Abstract
As modulators of dopamine availability and release in the brain, COMT and BDNF polymorphisms have demonstrated interactions on human cognition; however, the underlying neural mechanisms remain largely unknown. In this study, we aimed to investigate the interactions of COMT rs4680 and BDNF rs6265 on global functional connectivity density (gFCD) of the brain in 265 healthy young subjects. We found a significant COMT × BDNF interaction on the gFCD in the left frontal eye field (FEF), showing an inverted U‐shape modulation by the presumed dopamine signaling. This finding was consistently repeated in the gFCD analyses using other four connection thresholds. Our findings reveal a COMT × BDNF interaction on the FCD in the left FEF, which may be helpful for understanding the neural mechanisms of the COMT × BDNF interactions on the FEF‐related cognitive functions. Hum Brain Mapp 37:2468–2478, 2016. © 2016 Wiley Periodicals, Inc.
Keywords: COMT, BDNF, functional connectivity density, dopamine, attention
Abbreviations
- AG
Angular gyrus
- ANCOVA
Analysis of covariance
- BDNF
Brain‐derived neurotrophic factor
- BRAVO
Brain volume
- COMT
Catechol‐O‐methyltransferase
- FA
Flip angle
- FCD
Functional connectivity density
- FD
Framewise displacement
- FEF
Frontal eye field
- FOV
Field of view
- MCC
Mid‐cingulate cortex
- MNI
Montreal Neurological Institute
- MOG
Middle occipital gyrus
- PFC
Prefrontal cortex
- pSTG
Posterior superior temporal gyrus
INTRODUCTION
Catechol‐O‐methyltransferase (COMT) catalyzes the degradation of synaptic dopamine in the brain. The COMT rs4680 polymorphism reduces enzymatic activity and increases synaptic dopamine concentration in the Met‐carriers [Mannisto and Kaakkola, 1999; Matsumoto et al., 2003; Tunbridge et al., 2006]. This effect is especially prominent in the prefrontal cortex (PFC) [Akil et al., 2003; Mannisto and Kaakkola, 1999; Seamans and Yang, 2004] due to the lack of dopamine transporter in this region [Lewis et al., 2001]. Brain‐derived neurotrophic factor (BDNF) regulates neuronal survival, differentiation and synaptic plasticity [Chao, 2003; Huang and Reichardt, 2001; Poo, 2001] and exhibits the highest expression in the PFC [Pezawas et al., 2004]. The BDNF rs6265 polymorphism affects the trafficking of BDNF and reduces BDNF release [Chen et al., 2004; Chen et al., 2006; Egan et al., 2003; Yeh et al., 2012], resulting reduced activity‐dependent dopamine release in the Met carriers [Pecina et al., 2014].
Both COMT rs4680 and BDNF rs6265 polymorphisms act on the dopamine system [Tunbridge et al., 2006; Pecina et al., 2014], modulate stress reaction and cognitive functions [Papaleo et al., 2008; Ren‐Patterson et al., 2005; Ursini et al., 2011], and involve in several psychiatric disorders [Nolan et al., 2004; Ho et al., 2007; Twamley et al., 2014]; these findings indicate a possible COMT × BDNF interaction on their external phenotypes. Indeed, significant COMT × BDNF interactions have been reported on boredom susceptibility of sensation seeking traits [Kang et al., 2010], implicit grammar learning [Witte et al., 2012], resilience [Kang et al., 2013], and cognitive performance [Das et al., 2014] in healthy subjects, and on symptoms and cognition in schizophrenia [Han et al., 2008], dysfunctional beliefs in obsessive‐compulsive disorder [Alonso et al., 2013], and anxiety sensitivity in panic disorder [Konishi et al., 2014]. To investigate the neural mechanisms underlying the COMT × BDNF interaction on these external phenotypes, a COMT × BDNF interaction has been found in paired associative stimulation‐induced plasticity in the motor cortex [Witte et al., 2012] and resting‐state functional connectivity (rsFC) between the ventral striatum and the anterior cingulate cortex [Wang et al., 2015]. However, these hypothesis‐driven analyses cannot provide us a full picture of the COMT × BDNF interaction on brain properties.
As a data‐driven method, the functional connectivity density (FCD) mapping can voxel‐wisely identify the intergroup differences in FCD [Tomasi and Volkow, 2010, 2011a, 2011b]. In this study, we aimed to identify the COMT × BDNF interaction on the global FCD (gFCD) in healthy young subjects. We predict that the PFC may show a COMT × BDNF interaction on the gFCD because both genetic variations have a large effect on the PFC [Mannisto and Kaakkola, 1999; Pezawas et al., 2004; Seamans and Yang, 2004].
MATERIALS AND METHODS
Subjects
A total of 323 right‐handed healthy young adults were recruited for this study. They were carefully screened to ensure that they had no history of psychiatric or neurological illness, psychiatric treatment, or drug or alcohol abuse and that they had no contraindications to MRI examination. Only Chinese Han populations were included to purify the sample. All subjects were strongly right‐handed according to the Chinese edition of the Edinburgh Handedness Inventory [Oldfield, 1971]. The study was approved by the Medical Research Ethics Committee of Tianjin Medical University, and all participants provided written informed consent. Fifty‐eight subjects were excluded from further analysis because of poor imaging quality (28 subjects) or genotyping failure (30 subjects). The remaining 265 healthy young adults (145 females and 120 males; mean age, 22.8 years; range, 18‐29 years) were ultimately included in the imaging analysis.
Genotyping
For each subject, the COMT rs4680 and BDNF rs6265 were genotyped using the polymerase chain reaction and ligation detection reaction [Thomas et al., 2004; Yi et al., 2009] with technical support from the Shanghai Biowing Applied Biotechnology Company. The detailed procedures for genotyping have been previously described [Wang et al., 2015].
Image Acquisition
MR images were acquired using a Signa HDx 3.0 tesla MR scanner (General Electric) with 8‐channel radio‐frequency coils. Tight but comfortable foam padding was used to minimize head motion, and ear plugs were used to reduce scanner noise. The resting‐state fMRI data were obtained using single‐shot echo planar imaging with the following parameters: repetition time (TR)/echo time (TE) = 2,000/30 ms; field of view (FOV) = 240 × 240 mm; matrix = 64 × 64; flip angle (FA) = 90°; slice thickness = 4 mm; no gap; 40 interleaved transverse slices; 180 volumes. During the fMRI scans, all subjects were instructed to keep their eyes closed, to relax and move as little as possible, to think of nothing in particular, and to not fall asleep. Sagittal 3D T1‐weighted images were acquired using a brain volume (BRAVO) sequence (TR/TE = 8.1/3.1 ms; inversion time = 450 ms; FA = 13°; FOV = 256 × 256 mm; matrix = 256 × 256; slice thickness = 1 mm; no gap; 176 sagittal slices). The same MRI datasets have been used to investigate the COMT × DRD2 interaction on FCD [Tian et al., 2013].
Data Preprocessing
The fMRI data were preprocessed using SPM8 (http://www.fil.ion.ucl.ac.uk/spm). The first 10 volumes for each subject were discarded to allow the signal to reach equilibrium and the participants to adapt to the scanning noise. The remaining 170 volumes were then corrected for the acquisition time delay between slices. The fMRI data from the finally included 265 subjects were within the defined motion thresholds (translational or rotational motion parameters lower than 2mm or 2°). We also calculated framewise displacement (FD), which indexes volume‐to‐volume changes in head position. These changes were obtained from the derivatives of the rigid‐body realignment estimates that are used to realign blood oxygen level‐dependent (BOLD) data during fMRI preprocessing [Power et al., 2012, 2013]. There was no main effect of each SNP and interaction on the FD (P > 0.05). The approach used to normalize these functional images included the following steps: (1) individual structural images were linear coregistered to the mean motion‐corrected functional image; (2) the transformed structural images were segmented into gray matter, white matter, and CSF, and gray matter was nonlinear coregistered to the Montreal Neurological Institute (MNI) space; and (3) the motion‐corrected functional volumes were spatially normalized to the MNI space using the parameters estimated during nonlinear coregistration. The functional images were then resampled into a voxel size of 3 × 3 × 3 mm3. After normalization, several nuisance covariates (six motion parameters and average BOLD signals of the ventricular and white matter) were regressed out from the data and the datasets were band‐pass filtered with frequency from 0.01 to 0.1 Hz.
gFCD Calculation
We calculated the gFCD of each voxel using the inhouse script that was written in the Linux platform according to the method described by Tomasi and Volkow [2011a, b]. The Pearson's linear correlation was used to calculate the rsFCs, and two voxels with a correlation coefficient > 0.6 were considered functionally connected. The calculation of the gFCD was restricted to voxels in the gray matter regions with a signal‐to‐noise ratio > 50% to minimize unwanted effects from susceptibility‐related signal‐loss artifacts [Tomasi and Volkow, 2010]. The gFCD at a given voxel x 0 was computed as the total number of rsFCs between x 0 and all other voxels. This calculation was repeated for all x 0 voxels in the brain. To increase the normality of the distribution, grand mean scaling of the gFCD was performed by dividing by the mean value of the qualified voxels of the whole brain. The gFCD maps were spatially smoothed with an 8 × 8 × 8 mm3 Gaussian kernel.
gFCD Analysis
A two‐way (COMT rs4680 and BDNF rs6265 genotypes) analysis of covariance (ANCOVA) was used to voxel‐wisely identify the main effect of COMT or BDNF and the COMT × BDNF interaction on the gFCD, controlling for the effects of age, gender and years of education. A corrected threshold of P < 0.005 was derived from a combined threshold of P < 0.005 for each voxel and a cluster size > 64 voxels which was determined by the AlphaSim program in the AFNI software (parameters: single voxel P = 0.005, 5,000 simulations, FWHM = 8 mm, cluster connection radius = 5 mm, with gray matter mask, http://afni.nimh.nih.gov/).
Validation Analysis
Because the threshold of Pearson correlation coefficient (r = 0.6) was arbitrarily selected, we also validated the reliability of our results using r = 0.4, 0.5, 0.7 and 0.8. In the validation analyses, we used the same ANCOVA model and corrected methods to identify the effects of the COMT rs4680 and BDNF rs6265 on the gFCD derived from the connection thresholds of r = 0.4, 0.5, 0.6, 0.7 and 0.8.
Connection Probability Maps
All the preprocessing steps were the same as the gFCD calculation. For each significant cluster under a certain connection threshold, we used this connection threshold to generate the rsFC map of this cluster for each subject. Based on the rsFC maps of this cluster of all subjects, we generated a connection probability map of this cluster under the connection threshold. The connection probability map may represent the connection pattern of this cluster at the connection threshold.
RESULTS
Demographic and Genetic Characteristics
The demographic data of these subjects are summarized in Table 1. The distributions of COMT rs4680 (129 Val/Val, 110 Met/Val, and 26 Met/Met) and BDNF rs6265 (83 Val/Val, 134 Met/Val, and 48 Met/Met) were in Hardy–Weinberg equilibrium (P > 0.05). Subjects who were either homozygous or heterozygous for the Met‐allele were merged into a group of Met‐allele carriers of COMT rs4680 or BDNF rs6265, because of the relatively low frequency of the Met homozygotes; this method has been used previously to address skewed genotypic distributions [Aguilera et al., 2008; Ettinger et al., 2008; Li et al., 2009; Taylor et al., 2007].
Table 1.
Demographic data of subjects
| Genotypic groups | n = 265 | Age (years) | Years of education | Gender (male : female) |
|---|---|---|---|---|
| COMT Met‐BDNF Met | 93 | 22.2 ± 2.2 | 15.2 ± 2.1 | 43:50 |
| COMT Met‐BDNF Val/Val | 43 | 23.6 ± 2.6 | 15.9 ± 2.2 | 21:22 |
| COMT Val/Val‐BDNF Met | 89 | 22.9 ± 2.4 | 15.9 ± 2.1 | 38:51 |
| COMT Val/Val‐BDNF Val/Val | 40 | 22.9 ± 2.6 | 15.8 ± 2.0 | 18:22 |
| P values | 0.017 | 0.119 | 0.920 |
Mean gFCD Maps of Genotypic Subgroups
Using a connection threshold of 0.6, the mean gFCD maps of the four genotypic subgroups are shown in Figure 1. The four subgroups had similar gFCD spatial distribution: the greatest in the posterior cingulate cortex, precuneus, and medial occipital cortex; the medium in the medial and dorsolateral PFC, and lateral parietal cortex; and the lowest in the sensorimotor cortex and anterior temporal cortex.
Figure 1.
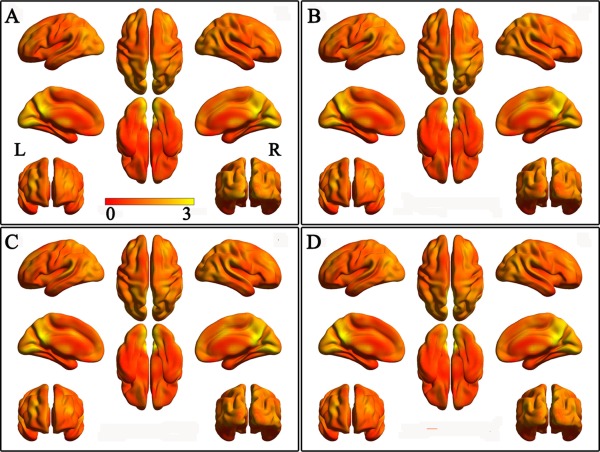
The mean gFCD maps in each of 4 groups with connectivity threshold of 0.6. A: COMT Met‐BDNF Val/Val subgroup; B: COMT Met‐BDNF Met subgroup; C: COMT Val/Val‐BDNF Val/Val subgroup; D: COMT Val/Val‐BDNF Met subgroup. [Color figure can be viewed in the online issue, which is available at http://wileyonlinelibrary.com.]
gFCD Analysis Based on the Recommended Connection Threshold
Based on the recommended connection threshold of 0.6 to calculate the gFCD map for each subject [Tomasi and Volkow, 2010], the genetic interactive effects on gFCD are shown in Figure 2. Although neither of the COMT rs4680 or the BDNF rs6265 showed a significant main effect on gFCD, we found a significant COMT × BDNF interaction (AlphaSim corrected, P < 0.005) on the gFCD in the left frontal eye field (FEF) (Fig. 2A), the location of which was confirmed by a meta‐analysis (Details see Supplementary Materials). The distribution of the gFCD in the left FEF across these genotypic subgroups was more likely an invert U‐shape according the presumed dopamine signaling from high to low (COMT Met‐BDNF Val/Val > COMT Val/Val‐BDNF Val/Val and COMT Met‐BDNF Met > COMT Val/Val‐BDNF Met). Specifically, the COMT Met‐BDNF Val/Val carriers had significant reduced gFCD than the COMT Val/Val‐BDNF Val/Val (P = 0.009) and COMT Met‐BDNF Met carriers (P = 0.012). The COMT Val/Val‐BDNF Met carriers had a trend towards gFCD reduction than the COMT Val/Val‐BDNF Val/Val (P = 0.091). There were no significant differences in gFCD between any other contrasts (Fig. 2B).
Figure 2.
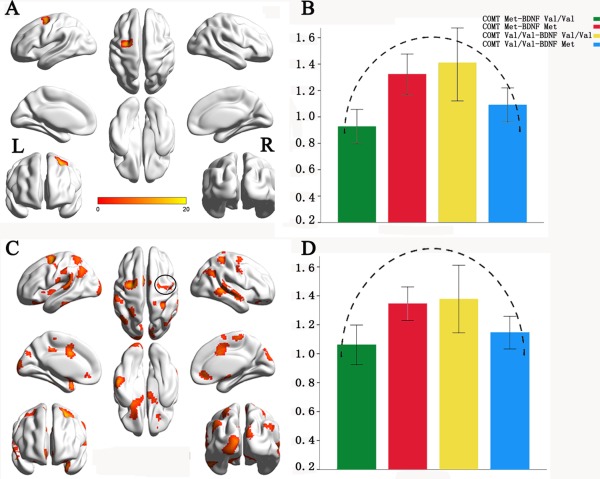
The COMT × BDNF interactions on global functional connectivity density (gFCD) with the connection threshold of 0.6. A: Brain region with significant interaction (AlphaSim corrected, P < 0.005). B: The modulation pattern of the FCD of the left FEF by the presumed dopamine signaling from high to low. C: Brain regions with significant interactions (AlphaSim corrected, P < 0.05). D: The modulation pattern of the FCD of the right FEF by the presumed dopamine signaling from high to low. [Color figure can be viewed in the online issue, which is available at http://wileyonlinelibrary.com.]
We also used a lenient statistical threshold (AlphaSim corrected, P< 0.05) to confirm whether the right FEF showed the similar interaction effect between COMT rs4680 and BDNF rs6265. The right FEF showed a COMT × BDNF interaction (Fig. 2C) and the gFCD distribution of the right FEF among four genotypic subgroups was similar with that of the left FEF that exhibited an invert U‐shape (Fig. 2D). We used a lenient statistical threshold (P < 0.05, uncorrected) to identify potential main effects of COMT and BDNF on gFCD. We found that multiple brain regions show main effects of COMT rs4680 and BDNF rs6265 on gFCD (Supporting Information Fig. S1).
Validation Analysis
To validate our results derived from the connection threshold of 0.6, we also repeated the gFCD analysis (AlphaSim corrected, P < 0.005) using the connection thresholds from 0.4 to 0.8 with a step of 0.1 (Fig. 3, Table 2). The left FEF consistently showed a significant COMT × BDNF interaction at all thresholds; the left angular gyrus (AG) exhibited an interaction effect at r = 0.4, 0.5 and 0.7; the left middle occipital gyrus (MOG) had an interaction at r = 0.4 and 0.5; the left posterior superior temporal gyrus (pSTG) showed an intraction at r = 0.4 and 0.8; and the right mid‐cingulate cortex (MCC) exhibited an interaction at r = 0.4. In these validation analyses, the distribution of the gFCD of these genotypic subgroups showed an invert U‐shape in the left FEF, the left pSTG and the right MCC and a U‐shape in the left AG and MOG (Fig. 3 and Supporting Information Fig. S2).
Figure 3.
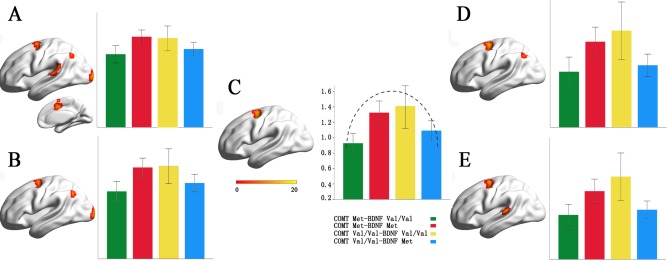
The COMT × BDNF interactions on gFCD with the connection thresholds ranged from 0.4 to 0.8. A: connection threshold = 0.4; B: connection threshold = 0.5; C: connection threshold = 0.6; D: connection threshold = 0.7; E: connection threshold = 0.8. [Color figure can be viewed in the online issue, which is available at http://wileyonlinelibrary.com.]
Table 2.
Brain areas with significant COMT × BDNF interactions on gFCD using different connection thresholds ranged from 0.4 to 0.8
| Connection thresholds | Brain regions | Peak F‐score | Cluster size (voxels) | MNI coordinates (x, y, z) |
|---|---|---|---|---|
| r = 0.4 | Left frontal eye field | 12.51 | 67 | −24, −9, 45 |
| Left angular gyrus | 14.30 | 96 | −39, −69, 54 | |
| Left middle occipital cortex | 15.15 | 100 | −21, −99, 6 | |
| Right mid‐cingulate cortex | 15.62 | 122 | 12, 6, 45 | |
| Left posterior superior temporal gyrus | 11.67 | 68 | −54, −27, 12 | |
| r = 0.5 | Left frontal eye field | 12.71 | 87 | −24, 0, 63 |
| Left angular cortex | 13.90 | 101 | −39, −69, 54 | |
| Left middle occipital gyrus | 13.52 | 121 | −21, −99 6 | |
| r = 0.6 | Left frontal eye field | 15.00 | 113 | −27, −3,54 |
| r = 0.7 | Left frontal eye field | 18.41 | 128 | −24, 0, 63 |
| Left angular cortex | 13.21 | 65 | −39, −72, 51 | |
| r = 0.8 | Left frontal eye field | 18.12 | 105 | −24, 0, 63 |
| Left posterior superior temporal cortex | 11.82 | 67 | −66, −33,12 |
Because we used a relatively strict statistical threshold (AlphaSim corrected, P < 0.05), we also re‐performed our analyses using a cluster‐corrected P < 0.05 threshold. Using this threshold, more brain regions showed significant interactions (Supporting Information Fig. S3). From Table 2, it seems that the strength of the interaction on FCD of the left FEF as a function of r values. We also tested correlations between the strengths (F values) of the interaction on gFCD of the left FEF and the connection thresholds of r. We found a significant correlation (r = 0.943, P = 0.016) (Supporting Information Fig. S4).
Connection Probability Maps
The connection probability maps of the significant clusters at different connection thresholds are shown in Figure 4 and Supporting Information Figure S5. At the same connection probability, brain regions exhibiting significant rsFCs with a certain cluster were largely different across connection thresholds. For example, at the same connection probability of 0.05, brain regions exhibiting rsFCs with the left FEF were gradually reduced at connection thresholds from 0.4 to 0.8 (Supporting Information Fig. S5). For several clusters with significant gFCD differences at a certain connection threshold, the connection probability maps may help to determine whether they are located in the same functional network. For example, under the connection threshold of r = 0.4, five clusters showed a significant interaction effect on gFCD (Fig. 3A). The 20% connection probability maps showed that they belong to three distinct functional networks (Fig. 4). Although four clusters mainly connected with the frontal, parietal and temporal cortices, the connection pattern of the left AG was completely different from those of the other three clusters. The left FEF, the right MCC and the left pSTG showed similar connectivity pattern and they were consistently included in the connection probability map of each of the three clusters, suggesting that they belong to the same functional network. The left MOG only connected with the occipital regions.
Figure 4.

The connection probability maps of each significant cluster at different connection thresholds. Columns 1–5 show connection probability maps of the left FEF, the left pSTG, the right MCC, the left AG and the left MOG. Rows 1–5 are connection probability maps at connection thresholds of 0.4–0.8.The blue areas are clusters with significant FCD differences, which have been used as seed regions for the calculation of functional connectivity. Abbreviation: AG, angular cortex; FEF, frontal eye field; MCC, mid‐cingulate cortex; MOC, middle occipital cortex; pSTG, posterior superior temporal gyrus. [Color figure can be viewed in the online issue, which is available at http://wileyonlinelibrary.com.]
The rsFC Difference Between Subgroups With High and Low gFCD of the Left FEF
We combined the COMT Met‐BDNF Val/Val and COMT Val/Val‐BDNF Met carriers into a low gFCD group and combined the COMT Val/Val‐BDNF Val/Val and COMT Met‐BDNF Met carriers into a high gFCD group. We then compared the rsFC difference of the left FEF between the two subgroups within a mask of the 20% connection probability map of the left FEF derived from a connection threshold of r = 0.6. Compared with the high gFCD group, the low gFCD group showed significantly lower rsFC between the left FEF (seed region) and the right FEF, the left parietal cortex, precentral gyrus and insular cortex (AlphaSim corrected, P < 0.005) (Fig. 5).
Figure 5.
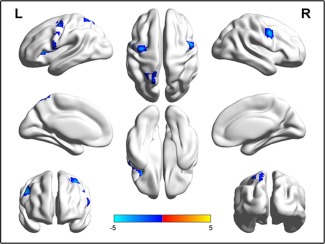
Connectivity differences of the left FEF between the high and low FCD subgroups (AlphaSim corrected, P < 0.005). The low FCD subgroup includes the COMT Met‐BDNF Val/Val and COMT Val/Val‐BDNF Met carriers and the high FCD subgroup includes the COMT Val/Val‐BDNF Val/Val and the COMT Met‐BDNF Met carriers. The blue color represents brain regions with significant lower functional connectivity of the left FEF in the low FCD subgroup than in the high FCD subgroup. [Color figure can be viewed in the online issue, which is available at http://wileyonlinelibrary.com.]
DISCUSSION
In the present study, we investigated the interaction of COMT rs4680 and BDNF rs6265 on the gFCD in healthy young adults. We found a significant COMT × BDNF interaction on the gFCD in the left FEF, suggesting an invert U‐shaped modulation by the presumed dopamine signaling. This finding was repeated in the gFCD analyses using other four connection thresholds.
Using a seed‐based rsFC analysis, one study has investigated COMT × BDNF interaction on the rsFC of the ventral striatum in healthy subjects [Wang et al., 2015]. The authors found a significant COMT × BDNF interaction on the rsFC between the ventral striatum and the PFC [Wang et al., 2015]. However, this hypothesis‐driven method cannot provide a complete picture of the COMT × BDNF interaction on the rsFCs of the whole brain. The FCD mapping is an ultra‐fast voxel‐wise data‐driven method that measures the number of rsFCs of a given voxel with all other voxels in the entire brain [Tomasi and Volkow, 2010, 2011a,b]. Greater gFCD values for particular voxels indicate that those voxels are functionally connected to a greater number of other brain voxels and suggest that those voxels play more important roles in the information processing. However, in the gFCD calculation, the connection threshold is arbitrarily selected by investigators. To validate the reproducibility of our findings, we repeated our gFCD analysis using the connection thresholds from 0.4 to 0.8 with a step of 0.1. Although results derived from different connection thresholds were not totally the same, the left FEF was the only region where the gFCD showed a significant COMT × BDNF interaction in all the five analyses. Moreover, the modulation patterns of the presumed dopamine signaling on the gFCD of the left FEF were very similar across different connection thresholds. These findings suggest that our results are robust to different thresholds.
The COMT is the main modulator of the synaptic dopamine concentration in the PFC due to the lack of dopamine transporter [Mannisto and Kaakkola, 1999; Seamans and Yang, 2004]. The BDNF exhibits the highest expression in the PFC [Pezawas et al., 2004] and involves in modulating activity‐dependent dopamine release [Pecina et al., 2014]. These may explain why we consistently found a significant COMT × BDNF interaction on the FCD in the left FEF, a brain region of the PFC.
Both COMT and BDNF are expected to modulate dopamine on their own and have exhibited their respective effects on rsFCs [Liu et al., 2010; Meyer et al., 2014; Wang et al., 2014]. In this study, the lack of significant main effects of COMT and BDNF on gFCD may be explained by the lack of enough power of the current sample to obtain significant main effects. This is confirmed by the finding of significant main effects at a lenient statistical threshold (P < 0.05, uncorrected). Similarly, the lack of power may also explain for the lack of significant COMT × BDNF interaction on the gFCD of the right FEF at the current statistical threshold. Future studies with large enough samples are needed to clarify these questions.
We found a linear correlation between the strengths of the interaction effect on gFCD of the left FEF and the connection thresholds of r. At the same connection probability, we also found that brain regions exhibiting rsFCs with the left FEF were gradually reduced at connection thresholds from 0.4 to 0.8. These findings suggest that brain regions with stronger rsFCs with the left FEF may make a greater contribution to the interaction effect on the gFCD of the left FEF than brain regions with weaker rsFCs with this region.
Although there is inconsistency across connection thresholds, four other clusters also show significant COMT × BDNF interaction. The connection probability maps showed that they belong to three distinct functional networks. The FEF, MCC and pSTG belong to the dorsal attention network (DAN); the AG belongs to an independent fronto‐parieto‐temporal network; and the MOG belongs to the visual network. Based on COMT Met and BDNF Val/Val carriers having higher synaptic dopamine concentration [Mannisto and Kaakkola, 1999; Pecina et al., 2014], we can generate genotypic subgroups with different levels of the presumed dopamine signaling (COMT Met‐BDNF Val/Val > COMT Val/Val‐BDNF Val/Val and COMT Met‐BDNF Met > COMT Val/Val‐BDNF Met). We found an invert U‐shape modulation of the presumed dopamine signaling on the FCD in the DAN regions and a U‐shape modulation in the other two clusters. These findings provide new evidence for the hypothesis of a network‐dependent modulation of the dopamine signaling on brain functional properties [Tian et al., 2013; Zhao et al., 2015].
Dopamine has been proposed to play an important role in the modulation of attention and has been linked to the integrity of the attention‐related networks, especially the DAN [Corbetta et al., 2008; Dang et al., 2012; Li et al., 2006; Nieoullon, 2002]. The DAN, involved in the endogenous goal‐driven attention orienting (top‐down) process, is hypothesized to modulate externally directed attention by amplifying or attenuating the saliency of relevant and irrelevant cues [Astafiev et al., 2003; Corbetta and Shulman, 2002; Fox et al., 2006; Giesbrecht et al., 2003; Hopfinger et al., 2000; Ptak and Schnider, 2010; Shulman et al., 2003]. For example, the prefrontal dopamine play a dominant role in the modulation of top‐down attention [Noudoost and Moore, 2011], and the top‐down selective visual attention was modulated by gene variants (i.e., COMT Val158Met) of the dopaminergic system [Schneider et al., 2015]. The administration of methylphenidate, a dopamine reuptake blocker, can enhance the activation in the DAN during visual attention and memory tasks [Muller et al., 2005; Tomasi et al., 2011]. In attention deficit hyperactivity disorder, most of available treatments target on the dopamine system [Solanto, 1998]. Destroying dopaminergic terminals in the frontal cortex in primates can impair the ability to acquire attentional sets [Crofts et al., 2001]. Our finding of the interaction of COMT rs4680 and BDNF rs6265 on FCD in the left FEF of the DAN suggests that they jointly modulate the functional connectivity of the DAN by acting on the dopamine system, which is also supported by our finding that the left FEF showed lower rsFC with several other regions of the DAN in the low gFCD group than in the high gFCD group.
There are several limitations in this study. Although this study included 265 subjects, this sample size is not large enough to identify significant main effects of COMT rs4680 and BDNF rs6265. Except for COMT and BDNF, several other genetic or environmental factors may also affect dopamine system and effect on the gFCD of the brain. These factors and their possible interactions should be further investigated. It has been reported that the use of multichannel array head coils may have an effect on the rsFC [Anteraper et al., 2013] and FCD [Tomasi et al., 2015]. In the present study, only an 8‐channel head coil was used, which may lower the sensitivity of our study to identify meaningful findings.
CONCLUSION
In this study, we used a data‐driven method to search brain regions that showed a significant interaction between COMT rs4680 and BDNF rs6265 on the gFCD in healthy young adults. We repeatedly found a significant COMT × BDNF interaction in the left FEF of the DAN. Moreover, we found that the COMT × BDNF interaction can be explained by an invert U‐shape modulation model of the dopamine. These findings may be helpful for understanding the neural mechanisms of the COMT × BDNF interactions on the FEF‐related attention functions.
Supporting information
Supporting Information
Disclosure: The authors have reported no conflicts of interest.
REFERENCES
- Aguilera M, Barrantes‐Vidal N, Arias B, Moya J, Villa H, Ibanez MI, Ruiperez MA, Ortet G, Fananas L (2008): Putative role of the COMT gene polymorphism (Val158Met) on verbal working memory functioning in a healthy population. Am J Med Genet 147B:898–902. [DOI] [PubMed] [Google Scholar]
- Akil M, Kolachana BS, Rothmond DA, Hyde TM, Weinberger DR, Kleinman JE (2003): Catechol‐O‐methyltransferase genotype and dopamine regulation in the human brain. J Neurosci 23:2008–2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso P, Lopez‐Sola C, Gratacos M, Fullana MA, Segalas C, Real E, Cardoner N, Soriano‐Mas C, Harrison BJ, Estivill X, Menchon JM (2013): The interaction between Comt and Bdnf variants influences obsessive‐compulsive‐related dysfunctional beliefs. J Anxiety Disorder 27:321–327. [DOI] [PubMed] [Google Scholar]
- Anteraper SA, Whitfield‐Gabrieli S, Keil B, Shannon S, Gabrieli JD, Triantafyllou C (2013): Exploring functional connectivity networks with multichannel brain array coils. Brain connectivity 3:302–315. [DOI] [PubMed] [Google Scholar]
- Astafiev SV, Shulman GL, Stanley CM, Snyder AZ, Van Essen DC, Corbetta M (2003): Functional organization of human intraparietal and frontal cortex for attending, looking, and pointing. J Neurosci 23:4689–4699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao MV (2003): Neurotrophins and their receptors: a convergence point for many signalling pathways. Nat Rev Neurosci 4:299–309. [DOI] [PubMed] [Google Scholar]
- Chen ZY, Patel PD, Sant G, Meng CX, Teng KK, Hempstead BL, Lee FS (2004): Variant brain‐derived neurotrophic factor (BDNF) (Met66) alters the intracellular trafficking and activity‐dependent secretion of wild‐type BDNF in neurosecretory cells and cortical neurons. J Neurosci 24:4401–4411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZY, Jing D, Bath KG, Ieraci A, Khan T, Siao CJ, Herrera DG, Toth M, Yang C, McEwen BS, Hempstead BL, Lee FS (2006): Genetic variant BDNF (Val66Met) polymorphism alters anxiety‐related behavior. Science 314:140–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbetta M, Shulman GL (2002): Control of goal‐directed and stimulus‐driven attention in the brain. Nat Rev Neurosci 3:201–215. [DOI] [PubMed] [Google Scholar]
- Corbetta M, Patel G, Shulman GL (2008): The reorienting system of the human brain: from environment to theory of mind. Neuron 58:306–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crofts HS, Dalley JW, Collins P, Van Denderen JC, Everitt BJ, Robbins TW, Roberts AC (2001): Differential effects of 6‐OHDA lesions of the frontal cortex and caudate nucleus on the ability to acquire an attentional set. Cereb Cortex 11:1015–1026. [DOI] [PubMed] [Google Scholar]
- Dang LC, O'Neil JP, Jagust WJ (2012): Dopamine supports coupling of attention‐related networks. J Neurosci 32:9582–9587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das D, Tan X, Bielak AA, Cherbuin N, Easteal S, Anstey KJ (2014): Cognitive ability, intraindividual variability, and common genetic variants of catechol‐O‐methyltransferase and brain‐derived neurotrophic factor: a longitudinal study in a population‐based sample of older adults. Psychol Aging 29:393–403. [DOI] [PubMed] [Google Scholar]
- Egan MF, Kojima M, Callicott JH, Goldberg TE, Kolachana BS, Bertolino A, Zaitsev E, Gold B, Goldman D, Dean M, Lu B, Weinberger DR (2003): The BDNF val66met polymorphism affects activity‐dependent secretion of BDNF and human memory and hippocampal function. Cell 112:257–269. [DOI] [PubMed] [Google Scholar]
- Ettinger U, Kumari V, Collier DA, Powell J, Luzi S, Michel TM, Zedomi O, Williams SC (2008): Catechol‐O‐methyltransferase (COMT) val158met genotype is associated with BOLD response as a function of task characteristic. Neuropsychopharmacology 33:3046–3057. [DOI] [PubMed] [Google Scholar]
- Fox MD, Corbetta M, Snyder AZ, Vincent JL, Raichle ME (2006): Spontaneous neuronal activity distinguishes human dorsal and ventral attention systems. Proc Natl Acad Sci U S A 103:10046–10051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giesbrecht B, Woldorff MG, Song AW, Mangun GR (2003): Neural mechanisms of top‐down control during spatial and feature attention. NeuroImage 19:496–512. [DOI] [PubMed] [Google Scholar]
- Han DH, Park DB, Choi TY, Joo SY, Lee MK, Park BR, Nishimura R, Chu CC, Renshaw PF (2008): Effects of brain‐derived neurotrophic factor‐catecholamine‐O‐methyltransferase gene interaction on schizophrenic symptoms. Neuroreport 19:1155–1158. [DOI] [PubMed] [Google Scholar]
- Ho BC, Andreasen NC, Dawson JD, Wassink TH (2007): Association between brain‐derived neurotrophic factor Val66Met gene polymorphism and progressive brain volume changes in schizophrenia. Am J Psychiatry 164:1890–1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopfinger JB, Buonocore MH, Mangun GR (2000): The neural mechanisms of top‐down attentional control. Nat Neurosci 3:284–291. [DOI] [PubMed] [Google Scholar]
- Huang EJ, Reichardt LF (2001): Neurotrophins: roles in neuronal development and function. Ann Rev Neurosci 24:677–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang JI, Song DH, Namkoong K, Kim SJ (2010): Interaction effects between COMT and BDNF polymorphisms on boredom susceptibility of sensation seeking traits. Psychiatry Res 178:132–136. [DOI] [PubMed] [Google Scholar]
- Kang JI, Kim SJ, Song YY, Namkoong K, An SK (2013): Genetic influence of COMT and BDNF gene polymorphisms on resilience in healthy college students. Neuropsychobiology 68:174–180. [DOI] [PubMed] [Google Scholar]
- Konishi Y, Tanii H, Otowa T, Sasaki T, Tochigi M, Umekage T, Motomura E, Shiroyama T, Kaiya H, Okazaki Y, Okada M (2014): Genexgenexgender interaction of BDNF and COMT genotypes associated with panic disorder. Prog Neuropsychopharmacol Biol Psychiatry 51:119–125. [DOI] [PubMed] [Google Scholar]
- Lewis DA, Melchitzky DS, Sesack SR, Whitehead RE, Auh S, Sampson A (2001): Dopamine transporter immunoreactivity in monkey cerebral cortex: regional, laminar, and ultrastructural localization. J Comp Neurol 432:119–136. [DOI] [PubMed] [Google Scholar]
- Li D, Sham PC, Owen MJ, He L (2006): Meta‐analysis shows significant association between dopamine system genes and attention deficit hyperactivity disorder (ADHD). Hum Mol Genet 15:2276–2284. [DOI] [PubMed] [Google Scholar]
- Li J, Yu C, Li Y, Liu B, Liu Y, Shu N, Song M, Zhou Y, Zhu W, Li K, Jiang T (2009): COMT val158met modulates association between brain white matter architecture and IQ. Am J Med Genet B Neuropsychiatr Genet 150B:375–380. [DOI] [PubMed] [Google Scholar]
- Liu B, Song M, Li J, Liu Y, Li K, Yu C, Jiang T (2010): Prefrontal‐related functional connectivities within the default network are modulated by COMT val158met in healthy young adults. J Neurosci 30:64–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannisto PT, Kaakkola S (1999): Catechol‐O‐methyltransferase (COMT): biochemistry, molecular biology, pharmacology, and clinical efficacy of the new selective COMT inhibitors. Pharmacol Rev 51:593–628. [PubMed] [Google Scholar]
- Matsumoto M, Weickert CS, Akil M, Lipska BK, Hyde TM, Herman MM, Kleinman JE, Weinberger DR (2003): Catechol O‐methyltransferase mRNA expression in human and rat brain: evidence for a role in cortical neuronal function. Neuroscience 116:127–137. [DOI] [PubMed] [Google Scholar]
- Meyer BM, Huemer J, Rabl U, Boubela RN, Kalcher K, Berger A, Banaschewski T, Barker G, Bokde A, Buchel C, Conrod P, Desrivieres S, Flor H, Frouin V, Gallinat J, Garavan H, Heinz A, Ittermann B, Jia T, Lathrop M, Martinot JL, Nees F, Rietschel M, Smolka MN, Bartova L, Popovic A, Scharinger C, Sitte HH, Steiner H, Friedrich MH, Kasper S, Perkmann T, Praschak‐Rieder N, Haslacher H, Esterbauer H, Moser E, Schumann G, Pezawas L (2014): Oppositional COMT Val158Met effects on resting state functional connectivity in adolescents and adults. Brain Struct Funct 221:103–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller U, Suckling J, Zelaya F, Honey G, Faessel H, Williams SC, Routledge C, Brown J, Robbins TW, Bullmore ET (2005): Plasma level‐dependent effects of methylphenidate on task‐related functional magnetic resonance imaging signal changes. Psychopharmacology 180:624–633. [DOI] [PubMed] [Google Scholar]
- Nieoullon A (2002): Dopamine and the regulation of cognition and attention. Prog Neurobiol 67:53–83. [DOI] [PubMed] [Google Scholar]
- Nolan KA, Bilder RM, Lachman HM, Volavka J (2004): Catechol O‐methyltransferase Val158Met polymorphism in schizophrenia: differential effects of Val and Met alleles on cognitive stability and flexibility. Am J Psychiatry 161:359–361. [DOI] [PubMed] [Google Scholar]
- Noudoost B, Moore T (2011): The role of neuromodulators in selective attention. Trend Cognit Sci 15:585–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldfield RC (1971): The assessment and analysis of handedness: the Edinburgh inventory. Neuropsychologia 9:97–113. [DOI] [PubMed] [Google Scholar]
- Papaleo F, Crawley JN, Song J, Lipska BK, Pickel J, Weinberger DR, Chen J (2008): Genetic dissection of the role of catechol‐O‐methyltransferase in cognition and stress reactivity in mice. J Neurosci 28:8709–8723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pecina M, Martinez‐Jauand M, Love T, Heffernan J, Montoya P, Hodgkinson C, Stohler CS, Goldman D, Zubieta JK (2014): Valence‐specific effects of BDNF Val66Met polymorphism on dopaminergic stress and reward processing in humans. J Neurosci 34:5874–5881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pezawas L, Verchinski BA, Mattay VS, Callicott JH, Kolachana BS, Straub RE, Egan MF, Meyer‐Lindenberg A, Weinberger DR (2004): The brain‐derived neurotrophic factor val66met polymorphism and variation in human cortical morphology. J Neurosci 24:10099–10102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poo MM (2001): Neurotrophins as synaptic modulators. Nat Rev Neurosci 2:24–32. [DOI] [PubMed] [Google Scholar]
- Power JD, Barnes KA, Snyder AZ, Schlaggar BL, Petersen SE (2012): Spurious but systematic correlations in functional connectivity MRI networks arise from subject motion. NeuroImage 59:2142–2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power JD, Barnes KA, Snyder AZ, Schlaggar BL, Petersen SE (2013): Steps toward optimizing motion artifact removal in functional connectivity MRI; A reply to Carp. NeuroImage 76:439–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptak R, Schnider A (2010): The dorsal attention network mediates orienting toward behaviorally relevant stimuli in spatial neglect. J Neurosci 30:12557–12565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren‐Patterson RF, Cochran LW, Holmes A, Sherrill S, Huang SJ, Tolliver T, Lesch KP, Lu B, Murphy DL (2005): Loss of brain‐derived neurotrophic factor gene allele exacerbates brain monoamine deficiencies and increases stress abnormalities of serotonin transporter knockout mice. J Neurosci Res 79:756–771. [DOI] [PubMed] [Google Scholar]
- Schneider KK, Schote AB, Meyer J, Frings C (2015): Genes of the dopaminergic system selectively modulate top‐down but not bottom‐up attention. Cognit Affect Behav Neurosci 15:104–116. [DOI] [PubMed] [Google Scholar]
- Seamans JK, Yang CR (2004): The principal features and mechanisms of dopamine modulation in the prefrontal cortex. Prog Neurobiol 74:1–58. [DOI] [PubMed] [Google Scholar]
- Shulman GL, McAvoy MP, Cowan MC, Astafiev SV, Tansy AP, d'Avossa G, Corbetta M (2003): Quantitative analysis of attention and detection signals during visual search. J Neurophysiol 90:3384–3397. [DOI] [PubMed] [Google Scholar]
- Solanto MV (1998): Neuropsychopharmacological mechanisms of stimulant drug action in attention‐deficit hyperactivity disorder: a review and integration. Behav Brain Res 94:127–152. [DOI] [PubMed] [Google Scholar]
- Taylor WD, Zuchner S, Payne ME, Messer DF, Doty TJ, MacFall JR, Beyer JL, Krishnan KR (2007): The COMT Val158Met polymorphism and temporal lobe morphometry in healthy adults. Psychiatry Res 155:173–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas G, Sinville R, Sutton S, Farquar H, Hammer RP, Soper SA, Cheng YW, Barany F (2004): Capillary and microelectrophoretic separations of ligase detection reaction products produced from low‐abundant point mutations in genomic DNA. Electrophoresis 25:1668–1677. [DOI] [PubMed] [Google Scholar]
- Tian T, Qin W, Liu B, Jiang T, Yu C (2013): Functional connectivity in healthy subjects is nonlinearly modulated by the COMT and DRD2 polymorphisms in a functional system‐dependent manner. J Neurosci 33:17519–17526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasi D, Volkow ND (2010): Functional connectivity density mapping. Proc Natl Acad Sci U S A 107:9885–9890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasi D, Volkow ND (2011a): Association between functional connectivity hubs and brain networks. Cereb Cortex 21:2003–2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasi D, Volkow ND (2011b): Functional connectivity hubs in the human brain. NeuroImage 57:908–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasi D, Volkow ND, Wang GJ, Wang R, Telang F, Caparelli EC, Wong C, Jayne M, Fowler JS (2011): Methylphenidate enhances brain activation and deactivation responses to visual attention and working memory tasks in healthy controls. NeuroImage 54:3101–3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasi D, Shokri‐Kojori E, Volkow ND (2015): High‐resolution functional connectivity density: hub locations, sensitivity, specificity, reproducibility, and reliability. Cereb Cortex (Epub ahead of print). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tunbridge EM, Harrison PJ, Weinberger DR (2006): Catechol‐o‐methyltransferase, cognition, and psychosis: Val158Met and beyond. Biol Psychiatry 60:141–151. [DOI] [PubMed] [Google Scholar]
- Twamley EW, Hua JP, Burton CZ, Vella L, Chinh K, Bilder RM, Kelsoe JR (2014): Effects of COMT genotype on cognitive ability and functional capacity in individuals with schizophrenia. Schizophr Res 159:114–117. [DOI] [PubMed] [Google Scholar]
- Ursini G, Bollati V, Fazio L, Porcelli A, Iacovelli L, Catalani A, Sinibaldi L, Gelao B, Romano R, Rampino A, Taurisano P, Mancini M, Di Giorgio A, Popolizio T, Baccarelli A, De Blasi A, Blasi G, Bertolino A (2011): Stress‐related methylation of the catechol‐O‐methyltransferase Val 158 allele predicts human prefrontal cognition and activity. J Neurosci 31:6692–6698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Zhang Y, Liu B, Long H, Yu C, Jiang T (2014): Dosage effects of BDNF Val66Met polymorphism on cortical surface area and functional connectivity. J Neurosci 34:2645–2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Liu B, Long H, Fan L, Li J, Zhang X, Qiu C, Yu C, Jiang T (2015): Epistatic interaction of BDNF and COMT on the frontostriatal system. Neuroscience 298:380–388. [DOI] [PubMed] [Google Scholar]
- Witte AV, Kurten J, Jansen S, Schirmacher A, Brand E, Sommer J, Floel A (2012): Interaction of BDNF and COMT polymorphisms on paired‐associative stimulation‐induced cortical plasticity. J Neurosci 32:4553–4561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh TK, Hu CY, Yeh TC, Lin PJ, Wu CH, Lee PL, Chang CY (2012): Association of polymorphisms in BDNF, MTHFR, and genes involved in the dopaminergic pathway with memory in a healthy Chinese population. Brain Cognit 80:282–289. [DOI] [PubMed] [Google Scholar]
- Yi P, Chen Z, Zhao Y, Guo J, Fu H, Zhou Y, Yu L, Li L (2009): PCR/LDR/capillary electrophoresis for detection of single‐nucleotide differences between fetal and maternal DNA in maternal plasma. Prenatal Diagn 29:217–222. [DOI] [PubMed] [Google Scholar]
- Zhao F, Zhang X, Qin W, Liu F, Wang Q, Xu Q, Wang J, Yu C (2015): Network‐Dependent Modulation of COMT and DRD2 Polymorphisms in Healthy Young Adults. Sci Rep 5:17996. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information


