Leptospirosis is one of the most widespread zoonoses caused by pathogenic Leptospira spp. In this study, we report that the LIC11966/ErpY-like lipoprotein is a surface-exposed outer membrane protein exclusively present in pathogenic species of Leptospira. The recombinant ErpY (rErpY)-like protein is recognized by the immunoglobulins of confirmed leptospirosis sera of diverse hosts (human, bovine, and canine), suggesting the expression of the native leptospiral surface protein during infection.
Keywords: Leptospira, lipoprotein, antibodies, ELISA, recombinant protein, extracellular matrix component, LIC11966, factor H, fibrinogen
ABSTRACT
Leptospirosis is one of the most widespread zoonoses caused by pathogenic Leptospira spp. In this study, we report that the LIC11966/ErpY-like lipoprotein is a surface-exposed outer membrane protein exclusively present in pathogenic species of Leptospira. The recombinant ErpY (rErpY)-like protein is recognized by the immunoglobulins of confirmed leptospirosis sera of diverse hosts (human, bovine, and canine), suggesting the expression of the native leptospiral surface protein during infection. Circular dichroism of pure rErpY-like protein showed the secondary structural integrity to be uncompromised during the purification process. Analysis of the rErpY-like protein by native polyacrylamide gel electrophoresis, chemical cross-linking, dynamic light scattering, and field emission transmission electron microscopy demonstrated it undergoes supramolecular assembly. The rErpY-like protein can bind to diverse host extracellular matrices, and it presented a saturable and strong binding affinity (dissociation constant [KD] of 70.45 ± 4.13 nM) to fibrinogen, a central host plasma component involved in blood clotting. In the presence of the rErpY-like supramolecule, thrombin-catalyzed fibrin clot formation is inhibited up to 7%, implying its role in inhibiting blood coagulation during Leptospira infection. In addition, binding of the rErpY-like supramolecule to complement factors H and I suggests the protein also contributes to Leptospira evading innate host defense during infection by inactivating alternative complement pathways. This study reveals that rErpY-like protein is functionally active in the supramolecular state and performs moonlighting activity under the given in vitro conditions.
INTRODUCTION
Leptospirosis, a widespread zoonosis, especially in the tropical regions of the world, is caused by the pathogenic species of the spirochete Leptospira (1). It is an infectious disease found in farmers, veterinarians, abattoir workers, and people working in close contact with animals, with more than one million cases reported each year (2). Leptospirosis is among the most underdiagnosed diseases owing to its wide range of symptoms, ranging from jaundice to renal failure (1). The most severe forms of leptospirosis are known as Weil’s syndrome, where pulmonary hemorrhage may result in mortality rates of up to 70% (3–5). The molecular understanding of pathogenicity and virulence of leptospires is still in the early stages. The origin of pathophysiological symptoms of leptospirosis and the severity of disease remain virtually unknown (6, 7).
The comprehensive interrogation of host-pathogen interplay targeting outer membrane proteins of Leptospira has been actively under study to understand its pathophysiology. However, to date, only a few virulence factors of Leptospira have been functionally characterized and well understood. It is now established that Leptospira adherence with the host cells, extracellular matrix, and plasma proteins contributes to bacterial dissemination and host immune evasion (8). Various pieces of evidence for the exploitation of host plasma proteins, like complement factors (9, 10), plasminogen (10, 11), ferritin (12), and fibrinogen (Fg) (13), by the leptospires have been reported. The advent of the whole-genome sequence of Leptospira established that a large share of genes represent putative proteins with no identified function or are exclusively present only in pathogenic species of Leptospira (14). Several such leptospiral proteins (13, 15, 16) have been reported to interact with human Fg and complement regulatory proteins. Such binding proteins benefit the bacteria in intervening thrombin-catalyzed clot formation or inhibiting complement activation, essential for successful establishment in the host and impeding the innate defense system. In a recent study, a protein annotated ErpY-like (LIC11966) in Leptospira was demonstrated to be an Fg-binding protein with diagnostic and subunit vaccine potential (17–19). The ErpY protein annotation originated from outer surface protein E/F-related proteins of another pathogenic spirochete, Borrelia burgdorferi (20). In the genus Borrelia, erp genes have been subdivided into three distinct gene families, ospE, ospF, and elp, where ErpY belongs to OspF protein family (21). These erp genes possess well-conserved leader polypeptide sequences and encode highly charged lipoproteins (large number of lysine and glutamate residues) localized to the bacterial outer surface (22). The first description of LIC11966 as an ErpY-like lipoprotein of Leptospira (17) was given due to its 26% sequence identity with ErpY of B. burgdorferi. The ErpY-like protein with no known function is conserved among pathogenic Leptospira spp., with up to 99% pairwise sequence identity. The evaluation of recombinant ErpY (rErpY)-like protein as a diagnostic antigen for leptospirosis has not been done extensively in bovines and canines to date. Moreover, being a conserved protein exclusively in pathogenic Leptospira, it may be associated with virulence. The present study deals with the characterization of a putative ErpY-like lipoprotein of Leptospira interrogans, emphasizing its subcellular location, diagnostic importance, and virulence activity.
RESULTS
In silico analysis of LIC11966/ErpY-like protein.
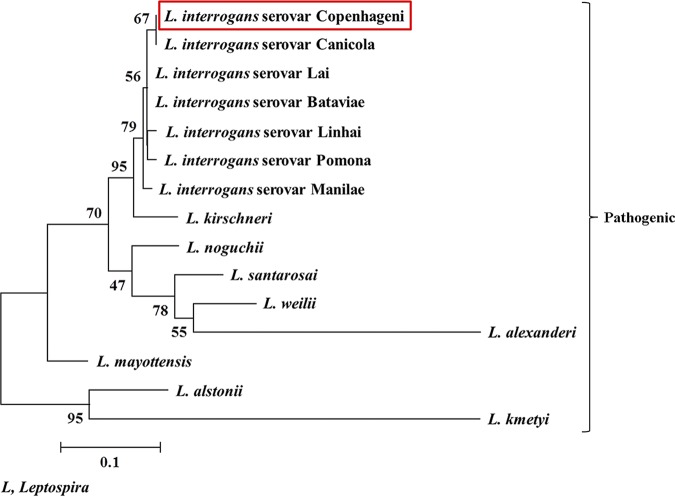
Bioinformatics analysis of LIC11966 using the SignalP 5.0 program (23) predicted a signal peptide with the cleavage site between the 22nd and 23rd residues at the N terminus. Also, the amino acid sequence of LIC11966 (159 residues) was analyzed manually to identify its signal peptide with the criteria set for spirochetal lipoproteins (24). The signal peptide cleavage site in LIC11966 lipoprotein by signal peptidase (Lsp) was consistent with the findings predicted through the SignalP 5.0 program. The signal peptide (22 residues) of LIC11966 fulfills all the criteria set for a spirochete protein to be categorized as a lipoprotein. The PSORT program (25) predicted LIC11966 to be localized more toward the periplasmic region than the outer membrane of Leptospira. The orthologs of LIC11966 were found to be highly conserved among the serovars of L. interrogans, with 99 to 100% sequence identity on BLAST search analysis. The phylogenetic tree constructed using the 14 representative leptospiral LIC11966 orthologs shows a high level of sequence conservation among pathogenic species of Leptospira and with the lowest sequence identity of 57% (Table 1 and Fig. 1).
TABLE 1.
Comparative analyses of the LIC11966/ErpY-like protein orthologs among Leptospira species
| Leptospira species (serovar) | Query coverage (%) | Sequence identity (%) | NCBI accession no. |
|---|---|---|---|
| L. interrogans (Canicola) | 100 | 100 | OCC30350.1 |
| L. interrogans (Lai) | 100 | 99 | NP_712120.1 |
| L. interrogans (Linhai) | 100 | 99 | AJR14687.1 |
| L. interrogans (Manilae) | 100 | 99 | EYU63405.1 |
| L. interrogans (Bataviae) | 100 | 99 | OAM75663.1 |
| L. interrogans (Pomona) | 100 | 99 | EMI70432.1 |
| L. kirschneri | 100 | 92 | WP_004755938.1 |
| L. noguchii | 100 | 85 | WP_002151500.1 |
| L. mayottensis | 100 | 79 | WP_117340238.1 |
| L. santarosai | 100 | 78 | WP_004476249.1 |
| L. weilii | 98 | 80 | WP_002622075.1 |
| L. alstonii | 98 | 71 | WP_020772825.1 |
| L. kmetyi | 98 | 57 | WP_040912836.1 |
| L. alexanderi | 76 | 64 | WP_078128751.1 |
FIG 1.
Phylogenetic analysis of Leptospira spp. based on the amino acid sequence of LIC11966/ErpY-like protein of L. interrogans serovar Copenhageni by the maximum likelihood method. The amino acid sequence of ErpY-like protein was retrieved from the NCBI protein database, and a total of 14 orthologs of ErpY-like protein were retrieved through NCBI protein BLAST. The obtained sequences were aligned, and the phylogenetic tree was constructed using the MEGA, version 7.0.26, program. The tree with the highest log likelihood (−987.90), inferred following 1,000 bootstrap replications, is shown where a bootstrap value of greater than 50 indicates the reliability of the data. The tree was drawn to scale, with branch lengths measured in the number of substitutions per site. The resulting phylogram shows that ErpY-like protein (red box) is exclusively present in pathogenic species of Leptospira with a high level of sequence conservation.
ErpY-like protein is found exclusively in the pathogenic Leptospira genome and is a predominant outer membrane surface-exposed protein.
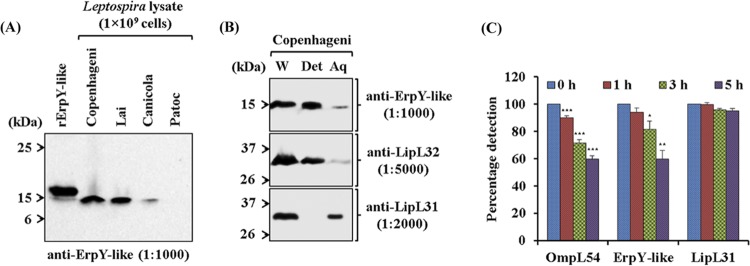
Immunoblot assay using anti-ErpY-like antibody specifically detected the native ErpY-like protein (∼15 kDa) in the lysates (1 × 109 cells) of pathogenic L. interrogans serovars (Copenhageni, Lai, and Canicola) (Fig. 2A). However, there was no detection of native ErpY-like protein in the lysate (1 × 109 cells) of nonpathogenic Leptospira biflexa serovar Patoc. Immunoblot assay detected the rErpY-like (∼18 kDa) protein at relatively larger molecular size than the native ErpY-like (∼15 kDa) protein due to the presence of an additional N-terminal vector fusion of 2.5 kDa in recombinant protein (Fig. 2A). These results suggest that the in vitro-cultured Leptospira (IVCL) expresses the native ErpY-like protein and shares an immunogenic epitope with the recombinant protein.
FIG 2.
Characterization of the recombinant Leptospira ErpY-like protein. (A) Immunoblot analysis of recombinant and native ErpY-like antigen using anti-ErpY-like protein antibody. Laboratory animal (mouse)-generated polyclonal antisera (1:1,000) after immunization with rErpY-like protein can detect both the native ErpY-like antigen in the pathogenic Leptospira (L. interrogans serovar Copenhageni, Lai, or Canicola) and the purified rErpY-like antigen (200 ng). The lysates of nonpathogenic L. biflexa serovar Patoc were used as a negative control in the immunoblot. (B) Phase partitioning of whole-cell protein of L. interrogans serovar Copenhageni using Triton X-114 and its immunoblot with the anti-rErpY-like antibody. Live leptospires were subjected to 1% Triton X-114 to partition proteins into detergent (Det) and aqueous-phase (Aq) fractions. Phase-partitioned proteins were resolved on 12% SDS-polyacrylamide gel, electroblotted on nitrocellulose membrane, and probed with anti-ErpY-like (1:1,000), anti-LipL32 (1:5,000), or anti-LipL31 (1:2,000) antibody. (Top) The ErpY-like protein was detected (15 kDa) predominantly in the detergent phase. LipL32, a known outer membrane protein of Leptospira, was detected predominantly in the detergent phase (middle), whereas a leptospiral cytoplasmic membrane protein, LipL31, was detected in the aqueous phase (bottom). Whole-cell lysate of L. interrogans serovar Copenhageni (W) was used in the immunoblot as a marker for ErpY-like protein, LipL32, and LipL31. (C) Protease accessibility assay of L. interrogans serovar Copenhageni using proteinase K (PK). Live leptospire suspensions (5 × 108 cells per suspension) were incubated with 5 μg of PK at the indicated time points. The suspensions were harvested, washed, resuspended in PBS, and coated on a microtiter plate (5 × 107 cells per well). In ELISA, the reactivity of ErpY-like protein with its antiserum showed a perpetual decrease in signal from 0 to 5 h of PK treatment. A similar trend of reduction in signal was observed for OmpL54, a known surface-exposed outer membrane protein of Leptospira. The serological detection of antigen LipL31, a known cytoplasmic membrane protein of Leptospira, demonstrates the uncompromised integrity of leptospire inner membrane during treatment with PK. Error bars represent SD from three replicates and are indicative of two independent experiments. Statistical analysis was performed using Student's t test by comparing the signals obtained for 0 h and other time points of treatment with proteinase K (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
To determine the ErpY-like protein subcellular location, detergent extraction and phase partition of L. interrogans serovar Copenhageni were performed using Triton X-114. Immunoblot assay of the phase-partitioned fractions of Leptospira detected the ErpY-like protein predominantly in the hydrophobic detergent phase, indicating that the ErpY-like protein is an outer membrane protein (Fig. 2B). To validate the subcellular localization experiment in Leptospira using Triton X-114, known LipL32 and LipL31 proteins, located in the outer (26) and cytoplasmic membrane (27), respectively, were probed with specific antibodies in the phase-partitioned fraction. The assessment of ErpY-like protein as a surface-exposed protein of Leptospira also was executed by protease accessibility assay using proteinase K (PK) treatment at different time intervals on live spirochetes. At a range of 1 to 5 h of proteinase treatment on live spirochetes, a gradual decrease in the absorbance for detection of ErpY-like protein in the enzyme-linked immunosorbent assay (ELISA) suggested it was a surface-exposed protein of Leptospira (Fig. 2C). Moreover, a known leptospiral surface-exposed protein, OmpL54, and a cytoplasmic membrane protein, LipL31 (28), were used as controls to validate the protease accessibility assay in Leptospira. During the various intervals of proteinase treatment on leptospires, an insignificant change in the serological detection of LipL31 indicates that the cellular integrity of the Leptospira inner membrane was uncompromised (Fig. 2C). On the other hand, the reduction in absorbance for detecting OmpL54 after proteinase treatment validates the quality of the protease accessibility assay performed in this study and agrees with results published elsewhere (28).
Recognition of rErpY-like protein by serum obtained during natural Leptospira infection.
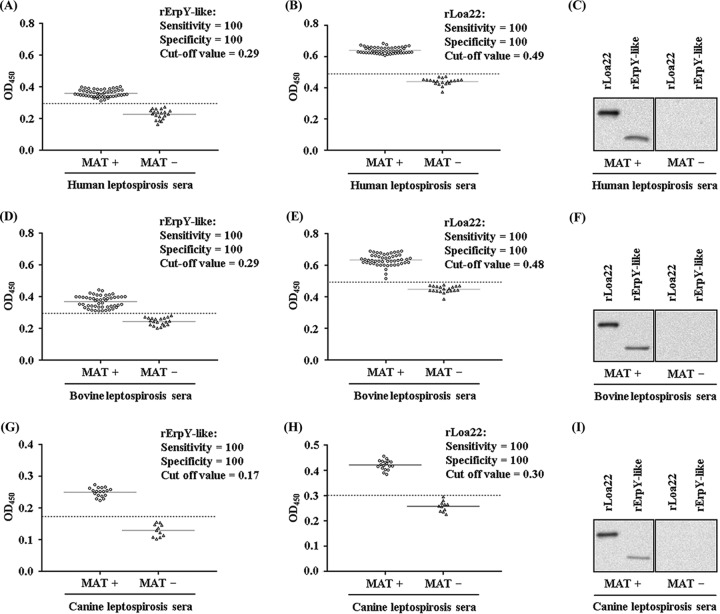
The naturally infected hosts of Leptospira species were evaluated for their ability to recognize rErpY-like protein as an antigen. Indirect ELISA was conducted for comparing the recognition of rErpY-like antigen with sera of broad host range (human, bovine, and canine) that tested positive for leptospirosis by the microscopic agglutination test (MAT). As a control, serum samples of each host (human, bovine, and canine) that tested negative for leptospirosis by the MAT were included for assessment of recognition of rErpY-like antigen. The antigen recombinant Loa22 (rLoa22), a known serological diagnostic candidate of Leptospira (29), was added to validate the MAT results and compare the sensitivity/specificity of rErpY-like protein as a potential antigen. To further substantiate our assessment, immunoblots were performed with the pooled serum samples of the host (human/bovine/canine) that tested positive or negative for leptospirosis by MAT. The calculated sensitivity and specificity of the indirect ELISA to recognize antigens (rErpY-like and rLoa22) with human serum were both 100% (Fig. 3A and B). However, the measured cutoff value of absorbance for detecting rErpY-like antigen (0.29) was lower than that for rLoa22 (0.49) in humans naturally infected with Leptospira (Table 2). On the same note, immunoblots performed with MAT-positive pooled human serum could detect rLoa22 and rErpY-like antigens, unlike the MAT-negative pooled serum for leptospirosis (Fig. 3C). It is now well established that Leptospira species have a wide host range where they can sustain themselves, and these hosts respond immunologically in diverse ways during leptospirosis; therefore, it was interesting to evaluate the reactivity of immunoglobulins from sera of infected hosts to rErpY-like protein. Under similar experimental conditions, immunoassay was performed with the bovine serum samples that tested positive or negative for leptospirosis by MAT. In a trend the same as that of human serum for detecting antigens (rErpY-like and rLoa22) in this study, the bovine leptospirosis serum also scored 100% sensitivity and specificity (Fig. 3D and E). In agreement with the ELISA results, immunoblotting performed with the pooled bovine serum (MAT positive) detected rLoa22 and rErpY-like antigen (Fig. 3F). The serum samples that were MAT positive for leptospirosis in canines also detected antigens (rErpY-like and rLoa22) with specificity and sensitivity of 100% (Fig. 3G and H). Among the tested sera of various hosts in this study, canine serum used for serological detection of leptospirosis demonstrated the lowest cutoff to detect both rErpY-like antigen (0.17) and rLoa22 (0.30) (Table 2). Nevertheless, immunoblotting with canine serum samples (MAT positive) could detect rLoa22 and rErpY-like antigen at an adequate level compared to that of MAT-negative serum samples (Fig. 3I). The obtained data for detecting rErpY-like antigen by ELISA were compared with those for the MAT using Cohen’s kappa coefficient value. The calculated kappa value (kappa, 1.0) for detecting leptospirosis in the serum samples of the different hosts (human, bovine, and canine) can be interpreted as very good strength of agreement.
FIG 3.
Leptospirosis-positive sera of various hosts recognize recombinant ErpY-like protein. ELISA was performed with two antigens (rErpY-like and rLoa22) independently to determine the reactivity of human, bovine, and canine sera testing MAT positive or negative for leptospirosis. Serum sample reactivity against the two antigens was recorded in triplicate. The mean absorbance values obtained from each serum sample are demarcated as empty circles or as empty triangles for leptospirosis MAT-positive or -negative samples, respectively. Solid black horizontal lines represent the means for each group. The cutoff value (dashed black horizontal lines) of the assay was derived from the mean absorbance values obtained from leptospirosis MAT-negative samples (control group) plus two SD for the antigen used. (A) ELISA to detect rErpY-like antigen (400 ng) using MAT-positive (n = 50) and -negative (n = 20) serum samples of human (1:100) for leptospirosis. Infected human sera recognized the rErpY-like protein with 100% sensitivity and specificity. OD450, optical density at 450 nm. (B) ELISA to detect rLoa22 (400 ng) antigen using MAT-positive and -negative human serum samples for leptospirosis. Infected human sera recognized the rLoa22 antigen with 100% sensitivity and specificity. (C) Immunoblot of antigens (rErpY-like and rLoa22) using the pooled human serum. (Left) Pooled human serum testing MAT positive for leptospirosis recognized both antigens. (Right) In contrast, there was no recognition of both antigens using a pooled human serum testing negative for leptospirosis. (D) ELISA to detect rErpY-like antigen using bovine serum samples testing MAT positive (n = 50) or negative (n = 20) for leptospirosis. (E) ELISA to detect rLoa22 antigen using bovine serum samples testing MAT positive or negative for leptospirosis. (F) Immunoblot using pooled bovine serum testing MAT positive or negative for leptospirosis. (G) ELISA to detect rErpY-like antigen using canine serum samples testing MAT positive (n = 18) or negative (n = 18) for leptospirosis. (H) ELISA to detect rLoa22 antigen using canine serum samples testing MAT positive or negative for leptospirosis. (I) Immunoblot using pooled canine serum testing MAT positive or negative for leptospirosis.
TABLE 2.
Measured ELISA data for the recognition of antigens (rErpY-like protein and rLoa22) using sera of hosts (human, bovine, and canine) testing positive or negative for leptospirosis by MAT
| Host and MAT serum result (no. of samples) | Recombinant protein | OD (means ± SD) | Cutoff (means + 2 SD) | Sensitivity (%) | Specificity (%) |
|---|---|---|---|---|---|
| Human | |||||
| Positive (50) | rErpY-like | 0.3587 ± 0.0230 | _ | 100 | _ |
| rLoa22 | 0.6339 ± 0.0360 | _ | 100 | _ | |
| Negative (20) | rErpY-like | 0.2268 ± 0.0308 | 0.2883 | _ | 100 |
| rLoa22 | 0.4478 ± 0.0212 | 0.4903 | _ | 100 | |
| Bovine | |||||
| Positive (50) | rErpY-like | 0.3684 ± 0.0373 | _ | 100 | _ |
| rLoa22 | 0.6404 ± 0.0194 | _ | 100 | _ | |
| Negative (20) | rErpY-like | 0.2431 ± 0.0236 | 0.2902 | _ | 100 |
| rLoa22 | 0.4401 ± 0.0223 | 0.4846 | _ | 100 | |
| Canine | |||||
| Positive (18) | rErpY-like | 0.2497 ± 0.0143 | _ | 100 | _ |
| rLoa22 | 0.4215 ± 0.0191 | _ | 100 | _ | |
| Negative (11) | rErpY-like | 0.1291 ± 0.0198 | 0.1687 | _ | 100 |
| rLoa22 | 0.2578 ± 0.0207 | 0.2992 | _ | 100 |
Interaction of rErpY-like protein with host ECM and plasma components.
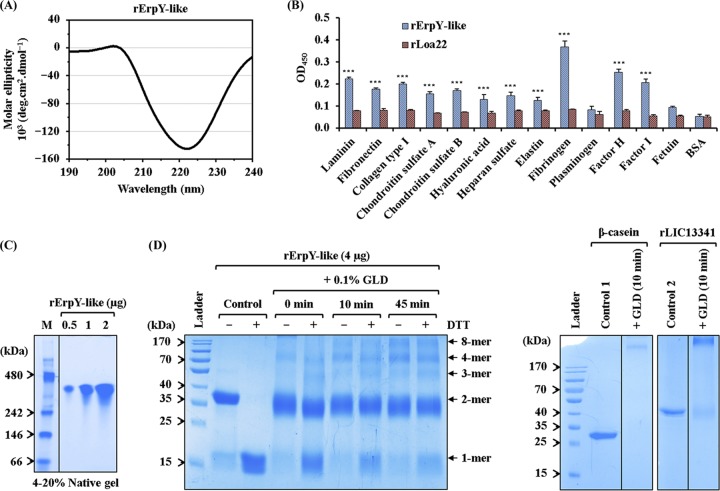
Several leptospiral surface proteins have been established to mediate pathogen colonization and dissemination in host tissues by adhering to extracellular matrix (ECM) proteins (13, 30). Moreover, Leptospira binding to host-specific plasma components through its surface-exposed membrane proteins may cause an imbalance in physiological processes like blood coagulation and complement activation cascade in the host (31). Before attempting to understand the role of ErpY-like protein as an outer membrane protein of Leptospira, we first evaluated the secondary structure integrity of rErpY-like protein by circular dichroism (CD) spectroscopy (Fig. 4A). The obtained experimental CD spectral data for rErpY-like protein using the K2D2 program (32) predicted 87.6% α-helix and 0.5% β-strand. The secondary structure prediction for native ErpY-like protein using the PSIPRED program (34) was close to the experimental CD data, indicating its suitability for downstream biochemical studies. Using ELISA, we evaluated the ability of rErpY-like protein to bind to a spectrum of host ECM and plasma components. Among all the host ligands (laminin, fibronectin, collagen type I, chondroitin sulfate A and B, hyaluronic acid, heparan sulfate, elastin, fibrinogen, plasminogen, factor H, and factor I) examined for interaction with rErpY-like protein using ELISA, a higher absorbance for binding to Fg, complement factor H (CFH), and factor I (CFI) was evident (Fig. 4B). This binding study of rErpY-like protein to ECM leads us to hypothesize that during natural Leptospira infection, ErpY-like protein plays a moonlighting role in disrupting blood coagulation processes and evading complement-mediated lysis of Leptospira.
FIG 4.
Recombinant ErpY-like protein binds to host extracellular matrix and plasma components. (A) Far-UV (190 to 240 nm) CD spectra of rErpY-like protein. The analyzed CD spectral data show the predominance of α-helix (87.6%), which is in agreement with the theoretically predicted secondary structure of the rErpY-like protein. The spectra are represented as averages from three scans with a scanning speed of 100 nm min−1. (B) Interaction between rErpY-like protein and commercial host ECM components. The binding of rErpY-like protein to ECM components (laminin, fibronectin, collagen type I, chondroitin sulfate A and B, hyaluronic acid, heparan sulfate, and elastin) or plasma components (fibrinogen, plasminogen, factor H, and factor I) was analyzed through ELISA. The plasma components fetuin (highly glycosylated protein) and bovine serum albumin fraction V (nonglycosylated protein) were included as negative controls for ligands. rLoa22, which is known to interact moderately with host ECM components, was included in the assay as a negative-control protein. Protein-specific antisera (anti-ErpY-like protein or anti-Loa22 antibody) were used to measure the interaction of recombinant proteins with host ligands. The rErpY-like protein was recorded to interact preferentially with plasma fibrinogen, followed by factor H and other host ligands. The bar represents mean absorbance values (450 nm) for a ligand. Error bars are indicative of SD from two independent experiments, each performed in triplicate. For statistical analyses, the binding of rErpY-like protein with host ligands was compared to that with fetuin or BSA by the two-tailed t test (***, P < 0.001). (C) Pure rErpY-like protein forms a supramolecule. The purified rErpY-like protein (0.5, 1, and 2 μg) was resolved on a 4 to 20% native-PAGE and stained with Coomassie blue. The standard native molecular weight marker (M) was run in lane 1. The rErpY-like protein resolved at around 300 kDa. The gel in this figure has been spliced for labeling purposes. (D) Denaturing polyacrylamide gel electrophoresis of rErpY-like protein after glutaraldehyde (GLD) cross-linking. Chemical cross-linking of rErpY-like protein with glutaraldehyde (0 to 45 min) shows multiple higher-molecular-weight bands when resolved in the presence (+) or absence (−) of DTT. In the panel on the right, the cross-linked products of two control proteins, β-casein and rLIC13341, demonstrate only one band of larger size.
Interestingly, the binding of the rErpY-like protein to another plasma component like plasminogen was statistically insignificant compared to that of the ligands fetuin and bovine serum albumin (BSA; negative control) (Fig. 4B). Moreover, a negative-control protein, rLoa22, used in the assay showed moderate binding with all the examined ECM and plasma components of the host (Fig. 4B) and agrees with an earlier report (35). The moderate binding of rLoa22 to ECM components validates the experimental procedures and the quality of ligands used in this assay. Binding of the rErpY-like protein to host plasma components like CFH/I and Fg prompted us to evaluate if rErpY-like protein tends to undergo supramolecular assembly. To determine the self-assembly property of rErpY-like protein, the purified recombinant protein was resolved on native-PAGE and stained with Coomassie blue. In the native-PAGE gel, the proteins migrate according to not only their size but also their shape and hydrodynamic properties, and therefore the results may not represent the actual size of the protein. The apparent molecular weight of rErpY-like protein resolved on native-PAGE was found to be around 300 kDa (Fig. 4C). The ability of the rErpY-like protein to form supramolecules, as evidenced by native-PAGE images, encouraged us to find whether chemical cross-linking with glutaraldehyde could detect the transient assembly of rErpY-like protein subunits on denaturing polyacrylamide gel. As expected, multiple transient bands of cross-linked rErpY-like protein corresponding to 1-, 2-, 3-, 4-, and 8-mers were evident on SDS-PAGE stained with Coomassie blue until 45 min of the cross-linking reaction (Fig. 4D). In contrast, cross-linked products of two control proteins, β-casein and recombinant protein LIC13341 of Leptospira, previously studied in our laboratory (26), show only one transient cross-linked band (Fig. 4D, right). Among all the multiple bands visualized, the 2-mer band of rErpY-like protein appeared in the majority of bands when resolved in the absence of the reducing agent dithiothreitol (DTT). We further looked into the hydrodynamic size of the supramolecule rErpY-like protein by dynamic light scattering, as the 12% SDS-PAGE cannot resolve the cross-linked product of more than 170 to 200 kDa.
DLS and FETEM of rErpY-like protein.
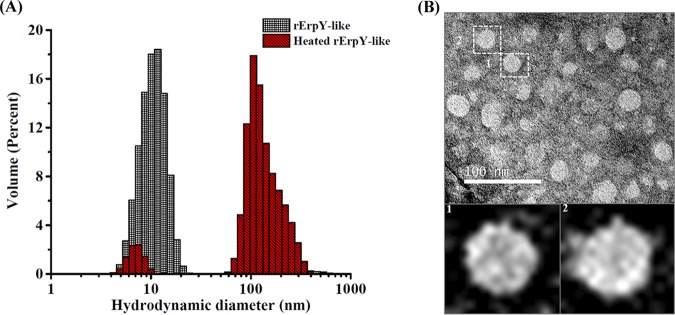
Dynamic light scattering (DLS) was performed to determine the size distribution profile of the purified rErpY-like oligomeric protein or its heat-denatured form in solution. The size distribution profile of purified rErpY-like protein using DLS demonstrated a homogenous distribution of a single major population species with a mean hydrodynamic diameter (DH) of 10.88 ± 5.18 nm and with an estimated molecular weight of 177 ± 84.3 kDa (Table 3). On the other hand, the heat-denatured rErpY-like protein demonstrated a heterogeneous size distribution, composed of a significant population with a DH of 163.6 ± 57.92 nm and a minor population with a mean DH of 6.94 ± 1.03 nm. Such a shift in size distribution profile of heat-denatured rErpY-like protein suggests rErpY-like protein has aggregated, whereas the pure rErpY-like protein in solution tends to form a supramolecule with homogenous size distribution (Fig. 5A). The values of particle polydispersity, polydispersity index, count rate, volume, percent intensity, and estimated MW (molecular weight) of major peaks of rErpY-like protein are shown in Table 3. The supramolecule rErpY-like protein topography was also imaged using field emission transmission electron microscopy (FETEM). A negative-stain FETEM image of the rErpY-like supramolecule demonstrates the homogenous distribution of the particles with identical spherical shape and size (∼20 nm diameter) throughout the grid (Fig. 5B).
TABLE 3.
Dynamic light scattering data of rErpY-like protein
| Sample | Particle Pda (nm) | Pd index | Count rate (cps) | Vol % (mass %) | Intensity (%) | Estimated MWb (kDa, means ± SD) |
|---|---|---|---|---|---|---|
| rErpY-like | 5.348 | 0.282 | 232,700 | 99 | 89.7 | 177 ± 84.3 |
| Heat-treated rErpY-like | 70.38 | 0.33 | 533,300 | 91.4 | 92.6 | 1.01 × 105 ± 1.49 × 104 |
Pd, polydispersity.
Data shown are for the major peaks.
FIG 5.
Analyses of rErpY-like supramolecule by DLS and FETEM. (A) Size distribution profile of purified rErpY-like protein by dynamic light scattering. The purified rErpY-like protein shows a single major peak with an average hydrodynamic diameter of 10.88 ± 5.18 nm, whereas the heat-denatured protein shows a shift in the size distribution of the major peak with an average diameter of 163.6 ± 57.92 nm and a minor peak corresponding to the nonaggregated rErpY-like protein (red striped bars). The representative size distribution data of the 30 scans are presented here. (B) Negative-stain FETEM image of rErpY-like protein. FETEM image demonstrates the self-assembly of the rErpY-like protein monomer into spherical particles of nearly uniform shape and size (∼20-nm diameter) throughout the grid. The sample image was acquired at ×40,000 magnification. Micrograph scale bar, 100 nm. For clarity, particle 1 and 2 magnified images are shown.
Characterization of rErpY-like protein interaction with human fibrinogen.
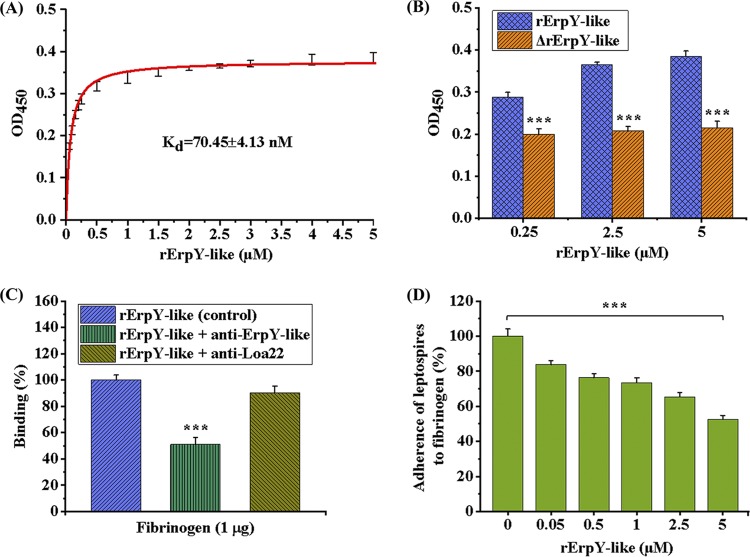
The optimal binding of the rErpY-like protein to human Fg was evaluated on a quantitative basis through indirect ELISA. A dose-dependent and saturable binding curve was obtained when increasing concentrations of the rErpY-like protein (0 to 5 μM) were allowed to bind to a fixed amount of human Fg (1 μg) onto a microtiter plate. The rErpY-like protein binding to Fg ligand achieved saturation at 5 μM concentration of recombinant protein, demonstrating the interaction was specific (Fig. 6A). The value of calculated dissociation constant (KD) of rErpY-like protein binding to ligand Fg was 70.45 ± 4.13 nM (Fig. 6A). To determine if the structural epitopes of rErpY-like protein were critical for the binding to Fg, the recombinant protein was heat denatured for 10 min at 95°C prior to its binding to the ligand. Severe reduction in binding of the heat-denatured rErpY-like protein at different concentrations to Fg was observed, in contrast to results with the structurally intact rErpY-like protein (Fig. 6B). The binding assay of the structurally intact or heat-denatured rErpY-like protein to Fg along with the DLS data suggests that ligand binding is dependent on both structural conformation and assembly of supramolecule subunits of recombinant protein. Also, the role of immunogenic epitopes of rErpY-like protein in interaction with host Fg was assessed by allowing the rErpY-like protein to bind with its specific immunoglobulins (serum-containing anti-rErpY-like) before determining the binding reaction to Fg. The specific immunoglobulins inhibited (51%) the interaction of rErpY-like protein to ligand Fg, unlike the control, where no serum was used (Fig. 6C). The serum anti-Loa22 utilized as a source of nonspecific antibody demonstrated insignificant inhibition (5%) of rErpY-like protein binding to Fg (Fig. 6C), suggesting that immunogenic epitopes of rErpY-like protein are essential in the binding to Fg.
FIG 6.
Binding characterization of the rErpY-like protein to human fibrinogen. (A) Dose-dependent binding of the rErpY-like protein to human fibrinogen. Fibrinogen (1 μg/well)-coated microtiter plates were incubated with increasing concentrations of rErpY-like protein (0.05 to 5 μM), and the ligand interaction was detected by anti-ErpY-like antibody (1:1,000). The mean absorbance values of rErpY-like protein binding to fibrinogen as a function of rErpY-like protein concentration are shown. Binding saturation was recorded at 5 μM rErpY-like protein with a dissociation constant (KD) of 70.45 ± 4.13 nM. (B) Heat-denatured rErpY-like protein fails to bind human fibrinogen. The rErpY-like protein was heat denatured for 10 min at 95°C. The untreated (control) or heat-denatured rErpY-like protein (0.25 to 5 μM) was allowed to interact with fibrinogen-coated microtiter plates. In contrast to dose-dependent binding of the untreated rErpY-like protein to fibrinogen, no significant change in binding to fibrinogen was observed with increasing concentrations of heat-denatured rErpY-like (ΔrErpY-like) protein. (C) Specific antibody-mediated inhibition of rErpY-like antigen binding to human fibrinogen. The rErpY-like protein was incubated individually with the antibody against rErpY-like/rLoa22/blank before its interaction with fibrinogen-coated microtiter plates. The specific antibody-antigen complex generation showed a reduction in interaction with fibrinogen (51%) relative to the control with no antibody supplementation. In contrast, a relative reduction of 5% was observed when serum against rLoa22 antigen was used as a nonspecific antibody control. (D) Presence of rErpY-like protein inhibits live leptospire adherence to human fibrinogen. Adhesion of leptospires to fibrinogen was reduced in the presence of an increasing concentration of rErpY-like protein. Fibrinogen-bound leptospires were quantified by anti-Loa22 antibody, and the absorbance obtained in the absence of rErpY-like protein was measured as 100%. The live leptospire adherence to fibrinogen was reduced to 53% in the presence of a saturated concentration (5 μM) of rErpY-like protein. The binding measurements were performed in three replicates, and the data shown are representative of two independent experiments. Statistical analyses were performed by the two-tailed t test (***, P < 0.001).
We were interested in evaluating the effect of rErpY-like protein on adherence of live Leptospira organisms, expressing the native ErpY-like surface protein, to Fg. Indirect ELISA quantified the impact of the presence of rErpY-like protein on adherence of leptospires to Fg. Live leptospires were allowed to competitively adhere to Fg preincubated with different concentrations of rErpY-like protein (0 to 5 μM). Fibrinogen-bound leptospires were quantified using antiserum against Loa22, a predominant surface-exposed leptospiral membrane protein (36). A dose-dependent reduction in the number of leptospires adhering to Fg preincubated with increasing rErpY-like protein concentrations indicates specific interaction of ErpY-like protein with the Fg. It turned out that only 53% of leptospires could bind to Fg in the presence of saturable binding concentration (5 μM) of the rErpY-like protein (Fig. 6D).
Inhibition of fibrin clot formation catalyzed by thrombin.
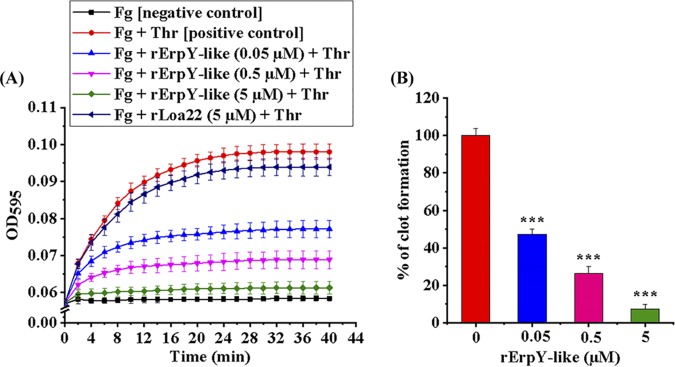
The binding ability of the rErpY-like protein to plasma component Fg prompted us to address whether the presence of rErpY-like protein can interfere in the thrombin-catalyzed conversion of soluble fibrinogen into insoluble fibrin, a physiological reaction essential for initiating blood clot formation. The enzymatic conversion of soluble fibrinogen into insoluble fibrin can be detected by measuring the increase in absorbance (595 nm). Therefore, the kinetics of fibrin formation from fibrinogen, catalyzed by thrombin in the presence or absence of increasing concentrations (0 to 5 μM) of rErpY-like protein, was measured (Fig. 7A). The thrombin-catalyzed clot formation in the absence of rErpY-like protein demonstrated maximal clotting (100%; positive control), whereas the fibrinogen clot reaction with no added thrombin showed a lack of any measurable clotting (negative control). rLoa22, a known protein of Leptospira with moderate ability to bind to Fg, was used as another control in thrombin-catalyzed clot formation. The kinetics of clot formation in the presence of rLoa22 (5 μM) was similar to that of the positive control of the assay (Fig. 7A). On the other hand, the kinetics of clot formation regularly decreased (47 to 7%) in the presence of an increasing concentration of rErpY-like protein (0.05 to 5 μM) (Fig. 7B).
FIG 7.
Inhibition of thrombin-catalyzed fibrin clot formation in the presence of rErpY-like protein. (A) Recombinant ErpY-like protein dose-dependent inhibition of fibrin clot reaction. Increasing concentrations of rErpY-like protein (0.05 to 5 μM) were preincubated with human fibrinogen (Fg; 1 mg ml−1) in a microtiter plate for 2 h at 37°C before initiating the thrombin (Thr; 10 mU/well)-catalyzed fibrin clot formation. The clotting reaction in the well containing fibrinogen plus thrombin is taken as a positive control, while the reaction mixture containing only fibrinogen is the negative control. rLoa22 was used to check the specificity of the clot inhibition assay. The absorbance obtained during the thrombin-catalyzed fibrin clot reaction was measured every 2 min for 40 min. The recorded absorbance (595 nm) versus time (min) plot shows inhibition of thrombin-catalyzed fibrin clot formation from fibrinogen in the presence of rErpY-like protein. (B) Reduction in the percentage of fibrin clot formation in the presence of rErpY-like protein. The absorbance obtained during fibrin clot formation in the positive control of the assay shown in panel A was taken as 100%. Fibrin clot formation at a saturation time point (40 min) in the presence of an increasing concentration of rErpY-like protein was calculated as a percentage. The thrombin-catalyzed fibrin clot formation was reduced to 7% in the presence of a saturated concentration of rErpY-like protein (5 μM). The measurements were performed in triplicates, and the data shown are representative of two independent experiments. Statistical analysis was performed by the two-tailed t test (***, P < 0.001).
Leptospira rErpY-like protein is a complement factor H binding protein.
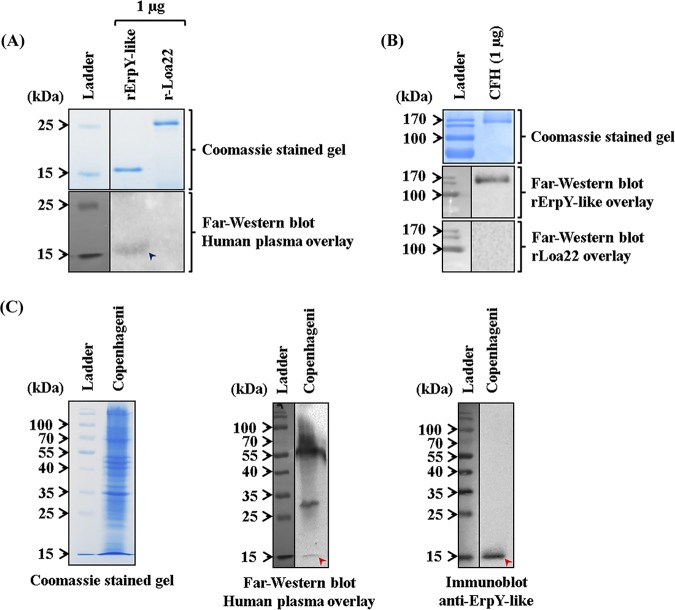
It is a known fact that few pathogens can evade the host’s innate immune responses more efficiently by complement inactivation, thereby being able to survive and enhance host colonization. The alternative pathway of complement activation is an essential arm of vertebrate innate immunity, which rapidly clears susceptible microorganisms from the host in the absence of antibody (37). In this study, we observed that CFH and CFI show relatively higher interactions with rErpY-like protein than plasminogen, another soluble plasma component. Interestingly, CFH is a soluble glycoprotein present in normal plasma at a concentration of 0.3 to 0.5 mg ml−1 and is also a cofactor for CFI, a serine protease of the alternative pathway of complementation (37). Therefore, it was interesting to determine if the recombinant protein and native ErpY-like protein of Leptospira could bind to the CFH present in normal human plasma. To address this, we performed far-Western blotting of rErpY-like protein as the bait protein on nitrocellulose membrane and human plasma (10%) as the prey protein component. The rErpY-like protein (18 kDa) could bind to the CFH (prey protein) present in the human plasma overlay and when probed with an anti-CFH antibody (Fig. 8A, bottom). In contrast, the same amount of rLoa22 failed to bind CFH during the overlay experiment (Fig. 8A, bottom). Similarly, human CFH was used as bait and rErpY-like protein as prey to identify the protein-ligand interaction through far-Western blotting. Human CFH was resolved on SDS-PAGE (Fig. 8B, top) and electroblotted, and the membrane was overlaid with recombinant proteins. It was evident that rErpY-like protein showed interaction with CFH (∼150 kDa) (Fig. 8B, middle), whereas an equivalent amount of rLoa22 did not show any interaction with CFH (Fig. 8B, bottom). The whole-cell lysate of L. interrogans serovar Copenhageni used as bait could bind to the CFH component present in human plasma. Detection at approximately 15 kDa in the far-Western blot probably corresponds to the native ErpY-like protein of Leptospira (Fig. 8C, middle). Additionally, two other Leptospira proteins (∼50 and 30 kDa) were detected (Fig. 8C, middle), in agreement with a previous report (38). Detection of the native protein in Leptospira lysate using anti-ErpY-like antibody (Fig. 8C, right) indicates that the CFH-bound protein detected at approximately 15 kDa in the far-Western blot (Fig. 8C, middle) is, in fact, the ErpY-like protein of Leptospira. The binding of rErpY-like protein (18 kDa) to CFH by far-Western blotting was in agreement with the binding ability of native ErpY-like protein (15 kDa) of Leptospira.
FIG 8.
Recombinant and native ErpY-like protein interacts with complement factor H present in human plasma. (A) Comparison of far-Western and Coomassie blue-stained gel of rErpY-like and rLoa22 proteins. (Top) The recombinant proteins (rErpY-like protein and rLoa22) resolved on SDS-polyacrylamide gel were stained with Coomassie blue to demonstrate the equal loading of the recombinant proteins. An equivalent amount of recombinant protein was electroblotted onto the nitrocellulose membrane to perform far-Western blotting with anti-CFH antibody after overlaying it with 10% human plasma. (Bottom) Far-Western blot demonstrated the rErpY-like protein (∼18 kDa, blue arrowhead) interaction with the CFH component of human plasma, whereas rLoa22 (negative control) did not show any interaction with CFH. (B) Comparison of far-Western and Coomassie blue-stained gel of CFH. (Top) The human CFH (1 μg) of 150 kDa was resolved on SDS-polyacrylamide gel and stained with Coomassie blue. An equivalent amount of CFH was electroblotted onto nitrocellulose membrane and overlaid with 100 ng/μl of rErpY-like protein (middle) or rLoa22 (bottom). Far-Western blot demonstrated the rErpY-like protein interaction with CFH (∼150 kDa), whereas rLoa22 (negative control) did not show any interaction with CFH. (C) Comparison of far-Western blot and Coomassie blue-stained gel of whole-cell lysate of Leptospira. The whole-cell lysate of L. interrogans serovar Copenhageni was resolved on SDS-polyacrylamide gel for Coomassie blue staining (left) and far-Western blotting (middle). Detection at approximately 15 kDa (red arrowhead) demonstrates the possible interaction of native ErpY-like protein of Leptospira with CFH present in human plasma (middle). (Right) Detection of the native protein (15 kDa, red arrowhead) in Leptospira lysate using anti-ErpY-like antibody. The gels in this figure have been spliced for labeling purposes.
DISCUSSION
To date, the role of ErpY-like protein in Leptospira pathogenesis has not been known; however, it is a potential vaccine and diagnostic candidate for leptospirosis in the specific host (18, 19). We demonstrate that ErpY-like protein of Leptospira is a surface-exposed outer membrane lipoprotein, and its recombinant form of antigen can be implemented in serological diagnosis of leptospirosis in humans, bovines, and canines. The serological diagnosis using rErpY-like protein in this study is in agreement with the previous reports for detecting leptospirosis in humans (17), swine (18), and rabbits (17). This serological study, in agreement with the other reports, establishes that the expression of ErpY-like protein during Leptospira natural infection evokes an immune response in a broad host range, including bovines and canines. In this study, we also compared the diagnostic potential of rErpY-like antigen with that of rLoa22, a known diagnostic antigen. The diverse Leptospira hosts (human, bovine, and canine) positive for leptospirosis by MAT demonstrated serological reactivity with rLoa22 superior to that of rErpY-like antigen in terms of the ELISA cutoff value. Nevertheless, the immunoblotting performed on these two antigens with the pooled sera of diverse hosts did not find any difference in serological reactivity.
A study done on golden Syrian hamsters infected with L. interrogans serovar Pomona failed to detect ErpY-like protein expression in the liver samples using an immunofluorescence technique; in contrast, human serum positive for leptospirosis showed reactivity to rErpY-like protein using ELISA (17). Failure of polyclonal antibody reactivity to ErpY-like antigen in a hamster model may be due to either the low expression of antigen in L. interrogans serovar Pomona or the low immune response in hamster against ErpY-like protein. Moreover, the expression of ErpY-like protein in L. interrogans serovar Pomona under in vitro growth has been reported to be modulated by iron limitation and the presence of serum (17). Interestingly, an attenuated strain of L. interrogans serovar Manilae (M1352; defective in lipopolysaccharide biosynthesis) immunized to hamsters conferred protection against leptospirosis and demonstrated serological recognition of rLA1939 (ErpY-like protein ortholog) (39). In a follow-up of a similar study, the rErpY-like protein was shown to protect hamsters (62.5%) against mortality but not against tissue lesions developed during laboratory-generated Leptospira infection (19).
Pathogenic species of Leptospira exclusively express the ErpY-like protein; hence, we speculate that it is one of the virulence factors required during the pathogenesis of Leptospira. Biophysical interaction studies reported earlier have demonstrated that Erp proteins in spirochetes bind with host Fg and CFH (19, 22). Moreover, bacterial adhesion to host extracellular matrix (ECM) components or plasma protein is assumed to be an indispensable step in the pathogenesis of spirochete infection. In accordance with this, the ErpY protein of B. burgdorferi has been demonstrated as a binding partner to factor H complement regulating protein from rodents, dogs, and cats (22). The Leptospira ErpY-like protein, a predominant outer membrane protein, has 26% identity to ErpY protein of Borrelia. Therefore, we evaluated whether the secondary structure of the recombinant protein is retained in the purified rErpY-like protein before pursuing any interaction study with host extracellular and plasma protein components. The CD spectra of rErpY-like protein suggested the secondary structure of the purified protein is retained. The interaction of the rErpY-like protein with the host ECM component was compared with the interaction of another Leptospira protein (rLoa22) known to have a low/moderate binding property to ECM components as described before by our laboratory (26). In this study, the assay of binding of rErpY-like protein to host ECM or plasma component indicates it facilitates Leptospira in adhesion. Interestingly, the rErpY-like protein showed a higher preference for specific plasma proteins (Fg, CFH, and CFI). Such host plasma protein components are known to form oligomeric structures (40–42). This raised our interest in determining if ErpY-like protein also undergoes supramolecular assembly. Native gel electrophoresis, chemical cross-linking, DLS, and transmission electron microscopy imaging of rErpY-like protein demonstrated the tendency of this protein to oligomerize in solution, and further comprehensive analysis of such protein structure is warranted.
The plasma component Fg plays a central role in coagulation and thrombosis in host blood. Several important pathogens, including spirochetes, exploit host Fg through a diverse mechanism to enhance its survival and colonization in the host (43–46). The Fg-binding protein also contributes to increasing the virulence of pathogens during wound infection (47). In Leptospira, to date multiple Fg-binding proteins, viz, LigA, LigB (16, 48), OmpL37 (49), rLIC12238 (50), Lsa33, Lsa25 (51), Lsa30, OmpL1, rLIC11360, and rLIC11975 (13), with different binding affinities to Fg, have been established. Also, many such recombinant proteins along with the live virulent L. interrogans strains have been demonstrated to have a direct role in inhibiting thrombin-catalyzed fibrin clot formation (13, 16, 48). Such a large repertoire of Fg-binding proteins encoded by virulent Leptospira genomes is discrete in nature. However, in this study, we demonstrate that the binding of the rErpY-like protein to Fg biochemically is dose dependent, saturable, and specific in nature. The measured KD value (70.45 ± 4.13 nM) of rErpY-like protein to Fg demonstrated a stronger affinity relative to that of other reported Fg-binding proteins, like rLIC12238 and Lsa33 (733 ± 277 to 128 ± 90 nM) of Leptospira. Moreover, in the presence of rErpY-like protein, the thrombin-catalyzed fibrin clot formation from fibrinogen and the binding of live L. interrogans to Fg were inhibited in a dose-dependent manner. Thus, Leptospira species, in agreement with the other reports and like other pathogens, encode multiple Fg-binding proteins (43, 52), including the rErpY-like protein that may enhance its survival, colonization, and dissemination in the host during infection.
The activation of the alternative complement pathway in a host leads to C3b opsonization on the surfaces of invading microbes that may result in bacterial destruction (53). However, such complement activation is not deleterious to host cells due to the presence of regulatory CFH that coats the host cell surface and either thwarts C3b opsonization on the cell surface or promotes degradation of bound C3b with the help of CFI. Inspired by the vertebrate host cell of self-defense from its complement-mediated killing, several pathogens, including Leptospira, have evolved a mechanism of hijacking CFH to evade complement-mediated execution (31, 38, 54–57). It has been demonstrated that pathogenic Leptospira possesses the capacity to recruit and bind not only the host CFH but also factor H-like 1 (FHL-1), factor H-related 1 (FHR-1), and C4b binding protein (C4BP) to avoid complement-mediated killing (9, 38, 58, 59). As multiple outer membrane proteins of Leptospira that can adhere to complement regulator FH have already been established, the precise contributory role of ErpY-like protein for alternative complement system inactivation requires further evaluation.
In this study, we have identified a conserved immunogenic ErpY-like lipoprotein localized on the outer membrane of pathogenic L. interrogans. The rErpY-like protein of Leptospira that tends to form a supramolecule structure can be serologically detected by antibodies present in the serum of humans, bovines, and canines infected naturally with Leptospira. Our experimental analyses suggest rErpY-like protein interferes with fibrin clot formation and in the evasion of complement-mediated killing by binding to CFH and CFI. Further investigation on ErpY-like protein may give its exact role in evading complement-mediated execution and inhibition of blood clot during infection. Moreover, deciphering the structure of the rErpY-like protein as a supramolecule may provide better insights into its molecular interaction with host plasma components. The characterization of novel diagnostic rErpY-like protein as an antigen for diagnosing leptospirosis and its possible role as an adhesin to diverse host ECM components may help in developing new strategies for disease intervention.
MATERIALS AND METHODS
Bacterial strains, culture media, and serum samples.
The pathogenic Leptospira reference strains (L. interrogans serovar Copenhageni strain Fiocruz L1-130, L. interrogans serovar Lai strain Lai, and L. interrogans serovar Canicola strain Hond Utrecht IV) and a nonpathogenic Leptospira strain (L. biflexa serovar Patoc strain Patoc 1) were grown as described before (26). Serum samples positive or negative for leptospirosis in a diverse host (human, bovine, and canine) by microscopic agglutination test (MAT) were obtained from the Indian Council of Agricultural Research-National Institute of Veterinary Epidemiology and Disease Informatics (ICAR-NIVEDI), Bengaluru, Karnataka, India. The serum samples were screened for MAT using the methodology described before (26). The Escherichia coli strain DH5α or BL21(DE3) was grown at 37°C in Luria-Bertani (LB) broth medium (M1245; HiMedia) or LB agar (M1151; HiMedia) with or without ampicillin (61314; SRL) or kanamycin (99311; SRL) at a concentration of 100 μg ml−1 for gene cloning and protein expression studies.
In silico analysis of protein.
The LIC11966/ErpY-like protein (new locus tag, LIC_RS10040) is annotated as a hypothetical protein of L. interrogans serovar Copenhageni by the NCBI (National Center for Biotechnology Information) database. The amino acid sequence of the ErpY-like protein was retrieved using its reference sequence identifier (WP_000716642.1) from the NCBI protein database and was analyzed to identify the signal peptide as reported previously (60). NCBI protein BLAST (basic local alignment search tool) was used to identify orthologs of ErpY-like protein among Leptospira species. Phylogenetic analysis of Leptospira was performed based on ErpY-like protein as described previously (26). Analysis of theoretical secondary structures of the recombinant protein was performed using the web server PSIPRED, version 3.3 (61).
Cloning, protein overexpression, purification, and polyclonal antibody generation.
Genomic DNA (gDNA) was isolated from Leptospira as described previously by our laboratory (26). All of the enzymes used for DNA manipulations were purchased from New England Biolabs or Thermo Fisher Scientific. The coding sequence (CDS) of the erpY-like gene without its signal peptide coding sequence was PCR amplified (414 bp) from gDNA using gene-specific primers erpY-like forward, 5’CTAGCTAGCTGCAAACAAGATCCAGTAGAT3’ (NheI indicated in italics), and reverse, 5’CCGCTCGAGTTATTGAGAAGCGTATTCTTTC3’ (XhoI indicated in italics). The pTZ57R/T plasmid construct (K1214; Thermo Fisher Scientific) was used to facilitate the erpY-like gene cloning into a pET28a expression vector (69864; Novagen). The sequencing of the cloned DNA insert in pET28a was done by Eurofins Genomics India, Pvt. Ltd., Bengaluru, India. Induction of expression and purification of the recombinant ErpY-like (rErpY-like; 17.8 kDa) protein in the E. coli BL21(DE3) strain were done as described previously (62). Similarly, the recombinant LIC10191/Loa22 (rLoa22), used in this study, was purified as described elsewhere (63). Female BALB/c mice (n = 5), 4 to 6 weeks old, were immunized with recombinant protein for the generation of specific polyclonal antibodies as described before (26). The immunization experiments were performed at the Department of Microbiology, College of Veterinary Science, Assam Agricultural University, Guwahati, India, after approval by the Institutional Animal Ethics Committee (approval no. 770/ac/CPCSEA/FVSC/AAU/IAEC/13-14). The antibodies against OmpL54, LipL31, and LipL32, raised in rabbits, were a generous gift from David Haake (Division of Infectious Diseases, UCLA, Los Angeles, CA, USA).
CD spectroscopy.
A circular dichroism (CD) spectropolarimeter (model J-815; JASCO, Japan) was used to measure the molar ellipticity (Ф) of rErpY-like protein (in terms of degrees centimeter square per decimole) as described previously by our laboratory (26).
Chemical cross-linking of rErpY-like protein of Leptospira.
Purified rErpY-like protein (4 μg per reaction mixture) was cross-linked with glutaraldehyde solution (0.1%) in a cross-linking buffer (50 mM phosphate buffer, 100 mM KCl, 5 mM MgCl2, 5% glycerol; pH 7.6) at room temperature. The reactions were terminated at various time intervals (0 to 45 min), resolved on 12% SDS-PAGE with or without the reducing agent dithiothreitol (DTT), and visualized by Coomassie blue staining as described previously (64).
Native-PAGE of rErpY-like protein.
Various amounts of pure rErpY-like protein (0.5 to 2 μg) were mixed with 3× native sample buffer (240 mM Tris-HCl, 30% glycerol, 0.03% bromophenol blue; pH 6.8). The rErpY-like protein subunit assembly was analyzed on a 4 to 20% gradient gel (catalog no. 456-1096; Mini-Protean; Bio-Rad) after resolving for 2 h at 120 V. The resolved proteins in native gradient gel were visualized with Coomassie blue stain, and the size was estimated with standard protein markers (928387; Invitrogen) of native-PAGE.
DLS.
Untreated or heat-denatured (30 min at 100°C) rErpY-like protein (0.5 μg μl−1), after overnight incubation at 4°C in a phosphate buffer (50 mM sodium phosphate buffer, pH 7.4), was used in DLS experiments to record scattering as described previously (64). A total of thirty autocorrelation functions were recorded for each of the protein samples, and intensity-weighted hydrodynamic diameters were determined. The profile corresponding to average sizes is reported and discussed. The hydrodynamic diameter and molecular weight of rErpY-like protein were generated assuming a globular conformation model using the Malvern Zetasizer software.
FETEM.
The rErpY-like protein sample (5 μl of 40 ng μl−1) was drop-cast on a UV-irradiated carbon-coated grid (number AGS147-4H; Agar Scientific), adsorbed, and stained with uranyl acetate solution for field emission transmission electron microscopy (FETEM; JEOL-JEM-2100F) as described previously (65). Images were acquired at an instrumental magnification of ×40,000 and 200 keV. Selective particles of the image were enlarged using the MS Office image viewer tool for clarity.
Triton X-114 extraction and phase partitioning of membrane proteins.
A viable culture of L. interrogans serovar Copenhageni (5 × 109 cells) in mid-log phase (7 days old) was harvested, washed with phosphate-buffered saline (PBS), and subjected to detergent (Triton X-114) extraction and phase partitioning as described previously by our laboratory (26). Briefly, the whole lysates and the phase-partitioned fractions were resolved on SDS-polyacrylamide gel and then subjected to immunoblot analyses using anti-ErpY-like (1:1,000), anti-LipL32 (1:5,000), or anti-LipL31 (1:2,000) antibody.
Immunoblot assays.
The spirochete whole-cell lysate, detergent-mediated phase-partitioned fraction, and recombinant proteins (rErpY-like and rLoa22) were immunoblotted as described before by our laboratory (26). The membranes were probed for 2 h at room temperature with specific polyclonal antisera (anti-ErpY-like at 1:1,000, anti-Loa22 at 1:1,000, anti-LipL32 at 1:5,000, and anti-LipL31 at 1:2,000) as primary antibodies against the recombinant proteins or with the pooled sera (1:400) of the respective host (human, bovine, and/or canine) testing MAT positive/negative for leptospirosis. The horseradish peroxidase (HRP)-conjugated secondary antibodies (goat anti-mouse IgG, goat anti-rabbit IgG, goat anti-human IgG, rabbit anti-bovine IgG, and rabbit anti-dog IgG) were used (1:5,000) to probe bound primary antibodies.
Protease accessibility assay.
A viable culture of L. interrogans serovar Copenhageni (2.5 × 109 cells) in mid-log phase was harvested, washed, and resuspended in 5 ml of PBS. Each 1-ml suspension of live leptospires (5 × 108 cells) was incubated with 5 μg of proteinase K (PK; 36331; SRL) for 0, 1, 3, or 5 h before the inhibition of PK activity by 1 mM phenylmethylsulfonyl fluoride (PMSF; RM1592; HiMedia) as described previously by our laboratory (26). The bound bacteria in the microtiter plate were probed individually with anti-OmpL54 (1:50), anti-ErpY-like (1:1,000), or anti-LipL31 (1:1,000) antibody. The HRP-conjugated secondary antibodies (anti-mouse and anti-rabbit, 1:5,000) were added to detect bound primary antibodies. The absorbance values obtained were analyzed and plotted as described before (26).
Binding of recombinant protein to host ligands.
Binding of the rErpY-like protein to the host ligands, ECM, or plasma components was analyzed by indirect ELISA (enzyme-linked immunosorbent assay) as described previously (26). The ECM components selected in this study are laminin (L2020; Sigma), collagen type I (C9791 [Sigma] and 08-115 [Merck]), chondroitin sulfate A and B (C9819 and C3788; Sigma), elastin (E1625; Sigma), hyaluronic acid (H7630; Sigma), and heparan sulfate (H7640; Sigma). The host plasma components used in this study are fibronectin (F4759; Sigma), fibrinogen (Fg, F3879; Sigma), plasminogen (P7999; Sigma), complement factor H (CFH; C5813; Sigma), and complement factor I (CFI; C5938; Sigma). Additionally, the glycoprotein fetuin (P6042; New England Biolabs) and bovine serum albumin, fraction V (BSA; 97350; SRL), are included as control ligands. Briefly, 1 μg of each host ligand in PBS was coated, and then each well was blocked with BSA. The recombinant proteins (rErpY-like and rLoa22) were added individually in each ligand-coated well, and binding was determined by specific primary (anti-ErpY-like/anti-Loa22 antibody) and secondary (anti-mouse or anti-rabbit antibody) antibody.
Serological assay using sera testing MAT positive or negative for leptospirosis.
The sera testing MAT positive or negative for leptospirosis (human, n = 50 positive and 20 negative; bovine, n = 50 positive and 20 negative; canine, n = 18 positive and 11 negative) were used to check immunoglobulin reactivity to recombinant antigens (rErpY-like and rLoa22; 400 ng each) on microtiter plates by ELISA. The percent sensitivity and specificity of the assay were calculated as described previously (26). The absorbance of MAT-negative serum samples of each host group for each antigen was used as a control in ELISA to calculate the background cutoff value (means + 2 standard deviations [SD]).
Dose-response curve of rErpY-like protein binding to fibrinogen.
The rErpY-like protein (0.05 to 5 μM) was added in increasing concentrations into Fg-coated microtiter wells and was probed with specific primary and secondary antibodies as described in “Binding of recombinant protein to host ligands,” above. The obtained ELISA data were plotted in the OriginLab software to calculate the dissociation constant (KD) through a fitted Michaelis-Menten curve. Similarly, increasing concentrations (0.25 to 5 μM) of rErpY-like protein (control) or its heat-denatured form (95°C, 10 min) were used to measure the effect of heat denaturation of rErpY-like protein in binding to human fibrinogen.
Antibody inhibition assay.
A microtiter plate was coated with Fg and blocked with BSA. The rErpY-like protein (1 μg) was preincubated with specific anti-ErpY-like antibody or nonspecific anti-Loa22 antibody (1:100) in PBS with Tween 20 (PBS-T) (0.05%) for 1 h as described previously (13). The preincubated antibody-blocked rErpY-like protein (1 μg) complex and the pure rErpY-like protein (1 μg) were added independently into Fg-coated wells in triplicates and incubated for 2 h. After being washed, the wells were probed with monoclonal anti-His antibody (1:1,000) (05-949; Merck). Further analysis of protein interaction to the Fg was performed as described in “Binding of recombinant protein to host ligands,” above. The obtained ELISA data of the antibody inhibition assay were plotted in relative percentages, presuming the binding of the pure rErpY-like protein to Fg to be 100%.
Inhibition of live leptospire adherence to fibrinogen in the presence of rErpY-like protein.
Increasing concentrations of rErpY-like protein (0.05 to 5 μM) were added separately in triplicates into Fg-coated wells. After three washings, live L. interrogans serovar Copenhageni (4 × 107) cells were incubated into each well for 90 min. Each well was washed with PBS-T (0.05%) before the addition of anti-Loa22 antibody (1:1,000). After 2 h of incubation, secondary antibody (1:5,000) was used to detect inhibition of leptospire binding as described before by our laboratory (26).
Fibrin clot inhibition assay.
Inhibition of thrombin-catalyzed fibrin clot formation from Fg in the presence of recombinant proteins of Leptospira was performed as described previously (13). Briefly, increasing concentrations of rErpY-like protein (0.05 to 5 μM) and the single highest concentration of rLoa22 (5 μM) were added independently to soluble Fg (1 mg ml−1) in PBS (300 μl). An aliquot (90 μl) of these complex solutions was added onto a microtiter plate in triplicates. Thrombin (0.1 U/10 μl) (69671-3; Merck) was added to each well, and every 2 min the absorbance (595 nm) was recorded for 40 min at 25°C. A graph of absorbance (595 nm) versus time (in minutes) was plotted in the OriginLab software. The relative percentage of fibrin clot formation in the presence of rErpY-like protein was calculated presuming the absorbance obtained in the positive control of the assay to be 100%.
Far-Western blotting.
The whole-cell lysate of L. interrogans serovar Copenhageni, the recombinant proteins (rErpY-like protein and rLoa22, 1 μg each), and CFH (1 μg) were subjected to SDS-PAGE (12%) and far-Western blotting. The electroblotted membrane was overlaid with 10% human plasma (P9523; Sigma) as a source of CFH or 100 ng/μl of recombinant protein for 90 min after blocking with 5% nonfat dry milk for 2 h and was subsequently probed with specific primary antibody (1:1,000) of human CFH (anti-CFH antibody; SAB1401172; Sigma) or anti-ErpY-like/Loa22 (1:1,000) antibody and the HRP-conjugated secondary IgG (1:5,000).
Statistical analysis.
All data points in graphs are representative of the means ± SD. Two-tailed Student’s paired t test was used to determine the significance of the differences between the means. P values between 0.05 and 0.01 (*), 0.001 and 0.01 (**), and less than 0.0001 (***) were considered statistically significant, very significant, and extremely significant, respectively. Two independent experiments were performed, each one in duplicate or triplicate as described above. Statistical agreement between ELISA and MAT was determined using Cohen’s kappa coefficient as described previously (26).
ACKNOWLEDGMENTS
We gratefully acknowledge ICMR, Port Blair, India, for providing the Leptospira strains and Nitin Chaudhary, Department of Biosciences and Bioengineering (BSBE), Indian Institute of Technology, Guwahati (IIT Guwahati), for providing help in recording and analyzing the DLS experiments. We appreciate the efforts made by Shankar Prasad Kanaujia (BSBE) in refining the manuscript. We acknowledge the central instruments facility, IIT Guwahati, for electron microscopy. We also thank the Department of Microbiology, College of Veterinary Science, Guwahati, for generating polyclonal antibodies.
This work was financially supported by the Department of Science and Technology (DST), Science and Engineering Research Board (SERB), and ICMR Government of India, bearing project numbers SERB/EMR/2015/000255 and Leptos/10/2013-ECD-I.
We have no conflicts of interest to declare.
REFERENCES
- 1.Bharti AR, Nally JE, Ricaldi JN, Matthias MA, Diaz MM, Lovett MA, Levett PN, Gilman RH, Willig MR, Gotuzzo E, Vinetz JM. 2003. Leptospirosis: a zoonotic disease of global importance. Lancet Infect Dis 3:757–771. doi: 10.1016/S1473-3099(03)00830-2. [DOI] [PubMed] [Google Scholar]
- 2.Costa F, Hagan JE, Calcagno J, Kane M, Torgerson P, Martinez-Silveira MS, Stein C, Abela-Ridder B, Ko AI. 2015. Global morbidity and mortality of leptospirosis: a systematic review. PLoS Negl Trop Dis 9:e0003898. doi: 10.1371/journal.pntd.0003898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nicodemo AC, Duarte MIS, Alves VA, Takakura CF, Santos RT, Nicodemo EL. 1997. Lung lesions in human leptospirosis: microscopic, immunohistochemical, and ultrastructural features related to thrombocytopenia. Am J Trop Med Hyg 56:181–187. doi: 10.4269/ajtmh.1997.56.181. [DOI] [PubMed] [Google Scholar]
- 4.Marchiori SE, Netto B, Tavares W. 1992. Pulmonary compromise in leptospirosis. Rev Soc Bras Med Trop 25:21–30. doi: 10.1590/S0037-86821992000100004. [DOI] [PubMed] [Google Scholar]
- 5.Segura ER, Ganoza CA, Campos K, Ricaldi JN, Torres S, Silva H, Cespedes MJ, Matthias MA, Swancutt MA, Linan RL, Gotuzzo E, Guerra H, Gilman RH, Vinetz JM. 2005. Clinical spectrum of pulmonary involvement in leptospirosis in a region of endemicity, with quantification of leptospiral burden. Clin Infect Dis 40:343–351. doi: 10.1086/427110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vieira ML, Naudin C, Mörgelin M, Romero EC, Nascimento ALT, Herwald H. 2016. Modulation of hemostatic and inflammatory responses by Leptospira spp. PLoS Negl Trop Dis 10:e0004713. doi: 10.1371/journal.pntd.0004713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Evangelista KV, Coburn J. 2010. Leptospira as an emerging pathogen: a review of its biology, pathogenesis and host immune responses. Future Microbiol 5:1413–1425. doi: 10.2217/fmb.10.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Evangelista KV, Hahn B, Wunder EA Jr, Ko AI, Haake DA, Coburn J. 2014. Identification of cell-binding adhesins of Leptospira interrogans. PLoS Negl Trop Dis 8:e3215. doi: 10.1371/journal.pntd.0003215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Castiblanco-Valencia MM, Fraga TR, Silva LB, Monaris D, Abreu PAE, Strobel S, Józsi M, Isaac L, Barbosa AS. 2012. Leptospiral immunoglobulin-like proteins interact with human complement regulators factor H, FHL-1, FHR-1, and C4BP. J Infect Dis 205:995–1004. doi: 10.1093/infdis/jir875. [DOI] [PubMed] [Google Scholar]
- 10.Cavenague MF, Teixeira AF, Filho SA, Souza GO, Vasconcellos SA, Heinemann MB, Nascimento AL. 2019. Characterization of a novel protein of Leptospira interrogans exhibiting plasminogen, vitronectin and complement binding properties. Int J Med Microbiol 309:116. doi: 10.1016/j.ijmm.2018.12.005. [DOI] [PubMed] [Google Scholar]
- 11.Santos JV, Pereira PR, Fernandes LG, Siqueira GH, de Souza GO, Souza Filho A, Vasconcellos SA, Heinemann MB, Chapola EG, Nascimento AL. 2018. Binding of human plasminogen by the lipoprotein LipL46 of Leptospira interrogans. Mol Cell Probes 37:12–21. doi: 10.1016/j.mcp.2017.10.004. [DOI] [PubMed] [Google Scholar]
- 12.Lo M, Murray GL, Khoo CA, Haake DA, Zuerner RL, Adler B. 2010. Transcriptional response of Leptospira interrogans to iron limitation and characterization of a PerR homolog. Infect Immun 78:4850–4859. doi: 10.1128/IAI.00435-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oliveira R, Domingos RF, Siqueira GH, Fernandes LG, Souza NM, Vieira ML, de Morais ZM, Vasconcellos SA, Nascimento AL. 2013. Adhesins of Leptospira interrogans mediate the interaction to fibrinogen and inhibit fibrin clot formation in vitro. PLoS Negl Trop Dis 7:e2396. doi: 10.1371/journal.pntd.0002396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Picardeau M. 2015. Genomics, proteomics, and genetics of leptospira, p 43–63. In Adler B. (ed), Leptospira and leptospirosis. Springer, New York, NY. [DOI] [PubMed] [Google Scholar]
- 15.Siqueira GH, Teixeira AF, Fernandes LG, de Souza GO, Kirchgatter K, Romero EC, Vasconcellos SA, Vieira ML, Nascimento ALT. 2015. The recombinant LIC10508 is a plasma fibronectin, plasminogen, fibrinogen and C4BP-binding protein of Leptospira interrogans. FEMS Pathog Dis 74:ftv118. doi: 10.1093/femspd/ftv118. [DOI] [PubMed] [Google Scholar]
- 16.Lin YP, McDonough SP, Sharma Y, Chang YF. 2011. Leptospira immunoglobulin‐like protein B (LigB) binding to the C‐terminal fibrinogen αC domain inhibits fibrin clot formation, platelet adhesion and aggregation. Mol Microbiol 79:1063–1076. doi: 10.1111/j.1365-2958.2010.07510.x. [DOI] [PubMed] [Google Scholar]
- 17.Eshghi A, Cullen PA, Cowen L, Zuerner RL, Cameron CE. 2009. Global proteome analysis of Leptospira interrogans. J Proteome Res 8:4564–4578. doi: 10.1021/pr9004597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Padilha BCR, Simão HQ, Oliveira TL, Hartwig DD. 2019. The use of ErpY-like recombinant protein from Leptospira interrogans in the development of an immunodiagnostic test for swine leptospirosis. Acta Tropica 193:31. doi: 10.1016/j.actatropica.2019.01.027. [DOI] [PubMed] [Google Scholar]
- 19.Oliveira TL, Schuch RA, Inda GR, Roloff BC, Neto ACPS, Amaral M, Dellagostin OA, Hartwig DD. 2018. LemA and Erp Y-like recombinant proteins from Leptospira interrogans protect hamsters from challenge using AddaVax as adjuvant. Vaccine 36:2574–2580. doi: 10.1016/j.vaccine.2018.03.078. [DOI] [PubMed] [Google Scholar]
- 20.Stevenson B, Zuckert W, Akins DR. 2000. Repetition, conservation, and variation: the multiple cp32 plasmids of Borrelia species. J Mol Microbiol Biotechnol 2:411–422. [PubMed] [Google Scholar]
- 21.Akins DR, Caimano MJ, Yang X, Cerna F, Norgard MV, Radolf JD. 1999. Molecular and evolutionary analysis of Borrelia burgdorferi 297 circular plasmid-encoded lipoproteins with OspE-and OspF-like leader peptides. Infect Immun 67:1526–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stevenson B, El-Hage N, Hines MA, Miller JC, Babb K. 2002. Differential binding of host complement inhibitor factor H by Borrelia burgdorferi Erp surface proteins: a possible mechanism underlying the expansive host range of Lyme disease spirochetes. Infect Immun 70:491–497. doi: 10.1128/IAI.70.2.491-497.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Almagro Armenteros JJ, Tsirigos KD, Sønderby CK, Petersen TN, Winther O, Brunak S, von Heijne G, Nielsen H. 2019. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat Biotechnol 37:420–423. doi: 10.1038/s41587-019-0036-z. [DOI] [PubMed] [Google Scholar]
- 24.Setubal JC, Reis M, Matsunaga J, Haake DA. 2006. Lipoprotein computational prediction in spirochaetal genomes. Microbiology 152:113. doi: 10.1099/mic.0.28317-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakai K, Horton P. 1999. PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem Sci 24:34–36. doi: 10.1016/S0968-0004(98)01336-X. [DOI] [PubMed] [Google Scholar]
- 26.Ghosh KK, Prakash A, Shrivastav P, Balamurugan V, Kumar M. 2018. Evaluation of a novel outer membrane surface-exposed protein, LIC13341 of Leptospira, as an adhesin and serodiagnostic candidate marker for leptospirosis. Microbiology 164:1023–1037. doi: 10.1099/mic.0.000685. [DOI] [PubMed] [Google Scholar]
- 27.Abreu PA, Seguro AC, Canale D, da Silva AMG, do L, Matos RB, Gotti TB, Monaris D, de Jesus DA, Vasconcellos SA, de Brito T. 2017. Lp25 membrane protein from pathogenic Leptospira spp. is associated with rhabdomyolysis and oliguric acute kidney injury in a guinea pig model of leptospirosis. PLoS Negl Trop Dis 11:e0005615. doi: 10.1371/journal.pntd.0005615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pinne M, Haake DA. 2009. A comprehensive approach to identification of surface-exposed, outer membrane-spanning proteins of Leptospira interrogans. PLoS One 4:e6071. doi: 10.1371/journal.pone.0006071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chalayon P, Chanket P, Boonchawalit T, Chattanadee S, Srimanote P, Kalambaheti T. 2011. Leptospirosis serodiagnosis by ELISA based on recombinant outer membrane protein. Trans R Soc Trop Med Hyg 105:289–297. doi: 10.1016/j.trstmh.2011.01.008. [DOI] [PubMed] [Google Scholar]
- 30.Atzingen MV, Barbosa AS, De Brito T, Vasconcellos SA, de Morais ZM, Lima DM, Abreu PA, Nascimento AL. 2008. Lsa21, a novel leptospiral protein binding adhesive matrix molecules and present during human infection. BMC Microbiol 8:70. doi: 10.1186/1471-2180-8-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Siqueira GH, Atzingen MV, de Souza GO, Vasconcellos SA, Nascimento AL. 2016. Leptospira interrogans Lsa23 protein recruits plasminogen, factor H and C4BP from normal human serum and mediates C3b and C4b degradation. Microbiology 162:295–308. doi: 10.1099/mic.0.000217. [DOI] [PubMed] [Google Scholar]
- 32.Perez-Iratxeta C, Andrade-Navarro MA. 2008. K2D2: estimation of protein secondary structure from circular dichroism spectra. BMC Struct Biol 8:25. doi: 10.1186/1472-6807-8-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reference deleted.
- 34.Buchan DW, Minneci F, Nugent TC, Bryson K, Jones DT. 2013. Scalable web services for the PSIPRED protein analysis workbench. Nucleic Acids Res 41:W349–W357. doi: 10.1093/nar/gkt381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barbosa AS, Abreu PA, Neves FO, Atzingen MV, Watanabe MM, Vieira ML, Morais ZM, Vasconcellos SA, Nascimento AL. 2006. A newly identified leptospiral adhesin mediates attachment to laminin. Infect Immun 74:6356–6364. doi: 10.1128/IAI.00460-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Malmström J, Beck M, Schmidt A, Lange V, Deutsch EW, Aebersold R. 2009. Proteome-wide cellular protein concentrations of the human pathogen Leptospira interrogans. Nature 460:762. doi: 10.1038/nature08184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brissette CA, Cooley AE, Burns LH, Riley SP, Verma A, Woodman ME, Bykowski T, Stevenson B. 2008. Lyme borreliosis spirochete Erp proteins, their known host ligands, and potential roles in mammalian infection. Int J Med Microbiol 298:257–267. doi: 10.1016/j.ijmm.2007.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Verma A, Hellwage J, Artiushin S, Zipfel PF, Kraiczy P, Timoney JF, Stevenson B. 2006. LfhA, a novel factor H-binding protein of Leptospira interrogans. Infect Immun 74:2659–2666. doi: 10.1128/IAI.74.5.2659-2666.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Srikram A, Zhang K, Bartpho T, Lo M, Hoke DE, Sermswan RW, Adler B, Murray GL. 2011. Cross-protective immunity against leptospirosis elicited by a live, attenuated lipopolysaccharide mutant. J Infect Dis 203:870–879. doi: 10.1093/infdis/jiq127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Perkins SJ, Nealis AS, Sim RB. 1991. Oligomeric domain structure of human complement factor H by X-ray and neutron solution scattering. Biochemistry 30:2847–2857. doi: 10.1021/bi00225a017. [DOI] [PubMed] [Google Scholar]
- 41.Goldberger G, Bruns G, Rits M, Edge M, Kwiatkowski D. 1987. Human complement factor I: analysis of cDNA-derived primary structure and assignment of its gene to chromosome 4. J Biol Chem 262:10065–10071. [PubMed] [Google Scholar]
- 42.Mosesson M. 2005. Fibrinogen and fibrin structure and functions. J Thromb Haemost 3:1894–1904. doi: 10.1111/j.1538-7836.2005.01365.x. [DOI] [PubMed] [Google Scholar]
- 43.Rivera J, Vannakambadi G, Höök M, Speziale P. 2007. Fibrinogen-binding proteins of Gram-positive bacteria. Thromb Haemost 98:503–511. doi: 10.1160/TH07-03-0233. [DOI] [PubMed] [Google Scholar]
- 44.Houston S, Hof R, Francescutti T, Hawkes A, Boulanger MJ, Cameron CE. 2011. Bifunctional role of the Treponema pallidum extracellular matrix binding adhesin Tp0751. Infect Immun 79:1386–1398. doi: 10.1128/IAI.01083-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Haapasalo M, Müller K, Uitto V, Leung W, McBride B. 1992. Characterization, cloning, and binding properties of the major 53-kilodalton Treponema denticola surface antigen. Infect Immun 60:2058–2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Flick MJ, Du X, Prasad JM, Raghu H, Palumbo JS, Smeds E, Höök M, Degen JL. 2013. Genetic elimination of the binding motif on fibrinogen for the S. aureus virulence factor ClfA improves host survival in septicemia. Blood 121:1783–1794. doi: 10.1182/blood-2012-09-453894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shannon O, Flock J-I. 2004. Extracellular fibrinogen binding protein, Efb, from Staphylococcus aureus binds to platelets and inhibits platelet aggregation. Thromb Haemost 91:779–789. doi: 10.1160/TH03-05-0287. [DOI] [PubMed] [Google Scholar]
- 48.Choy HA, Kelley MM, Croda J, Matsunaga J, Babbitt JT, Ko AI, Picardeau M, Haake DA. 2011. The multifunctional LigB adhesin binds homeostatic proteins with potential roles in cutaneous infection by pathogenic Leptospira interrogans. PLoS One 6:e16879. doi: 10.1371/journal.pone.0016879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pinne M, Choy HA, Haake DA. 2010. The OmpL37 surface-exposed protein is expressed by pathogenic Leptospira during infection and binds skin and vascular elastin. PLoS Negl Trop Dis 4:e815. doi: 10.1371/journal.pntd.0000815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vieira ML, Atzingen MV, Oliveira TR, Oliveira R, Andrade DM, Vasconcellos SA, Nascimento AL. 2010. In vitro identification of novel plasminogen-binding receptors of the pathogen Leptospira interrogans. PLoS One 5:e11259. doi: 10.1371/journal.pone.0011259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Domingos RF, Vieira ML, Romero EC, Gonçales AP, de Morais ZM, Vasconcellos SA, Nascimento AL. 2012. Features of two proteins of Leptospira interrogans with potential role in host-pathogen interactions. BMC Microbiol 12:50. doi: 10.1186/1471-2180-12-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sun H. 2006. The interaction between pathogens and the host coagulation system. Physiology 21:281–288. doi: 10.1152/physiol.00059.2005. [DOI] [PubMed] [Google Scholar]
- 53.Janeway CA Jr, Travers P, Walport M, Shlomchik MJ. 2001. The complement system and innate immunity Immunobiology: the immune system in health and disease, 5th ed Garland Science, New York, NY. [Google Scholar]
- 54.Joiner KA. 1988. Complement evasion by bacteria and parasites. Annu Rev Microbiol 42:201–230. doi: 10.1146/annurev.mi.42.100188.001221. [DOI] [PubMed] [Google Scholar]
- 55.Lindahl G, Sjöbring U, Johnsson E. 2000. Human complement regulators: a major target for pathogenic microorganisms. Curr Opin Immunol 12:44–51. doi: 10.1016/S0952-7915(99)00049-7. [DOI] [PubMed] [Google Scholar]
- 56.da Silva LB, Miragaia LS, Breda LCD, Abe CM, Schmidt MCB, Moro AM, Monaris D, Conde JN, Józsi M, Isaac L, Abreu PAE, Barbosa AS. 2015. Pathogenic Leptospira species acquire factor H and vitronectin via the surface protein LcpA. Infect Immun 83:888–897. doi: 10.1128/IAI.02844-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fraga TR, Isaac L, Barbosa AS. 2016. Complement evasion by pathogenic Leptospira. Front Immunol 7:623. doi: 10.3389/fimmu.2016.00623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Barbosa AS, Abreu PA, Vasconcellos SA, Morais ZM, Gonçales AP, Silva AS, Daha MR, Isaac L. 2009. Immune evasion of leptospira species by acquisition of human complement regulator C4BP. Infect Immun 77:1137–1143. doi: 10.1128/IAI.01310-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Meri T, Murgia R, Stefanel P, Meri S, Cinco M. 2005. Regulation of complement activation at the C3-level by serum resistant leptospires. Microb Pathog 39:139–147. doi: 10.1016/j.micpath.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 60.Cullen PA, Haake DA, Adler B. 2004. Outer membrane proteins of pathogenic spirochetes. FEMS Microbiol Rev 28:291–318. doi: 10.1016/j.femsre.2003.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jones DT. 1999. Protein secondary structure prediction based on position-specific scoring matrices. J Mol Biol 292:195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- 62.Dixit B, Ghosh KK, Fernandes G, Kumar P, Gogoi P, Kumar M. 2016. Dual nuclease activity of a Cas2 protein in CRISPR-Cas subtype I-B of Leptospira interrogans. FEBS Lett 590:1002–1016. doi: 10.1002/1873-3468.12124. [DOI] [PubMed] [Google Scholar]
- 63.Ghosh KK, Prakash A, Balamurugan V, Kumar M. 2018. Catecholamine-modulated novel surface-exposed adhesin LIC20035 of Leptospira spp. binds host extracellular matrix components and is recognized by the host during infection. Appl Environ Microbiol 84:e02360-17. doi: 10.1128/AEM.02360-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dhara A, Hussain MS, Datta D, Kumar M. 2019. Insights to the assembly of a functionally active leptospiral ClpP1P2 protease complex along with its ATPase chaperone ClpX. ACS Omega 4:12880–12895. doi: 10.1021/acsomega.9b00399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lin M-H, Chang Y-C, Hsiao C-D, Huang S-H, Wang M-S, Ko Y-C, Yang C-W, Sun Y-J. 2013. LipL41, a hemin binding protein from Leptospira santarosai serovar Shermani. PLoS One 8:e83246. doi: 10.1371/journal.pone.0083246. [DOI] [PMC free article] [PubMed] [Google Scholar]