Abstract
PICALM and CLU are two major risk genes of late‐onset Alzheimer's disease (LOAD), and there is strong molecular evidence suggesting their interaction on amyloid‐beta deposition, hence finding functional dependency between their risk genotypes may lead to better understanding of their roles in LOAD development and greater clinical utility. In this study, we mainly investigated interaction effects of risk loci PICALM rs3581179 and CLU rs11136000 on hippocampal degeneration in both young and elderly adults in order to understand their neural mechanism on aging process, which may help identify robust biomarkers for early diagnosis and intervention. Besides volume we also assessed hippocampal shape phenotypes derived from diffeomorphic metric mapping and nonlinear dimensionality reduction. In elderly individuals (75.6 ± 6.7 years) significant interaction effects existed on hippocampal volume (P < 0.001), whereas in young healthy adults (19.4 ± 1.1 years) such effects existed on a shape phenotype (P = 0.01) indicating significant variation at hippocampal head and tail that mirror most AD vulnerable regions. Voxel‐wise analysis also pointed to the same regions but lacked statistical power. In both cohorts, PICALM protective genotype AA only exhibited protective effects on hippocampal degeneration and cognitive performance when combined with CLU protective T allele, but adverse effects with CLU risk CC. This study revealed novel PICALM and CLU interaction effects on hippocampal degeneration along aging, and validated effectiveness of diffeomorphometry in imaging genetics study. Hum Brain Mapp 37:2419–2430, 2016. © 2016 Wiley Periodicals, Inc.
Keywords: PICALM, CLU, interaction, hippocampus, Alzheimer's disease
INTRODUCTION
Late‐onset Alzheimer's disease (LOAD) is a neurodegenerative disorder that has no effective treatment so far. There have been increasing needs to develop early intervention strategies that can slow down or stop the progression of the disease [Erk et al., 2011; Filippini et al., 2009; Zhang et al., 2014, 2015]. Because LOAD is highly heritable [Gatz et al., 2006], risk genetic variants can be used as robust biomarkers to identify at‐risk population who are likely to develop LOAD and benefit from early intervention. Emerging evidence has shown that major risk genes will interplay to modulate the brain structure and cognitive performance [Morgen et al., 2014; Zhang et al., 2014], hence finding dependence of one gene on another will contribute to more accurate identification of at‐risk population. In this study, we focus on two risk genes identified through genome‐wide association studies (GWAS), namely clusterin/apolipoprotein J protein gene (CLU) and phosphatidylinositol‐binding clathrin assembly protein gene (PICALM) [Corneveaux et al., 2010; Harold et al., 2009; Lambert et al., 2013]. Clusterin and PICALM may play reverse roles in amyloid‐beta (Aβ) deposition which is a hallmark of AD pathology: clusterin drives Aβ away from brain [Cirrito et al., 2008] whereas PICALM promotes Aβ deposition [Ehrlich et al., 2004; McMahon and Boucrot 2011], a strong evidence suggesting existence of an interaction between them. However, whether risk variants in the two genes interact to modulate brain structure and disease risk remains to be identified.
Many neuroimaging genetics studies towards understanding neural mechanism of CLU and PICALM were focused on their individual effects on neuroimaging and neuropsychological measurements [Biffi et al., 2010; Erk et al., 2011; Furney et al., 2011; Melville et al., 2012], whereas their interaction effects have been rarely studied. One study reported CLU and PICALM interacted to modulate resting‐state functional MRI (rs‐fMRI) connectivity of hippocampus in Chinese Han young healthy adults [Zhang et al., 2014], suggesting impact of the two genes on hippocampal structure, but no main or interaction effects of the two genes were observed on hippocampal volume [Bralten et al., 2011; Zhang et al., 2014]. Such ambiguity between different imaging modalities may arise from the limitation of gross volume measure which lacks ability to encode regional variability to reflect dynamic genetic effects. The commonly used voxel‐wise analysis is able to localize anatomical variability, but it is underpowered to detect small genetic effects due to heavy multiple test burden.
To overcome limitation of traditional morphometric analysis approaches, we employed diffeomorphic metric mapping [Yang et al., 2015] and nonlinear dimensionality reduction approach to derive compact hippocampal shape descriptors as a strategy to investigate effects of risk loci CLU rs11136000 and PICALM rs3851179 that were the most reported loci in GWAS [Corneveaux et al., 2010; Harold et al., 2009; Lambert et al., 2013] and imaging genetics studies [Biffi et al., 2010; Erk et al., 2011; Furney et al., 2011; Melville et al., 2012; Zhang et al., 2014]. Diffeomorphic metric based shape analysis had the ability to capture nonlinearity of the shape space [Miller et al., 2009; Yang et al., 2012] and may yield shape descriptors proximal to gene functions, hence increasing the detection power. Besides neuroimaging traits, we also analysed effects of the two risk loci on clinical diagnosis and memory performance in order to find the correspondence between imaging and function.
We performed analysis in both elderly and young healthy adults with the aim to understand the evolutional pattern of the genetic effects on the aging process. Ideally a consistent pattern across early and later life can provide strong basis for early intervention and disease progress monitoring. Towards this goal, we first performed analysis in elderly subjects (55–90 years) mixed with healthy controls and AD patients in order to find CLU and PICALM effects on hippocampal degeneration and disease risk, then we verified whether such effects could be traced back to young healthy adults. Elderly subjects were drawn from Alzheimer's Disease Neuroimaging Initiative (ADNI) study, and young healthy subjects from Chinese Han population. Effects of CLU and PICALM in the two cohorts are supposed to be comparable because their link to LOAD has been replicated in different ethnic groups, including Caucasian population and Chinese Han population [Carrasquillo et al., 2010; Chen et al., 2012; Corneveaux et al., 2010; Harold et al., 2009; Lee et al., 2011].
MATERIALS AND METHODS
Young Healthy Subjects
We recruited 360 young healthy Chinese university students (186 males and 174 females; mean age = 19.41 ± 1.09 years, range = 17–24 years; school education = 12.33 ± 0.80 years, range = 10–16 years]. The study was approved by the Ethics Committee of School of Life Science and Technology at University of Electronic Science and Technology of China, and all participants gave written informed consent. All of the participants were carefully screened to exclude individuals with a history of neurological or psychological diseases in the subjects or their third‐degree relatives, psychiatric treatment, drug or alcohol abuse, traumatic brain injury, or visible brain lesions on conventional MRI. All subjects were examined using the Chinese Revised Wechsler Adult Intelligence Scale (WAIS‐RC). 321 subjects passing all stages of quality control procedures were included in this study.
ADNI Study
Data used in the preparation of this article were obtained from the Alzheimer's Disease Neuroimaging Initiative (ADNI) database (http://www.adni.loni.usc.edu). The ADNI was launched in 2003 by the National Institute on Aging (NIA), the National Institute of Biomedical Imaging and Bioengineering (NIBIB), the Food and Drug Administration (FDA), private pharmaceutical companies and nonprofit organizations, as a $60 million, 5‐year public–private partnership. The primary goal of ADNI has been to test whether serial magnetic resonance imaging (MRI), positron emission tomography (PET), other biological markers, and clinical and neuropsychological assessment can be combined to measure the progression of mild cognitive impairment (MCI) and early Alzheimer's disease (AD). Determination of sensitive and specific markers of very early AD progression is intended to aid researchers and clinicians to develop new treatments and monitor their effectiveness, as well as lessen the time and cost of clinical trials.
The Principal Investigator of this initiative is Michael W. Weiner, MD, VA Medical Center and University of California—San Francisco. ADNI is the result of efforts of many co‐investigators from a broad range of academic institutions and private corporations, and subjects have been recruited from over 50 sites across the U.S. and Canada. The initial goal of ADNI was to recruit 800 subjects but ADNI has been followed by ADNI‐GO and ADNI‐2. To date these three protocols have recruited over 1500 adults, ages 55–90, to participate in the research, consisting of cognitively normal older individuals, people with early or late MCI, and people with early AD. The follow up duration of each group is specified in the protocols for ADNI‐1, ADNI‐2, and ADNI‐GO. Subjects originally recruited for ADNI‐1 and ADNI‐GO had the option to be followed in ADNI‐2. For up‐to‐date information, see http://www.adni-info.org.
Baseline data of ADNI1 were used in this study. 818 subjects were genotyped as part of the ADNI‐1 study. But to reduce population stratification effects, we only included 699 subjects of European ancestry clustered with CEU samples in HapMap phase3 data in this study, containing 194 controls, 337 MCIs and 168 ADs.
Genotyping
Ethylene diamine tetraacetic acid (EDTA) anticoagulated venous blood samples were collected from all Chinese young healthy individuals. Genomic DNA was extracted from whole blood using the EZgene Blood gDNA Miniprep Kit (Biomiga, San Diego, CA) according to the manufacturer's recommendations. Genotype data for CLU (rs11136000) and PICALM (rs3851179) and APOE (rs429358 and rs7412) were obtained using the standard Illumina genotyping protocol (Illumina).
ADNI samples were genotyped with Human610‐Quad BeadChip, which was described in detail elsewhere [Saykin et al., 2010]. When analysing population structure using genome‐wide data, we adopted the following quality control criteria: genotype call rate >98%, significant deviation from Hardy–Weinberg equilibrium (HWE) P > 10−6 and minor allele frequency >0.02. After QC procedure, 522 236 SNPs remained, and the maximum genotyping missing rate per subject was 0.102.
MRI Data Acquisition
MRI scans of Chinese young healthy subjects were performed on a MR750 3.0 T magnetic resonance scanner (GE Healthcare). Resting‐state functional imaging data were acquired using a gradient‐echo echo‐planar‐imaging (GRE‐EPI) sequence with the following parameters: repetition time (TR) = 2,000 ms, echo time (TE) = 30 ms, field of view (FOV) = 240 × 240 mm2, matrix = 64 × 64, flip angle = 90°, voxel size = 3.75 × 3.75 × 4.0 mm3, 39 slices, and 255 volumes. High‐resolution 3D T1‐weighted brain volume (BRAVO) MRI sequence was subsequently performed with the following parameters: TR = 8.16 ms, TE = 3.18 ms, flip angle = 7°, FOV = 256 × 256 mm2, voxel size = 1 × 1 × 1 mm3, and 188 slices. Before the scanning, all subjects were informed that they should move as little as possible, keep their eyes closed, think of nothing in particular and avoid falling asleep. Then, subjects were asked whether they fell asleep during and after the scanning to confirm that the included subjects did not fall asleep. ADNI MRI data acquisition was described in details elsewhere (http://www.adni.loni.usc.edu/about/centers-cores/mri-core/).
Memory Test
Individual working memory (WM) capacity in Chinese young healthy subjects was evaluated using the n‐back task, which has been widely used in previous studies on WM [Owen et al., 2005]. This task was performed on a computer in a quiet room outside the MRI scanner before performing magnetic resonance scanning for each subject, and the data were evaluated using E‐Prime, Version 2.0 (http://www.pstnet.com/eprime.cfm) as described in previous study [Liu et al., 2014]. Episodic memory performance in elderly was measured by MMSE (Mini‐Mental State Examination) score available in ADNI.
Hippocampal Segmentation
The hippocampal images of elderly individuals were semiautomated segmentations provided by ADNI, using high‐dimensional brain mapping tool SNT which was commercially available from Medtronic Surgical Navigation Technologies (Louisville, CO). SNT hippocampal volumetry has been previously validated on the normal aging, MCI and AD subjects [Hsu et al., 2002]. It first used 22 control points manually placed on the individual brain MRI as local landmarks. Fluid image transformation was then used to match the individual brains to a template brain [Christensen et al., 1997]. The segmentations were manually edited by qualified reviewers if the boundaries delineated by SNT were not accurate.
Hippocampal segmentation of young adult brains adopted multiatlases segmentation using joint label fusion [Wang et al., 2013] which achieved the state‐of‐the‐art segmentation accuracy. To be consistent with ADNI hippocampal segmentation, atlases were chosen from ADNI brains closest to young adult brains. Specifically, five ADNI brains closest to the center of young adult brains were selected as templates. The main steps of this pipeline are as follows: (1) Affine registration was first used to align template to subject's T1 image; (2) ROIs of the subject and aligned template were segmented and aligned again by affine transformation; (3) Affine‐aligned ROIs were then registered by nonlinear image registration implemented using ANTS software tools (http://stnava.github.io/ANTs/); (4) Hippocampal mask of template was then transformed to subject space by composing affine and nonlinear transformations; (5) Multi‐atlas segmentations were fused by minimizing the total expectation of labeling error, which not only considered intensity similarity between atlases and target, but also between atlases [Wang et al., 2013); (6) Manual checking was followed to correct those segmentations with minor errors, e.g., spikes, and remove those with obvious errors. Segmentation accuracy was measured by.
Hippocampal Shape Analysis
Hippocampal surfaces generated from the segmentations were smoothed using a low pass filter [Taubin, 1995]. Landmarks on the hippocampal surface were identified using 'blended intrinsic maps' [Kim et al., 2011] which provided low‐distortion mapping for pairs of surfaces with large deformations. One subject from each cohort was chosen to create hippocampal templates. Since the cloud of dense landmarks was used to represent hippocampal shapes, it was sufficient to characterize the shape variations regardless of the landmark locations, so we just chose the shapes whose mesh points were relatively evenly spaced as the template. For ADNI subjects, there were 1,103 mesh points on the left template, and 1,077 points on the right. For young subjects, there were 1,177 mesh points on the left template and 1,242 on the right. These surface points were mapped to the remaining surfaces by the method described above. Manual checking was followed to remove hippocampi with nonsmooth mappings, e.g., mesh folding or joining of distant regions. 483 ADNI subjects and 336 young adults passing all stages of hippocampal segmentation and landmark identification were included in shape analysis in the next.
Landmarked shapes were rigidly aligned to the template and a new template was generated by averaging the aligned shapes. Diffeomorphic metric mappings based on stationary velocity field parameterization [Yang et al., 2015] were then performed between the new template and rigidly aligned shapes. In this approach, shape metric was defined as the length of the shortest Lie group exponential path in Riemannian manifold of diffeomorphisms connecting two shapes. To construct a pairwise shape distance matrix, we computed distances between any two shapes using their second order approximation which used only their mapping information relative to the template [Yang et al., 2015], thus avoiding very high computational loads of pairwise diffeomorphic metric mappings. The second order approximation has been shown to improve significantly over the first order one, and achieve high accuracy in hippocampal shape metric estimation [Yang et al., 2015]. Sum of the left and right hippocampal shape metric matrices was projected onto a low‐dimensional Euclidean space using multidimensional scaling (MDS) that preserved the relationship of any two shapes described in the pair‐wise metric matrix. MDS components were ordered according to their variance, with the first one having the largest variance. However, selecting intrinsic dimensions of the shape space is a well‐known open question. One of the most commonly used method is to plot the eigenvalues of the kernel matrix stemmed from the distance or covariance matrix in descending order (called scree plot) and look for a “big gap” or “elbow” in such a graph [Jolliffe, 2002]. Though this approach is somewhat ad hoc, it has been shown as an effective approach in practice [Zhu and Ghodsi, 2006], so we adopted it to select shape components in this study.
Statistical Analysis
Hardy‐Weinberg equilibrium between expected and observed genotype distributions were tested in PLINK [Purcell et al., 2007] (version 1.9). Effects of gender and handedness were tested by χ2 test. Genetic association with hippocampal phenotypes adopted general linear regression model including minor allele dosages of CLU and PICALM loci and their interaction term. For elderly subjects, covariates included age, sex, education years, handness, total intracranial volume, APOE ε4 dosage, and the first four MDS components of genetic data merged with HapMap3 data. In young healthy adults, the covariates included age, sex, education years, handness, brain volume and APOE ε4 dosage. Interaction effect was evaluated by P value of the regression coefficient for the interaction term, and main effects were evaluated without the interaction term. All the regression analysis and χ 2 test were performed in Matlab software.
To observe the shape variation pattern each shape component characterized, the subjects were divided into two groups with coordinates above or below the mean value for each component, namely mean (+) and mean (−) groups, and two sample t tests were performed at each surface vertex using as phenotype the shape displacement relative to template which was projected onto the normal direction. The P value of each vertex was corrected using Gaussian random field theory [Taylor and Worsley, 2007; Worsley et al., 1999]. The tests were performed using software package SurfStat (http://www.math.mcgill.ca/keith/surfstat/).
RESULTS
Hippocampal Segmentation and Landmark Identification Results
For multiatlas hippocampal segmentation, dice measurement for the leave‐one out crossvalidation of the five ADNI atlases was 0.879 ± 0.037 (L/R). 24 out of 360 young subjects were observed with obvious hippocampal segmentation errors, e.g., spikes, and removed from further analysis.
The hippocampal landmark correspondence of two elderly subjects identified through blended intrinsic maps was shown in Figure 1 by fringe patterns and color‐coded landmarks. As was shown, the correspondence agreed with manual verification well. Based on the surfaces constructed from landmarks, only two young subjects were observed with obvious abnormal landmark identification such as significant mesh folding, and none of the elderly subjects were observed with obvious abnormality. To quantify the identification error, the hippocampal surfaces were deformed by diffeomorphic transformation, so the ground truth of landmark correspondence between original and deformed ones was known. Ten pairs of randomly chosen surfaces were generated. The average distance of the landmarks identified by blended intrinsic map to the ground truth was 0.16 ± 0.076 mm, much smaller than the average distance of neighbour landmarks (∼1 mm).
Figure 1.
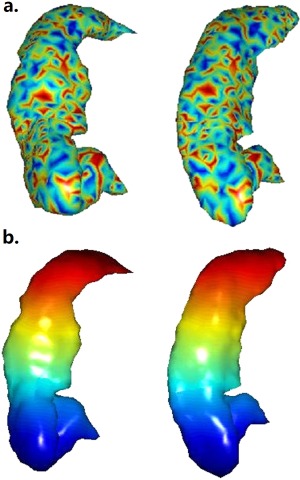
Hippocampal landmark correspondence identified by blended intrinsic maps. (a) Fringe pattern at similar locations indicated correspondence, (b) landmarks were color‐coded.
Hippocampal Shape Analysis Results
Diffeomorphic metric based analysis was performed in 483 elderly subjects and 336 young adults separately. Scree plots for MDS components were shown in Figure 2. For elderly subjects, there was “big gap” between the second and third components (see Fig. 2a), which was not observed between rest components, so the first two were retained for further analysis. The first shape component had high correlation with bilateral hippocampal volume (r = 0.915, P < 10−6), as well as significant correlation with age factor (r = 0.22, P < 10−6), and the first two components both had significant correlation with diagnosis (P < 0.001 for all). For young subjects, there was “big gap” between the third and fourth components (Fig. 2b), so the first three were retained for further analysis, and they all significantly associated with volume (r = 0.7, 0.33, and 0.41, respectively, P < 10−6 for all).
Figure 2.
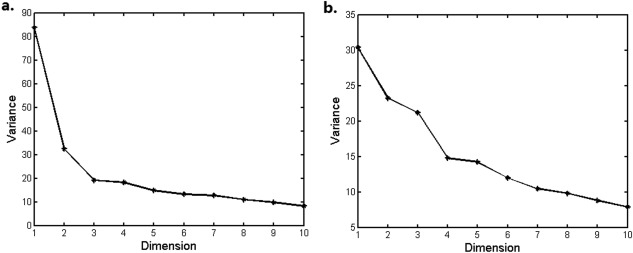
Scree plots of MDS components of hippocampal shapes. (a) Elderly subjects, (b) young adults.
Genetic Association Results
After quality control, 421 elderly subjects and 321 young adults were involved in imaging genetics study. Characteristics of these subjects were shown in Table 1. Genotype distributions for the CLU and PICALM risk loci were in Hardy–Weinberg equilibrium (P > 0.05) in both elderly and young subjects, and there were no significant age, sex, and education difference between genotypes (P > 0.05).
Table 1.
Characteristics of ADNI subjects (N = 421) and Chinese Han young healthy adults (N = 321) involved in the imaging genetics study; age and education years were denoted by ‘Mean ± SD’
| ADNI subjects | Chinese young healthy adults | |
|---|---|---|
| Diagnosis | 116 controls, 215 MCI, 90 AD | All normal |
| Sex | 247M/174F | 163M/158F |
| Age | 75.6 ± 6.7 years | 19.4 ± 1.1 years |
| Education | 15.6 ± 3.1 years | 12.3 ± 0.8 years |
| CLU genotype status | 160 CC/194 CT/67 TT | 205 CC/96 CT/20 TT |
| PICALM genotype status | 181 GG/186 GA/54 AA | 120 GG/154 GA/47 AA |
PICALM–CLU effects in elderly
In 421 mixed elderly subjects, CLU and PICALM showed significant interaction effects on the bilateral hippocampal volume (P = 9.7 × 10−4) as well as the first hippocampal shape component (P = 0.002), but not on the second component (P > 0.05). In CLU risk C allele homozygotes, PICALM protective AA genotype exhibited lower mean hippocampal volume than risk allele G carriers, whereas this effect was reversed in CLU protective T allele carriers (see Fig. 3a). PICALM rs3851179 also had nominally significant main effect on hippocampal volume (P = 0.05), but CLU rs11136000 did not (P = 0.47), consistent with previous finding (Biffi et al., 2010).
Figure 3.
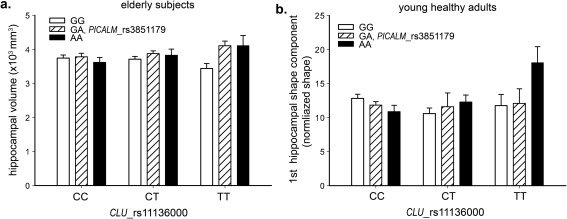
Hippocampal phenotypes for PICALM and CLU genotypes. (a) Hippocampal volume in 421 elderly subjects, (b) the 2nd hippocampal shape component in young healthy adults. Error bars were denoted by 'mean ± SEM'.
We further performed analysis in specific diagnostic groups. In each of the three groups PICALM AA genotype in CLU TT group exhibited obviously higher mean hippocampal volume than GG homozygotes, whereas in CLU risk CC group, such effects were weaker or reversed (see Fig. 4). Statistically, the interaction effects were significant in healthy controls (P = 0.047, n = 116), but not in MCIs (P = 0.17, n = 215) and AD patients (P = 0.11, n = 90). Main effects of PICALM rs3851179 were marginally significant in AD patients (P = 0.056), but not in other two groups. When three groups were merged and diagnosis used as covariate, significant PICALM–CLU interaction still remained on hippocampal volume (P = 0.017), further demonstrating the effects were not confounded by disease. In healthy elderly (n = 116) 1.01% of the hippocampal volume variance was explained by the interaction effects of the two variants after adjusting for other covariates, as opposed to 0.03% by just their main effects.
Figure 4.

Hippocampal volumes for PICALM and CLU genotypes in three diagnostic groups of elderly subjects. Error bars were denoted by 'mean ± SEM'.
With regard to cognitive functions, PICALM protective AA genotype exhibited a higher AD ratio and lower mean adjusted MMSE score (after adjusting for covariates) than risk G carriers in the CLU risk CC group, which was reversed in CLU protective T carriers (Table 2). This pattern reflected the imaging findings, but statistical significance was not reached.
Table 2.
AD ratios, adjusted MMSE score and WM test for PICALM genotypes with CLU CC genotype or T carriers in elderly or young subjects; age and residuals were denoted by ‘Mean ± SD’
| PICALM genotype | Elderly subjects (N = 699) | Young healthy subjects (N = 321) | ||||
|---|---|---|---|---|---|---|
| Age | AD ratio | MMSE residual | Age | Working memory test (2‐back condition) | ||
| CLU CC | GG | 74.5 ± 6.8 | 22.9% | −0.18 ± 0.16 | 19.5 ± 1.1 | 0.073 ± 0.14 |
| GA | 75.7 ± 6.8 | 19.7% | 0.16 ± 0.15 | 19.3 ± 1.2 | −0.096 ± 1.35 | |
| AA | 76.0 ± 6.7 | 26.3% | −0.48 ± 0.28 | 19.4 ± 1.1 | −0.25 ± 1.87 | |
| CLU CT/TT | GG | 75.5 ± 6.8 | 23.9% | 0.027 ± 0.13 | 19.0 ± 1.0 | 0.087 ± 0.1 |
| GA | 75.4 ± 6.8 | 25.5% | 0.002 ± 0.12 | 19.5 ± 0.9 | 0.109 ± 0.13 | |
| AA | 76.3 ± 6.2 | 19.6% | 0.26 ± 0.23 | 19.1 ± 0.9 | 0.114 ± 0.09 | |
PICALM–CLU effects in young healthy adults
We next tested the interaction effects of the two loci on bilateral hippocampal volume as well as the first three shape components in 321 young healthy subjects. The analysis revealed that a significant interaction existed on the second hippocampal shape component (P = 0.01), demonstrating the same interaction pattern as that observed in the elderly cohort (Fig. 3b). Even corrected for multiple tests, this effect was still significant (adjusted P = 0.04). Main effect terms of the two variants in the full model were significant with P = 0.01 and 0.02 for PICALM and CLU, respectively, though their marginal main effects were nonsignificant when interaction term was not presented. As demonstrated in Figure 5a, the second component characterized significant outward deformations at head, tail, and fimbria regions for mean (+) group relative to mean (−) group, reflecting significant local volume variations at these sites. In contrast, the first shape component with high correlation with bilateral volume (r = 0.7, P < 10−12) indicated significant outward variation from head to tail (Fig. 5b), which was similar to that in elderly (Fig. 5c), but no interaction effects existed on this component in young healthy adults (P = 0.53). Based on this observation, we could conclude that the PICALM AA genotype tended to result in greater volume loss in the head and tail regions of the hippocampus than observed for the GG genotype in the CLU risk CC group, versus a smaller one than for the GG genotype in CLU protective T carriers.
Figure 5.

Significant outward deformation of mean (+) group relative to mean (−) group for hippocampal shape components. (a) Second shape component in young adults; (b) 1st shape component in young adults; (c) 1st shape component in elderly subjects. M: medial; P: posterior. [Color figure can be viewed in the online issue, which is available at http://wileyonlinelibrary.com.]
We also performed voxel‐wise analysis of the CLU–PICALM interaction using the landmark displacement vector as phenotype. The strongest interaction effects again appeared at the head, tail, and fimbria regions (see Fig. 6), consistent to the imaging pattern characterized by the second shape component (Fig. 5a). But none of these effects achieved statistical significance (P < 0.05) after multiple test correction, demonstrating the necessity of dimensionality reduction.
Figure 6.
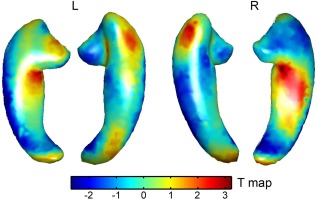
Voxel‐wise analysis of PICALM–CLU interaction using landmark displacement vectors as phenotypes. T maps of the interaction term are shown.
In relation to the imaging interaction pattern, the PICALM protective AA genotype demonstrated a higher mean WM (2‐back) test score (after adjustment) than observed for the risk G allele in CLU protective T carriers, with an adverse effect being recorded in the CLU risk CC group (Table 2), although statistical significance was not reached.
DISCUSSION
The main findings of this study are that common LOAD risk loci in CLU and PICALM exhibited significant interaction effects on hippocampal morphology in both young healthy adults and elderly individuals. The interaction effects in young healthy adults were mainly manifested at the most AD vulnerable regions, e.g., CA1 and subiculum [Hyman et al., 1984; Van Hoesen and Hyman, 1990], but this effect was cumulative and spread to the whole volume with aging, which finally led to differential disease risk. In both elderly and young subjects, the PICALM LOAD protective genotype AA was only protective against hippocampal neurodegeneration and memory deficits in association with the CLU protective T allele, whereas adverse effects were observed for the CLU risk CC genotype. This genetic interaction pattern, together with neuroimaging and neuropsychological measures, provides robust biomarkers for at‐risk population identification and disease progression monitoring.
Interestingly, this study was the first to observe the significant impact of CLU and PICALM on structural MRI phenotypes in young healthy adults, showing the power of this shape analysis approach in genetic association studies. Voxel‐wise genetic association revealed similar interaction pattern in the case of the hippocampus surface but was underpowered. The metric based analysis also showed greater power than the PCA based analysis. Previous imaging genetics studies have not found significant effects of major LOAD risk genes including APOE, CLU, and PICALM on hippocampal volume in cognitive normal adults aged from 20 to 50 [Jack et al., 2015; Zhang et al., 2014), whereas our finding showed that these genetic effects may exist in subregions of this brain structure, and that shape analysis can capture such dynamic effects. The second shape component indicated that a subgroup of young healthy adults exhibited more atrophy in the hippocampal head and tail regions, with the interaction of CLU and PICALM explaining 1.79% of this variance. In terms of only the two loci considered, this effect was significant and understandable given that a single locus can usually explain up to 0.5% of the imaging variance [Hibar et al., 2015], demonstrating significant impact of the two genes in hippocampal development.
Our structural findings also corroborate the results of a previous fMRI study which reported a CLU–PICALM interaction on resting‐state functional connectivity of the hippocampus in an independent Chinese young healthy cohort and indicated that PICALM protective AA genotype with CLU risk CC homozygotes would suffer hyperactivity of hippocampus that may predict faster cognitive decline in the future [Zhang et al., 2014], whereas our data further identified structural basis of this functional finding by showing that this genotype combination exhibited the heaviest degeneration in hippocampal head and tail regions, which may facilitate the development of new therapeutic approaches. Our study also substantially extends previous imaging studies of CLU and PICALM in the elderly [Biffi et al., 2010; Furney et al., 2011; Melville et al., 2012] by showing that their interaction can significantly increase the power in explaining hippocampal volume variation.
The molecular mechanism underlying the effect of CLU and PICALM interaction on neurodegeneration may involve their reverse modulation of Aβ levels that affect the extent of neuronal loss. Clusterin (apopoliprotein J) mediates the clearance of Aβ by enhancing endocytosis [Cirrito et al., 2008] and binding Aβ into an insoluble form [DeMattos et al., 2004; Nuutinen et al., 2009], whereas PICALM promotes Aβ generation via clathrin‐mediated endocytosis [Ehrlich et al., 2004; McMahon and Boucrot, 2011]. Moreover, clusterin is an inducible lipoprotein, the expression of which can be greatly increased under AD pathological conditions to promote the clearance of Aβ [Bertrand et al., 1995; Nuutinen et al., 2007]. This protective mechanism may explain why the interaction of the PICALM risk GG genotype with the CLU risk CC genotype results in less neurodegeneration than that observed in the case of PICALM protective AA and CLU CC. In contrast, the effect of the interaction between APOE and PICALM reported in a previous study was quite different, showing that the PICALM risk GG genotype in APOE risk ε4 carriers produced the lowest gray matter volume and memory test scores [Morgen et al., 2014]. One explanation for this is that apoE is not an inducible protein although it has similar functions to clusterin/apoJ in terms of Aβ clearance in the brain [DeMattos et al., 2004], and its low level expression is a marker of AD pathology. Together with previous studies, our findings contribute to a more profound understanding of the complex roles of major LOAD risk genes underlying AD.
In conclusion, the hippocampal shape features derived from the diffeomorphic metric‐based shape analysis have led to the identification of significant CLU‐PICALM interaction effects on hippocampal morphology in young healthy adults, which were not identified by volume measurement and voxel‐wise analysis. The consistent genetic interaction pattern on imaging and cognitive measures across young and elderly adults suggested robust biomarkers for the early diagnosis and treatment of AD. It showed that the interaction analysis could significantly improve the clinical outcome of PICALM and CLU in explaining brain development and predicting LOAD risk, and this study may prompt further interaction analysis of more LOAD risk genes. We can also apply this diffeomorphometry approach to other brain structures to discover novel neural mechanism of risk genes of LOAD and other mental diseases.
ACKNOWLEDGMENTS
ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: Alzheimer's Association; Alzheimer's Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc.; Biogen Idec Inc.; Bristol‐Myers Squibb Company; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; EuroImmun; F. Hoffmann‐La Roche Ltd and its affiliated company Genentech, Inc.; Fujirebio; GE Healthcare; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research and Development, LLC.; Johnson and Johnson Pharmaceutical Research and Development LLC.; Medpace, Inc.; Merck and Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Synarc Inc.; and Takeda Pharmaceutical Company. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (http://www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer's Disease Cooperative Study at the University of California, San Diego. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California. Conflict of interest: The authors declare no competing financial interests.
Disclosure: The authors declare no competing financial interests.
REFERENCES
- Bertrand P, Poirier J, Oda T, Finch CE, Pasinetti GM (1995): Association of apolipoprotein E genotype with brain levels of apolipoprotein E and apolipoprotein J (clusterin) in Alzheimer disease. Brain Res Mol Brain Res 33:174–178. [DOI] [PubMed] [Google Scholar]
- Biffi A, Anderson CD, Desikan RS, Sabuncu M, Cortellini L, Schmansky N, Salat D, Rosand J, Alzheimer's Disease Neuroimaging (2010): Genetic variation and neuroimaging measures in Alzheimer disease. Arch Neurol 67:677–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bralten J, Arias‐Vasquez A, Makkinje R, Veltman JA, Brunner HG, Fernandez G, Rijpkema M, Franke B (2011): Association of the Alzheimer's gene SORL1 with hippocampal volume in young, healthy adults. Am J Psychiatry 168:1083–1089. [DOI] [PubMed] [Google Scholar]
- Carrasquillo MM, Belbin O, Hunter TA, Ma L, Bisceglio GD, Zou F, Crook JE, Pankratz VS, Dickson DW, Graff‐Radford NR, Petersen RC, Morgan K, Younkin SG (2010): Replication of CLU, CR1, and PICALM associations with Alzheimer disease. Arch Neurol 67:961–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LH, Kao PY, Fan YH, Ho DT, Chan CS, Yik PY, Ha JC, Chu LW, Song YQ (2012): Polymorphisms of CR1, CLU and PICALM confer susceptibility of Alzheimer's disease in a southern Chinese population. Neurobiol Aging 33:210 e1–210 e7. [DOI] [PubMed] [Google Scholar]
- Christensen GE, Joshi SC, Miller MI (1997): Volumetric transformation of brain anatomy. IEEE Trans Med Imaging 16:864–877. [DOI] [PubMed] [Google Scholar]
- Cirrito JR, Kang JE, Lee J, Stewart FR, Verges DK, Silverio LM, Bu G, Mennerick S, Holtzman DM (2008): Endocytosis is required for synaptic activity‐dependent release of amyloid‐beta in vivo . Neuron 58:42–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corneveaux JJ, Myers AJ, Allen AN, Pruzin JJ, Ramirez M, Engel A, Nalls MA, Chen K, Lee W, Chewning K, Villa SE, Meechoovet HB, Gerber JD, Frost D, Benson HL, O'Reilly S, Chibnik LB, Shulman JM, Singleton AB, Craig DW, Van Keuren‐Jensen KR, Dunckley T, Bennett DA, De Jager PL, Heward C, Hardy J, Reiman EM, Huentelman MJ (2010): Association of CR1, CLU and PICALM with Alzheimer's disease in a cohort of clinically characterized and neuropathologically verified individuals. Hum Mol Genet 19:3295–3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMattos RB, Cirrito JR, Parsadanian M, May PC, O'Dell MA, Taylor JW, Harmony JA, Aronow BJ, Bales KR, Paul SM, Holtzman DM (2004): ApoE and clusterin cooperatively suppress Abeta levels and deposition: evidence that ApoE regulates extracellular Abeta metabolism in vivo . Neuron 41:193–202. [DOI] [PubMed] [Google Scholar]
- Ehrlich M, Boll W, Van Oijen A, Hariharan R, Chandran K, Nibert ML, Kirchhausen T (2004): Endocytosis by random initiation and stabilization of clathrin‐coated pits. Cell 118:591–605. [DOI] [PubMed] [Google Scholar]
- Erk S, Meyer‐Lindenberg A, Opitz von Boberfeld C, Esslinger C, Schnell K, Kirsch P, Mattheisen M, Muhleisen TW, Cichon S, Witt SH, Rietschel M, Nothen MM, Walter H (2011): Hippocampal function in healthy carriers of the CLU Alzheimer's disease risk variant. J Neurosci 31:18180–18184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippini N, MacIntosh BJ, Hough MG, Goodwin GM, Frisoni GB, Smith SM, Matthews PM, Beckmann CF, Mackay CE (2009): Distinct patterns of brain activity in young carriers of the APOE‐epsilon4 allele. Proc Natl Acad Sci USA 106:7209–7214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furney SJ, Simmons A, Breen G, Pedroso I, Lunnon K, Proitsi P, Hodges A, Powell J, Wahlund LO, Kloszewska I, Mecocci P, Soininen H, Tsolaki M, Vellas B, Spenger C, Lathrop M, Shen L, Kim S, Saykin AJ, Weiner MW, Lovestone S, Alzheimer's Disease Neuroimaging Initiative, AddNeuroMed Consortium (2011): Genome‐wide association with MRI atrophy measures as a quantitative trait locus for Alzheimer's disease. Mol Psychiatry 16:1130–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatz M, Reynolds CA, Fratiglioni L, Johansson B, Mortimer JA, Berg S, Fiske A, Pedersen NL (2006): Role of genes and environments for explaining Alzheimer disease. Arch Gen Psychiatry 63:168–174. [DOI] [PubMed] [Google Scholar]
- Harold D, Abraham R, Hollingworth P, Sims R, Gerrish A, Hamshere ML, Pahwa JS, Moskvina V, Dowzell K, Williams A, Jones N, Thomas C, Stretton A, Morgan AR, Lovestone S, Powell J, Proitsi P, Lupton MK, Brayne C, Rubinsztein DC, Gill M, Lawlor B, Lynch A, Morgan K, Brown KS, Passmore PA, Craig D, McGuinness B, Todd S, Holmes C, Mann D, Smith AD, Love S, Kehoe PG, Hardy J, Mead S, Fox N, Rossor M, Collinge J, Maier W, Jessen F, Schurmann B, Heun R, van den Bussche H, Heuser I, Kornhuber J, Wiltfang J, Dichgans M, Frolich L, Hampel H, Hull M, Rujescu D, Goate AM, Kauwe JS, Cruchaga C, Nowotny P, Morris JC, Mayo K, Sleegers K, Bettens K, Engelborghs S, De Deyn PP, Van Broeckhoven C, Livingston G, Bass NJ, Gurling H, McQuillin A, Gwilliam R, Deloukas P, Al‐Chalabi A, Shaw CE, Tsolaki M, Singleton AB, Guerreiro R, Muhleisen TW, Nothen MM, Moebus S, Jockel KH, Klopp N, Wichmann HE, Carrasquillo MM, Pankratz VS, Younkin SG, Holmans PA, O'Donovan M, Owen MJ, Williams J (2009): Genome‐wide association study identifies variants at CLU and PICALM associated with Alzheimer's disease. Nat Genet 41:1088–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibar DP, Stein JL, Renteria ME, Arias‐Vasquez A, Desrivieres S, Jahanshad N, Toro R, Wittfeld K, Abramovic L, Andersson M, Aribisala BS, Armstrong NJ, Bernard M, Bohlken MM, Boks MP, Bralten J, Brown AA, Chakravarty MM, Chen Q, Ching CR, Cuellar‐Partida G, den Braber A, Giddaluru S, Goldman AL, Grimm O, Guadalupe T, Hass J, Woldehawariat G, Holmes AJ, Hoogman M, Janowitz D, Jia T, Kim S, Klein M, Kraemer B, Lee PH, Olde Loohuis LM, Luciano M, Macare C, Mather KA, Mattheisen M, Milaneschi Y, Nho K, Papmeyer M, Ramasamy A, Risacher SL, Roiz‐Santianez R, Rose EJ, Salami A, Samann PG, Schmaal L, Schork AJ, Shin J, Strike LT, Teumer A, van Donkelaar MM, van Eijk KR, Walters RK, Westlye LT, Whelan CD, Winkler AM, Zwiers MP, Alhusaini S, Athanasiu L, Ehrlich S, Hakobjan MM, Hartberg CB, Haukvik UK, Heister AJ, Hoehn D, Kasperaviciute D, Liewald DC, Lopez LM, Makkinje RR, Matarin M, Naber MA, McKay DR, Needham M, Nugent AC, Putz B, Royle NA, Shen L, Sprooten E, Trabzuni D, van der Marel SS, van Hulzen KJ, Walton E, Wolf C, Almasy L, Ames D, Arepalli S, Assareh AA, Bastin ME, Brodaty H, Bulayeva KB, Carless MA, Cichon S, Corvin A, Curran JE, Czisch M, de Zubicaray GI, Dillman A, Duggirala R, Dyer TD, Erk S, Fedko IO, Ferrucci L, Foroud TM, Fox PT, Fukunaga M, Gibbs JR, Goring HH, Green RC, Guelfi S, Hansell NK, Hartman CA, Hegenscheid K, Heinz A, Hernandez DG, Heslenfeld DJ, Hoekstra PJ, Holsboer F, Homuth G, Hottenga JJ, Ikeda M, Jack CR, Jr Jenkinson M, Johnson R, Kanai R, Keil M, Kent JW, Jr Kochunov P, Kwok JB, Lawrie SM, Liu X, Longo DL, McMahon KL, Meisenzahl E, Melle I, Mohnke S, Montgomery GW, Mostert JC, Muhleisen TW, Nalls MA, Nichols TE, Nilsson LG, Nothen MM, Ohi K, Olvera RL, Perez‐Iglesias R, Pike GB, Potkin SG, Reinvang I, Reppermund S, Rietschel M, Romanczuk‐Seiferth N, Rosen GD, Rujescu D, Schnell K, Schofield PR, Smith C, Steen VM, Sussmann JE, Thalamuthu A, Toga AW, Traynor BJ, Troncoso J, Turner JA, Valdes Hernandez MC, van't Ent D, van der Brug M, van der Wee NJ, van Tol MJ, Veltman DJ, Wassink TH, Westman E, Zielke RH, Zonderman AB, Ashbrook DG, Hager R, Lu L, McMahon FJ, Morris DW, Williams RW, Brunner HG, Buckner RL, Buitelaar JK, Cahn W, Calhoun VD, Cavalleri GL, Crespo‐Facorro B, Dale AM, Davies GE, Delanty N, Depondt C, Djurovic S, Drevets WC, Espeseth T, Gollub RL, Ho BC, Hoffmann W, Hosten N, Kahn RS, Le Hellard S, Meyer‐Lindenberg A, Muller‐Myhsok B, Nauck M, Nyberg L, Pandolfo M, Penninx BW, Roffman JL, Sisodiya SM, Smoller JW, van Bokhoven H, van Haren NE, Volzke H, Walter H, Weiner MW, Wen W, White T, Agartz I, Andreassen OA, Blangero J, Boomsma DI, Brouwer RM, Cannon DM, Cookson MR, de Geus EJ, Deary IJ, Donohoe G, Fernandez G, Fisher SE, Francks C, Glahn DC, Grabe HJ, Gruber O, Hardy J, Hashimoto R, Hulshoff Pol HE, Jonsson EG, Kloszewska I, Lovestone S, Mattay VS, Mecocci P, McDonald C, McIntosh AM, Ophoff RA, Paus T, Pausova Z, Ryten M, Sachdev PS, Saykin AJ, Simmons A, Singleton A, Soininen H, Wardlaw JM, Weale ME, Weinberger DR, Adams HH, Launer LJ, Seiler S, Schmidt R, Chauhan G, Satizabal CL, Becker JT, Yanek L, van der Lee SJ, Ebling M, Fischl B, Longstreth WT, Jr Greve D, Schmidt H, Nyquist P, Vinke LN, van Duijn CM, Xue L, Mazoyer B, Bis JC, Gudnason V, Seshadri S, Ikram MA, Alzheimer's Disease Neuroimaging I, Consortium C, Epigen Imagen Sys Martin NG, Wright MJ, Schumann G, Franke B, Thompson PM, Medland SE (2015): Common genetic variants influence human subcortical brain structures. Nature 520:224–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu YY, Schuff N, Du AT, Mark K, Zhu X, Hardin D, Weiner MW (2002): Comparison of automated and manual MRI volumetry of hippocampus in normal aging and dementia. J Magn Reson Imaging 16:305–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman BT, Van Hoesen GW, Damasio AR, Barnes CL (1984): Alzheimer's disease: Cell‐specific pathology isolates the hippocampal formation. Science 225:1168–1170. [DOI] [PubMed] [Google Scholar]
- Jack CRJ, Wiste HJ, Weigand SD, Knopman DS, Vemuri P, Mielke MM, Lowe V, Senjem ML, Gunter JL, Machulda MM, Gregg BE, Pankratz VS, Rocca WA, Petersen RC (2015): Age, sex, and APOE epsilon4 effects on memory, brain structure, and beta‐amyloid across the adult life span. JAMA Neurol 72:511–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolliffe IT (2002): Principal Component Analysis. New York: Springer; pp 487. [Google Scholar]
- Kim VG, Lipman Y, Funkhouser T (2011): Blended intrinsic maps. ACM Trans Graphics 30:1–12. [Google Scholar]
- Lambert JC, Ibrahim‐Verbaas CA, Harold D, Naj AC, Sims R, Bellenguez C, DeStafano AL, Bis JC, Beecham GW, Grenier‐Boley B, Russo G, Thorton‐Wells TA, Jones N, Smith AV, Chouraki V, Thomas C, Ikram MA, Zelenika D, Vardarajan BN, Kamatani Y, Lin CF, Gerrish A, Schmidt H, Kunkle B, Dunstan ML, Ruiz A, Bihoreau MT, Choi SH, Reitz C, Pasquier F, Cruchaga C, Craig D, Amin N, Berr C, Lopez OL, De Jager PL, Deramecourt V, Johnston JA, Evans D, Lovestone S, Letenneur L, Moron FJ, Rubinsztein DC, Eiriksdottir G, Sleegers K, Goate AM, Fievet N, Huentelman MW, Gill M, Brown K, Kamboh MI, Keller L, Barberger‐Gateau P, McGuiness B, Larson EB, Green R, Myers AJ, Dufouil C, Todd S, Wallon D, Love S, Rogaeva E, Gallacher J, St George‐Hyslop P, Clarimon J, Lleo A, Bayer A, Tsuang DW, Yu L, Tsolaki M, Bossu P, Spalletta G, Proitsi P, Collinge J, Sorbi S, Sanchez‐Garcia F, Fox NC, Hardy J, Deniz Naranjo MC, Bosco P, Clarke R, Brayne C, Galimberti D, Mancuso M, Matthews F, European Alzheimer's Disease I, Genetic Environmental Risk in Alzheimer's D, Alzheimer's Disease Genetic C, Cohorts for H, Aging Research in Genomic E, Moebus S, Mecocci P, Del Zompo M, Maier W, Hampel H, Pilotto A, Bullido M, Panza F, Caffarra P, Nacmias B, Gilbert JR, Mayhaus M, Lannefelt L, Hakonarson H, Pichler S, Carrasquillo MM, Ingelsson M, Beekly D, Alvarez V, Zou F, Valladares O, Younkin SG, Coto E, Hamilton‐Nelson KL, Gu W, Razquin C, Pastor P, Mateo I, Owen MJ, Faber KM, Jonsson PV, Combarros O, O'Donovan MC, Cantwell LB, Soininen H, Blacker D, Mead S, Mosley TH, Jr Bennett DA, Harris TB, Fratiglioni L, Holmes C, de Bruijn RF, Passmore P, Montine TJ, Bettens K, Rotter JI, Brice A, Morgan K, Foroud TM, Kukull WA, Hannequin D, Powell JF, Nalls MA, Ritchie K, Lunetta KL, Kauwe JS, Boerwinkle E, Riemenschneider M, Boada M, Hiltuenen M, Martin ER, Schmidt R, Rujescu D, Wang LS, Dartigues JF, Mayeux R, Tzourio C, Hofman A, Nothen MM, Graff C, Psaty BM, Jones L, Haines JL, Holmans PA, Lathrop M, Pericak‐Vance MA, Launer LJ, Farrer LA, van Duijn CM, Van Broeckhoven C, Moskvina V, Seshadri S, Williams J, Schellenberg GD, Amouyel P (2013): Meta‐analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer's disease. Nat Genet 45:1452–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Cheng R, Barral S, Reitz C, Medrano M, Lantigua R, Jimenez‐Velazquez IZ, Rogaeva E, St George‐Hyslop PH, Mayeux R (2011): Identification of novel loci for Alzheimer disease and replication of CLU, PICALM, and BIN1 in Caribbean Hispanic individuals. Arch Neurol 68:320–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Zhang X, Hou B, Li J, Qiu C, Qin W, Yu C, Jiang T (2014): The impact of MIR137 on dorsolateral prefrontal‐hippocampal functional connectivity in healthy subjects. Neuropsychopharmacology 39:2153–2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon HT, Boucrot E (2011): Molecular mechanism and physiological functions of clathrin‐mediated endocytosis. Nat Rev Mol Cell Biol 12:517–533. [DOI] [PubMed] [Google Scholar]
- Melville SA, Buros J, Parrado AR, Vardarajan B, Logue MW, Shen L, Risacher SL, Kim S, Jun G, DeCarli C, Lunetta KL, Baldwin CT, Saykin AJ, Farrer LA, Alzheimer's Disease Neuroimaging Initiative (2012): Multiple loci influencing hippocampal degeneration identified by genome scan. Ann Neurol 72:65–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MI, Priebe CE, Qiu A, Fischl B, Kolasny A, Brown T, Park Y, Ratnanather JT, Busa E, Jovicich J, Yu P, Dickerson BC, Buckner RL, Morphometry B (2009): Collaborative computational anatomy: An MRI morphometry study of the human brain via diffeomorphic metric mapping. Hum Brain Mapp 30:2132–2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgen K, Ramirez A, Frolich L, Tost H, Plichta MM, Kolsch H, Rakebrandt F, Rienhoff O, Jessen F, Peters O, Jahn H, Luckhaus C, Hull M, Gertz HJ, Schroder J, Hampel H, Teipel SJ, Pantel J, Heuser I, Wiltfang J, Ruther E, Kornhuber J, Maier W, Meyer‐Lindenberg A (2014): Genetic interaction of PICALM and APOE is associated with brain atrophy and cognitive impairment in Alzheimer's disease. Alzheimers Dement 10:S269–S276. [DOI] [PubMed] [Google Scholar]
- Nuutinen T, Huuskonen J, Suuronen T, Ojala J, Miettinen R, Salminen A (2007): Amyloid‐beta 1‐42 induced endocytosis and clusterin/apoJ protein accumulation in cultured human astrocytes. Neurochem Int 50:540–547. [DOI] [PubMed] [Google Scholar]
- Nuutinen T, Suuronen T, Kauppinen A, Salminen A (2009): Clusterin: A forgotten player in Alzheimer's disease. Brain Res Rev 61:89–104. [DOI] [PubMed] [Google Scholar]
- Owen AM, McMillan KM, Laird AR, Bullmore E (2005): N‐back working memory paradigm: A meta‐analysis of normative functional neuroimaging studies. Hum Brain Mapp 25:46–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd‐Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC (2007): PLINK: A tool set for whole‐genome association and population‐based linkage analyses. Am J Hum Genet 81:559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saykin AJ, Shen L, Foroud TM, Potkin SG, Swaminathan S, Kim S, Risacher SL, Nho K, Huentelman MJ, Craig DW, Thompson PM, Stein JL, Moore JH, Farrer LA, Green RC, Bertram L, Jack CR, Jr Weiner MW, Alzheimer's Disease Neuroimaging Initiative (2010): Alzheimer's Disease Neuroimaging Initiative biomarkers as quantitative phenotypes: Genetics core aims, progress, and plans. Alzheimers Dement 6:265–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taubin G (1995) Curve and surface smoothing without shrinkage. Proceedings of the Fifth International Conference on Computer Vision, Cambridge, Massachusetts. pp 852–857.
- Taylor JE, Worsley KJ (2007): Detecting sparse signals in random fields, with an application to brain mapping. J Am Stat Assoc 102:913–928. [Google Scholar]
- Van Hoesen GW, Hyman BT (1990): Hippocampal formation: anatomy and the patterns of pathology in Alzheimer's disease. Prog Brain Res 83:445–457. [DOI] [PubMed] [Google Scholar]
- Wang HZ, Suh JW, Das SR, Pluta JB, Craige C, Yushkevich PA (2013): Multi‐atlas segmentation with joint label fusion. IEEE Trans Pattern Anal Mach Intell 35:611–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worsley KJ, Andermann M, Koulis T, MacDonald D, Evans AC (1999): Detecting changes in nonisotropic images. Hum Brain Mapp 8:98–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Tan MZ, Qiu A (2012): CSF and brain structural imaging markers of the Alzheimer's pathological cascade. PLoS One 7:e47406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XF, Li YH, Reutens D, Jiang TZ (2015): Diffeomorphic metric landmark mapping using stationary velocity field parameterization. Int J Comput Vis 115:69–86. [Google Scholar]
- Zhang P, Qin W, Wang D, Liu B, Zhang Y, Jiang T, Yu C (2014): Impacts of PICALM and CLU variants associated with Alzheimer's disease on the functional connectivity of the hippocampus in healthy young adults. Brain Struct Funct 220:1463–1475. [DOI] [PubMed] [Google Scholar]
- Zhang X, Yu JT, Li J, Wang C, Tan L, Liu B, Jiang T (2015): Bridging integrator 1 (BIN1) genotype effects on working memory, hippocampal volume, and functional connectivity in young healthy individuals. Neuropsychopharmacology 40:1794–1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu M, Ghodsi A (2006): Automatic dimensionality selection from the scree plot via the use of profile likelihood. Comput Stat Data Anal 51:918–930. [Google Scholar]


