Abstract
Tuberous sclerosis complex (TSC) is a genetic disorder caused by mutations in TSC1 or TSC2. Patients frequently have epilepsy, autism spectrum disorder, and/or intellectual disability, as well as other systemic manifestations. In this study, we differentiated human induced pluripotent stem cells (iPSCs) from a female patient with TSC with one or two mutations in TSC2 into neurons using induced expression of NGN2 to examine neuronal dysregulation associated with the neurological symptoms in TSC. Using this method, neuronal differentiation was comparable between the three genotypes of iPSCs. We observed that TSC2+/− neurons show mTOR complex 1 (mTORC1) hyperactivation and associated increased cell body size and process outgrowth, as well as exacerbation of the abnormalities by loss of the second allele of TSC2 in TSC2−/− neurons. Interestingly, iPSC-derived neurons with either a single or biallelic mutation in TSC2 demonstrated hypersynchrony and downregulation of FMRP targets. However, only neurons with biallelic mutations of TSC2 demonstrated hyperactivity and transcriptional dysregulation observed in cortical tubers. These data demonstrate that loss of one allele of TSC2 is sufficient to cause some morphological and physiological changes in human neurons but that biallelic mutations in TSC2 are necessary to induce gene expression dysregulation present in cortical tubers. Finally, we found that treatment of iPSC-derived neurons with rapamycin reduced neuronal activity and partially reversed gene expression abnormalities, demonstrating that mTOR dysregulation contributes to both phenotypes. Therefore, biallelic mutations in TSC2 and associated molecular dysfunction, including mTOR hyperactivation, may play a role in the development of cortical tubers.
SIGNIFICANCE STATEMENT In this study, we examined neurons derived from induced pluripotent stem cells with two, one, or no functional TSC2 (tuberous sclerosis complex 2) alleles and found that loss of one or both alleles of TSC2 results in mTORC1 hyperactivation and specific neuronal abnormalities. However, only biallelic mutations in TSC2 resulted in elevated neuronal activity and upregulation of cell adhesion genes that is also observed in cortical tubers. These data suggest that loss of heterozygosity of TSC1 or TSC2 may play an important role in the development of cortical tubers, and potentially epilepsy, in patients with TSC.
Keywords: cortical tuber, FMRP, iPSC, mTOR, TSC2, tuberous sclerosis
Introduction
Tuberous sclerosis complex (TSC) is a neurodevelopmental disorder that is associated with the development of benign tumors or hamartomas in multiple organs. It is caused by mutations in either TSC1 or TSC2, which encode the TSC1 and TSC2 proteins that with TBC1D7 form a complex that inhibits mTOR complex 1 (mTORC1) through inactivation of RHEB, which is a major mTORC1 activator (Lipton and Sahin, 2014; Switon et al., 2017). Although TSC2 contains the GTPase function that inactivates RHEB, both TSC1 and TSC2 are required for the stability of the complex, which explains why patients with either gene mutation have similar clinical characteristics. Most studies have demonstrated that several characteristics of TSC are caused by disinhibition of mTORC1, and mTORC1 inhibitors such as sirolimus and everolimus have been shown to be effective for several manifestations of TSC (Bissler et al., 2013; Franz et al., 2013; French et al., 2016). However, many questions regarding disease mechanisms in TSC remain.
Patients with TSC frequently have heterozygous loss of function variants in either TSC1 or TSC2, but most animal models with heterozygous mutations demonstrate subtle or no symptoms (Uhlmann et al., 2002; Goorden et al., 2007; Ehninger et al., 2008; Yuan et al., 2012). Several benign tumors observed in TSC such as subependymal giant cell astrocytomas and angiomyolipomas are associated with somatic mutations in the second allele of either TSC1 or TSC2 or loss of heterozygosity (LOH), resulting in near or complete absence of the protein (Henske et al., 1996; Chan et al., 2004; Giannikou et al., 2016). Indeed, animal models with biallelic or homozygous mutations commonly recapitulate features of TSC (Uhlmann et al., 2002; Meikle et al., 2007; Way et al., 2009; Yuan et al., 2012). These data suggest that LOH may play an important role in the pathogenesis of TSC. However, cortical tubers are the most common neuropathological finding in TSC and show much lower rates of LOH (<35%) (Qin et al., 2010; Martin et al., 2017). In addition, cortical tubers are often associated with seizures in patients with TSC who have refractory epilepsy, suggesting that these lesions contribute to the neurological aspects of the disorder (Major et al., 2009; Mohamed et al., 2012). Given their low rates of LOH, it is possible that haploinsufficiency of either TSC1 or TSC2 is able to engage the pathogenic mechanisms responsible for the development of cortical tubers or that the number of cells with LOH is below the current threshold for detection.
We have previously used induced pluripotent stem cells (iPSCs) from patients with TSC and differentiated them into cerebellar Purkinje cells (PCs). We observed mTORC1 disinhibition and other neuronal abnormalities in PCs with heterozygous mutations in TSC2 (Sundberg et al., 2018). In addition, another study that used embryonic stem cells (ESCs) with an engineered deletion of TSC2 observed similar, albeit less pronounced, dysregulation in neurons derived from ESCs with heterozygous TSC2 mutations (Costa et al., 2016). Interestingly, other studies of iPSC-derived neurons from patients with TSC have also observed abnormalities in neurons derived from iPSCs with heterozygous mutations (Li et al., 2017; Zucco et al., 2018; Nadadhur et al., 2019). However, these studies have observed multiple conflicting findings in characteristics such as neurite outgrowth and neuronal activity.
One limitation of the prior studies was that the differentiation protocols used led to mixed cultures with varying numbers of neurons and glia between genotypes. These approaches identified alterations in proliferation and cell fate but may have also introduced substantial variability, contributing to the inconsistencies between studies. Therefore, we chose to use a transcription factor-based approach that drives differentiation of excitatory, cortical neurons and results in relatively homogeneous cultures (Zhang et al., 2013). This approach allowed us to interrogate the effects of TSC2 gene dosage in human neurons that are independent of its effects on differentiation. We then used a combination of techniques to characterize the alterations caused by dysfunction of the TSC complex in differentiated neurons.
Materials and Methods
iPSC lines and neuronal differentiation.
The subjects enrolled in this study were recruited through Boston Children's Hospital, and the protocol was approved by Boston Children's Hospital institutional review board (P00008224). Informed consent was obtained from all participants and/or their parents as appropriate. Characteristics of the subjects involved and the methods used for reprogramming have been described previously (Ebrahimi-Fakhari et al., 2016; Sundberg et al., 2018). Briefly, the TSC2+/− line was derived from a female patient with TSC and the control, TSC2+/+ line was derived from her father. The third line with biallelic mutations in TSC2 was created using gene editing in the TSC2+/− line.
For neuronal differentiation, we used a protocol previously published from Sudhof and colleagues using induced NGN2 expression with minor changes (Zhang et al., 2013). Human iPSCs were dissociated into single cells with Accutase (catalog #AT 104–500; Innovative Cell Technology) and seeded onto Geltrex-coated plates (catalog #A1413301; Thermo Fisher Scientific). On differentiation day −1, iPSCs were then infected with concentrated rtTA-, EGFP-, and NGN2-expressing lentiviruses in presence of polybrene (8 μg/ml, catalog #TR-1003-G; Sigma-Aldrich, RRID: Addgene_52047) overnight. The next day, NGN2 expression was induced using doxycycline (2 μg/ml, catalog #324385–1GM; Millipore), and infected cells were then selected using puromycin (1 μg/ml, catalog #ant-pr-1; Invitrogen) for up to 48 h. Growth factors BDNF (10 ng/ml, catalog #450–02; Peprotech) and NT3 (10 ng/ml, catalog #450–03; Peprotech), and laminin (0.2 mg/L, catalog #23017–015; Thermo Fisher Scientific) were added in N2 medium for the first 2 d. Cells were then fed with BDNF (10 ng/ml), NT3 (10 ng/ml), laminin (0.2 mg/L), doxycycline (2 μg/ml), and Ara-C (2 μm, catalog #C1768; Sigma-Aldrich) in B27 media and fed every other day until differentiation day 6. Cells were then dissociated with papain (catalog #LK003178; Worthington) supplemented with DNase (catalog #LK003172; Worthington) and replated with or without human iPSC-derived astrocytes (Astro.4U; Ncardia) and cultured for various durations until they were assayed as indicated below.
Immunocytochemistry and automated microscopy.
Six days after NGN2 induction with doxycycline, iPSC-derived neurons were either replated with or without astrocytes in black 96 well, flat glass bottom plates (catalog #655090; Grenier) coated with PDL (100 μg/ml, catalog #P6407; Sigma-Aldrich) overnight followed by laminin (5 μg/ml) overnight. For short-term culture, 10,000 neurons were plated into each well, and cells were fixed 24 h after replating. For long-term culture, 20,000 neurons and 3000 astrocytes were plated into each well, and cells were maintained for 18 d in vitro before fixation. For both time points, cells were fixed with 4% PFA (catalog #15686; Electron Microscopy Sciences) and then blocked with 5% normal goat serum (catalog #G9023–10ML; Sigma-Aldrich) and 0.1% Triton-X (catalog #AC215682500; Thermo Fisher Scientific) for 1 h. Cells were incubated with primary antibody in 0.5% normal goat serum overnight at 4C. Primary antibodies included anti-HuC/HuD (1:250, 16A11, catalog #A-21271; Thermo Fisher Scientific, RRID:AB_221448), anti-TUBB3 (1:500, catalog #AB9354 Millipore, RRID:AB_570918; 1:500, catalog #ab18207 Abcam, RRID:AB_444319), anti-MAP2 (1:500, catalog #ab32454 Abcam, RRID:AB_776174), anti-PSD95 (1:50, K28/43; NeuroMab, RRID:AB_2292909), anti-Synapsin-1 (1:500, catalog #AB1543P; Millipore, RRID:AB_90757), anti-VGLUT1 (1:1000, 135 302 Synaptic Systems, RRID:AB_887875), anti-NANOG (1:250, catalog #ab109250; Abcam, RRID:AB_10863442), anti-OCT4 (1:250, catalog #ab181557; Abcam, RRID:AB_2687916), anti-GFAP (1:200, catalog #173–002; Synaptic Systems, RRID:AB_887720), anti-TBR1 (1:200, catalog #ab31940; Abcam, RRID:AB_2200219), CTIP2 (1:400, catalog #ab18465; Abcam, RRID:AB_2064130), or anti-Ki67 (1:500, catalog #ab15580; Abcam, RRID:AB_443209). Cells were washed three times with PBS, and then incubated with fluorophore-conjugated secondary antibodies (Invitrogen) for 1 h at room temperature. Cells were then washed three times with PBS and counterstained with DAPI or Hoechst 33342 (4 μg/ml for 5 min; Invitrogen). Cells were then maintained in PBS with 0.01% sodium azide (catalog #S2002; Sigma-Aldrich) or mounted onto glass slides (Fisher Scientific) until imaging. Plates were imaged and analyzed using automated high content imaging platforms, including ArrayScan XTI (Thermo Fisher Scientific) and ImageXpress Micro Confocal (Molecular Devices) systems.
Western blotting.
Fourteen days after NGN2 induction with doxycycline, protein from iPSC-derived neurons without astrocytes was collected. Protein lysates were collected in RIPA buffer (catalog #sc-24948; Santa Cruz Biotechnology) with phosphatase and protease inhibitors and spun at 16,000 g to clarify the lysate. Protein concentration was determined using the BCA assay (catalog #A53226; Thermo Fisher Scientific). Laemmle buffer was added to protein lysates, and lysates were boiled at 95°C for 5 min. Equal masses were loaded onto either 4–20% gradient or 10% acrylamide gels, and gels were transferred to PVDF membranes (Immobilon-FL) using a semidry transfer apparatus (Bio-Rad). Total protein was determined using the REVERT stain (LI-COR) and imaged using a fluorescence imager (LI-COR). Total protein stain was washed off, and membranes were blocked with Odyssey blocking buffer for 1 h at room temperature. Membranes were then incubated in primary antibody overnight at 4°C. Primary antibodies included anti-TSC2 (1:1000; Cell Signaling Technology, RRID:AB_10547134), anti-pS6 S240/244 (1:1000; Cell Signaling Technology, RRID:AB_10694233), and anti-S6 (1:200; Santa Cruz Biotechnology, RRID:AB_1129205), anti-pS65 4EBP1 (1:1000, Cell Signaling, RRID:AB_330947), anti-4EBP1 (1:1000, Cell Signaling, RRID:AB_331692), anti-S473 AKT (1:1000, Cell Signaling, RRID:AB_2315049), and anti-AKT (1:1000, Cell Signaling, RRID:AB_915783). Membranes were then washed three times with TBS-T, and then incubated in secondary antibody (LI-COR) for 1 h at room temperature. Membranes were then washed three times with TBS-T and once with DI water, and membranes were imaged using a fluorescent imager (LI-COR). Quantification of Western blots was performed using ImageStudio (LI-COR). Differences between genotypes were determined using an ANOVA with Dunnett's multiple-comparisons test.
ActivSignal pathway proteomics profiling.
Six days after doxycycline induction of NGN2, neurons were replated without astrocytes and cultured for 24 h in black 96 well, flat bottom plates. Then, 10,000 neurons were plated into each PDL/laminin-coated well. Cells were fixed with 4% PFA and kept in PBS for immunopaired antibody detection for signaling pathway profiling, which was performed at ActivSignal. Quantified values between different lines were compared using a t test, and correction for multiple comparisons was performed using a false discovery rate (FDR).
Multielectrode array.
Forty-eight well CytoView MEA plates (M768-tMEA-48B; Axion Biosystems) were coated with 0.1% PEI (catalog #408727; Sigma-Aldrich) at room temperature for 1 h, washed with sterile water, and dried overnight. Six days after doxycycline induction of NGN2, iPSC-derived neurons were dissociated as above, and 50,000 iPSC-derived neurons and 7500 human iPSC-derived astrocytes were resuspended in 7 μl of media supplemented with Y-27632 (catalog #10005583; Cayman) and laminin (10 μg/ml) and were manually spotted in each well. Spontaneous network activity was then recorded with a Maestro Original MEA System (Axion Biosystems) every other day for 25 d. Data were sampled at 12.5 kHz, digitized, and analyzed using Axion Integrated Studio software with a 200 Hz high-pass and 3000 Hz low-pass filter with an adaptive spike detection threshold set at 6 times the SD for each electrode with 1 s binning. The burst detector was set to detect network burst, with maximum 100 ms interspike interval, minimum 50 spikes, and minimum 35% participating electrodes. The window size for synchrony parameters is set at 20 ms. After recording, the data were analyzed using the Neurostatistics compiler (Axion). These summary files were collated and analyzed using custom R scripts. For statistical analysis of MEA data, the values of each parameter were fitted to a sigmoidal curve with three coefficients (midpoint, slope, and plateau) using nonlinear least-squares estimates. These coefficients between different genotypes were then compared using a Z test (Clogg et al., 1995).
For rapamycin treatment, iPSC-derived neurons were prepared and plated in the same manner. On day 14 of differentiation, either 20 nm rapamycin or the same volume of DMSO was added to the media and to all subsequent media changes, until day 31 of differentiation when it was removed. Recordings and data analysis were performed as above.
Transcriptional profiling.
NGN2-induced neurons were cultured for 14 d and total RNA was isolated (catalog #740971.50; Takara Bio) and converted into cDNA using SuperScript IV VILO (catalog #11756050; Thermo Fisher Scientific). Libraries were created and amplified using the Ion AmpliSeq Transcriptome Human Gene Expression Kit (catalog #A31446; Thermo Fisher Scientific) using automated Ion Chef instrument and then sequenced on an IonTorrent S5 Sequencer (Thermo Fisher Scientific). Gene expression was quantified using the Torrent Suite. The data were imported into R, log2 transformed, and genes with low expression (mean expression <2) were removed. Differential expression analysis was performed using LIMMA (Law et al., 2014) and the p-values were corrected for multiple comparisons using FDR. Weighted gene coexpression network analysis (WGCNA) was performed as described previously (Zhang and Horvath, 2005; Sundberg et al., 2018). Briefly, a correlation matrix of all expressed genes was calculated and then taken to the power of 6 to approximate scale free topology, resulting in an adjacency matrix. A topological overlap matrix was calculated from the adjacency matrix, and then hierarchical clustering was performed. Modules were identified using the dynamic tree cut algorithm. Module eigengenes (MEs) were determined by obtaining the first principal component of the scaled expression data within a module. Module eigengene-based connectivity (kME) was determined by calculating the correlation between the expression of a gene and the ME (Dong and Horvath, 2007).
For analysis of gene expression within cortical tubers, raw sequencing data were obtained for all samples. These sequencing data were mapped to the human genome (Ensembl 92) using STAR (Dobin et al., 2013). The transcriptome was then assembled and quantified using Cufflinks. Normalized gene-level expression data were then imported into R, and LIMMA was used to calculate differential expression. p-values were corrected for multiple comparisons using an FDR.
Gene ontology analysis was performed using DAVID (https://david.ncifcrf.gov). Filtered categories within biological process, cellular compartment, and molecular function were tested for enrichment.
For qPCR, RNA was isolated as above and reverse transcribed into cDNA. PCRs were setup using 2× SYBR green master mix and specific primers for each target. Relative target abundance was determined using a QuantStudio thermocycler. GAPDH was used as a loading control for each gene. Fold changes were calculated using the ΔΔCt method, and statistical significance was calculated using a Wilcoxon rank-sum test between the ΔCt values. Correction for multiple comparisons was performed using a Bonferroni correction.
For rapamycin treatment, iPSCs were differentiated into neurons as above. Either 20 nm rapamycin or the same volume of DMSO was added to the medium beginning on day 7 of differentiation and treatments were continued until day 14, when cells were collected for RNA isolation as above. cDNA synthesis, qPCR, and data analysis were performed as above.
Statistical tests.
For automated image analysis, Western blot, and qPCR results, the data were imported into Prism and the indicated statistical tests were performed. For MEA results, the data were imported into R. The NLS function was used for fitting to a generic sigmoidal equation. The calculated coefficients were compared using custom scripts that implements a Z test for coefficients described previously (Clogg et al., 1995). Gene expression data and RNA sequencing data were analyzed as described above and imported into R. Analysis was performed with the LIMMA and WGCNA packages as described above.
Results
Characterization of iPSC-derived neurons with TSC2 mutations
We used three iPSC lines for this study that we have previously demonstrated to be pluripotent and without karyotypic abnormalities (Sundberg et al., 2018). The first line was derived from a patient with TSC due to an 18bp in-frame deletion in exon 40 of TSC2 (c.5238_5255del p.H1746_R1751del, TSC2+/−). The second line was derived from the patient line and had a frameshift mutation induced in the second allele of TSC2 using TALEN technology (TSC2−/−), modeling LOH that is observed in TSC. Finally, a third line was derived from the unaffected parent of the patient, and this line was used as a control (TSC2+/+).
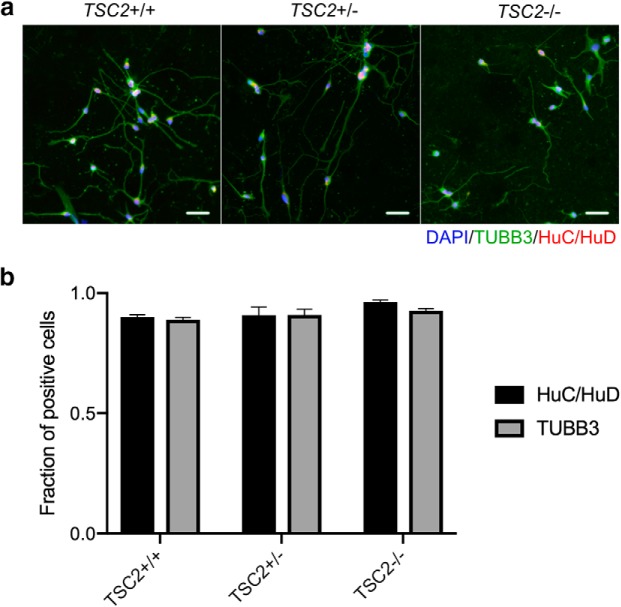
For this study, we used a transcription factor-based differentiation protocol that uses induced expression of NGN2 to directly differentiate iPSCs into cortical neurons (Zhang et al., 2013). iPSCs with all three TSC2 genotypes were transduced with lentiviral vectors that encode NGN2 and EGFP under the tetracycline promoter, as well as the puromycin resistance gene for selection. iPSCs were grown under feeder-free conditions, and on day 0 of differentiation, doxycycline was added to induce differentiation. Seven days after doxycycline induction of NGN2, we observed that ∼90% of cells demonstrated expression of HuC/HuD and β-tubulin III, consistent with immature neurons that have committed to neuronal fate (Fig. 1a). In addition, we observed similar rates of the expression of both markers, demonstrating that neuronal differentiation was similar for all three genotypes using this protocol (Fig. 1b). We did not observe cells that were positive for pluripotency markers such as OCT4 or NANOG or markers of glia such as GFAP, indicative of efficient neuronal differentiation (Fig. 1-1). In addition, we performed staining for Ki67 to determine whether there was another nonpluripotent stem cell population, and we observed few Ki67-positive cells, but the majority of these cells were also positive for β-tubulin III (Fig. 1-2).
Figure 1.
Differentiation of iPSCs into neurons. iPSCs from a patient with TSC due to a mutation in TSC2 (TSC2+/−), a line derived from the patient line with LOH of TSC2 (TSC2−/−), and an unaffected, related individual (TSC2+/+) were transduced with a lentivirus encoding the transcription factor NGN2 under a tetracycline inducible promoter as described previously (Zhang et al., 2013). Cells were then treated with doxycycline to induce expression of NGN2, and we assayed markers of neuronal differentiation. a, Seven days after doxycycline, we observed markers of neuronal commitment (HuC/HuD) and immature neurons (tubulin-B3/TUJ1) in all three lines. Scale bars, 50 μm. b, Quantification of the fraction of cells from all three lines that were positive for either HuC/HuD or TUBB3 7 d after doxycycline treatment using automated microscopy. There were no significant differences between genotypes. Data represent 25 images from each well across five independent wells from one differentiation (p > 0.05, ANOVA). Bar plot shows mean ± SEM. Figure 1-1) shows the lack of expression of the glial marker GFAP and pluripotency markers, NANOG and OCT4, corroborating efficient neuronal differentiation. (Figure 1-2) shows scarce expression of Ki67 that frequently colocalizes with TUBB3.
iPSC differentiation into neurons is highly efficient. iPSCs with three different genotypes of TSC2 (TSC2+/+, TSC2+/-, and TSC2-/-) were differentiated into neurons using induced expression of NGN2. a) Seven days after doxycycline, we did not observe expression of glial markers such as GFAP in any of the three lines. In addition, we did not observe markers of undifferentiated pluripotent cells such as b) NANOG or c) OCT4 in any of the three lines. Scale bars=50μm. Download Figure 1-1, TIF file (6MB, tif)
Dividing cell populations in neuronal cultures. iPSCs with three different genotypes of TSC2 (TSC2+/+, TSC2+/-, and TSC2-/-) were differentiated into neurons using induced expression of NGN2. a) Seven days after doxycycline, we evaluated for other non-pluripotent stem cells using immunostaining for Ki67. We observed a small fraction of Ki67-positive cells in cultures from all three genotypes, but the majority of these cells were also positive for TUBB3, consistent with a neuronal identity. Scale bars=50μm. Download Figure 1-2, TIF file (1.4MB, tif)
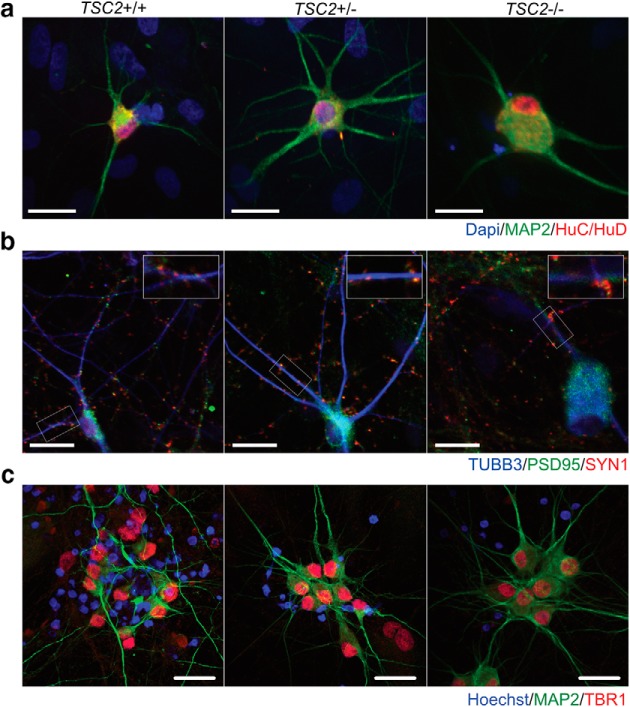
We then cultured these differentiating cells in the presence of astrocytes for three and a half weeks to facilitate neuronal maturation and synapse formation. We observed that these cells demonstrated expression of HuC/HuD and the mature neuronal marker, MAP2 (Fig. 2a). In addition, processes from these cells demonstrated expression of PSD95 and synapsin-1 in close apposition (Fig. 2b), consistent with formation of functional synapses. Finally, these neurons were positive for TBR1 and CTIP2 expression (Fig. 2c; Fig. 2-1), markers of developing lower-layer cortical pyramidal neurons (Saito et al., 2011). We observed the expression of each of these markers in all three genotypes, demonstrating that iPSCs are capable of differentiating into neurons regardless of the allelic dosage of TSC2 under these conditions.
Figure 2.
Characterization of iPSC-derived neurons. iPSCs with inducible NGN2 were treated with doxycycline and replated with astrocytes after six days to promote maturation. Neuronal and synaptic marker expression was assayed at 24 d after initial doxycycline treatment. a, Neurons from all three lines displayed expression of HuC/HuD and MAP2. b, Synaptic markers PSD-95 and Synapsin-1 demonstrate coclustering along neurites labeled by TUBB3, consistent with formation of functional synapses. Scale bars, 25 μm. c, TBR1, a marker of early lower layer cortical neurons, was expressed in iPSC-derived neurons from all three genotypes, suggestive of a pyramidal neuron fate. Scale bars, 20 μm. (Figure 2-1) shows the expression of CTIP2, another marker of early lower layer cortical neurons in iPSC-derived neurons.
Characterization of iPSC-derived neurons. iPSCs with inducible NGN2 were treated with doxycycline and re-plated with astrocytes after six days to promote maturation. CTIP2, a marker of early lower layer cortical neurons, was also expressed in iPSC-derived neurons, suggestive of a pyramidal neuron fate. Scale bars=20μm. Download Figure 2-1, TIF file (1.4MB, tif)
mTOR activity in iPSC-derived neurons
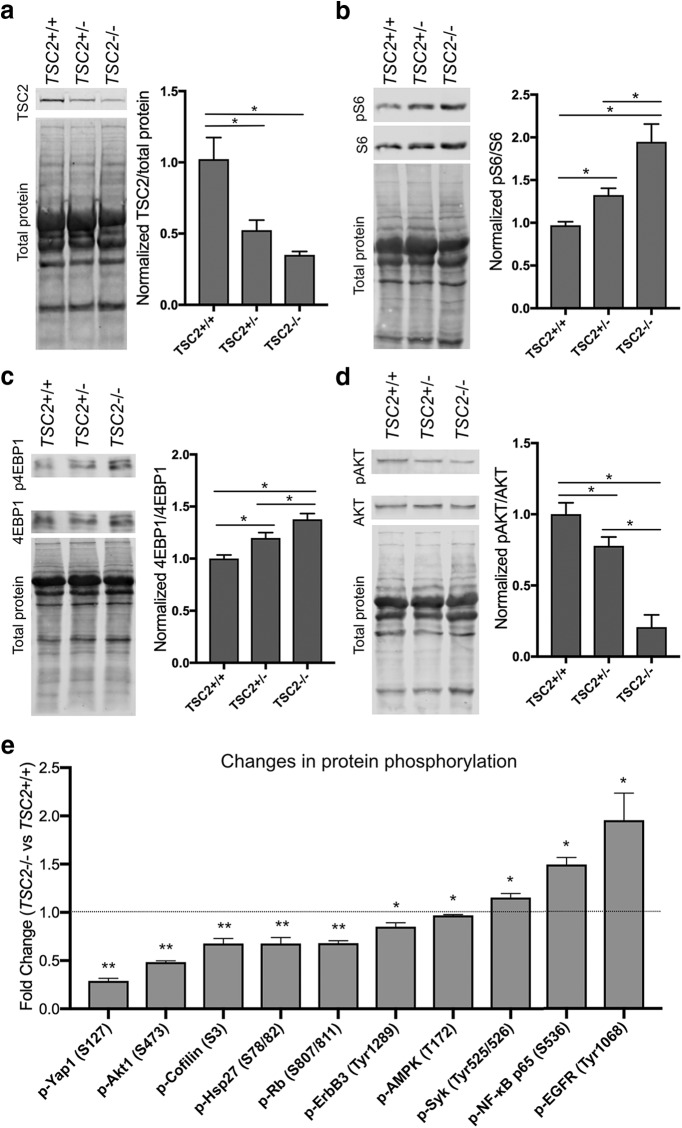
To assay TSC2 levels and mTOR activity, protein lysates were collected from iPSC-derived neurons 14 d after doxycycline induction of NGN2 from all three TSC2 genotypes. We then performed Western blots for TSC2, and we found that there was a significant reduction in TSC2 levels in both the TSC2+/− and TSC2−/− neurons compared with TSC2+/+ neurons (Fig. 3a). Interestingly, the reduction in TSC2 levels between TSC2+/− and TSC2−/− was not significant, and there was a small amount of residual TSC2 in the TSC2−/− neurons, likely due to persistent protein expression from the p.H1746_R1751del allele, which is an in-frame deletion. To evaluate mTORC1 activation, we performed Western blots for the phosphorylation site of the well characterized mTORC1 target, ribosomal protein S6 S240/244 (p-S6). We found significantly increased levels of p-S6 in both the TSC2+/− and TSC2−/− neurons compared with the TSC2+/+ neurons, indicative of mTORC1 hyperactivation in both lines, and this ratio was significantly increased in the TSC2−/− line compared with the TSC2+/− line (Fig. 3b). In addition, we observed similar increases in another mTORC1 target, 4EBP1 phosphorylated at S65, in both the TSC2+/− and TSC2−/− neurons compared with the TSC2+/+ neurons (Fig. 3c). Previous functional studies have demonstrated that the p.H1746_R1751del variant of TSC2 is nonfunctional in mTORC1 regulation and destabilizes the TSC1-TSC2 complex (Hoogeveen-Westerveld et al., 2011), which explains the significant increase in mTORC1 activity without a significant reduction in TSC2 levels between the TSC2+/− and TSC2−/− neurons. TSC2 has also been shown to be involved mTORC2 mediated phosphorylation of AKT, and we observed significantly reduced phosphorylation of AKT at S473 in both the TSC2+/− and TSC2−/− neurons compared with TSC2+/+ neurons (Fig. 3d). These data demonstrate that loss of one or both alleles of TSC2 is sufficient to cause dysregulation of mTOR activity in human iPSC-derived neurons.
Figure 3.
mTORC1 hyperactivation in iPSC-derived neurons. a, Western blot of TSC2 from iPSC-derived neurons from all three iPSC lines at 14 d after NGN2 induction with doxycycline. Quantification of TSC2 levels from three separate differentiations of each line normalized to total protein (*p < 0.05; ANOVA with Dunnett's multiple-comparisons test). b, c, Western blot of mTORC1 targets (b) pS6 and (c) phosphorylated 4EBP1, from iPSC-derived neurons at 14 d after treatment with doxycycline. Quantification of pS6 and phosphorylated 4EBP1 levels from six separate differentiations of each line normalized to total S6 and 4EBP1 levels, respectively (*p < 0.05; ANOVA with Dunnett's multiple-comparisons test). d, Western blot of the mTORC2 target phosphorylated AKT from iPSC-derived neurons at 14 d after treatment with doxycycline. Quantification of phosphorylated AKT levels from six separate differentiations of each line normalized to total AKT levels (*p < 0.05; ANOVA with Dunnett's multiple-comparisons test). e, Evaluation of phosphoprotein levels using the ActivSignal platform. Only phosphorylation sites that were different between TSC2−/− and TSC2+/+ with an FDR<0.10 are shown in the figure (n = 3 wells; t test; *FDR < 0.1 **FDR < 0.05). The fold changes and p-values for all phosphoproteins are shown in (Figure 3-1). Bar plots show mean ± SEM.
Investigation of dysregulated signaling pathways due to loss of TSC2 using the ActivSignal platform. Three separate replicates of TSC2+/+ iPSC-derived neurons and TSC2-/- iPSC-derived neurons were analyzed using the ActivSignal platform. The first column identifies the protein that is being evaluated. The second column shows the fold change (TSC2-/- vs TSC2+/+). The third column shows the standard deviation. The fourth column shows the p-value calculated using a T-test, and the fifth column shows the correction for multiple comparisons using the FDR. Download Figure 3-1, XLSX file (53.9KB, xlsx)
Few mTORC1 targets have been validated in human neurons, and therefore, we performed a broad screen for dysregulated phosphoproteins in TSC2−/− neurons using the Immuno-Paired-Antibody Detection platform (ActivSignal). This assay provides quantitative data on the phosphorylation levels of 45 proteins per sample in parallel, assessing the activation or inhibition of various signaling pathways. TSC2+/+ and TSC2−/− neurons were assayed seven days after doxycycline induction of NGN2, and of the 45 phosphorylation sites evaluated using this platform, 41 were detected in both samples (Fig. 3-1). We then compared the level of phosphorylation between the TSC2+/+ and TSC2−/− neurons, and we found that 10 phosphorylation sites demonstrated a significant change between genotypes (FDR < 0.10) (Fig. 3e). One phosphorylation site that was decreased in TSC2−/− neurons was AKT S473, which is consistent with our Western blot data (Fig. 3d). Otherwise, we observed that YAP1, Cofilin, Hsp27, Rb, ErbB3, and Ampk demonstrated reduced phosphorylation, while Syk, NFκB p65, and EGFR showed elevated levels of phosphorylation. These data indicate widespread disruption in the phosphoproteome due to loss of a function TSC complex, which is likely due to a combination of direct and indirect effects of mTORC1 disinhibition.
Morphological alterations in TSC2-deficient neurons
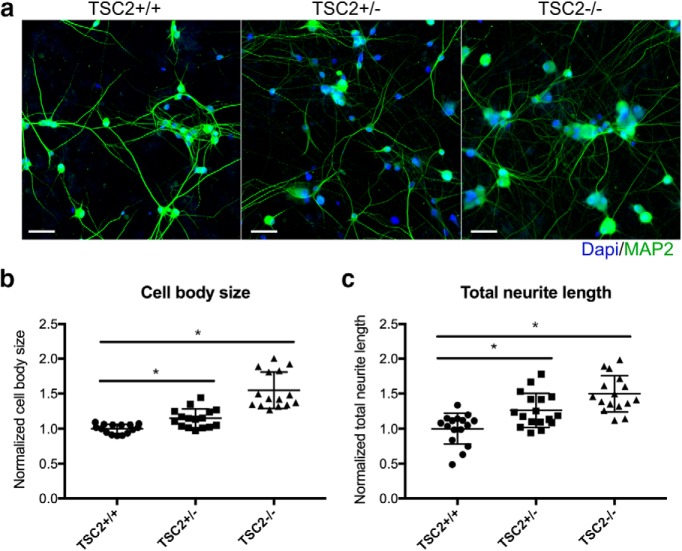
Animal models, neuropathological examinations of cortical tubers, and human neurons studied in vitro have demonstrated multiple cytological abnormalities, including enlarged soma size and changes in length and branching of neuronal processes in TSC2-deficient neurons. Six days after doxycycline induction of NGN2, we replated iPSC-derived neurons with human iPSC-derived astrocytes and allowed them to mature for two and a half weeks. We then stained these cells for MAP2 and used automated microscopy to capture multiple fields of view from each independent well for each genotype (Fig. 4a). These images were then analyzed using the Neuronal Profiler algorithm, which automatically segments cell bodies and neurites and quantifies these morphological features. We observed that TSC2+/− and TSC2−/− neurons demonstrated increased soma size compared with TSC2+/+ neurons at this time point (Fig. 4b). In addition, MAP2-positive dendrites demonstrated increased length in TSC2+/− and TSC2−/− neurons compared with TSC2+/+ neurons (Fig. 4c). These data demonstrate the TSC2+/− and TSC2−/− neurons display cytological abnormalities consistent with those previously described in cortical tubers and animal models.
Figure 4.
Increased soma size and dendritic growth in TSC2-deficient iPSC-derived neurons. iPSC-derived neurons from all three lines three and a half weeks after doxycycline treatment were stained with MAP2 to highlight neuronal cell bodies and dendrites. a, Images acquired with an automated microscopy system and analyzed using Neuronal Profiler. b, c, These plots demonstrate the (b) average cell body area of MAP2-positive cells and (c) average MAP2-positive neurite length from all three iPSC-derived cell lines. Data represent five independent wells across three separate differentiations, and each point represents the mean of 25 fields of view from a single well. Data were normalized to the mean value of the TSC2+/+ line for each differentiation. *p < 0.05; ANOVA with Dunnett's multiple-comparisons test.
Increased activity and synchrony in TSC2-deficient iPSC-derived neurons
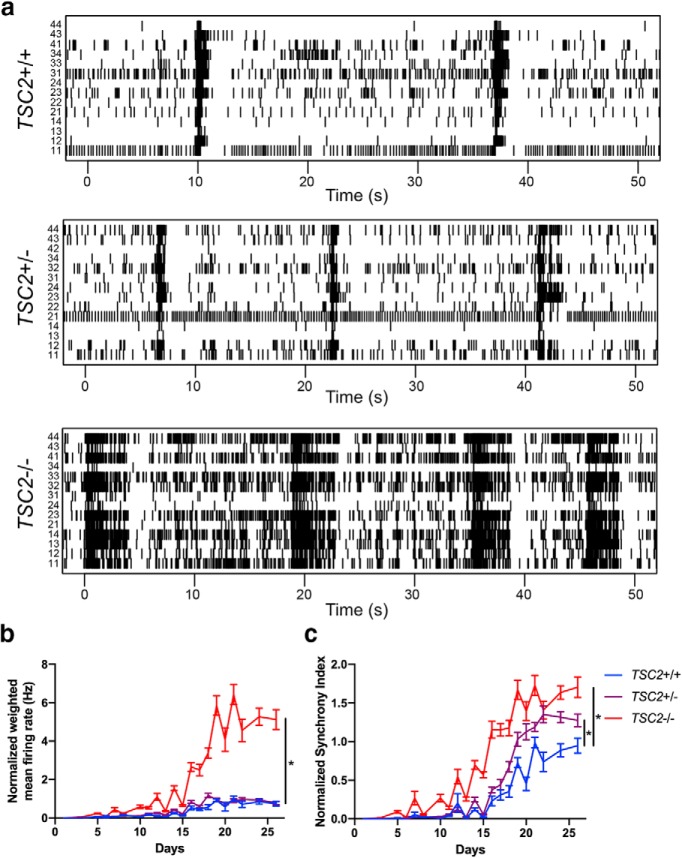
To investigate physiological alterations that may be present in TSC2-deficient human neurons, we used a multielectrode array (MEA), which allows extracellular recording of neuronal cultures through maturation. We differentiated iPSCs from the three cell lines into neurons in parallel and replated these neurons with human astrocytes on differentiation day six on PEI-coated MEA plates. This experiment was repeated three times using three independently differentiated neuronal batches. For each replicate, 16 wells were dedicated to each genotype with 16 electrodes per well. We recorded extracellular activity from each electrode for 5 min every 2–3 d for 25 d. We then used the Neuronal Statistics Compiler (Axion) to analyze these recordings. To ensure robustness of the data, we discarded any well that had <25% active electrodes. We observed that TSC2−/− neurons displayed increased activity compared with TSC2+/− and TSC2−/− neurons (Fig. 5a). We then analyzed the weighted mean firing rate, which adjusts the overall firing rate by taking the number of inactive electrodes into consideration. We normalized each differentiation to the maximal average weighted mean firing rate of the TSC2+/+ neurons and used nonlinear regression to fit the normalized weighted mean firing rate of each well to a sigmoidal curve (Fig. 5b). We then used a Z test to determine whether there was a significant difference in any of the coefficients from the regression analysis. Using this assay, TSC2−/− neurons demonstrated a significantly greater plateau weighted mean firing rate than the TSC2+/+ neurons. Interestingly, there was no significant difference between the plateau coefficients for the TSC2+/− and TSC2+/+ neurons. However, mid-point coefficients for both the TSC2−/− and TSC2+/− neurons were significantly less than the mid-point coefficient for the TSC2+/+ neurons. These data demonstrate that both TSC2+/− and TSC2−/− neurons become active at earlier time points than the TSC2+/+ neurons. However, the TSC2+/− neurons appear to be constrained in their overall activity and not different from TSC2+/+ neurons, whereas TSC2−/− neurons show dramatically elevated overall firing rates.
Figure 5.
Increased activity and synchrony of TSC2-deficient human neurons. iPSC-derived neurons were plated onto an MEA at six days after doxycycline treatment, and extracellular activity was monitored for the next 25 d. a, Raster plots of MEA recordings of neurons from all three lines. The numbers on the left side of the raster denote the electrode and time is represented on the x-axis. Each line represents a spike detected by the designated electrode. b, c, Normalized (b) weighted mean firing rate and (c) synchrony index of iPSC-derived neurons from all three lines throughout maturation. These data are compiled across three separate differentiations, each with 16 independent wells per genotype. Data are normalized to the mean maximum value for the TSC2+/+ neurons for each differentiation. Values for each well were fit to a sigmoidal curve, and differences between coefficients for each genotype were calculated using a Z test. *p < 0.01. Graph shows mean ± SEM.
Epilepsy is characterized by hypersynchronous neuronal discharges, and given the high prevalence of epilepsy in patients with TSC, we examined the synchrony index from each genotype, which is related to the cross-correlation between all pairs of electrodes within a well. We normalized each differentiation to the maximal average synchrony index of the TSC2+/+ neurons, and we used nonlinear regression to fit the normalized synchrony index of each well to a sigmoidal curve (Fig. 5c). We then used a Z test to determine whether where there was a significant difference in any of the coefficients from the regression analysis. We observed that both the TSC2+/− and TSC2−/− neurons demonstrated significantly greater plateau synchrony indices compared with TSC2+/+ neurons. In addition, we found that the mid-point coefficients for both the TSC2+/− and TSC2−/− neurons were significantly less than the mid-point coefficient of the TSC2+/+ neurons. These data demonstrate that both TSC2+/− and TSC2−/− neurons show increased synchronous neuronal activity and that this activity arises earlier compared with TSC2+/+ neurons.
Transcriptional profiling of TSC2-deficient iPSC-derived neurons
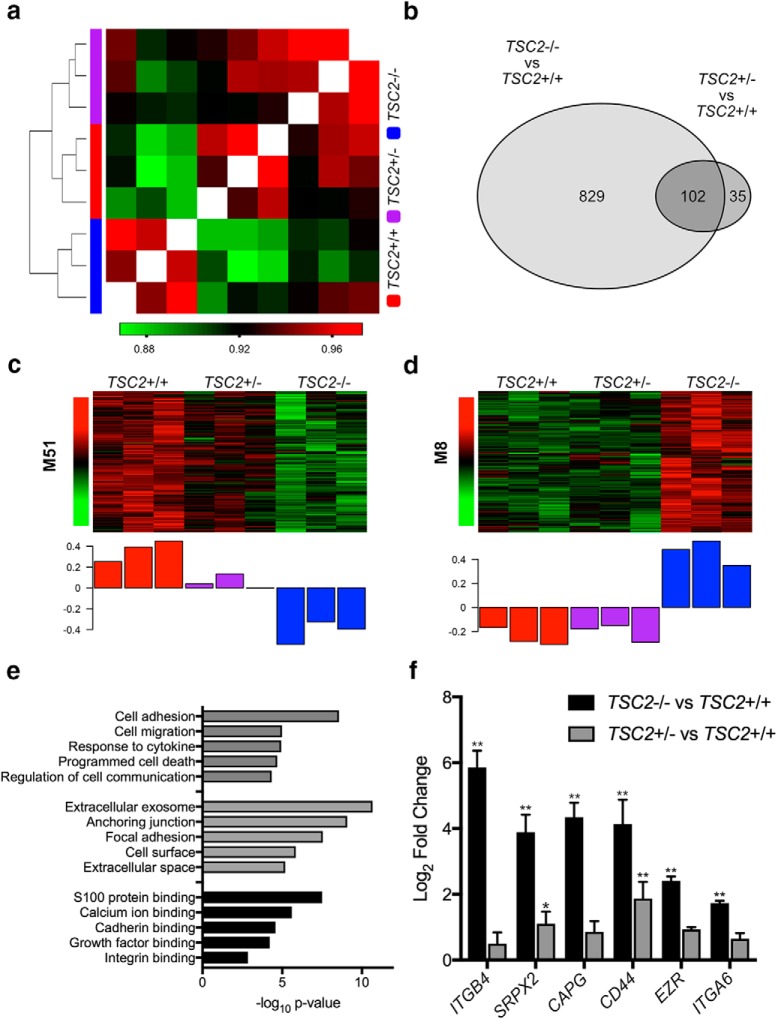
To investigate the mechanisms underlying the morphological and electrophysiological abnormalities that we observed in TSC2-deficient neurons, we performed transcriptional profiling of human neurons from all three genotypes. We differentiated iPSCs from all three lines and collected total RNA from iPSC-derived neurons on differentiation day 14. We then performed gene expression profiling from three independent differentiations of each line using targeted sequencing with the Ion AmpliSeq transcriptome human gene expression kit and the IonTorrent S5 Sequencer. Gene level expression data were quantified using the Torrent Suite software. After removal of genes with low expression, we compared the Pearson correlation coefficients between all nine samples, and we found that samples clustered based on allelic dose of TSC2 (Fig. 6a). Interestingly, gene expression in the TSC2+/− neurons appeared more similar to the TSC2+/+ neurons than the TSC2−/− neurons, despite the TSC2+/− and TSC2−/− neurons sharing an identical genetic background. These data demonstrate that biallelic mutations in TSC2 have a greater impact on neuronal gene expression than the loss of only one allele.
Figure 6.
Gene expression alterations in TSC2-deficient human neurons. Gene expression was assayed in iPSC-derived neurons from all three lines at 14 d after doxycycline treatment from three separate differentiations. a, Heat map demonstrating the Pearson correlation between all samples, where the value of the correlation is shown on the color bar below. Samples were ordered using hierarchical clustering and the dendrogram is shown to the left of the hea tmap. The samples are denoted by the colors on the left of the heat map, where red-TSC2+/+, purple-TSC2+/−, and blue-TSC2−/−. b, Venn diagram of differentially expressed genes between TSC2−/− and TSC2+/+ neurons and differentially expressed genes between TSC2+/− and TSC2+/+ neurons. Differentially expressed genes between TSC2+/− and TSC2+/+ neurons are shown in (Figure 6-1), and differentially expressed genes between TSC2−/− and TSC2+/+ neurons are shown in (Figure 6-2). c, Heat map demonstrating the scaled expression of each gene within the M51 module (rows) across all samples (columns), where red corresponds to higher relative expression and green denotes lower relative expression. The bar plot below represents the first principal component of expression within the module or the ME. Venn diagrams of overlap between genes within the M51 module and previously identified downregulated genes in TSC2-deficient cerebellar Purkinje cells, as well as FMRP target transcripts, are shown in (Figure 6-3). (Figure 6-4) shows a Western blot of FMRP in iPSC-derived neurons 14 d after doxycycline treatment, which shows no significant change of FMRP expression between the different lines. d, Heat map and ME of genes within the M8 module. (Figure 6-5) shows all the genes that are differentially expressed between cortical tubers and unaffected controls. Venn diagram of the overlap between the M8 module and genes upregulated in cortical tubers is shown in (Figure 6-6). e, Bar plot of gene ontology analysis of genes in M8 and upregulated in cortical tubers. The enriched gene ontology terms are denoted on the left and the x-axis represents −log10 of the p-value. f, Bar plot of qPCR performed in iPSC-derived neurons for six genes present in M8 and upregulated in cortical tubers. The y-axis represents log2 fold change calculated by the ΔΔCt method, and the gene names are shown along the x-axis. **p (Bonferroni correction) < 0.01, *p (Bonferroni correction) < 0.05, Wilcoxon's rank-sum test. Bar plot shows mean ± SEM.
Differentially expressed genes between TSC2+/- and TSC2+/+ neurons. The first column shows a unique identifier from the Ampliseq analysis pipeline. The second column shows the associated gene symbols, and the third column shows the associated full gene name. The fourth column shows the mean expression in TSC2+/+ neurons, while the fifth column shows the mean expression in TSC2+/- neurons. The sixth column shows the fold change between TSC2+/- and TSC2+/+ neurons, and the seventh and eighth columns show the p-value and false discovery rate, respectively. Download Figure 6-1, XLSX file (73.1KB, xlsx)
Differentially expressed genes between TSC2-/- and TSC2+/+ neurons. The first column shows a unique identifier from the Ampliseq analysis pipeline. The second column shows the associated gene symbols, and the third column shows the associated full gene name. The fourth column shows the mean expression in TSC2+/+ neurons, while the fifth column shows the mean expression in TSC2-/- neurons. The sixth column shows the fold change between TSC2-/- and TSC2+/+ neurons, and the seventh and eighth columns show the p-value and false discovery rate, respectively. Download Figure 6-2, XLSX file (176KB, xlsx)
Comparison of M51 co-expression module identified from gene expression data from iPSC-derived neurons with other datasets. a) This Venn diagram shows the overlap between the M51 co-expression modules that contains genes that are down-regulated concomitantly with loss of TSC2 alleles in iPSC-derived neurons and the M99 co-expression module that also contains genes whose down-regulation is related to loss of TSC2 alleles in iPSC-derived Purkinje cell neurons (Sundberg et al., 2018). b-d) These Venn diagrams shows the overlap between the M51 co-expression module and transcripts that were found to bind to FMRP (Brown et al., 2001; Darnell et al., 2011; Ascano et al., 2012). Download Figure 6-3, TIF file (432.7KB, tif)
Expression of FMRP in iPSC-derived cortical neurons. Western blot of FMRP from iPSC-derived neurons from all three iPSC lines at fourteen days after NGN2 induction with doxycycline. Quantification of FMRP levels from three separate differentiations of each line normalized to total protein shows no significant difference between the lines (ANOVA with Dunnett’s multiple comparison test). Download Figure 6-4, TIF file (174.2KB, tif)
Differentially expressed genes between cortical tubers and brain samples of unaffected individuals across two datasets (Martin et al., 2017; Mills et al., 2017). The first column shows the Ensembl transcript ID for the mapped gene, and the second column shows the associated gene symbol. The third column shows the fold change between the cortical tuber and samples from unaffected individuals. The fourth and fifth columns show the p-value and false discovery rate of the comparison between the cortical tubers and samples from unaffected individuals. Download Figure 6-5, XLSX file (194.5KB, xlsx)
Comparison of M8 co-expression module identified from gene expression data from iPSC-derived neurons with differentially expressed genes from cortical tubers. This Venn diagram shows the overlap between the M8 co-expression module and genes that are up-regulated in cortical tubers across two datasets (Martin et al., 2017; Mills et al., 2017). Download Figure 6-6, TIF file (75.8KB, tif)
Using LIMMA to calculate differential expression, we identified 931 genes that were differentially expressed between TSC2−/− and TSC2+/+ neurons (FDR < 0.05 and log2 FC > 0.5), of which 689 were upregulated and 242 were downregulated (Fig. 6-1). We also identified 137 genes that were differentially expressed between TSC2+/− and TSC2+/+ neurons, of which 94 were upregulated and 43 were downregulated (Fig. 6-2). We then compared these two lists of genes and found a highly significant overlap (p = 9.5e-109; Fig. 6b), demonstrating that loss of one allele of TSC2 results in a similar effect on gene expression as loss of both alleles of TSC2. However, LOH has a substantially larger effect on gene expression than the heterozygous mutation.
Previously, we had found that targets of the RNA binding protein, Fragile X Mental Retardation Protein (FMRP), were downregulated in a manner related to the number of functional TSC2 alleles (Sundberg et al., 2018). In this study, we performed an unsupervised network analysis using WGCNA on the transcriptional data from these iPSC-derived neurons. We identified 32 modules of coexpressed genes and examined the module eigengenes (MEs) for modules that demonstrated downregulation of genes related to the allelic dose of TSC2. The module with the greatest correlation between its ME and TSC2 genotype was M51 (r = 0.91; Fig. 6c). We compared the genes associated with M51 module (kME>0.5) to a group of coexpressed genes that we had found to be downregulated in Purkinje neurons (Sundberg et al., 2018), and we found a significant overlap between these two groups (p = 1.94e-11; hypergeometric probability; (Fig. 6-3). We then compared the M51 genes to previously published datasets of FMRP targets (Brown et al., 2001; Darnell et al., 2011; Ascano et al., 2012) and also found significant overlaps between these lists (Ascano p = 7.41e-61; Darnell p = 0.0025; Brown p = 0.0065; hypergeometric probability; (Fig. 6-3). These data independently corroborate our prior study and demonstrate that FMRP target transcripts are downregulated in multiple types of iPSC-derived neurons with loss of TSC2. We then examined the protein level of FMRP in iPSC-derived neurons from all three genotypes after 14 d of differentiation. However, we did not observe a significant change in the steady-state levels of FMRP (Fig. 6-4), suggesting that FMRP function may be dysregulated independent of its level of expression.
We also compared the gene expression dysregulation in these iPSC-derived neurons to cortical tubers that were surgically resected from patients with TSC. We obtained sequencing data from two large studies that compared cortical tubers to matched postmortem samples from individuals without TSC (Martin et al., 2017; Mills et al., 2017). Together, this dataset consists of 26 cortical tubers and 22 specimens from unaffected individuals. We then mapped both datasets to same version of the human genome and calculated differential expression between cortical tubers and control samples across both datasets using LIMMA. We identified 2222 genes that were differentially expressed between cortical tubers and samples from unaffected individuals where 1759 genes showed higher expression in cortical tubers and 463 genes showed higher expression in specimens from unaffected individuals (FDR < 0.05 and log2 FC > 0.5, (Fig. 6-5). We then investigated the coexpression modules in the iPSC-derived neurons to identify whether there were patterns of dysregulated genes that were also observed in cortical tubers. We found that genes associated with the M8 module (kME > 0.5) demonstrated a significant overlap with genes that were upregulated in cortical tubers (p = 6.19e-15; hypergeometric probability; (Fig. 6-6). Interestingly, the M8 module corresponds to genes that are upregulated in TSC2−/− neurons but show similar expression between TSC2+/− and TSC2+/+ neurons (Fig. 6d). These data indicate that gene expression dysregulation that is observed in TSC2−/− neurons is also present in cortical tubers, but these genes are not altered in TSC2+/− neurons.
To investigate the functions of the shared gene expression abnormalities between cortical tubers and TSC2−/− neurons, we used gene ontology analysis on the 149 genes that overlap between the two lists. This group was enriched in genes whose proteins are localized to the extracellular region, especially within exosomes (p = 2.17e-11; Fig. 6e). In addition, these genes are known to regulate cell adhesion (p = 2.77e-9) and cell migration (p = 1.04e-5). We then examined several genes that showed high connectivity within the M8 module and increased expression in cortical tubers using qPCR. We validated significant upregulation of six candidate genes in TSC2−/− neurons, including ITGB4, ITGA6, SRPX2, EZR, CD44, and CAPG (Fig. 6f). Interestingly, all six genes demonstrated a trend toward upregulation in TSC2+/− neurons, but only two genes (CD44 and SRPX2) were significantly differentially expressed compared with TSC2+/+ neurons. These data demonstrate that biallelic but not monoallelic loss of TSC2 in neurons results in a pattern of dysregulated of gene expression that can be identified in cortical tubers and is related to increased expression of genes associated with cellular adhesion and motility.
Inhibition of mTORC1 in TSC2-deficient iPSC-derived neurons
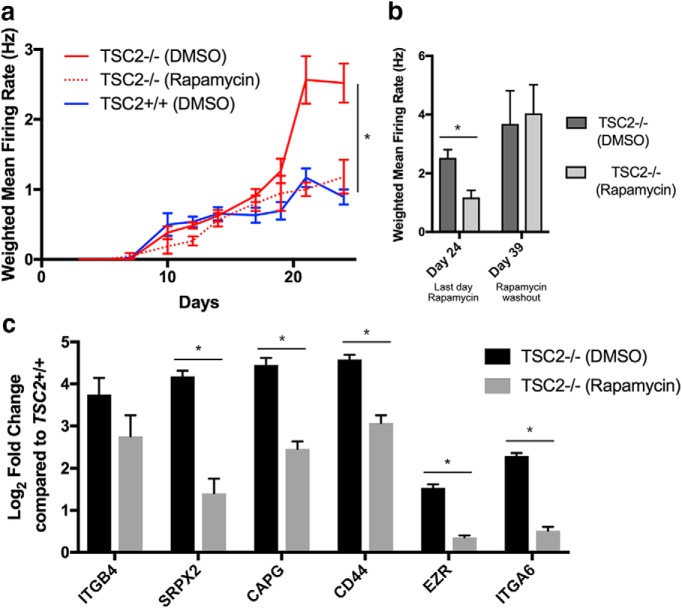
We then examined the contribution of mTORC1 hyperactivation to a subset of the abnormalities that we identified in TSC2-deficient iPSC-derived neurons. Initially, we performed MEA recordings of from iPSC-derived neurons from all three TSC2 genotypes, as described above. At day 7 of recording, we treated half of the neurons of each genotype with the mTORC1 inhibitor, rapamycin (20 nm), and half with DMSO as a vehicle control, and we continued recording network activity every other day. Consistent with our prior data, we observed that vehicle-treated TSC2−/− neurons showed significant hyperactivity compared with vehicle-treated TSC2+/+ neurons (Fig. 7a). Strikingly, rapamycin treatment reduced activity of TSC2−/− neurons to levels comparable to vehicle-treated TSC2+/+ neurons (Fig. 7a). There were no significant differences between the activity of either vehicle- or rapamycin-treated TSC2+/− and TSC2+/+ neurons (Fig. 7-1). After 24 d of recording, we stopped rapamycin and vehicle treatments and continued recording for 14 d. Although the activity in TSC2−/− neurons was significantly reduced with rapamycin treatment, these cells substantially increased their activity during the washout period (Fig. 7b). There was no significant difference between the activities of vehicle- and rapamycin-treated TSC2−/− neurons after the washout period (Fig. 7b). These data show that hyperactivity in TSC2−/− neurons is due to mTORC1 hyperactivation but that continued inhibition of mTORC1 is required to constrain network activity.
Figure 7.
mTORC1 inhibition reduces hyperactivity and gene expression changes in TSC2−/− neurons. a, Weighted mean firing rate of iPSC-derived neurons from TSC2−/− neurons treated with rapamycin or vehicle, as well as vehicle-treated TSC2+/+ neurons. Each point represents eight independent wells per treatment per genotype. Values for each well were fit to a sigmoidal curve, and differences between coefficients for each genotype were calculated using a Z test. *p < 0.01. Graph shows mean ± SEM. (Figure 7-1) shows the activity of rapamycin- and vehicle-treated neurons from all three TSC2 genotypes. b, Bar plot of the weighted mean firing rate of TSC2−/− neurons on the last day of rapamycin treatment (day 24) and after rapamycin washout (day 39). Vehicle-treated TSC2−/− neurons show increased activity compared with rapamycin-treated TSC2−/− neurons, but there is no difference between the two groups after rapamycin washout. (*p < 0.05, t test). c, Bar plot of qPCR assaying the expression of six genes from TSC2−/− neurons treated with rapamycin or vehicle compared with vehicle-treated TSC2+/+ neurons. The y-axis represents log2 fold change calculated by the ΔΔCt method, and the gene names are shown along the x-axis. *p < 0.05, Wilcoxon rank-sum test. Bar plot shows mean ± SEM.
mTORC1 inhibition reduces hyperactivity and gene expression changes in TSC2-/- neurons. a) Weighted mean firing rate of iPSC-derived neurons from TSC2+/+, TSC2+/-, and TSC2-/- neurons treated with rapamycin or vehicle. Each point represents 8 independent wells per treatment per genotype. Values for each well were fit to a sigmoidal curve, and differences between coefficients for each genotype were calculated using a Z-test. Graph shows mean ± SEM. Download Figure 7-1, TIF file (174KB, tif)
To examine the contribution of mTORC1 hyperactivation to the gene expression abnormalities that were also observed in cortical tubers, we treated iPSC-derived neurons with either 20 nm rapamycin or DMSO from day of differentiation 7 until RNA collection at day of differentiation 14. We then examined the expression of the six genes that we had confirmed were upregulated in TSC2−/− neurons. Interestingly, we found that 5 of 6 genes were significantly reduced by rapamycin in TSC2−/− neurons (Fig. 7c). However, all six genes were still expressed at higher levels in rapamycin-treated TSC2−/− neurons compared with vehicle-treated TSC2+/+ neurons (Fig. 7c), indicating that rapamycin only partially reversed the upregulation of these genes within this timeframe of treatment.
Discussion
TSC is a systemic disorder, but the neurological manifestations are among the most prevalent and cause substantial morbidity. We have used iPSCs with the full allelic series of TSC2 dosages to study the morphological and physiological characteristics of neurons derived from these cells. In addition, we have used a transcription factor-based method of neuronal differentiation that is both highly efficient and homogeneous. We found that this protocol was capable of resulting in high rates of neuronal differentiation in all three lines, and neurons derived using this protocol expressed mature neuronal and synaptic markers. We found that both TSC2+/− and TSC2−/− neurons demonstrated increased mTORC1 activation, cell body size, and neurite length compared with TSC2+/+ neurons. Using MEA analysis, we found that TSC2−/− neurons showed a dramatic increase in neuronal activity, while both the TSC2+/− and TSC2−/− neurons demonstrated earlier and greater synchrony. In addition, we found that TSC2−/− neurons displayed a pattern of gene expression dysregulation that was also present in cortical tubers. Finally, we demonstrate that mTORC1 inhibition is capable of reducing network activity in TSC2−/− neurons, but it does not fully reverse certain gene expression changes, and network hyperactivity returns when mTORC1 inhibition is discontinued.
In this study, induced expression of NGN2 led robustly to differentiation of neurons regardless of TSC2 genotype. However, prior studies have observed that loss of one or both alleles of TSC2 in stem cells results in impaired neuronal differentiation. Differentiation of ESCs with a homozygous mutation in TSC2 showed that there were fewer HuC/HuD+ cells at an early time point (Costa et al., 2016), and cerebral spheroids created from iPSCs with a homozygous mutation in TSC2 also showed reduced numbers of neurons (Blair et al., 2018). However, a study with iPSCs derived from TSC patients suggested that this effect was transient (Zucco et al., 2018). These studies have observed increased proliferation of TSC2-deficient neuroprogenitors (Li et al., 2017; Sundberg et al., 2018) due to hyperactivation of mTORC1, reduced activity of AKT, and increased STAT3 signaling (Grabole et al., 2016; Zucco et al., 2018). It is likely that we did not observe any differences in neuronal differentiation between the various TSC2 genotypes because induction of NGN2 bypasses the need for initial differentiation into neuroprogenitors before final differentiation into neurons, resulting in a differentiation program that appears to be unaffected by loss of TSC2.
Epilepsy is highly prevalent among patients with TSC, affecting up to 90% of individuals (Chu-Shore et al., 2010). Consistent with this phenotype, we found that neurons with biallelic TSC2 mutations show dramatic hyperactivity, whereas neurons with a single TSC2 mutation do not demonstrate a difference in overall neuronal activity. Several animal models with disruption of the TSC have shown that pyramidal neurons are hyperexcitable (Bateup et al., 2011, 2013; Abs et al., 2013; Lozovaya et al., 2014). However, neurons derived from human embryonic stem cells with a homozygous TSC2 mutation showed reduced excitability (Costa et al., 2016). One possible explanation is that this study used dual SMAD inhibition for patterning of neuroprogenitors before inducing terminal neuronal differentiation, whereas our study used induction of NGN2 in iPSCs to directly drive differentiation of excitatory neurons. Therefore, neurons that showed reduced activity due to loss of TSC2 may reflect the phenotype in a separate neuronal subtype. In fact, it has been demonstrated that iPSC-derived Purkinje neurons with TSC2 mutations are also less excitable (Sundberg et al., 2018). How TSC2 loss affects neuronal excitability in different ways in different neuronal subtypes is an open and important question. Another possibility is that the reduction in the neuronal population in the mixed cortical cultures, as indicated by fewer HuC/HuD+ cells (Costa et al., 2016), led to reduced or delayed network formation, neuron maturation and activity. One study used iPSCs from patients with either a TSC1 or TSC2 mutation found that these cells showed increased activity compared with unrelated, unaffected controls (Nadadhur et al., 2019). We did not observe a difference in overall activity levels between TSC2+/− and TSC2+/+ neurons, but the TSC2+/− neurons attained their peak activity slightly earlier than the TSC2+/+ neurons. Therefore, it is possible that this study evaluated neuronal activity during this window when there is a difference between heterozygous neurons and controls, and this window may be substantially elongated due to the slower differentiation methods.
Interestingly, we found that synchrony was increased and attained earlier in both TSC2+/− and TSC2−/−, suggesting an intrinsic abnormality in neuronal connectivity. Neuronal synchrony is thought to be mediated by underlying oscillations where neurons showing fluctuating excitability, leading to most neurons firing together during the more excitable window (Traub et al., 2004). A study using hippocampal slices in which Pten was deleted in a subset of neurons found that mutant cells were more easily recruited into epochs with a significant portion of active neurons, whereas the frequency of these events was not increased (Barrows et al., 2017). Thus, these data suggest that mTORC1 hyperactivation may be sufficient to increase synchrony, independent of overall network activity. One potential mechanism of increased synchrony in TSC2-deficient neurons is reduction of FMRP target transcripts, which we observed in both this and a prior study (Sundberg et al., 2018). Loss of Fmr1 in animal models has been demonstrated to cause hypersynchrony and increased connectivity within local cortical circuits (Contractor et al., 2015). Interestingly, we did not observe a significant decrease in FMRP protein levels in this study, which suggests that dysregulation of FMRP target transcripts may be due to altered function of FMRP due to loss of TSC2.
Cortical tubers are a pathological hallmark of TSC, and we found similar gene expression dysregulation in TSC2−/− neurons and cortical tubers. Although LOH has been suggested to play a role in the development of cortical tubers (Crino et al., 2010), studies have found second hit mutations in either TSC1 or TSC2 in <35% of samples (Qin et al., 2010; Martin et al., 2017). Therefore, despite cortical tubers consisting of mostly heterozygous cells, the pattern of gene expression dysregulation is similar to that of neurons with biallelic loss of TSC2. Although it is possible that neurons with LOH are driving the gene expression differences observed in cortical tubers, this explanation is probably less likely because of the scarcity of these cells in tubers. One potential mechanism involves non-cell autonomous signaling from cells with LOH that drives gene expression dysregulation in surrounding heterozygous cells. Interestingly, we found that upregulated genes were associated with exosomes, and it has been shown that exosomes from Tsc1-null cells can transform wild-type cells to have a similar phenotype (Patel et al., 2016). In addition, several of these genes have known roles in synapse formation, maturation, and maintenance. For example, CD44 is present at synapses in rat hippocampal neurons, and knockdown of CD44 led to reduced numbers of functional synapses (Roszkowska et al., 2016). ITGA6 and ITGB4 form a complex that is important for proper neurodevelopment (Georges-Labouesse et al., 1998). Finally, SRPX2 can promote synapse formation in cultured neurons, and it is known to be involved in Rolandic epilepsy and language disorders (Sia et al., 2013). Together, these data demonstrate that LOH of TSC2 leads to upregulation of several genes that promote synaptogenesis, which is also seen in cortical tubers, suggesting that these genes may play a role in the hyperexcitability present in cortical tubers.
To examine the mechanism underlying a subset of the identified neuronal abnormalities, we treated iPSC-derived neurons with the mTORC1 inhibitor rapamycin. We found that rapamycin prevented hyperactivity in TSC2−/− iPSC-derived neurons, but hyperactivity returned after discontinuation of treatment, suggesting that mTORC1 hyperactivation in differentiated TSC2−/− neurons is sufficient to cause hyperactivity. These data are consistent with a prior study that showed that deletion of Tsc1 in adult mice led to hyperexcitability (Abs et al., 2013). We also observed that rapamycin treatment partially rescued several of the gene expression changes that we had observed in TSC2−/− neurons. These data suggest that either initial mTORC1 dysregulation during differentiation or mTORC1-independent processes can cause these changes in gene expression. Interestingly, a phase III trial of everolimus for refractory epilepsy in TSC showed a significant reduction in seizures, but patients continued to have an average of 4.9 seizures per week on everolimus therapy (French et al., 2016). Therefore, it is possible that these residual or mTORC1-independent effects may be contributing to ongoing seizures in patients with TSC who have refractory epilepsy.
In this study, we found that a single mutation in TSC2 is capable of causing mTORC1 hyperactivation and increased synchrony in human iPSC-derived neurons. However, biallelic mutations in TSC2 were necessary to cause hyperactivity and gene expression alterations observed in cortical tubers. We observed comparable rates of neuronal differentiation between TSC2 genotypes using induced expression of NGN2, which suggests that the abnormalities that we observed in TSC2+/− and TSC2−/− neurons can be independent of alterations in development. This implies that ongoing neuronal dysregulation due to reduction in TSC2 may play a role in the neurological symptoms in TSC.
Footnotes
The Sahin laboratory has received grant funding from the U.S. Department of Defense (Grant W81XWH-15-1-0189) and the Nancy Lurie Marks Family Foundation related to this study. K.D.W. is funded by the National Institutes of Health (Grant R25NS070682). We thank David Kwiatkowski for critical reading of this manuscript. The lentiviral plasmids were gifts from Thomas Sudhof at Stanford University.
M. Sahin reports grant support from Novartis, Roche, Pfizer, Ipsen, LAM Therapeutics, and Quadrant Biosciences unrelated to this project and has served on scientific advisory boards for Sage, Roche, Celgene, and Takeda. The remaining authors declare no competing financial interests.
References
- Abs E, Goorden SM, Schreiber J, Overwater IE, Hoogeveen-Westerveld M, Bruinsma CF, Aganović E, Borgesius NZ, Nellist M, Elgersma Y (2013) TORC1-dependent epilepsy caused by acute biallelic Tsc1 deletion in adult mice. Ann Neurol 74:569–579. 10.1002/ana.23943 [DOI] [PubMed] [Google Scholar]
- Ascano M Jr, Mukherjee N, Bandaru P, Miller JB, Nusbaum JD, Corcoran DL, Langlois C, Munschauer M, Dewell S, Hafner M, Williams Z, Ohler U, Tuschl T (2012) FMRP targets distinct mRNA sequence elements to regulate protein expression. Nature 492:382–386. 10.1038/nature11737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrows CM, McCabe MP, Chen H, Swann JW, Weston MC (2017) PTEN loss increases the connectivity of fast synaptic motifs and functional connectivity in a developing hippocampal network. J Neurosci 37:8595–8611. 10.1523/JNEUROSCI.0878-17.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateup HS, Takasaki KT, Saulnier JL, Denefrio CL, Sabatini BL (2011) Loss of Tsc1 in vivo impairs hippocampal mGluR-LTD and increases excitatory synaptic function. J Neurosci 31:8862–8869. 10.1523/JNEUROSCI.1617-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateup HS, Johnson CA, Denefrio CL, Saulnier JL, Kornacker K, Sabatini BL (2013) Excitatory/inhibitory synaptic imbalance leads to hippocampal hyperexcitability in mouse models of tuberous sclerosis. Neuron 78:510–522. 10.1016/j.neuron.2013.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bissler JJ, Kingswood JC, Radzikowska E, Zonnenberg BA, Frost M, Belousova E, Sauter M, Nonomura N, Brakemeier S, de Vries PJ, Whittemore VH, Chen D, Sahmoud T, Shah G, Lincy J, Lebwohl D, Budde K (2013) Everolimus for angiomyolipoma associated with tuberous sclerosis complex or sporadic lymphangioleiomyomatosis (EXIST-2): a multicentre, randomised, double-blind, placebo-controlled trial. Lancet 381:817–824. 10.1016/S0140-6736(12)61767-X [DOI] [PubMed] [Google Scholar]
- Blair JD, Hockemeyer D, Bateup HS (2018) Genetically engineered human cortical spheroid models of tuberous sclerosis. Nat Med 24:1568–1578. 10.1038/s41591-018-0139-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown V, Jin P, Ceman S, Darnell JC, O'Donnell WT, Tenenbaum SA, Jin X, Feng Y, Wilkinson KD, Keene JD, Darnell RB, Warren ST (2001) Microarray identification of FMRP-associated brain mRNAs and altered mRNA translational profiles in fragile X syndrome. Cell 107:477–487. 10.1016/S0092-8674(01)00568-2 [DOI] [PubMed] [Google Scholar]
- Chan JA, Zhang H, Roberts PS, Jozwiak S, Wieslawa G, Lewin-Kowalik J, Kotulska K, Kwiatkowski DJ (2004) Pathogenesis of tuberous sclerosis subependymal giant cell astrocytomas: biallelic inactivation of TSC1 or TSC2 leads to mTOR activation. J Neuropathol Exp Neurol 63:1236–1242. 10.1093/jnen/63.12.1236 [DOI] [PubMed] [Google Scholar]
- Chu-Shore CJ, Major P, Camposano S, Muzykewicz D, Thiele EA (2010) The natural history of epilepsy in tuberous sclerosis complex. Epilepsia 51:1236–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clogg CC, Petkova E, Haritou A (1995) Statistical methods for comparing regression coefficients between models. Am J Sociol 100:1261–1293. 10.1086/230638 [DOI] [Google Scholar]
- Contractor A, Klyachko VA, Portera-Cailliau C (2015) Altered neuronal and circuit excitability in fragile X syndrome. Neuron 87:699–715. 10.1016/j.neuron.2015.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa V, et al. (2016) mTORC1 inhibition corrects neurodevelopmental and synaptic alterations in a human stem cell model of tuberous sclerosis. Cell Rep 15:86–95. 10.1016/j.celrep.2016.02.090 [DOI] [PubMed] [Google Scholar]
- Crino PB, Aronica E, Baltuch G, Nathanson KL (2010) Biallelic TSC gene inactivation in tuberous sclerosis complex. Neurology 74:1716–1723. 10.1212/WNL.0b013e3181e04325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell JC, Van Driesche SJ, Zhang C, Hung KY, Mele A, Fraser CE, Stone EF, Chen C, Fak JJ, Chi SW, Licatalosi DD, Richter JD, Darnell RB (2011) FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell 146:247–261. 10.1016/j.cell.2011.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR (2013) STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29:15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J, Horvath S (2007) Understanding network concepts in modules. BMC Syst Biol 1:24. 10.1186/1752-0509-1-24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebrahimi-Fakhari D, Saffari A, Wahlster L, DiNardo A, Turner D, Lewis TL Jr, Conrad C, Rothberg JM, Lipton JO, Kölker S, Hoffmann GF, Han MJ, Polleux F, Sahin M (2016) Impaired mitochondrial dynamics and mitophagy in neuronal models of tuberous sclerosis complex. Cell Rep 17:2162. 10.1016/j.celrep.2016.10.051 [DOI] [PubMed] [Google Scholar]
- Ehninger D, Han S, Shilyansky C, Zhou Y, Li W, Kwiatkowski DJ, Ramesh V, Silva AJ (2008) Reversal of learning deficits in a Tsc2+/− mouse model of tuberous sclerosis. Nat Med 14:843–848. 10.1038/nm1788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franz DN, Belousova E, Sparagana S, Bebin EM, Frost M, Kuperman R, Witt O, Kohrman MH, Flamini JR, Wu JY, Curatolo P, de Vries PJ, Whittemore VH, Thiele EA, Ford JP, Shah G, Cauwel H, Lebwohl D, Sahmoud T, Jozwiak S (2013) Efficacy and safety of everolimus for subependymal giant cell astrocytomas associated with tuberous sclerosis complex (EXIST-1): a multicentre, randomised, placebo-controlled phase 3 trial. Lancet 381:125–132. 10.1016/S0140-6736(12)61134-9 [DOI] [PubMed] [Google Scholar]
- French JA, Lawson JA, Yapici Z, Ikeda H, Polster T, Nabbout R, Curatolo P, de Vries PJ, Dlugos DJ, Berkowitz N, Voi M, Peyrard S, Pelov D, Franz DN (2016) Adjunctive everolimus therapy for treatment-resistant focal-onset seizures associated with tuberous sclerosis (EXIST-3): a phase 3, randomised, double-blind, placebo-controlled study. Lancet 388:2153–2163. 10.1016/S0140-6736(16)31419-2 [DOI] [PubMed] [Google Scholar]
- Georges-Labouesse E, Mark M, Messaddeq N, Gansmüller A (1998) Essential role of alpha 6 integrins in cortical and retinal lamination. Curr Biol 8:983–986. 10.1016/S0960-9822(98)70402-6 [DOI] [PubMed] [Google Scholar]
- Giannikou K, Malinowska IA, Pugh TJ, Yan R, Tseng YY, Oh C, Kim J, Tyburczy ME, Chekaluk Y, Liu Y, Alesi N, Finlay GA, Wu CL, Signoretti S, Meyerson M, Getz G, Boehm JS, Henske EP, Kwiatkowski DJ (2016) Whole exome sequencing identifies TSC1/TSC2 biallelic loss as the primary and sufficient driver event for renal angiomyolipoma development. PLoS Genet 12:e1006242. 10.1371/journal.pgen.1006242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goorden SM, van Woerden GM, van der Weerd L, Cheadle JP, Elgersma Y (2007) Cognitive deficits in Tsc1+/− mice in the absence of cerebral lesions and seizures. Ann Neurol 62:648–655. 10.1002/ana.21317 [DOI] [PubMed] [Google Scholar]
- Grabole N, Zhang JD, Aigner S, Ruderisch N, Costa V, Weber FC, Theron M, Berntenis N, Spleiss O, Ebeling M, Yeo GW, Jagasia R, Kiialainen A (2016) Genomic analysis of the molecular neuropathology of tuberous sclerosis using a human stem cell model. Genome Med 8:94. 10.1186/s13073-016-0347-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henske EP, Scheithauer BW, Short MP, Wollmann R, Nahmias J, Hornigold N, van Slegtenhorst M, Welsh CT, Kwiatkowski DJ (1996) Allelic loss is frequent in tuberous sclerosis kidney lesions but rare in brain lesions. Am J Hum Genet 59:400–406. [PMC free article] [PubMed] [Google Scholar]
- Hoogeveen-Westerveld M, et al. (2011) Functional assessment of variants in the TSC1 and TSC2 genes identified in individuals with tuberous sclerosis complex. Hum Mutat 32:424–435. 10.1002/humu.21451 [DOI] [PubMed] [Google Scholar]
- Law CW, Chen Y, Shi W, Smyth GK (2014) voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol 15:R29. 10.1186/gb-2014-15-2-r29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Cao J, Chen M, Li J, Sun Y, Zhang Y, Zhu Y, Wang L, Zhang C (2017) Abnormal neural progenitor cells differentiated from induced pluripotent stem cells partially mimicked development of TSC2 neurological abnormalities. Stem Cell Reports 8:883–893. 10.1016/j.stemcr.2017.02.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipton JO, Sahin M (2014) The neurology of mTOR. Neuron 84:275–291. 10.1016/j.neuron.2014.09.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozovaya N, et al. (2014) Selective suppression of excessive GluN2C expression rescues early epilepsy in a tuberous sclerosis murine model. Nat Commun 5:4563. 10.1038/ncomms5563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Major P, Rakowski S, Simon MV, Cheng ML, Eskandar E, Baron J, Leeman BA, Frosch MP, Thiele EA (2009) Are cortical tubers epileptogenic? evidence from electrocorticography. Epilepsia 50:147–154. 10.1111/j.1528-1167.2008.01814.x [DOI] [PubMed] [Google Scholar]
- Martin KR, Zhou W, Bowman MJ, Shih J, Au KS, Dittenhafer-Reed KE, Sisson KA, Koeman J, Weisenberger DJ, Cottingham SL, DeRoos ST, Devinsky O, Winn ME, Cherniack AD, Shen H, Northrup H, Krueger DA, MacKeigan JP (2017) The genomic landscape of tuberous sclerosis complex. Nat Commun 8:15816. 10.1038/ncomms15816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meikle L, Talos DM, Onda H, Pollizzi K, Rotenberg A, Sahin M, Jensen FE, Kwiatkowski DJ (2007) A mouse model of tuberous sclerosis: neuronal loss of Tsc1 causes dysplastic and ectopic neurons, reduced myelination, seizure activity, and limited survival. J Neurosci 27:5546–5558. 10.1523/JNEUROSCI.5540-06.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills JD, et al. (2017) Coding and small non-coding transcriptional landscape of tuberous sclerosis complex cortical tubers: implications for pathophysiology and treatment. Sci Rep 7:8089. 10.1038/s41598-017-06145-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohamed AR, Bailey CA, Freeman JL, Maixner W, Jackson GD, Harvey AS (2012) Intrinsic epileptogenicity of cortical tubers revealed by intracranial EEG monitoring. Neurology 79:2249–2257. 10.1212/WNL.0b013e3182768923 [DOI] [PubMed] [Google Scholar]
- Nadadhur AG, Alsaqati M, Gasparotto L, Cornelissen-Steijger P, van Hugte E, Dooves S, Harwood AJ, Heine VM (2019) Neuron-glia interactions increase neuronal phenotypes in tuberous sclerosis complex patient iPSC-derived models. Stem Cell Reports 12:42–56. 10.1016/j.stemcr.2018.11.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel B, Patel J, Cho JH, Manne S, Bonala S, Henske E, Roegiers F, Markiewski M, Karbowniczek M (2016) Exosomes mediate the acquisition of the disease phenotypes by cells with normal genome in tuberous sclerosis complex. Oncogene 35:3027–3036. 10.1038/onc.2015.358 [DOI] [PubMed] [Google Scholar]
- Qin W, Kozlowski P, Taillon BE, Bouffard P, Holmes AJ, Janne P, Camposano S, Thiele E, Franz D, Kwiatkowski DJ (2010) Ultra deep sequencing detects a low rate of mosaic mutations in tuberous sclerosis complex. Hum Genet 127:573–582. 10.1007/s00439-010-0801-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roszkowska M, Skupien A, Wójtowicz T, Konopka A, Gorlewicz A, Kisiel M, Bekisz M, Ruszczycki B, Dolezyczek H, Rejmak E, Knapska E, Mozrzymas JW, Wlodarczyk J, Wilczynski GM, Dzwonek J (2016) CD44: a novel synaptic cell adhesion molecule regulating structural and functional plasticity of dendritic spines. Mol Biol Cell 27:4055–4066. 10.1091/mbc.E16-06-0423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito T, Hanai S, Takashima S, Nakagawa E, Okazaki S, Inoue T, Miyata R, Hoshino K, Akashi T, Sasaki M, Goto Y, Hayashi M, Itoh M (2011) Neocortical layer formation of human developing brains and lissencephalies: consideration of layer-specific marker expression. Cereb Cortex 21:588–596. 10.1093/cercor/bhq125 [DOI] [PubMed] [Google Scholar]
- Sia GM, Clem RL, Huganir RL (2013) The human language-associated gene SRPX2 regulates synapse formation and vocalization in mice. Science 342:987–991. 10.1126/science.1245079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundberg M, Tochitsky I, Buchholz DE, Winden K, Kujala V, Kapur K, Cataltepe D, Turner D, Han MJ, Woolf CJ, Hatten ME, Sahin M (2018) Purkinje cells derived from TSC patients display hypoexcitability and synaptic deficits associated with reduced FMRP levels and reversed by rapamycin. Mol Psychiatry 23:2167–2183. 10.1038/s41380-018-0018-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Switon K, Kotulska K, Janusz-Kaminska A, Zmorzynska J, Jaworski J (2017) Molecular neurobiology of mTOR. Neuroscience 341:112–153. 10.1016/j.neuroscience.2016.11.017 [DOI] [PubMed] [Google Scholar]
- Traub RD, Bibbig A, LeBeau FE, Buhl EH, Whittington MA (2004) Cellular mechanisms of neuronal population oscillations in the hippocampus in vitro. Annu Rev Neurosci 27:247–278. 10.1146/annurev.neuro.27.070203.144303 [DOI] [PubMed] [Google Scholar]
- Uhlmann EJ, Wong M, Baldwin RL, Bajenaru ML, Onda H, Kwiatkowski DJ, Yamada K, Gutmann DH (2002) Astrocyte-specific TSC1 conditional knockout mice exhibit abnormal neuronal organization and seizures. Ann Neurol 52:285–296. 10.1002/ana.10283 [DOI] [PubMed] [Google Scholar]
- Way SW, McKenna J 3rd, Mietzsch U, Reith RM, Wu HC, Gambello MJ (2009) Loss of Tsc2 in radial glia models the brain pathology of tuberous sclerosis complex in the mouse. Hum Mol Genet 18:1252–1265. 10.1093/hmg/ddp025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan E, Tsai PT, Greene-Colozzi E, Sahin M, Kwiatkowski DJ, Malinowska IA (2012) Graded loss of tuberin in an allelic series of brain models of TSC correlates with survival, and biochemical, histological and behavioral features. Hum Mol Genet 21:4286–4300. 10.1093/hmg/dds262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Horvath S (2005) A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol 4:Article17. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Pak C, Han Y, Ahlenius H, Zhang Z, Chanda S, Marro S, Patzke C, Acuna C, Covy J, Xu W, Yang N, Danko T, Chen L, Wernig M, Südhof TC (2013) Rapid single-step induction of functional neurons from human pluripotent stem cells. Neuron 78:785–798. 10.1016/j.neuron.2013.05.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zucco AJ, Pozzo VD, Afinogenova A, Hart RP, Devinsky O, D'Arcangelo G (2018) Neural progenitors derived from tuberous sclerosis complex patients exhibit attenuated PI3K/AKT signaling and delayed neuronal differentiation. Mol Cell Neurosci 92:149–163. 10.1016/j.mcn.2018.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
iPSC differentiation into neurons is highly efficient. iPSCs with three different genotypes of TSC2 (TSC2+/+, TSC2+/-, and TSC2-/-) were differentiated into neurons using induced expression of NGN2. a) Seven days after doxycycline, we did not observe expression of glial markers such as GFAP in any of the three lines. In addition, we did not observe markers of undifferentiated pluripotent cells such as b) NANOG or c) OCT4 in any of the three lines. Scale bars=50μm. Download Figure 1-1, TIF file (6MB, tif)
Dividing cell populations in neuronal cultures. iPSCs with three different genotypes of TSC2 (TSC2+/+, TSC2+/-, and TSC2-/-) were differentiated into neurons using induced expression of NGN2. a) Seven days after doxycycline, we evaluated for other non-pluripotent stem cells using immunostaining for Ki67. We observed a small fraction of Ki67-positive cells in cultures from all three genotypes, but the majority of these cells were also positive for TUBB3, consistent with a neuronal identity. Scale bars=50μm. Download Figure 1-2, TIF file (1.4MB, tif)
Characterization of iPSC-derived neurons. iPSCs with inducible NGN2 were treated with doxycycline and re-plated with astrocytes after six days to promote maturation. CTIP2, a marker of early lower layer cortical neurons, was also expressed in iPSC-derived neurons, suggestive of a pyramidal neuron fate. Scale bars=20μm. Download Figure 2-1, TIF file (1.4MB, tif)
Investigation of dysregulated signaling pathways due to loss of TSC2 using the ActivSignal platform. Three separate replicates of TSC2+/+ iPSC-derived neurons and TSC2-/- iPSC-derived neurons were analyzed using the ActivSignal platform. The first column identifies the protein that is being evaluated. The second column shows the fold change (TSC2-/- vs TSC2+/+). The third column shows the standard deviation. The fourth column shows the p-value calculated using a T-test, and the fifth column shows the correction for multiple comparisons using the FDR. Download Figure 3-1, XLSX file (53.9KB, xlsx)
Differentially expressed genes between TSC2+/- and TSC2+/+ neurons. The first column shows a unique identifier from the Ampliseq analysis pipeline. The second column shows the associated gene symbols, and the third column shows the associated full gene name. The fourth column shows the mean expression in TSC2+/+ neurons, while the fifth column shows the mean expression in TSC2+/- neurons. The sixth column shows the fold change between TSC2+/- and TSC2+/+ neurons, and the seventh and eighth columns show the p-value and false discovery rate, respectively. Download Figure 6-1, XLSX file (73.1KB, xlsx)
Differentially expressed genes between TSC2-/- and TSC2+/+ neurons. The first column shows a unique identifier from the Ampliseq analysis pipeline. The second column shows the associated gene symbols, and the third column shows the associated full gene name. The fourth column shows the mean expression in TSC2+/+ neurons, while the fifth column shows the mean expression in TSC2-/- neurons. The sixth column shows the fold change between TSC2-/- and TSC2+/+ neurons, and the seventh and eighth columns show the p-value and false discovery rate, respectively. Download Figure 6-2, XLSX file (176KB, xlsx)
Comparison of M51 co-expression module identified from gene expression data from iPSC-derived neurons with other datasets. a) This Venn diagram shows the overlap between the M51 co-expression modules that contains genes that are down-regulated concomitantly with loss of TSC2 alleles in iPSC-derived neurons and the M99 co-expression module that also contains genes whose down-regulation is related to loss of TSC2 alleles in iPSC-derived Purkinje cell neurons (Sundberg et al., 2018). b-d) These Venn diagrams shows the overlap between the M51 co-expression module and transcripts that were found to bind to FMRP (Brown et al., 2001; Darnell et al., 2011; Ascano et al., 2012). Download Figure 6-3, TIF file (432.7KB, tif)
Expression of FMRP in iPSC-derived cortical neurons. Western blot of FMRP from iPSC-derived neurons from all three iPSC lines at fourteen days after NGN2 induction with doxycycline. Quantification of FMRP levels from three separate differentiations of each line normalized to total protein shows no significant difference between the lines (ANOVA with Dunnett’s multiple comparison test). Download Figure 6-4, TIF file (174.2KB, tif)
Differentially expressed genes between cortical tubers and brain samples of unaffected individuals across two datasets (Martin et al., 2017; Mills et al., 2017). The first column shows the Ensembl transcript ID for the mapped gene, and the second column shows the associated gene symbol. The third column shows the fold change between the cortical tuber and samples from unaffected individuals. The fourth and fifth columns show the p-value and false discovery rate of the comparison between the cortical tubers and samples from unaffected individuals. Download Figure 6-5, XLSX file (194.5KB, xlsx)
Comparison of M8 co-expression module identified from gene expression data from iPSC-derived neurons with differentially expressed genes from cortical tubers. This Venn diagram shows the overlap between the M8 co-expression module and genes that are up-regulated in cortical tubers across two datasets (Martin et al., 2017; Mills et al., 2017). Download Figure 6-6, TIF file (75.8KB, tif)
mTORC1 inhibition reduces hyperactivity and gene expression changes in TSC2-/- neurons. a) Weighted mean firing rate of iPSC-derived neurons from TSC2+/+, TSC2+/-, and TSC2-/- neurons treated with rapamycin or vehicle. Each point represents 8 independent wells per treatment per genotype. Values for each well were fit to a sigmoidal curve, and differences between coefficients for each genotype were calculated using a Z-test. Graph shows mean ± SEM. Download Figure 7-1, TIF file (174KB, tif)