Abstract
This study provides first data about the spatial variability of fMRI sensorimotor localizations when investigating the same subjects at different fMRI sites. Results are comparable to a previous patient study. We found a median between‐site variability of about 6 mm independent of task (motor or sensory) and experimental standardization (high or low). An intraclass correlation coefficient analysis using data quality measures indicated a major influence of the fMRI site on variability. In accordance with this, within‐site localization variability was considerably lower (about 3 mm). We conclude that the fMRI site is a considerable confound for localization of brain activity. However, when performed by experienced clinical fMRI experts, brain pathology does not seem to have a relevant impact on the reliability of fMRI localizations. Hum Brain Mapp 37:2151–2160, 2016. © 2016 Wiley Periodicals, Inc.
Keywords: FMRI, motor cortex, somatosensory cortex, multicenter study, variability
Abbreviations
- CNR
Contrast‐to‐noise ratio
- COM
Center of mass
- ED
Euclidian distance
- EPI
Echo planar imaging
- FWE
Family‐wise error
- ICC
Intraclass correlation coefficient
- PSC
Percent signal change
- ROI
Region of interest
- SFNR
Signal‐to‐fluctuation‐noise‐ratio
- SPM
Statistical parametric mapping
- TE
Echo time
- TR
Repetition time
INTRODUCTION
Because of its widespread availability, safety and flexibility, fMRI has become a standard research tool in neuroimaging and clinical application is growing [Orringer et al., 2013; Stippich, 2015]. Within the current upsurge of the “Big Data” philosophy, multicenter studies can enhance statistical power by measuring a large sample of subjects. However, fMRI measurements are susceptible to many potential sources of variability, which subsequently can change the statistical significance or the spatial localization of activations [Handwerker et al., 2012]. Several fMRI studies have investigated variability in a traveling subjects design across different sites [Bosnell et al., 2008; Brown et al., 2011; Casey et al., 1998; Costafreda et al., 2007; Forsyth et al., 2014; Friedman et al., 2006a; Friedman et al., 2006b, 2007; Gee et al., 2015; Gountouna et al., 2010; Gradin et al., 2010; Suckling et al., 2008; Sutton et al., 2008; Yendiki et al., 2010; Zou et al., 2005]. The FBIRN collaboration showed that field strength, number of subjects, and k‐space recording (raster, spiral, or dualecho) significantly influenced reproducibility and that higher field strength (3T and 4) yielded in better reproducibility than 1.5T [Zou et al., 2005]. Furthermore, they demonstrated that inter‐scanner variability can be reduced by controlling for signal‐to‐fluctuation‐noise‐ratio (SFNR) and that smoothness equalization reduces scanner to scanner variation [Friedman et al., 2006a; Friedman et al., 2006b]. They also found higher within‐site than between‐site reproducibility for percent signal change (PSC) and contrast‐to‐noise (CNR) median values, but demonstrated that by adjusting the data in multiple steps (increasing the size of the region of interest (ROI), adjusting for smoothness differences, inclusion of additional runs) reproducibility between sites can be improved [Friedman et al., 2007].
Other studies reported greater variation related to subject than to site using motor tasks, working memory and emotional processing tasks [Bosnell et al., 2008; Brown et al., 2011; Forsyth et al., 2014; Gee et al., 2015; Gradin et al., 2010; Suckling et al., 2008; Yendiki et al., 2010].
For clinical fMRI applications the most important issue is the spatial localization of brain activity. The functional localization of motor, language or memory activity is often decisive for further surgical procedures [Sartor and Stippich, 2007]. For this reason, we recently performed a clinical study investigating the reliability of fMRI localizations in brain tumor patients with considerably distorted brains [Wurnig et al., 2013]. When investigating the same patients at different fMRI sites, the spatial localization of brain activity varied between sites by about 6 mm but reached 16.5 mm in the worst patient.
To judge the influence of brain pathology on these results, we here present a study with healthy subjects using an identical experimental design. Although several aspects of between‐site fMRI variability have already been investigated, data about spatial localization (peak activation shifts) are currently missing. We compared a highly standardized somatosensory and a non‐standardized motor task using various localization measures and signal quality indices. Traveling subjects were scanned twice on consecutive days at five participating sites to evaluate both between‐site and within‐site variability.
MATERIAL AND METHODS
The general methods including tasks, experimental setup, imaging parameters and principal data analysis were the same as in Wurnig et al. [2013].
The study was approved by the local ethics committees. 18 healthy subjects with no prior history of neurological diseases or MR contraindications were scanned twice on consecutive days at five MR sites, except for the motor task at site 5, which was measured only once on the first day for organizational reasons. One subject (male, age 27) was excluded because of poor data quality and missing data. From the other 17 subjects, seven measurements of the somatosensory task and five measurements of the motor task were excluded because of extensive artifacts or missing data. Analysis included therefore 17 subjects (12 male, five female, mean age 32.1), 163/170 measurements for the somatosensory task and 148/163 measurements for the motor task. The median time interval between site visits was 51.5 days (range 11.5–190.5) and the order of visits is depicted in Table 1. Data acquisitions were performed within the same time periods as the patient investigations [Wurnig et al., 2013] and with the same experimental teams.
Table 1.
Demographics and order of visits of subjects
| Subject | Sex | Age | Site 1 | Site 2 | Site 3 | Site 4 | Site 5 |
|---|---|---|---|---|---|---|---|
| 1 | m | 23 | 1 | 5 | 2 | 4 | 3 |
| 2 | m | 51 | 1 | 5 | 2 | 4 | 3 |
| 3 | m | 64 | 1 | 5 | 2 | 4 | 3 |
| 4 | m | 28 | 2 | 5 | 1 | 4 | 3 |
| 5 | m | 26 | 2 | 5 | 1 | 4 | 3 |
| 6 | m | 25 | 1 | 5 | 2 | 3 | 4 |
| 7 | m | 36 | 2 | 5 | 1 | 3 | 4 |
| 8 | m | 22 | 1 | 5 | 2 | 3 | 4 |
| 9 | f | 24 | 1 | 5 | 2 | 3 | 4 |
| 10 | f | 29 | 2 | 5 | 1 | 3 | 4 |
| 11 | m | 27 | 2 | 5 | 1 | 4 | 3 |
| 12 | m | 26 | 2 | 3 | 1 | 4 | 5 |
| 13 | m | 29 | 5 | 3 | 4 | 1 | 2 |
| 14 | f | 24 | 2 | 4 | 1 | 3 | 5 |
| 15 | f | 26 | 4 | 1 | 5 | 3 | 2 |
| 16 | f | 23 | 5 | 1 | 4 | 3 | 2 |
| 17 | m | 31 | 3 | 5 | 4 | 1 | 2 |
Participating sites and scanner specification were:
Department of Neurology and MR Center of Excellence, Medical University of Vienna, Austria. Scanner specification: 3‐T Medspec (Bruker Medical, Ettlingen, Germany) in 12 subjects, 3‐T Magnetom Trio (Siemens, Erlangen, Germany) in five subjects.
Department of Neurology, Christian‐Doppler‐Clinic, Paracelsus Medical University, Salzburg, Austria. Scanner specification: 1.5‐T Gyroscan ACS‐NT Powertrak 6000 (Philips Healthcare, Best, the Netherlands) in five subjects; 3‐T Achieva (Philips Healthcare) in 12 subjects.
Department of Radiology, Medical University of Innsbruck, Austria. Scanner specification: 1.5‐T Magnetom Sonata (Siemens, Erlangen, Germany)
Institute for Diagnostic Radiology, Stiftungsklinikum Mittelrhein, Koblenz, Germany. Scanner specification: Philips, Eindhofen Intera Achieva 3Tesla TX (Release 3.2.1.)
Department of Neurology, Heinrich‐Heine‐University Düsseldorf, Düsseldorf, Germany. Scanner specifications: TIM Trio 3.0 Tesla MRI scanner (Siemens, Erlangen, Germany)
Data Acquisition and Tasks
Somatosensory task
Subjects were measured using a highly standardized somatosensory task consisting of 10 runs of 2:30min duration each (blocked design, three activation, four rest periods, 20s duration each). The paradigm consisted of stimulation of fingers II and III of the right hand with a highly standardized vibrotactile stimulus [Gallasch et al., 2010]. The stimulator is MR compatible and all electromechanical parts of the stimulator were located outside the scanning room. The stimulation itself was applied through finger cuffs with standardized oscillation impulses (1–20 Hz) using compressed air. For each activation period of the somatosensory task a pseudorandom stimulation was used. Each stimulation program consisted of four blocks of differing stimulation frequencies (between 1 and 9 Hz) and differing durations (between 3 and 11s).
The following fMRI sequence parameters for the somatosensory task were standardized across all sites: Echo planar imaging (EPI) sequences, repetition time (TR) 4,000 ms, flip angle 90°, field of view 230 × 230 mm, 30 slices (3 mm thickness, no gap) were obtained parallel to the anterior commissure ‐ posterior commissure plane. Echo time (TE; 30‐55 ms) and voxel size (1.6 × 1.6 to 2.4 × 2.4 mm) slightly varied for locally optimized blood‐oxygen‐level–dependent signal.
Motor task
For the non‐standardized motor task each site used their site‐specific task and setup to locate brain activity evoked by right hand movements.
Site 1: Fist clenching of the right hand, self‐paced at a frequency of approximately 1 Hz, seven to eight runs, EPI sequences, RT 4,000 ms, TE 55 ms, flip angle 90°, field of view 230 × 230 mm, matrix size 96 × 96, in plane resolution 2.4 × 2.4 mm. 30 slices (3 mm thickness, no gap) obtained parallel to the anterior commissure‐posterior commissure plane.
Site 2: Fist clenching of the right hand, EPI sequences blocked design, eight activation, nine rest periods. TR 4,000 ms, TE 45 ms, flip angle 90°, two runs, either an in plane resolution of 1.6 × 1.6 mm, matrix size of 144 × 144, or an in plane resolution of 1.8 × 1 × 8 mm and a matrix size of 128 × 128; 30 slices of 3 mm thickness (no gap).
Site 3: Fist clenching of the right hand, one run, blocked design, three activation, four rest periods, EPI sequences, TR 4,000 ms, TE 66 ms, flip angle 90°, field of view 230 × 230 mm, matrix size 96 × 96, in plane resolution 2.4 × 2.4 mm. 30 slices (3 mm thickness, no gap).
Site 4: Fist clenching of the right hand, one run, three activation, four rest periods, EPI sequences, TR 4,000 ms, TE 35 ms, flip angle 90°, field of view 230 × 230 mm, matrix size 112 × 112, voxel size 2.05 × 2.05 mm, 30 slices (3 mm thickness, no gap).
Site 5: Sequential finger opposition‐paradigm (order: 5th, 3rd, 4th and 2nd finger versus the 1st [Kleiser et al., 2005]. Standard EPI sequences with TR 2,000 ms, TE 30 ms, flip angle 90°, spatial resolution of 1.5 × 1.5 × 3.0 mm³, 44 slices with a matrix of 128 × 128 pixels.
Data Analysis
Data was converted into the NIFTI data format after collection from participating sites and was visually checked using the SPM8 Display option (Statistical Parametric Mapping, Wellcome Department of Imaging Neuroscience, London, UK) for artifacts. Analysis of fMRI data was done with SPM8 and local scripts. For all analyses a ROI was manually drawn using the software MRIcron (http://www.mccauslandcenter.sc.edu/mricro/mricron/) and encompassing the primary sensorimotor cortex for each subject. The extent of the ROI was defined anatomically (compare [Beisteiner et al., 2011]).
No slice timing correction was performed. For head‐motion control, realignment of all volumes to the first volume of each measurement was done. Default settings were used except for the factors “Quality: 1” and “Separation: 2” to improve the quality of the realignment. For our study goal “definition of spatial localization of brain activity” a critical issue is to minimize postprocessing related data manipulations, which could affect the final localization results. Therefore, no normalization to a standard space was performed. Instead, coregistration of all volumes of a single site to the first volume of the same reference site (site 2) was done using SPM's Coregistration (Estimate and Reslice) option. This step resulted in identical subject‐specific brain positions over all five sites and ensured comparability with the previously published data in patients [Wurnig et al., 2013]. Since no normalization to MNI space was done, the expression “identical brain positions” used in this manuscript refers to the individual EPI space of the reference site. Successful registration was checked visually using the Display option in SPM8. Data was smoothed by a Gaussian kernel with a full width at half maximum of double voxel size.
Individual SPM t‐maps were calculated using a general linear model with default settings. Further statistical analyses were done at a commonly used threshold [family‐wise error (FWE) P < 0.05], but without cluster threshold.
Quantification of Spatial Variability of Functional MR Imaging Localizations
To define individual sensorimotor fMRI localizations, the center of mass (COM) of active voxels within the ROI was defined using in‐house MATLAB scripts. The COM coordinate of each activation cluster (j) was calculated as the sum over all voxel coordinates (X i) divided by the number of voxels (n j) where each voxel is weighted with the SPM t‐value (t i) [Fesl et al., 2008]. Only voxels above the threshold of FWE P < 0.05 in the ROI were included.
The COM represents the most likely position for task‐specific functional tissue and is commonly used in somatotopic studies to quantify localization differences [Cunningham et al., 2013]. To quantify spatial between‐site variability, the median Euclidian Distance (ED) of COMs between the first five measurements of each subject was calculated as follows (visit 1 at the five sites):
The Euclidian Distances were calculated separately for each task and each subject for every possible two‐measurement comparison (10 possible ED values per subject and task—however, only measurements generating suprathreshold voxels (FWE P < 0.05) were considered).
The median of these values was taken as median between‐site distance (per subject and task) and used for further statistical processing.
To quantify spatial within‐site variability, the median ED of COMs between visit 1 and visit 2 was calculated as follows:
The Euclidian Distances between visit 1 and visit 2 were calculated separately for each task, each site and each subject except for the motor task at site 5 where the motor task was measured only at visit 1 (somatosensory task: five possible ED values per subject and task, motor task: four possible ED values per subject and task—however, only measurements generating suprathreshold voxels (FWE P < 0.05) were considered)
The median of these values was taken as median within‐site distance (per subject and task) and used for further statistical processing.
Results were compared using the Wilcoxon rank‐sum test. Bonferroni correction was applied to account for multiple comparison testing.
Additionally, a subgroup analysis was performed with the results of visit 1 of sites 1, 2, and 3 to be able to compare our healthy subject results with the results of our previous patient study [Wurnig et al., 2013]. The comparison with the patient data was carried out for each task separately, using the Mann‐Whitney U test for independent samples.
Data Quality Measures
Calculation of PSC, CNR and motion parameters
Calculation of PSC and CNR was based on significantly active voxels (FWE P < 0.05) and was done using local scripts as described in Beisteiner et al. [2011] and Geissler et al. [2014].
PSC was calculated as ΔS/S OFF where ΔS is defined as S ON‐S OFF, and S ON is the mean absolute signal of all time points within the ON (activation) phases in the blocked design and S OFF is the mean of all time points within OFF (rest) phases, respectively. For calculation of S OFF and S ON the blocked design was shifted by five seconds considering the delayed BOLD response. PSC was calculated for each voxel in the ROI of each run and then averaged to compute median PSC values for each center, subject and task.
CNR was calculated as ΔS CNR/σ t‐noise. ΔSCNR is the averaged voxel‐wise signal change in the temporally smoothed original signal and defined as mean value of all time points within the ON (activation) phase minus the mean value of all time points within the OFF (rest) phase of the blocked design. A Savitzky–Golay filter with a polynomial order of 2 and length 5 was used for smoothing. σ t‐noise is defined as the standard deviation of the difference between the original and smoothed signals. Median CNR values were calculated as described for PSC.
Motion parameters of each measurement were obtained from SPM's realignment procedure. The median spatial difference between every two consecutive volumes in each spatial direction was calculated and subsequently median translatory and rotatory motion per subject, center and task computed.
Statistical analysis
CNR, PSC and motion parameters were analyzed separately for the two tasks, using a random‐effect univariate ANOVA (main effects and two‐way interactions) to test for differences between sites, subjects and measurements and their interactions. Additionally, CNR, PCS and motion parameters were compared between tasks using the Wilcoxon rank‐sum test, with a P‐value of P < 0.0125 for significance after Bonferroni correction.
Reliability measures can be calculated using variance components estimates to assess the consistency of observations. We calculated variance components and associated intraclass correlation coefficients (ICC) values in the notion of Shrout and Fleiss [1979]. There are several types of ICC, but in the context of multicenter fMRI studies, where sites are not independent and the goal is to define the extent of the reliability of the data, the absolute level ICC (ICC 2,1) in the nomenclature of Shrout and Fleiss [1979] was mostly used. The ICC ranges from 0 to 1 and if it is large the absolute agreement of each rater (in this case fMRI site) would be high, suggesting that variability due to the site factor is low.
We adopted the criteria of Cicchetti [2001] to interpret ICCs: poor (<0.40), fair (0.41–0.59), good (0.60–0.74), and excellent (above 0.75).
For ICC computations variance components estimates were calculated for the median CNR and PSC values as well as median beta regressors of each measurement using the Varcomp routine (restricted Maximum Likelihood Estimation method) implemented in SPSS 22. Between‐site and within‐site ICCs were computed for all sites following the description of Friedman et al. [2007].
The model was defined as:
where Y ijk denotes the dependent measure for subject i, visit k and fMRI site j. Subject, site and visit were considered as random effects.
ICCs were subsequently calculated as:
Vd_subject would indicate the variance due to the subject effect, Vd_site the variance due to the site and Vd_subject‐by‐site the interaction of the two. ICC within‐site (test‐retest reliability) would be an ICC (1,1) and ICC between‐site an ICC (2,1) according to the definition of Shrout and Fleiss [1979] adapted to the more complex design as in Friedman et al. [2007].
RESULTS
Measurements Showing Activation Above Threshold
Choosing a threshold of FEW P < 0.05, we found activation within the ROI in 91.2% of measurements for the motor task and in 49.1% for the somatosensory task. When lowering the threshold to uncorrected P < 0.01, 99.3% measurements of the motor task and 93.3% of the somatosensory task showed activations.
Quantification of Spatial Variability of Functional MR Imaging Localizations
P‐values of comparisons are shown in Table 2. Table 3 summarizes the findings of spatial variability. Only voxels above the threshold of FEW P < 0.05 were included.
Table 2.
P‐values for COM variability comparisons using the Wilcoxon rank‐sum test
| Wilcoxon rank sum test | Pair 1 | Pair 2 | P‐values |
|---|---|---|---|
| Within‐site COM | Motor | Somatosensory | P = 284 |
| Between‐site COM | Motor | Somatosensory | P = 111 |
| Somatosensory task | Within‐site | Between‐site | P = 0.048 |
| Motor task | Within‐site | Between‐site | P < 0.001 |
P‐values are corrected for multiple comparisons using Bonferroni corrections (significance level is P ≤ 0.0125). The COM variabilities do not differ between tasks. However, COM variabilities are larger for the Between‐site comparison (motor task, bold P‐value).
Table 3.
Results of spatial variability based on the COM localizations
| Somatosensory task | Motor task | |||
|---|---|---|---|---|
| Between‐sites | Within‐sites | Between‐sites | Within‐sites | |
| Median ED‐COM | 5.9 mm | 3.9 mm | 5.9 mm | 1.8 mm |
| IQR | 4.8 mm | 1.9 mm | 5.0 mm | 2.4 mm |
| Maximum | 14.2 mm | 15.1 mm | 29.7 mm | 12.8 mm |
| Minimum | 0 mm | 0 mm | 1.6 mm | 0 mm |
ED‐COM denotes Euclidian distances of center of mass, IQR denotes interquartile range.
We found a median between‐site variability of 5.9 mm (IQR = 4.8 mm) for the somatosensory task and also 5.9 mm (IQR = 5 mm) for the motor task (median over all subjects). Maximum differences were 14.2 mm and 29.7 mm, respectively. We found a median spatial within‐site variability of 3.9 mm (IQR = 1.9 mm) for the somatosensory task and 1.8 mm (IQR = 2.4 mm) for the motor task (median over all subjects). Maximum differences were 15.1 mm for the somatosensory task and 12.8 mm for the motor task. Differences between the tasks were not significant.
The spatial variability of the motor task was significantly lower within sites than between sites (P < 0.001, Wilcoxon rank‐sum test), no statistically significant difference was found for the somatosensory task after Bonferroni correction although a trend toward significance (P = 0.048) was observed (Table 2). Figure 1 shows an example of the COM variability for a single subject.
Figure 1.
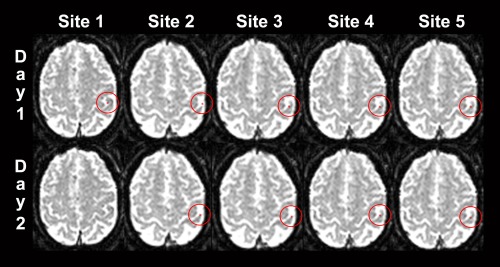
Example of the somatosensory variability of functional localizations (center of mass COM = red voxel). Data of a single subject with median variability of 3.9 mm (between‐site) and 2.4 mm (within‐site). The COM is projected on the respective axial functional echo‐planar image of the reference site (left is right). The COM position is shown for the five sites and the two measurements at each site (day 1 and day 2). Note that the COM‐slices and the COM‐within‐slice‐positions change only slightly. At site 1, day 2 no data survived thresholding.
Data Quality Measures
CNR, PSC and motion parameters
Median CNR and PSC values were significantly higher for the motor task than for the somatosensory task (P < 0.0001). Rotatory motion (P < 0.0001) and translatory motion (P = 0.008) was larger with the motor task. Table 4 shows median values of CNR, PSC and motion parameters for both tasks.
Table 4.
Descriptive results of contrast‐to‐noise ratio (CNR), percent signal change (PSC), and motion parameters
| Median | Interquartile Range | Maximum/Minimum | ||
|---|---|---|---|---|
| CNR | Somatosensory | 1.23 | 0.41 | 2.63/0.65 |
| Motor | 3.17 | 1.37 | 5.57/1.46 | |
| PSC | Somatosensory | 0.70 | 0.43 | 2.51/0.32 |
| Motor | 2.09 | 1.01 | 4.96/0.88 | |
| Translatory motion | Somatosensory | 0.0444 | 0.0357 | 0.1959/0.01394 |
| Motor | 0.0500 | 0.0572 | 0.2456/0.0154 | |
| Rotatory motion | Somatosensory | 0.0004 | 0.0002 | 0.00167/0.0002 |
| Motor | 0.0005 | 0.0003 | 0.0016/0.0002 |
CNR is shown in arbitrary units, PSC in percent, rotatory motion in rad and translatory motion as median distance in mm between two subsequent volumes.
ANOVA results
Details are listed in Table 5. Overall, the factor site and subject‐by site interaction showed the largest influence on CNR, PSC and motion parameters.
Table 5.
P‐values for significant factors in ANOVA results
| Motor task | Somatosensory task | |||||||
|---|---|---|---|---|---|---|---|---|
| All sites | 3T sites | All sites | 3T sites | |||||
| CNR | PSC | CNR | PSC | CNR | PSC | CNR | PSC | |
| Subject | 0.005 | |||||||
| Site | 0.000 | 0.000 | 0.000 | 0.007 | 0.001 | |||
| Visit | ||||||||
| Subject*site | 0.000 | 0.000 | 0.012 | 0.047 | ||||
| Subject*visit | ||||||||
| Site*visit | ||||||||
CNR and PSC values were analyzed separately for the two tasks, using a random‐effects univariate ANOVA (main effects and two‐way interactions). Boxes with P‐values indicate significance of the respective effect (subject, site or visit) or interaction. Blank boxes indicate lack of significance.
ICCs results
ICCs (Table 6) were excellent for within‐site comparisons with the exception of an untypically low within‐site ICC for CNR sensory. Between‐site ICCs were poor for both tasks. This indicates a large contribution of the factor site to between‐site variability but—as expectable—a low contribution of the factor site to within‐site variability.
Table 6.
Variance component estimates and ICCs for median CNR, PSC and Beta values
| Variance component estimates | Intraclass correlation coefficients | ||||||
|---|---|---|---|---|---|---|---|
| Subject | Site | Visit | Site*subject | Unexplained | ICC‐between‐site | ICC within‐site | |
| CNR Motor | 0.037 | 0.696 | 0.009 | 0.162 | 0.101 | 0.04 | 0.89 |
| CNR Sensory | 0.036 | 0.005 | 0.000 | 0.023 | 0.52 | 0.06 | 0.11 |
| PSC Motor | 0.091 | 0.280 | 0.000 | 0.145 | 0.098 | 0.15 | 0.84 |
| PSC Sensory | 0.031 | 0.063 | 0.002 | 0.059 | 0.026 | 0.17 | 0.85 |
| Beta Motor | 0.373 | 4.846 | 0.000 | 1.252 | 0.551 | 0.05 | 0.92 |
| Beta Sensory | 0.039 | 0.626 | 0.002 | 0.314 | 0.086 | 0.04 | 0.92 |
For ICC computations variance component estimates were calculated for the median CNR and PSC values as well as median beta regressors of each measurement. Values in the table show the respective estimates for each of the factors that were used to calculate between‐site and within‐site ICCs (see methods section for details). The obtained ICC values range from 0 to 1 with higher values signifying better reproducibility (agreement) and accordingly lower variability.
Comparison with previously published patients data
Comparison with our previously published data of 15 tumor patients [Wurnig et al., 2013], which were referred to presurgical evaluation and measured in the same traveling study design, except that only one measurement at three sites (sites 1, 2 and 3) was performed, showed similar results of spatial variability (COM analysis).
The subgroup analysis where we included only visit 1 of the three sites of the patient study, showed a median variability of 5.4 mm for the motor task (patients 5.7 mm) and 7.5 mm for the somatosensory task (patients 5.8 mm). Statistical comparisons between patient and subject data were non‐significant (P = 0.669 for the motor task and P = 0.109 for the somatosensory task, Mann‐Whitney U test). Site ICCs for beta‐values were poor in patients and subjects indicating a relevant influence of the factor site on localization variability (Table 7).
Table 7.
Comparison of the healthy subject data and our previously published results of tumor patients [Wurnig et al., 2013]
| Task | Motor | Somatosensory | ||
|---|---|---|---|---|
| Subjects | Patients | Subjects | Patients | |
| Percent activations (unc. P < 0.01) | 99.3% | 97.8% | 93.3% | 88.9% |
| Percent activations (FWE P < 0.05) | 91.2% (96.1) | 88.9% | 49.1% (49%) | 64.4% |
| Between site ED for COM | 5.9 mm (5.4) | 5.7 mm | 5.9 mm (7.5mm) | 5.8 mm |
| Max. ED for COM | 29.7 mm (18.7) | 16.5 mm | 14.2 mm (14.2) | 12 mm |
| Between‐site ICC (beta values) | 0.05 (0.02) | 0.23 | 0.04 (0.08) | 0.20 |
Between‐site Euclidian Distances (ED), maximum ED and ICC comparison were done at a threshold of FWE P < 0.05. ICC values are provided for beta values only, since ICCs for CNR and PSC were not calculated in patients. Values in brackets give healthy subject data resulting from visit 1 at the three patient sites.
DISCUSSION
In this study we investigated between‐site and within‐site variability of fMRI localizations. Our major finding was that compared to within‐site variability, the between‐site variability of fMRI localizations is considerably larger and ICC values are lower. This indicates a relevant influence of the local fMRI team and fMRI technology, which is important for multicenter studies. Most important factors for additional variability probably are hardware differences and different handling of subjects. Although median variability was low in both analyses (between‐ and within‐site), maximum differences ranged up to 3 cm with the non‐standardized motor task. This could be a critical value, e.g., in presurgical applications and indicates that some task standardization is beneficial.
The poor within‐site ICC for CNR with the somatosensory task is probably due to our stringent threshold (FEW P < 0.05), which generated significant activity in only 49.1% of the measurements—due to application of a rather weak stimulus. Analysis at a lower threshold (uncorrected P < 0.01) did show similar within‐site ICCs as seen for PSC and beta values (data with the authors).
Concerning the relationship of our data to our previously published data in tumor patients (Wurnig et al., 2013) using a similar design, the between‐site spatial variability in healthy subjects did not differ significantly from that of the brain tumor patients. This indicates that in the hands of experienced clinical fMRI experts brain pathology does not influence the quality of fMRI localizations to a relevant extent.
To our knowledge, there is no previous study comparing fMRI localizations with peak activation measures at different sites. Regarding spatial differences for repeated measurements using a finger tapping task at a single site, Marshall et al. [2004] reported COM differences with a standard deviation of about 3 mm at each coordinate at the group level. Rombouts et al. [1998] reported spatial differences of visual brain activation of COMs in a single center test‐retest study with three measurements to be 2.63 and 3.96 mm, respectively. Here—with minimized postprocessing influences (no brain normalizations)—we found median within‐site differences in the range of 1.8 to 3.9 mm.
With regard to the ICC results, previous studies using partly different measures and investigating a motor task [Bosnell et al., 2008; Friedman et al., 2007; Gountouna et al., 2010] or cognitive data [Brown et al., 2011; Forsyth et al., 2014; Gee et al., 2015; Gradin et al., 2010] showed lower between‐site variability. This might be explained by the low standardization in our study regarding site and hardware differences—deliberately chosen to generate a realistic picture of the functional localization landscape. In addition, with the non‐standardized motor task, the different number of runs or experimental setups used by the different sites is an important factor. Our within‐site ICCs values for the motor task indicate a high performance stability of the local clinical fMRI teams. The values are in the upper range of those reported in the literature [Aron et al., 2006; Kong et al., 2007; Quiton et al., 2014]. Within‐site ICCs for a somatosensory task have not previously been reported.
Concerning possible limitations of our study, standardization regarding the site factor was deliberately low, since we wanted to compare data achieved under real life clinical conditions [Wurnig et al., 2013]. For a realistic judgment of between‐site localization variability, differing hardware and experimental procedures have to be considered. In addition, patient fMRI requests are typically based on organizational criteria but not scanner issues. Concerning our database, we had to exclude one subject and 12 measurements either due to extensive artifacts or missing data and at site 5 there was only one data set for the motor task. However, our available test‐retest ICCs were high and a reanalysis with complete exclusion of site 5 did not substantially change the results.
The voxel size in our study varied between sites due to differences in the locally optimized acquisition schemes. Although an influence on variability results cannot be ruled out, its effect on COM calculation in smoothed data should be small. The rational behind this approach was to generate “real world” data, which requires acceptance of local voxel size choices and standard data analysis procedures like smoothing. Data smoothing prior to statistical analysis likely reduced the variability of our results.
In this study the between‐site intervals were not standardized for the five sites (in contrast to the within‐site intervals, which always were one day). Therefore, non‐stationarity at sensorimotor areas has to be considered as a possible confound.
CONCLUSION
In conclusion, our results suggest that the factor site is a considerable confound for localization of brain activity. Therefore, care must be taken, when pooling and interpreting fMRI results of different study sites, particularly when different scanners, field strengths or experimental setups are used. Comparing fMRI results from a single site is much more reliable. Importantly, when performed by experienced clinical fMRI experts, brain pathology does not seem to have a major impact on the reliability of fMRI localizations in the sensorimotor cortex.
REFERENCES
- Aron AR, Gluck MA, Poldrack RA (2006): Long‐term test‐retest reliability of functional MRI in a classification learning task. Neuroimage 29:1000–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beisteiner R, Robinson S, Wurnig M, Hilbert M, Merksa K, Rath J, Höllinger I, Klinger N, Marosi C, Trattnig S, Geissler A (2011): Clinical fMRI: Evidence for a 7T benefit over 3T. Neuroimage 57:1015–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosnell R, Wegner C, Kincses ZT, Korteweg T, Agosta F, Ciccarelli O, De Stefano N, Gass A, Hirsch J, Johansen‐Berg H, Kappos L, Barkhof F, Mancini L, Manfredonia F, Marino S, Miller DH, Montalban X, Palace J, Rocca M, Enzinger C, Ropele S, Rovira A, Smith S, Thompson A, Thornton J, Yousry T, Whitcher B, Filippi M, Matthews PM (2008): Reproducibility of fMRI in the clinical setting: Implications for trial designs. Neuroimage 42:603–610. [DOI] [PubMed] [Google Scholar]
- Brown GG, Mathalon DH, Stern H, Ford J, Mueller B, Greve DN, McCarthy G, Voyvodic J, Glover G, Diaz M, Yetter E, Ozyurt IB, Jorgensen KW, Wible CG, Turner JA, Thompson WK Potkin SG, Function Biomedical Informatics Research Network (2011): Multisite reliability of cognitive BOLD data. Neuroimage 54:2163–2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casey BJ, Cohen JD, O'Craven K, Davidson RJ, Irwin W, Nelson CA, Noll DC, Hu X, Lowe MJ, Rosen BR, Truwitt CL, Turski PA (1998): Reproducibility of fMRI results across four institutions using a spatial working memory task. Neuroimage 8:249–261. [DOI] [PubMed] [Google Scholar]
- Cicchetti DV (2001): The precision of reliability and validity estimates re‐visited: Distinguishing between clinical and statistical significance of sample size requirements. J Clin Exp Neuropsychol 23:695–700. [DOI] [PubMed] [Google Scholar]
- Costafreda SG, Brammer MJ, Vêncio RZN, Mourão ML, Portela LAP, de Castro CC, Giampietro VP, Amaro E (2007): Multisite fMRI reproducibility of a motor task using identical MR systems. J Magn Reson Imaging 26:1122–1126. [DOI] [PubMed] [Google Scholar]
- Cunningham DA, Machado A, Yue GH, Carey JR, Plow EB (2013): Functional somatotopy revealed across multiple cortical regions using a model of complex motor task. Brain Res 1531:25–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fesl G, Braun B, Rau S, Wiesmann M, Ruge M, Bruhns P, Linn J, Stephan T, Ilmberger J, Tonn J‐C, Brückmann H (2008): Is the center of mass (COM) a reliable parameter for the localization of brain function in fMRI? Eur Radiol 18:1031–1037. [DOI] [PubMed] [Google Scholar]
- Forsyth JK, McEwen SC, Gee DG, Bearden CE, Addington J, Goodyear B, Cadenhead KS, Mirzakhanian H, Cornblatt BA, Olvet DM, Mathalon DH, McGlashan TH, Perkins DO, Belger A, Seidman LJ, Thermenos HW, Tsuang MT, van Erp TGM, Walker EF, Hamann S, Woods SW, Qiu M, Cannon TD (2014): Reliability of functional magnetic resonance imaging activation during working memory in a multi‐site study: Analysis from the North American Prodrome Longitudinal Study. Neuroimage 97:41–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman L, Glover GH, Fbirn Consortium (2006a): Reducing interscanner variability of activation in a multicenter fMRI study: Controlling for signal‐to‐fluctuation‐noise‐ratio (SFNR) differences. Neuroimage 33:471–481. [DOI] [PubMed] [Google Scholar]
- Friedman L, Glover GH, Krenz D Magnotta V, FIRST BIRN (2006b): Reducing inter‐scanner variability of activation in a multicenter fMRI study: Role of smoothness equalization. Neuroimage 32:1656–1668. [DOI] [PubMed] [Google Scholar]
- Friedman L, Stern H, Brown GG, Mathalon DH, Turner J, Glover GH, Gollub RL, Lauriello J, Lim KO, Cannon T, Greve DN, Bockholt HJ, Belger A, Mueller B, Doty MJ, He J, Wells W, Smyth P, Pieper S, Kim S, Kubicki M, Vangel M, Potkin SG (2007): Test‐retest and between‐site reliability in a multicenter fMRI study. Hum Brain Mapp 29:958–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallasch E, Fend M, Rafolt D, Nardone R, Kunz A, Kronbichler M, Beisteiner R, Golaszewski S (2010): Cuff‐type pneumatic stimulator for studying somatosensory evoked responses with fMRI. Neuroimage 50:1067–1073. [DOI] [PubMed] [Google Scholar]
- Gee DG, McEwen SC, Forsyth JK, Haut KM, Bearden CE, Addington J, Goodyear B, Cadenhead KS, Mirzakhanian H, Cornblatt BA, Olvet D, Mathalon DH, McGlashan TH, Perkins DO, Belger A, Seidman LJ, Thermenos H, Tsuang MT, van Erp TGM, Walker EF, Hamann S, Woods SW, Constable T, Cannon TD (2015): Reliability of an fMRI paradigm for emotional processing in a multisite longitudinal study. Hum Brain Mapp 36:2558–2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geissler A, Matt E, Fischmeister F, Wurnig M, Dymerska B, Knosp E, Feucht M, Trattnig S, Auff E, Fitch WT, Robinson S, Beisteiner R (2014): Differential functional benefits of ultra highfield MR systems within the language network. Neuroimage 103:163–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gountouna V‐E, Job DE, McIntosh AM, Moorhead TWJ, Lymer GKL, Whalley HC, Hall J, Waiter GD, Brennan D, McGonigle DJ, Ahearn TS, Cavanagh J, Condon B, Hadley DM, Marshall I, Murray AD, Steele JD, Wardlaw JM, Lawrie SM (2010): Functional Magnetic Resonance Imaging (fMRI) reproducibility and variance components across visits and scanning sites with a finger tapping task. Neuroimage 49:552–560. [DOI] [PubMed] [Google Scholar]
- Gradin V, Gountouna V‐E, Waiter G, Ahearn TS, Brennan D, Condon B, Marshall I, McGonigle DJ, Murray AD, Whalley H, Cavanagh J, Hadley D, Lymer K, McIntosh A, Moorhead TW, Job D, Wardlaw J, Lawrie SM, Steele JD (2010): Between‐ and within‐scanner variability in the CaliBrain study n‐back cognitive task. Psychiatry Res 184:86–95. [DOI] [PubMed] [Google Scholar]
- Handwerker DA, Gonzalez‐Castillo J, D'Esposito M, Bandettini PA (2012): The continuing challenge of understanding and modeling hemodynamic variation in fMRI. Neuroimage 62:1017–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleiser R, Wittsack H‐J, Bütefisch CM, Jörgens S, Seitz RJ (2005): Functional activation within the PI‐DWI mismatch region in recovery from ischemic stroke: Preliminary observations. Neuroimage 24:515–523. [DOI] [PubMed] [Google Scholar]
- Kong J, Gollub RL, Webb JM, Kong J‐T, Vangel MG, Kwong K (2007): Test‐retest study of fMRI signal change evoked by electroacupuncture stimulation. Neuroimage 34:1171–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall I, Simonotto E, Deary IJ, Maclullich A, Ebmeier KP, Rose EJ, Wardlaw JM, Goddard N, Chappell FM (2004): Repeatability of motor and working‐memory tasks in healthy older volunteers: Assessment at functional MR imaging. Radiology 233:868–877. [DOI] [PubMed] [Google Scholar]
- Orringer D, Vago D, Golby A (2013): Clinical Applications and Future Directions of Functional MRI. Semin Neurol 32:466–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quiton RL, Keaser ML, Zhuo J, Gullapalli RP, Greenspan JD (2014): Intersession reliability of fMRI activation for heat pain and motor tasks. Neuroimage Clin 5:309–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rombouts SA, Barkhof F, Hoogenraad FG, Sprenger M, Scheltens P (1998): Within‐subject reproducibility of visual activation patterns with functional magnetic resonance imaging using multislice echo planar imaging. Magn Reson Imaging 16:105–113. [DOI] [PubMed] [Google Scholar]
- Sartor K, Stippich C (2007): Clinical Functional MRI. Berlin, Heidelberg: Springer Science & Business Media. [Google Scholar]
- Shrout PE, Fleiss JL (1979): Intraclass correlations: Uses in assessing rater reliability. Psychol Bull 86:420–428. [DOI] [PubMed] [Google Scholar]
- Stippich C, editor (2015): Clinical Functional MRI, 2nd ed Berlin, Heidelberg: Springer Berlin Heidelberg. [Google Scholar]
- Suckling J, Ohlssen D, Andrew C, Johnson G, Williams SCR, Graves M, Chen C‐H, Spiegelhalter D, Bullmore E (2008): Components of variance in a multicentre functional MRI study and implications for calculation of statistical power. Hum Brain Mapp 29:1111–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton BP, Goh J, Hebrank A, Welsh RC, Chee MWL, Park DC (2008): Investigation and validation of intersite fMRI studies using the same imaging hardware. J Magn Reson Imaging 28:21–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurnig MC, Rath J, Klinger N, Höllinger I, Geissler A, Fischmeister FP, Aichhorn M, Foki T, Kronbichler M, Nickel J, Siedentopf C, Staffen W, Verius M, Golaszewski S, Koppelstätter F, Knosp E, Auff E, Felber S, Seitz RJ, Beisteiner R (2013): Variability of clinical functional MR imaging results: A multicenter study. Radiology 268:521–531. [DOI] [PubMed] [Google Scholar]
- Yendiki A, Greve DN, Wallace S, Vangel M, Bockholt J, Mueller BA, Magnotta V, Andreasen N, Manoach DS, Gollub RL (2010): Multi‐site characterization of an fMRI working memory paradigm: Reliability of activation indices. Neuroimage 53:119–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou KH, Greve DN, Wang M, Pieper SD, Warfield SK, White NS, Manandhar S, Brown GG, Vangel MG, Kikinis R Wells WM, FIRST BIRN Research Group (2005): Reproducibility of functional MR imaging: Preliminary results of prospective multi‐institutional study performed by Biomedical Informatics Research Network. Radiology 237:781–789. [DOI] [PMC free article] [PubMed] [Google Scholar]


