Abstract
With Wolbachia-based arbovirus control programs being scaled and operationalised around the world, cost effective and reliable detection of Wolbachia in field samples and laboratory stocks is essential for quality control. Here we validate a modified loop-mediated isothermal amplification (LAMP) assay for routine scoring of Wolbachia in mosquitoes from laboratory cultures and the field, applicable to any setting. We show that this assay is a rapid and robust method for highly sensitive and specific detection of wAlbB Wolbachia infection within Aedes aegypti under a variety of conditions. We test the quantitative nature of the assay by evaluating pooled mixtures of Wolbachia-infected and uninfected mosquitoes and show that it is capable of estimating infection frequencies, potentially circumventing the need to perform large-scale individual analysis for wAlbB infection status in the course of field monitoring. These results indicate that LAMP assays are useful for routine screening particularly under field conditions away from laboratory facilities.
Introduction
Wolbachia releases are being undertaken in Aedes aegypti populations both for replacing existing populations with mosquitoes that have a reduced ability to transmit dengue [1, 2] and other arboviruses, and for suppressing mosquito populations directly due to sterility generated by males infected with Wolbachia [3]. A challenge in implementing Wolbachia-based strategies is that a high level of quality control is required for release success. This includes ensuring that source mosquito cultures used for releases remain infected by Wolbachia. It is also important to track infection status in release areas (particularly with releases aimed at replacing existing populations by Wolbachia-infected populations) given that not all releases are successful and periodic interventions may be needed for others [4, 5]. Successful replacement is dependent on Wolbachia frequencies in populations exceeding an unstable equilibrium point [1] and potential spread from a release site [6].
Wolbachia monitoring has been undertaken with a variety of approaches including staining, electron microscopy and PCR with Wolbachia-specific primers. For releases where high throughput is desirable, Wolbachia detection is currently achieved through qPCR fluorescence-based approaches such as RT/HRM (real time PCR/high resolution melt) such as described by Lee et al. [7] which achieves relative quantification of Wolbachia on a Roche 480 LightCycler® system allowing samples to be scored in 384-well plates. While this method is efficient and can be used to detect multiple Wolbachia strains along with identification of mosquito vectors of disease, it requires expertise and laboratory facilities that are well beyond what is readily available in many developing nations. Each assay also requires a substantial amount of time (around 1.5 h until results are available) as well as a dedicated qPCR machine.
Loop-mediated isothermal amplification (LAMP) is a powerful DNA amplification technique, enabling the detection of trace elements of DNA with high rapidity, sensitivity and accuracy [8]. It involves isothermal amplification through the interaction of four to six primers with up to eight target sites. When combined with a polymerase with high displacement activity, two outer primers assist two compound inner primers to form alternating loop structures within the DNA, providing self-primed single-stranded substrates for further inner primer interaction and replication. This process can be further accelerated by the addition of loop primers also targeted to the exposed single-stranded region of the loop [9].
This technique has found widespread application primarily in the diagnosis and monitoring of infectious diseases such as malaria [10, 11], West Nile virus [12, 13] and dengue [14]. LAMP has also been applied in other contexts including agriculture, quarantine, forensics, and environmental monitoring through DNA [15]. In all these contexts, the LAMP technique allows for fast and accurate assays that can be deployed with equivalent sensitivity to traditional PCR methods, but often with much cheaper costs and less technically-demanding deployment–ideal for field contexts and settings where laboratory expertise is limited.
Three LAMP assays have thus far been published for Wolbachia monitoring [16–18]. Gonçalves et al. (2014) targeted Wolbachia’s 16S ribosomal protein sequence, and amplified Wolbachia across multiple strains, including wAlbB, wMel, and wMelPop. A study applying this assay to field samples in Malaysia found that it compared favourably with a standard PCR method for Wolbachia detection in Aedes albopictus mosquitoes, detecting a higher infected rate than observed with PCR [19]. However, it should be noted that this study used an experimental design that did not include the loop primers, which Gonçalves et al. (2014) consider essential to ensure specificity of the assay.
A third Wolbachia paper using LAMP [18] independently targeted the 16S ribosomal protein sequence. As in the above Wolbachia assay [16], a range of Wolbachia strains were targeted and detected in Ae. albopictus and Ae. aegypti. However, an additional assay was also developed specific to wAlbB (ordinarily present within Ae. albopictus) and wPip strain Wolbachia surface proteins (wsp) [17]. This assay originally involved one loop primer and a specific detection method with an additional oligomeric probe designed for presence/absence discrimination of fluorescence by eye. The specific nature of this assay makes it a promising target for adaptation to monitor Ae. aegypti populations transinfected with wAlbB for disease control.
One important development of LAMP techniques has been to quantitatively assess targets (qLAMP) [20]. These developments have led to stable linear determinations of products across as many as nine orders of magnitude concentration over a wide range of human and agricultural pathogens [20–23]. Such quantitative assessments could be useful for Wolbachia monitoring, because a key feature of release success is the frequency of the endosymbiont in field populations. Currently in releases, the Wolbachia status of hundreds of mosquitoes is determined at a centralized facility using expensive equipment [1, 6]. In contrast, qLAMP conducted on pools of mosquitoes has the potential to provide rapid and cost-effective estimates of local Wolbachia frequencies. When implemented on a device such as the Genie® III, qLAMP assays require minimal training and are highly portable, reducing the load on centralised monitoring laboratories.
Accordingly, the aims of this research were to (a) adapt and extend the assay of Bhadra et al. [17] using a wAlbB wsp primer set as well as an Ae. aegypti ITS1 primer set [24] for efficient and specific detection of wAlbB-infected Ae. aegypti mosquitoes in the context of control efforts including those in sub-optimal conditions, (b) compare this method to established qPCR monitoring methods on samples taken from field locations, and (c) develop a quantitative form of the assay for use on pooled mosquito extractions to determine relative wAlbB frequencies. The results are expected to be applicable to a variety of projects involving Wolbachia within health and agricultural contexts.
Methods
Aedes aegypti samples
Laboratory colonies
Aedes aegypti were primarily derived from three laboratory colonies: (a) a wAlbB-infected colony with a wAlbB strain originating from Ae. albopictus [25], (b) an uninfected colony originating from wild populations in Cairns, Australia, and (c) a wMel-infected colony with a wMel strain originating from Drosophila melanogaster [26]. The three laboratory colonies of Ae. aegypti were crossed to mosquitoes of a common Australian background reared in an identical manner to each other, i.e. at 26°C with food provided ad libitum. Adults were sacrificed and stored in absolute ethanol before DNA extraction.
Alternate rearing and storage conditions
Aedes aegypti are exposed to a range of environmental conditions in nature, including heat stress and resource competition, which can produce adults of various sizes and with different Wolbachia loads [27]. We altered rearing conditions to simulate several scenarios affecting size and Wolbachia density. Small wAlbB-infected adults were produced according to Callahan et al. [28] by providing larvae with food ad libitum for 3 d and then depriving larvae of food until adulthood. Heat-stressed adults were generated according to Ross et al. [27] by holding eggs at a cyclical temperature regime of 30–40°C for one week. Eggs were then hatched and reared under standard conditions to produce adults with a reduced Wolbachia density. Field-collected adults are of variable age and may have taken a blood meal; we therefore tested 30 d old adults and 7 d old females that were stored in absolute ethanol at -20°C either immediately or 24 h after feeding on a human volunteer. Six mosquitoes were tested per treatment for these experiments.
We additionally tested the ability of the LAMP assay to detect wAlbB when mosquitoes were stored under suboptimal conditions that may be experienced during field sampling. wAlbB-infected adults reared in the laboratory were killed by shaking and stored for 1, 2, 3, 5, 10, 20 and 30 d at 26°C or for 10 d at 37°C in open air before storage in absolute ethanol at -20°C. Dead wAlbB-infected adults were also stored in water at 26°C for 3 d before transfer to ethanol and -20°C–simulating adults found floating in ovitraps as part of field collections. Six mosquitoes were tested for each scenario, except for 30 d at 26°C and 10 d at 37°C. As these represent the two most extreme scenarios, we tested 18 mosquitoes for each one.
Malaysian field samples
Malaysian samples were collected from three locations (researchers blinded as to origin) by staff from the Institute for Medical Research, Kuala Lumpur (https://www.imr.gov.my), and stored at -20°C in absolute ethanol before extraction. Mosquitoes were collected using ovitraps from two sites in Kuala Lumpur where the release of wAlbB-infected mosquitoes with a strain described by Ant et al. [29] is currently underway [2], as well as one control site where mosquitoes with Wolbachia have not been released. Mosquitoes were reared under standard conditions and sacrificed as adults for extraction. DNA was extracted from 24 mosquitoes from each field location for further analysis.
DNA extraction
For most experiments, individual mosquitoes were extracted by placing them in 200 μL of 0.3 M KOH and incubating tubes at 95°C for five minutes. This is the KOH concentration recommended for GeneWorks’ Lyse&Lamp reaction buffer for use on the Genie® III.
For pooled extractions, the quantity of KOH was increased–thus, a pool of 99 uninfected and one wAlbB-infected Ae. aegypti was extracted in 9 mL 0.3 M KOH, with aliquots taken before (i.e. negative control) and after the addition of the lone infected mosquito.
Our standard Wolbachia qPCR (LightCycler®) methods do not involve KOH. For comparisons between LAMP (Genie® III) and standard qPCR, genomic DNA was extracted using 250 μL of 5% Chelex® 100 Resin (Bio-Rad laboratories, Hercules CA) and 3 μL of Proteinase K (20 mg/ mL) (Roche Diagnostics Australia Pty. Ltd., Castle Hill New South Wales, Australia) solution. Tubes were incubated for 30 minutes at 65°C then for 10 minutes at 90°C. Following Chelex® extraction, an equivalent volume of 0.6 M KOH (Chem-Supply, Gillman, SA, Australia) was added to aliquots taken from each individual to produce final concentrations of 0.3 M KOH for analyses on the Genie® III, with an unadjusted aliquot being used for the qPCR assay.
qPCR assays
Aedes aegypti from the field sampling were tested for Wolbachia infection with qPCR according to Lee et al. [7] via the Roche LightCycler® 480. Three primer sets were used to amplify markers to confirm quality of mosquito DNA, the Ae. aegypti species and the presence or absence of the wAlbB infection. Crossing point (Cp) values of three consistent replicate runs were averaged to produce the final result. Differences in Cp values between the Ae. aegypti and wAlbB markers were transformed by 2n to produce relative Wolbachia density measures.
LAMP assays
LAMP primers for the wAlbB wsp sequence were derived from Bhadra et al. [17]. However, their OSD probe for wsp was replaced by an additional loop primer to increase detection speed (see S1 Table). LAMP primers for the Ae. aegypti ITS1 gene were taken from Schenkel et al. [24]. Primers were manufactured according to our specifications by Integrated DNA Technologies Inc. (Coralville, IA, USA) under the standard desalting purification process. Two alternative versions of each primer set were prepared to modify speed characteristics–a five primer and a six primer set. The wsp 5-primer set was identical to the original Bhadra primers, whereas the Ae. aegypti ITS1 5-primer set was constructed by removing the forward loop primer.
Typical LAMP reactions were conducted on a Genie® III machine (OptiGene Limited, Horsham UK)) according to GeneWorks’ Lyse&Lamp instructions, using their proprietary ISO-001-LNL Lyse&Lamp buffers. They involved combining 5 μL of a 20-fold dilution of extracted DNA with 20 μL master-mix, itself consisting of 15 μL Lyse&Lamp buffer, as well as enough of each LAMP primer to produce final concentrations of 20 pM FIP & BIP, 10 pM of each loop primer, and 5 pM each F3 & B3 respectively (in a final reaction volume of 25 μL).
Reactions were incubated at 65°C for 20–30 minutes. The Genie® III machine maintains real-time fluorescence detection throughout the incubation. Following amplification, an annealing curve analysis was conducted by reducing temperature by increments from 97 to 78°C in order to confirm the specificity of the amplified products.
Quantitative validation & frequency curves
To evaluate the quantitative efficacy of the LAMP primers, three wAlbB-infected Ae. aegypti KOH DNA extractions were first quantified using a Qubit 2.0 fluorometer (Thermo Fisher Scientific), then diluted up to 5,000-fold to form a standard concentration curve for estimating the concentration of the products of the Ae. aegypti ITS1 and wAlbB wsp LAMP primer sets. LAMP reactions were then run across this curve for both wAlbB and Ae. aegypti primer sets, comparing peak amplification time (Tp, min) with the log of relative concentration. Following visual inspection, regressions were carried out over the linear sections of that curve using the R function lm.
To investigate the ability of qLAMP to detect the relative frequencies of wAlbB-infected mosquitoes within a population, pooled DNA mixtures of both wAlbB-infected and uninfected individuals were created using laboratory-reared Ae. aegypti. Each pool was constructed to contain an equal volume of the DNA extract of twenty mosquitoes of mixed sex, but with different numbers of infected and uninfected mosquitoes. Eight frequency levels were constructed (infected/total): 0/20, 1/20, 3/20, 5/20, 8/20, 11/20, 15/20 20/20, with three separately generated pools at each frequency.
To further investigate the sensitivity of wAlbB detection in large pools of individuals where very few are infected, an additional pooled mixture was created by combining a single wAlbB-infected individual with 99 uninfected in the same extraction.
LAMP reactions targeting both wAlbB wsp and Ae. aegypti ITS1 regions were then run on these pools, using the previously described concentration regressions to derive relative concentrations of Wolbachia and Ae. aegypti DNA for each pool. The wAlbB wsp concentrations were then adjusted for overall mosquito DNA concentration based on Ae. aegypti ITS1 results–i.e. through calculating the ratio of Wolbachia concentration to Ae. aegypti concentration. Visual inspection and regressions were performed on the resulting frequency estimates, testing for goodness of fit with the known frequencies within each sample.
Results and discussion
Primer validation and characterization
The Ae. aegypti ITS1 primers from Schenkel et al. [24] exhibited a stable annealing point of 92.7°C (S.D. 0.11°C). With fresh reagents (i.e. isothermal buffer within two weeks of resuspension important as buffer decline slows amplification times, biasing quantitative assays), the use of all six primers produced positive detections with peak amplification times (Tp) ranging between 6 and 12 minutes over a 5,000-fold concentration gradient (S1 Fig).
The adapted wAlbB wsp LAMP primers from Bhadra et al. [17] (with an additional loop primer, i.e. with 6 primers) successfully amplified DNA from wAlbB-infected laboratory mosquitoes up to ten minutes faster than the Bhadra et al. primers alone, with a stable annealing point of 83.9°C (S.D. 0.17°C). With fresh reagents, these updated primers produced positive detections with Tp values ranging between 7 and 12 minutes over a 5,000-fold concentration gradient (S1 Fig). This primer was sensitive enough to detect the presence of wAlbB-infection in a 5,000-fold dilution of a KOH-extracted DNA from an Ae. aegypti individual. We also tested the assay’s sensitivity to the presence of infected individuals amongst large pools of uninfected individuals. We could detect a single wAlbB-infected Ae. aegypti among 99 uninfected mosquitoes with a Tp of 10 minutes–well within the quantitative bounds for this primer set (see S1 Fig).
To investigate the specificity of the modified wAlbB wsp LAMP primers, we challenged them with wMel-infected Ae. aegypti. No amplification was seen over a 30-min period in six wMel-infected samples.
When compared with the original 5-primer wAlbB wsp LAMP assay [17], the modified 6-primer assay was substantially faster (compare the concentration curve for 5-primer LAMP in Fig 1B with the curve for 6-primer (modified) LAMP in S1 Fig). This is expressed by a five-minute difference in intercept values for the respective regressions. The modified LAMP primer set we have developed thus represents a rapid, specific, and highly sensitive assay for wAlbB detection in Ae. aegypti mosquitoes.
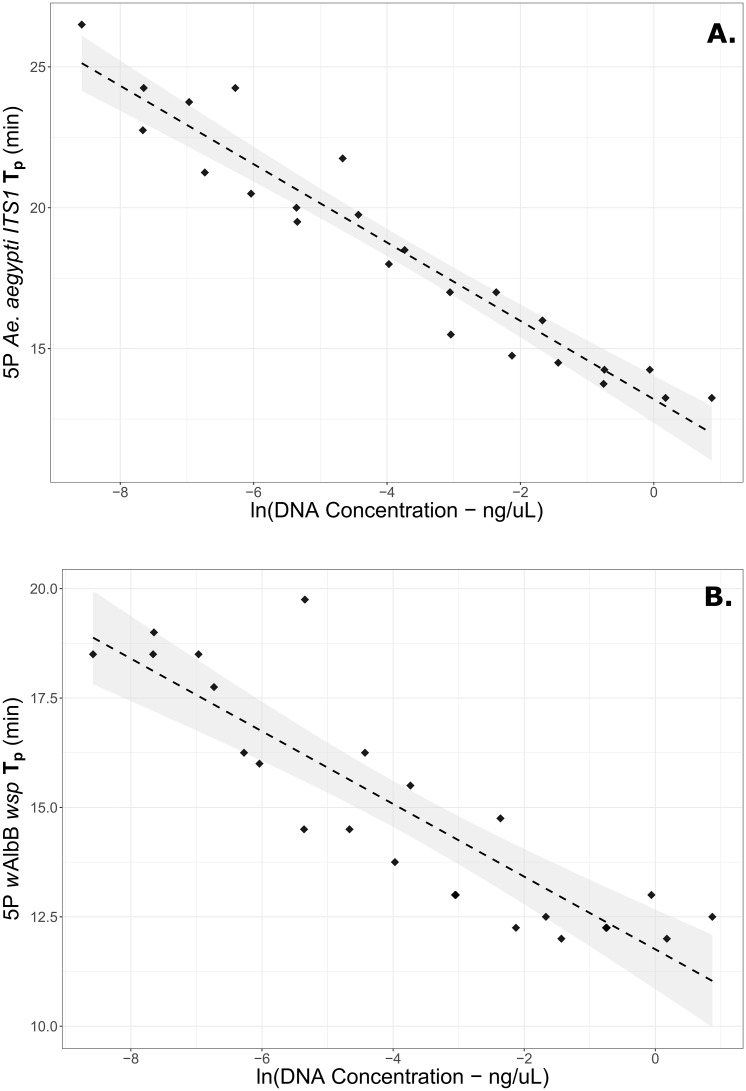
Fig 1. Concentration curves for (A) Ae. aegypti ITS1 & (B) wAlbB wsp LAMP 5-primer sets (i.e. the forward loop primer of each has been removed).
The horizontal axis shows the natural log of overall extracted DNA in the samples, while the vertical axis shows the peak amplification time (Tp) for each primer set under fresh reagent conditions. Linear regression lines are shown with 95% confidence intervals shaded.
Development of qLAMP for wAlbB in Ae. aegypti
Following KOH extractions, resulting DNA concentrations of three wAlbB individuals were first quantified using Qubit, then the samples were diluted up to 5,000-fold to form a standard concentration curve for measuring the Ae. aegypti ITS1 & wAlbB wsp LAMP primer sets. Concentration was highly correlated with amplification time under 6-primer conditions for each assay (S1 Fig); however, for highly precise quantitation these reactions were deemed too fast for the 15-second Tp resolution of the current Genie® III software. Accordingly, we repeated the curves on 5-primer LAMP sets, i.e. where the forward loop primer of each group had been removed (Fig 1).
A regression of 5-primer Ae. aegypti ITS1 Tp against the natural logarithm of overall DNA concentration was found to be highly significant (p = 1.2 e-13) with an adjusted R2 of 0.918 and a regression coefficient of -1.391 (S.E. 0.086). The regression of the original 5-primer wAlbB wsp Tp against the natural logarithm of overall DNA concentration was also highly significant (p = 1.09 e-08) with an adjusted R2 of 0.770 and a regression coefficient of -0.8306 (S.E. 0.094). The reduced R2 of wAlbB relative to Ae. aegypti may partly reflect variable concentrations of wAlbB within the three mosquitoes used for the curve.
Application of qLAMP to pooled DNA
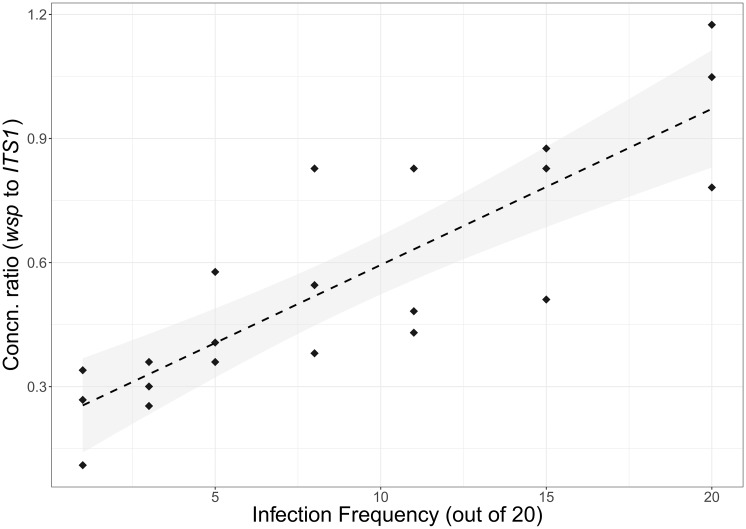
The association of wAlbB wsp to Ae. aegypti ITS1 concentration ratios to known frequency of the infection in DNA from pools of 20 mosquitoes is shown in Fig 2. The regression was highly significant (p = 1.10e-06) with a regression coefficient of 0.0377 (S.E. 0.0053) and an adjusted R2 of 0.7098 –higher than the R2 of 0.6091 for a regression of wAlbB concentration alone. These patterns suggest that the approach is sufficient for differentiation of Ae. aegypti with low, medium, or high wAlbB infection frequency.
Fig 2. Ratio of measured wAlbB concentrations to Ae. aegypti concentrations, compared to the actual infection frequency for pools of 20 mosquitoes with differing numbers of wAlbB-infected.
The linear regression line is shown with 95% confidence intervals shaded.
Effectiveness under conditions of stress or poor storage
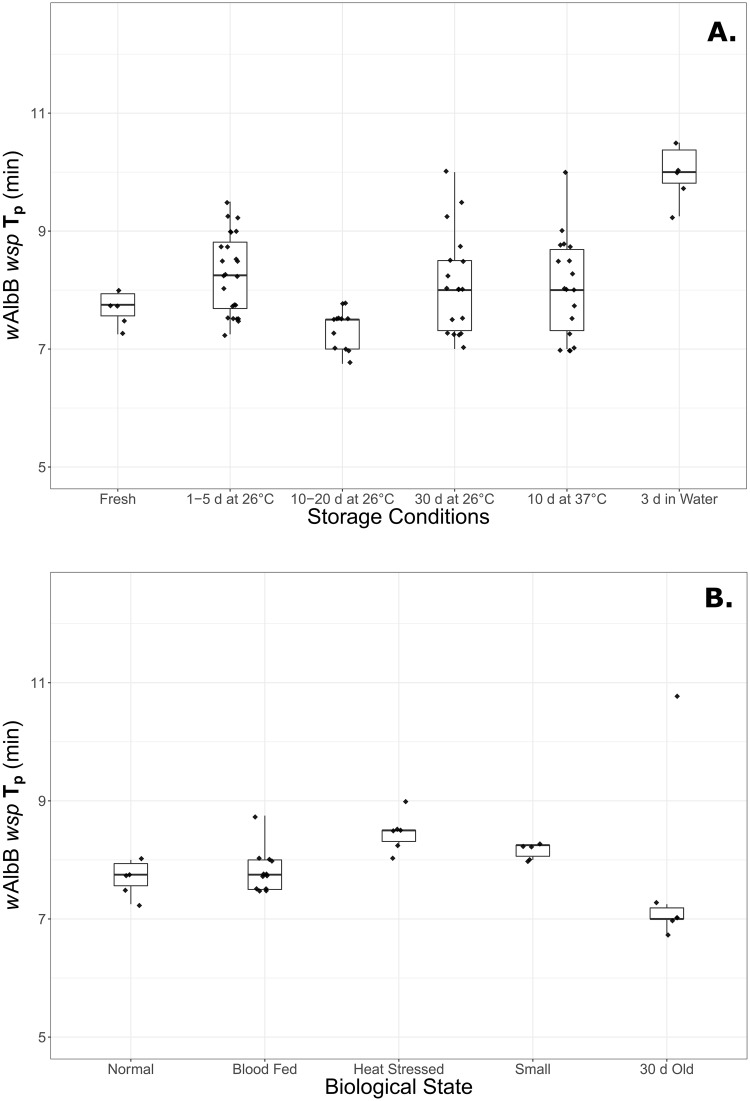
The wAlbB infection was readily detected when dead mosquitoes were stored in air for up to 30 d at 26°C and 10 d at 37°C–an improvement on the original Bhadra assay, which detected Wolbachia in only 40% of individuals stored for a week or more at 4°C, and failed to detect Wolbachia for individuals scored for a week at 37°C [17]. We obtained rapid amplification times for all storage conditions, though times for adults stored in water for 3 d were somewhat impacted (Fig 3A). Reliable detection of wAlbB was also achieved for adults that were aged, blood-fed, heat stressed or nutritionally stressed during development (Fig 3B). The modified LAMP assay we have developed is therefore robust to both poor storage conditions and low titre of Wolbachia. These features are valuable for field contexts such as the continued monitoring of Wolbachia releases, as the assay will reliably detect infections in low-titre situations and cope well with samples that have undergone degradation before DNA extraction. Our results suggest wAlbB will still be detectable where mosquito bodies have dried out in traps for extended periods (common in adult traps) or where they have been floating in water for some time (common in ovitraps).
Fig 3. Box plots of wAlbB peak amplification times (Tp) under differing storage conditions or biological states.
(A) Fresh samples compared to varying lengths of dry storage at 26°C, 10 days at 37°C, and 3 days in water. (B) Mosquitoes under standard rearing conditions compared to mosquitoes after blood feeding, those undergoing life histories of heat or resource stress, and aged mosquitoes.
Comparison with established PCR surveillance methods on Malaysian field samples
Over a series of LAMP processes, 24 individuals from each of three Malaysian locations (blindly scored as A, B and C) were tested by both LAMP and LightCycler for presence/absence of Wolbachia. For the LAMP results, 22/24 from location A were found to be infected (individuals 4 and 17 absent), and 22/24 from B (individuals 4 and 20 absent). All individuals of location C–which turned out to be a control site where no Wolbachia releases have occurred–were scored as uninfected, failing to amplify under LAMP conditions. All results were consistent with those of the qPCR assays (Table 1).
Table 1. Relative performance of qPCR and LAMP assays for wAlbB detection in mosquitoes sourced from three sites in Malaysia.
| Sample | N | Status | Infected (qPCR) | Infected (LAMP) | % difference |
|---|---|---|---|---|---|
| A | 24 | wAlbB release | 22 | 22 | 0 |
| B | 24 | wAlbB release | 22 | 22 | 0 |
| C | 24 | No release | 0 | 0 | 0 |
When tested with the Genie® III system, the LAMP assays developed in this paper provide a rapid, accurate and robust means of ascertaining wAlbB infection status of mosquitoes in field locations without the necessity for complex preparation or the use of developed laboratory facilities. LAMP assays can ascertain wAlbB presence/absence in pooled or individual Ae. aegypti mosquitoes in under 20 minutes’ amplification time under a wide variety of environmental or storage conditions, and they can detect infections of 1% or lower in pooled samples, making them suitable for a variety of applications such as rapid monitoring of areas peripheral to a local Wolbachia release when monitoring infection spread. Applied to individuals, this wAlbB assay shows a similar detection sensitivity and specificity as established qPCR monitoring.
While not sufficiently precise to determine the exact frequency of Wolbachia in a sample, this assay when coupled with the Ae. aegypti ITS1 primer set enables approximate quantification of infection frequency within a sample. This has clear applications for monitoring Wolbachia deployment for disease control. While an evaluation of release success requires an estimate of Wolbachia frequency, approximate estimates may be adequate for many purposes when combined with selective use of more accurate assays. Wolbachia could first be characterized as being absent, or as having a low, intermediate or high frequency at a site using a mobile, fast-acting, method as described here. Where an approximate determination of Wolbachia frequencies suggest ongoing issues with a release (e.g. where frequencies remain intermediate despite ongoing releases), other more accurate estimates could be obtained. Such a two-tiered monitoring scheme would assist with the scaling of Wolbachia releases during an operational phase.
The potential of quantitative LAMP for monitoring and control operations remains to be fully exploited. Until this point, most qLAMP assays have focused on ascertaining the concentrations of pathogens within specific infection contexts (e.g. 14, 21, 30, 31), although studies have considered microorganismal eDNA [14], pathogenic viruses in water [30] and forensic applications [31]. Our study reveals another application–ascertaining the approximate frequency of individuals infected with a symbiotic bacterium in disease-related releases. Other frequency-based applications could include monitoring the spread of disease resistance alleles, infections, and other traits distinguishable by DNA sequence on large scales using pooled DNA. These applications are further supported by the low cost and fast run-times of the assay. Our typical expense for a single individual extracted in 200μL 0.3M KOH run as part of an 8-strip LAMP reaction with the 6-primer wAlbB assay is approximately $3.30 AUD per sample (for comparison, our qPCR assay for Wolbachia in Ae. aegypti is approximately $1.04 per sample—$3.12 with three replicates). Additionally, with the use of the Genie® III machine and 5-minute KOH boil in a field context, time from commencing DNA extraction to determination of Wolbachia status can realistically be as short as an hour. The highly specific nature of a LAMP assay, owing to its four to six primers required, means it can be easily adapted to diverse targets, provided the primer design is conducted according to best practice.
Supporting information
Top. Wolbachia wAlbB wsp LAMP assay (6-primer) [17]. Bottom. Aedes aegypti ITS1 LAMP assay (6-primer) [24].
(DOCX)
Horizontal axes show the natural log of overall extracted DNA in the samples, while vertical axes show the peak amplification time (Tp) for each primer set under fresh reagent conditions. Linear regression lines are shown with 95% confidence intervals shaded. A regression of 6-primer Ae. aegypti ITS1 Tp against the natural logarithm of overall DNA concentration was found to be highly significant (p<2e-16) with an adjusted R2 of 0.919 and a regression coefficient of -0.580 (S.E. 0.029). The regression of 6-primer wAlbB wsp Tp against the natural logarithm of overall DNA concentration was also highly significant (p = 1.27e-13), with an adjusted R2 of 0.800 and a regression coefficient of -0.423 (S.E. 0.036). The reduced R2 value of wAlbB relative to Ae. aegypti may partly reflect variable concentrations of wAlbB within the three mosquitoes used for points along the curve.
(TIF)
Acknowledgments
Malaysian Ae. aegypti samples were supplied by Dr. Nazni W. Ahmad of the Institute for Medical Research, a division of the Ministry of Health, Malaysia.
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
AH received a grant (# 108508) from the Wellcome Trust (https://wellcome.ac.uk/). AH received a grant (Program Grant # 1132412) from the National Health and Medical Research Council (AU), (https://www.nhmrc.gov.au/). AH received a grant (Fellowship Grant # 1118640) from the National Health and Medical Research Council (AU), (https://www.nhmrc.gov.au/). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Hoffmann AA, Montgomery BL, Popovici J, Iturbe-Ormaetxe I, Johnson PH, Muzzi F, et al. Successful establishment of Wolbachia in Aedes populations to suppress dengue transmission. Nature. 2011;476(7361):454–7. http://www.nature.com/nature/journal/v476/n7361/abs/nature10356.html#supplementary-information [DOI] [PubMed] [Google Scholar]
- 2.Nazni WA, Hoffmann AA, Afizah AN, Cheong YL, Mancini MV, Golding N, et al. Establishment of Wolbachia strain wAlbB in Malaysian populations of Aedes aegypti for dengue control. bioRxiv. 2019:775965 10.1101/775965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bourtzis K, Dobson SL, Xi Z, Rasgon JL, Calvitti M, Moreira LA, et al. Harnessing mosquito-Wolbachia symbiosis for vector and disease control. Acta Tropica. 2014;132:S150–S63. 10.1016/j.actatropica.2013.11.004 [DOI] [PubMed] [Google Scholar]
- 4.Garcia GdA, Sylvestre G, Aguiar R, da Costa GB, Martins AJ, Lima JBP, et al. Matching the genetics of released and local Aedes aegypti populations is critical to assure Wolbachia invasion. PLOS Neglected Tropical Diseases. 2019;13(1):e0007023 10.1371/journal.pntd.0007023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nguyen TH, Le Nguyen H, Nguyen TY, Vu SN, Tran ND, Le TN, et al. Field evaluation of the establishment potential of wMelPop Wolbachia in Australia and Vietnam for dengue control. Parasites Vectors. 2015;8:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schmidt TL, Barton NH, Rasic G, Turley AP, Montgomery BL, Iturbe-Ormaetxe I, et al. Local introduction and heterogeneous spatial spread of dengue-suppressing Wolbachia through an urban population of Aedes aegypti. PLoS Biology. 2017;15(5):e2001894 10.1371/journal.pbio.2001894 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee SF, White VL, Weeks AR, Hoffmann AA, Endersby NM. High-throughput PCR assays to monitor Wolbachia infection in the dengue mosquito (Aedes aegypti) and Drosophila simulans. Applied and Environmental Microbiology. 2012;78(13):4740–3. 10.1128/AEM.00069-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N, et al. Loop-mediated isothermal amplification of DNA. Nucleic Acids Research. 2000;28(12):7 10.1093/nar/28.12.e63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nagamine K, Hase T, Notomi T. Accelerated reaction by loop-mediated isothermal amplification using loop primers. Molecular and Cellular Probes. 2002;16(3):223–9. 10.1006/mcpr.2002.0415 [DOI] [PubMed] [Google Scholar]
- 10.Lucchi NW, Demas A, Narayanan J, Sumari D, Kabanywanyi A, Kachur SP, et al. Real-Time Fluorescence Loop Mediated Isothermal Amplification for the diagnosis of malaria. PLoS One. 2010;5(10):e13733 10.1371/journal.pone.0013733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lucchi NW, Ndiaye D, Britton S, Udhayakumar V. Expanding the malaria molecular diagnostic options: opportunities and challenges for loop-mediated isothermal amplification tests for malaria control and elimination. Expert Review of Molecular Diagnostics. 2018;18(2):195–203. 10.1080/14737159.2018.1431529 [DOI] [PubMed] [Google Scholar]
- 12.Parida M, Posadas G, Inoue S, Hasebe F, Morita K. Real-Time Reverse Transcription Loop-Mediated Isothermal Amplification for rapid detection of West Nile Virus. Journal of Clinical Microbiology. 2004;42(1):257–63. 10.1128/JCM.42.1.257-263.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wheeler SS, Ball CS, Langevin SA, Fang Y, Coffey LL, Meagher RJ. Surveillance for Western Equine Encephalitis, St. Louis Encephalitis, and West Nile Viruses using Reverse Transcription Loop-Mediated Isothermal Amplification. PLoS One. 2016;11(1):e0147962 Epub 2016/01/26. 10.1371/journal.pone.0147962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dauner AL, Mitra I, Gilliland T Jr., Seales S, Pal S, Yang SC, et al. Development of a pan-serotype reverse transcription loop-mediated isothermal amplification assay for the detection of dengue virus. Diagnostic Microbiology and Infectious Disease. 2015;83(1):30–6. Epub 2015/06/03. 10.1016/j.diagmicrobio.2015.05.004 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee PLM. DNA amplification in the field: move over PCR, here comes LAMP. Molecular Ecology Resources. 2017;17(2):138–41. 10.1111/1755-0998.12548 [DOI] [PubMed] [Google Scholar]
- 16.Gonçalves DD, Cassimiro APA, de Oliveira CD, Rodrigues NB, Moreira LA. Wolbachia detection in insects through LAMP: loop mediated isothermal amplification. Parasites Vectors. 2014;7 10.1186/1756-3305-7-228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bhadra S, Riedel TE, Saldana MA, Hegde S, Pederson N, Hughes GL, et al. Direct nucleic acid analysis of mosquitoes for high fidelity species identification and detection of Wolbachia using a cellphone. PLOS Neglected Tropical Diseases. 2018;12(8):e0006671 Epub 2018/08/31. 10.1371/journal.pntd.0006671 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kulkarni A, Yu W, Jiang J, Sanchez C, Karna AK, Martinez KJ, et al. Wolbachia pipientis occurs in Aedes aegypti populations in New Mexico and Florida, USA. Ecology and Evolution. 2019;9(10):6148–56. 10.1002/ece3.5198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Noor-Shazleen-Husnie MM, Emelia O, Ahmad-Firdaus MS, Zainol-Ariffin P, Aishah-Hani A. Detection of Wolbachia in wild mosquito populations from selected areas in Peninsular Malaysia by loop-mediated isothermal amplification (LAMP) technique. Tropical Biomedicine. 2018;35(2):330–46. [PubMed] [Google Scholar]
- 20.Aoi Y, Hosogai M, Tsuneda S. Real-time quantitative LAMP (loop-mediated isothermal amplification of DNA) as a simple method for monitoring ammonia-oxidizing bacteria. Journal of Biotechnology. 2006;125(4):484–91. Epub 2006/06/23. 10.1016/j.jbiotec.2006.04.007 . [DOI] [PubMed] [Google Scholar]
- 21.Fujiwara-Nagata E, Eguchi M. Development and evaluation of a loop-mediated isothermal amplification assay for rapid and simple detection of Flavobacterium psychrophilum. Journal of Fish Diseases. 2009;32(10):873–81. Epub 2009/06/09. 10.1111/j.1365-2761.2009.01066.x . [DOI] [PubMed] [Google Scholar]
- 22.Chen BJ, Mani V, Huang ST, Hu YC, Shan HP. Bisintercalating DNA redox reporters for real-time electrochemical qLAMP. Biosensors and Bioelectronics. 2019;129:277–83. Epub 2018/09/30. 10.1016/j.bios.2018.09.056 . [DOI] [PubMed] [Google Scholar]
- 23.Su Y, Huang S, Hong L, Zou D, Tang Y, Chao S, et al. Establishment of the molecular beacon-loop-mediated isothermal amplification method for the rapid detection of Porphyromonas gingivalis. Journal of Microbiological Methods. 2019;160:68–72. Epub 2019/03/30. 10.1016/j.mimet.2019.01.013 . [DOI] [PubMed] [Google Scholar]
- 24.Schenkel CD, Kamber T, Schaffner F, Mathis A, Silaghi C. Loop-mediated isothermal amplification (LAMP) for the identification of invasive Aedes mosquito species. Medical and Veterinary Entomology. 2019;33(3):345–51. 10.1111/mve.12366 [DOI] [PubMed] [Google Scholar]
- 25.Xi Z, Khoo CC, Dobson SL. Wolbachia establishment and invasion in an Aedes aegypti laboratory population. Science (New York, NY). 2005;310(5746):326–8. Epub 2005/10/15. 10.1126/science.1117607 . [DOI] [PubMed] [Google Scholar]
- 26.Walker T, Johnson PH, Moreira LA, Iturbe-Ormaetxe I, Frentiu FD, McMeniman CJ, et al. The wMel Wolbachia strain blocks dengue and invades caged Aedes aegypti populations. Nature. 2011;476(7361):450–3. http://www.nature.com/nature/journal/v476/n7361/abs/nature10355.html#supplementary-information [DOI] [PubMed] [Google Scholar]
- 27.Ross PA, Ritchie SA, Axford JK, Hoffmann AA. Loss of cytoplasmic incompatibility in Wolbachia-infected Aedes aegypti under field conditions. PLOS Neglected Tropical Diseases. 2019;13(4):e0007357 10.1371/journal.pntd.0007357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Callahan AG, Ross PA, Hoffmann AA. Small females prefer small males: size assortative mating in Aedes aegypti mosquitoes. Parasites Vectors. 2018;11:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ant TH, Herd CS, Geoghegan V, Hoffmann AA, Sinkins SP. The Wolbachia strain wAu provides highly efficient virus transmission blocking in Aedes aegypti. PLoS Pathogens. 2018;14(1):19 10.1371/journal.ppat.1006815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang X, Lin X, Urmann K, Li L, Xie X, Jiang S, et al. Smartphone-based in-gel Loop-Mediated Isothermal Amplification (gLAMP) system enables rapid Coliphage MS2 quantification in environmental waters. Environmental Science & Technology. 2018;52(11):6399–407. Epub 2018/05/09. 10.1021/acs.est.8b00241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kitamura M, Kubo S, Tanaka J, Adachi T. Rapid screening method for male DNA by using the loop-mediated isothermal amplification assay. International Journal of Legal Medicine. 2018;132(4):975–81. Epub 2017/08/15. 10.1007/s00414-017-1661-z . [DOI] [PubMed] [Google Scholar]