuroimage 28:720–737. [DOI] [PubMed] [Google Scholar]
- Norden AD, Blumenfeld H (2002): The role of subcortical structures in human epilepsy. Epilepsy Behav 3:219–231. [DOI] [PubMed] [Google Scholar]
- Pulsipher DT, Seidenberg M, Guidotti L, Tuchscherer VN, Morton J, Sheth RD, Hermann B (2009): Thalamofrontal circuitry and executive dysfunction in recent‐onset juvenile myoclonic epilepsy. Epilepsia 50:1210–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raichle ME, MacLeod AM, Snyder AZ, Powers WJ, Gusnard DA, Shulman GL (2001): A default mode of brain function. Proc Natl Acad Sci USA 98:676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reshef DN, Reshef YA, Finucane HK, Grossman SR, McVean G, Turnbaugh PJ, Lander ES, Mitzenmacher M, Sabeti PC (2011): Detecting novel associations in large data sets. Science 334:1518–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson S, Basso G, Soldati N, Sailer U, Jovicich J, Bruzzone L, Kryspin‐Exner I, Bauer H, Moser E (2009): A resting state network in the motor control circuit of the basal ganglia. BMC Neurosci 10:137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rombouts SA, Damoiseaux JS, Goekoop R, Barkhof F, Scheltens P, Smith SM, Beckmann CF (2009): Model‐free group analysis shows altered BOLD FMRI networks in dementia. Hum Brain Mapp 30:256–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakoglu U, Pearlson GD, Kiehl KA, Wang YM, Michael AM, Calhoun VD (2010): A method for evaluating dynamic functional network connectivity and task‐modulation: Application to schizophrenia. Magma 23:351–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeley WW, Menon V, Schatzberg AF, Keller J, Glover GH, Kenna H, Reiss AL, Greicius MD (2007): Dissociable intrinsic connectivity networks for salience processing and executive control. J Neurosci 27:2349–2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SM, Fox PT, Miller KL, Glahn DC, Fox PM, Mackay CE, Filippini N, Watkins KE, Toro R, Laird AR, Beckmann CF (2009): Correspondence of the brain's functional architecture during activation and rest. Proc Natl Acad Sci USA 106:13040–13045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer SS (2002): Neural networks in human epilepsy: Evidence of and implications for treatment. Epilepsia 43:219–227. [DOI] [PubMed] [Google Scholar]
- Sridharan D, Levitin DJ, Menon V (2008): A critical role for the right fronto‐insular cortex in switching between central‐executive and default‐mode networks. Proc Natl Acad Sci USA 105:12569–12574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terry JR, Benjamin O, Richardson MP (2012): Seizure generation: The role of nodes and networks. Epilepsia 53:e166–e169. [DOI] [PubMed] [Google Scholar]
- Uddin LQ (2015): Salience processing and insular cortical function and dysfunction. Nat Rev Neurosci 16:55–61. [DOI] [PubMed] [Google Scholar]
- Wolf P, Yacubian EM, Avanzini G, Sander T, Schmitz B, Wandschneider B, Koepp M (2015): Juvenile myoclonic epilepsy: A system disorder of the brain. Epilepsy Res 114:2–12. [DOI] [PubMed] [Google Scholar]
- Xue K, Luo C, Zhang D, Yang T, Li J, Gong D, Chen L, Medina YI, Gotman J, Zhou D, Yao D (2014): Diffusion tensor tractography reveals disrupted structural connectivity in childhood absence epilepsy. Epilepsy Res 108:125–138. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Liao W, Wang Z, Xu Q, Yang F, Mantini D, Jiao Q, Tian L, Liu Y, Lu G (2014): Epileptic discharges specifically affect intrinsic connectivity networks during absence seizures. J Neurol Sci 336:138–145. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Xu Q, Liao W, Wang Z, Li Q, Yang F, Zhang Z, Liu Y, Lu G (2015): Pathological uncoupling between amplitude and connectivity of brain fluctuations in epilepsy. Hum Brain Mapp 36:2756–2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information


