Supplemental Digital Content is available in the text
Keywords: CNR, depressive disorder, rs1049353, rs2501432, triplet repeat polymorphism
Abstract
Studies investigating the association between gene variants and depression susceptibility found inconsistent data. The present study aimed to clarify whether CNR1rs1049353, CNR1 AAT triplet repeat, and CNR2rs2501432 polymorphisms confer higher risk for depressive disorder.
Literature from PubMed, Medline, Embase, Scopus, Cochrance Library, and Wanfang databases was searched (up to August 20, 2018). Seven case–control studies with various comorbidities were eligible. We targeted CNR single-nucleotide polymorphisms (SNPs) that have been reported by 2 or more studies to be involved in the current meta-analysis, resulting in a final list of 3 SNPs: CNR1rs1049353, CNR1 AAT triplet repeat polymorphism, and CNR2rs2501432. Odds ratios (ORs) and 95% confidence intervals (CIs) for allele and homozygote comparisons, dominant and recessive models, and triplet repeat polymorphism ((AAT)n≥5, ≥5 vs (AAT)n<5, <5 or <5, ≥5) were assessed using a random effect model as measures of association. Heterogeneity among included studies was analyzed using sensitivity test. Publication bias was also explored by Egger and rank correlation test.
overall, no significant association was found between depression and CNR1rs1049353 (G vs A: OR [95% CI] = 1.09 [0.61–1.95]; GG vs AA: 1.29 [0.73–2.26]; GG vs GA+AA: 1.10 [0.57–2.10]; GG+GA vs AA: 1.25 [0.72–2.18]; and AAT triplet repeat polymorphism ((AAT)n≥5, ≥5 vs (AAT)n<5, <5 or <5, ≥5): 1.92 [0.59–6.27]. In contrast, a significant association between CNR2rs2501432 and depression was detected, and the ORs and 95% CIs are as follows: allele contrast (OR = 1.39, 95% CI = [1.12–1.72], P = .003); homozygous (OR = 2.19, 95% CI = [1.34–3.59], P = .002); dominant (OR = 1.93,95% CI = [1.23–3.04], P = .005); and recessive (OR = 1.41, 95% CI = [1.04–1.92], P = .03).
This meta-analysis revealed that CNR1rs1049353 or AAT triplet repeat polymorphism had no association with susceptibility to depression, while CNR2rs2501432 polymorphism was a remarkable mark for depression patients.
1. Introduction
Depression is one of the most common psychiatric disorder with significant heritable components, and it is responsible for more “years lost” to disability than any other condition.[1] It affects an estimated 8% of person in the United States and accounts for more than $210 billion in healthcare costs annually.[2,3] The symptom of depression presents not only in mental disorders such as major depression (MD) and bipolar disorder (BD), but also in other chronic somatic diseases like rheumatoid arthritis, Parkinson disease (PD), and premenstrual dysphoric disorder (PMDD).[4–6] Recently, a large body of data has shown that patients diagnosed with depression have higher levels of CNRs (CNR1 and CNR2) in their neuroanatomical structures and circuits including the prefrontal cortex, hippocampus, amygdala, hypothalamus, and forebrain monoaminergic circuits.[7–9] Furthermore, mice lacking CNR1 can represent a validate and appropriate model to evaluate depressive-like disorders in animals,[10] and results from experimental research also found that the blockade of endocannabinoid system is a risk factor in the pathogenesis of depression as well as anxiety disorders.[11] Genetic variants of CNRs can alter the gene transcription and thereby protein expression and biologic function of these cannabinoid receptors.[12] The underlying mechanisms of the mood-regulating properties of CNRs signaling remain unclear.
Several studies have reported associations between genetic variants of the CNRs and susceptibility to depression in general population. The results of association between CNRs and predisposition to depressive disorder are contradicting. In a study of methadone-maintained outpatients, Icick et al[13] found that patients with at least 1 copy of the C allele of CNR1rs2023239 (including both CT heterozygous and CC homozygous subjects) had a lower prevalence of lifetime MD. Nevertheless, a study investigated the relation between CNR1 microsatellite polymorphism and mood disorders (i.e., depressive disorders and BDs) reported that this genetic variant was not likely to be involved in the pathogenesis or in the psychotic symptoms of mood disorders,[14] and similarly, Mitjans et al[15] did not showed any significant association between CNR1rs1049353 and major depressive disorder risk in their research. CNR2 is widely distributed in the brain and plays broad roles in mental diseases as well, including depression, drug addiction, and schizophrenia.[7] In Japanese alcoholic and depressed subjects, there was high incidence of CNR2 Q63R gene polymorphism.[16] Another study found a significant association between CNR2 524C>A (leu133 Ile) polymorphism and BD.[17]
The aim of this meta-analysis was to evaluate the main effects of CNR1rs1049353, CNR1 triplet repeat polymorphism, and CNR2rs2501432 pertaining to depressive disorders and to test for their consistency in association with the disease and heterogeneity in the effect size across studies.
2. Materials and methods
2.1. Search strategy
A systematic literature search was performed in databases of PubMed, Embase, Medline, Cochrance Library, Scopus, and Wanfang for studies which have investigated the relation between endocannabinoid genes polymorphisms and depression (up to August 20, 2018). Two independent researchers used the following keywords: (“mood disorders” OR “depressive disorder” OR “depressive episode” OR “depression” OR “Major depression” or “MD” OR “Bipolar disorder” OR “BD”) AND (“genetic polymorphism” OR “genetic variation” OR “genetic variant” OR “polymorphism” OR “variant”) AND (“cannabinoid receptors” OR “CB” OR “CB1” OR “CB2” OR “FAAH”) to retrieve related articles. To maximize the number of identified papers, the reference lists of eligible articles were also considered by manual searching. We did not put any restrictions on publication date, publication type, sample origin, country, or language to avoid any potential publication bias.
As this study is a meta-analysis of previously published studies, the ethical approval and patient consent are not required.
2.2. Inclusion and exclusion criteria
The inclusion criteria for this meta-analysis were as follows: human case–control design; patients diagnosed with depressive disorder by adequate diagnostic tool; providing adequate summary statistics on endocannabinoid genes polymorphisms for calculating the value of odds ratio (OR) and 95% confidence interval (CI); original papers; and investigating the relation between endocannabinoid gene polymorphisms and depression.
The exclusion criteria were: articles reporting overlapping data and studies with duplicated data.
2.3. Data extraction
In accordance with the PRISMA (Preferred reporting items for systematic reviews and meta-analyses) guidance, 2 independent investigators (XK and QM) identified the eligible studies and extracted reusable information. The following data from individual study were extracted whenever available: 1st author, publication year, number of cases and controls, geographic location, diagnostic tools for depression, genotyping method, frequencies of genotype and/or alleles in case and control groups, and P-value for Hardy–Weinberg equilibrium. Any discrepancies would be settled by a 3rd investigator (XL).
2.4. Statistical analysis
We used Stata 14.0 software (Stata Corporation, College Station, TX) and Review Manager (RevMan) 5.3 program (Copenhagen: The Nordic Cochrane Centre, the Cochrane Collaboration, 2014) to carry out all statistical procedures. The pooled OR and 95% CIs were utilized as measurements to estimate the relationship between CNRs (CNR1/2) polymorphisms and their respective risks for susceptibility to depression, under allele contrast (CNR1: G vs A; CNR2: A vs G), homozygote comparisons (CNR1:GG vs AA; CNR2: AA vs GG), and dominant (CNR1: GG vs GA+AA; CNR2: AA vs GA+GG) and recessive (CNR1:GG +GA vs AA; CNR2: AA+GA vs GG) models. Statistical heterogeneity among studies was assessed by the Q-test (meaningful value of P < .05), alongside the degree of inconsistency presented as the I2 index where I2 of 0% to 25% = low heterogeneity, 25% to 50% = moderate heterogeneity, 50% to 70% = large heterogeneity, and 75% to 100% = considerable heterogeneity. A random effect model was used to evaluate the pooled ORs and 95% CIs, as heterogeneity was found with P < .01 or I2 > 50%. Sensitivity analysis was conducted with all the comparisons using leave-one-out method through sequential omission of individual study to evaluate the effect on the remaining effect size estimates and subsequent I2 statistic for heterogeneity. Potential publication bias was examined by means of Egger linear regression test, rank correction test, and funnel plots.
3. Results
3.1. Study selection and characteristics
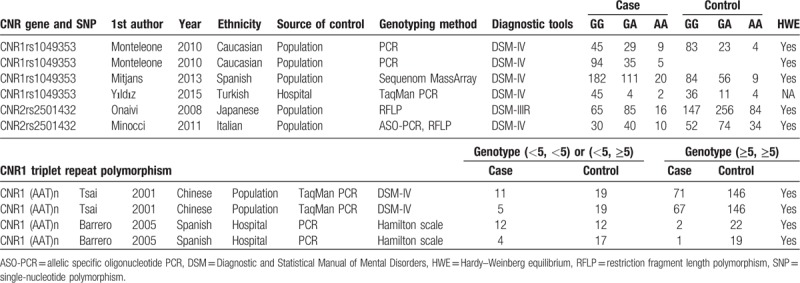
We can see the literature search and publication selection process from the flow diagram (Fig. 1). A total of 1063 potential papers were identified from 6 databases, and 187 of them were discarded owing to duplicating. After scrutinizing the titles and abstracts of remaining 876 records, articles that are not about endocannabinoid gene variants or depressive disorders (n = 711) or did not have abstracts (n = 9) were removed. Furthermore, reviews (n = 103), meta-analyses (n = 7) were omitted, or patents and book chapters (n = 10), and 36 potentially relevant studies were included in the full-text reviewing step. During the final step 29 papers were excluded for the following reasons: the full text was not available; studies that did not evaluate the association between endocannabinoid gene polymorphism and depression; they were not case–control designs; they did not provide worthy data about gene alleles or genotypes of endocannabinoid gene. Any endocannabinoid gene single-nucleotide polymorphisms (SNPs) that were reported by <2 studies were omitted. Eventually, 7 studies (with 827 depression people and 917 controls) performed between 2001 and 2015 were chosen in our meta-analysis, covering 3 gene SNPs (CNR1rs1049353, CNR1 AAT triplet repeat polymorphism, and CNR2rs2501432). Characteristics of the included studies are summarized in Table 1. Among these studies, 2 were carried out among Spanish, and each 1 among Chinese, Italian, Turkish, Caucasian, and Japanese. To establish the presence of depression, mean values of Diagnostic and Statistical Manual of Mental Disorders (DSM)-IV, DSM-IIIR, or Hamilton scale were used.
Figure 1.
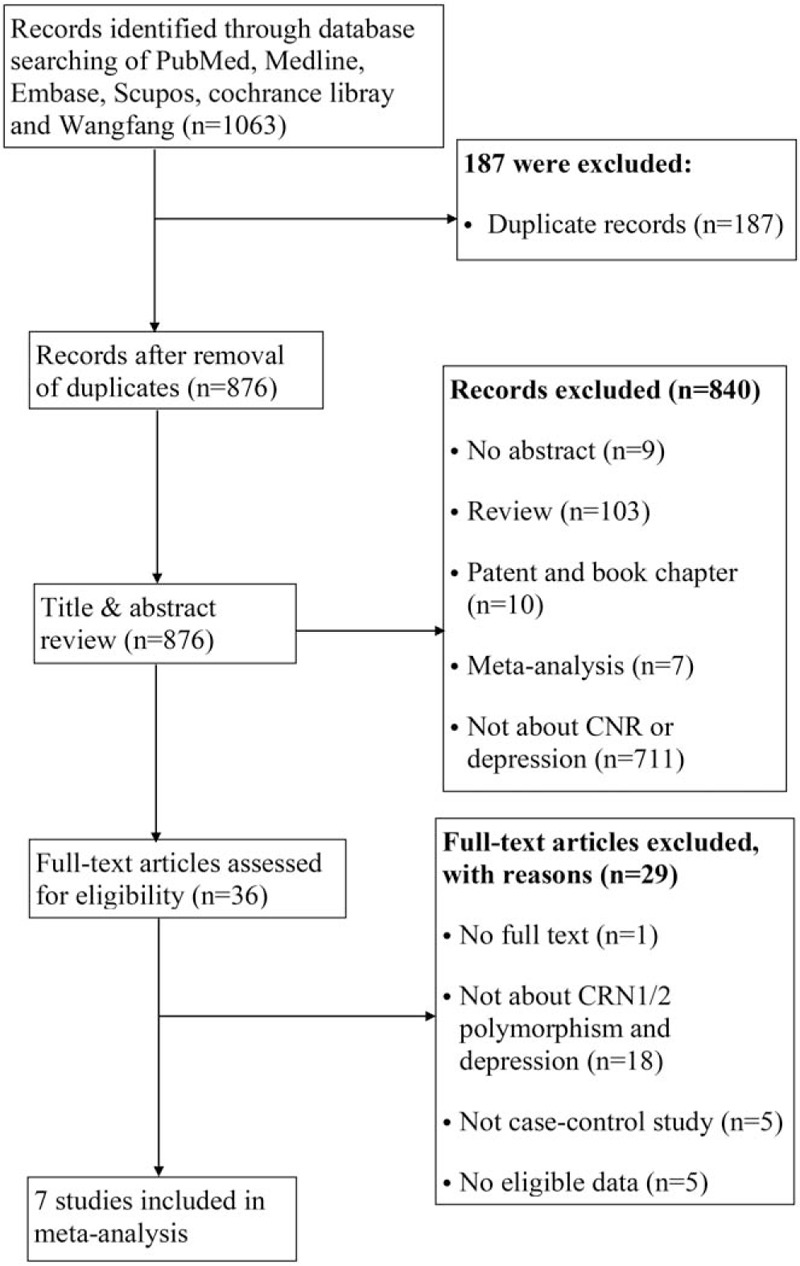
Preferred reporting items for systematic reviews and meta-analyses flow diagram.
Table 1.
Characteristics of individual studies estimating relation between CNR polymorphisms and depression.
3.2. Meta-analysis
3.2.1. CNR1rs1049353 and depressive disorder
Four studies were selected for CNR1rs1049353 gene polymorphism to calculate the combined OR (Figs. 2–6). There was no significant association between this polymorphism and predisposition to depression in any of the models we used. The OR and 95% CI for each model were as follows: 1.09 [0.61–1.95] in the allele comparison model (G vs A); 1.29 [0.73–2.26] in the homozygote comparison model (GG vs AA); 1.10 [0.57–2.10] in the dominant genetic model (GG vs GA+AA); 1.25 [0.72–2.18] in the recessive genetic model (GG vs GA+AA) and 1.92 [0.59–6.27] in triplet repeat polymorphism ((AAT)n≥5, ≥5 vs (AAT)n<5, <5 or <5, ≥5). Heterogeneity presented among studies in different genetic models was diverse and the data are detailed in Figures 2–6.
Figure 2.
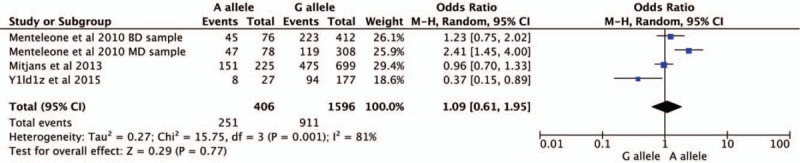
Forest plot of CNR1rs1049353 polymorphism and depressive disorder for allele model. CI = confidence interval.
Figure 6.
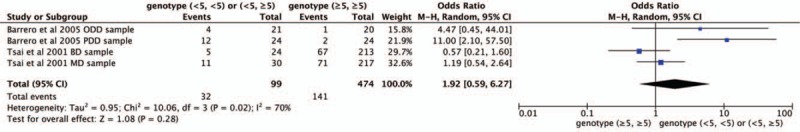
Forest plot of CNR1rs1049353 (AAT)n triple repeat polymorphism and depression. CI = confidence interval.
Figure 3.
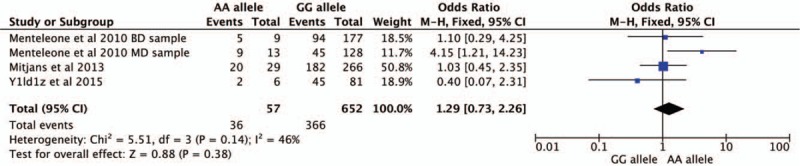
Forest plot of CNR1rs1049353 polymorphism and depression under homozygous model. CI = confidence interval.
Figure 4.
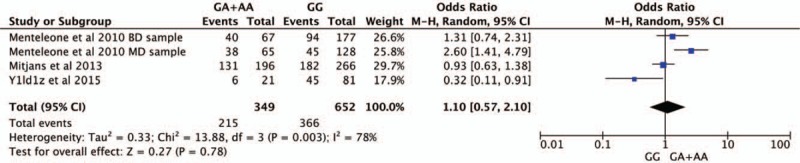
Forest plot of CNR1rs1049353 polymorphism and depression in dominant model. CI = confidence interval.
Figure 5.
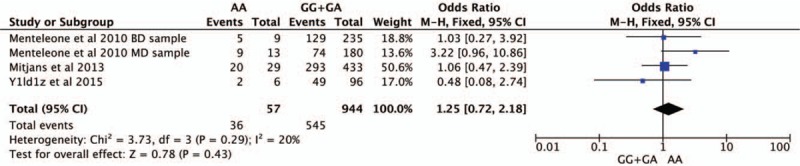
Forest plot of CNR1rs1049353 polymorphism and depressive disorder in recessive model. CI = confidence interval.
3.2.2. CNR2rs2501432 and depressive disorder
Our meta-analysis revealed an overall significant association between CNR2rs2501432 and depressive disorder susceptibility under all genetic models: allele contrast (OR = 1.39, 95% CI = [1.12–1.72], P = .003); homozygous (OR = 2.19, 95% CI = [1.34–3.59], P = .002); dominant (OR = 1.93, 95% CI = [1.23–3.04], P = .005); and recessive (OR = 1.41, 95% CI = [1.04–1.92], P = .03). Among all aforementioned genetic models, the homozygous model presented the strongest association with depressive disorders. The pooled effect-size estimates are demonstrated in Figure 7. There was no considerable heterogeneity detected between studies included in the analysis (I2 = 0%; P > .05).
Figure 7.
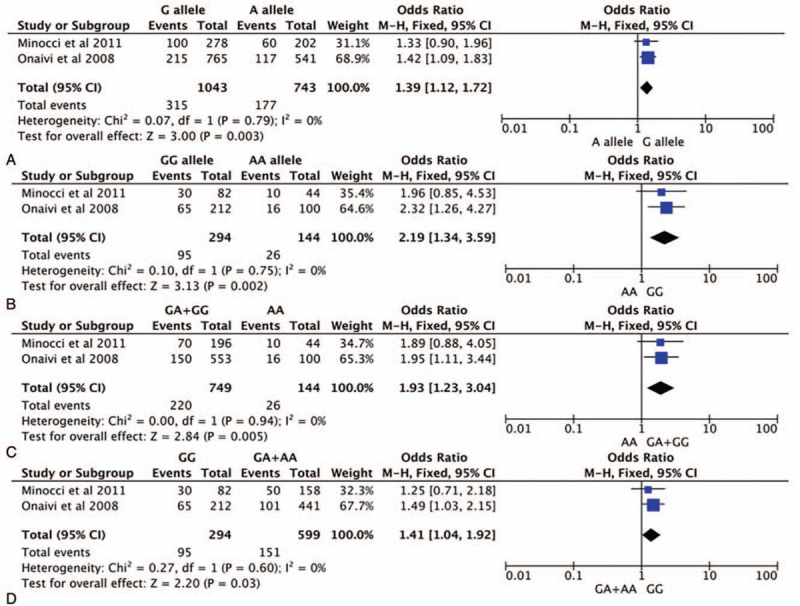
Forest plot of CNR2rs2501432 polymorphism and depression (A allele vs G allele), (B) AA vs GG, (C) AA vs GA+GG, and (D) AA+GA vs GG. CI = confidence interval.
3.3. Publication bias
The results of Egger test and rank correlation test did not present any evidence of obvious publication bias for each CNR1rs1049353 genetic models, and Table 2 includes the publication bias assessment method with its respective P-value for each test. In addition, the graphical representations of publication bias under homozygous and recessive model for CNR1rs1049353 SNP are shown below (Figs. 8 and 9). With respect to CNRrs2501432, publication bias was not estimated owing to insufficient data (<3 studies).
Table 2.
Evaluation of publication bias for CNR1 rs1049353.
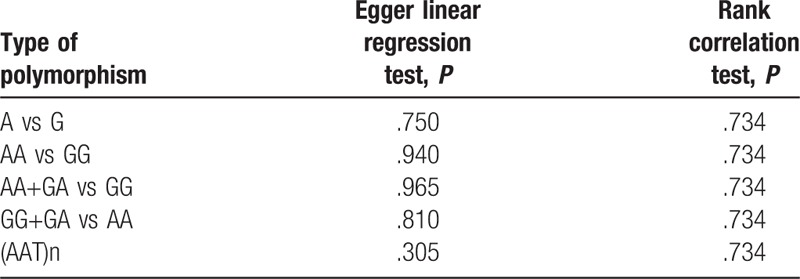
Figure 8.
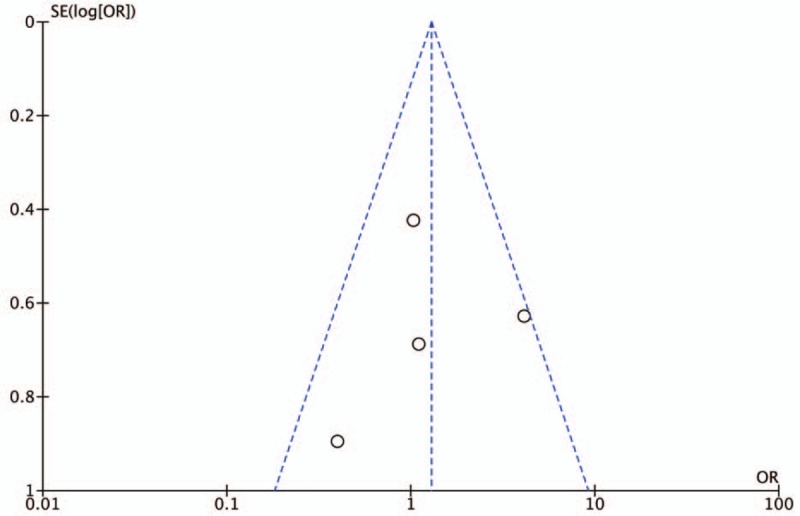
Funnel plot of publication bias for CNR1rs1049353 AA vs GG model. OR = odds ratio. SE = standard error.
Figure 9.
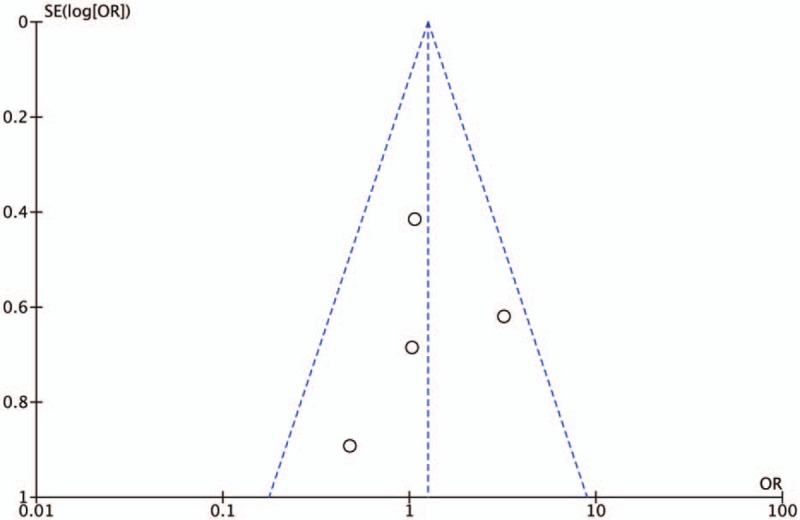
Funnel plot of publication bias for AA vs GA+GG model. OR = odds ratio. SE = standard error.
3.4. Sensitivity analysis
We performed sensitivity analysis in CNR1rs11049323 SNP and CNR1 AAT triplet repeat polymorphism to validate the robustness and reproducibility of our results using the leave-one-out approach by omitting 1 study at a time and repeating the meta-analysis, for all genetic models. During each study's deletion, the pooled effect size estimates did not vary dramatically. The P statistic demonstrated nonsignificant values ranging from 0.13 to 0.99. On the contrary, the omission of MD sample study altered the heterogeneity in allele contrast, homozygous, dominant, and recessive comparison models, and our observed heterogeneity changed by the deletion of PDD sample study in (AAT)n triple repeated genotype (Supplementary Table S1).
4. Discussion
In the present study, we evaluated the contribution of 3 CNR SNPs to the susceptibility to depression. Each genetic model of CNR1rs1049353 or CNR1 (AAT)n triplet repeat polymorphism had no association with increased depression risk, which included allele and homozygote comparisons, dominant and recessive models, and triplet repeat polymorphism ((AAT)n≥5, ≥5 vs (AAT)n<5, <5 or <5, ≥5) model. In contrast, a significant correlation was found between all four genetic models of CNR2rs2501432 SNPs and depression. To our best knowledge, this is the 1st meta-analysis to assess the relation between CNR1 and CNR2 polymorphism and depressive disorder susceptibility by combining all available literature up to date. Specially, 2 commonly interrogated SNPs in CNR with all possible genetic models, including allele and homozygote comparisons, dominant and recessive models, and (AAT)n triplet repeat polymorphism were analyzed across 7 case–control studies which covered depressive disorders regardless of their original diseases included major depressive disorder, BD, PMDDs, PDs, and osteoarthritis.
Giving the fact that the expression of CNRs is increased in brain of depressive patients, multiple studies have presented controversial results concerning the association of CNRs polymorphism with depression. Tsai et al[14] were the 1st to report the role of CNR1 gene triplet repeat polymorphism in depression. They demonstrated that the triplet repeat polymorphism in the promoter region of the CNR1 gene was not likely to be involved in the pathogenesis or in the psychotic symptoms of mood disorders (P = .830). In addition, a finding from Yildiz et al[18] did not support a major role of CNR1rs1040353 polymorphism in contributing to susceptibility to PMDD. Similarly, the results from our meta-analysis also illustrated no significant association between either CNR1rs1049353 SNP with all 4 genetic models or CNR1 (AAT)n triplet repeat polymorphism. However, some other studies found meaningful association of CNRs polymorphism with depressive disorders. Monteleone et al[19] observed that the distribution of CNR1rs1049353 genotypes and alleles significantly differed between depressive people and healthy controls (P = .01 for genotypes and P = .001 for alleles), with major depressive people showing a higher frequency of both AG, GG genotypes, and A allele as compared to healthy controls. Furthermore, Barrero et al[20] considered that in patients with PD, the presence of 2 long alleles with more than 5 repeated AAT trinucleotides in the CNR1 gene, was associated with a reduced prevalence of depression (P = .003). However, they did not find statistically significant association in osteoarthritis patients with depressive symptom. Many reasons might contribute to the conflicting results. First of all, we thought it would be worthwhile to address depressive disorder as a whole, so our meta-analysis included depressive patients with different comorbidities. Considering the polygenic effect on depressive disorders, the genetic factors that have impacted on the original diseases could share their contributions to depression as well.[21] Additionally, we could not do subgroup analyses on ethnicity, gender, age, and smoke status, owing to insufficient data, since there are experiments showed that these factors may have influence on depression.[22,23] However, each of our studies included in the meta-analysis selected matched subjects as controls, which might be helpful for this consideration. In future, large number of case–control studies could provide more reliable evidence for the role of these polymorphisms with respect to susceptibility to depression.
On the contrary, each genetic model of CNR2rs variants presented noticeable association with depression risk and the heterogeneity of these models was not significant. Which is consistent with the results reported by Onaivi and his coworkers.[16] They found that CNR2rs2501432 SNP has significant association with Japanese depressive patients (P = .007), and the proportion of subject with an GG genotype was significant higher in depression group than that in controls (P = .01). Also, the results from this meta-analysis are complemented by other converting evidence showing this marker may participate in human cognitive function, particularly relating to the area of depression-associated brain dysfunction.[7] Previous studies have confirmed that CNR2 protein is expressed in immune tissues, and it has been identified in mammalian brain as well. Studies have reported the involvement of CNR2 in alcoholism, depression, and drug abuse in mammalian, which provides the evidence of CNR2 in mammalian central nerve system. In human, a number of polymorphisms in CNR2 gene have been linked to diseases including pain, autoimmune disorders, and depression.[24] As many genetic variants play various roles in depression, variation in CNR2 rs250 may be a risk factor in depression in human, and targeting the CNR2 ligands could be a possibility for depression and its comorbidity therapy.
However, according to the study of Minocci and his colleagues,[17] the CNR2 524C>A has significant association with BD, while no statistically significant difference was detected between CNR2rs2501432 and BD. This inconsistency might come from the potential interactions of genetic-genetic, genetic-environmental, and different CNR2 SNPs during the development of depression. For instance, haplotype distribution of CNR plays important roles in depression is a MD etiology and clinical response to citalopram treatment.[15] Because of the number of study that reported this SNP was not >3, we did not perform publication bias test of this SNP.
Further publication bias test and sensitivity analysis for CNR1 provide additional support for the robustness of our results. Although heterogeneity existed in the analysis, results from sensitivity analysis indicates study included major depressive disorder patients (for CNR1rs1049353) and study investigated depression in PD (for CNR1 AAT triplet repeat polymorphism) played an important role contributing to this variance observed between studies. However, we only had one paper reporting each of the 2 diseases, a subgroup analysis based on which could not be performed.
Although our meta-analysis did not find a significant association for CNR1rs1049353 or CNR1 AAT triplet repeat polymorphism, which is consistent with the general trend form various depression studies, suggesting such an effect can be contributed to the polygenic nature of the disorder, this view believe that individual gene just make up a modest effect, but collectively, they have a cumulative effect on the likelihood of developing the disease.[25] Nevertheless, this generalization is constrained by the small number of studies available on these 2 variants, thus should be treated with caution.
As with all meta-analyses, several limitations merit consideration in our study. First of all, there is a lack of broader coverage on the investigation of other CNR polymorphisms related to depressive disorder risk. Second, in view of the relatively small sample size and the limited study number, the power used to detect the real difference between cases and controls may not be strong enough. Finally, significant heterogeneity that has been found in different comparison of CNR1 gene polymorphisms, which could be owing to the fact that the analysis encompasses few studies in the analysis or due to insufficient data that hindered further subgroup analysis on gender, comorbidity, gender, and smoking.
5. Conclusion
Our meta-analysis indicates significant association of CNR2rs2501432 with depressive disorders in general, while the existing literature does not suggest the relation of CNR1rs1049353 SNP or the length of CNR1 AAT triplet repeats polymorphism with depression. However, owing to the scarcity in the reusable literature focused on these variants, more large studies of wider coverage are needed to evaluate the overall effect of CNRs in depressive disorder as a whole, which in turn can potentially help to develop promising therapeutic targets and improve the treatment and functional outcome for depression.
Author contributions
Conceptualization: Xiangjuan Kong, Qingshan Miao.
Data curation: Xiangjuan Kong, Qingshan Miao.
Formal analysis: Qingshan Miao, Xiaozi Lu.
Investigation: Xiangjuan Kong, Zeng Zhang.
Software: Min Chen, Jinxiang Zhang.
Supervision: Jinguo Zhai.
Validation: Jinguo Zhai, Xiangjuan Kong, Qingshan Miao.
Writing – original draft: Xiangjuan Kong, Qingshan Miao.
Writing – review & editing: Jinguo Zhai.
Supplementary Material
Footnotes
Abbreviations: ASO-PCR = allelic specific oligonucleotide PCR, BD = bipolar disorder, CI = confidence interval, DSM = Diagnostic and Statistical Manual of Mental disorders, MD = major depression, OR = odds ratio, PD = Parkinson disease, PMDD = premenstrual dysphoric disorder, RFLP = restriction fragment length polymorphism, SNP = single-nucleotide polymorphism.
How to cite this article: Kong X, Miao Q, Lu X, Zhang Z, Chen M, Zhang J, Zhai J. The association of endocannabinoid receptor genes (CNR1 and CNR2) polymorphisms with depression. Medicine. 2019;98:46(e17403).
XK and QM contributed equally to the work.
The authors have no funding and conflicts of interest to disclose.
Supplemental Digital Content is available for this article.
References
- [1].Smith K. Mental health: a world of depression. Nature 2014;515:181. [DOI] [PubMed] [Google Scholar]
- [2].Maurer DM, Raymond TJ, Davis BN. Depression: screening and diagnosis. Am Fam Physician 2018;98:508–15. [PubMed] [Google Scholar]
- [3].Bhattacharjee S, Goldstone L, Vadiei N, et al. Depression screening patterns, predictors, and trends among adults without a depression diagnosis in ambulatory settings in the United States. Psychiatr Serv 2018;69:1098–100. [DOI] [PubMed] [Google Scholar]
- [4].Kwiatkowska B, Klak A, Maslinska M, et al. Factors of depression among patients with rheumatoid arthritis. Reumatologia 2018;56:219–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Kay DB, Tanner JJ, Bowers D. Sleep disturbances and depression severity in patients with Parkinson's disease. Brain Behav 2018;8:e00967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Acikgoz A, Dayi A, Binbay T. Prevalence of premenstrual syndrome and its relationship to depressive symptoms in first-year university students. Saudi Med J 2017;38:1125–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Onaivi ES. Neuropsychobiological evidence for the functional presence and expression of cannabinoid CB2 receptors in the brain. Neuropsychobiology 2006;54:231–46. [DOI] [PubMed] [Google Scholar]
- [8].Ishiguro H, Horiuchi Y, Tabata K, et al. Cannabinoid CB2 receptor gene and environmental interaction in the development of psychiatric disorders. Molecules 2018;23: [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].de Oliveira RW, Oliveira CL, Guimaraes FS, et al. Cannabinoid signalling in embryonic and adult neurogenesis: possible implications for psychiatric and neurological disorders. Acta Neuropsychiatr 2019;31:1–6. [DOI] [PubMed] [Google Scholar]
- [10].Valverde O, Torrens M. CB1 receptor-deficient mice as a model for depression. Neuroscience 2012;204:193–206. [DOI] [PubMed] [Google Scholar]
- [11].Patel S, Hillard CJ. Role of endocannabinoid signaling in anxiety and depression. Curr Top Behav Neurosci 2009;1:347–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Yao Y, Xu Y, Zhao J, et al. Detection of significant association between variants in cannabinoid receptor 1 gene (CNR1) and personality in African-American population. Front Genet 2018;9:199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Icick R, Peoc’h K, Karsinti E, et al. A cannabinoid receptor 1 polymorphism is protective against major depressive disorder in methadone-maintained outpatients. Am J Addict 2015;24:613–20. [DOI] [PubMed] [Google Scholar]
- [14].Tsai SJ, Wang YC, Hong CJ. Association study between cannabinoid receptor gene (CNR1) and pathogenesis and psychotic symptoms of mood disorders. Am J Med Genet 2001;105:219–21. [DOI] [PubMed] [Google Scholar]
- [15].Mitjans M, Serretti A, Fabbri C, et al. Screening genetic variability at the CNR1 gene in both major depression etiology and clinical response to citalopram treatment. Psychopharmacology (Berl) 2013;227:509–19. [DOI] [PubMed] [Google Scholar]
- [16].Onaivi ES, Ishiguro H, Gong JP, et al. Brain neuronal CB2 cannabinoid receptors in drug abuse and depression: from mice to human subjects. PLoS One 2008;3:e1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Minocci D, Massei J, Martino A, et al. Genetic association between bipolar disorder and 524A>C (Leu133Ile) polymorphism of CNR2 gene, encoding for CB2 cannabinoid receptor. J Affect Disord 2011;134:427–30. [DOI] [PubMed] [Google Scholar]
- [18].Yildiz M, Vural M, Erdal ME, et al. Lack of association of DRD3 and CNR1 polymorphisms with premenstrual dysphoric disorders. Iran J Reprod Med 2015;13:221–6. [PMC free article] [PubMed] [Google Scholar]
- [19].Monteleone P, Bifulco M, Maina G, et al. Investigation of CNR1 and FAAH endocannabinoid gene polymorphisms in bipolar disorder and major depression. Pharmacol Res 2010;61:400–4. [DOI] [PubMed] [Google Scholar]
- [20].Barrero FJ, Ampuero I, Morales B, et al. Depression in Parkinson's disease is related to a genetic polymorphism of the cannabinoid receptor gene (CNR1). Pharmacogenomics J 2005;5:135–41. [DOI] [PubMed] [Google Scholar]
- [21].Zivin K, Yosef M, Miller EM, et al. Associations between depression and all-cause and cause-specific risk of death: a retrospective cohort study in the Veterans Health Administration. J Psychosom Res 2015;78:324–31. [DOI] [PubMed] [Google Scholar]
- [22].Tjora T, Hetland J, Aaro LE, et al. The association between smoking and depression from adolescence to adulthood. Addiction 2014;109:1022–30. [DOI] [PubMed] [Google Scholar]
- [23].Karger A. Gender differences in depression [in German]. Bundesgesundheitsblatt Gesundheitsforschung Gesundheitsschutz 2014;57:1092–8. [DOI] [PubMed] [Google Scholar]
- [24].Dhopeshwarkar A, Mackie K. CB2 cannabinoid receptors as a therapeutic target--what does the future hold? Mol Pharmacol 2014;86:430–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Cannon TD, Keller MC. Endophenotypes in the genetic analyses of mental disorders. Annu Rev Clin Psychol 2006;2:267–90. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


