The Porphyromonas gingivalis strain ATCC 33277 (33277) and 381 genomes are nearly identical. However, strain 33277 displays a significantly diminished capacity to stimulate host cell Toll-like receptor 2 (TLR2)-dependent signaling and interleukin-1β (IL-1β) production relative to 381, suggesting that there are strain-specific differences in one or more bacterial immune-modulatory factors.
KEYWORDS: Porphyromonas gingivalis, genomics, inflammation, strain diversity, virulence factors
ABSTRACT
The Porphyromonas gingivalis strain ATCC 33277 (33277) and 381 genomes are nearly identical. However, strain 33277 displays a significantly diminished capacity to stimulate host cell Toll-like receptor 2 (TLR2)-dependent signaling and interleukin-1β (IL-1β) production relative to 381, suggesting that there are strain-specific differences in one or more bacterial immune-modulatory factors. Genomic sequencing identified a single nucleotide polymorphism in the 33277 fimB allele (A→T), creating a premature stop codon in the 33277 fimB open reading frame relative to the 381 fimB allele. Gene exchange experiments established that the 33277 fimB allele reduces the immune-stimulatory capacity of this strain. Transcriptome comparisons revealed that multiple genes related to carboxy-terminal domain (CTD) family proteins, including the gingipains, were upregulated in 33277 relative to 381. A gingipain substrate degradation assay demonstrated that cell surface gingipain activity is higher in 33277, and an isogenic mutant strain deficient for the gingipains exhibited an increased ability to induce TLR2 signaling and IL-1β production. Furthermore, 33277 and 381 mutant strains lacking CTD cell surface proteins were more immune-stimulatory than the parental wild-type strains, consistent with an immune-suppressive role for the gingipains. Our data show that the combination of an intact fimB allele and limited cell surface gingipain activity in P. gingivalis 381 renders this strain more immune-stimulatory. Conversely, a defective fimB allele and high-level cell surface gingipain activity reduce the capacity of P. gingivalis 33277 to stimulate host cell innate immune responses. In summary, genomic and transcriptomic comparisons identified key virulence characteristics that confer divergent host cell innate immune responses to these highly related P. gingivalis strains.
INTRODUCTION
Porphyromonas gingivalis is considered to be a keystone pathogen that is involved in the development of chronic periodontal disease (1–4), and interest in this microorganism includes its potential roles in other important chronic inflammatory disorders, including cardiovascular disease, rheumatoid arthritis and Alzheimer’s disease (5–7). P. gingivalis can modulate and dampen the ability of the host innate immune receptors known as Toll-like receptors (TLRs) to orchestrate proinflammatory responses aimed at controlling Gram-negative bacterial infections (6, 8, 9). We and our collaborators have previously shown that P. gingivalis employs lipid A phosphatases and a lipid A deacylase to evade host TLR4 recognition of its lipopolysaccharide (LPS), thus contributing to its ability to survive in macrophages, disseminate systemically, and exacerbate atherosclerosis in a murine model (10–12). P. gingivalis also elicits many of its pathogenic effects through the action of cell surface lipoprotein-dependent and fimbria-dependent interactions with host cell TLR2 signaling pathways (13–16). In addition, P. gingivalis gingipains promote TLR2-C5aR cross talk to selectively reconfigure neutrophil TLR2 responses to bacterial ligands. This gingipain-dependent mechanism is an important paradigm underlying the bacterium’s ability to persist in the periodontium, promoting both dysbiosis and chronic inflammation (8, 17).
Modulation of inflammasome activation is recognized as a key aspect of the innate immune response targeted by bacterial pathogens to disrupt host resolution of bacterial infection (18, 19). Inflammasomes are intracellular multiprotein complexes that sense a variety of microbial immunostimulatory molecules, including LPS, lipoproteins, and flagellin, to produce interleukin-1β (IL-1β) and IL-18 as major inflammatory mediators (20). Secreted IL-1β exerts multiple actions to combat bacterial infections, including stimulation of neutrophil recruitment and cytokine and chemokine production (21), and increases in IL-1β levels are associated with both periodontal disease and cardiovascular disease (22, 23). Inflammasome-dependent IL-1β production triggered by Gram-negative bacteria such as P. gingivalis requires a priming step involving the activation of TLR2 to elicit pro-IL-1β synthesis (20, 22). Subsequently, intracellular sensing of microbial factors via Nod-like receptor 3 results in the production and secretion of mature IL-1β (20, 22).
P. gingivalis’ capacities to invade host cells and to manipulate host inflammatory mediators are generally considered to be related to its potential to promote chronic inflammatory disease (2, 6, 9, 24). All strains of P. gingivalis express multiple immunomodulatory virulence factors, including fimbriae, LPS, gingipain proteases, and RagA-RagB antigens (9, 17, 25). However, it is presently unclear if one or more of these factors displays a dominant role in determining the ability of a particular strain to promote disease. Genomic alterations referred to as “pathogenicity islands” that occur between significantly divergent strains have been proposed to determine strain-specific disease association (26–28). For example, strain W83 expresses capsular polysaccharides, fimbrial variants, and RagA-RagB variants that are absent or divergent from those found in strain 33277 (25, 27, 29, 30). Notably, W83 is an isolate from clinical periodontal disease and exacerbates vascular inflammation in animal models, whereas strain 33277 does not exacerbate vascular inflammation in animal models (31, 32). The 33277 and W83 strains diverge significantly, expressing distinct types of multiple virulence factors, including fimbriae, gingipains, and capsular polysaccharides (25, 33). This type of genetic sequence divergence complicates applying a direct comparison of these two strains to rapidly elucidate genetic factors associated with the distinct capacities of these strains to promote host inflammatory responses.
However, the genetically similar strains 33277 and 381 (26, 28, 34) exhibit pronounced differences in their capacities to interact with vascular endothelial cells and to promote systemic inflammation in animal models (32, 33, 35). In addition, we have observed that when 33277 and 381 are grown to stationary phase in a defined culture medium, they display distinct abilities to elicit IL-1β production and to engage TLR2. Consequently, we directly compared the 33277 and 381 genomic sequences and their transcriptomes to identify defined strain-specific, disease-associated elements. The aim of the present study was to identify the genetic factor or factors that confer the differential capacities of these strains to promote host cell innate immune responses and to provide novel insights into characteristics that control strain virulence for P. gingivalis.
RESULTS
P. gingivalis strains 33277 and 381 exhibit distinct capacities to stimulate TLR2 signaling and IL-1β production.
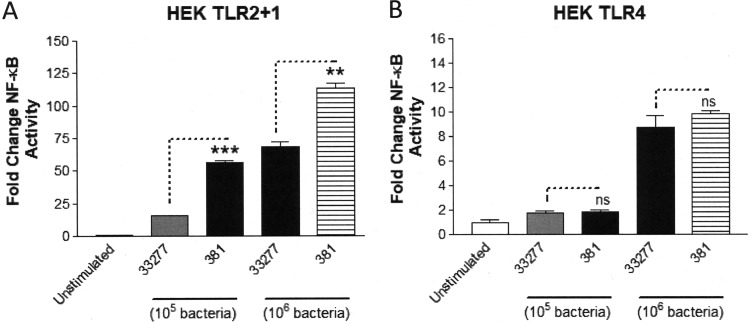
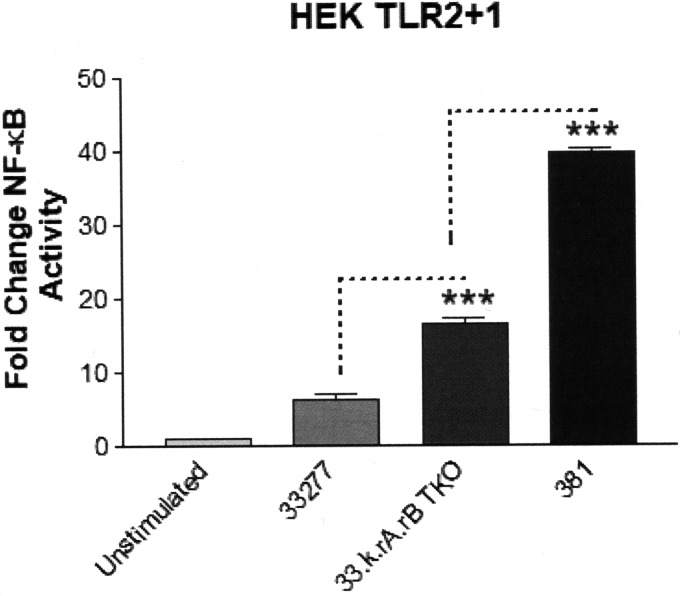
The TLR2/TLR1/CD14 (TLR2) signaling pathway represents a major target for P. gingivalis to promote immune-modulatory effects on host cells both in vitro and in vivo via cell-surface-associated, triacylated lipoproteins (13, 15, 36, 37). We investigated the distinct immune-stimulatory characteristics of strains 33277 and 381 to induce TLR2-dependent signaling since we previously observed that strain 381 elicited a more potent NF-κB response from HEK293 cells expressing TLR2 compared to strain 33277 (15). To determine if the differential activity was due to factors associated with the outer cell surface, we first washed the bacteria with growth medium to remove secreted factors and vesicle components. As shown in Fig. 1A, HEK cell TLR2-dependent signaling was more pronounced by strain 381 compared to strain 33277 at two different titers of bacteria. In contrast, both of the strains elicited reduced magnitudes of TLR4-dependent NF-κB activity relative to TLR2-dependent NF-κB activity and did not display different potencies as TLR4 agonists (Fig. 1B). These data are consistent with our experimental evidence demonstrating that P. gingivalis evades TLR4 signaling via lipid A remodeling (10, 12). Collectively, these data support the differential, strain-specific immune-stimulatory activities of P. gingivalis strains 381 and 33277 via TLR2 signaling.
FIG 1.
Strains 33277 and 381 differentially activate HEK cell TLR2-dependent, but not HEK cell TLR4-dependent, signaling. (A) HEK cells expressing TLR2 were stimulated with either 105 or 106 bacteria of both strains 33277 and 381. (B) HEK cells expressing TLR4 were stimulated with either 105 or 106 bacteria of both strains 33277 and 381. Fold change in NF-κB activation from bacterial stimulations is shown relative to the unstimulated control. Data are presented as means ± standard deviation (SD) from triplicate sample determinations and are representative of three independent experiments. Asterisks indicate statistical significance as determined by Student’s unpaired t test (**, P < 0.001; ***, P < 0.0001). ns, not statistically significant.
TLR2-mediated activation in human THP-1 macrophages is a key step for P. gingivalis-mediated inflammasome priming, and subsequent IL-1β production (22) and increased levels of IL-1β correlate with humans having periodontal disease and bearing P. gingivalis infections (22). We compared the abilities of the two strains to induce cytokine responses from differentiated THP-1 cells. Strain 381 exhibited a significantly more potent capacity to induce IL-1β production from THP-1 cells compared to strain 33277 (Fig. 2A). We observed a similar pattern of strain-specific immune stimulation with respect to IL-6 production in the THP-1 cells (Fig. 2B). The data presented in Fig. 2 demonstrate that P. gingivalis strains 33277 and 381 elicit distinct IL-1β and IL-6 responses in THP-1 cells, which correlate with the differential abilities of these to strains to activate TLR2 (Fig. 1).
FIG 2.
Strains 33277 and 381 exhibit distinct capacities to elicit innate immune responses in differentiated THP-1 cells. THP-1 cells were stimulated with strains 33277 and 381 (105 bacteria). (A) Fold change in IL-1β secretion following bacterial stimulations is shown relative to the unstimulated control. (B) Fold change in IL-6 secretion following bacterial stimulations is shown relative to the unstimulated control. Data are presented as means ± SD from triplicate sample determinations and are representative of three independent experiments. Asterisks indicate statistical significance as determined by Student’s unpaired t test (***, P < 0.0001).
Comparison of the strain 381 and 33277 genomes identifies a polymorphism in the fimB allele.
Based upon the similarity of the 33277 and 381 genomes (28, 34), we hypothesized that a limited number of gene polymorphisms or chromosomal rearrangements accounted for reducing the capacity of 33277 to stimulate host cell innate immune responses. We compared the genomes of our experimentally defined laboratory strains 381 and 33277 as an approach to elucidate the basis for their distinct immune-stimulatory capacities. We conducted a resequencing experiment and mapped duplicate read sets for each strain to the two published genomes. Single nucleotide polymorphisms (SNPs) that were found within the source genome that were possible due to sequencing errors or minor mutations from lab domestication over time were subtracted from the variable SNPS between the strains. We screened the coding regions in both genomes for polymorphisms that might be expected to alter cell surface structures or other key cellular factors (Table 1). The coding regions for six different genes were identified with high confidence as having authentic coding sequence polymorphisms between strains 33277 and 381. Notably, the 33277 fimB allele contains a previously identified polymorphism in its coding region that produces a premature stop codon in the fimB open reading frame (ORF) and potentially leading to the absence or truncation of the FimB component of the major fimbria in the cell surface of 33277 (38). Due to the stop codon present in the 33277 fimB coding region, the full-length fimB gene has been annotated as two separate coding sequences, PGN_0181 and PGN_0182. PGN_0181 encodes the N-terminal region of FimB, and PGN_0182 encodes the remaining C-terminal region of FimB. The fimB gene is part of the fimB to fimE gene cluster, and it is regulated independently from fimA, which encodes the major fimbrial structural subunit (39, 40). In contrast to fimA, fimB encodes a lipoprotein anchor component of the major fimbriae that restricts the length of fimbriae in P. gingivalis 381 and 33277 (38, 40). However, to the best of our knowledge the immune-modulatory capacity of FimB has not been reported. Given the prior evidence for the role of P, gingivalis fimbriae and lipoproteins in activating TLR2 (15, 41), and the high probability that 381 fimB allele encodes a cell surface lipoprotein (38, 40), we selected the fimB allele for further study as a potential novel determinant for the distinct immune-stimulatory capacities of strains 33277 and 381.
TABLE 1.
Coding region polymorphisms identified in the comparison of 33277 and 381 genomesa
| Coding sequence | Nucleotide polymorphism yielding the 381 change to 33277 | Outcome of polymorphism in 33277 protein |
|---|---|---|
| PGN_0076 (YWFC protein) | GCA→GTA | A→V |
| PGN_0181 (FimB) | AAA→TAA | Premature stop |
| PGN_0293 (RagA) | GAG→GTG; AGC→AAC; GCC→ACC | E→V; S→N; A→T |
| PGN_1085 (iron transport) | GTC→GGC | V→G |
| PGN_1328 (periplasmic binding) | C(8)→C(7) | Frameshift |
| PGN_1892 (phosphate transport) | AGT→TGT | S→C |
Gene numbers correspond to 33277 annotations.
The 33277 fimB allele contributes to its reduced immune-stimulatory capacity.
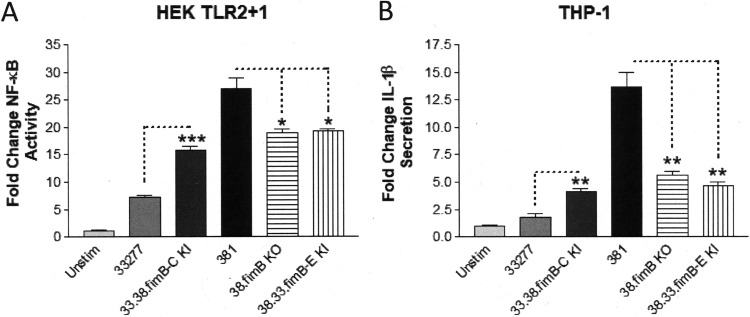
We examined the significance of the 33277 fimB polymorphism in modulating immune-modulatory activity by generating isogenic mutant strains in P. gingivalis 33277 and 381 containing alterations in their respective fimB loci. Notably, the entire 381 fimB to fimE gene cluster was toxic when cloned in Escherichia coli. Consequently, we were unable to construct a targeting vector bearing the intact 381 fimB to -E gene cluster for insertion into 33277. Instead, we created the mutant strain, 33.38.fimB-C KI (knock-in), which replaced the 33277 fimB to -C region with the native 381 fimB-C region at the expense of losing the fimD and fimE genes. For strain 381, we created the isogenic mutant strain, 38.fimB KO (knockout), which bears a deletion in the fimB gene while retaining fimC to -E. We also created strain 38.33.fimB-E KI, which contains a replacement of the native 381 fimB to -E gene cluster with the 33277 fimB to -E gene cluster, effectively replacing the 381 fimB gene with the 33277 fimB gene. In contrast to the 381 fimB to -E operon, the 33277 fimB to -E operon was not toxic in E. coli during recombinant plasmid propagation, suggesting that there is a functional distinction between these gene clusters in E. coli.
We first compared the abilities of the isogenic fimB mutant strains and respective parent strains to stimulate TLR2 activation in HEK293 cells (Fig. 3A). The 33277 isogenic mutant strain, 33.38.fimB-C KI, increased TLR2-dependent NF-κB activation significantly compared to the parental 33277 strain, although it did not stimulate TLR2 as potently as the 381 strain. Conversely, both 381 isogenic mutant strains, 38.fimB KO and 38.33.fimB-E KI, displayed significant reductions in their abilities to stimulate TLR2 compared to the parental 381 strain, although they were not reduced to the level displayed by the 33277 strain. These results indicate that the 381 fimB allele contributes to the ability of P. gingivalis to productively engage TLR2 and partially determines the distinct immune-stimulatory capacities of strains 33277 and 381. Furthermore, the immune-stimulatory capacity of FimB is likely independent from the main FimA structural unit since a 381 isogenic mutant strain bearing a deletion in the fimA allele did not exhibit a reduced capacity to stimulate TLR2 relative to the parental 381 strain (15).
FIG 3.
The distinct capacities of P. gingivalis strains 33277 and 381 to elicit immune-stimulatory responses are dependent on the fimB allele. (A) HEK cells expressing TLR2 were stimulated with the wild-type strains 33277 and 381 and the indicated isogenic fimB mutant strains (105 bacteria). Fold change in NF-κB activation from bacterial stimulations is shown relative to the unstimulated control. (B) Differentiated THP-1 cells were stimulated with wild-type strains 33277 and 381 and the indicated isogenic fimB mutant strains (105 bacteria). Fold change in IL-1β production from bacterial stimulations is shown relative to the unstimulated control. Data are presented as means ± SD from triplicate sample determinations and are representative of three independent experiments. Asterisks indicate statistical significance as determined by Student’s unpaired t test (*, P < 0.01; **, P < 0.001; ***, P < 0.0001).
We next tested the abilities of the wild-type and mutant strains to elicit IL-1β production in THP-1 cells (Fig. 3B). The strains displayed capacities to stimulate IL-1β secretion that were strikingly similar to the patterns observed for the TLR2 activation described above. Collectively, the TLR2 activation and IL-1β production results demonstrate that the intact 381 fimB allele performs a key role in conferring immune-stimulatory activity for this strain. These data also demonstrate that the truncated fimB gene, resulting from the 33277 fimB polymorphism in P. gingivalis 33277, contributes to the reduced immune-stimulatory activity of 33277. However, these data also indicate that the fimB gene is not the sole determinant of the strain-specific immune-modulatory characteristics, suggesting that additional genetic differences contribute to the distinct immune-stimulatory phenotypes of these strains.
Comparison of 33277 and 381 transcriptomes reveals multiple differences in gene expression patterns.
The genomic analyses of gene coding region polymorphisms identified a remarkably limited set of potential immune-modulatory molecules (Table 1), but it did not provide information on differences in gene expression levels between the strains. We decided to compare P. gingivalis strain transcriptomes to search for additional factors that could be differentially expressed between the strains and which could likely contribute to the distinct immune-modulatory phenotypes of these strains. We performed transcriptome sequencing (RNA-seq) analyses to compare the transcriptomes of 33277 and 381 strains grown to stationary phase (Table 2). (For complete strain transcriptome comparisons, see Table S1 in the supplemental material.) In this analysis, we focused our attention on genes that were expressed with a greater than 2-fold difference between the strains and which either represent outer cell membrane components or play a role in the expression of cell surface antigens. Using this approach, we identified a putative operon comprising three genes, PGN_1320, PGN_1318, and PGN_1317, which were much more highly expressed in 381 than the corresponding genes in 33277. Further examination of the genomic regions encompassing this predicted operon in both the 33277 and 381 strains revealed the presence of an insertion sequence (PGN_1319) in the 33277 genome that was absent in the corresponding position in the 381 genome (see Fig. S1 in the supplemental material). Based upon the pronounced increase in expression of this operon in strain 381 relative to strain 33277, and the presence of a putative outer membrane adhesin protein (PGN_1317), we selected this region for further functional analysis.
TABLE 2.
Select differentially expressed genes identified by comparison of the 33277 and 381 transcriptomesa
| Gene upregulated | Fold increase in RNA expression |
|---|---|
| Upregulated in 381 relative to 33277 | |
| PGN_1320 (permease) | 26.38 |
| PGN_1318 (ABC transporter) | 15.83 |
| PGN_1317 (gliding motility) | 11.20 |
| PGN_0288 (mfa2) | 4.18 |
| PGN_0287 (mfa1) | 3.70 |
| PGN_0289 (mfa3) | 3.57 |
| PGN_0180 (fimA) | 3.37 |
| PGN_0291 (mfa5) | 2.92 |
| PGN_0290 (mfa4) | 2.92 |
| PGN_0181 (fimB N terminus) | 1.97 |
| PGN_0183 (fimC) | 1.27 |
| PGN_0184 (fimD) | 1.19 |
| PGN_0182 (fimB C terminus) | 1.09 |
| Upregulated in 33277 relative to 381 | |
| PGN_1430 (HAE1 family) | 4.83 |
| PGN_1429 (conserved) | 4.29 |
| PGN_1841 (rpoA) | 4.08 |
| PGN_1843 (rpsK) | 3.97 |
| PGN_1842 (rpsD) | 3.90 |
| PGN_1904 (hagB) | 3.11 |
| PGN_1896 (wbaP) | 3.11 |
| PGN_1906 (hagC) | 2.99 |
| PGN_1877 (porW) | 2.55 |
| PGN_1054 (vimF) | 2.37 |
| PGN_1970 (rgpA) | 2.27 |
| PGN_0659 (HBP35) | 2.24 |
| PGN_1469 (dppIV) | 2.16 |
| PGN_0900 (prtT) | 2.14 |
| PGN_1466 (rgpB) | 2.11 |
| PGN_1728 (kgp) | 2.01 |
| PGN_0023 (porV) | 2.00 |
| PGN_0898 (PPAD) | 1.77 |
| PGN_0185 (fimE) | 1.22 |
Gene numbers correspond to 33277 annotations.
The PGN_1320-PGN_1317 operon does not contribute to the distinct immune-stimulatory characteristics of strains 33277 and 381.
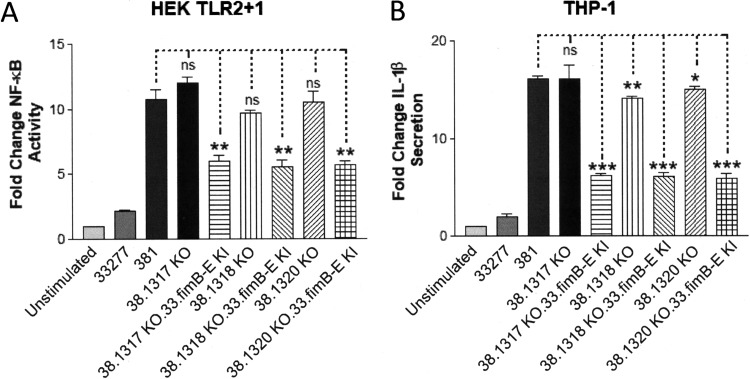
We constructed a series of isogenic mutant strains in the parental 381 strain that contained single deletions in the PGN_1320, PGN_1318, and PGN_1317 coding regions, designated 38.1320 KO, 38.1318 KO, and 38.1317 KO, respectively. We used these strains to test the possibility that the high expression level of one of these genes in strain 381 contributes to its increased immune-stimulatory capacity relative to strain 33277 (Fig. 4). We first compared the capacities of these mutant strains to activate TLR2 relative to the 33277 and 381 strains (Fig. 4A). These results indicated that none of the strains bearing PGN_1320, PGN_1318, or PGN_1317 appreciably altered the ability of the 381 strain to activate TLR2. However, as expected the introduction of the 33277 fimB allele into these strains (38.1320 KO.33.fimB-E KI, 38.1318 KO.33.fimB-E KI, and 38.1317 KO.33.fimB-E KI), led to reduced TLR2-dependent signaling.
FIG 4.
The immune-stimulatory capacity of strain 381 is not appreciably diminished in isogenic mutant strains bearing deletions in PGN_1320, PGN_1318, and PGN_1317 coding sequences. (A) HEK cells expressing TLR2 were stimulated with wild-type strains 33277 and 381 and the indicated isogenic mutant strains (105 bacteria). Fold change in NF-κB activation from bacterial stimulations is shown relative to the unstimulated control. (B) Differentiated THP-1 cells were stimulated with wild-type strains 33277 and 381 and the indicated isogenic mutant strains (105 bacteria). Fold change in IL-1β production from bacterial stimulations is shown relative to the unstimulated control. Data are presented as means ± SD from triplicate sample determinations and are representative of three independent experiments. Asterisks indicate statistical significance as determined by Student’s unpaired t test (*, P < 0.01; **, P < 0.001; ***, P < 0.0001). ns, not statistically significant.
We performed similar experiments in THP-1 cells to measure the capacities of the isogenic mutant strains to modulate IL-1β production (Fig. 4B). A similar pattern of immune-stimulatory activity was observed in the TLR2 assay. Based upon these data, we conclude that the highly expressed PGN_1320 to PGN_1317 operon in strain 381 does not appreciably contribute to the distinct immune-modulatory capacities of strains 381 and 33277. However, these experiments provide additional supporting evidence for the specific influence conferred by the fimB allele in mediating strain-specific host cell innate immune responses.
Given the apparent role of the fimB gene product in contributing to immune-stimulatory capacity of strain 381, we considered the possibility that the minor fimbrial components (e.g., mfa2) might also contribute to the immune-modulatory phenotype of this strain. Supporting this possibility, all of the genes encoding the minor fimbria subunits (mfa1 to mfa5) (42) were upregulated significantly (>3-fold) in the 381 strain relative to the 33277 strain (Table 2). In addition, the minor fimbriae have been reported to activate TLR2 (43). We constructed an isogenic 381 mutant strain that contained a deletion in the entire mfa1 to -5 gene cluster (38.mfa1-5 KO). We compared the immune-stimulatory capacity of this strain to those of the parental 381 strain and strain 33277 using both the TLR2 activation assay and the IL-1β production assay (see Fig. S2 in the supplemental material). Surprisingly, the removal of the gene cluster encoding the minor fimbrial subunits from 381 failed to reduce the strain’s immune-stimulatory capacity in our functional assays.
The data presented in Fig. 4 and Fig. S2 suggested to us that the distinct immune-stimulatory capacities exhibited by strains 381 and 33277 may not exclusively involve strain-specific activators that are preferentially expressed in strain 381. Instead, we considered the possibility that bacterial factors capable of suppressing innate immune responses may be more highly expressed in strain 33277, contributing to the diminished immune-stimulatory activity of this strain.
Strains 381 and 33277 display distinct cell surface gingipain activities.
The RNA-seq data in Table 2 indicate that multiple genes encoding factors associated with virulence were expressed more highly in strain 33277 compared to strain 381. Notably, several genes (wbaP, porW, vimF, and porV), which are associated with the expression of cell-surface-localized, carboxy-terminal domain (CTD)-containing proteins (44), were upregulated greater than 2-fold in strain 33277 (Table 2). In addition, genes encoding the gingipain proteases (kgp, rgpA, and rgpB), which also belong to the CTD family of proteins (44, 45), were upregulated in 33277. The gingipain cysteine proteases are critical for the ability of P. gingivalis to acquire nutrients and have well-established roles as modulators of host cell innate immune-responses in periodontal disease, including the ability to promote production of complement factor C5a or degrade host innate immune coreceptors such as CD14 (17, 46, 47).
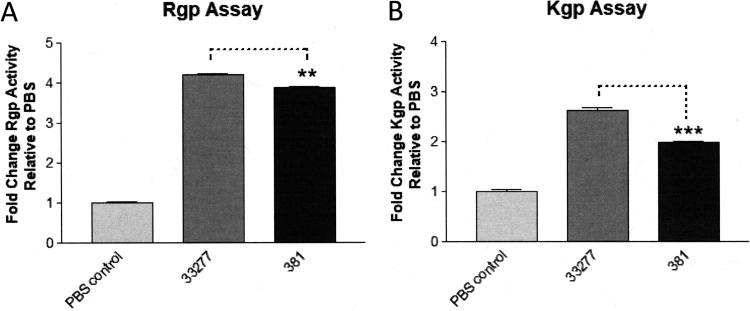
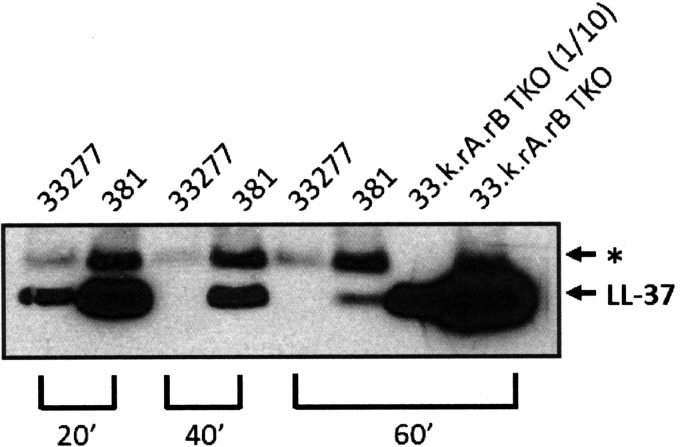
To investigate if these strains express distinct levels of cell surface gingipains, we measured arginine-dependent (Rgp) (Fig. 5A) and lysine-dependent (Kgp) gingipain activities (Fig. 5B) using a chromogenic substrate assay (48, 49). The results of these experiments indicate that both Rgp and Kgp gingipain activities are significantly reduced in strain 381 relative to strain 33277. We also compared the capacities of strains 33277 and 381 to degrade a defined gingipain substrate, the host defense peptide LL-37 (50). As shown in Fig. 6, Western blot analysis of the test samples confirmed that strain 33277 displays a substantially higher level of cell-surface-associated gingipain activity compared to strain 381, based upon its potent ability to degrade LL-37. An isogenic 33277 mutant strain bearing inactivating mutations in all three gingipain genes (33.k.rA.rB TKO) did not degrade the LL-37 substrate relative to the wild-type 33277 or the 381 strains confirmed the gingipain dependence of the LL-37 degradation assay. Examination of the 33.k.rA.rB TKO mutant using the chromogenic gingipain substrate assay confirmed that this strain has substantially reduced total gingipain activity compared to the wild-type P. gingivalis strains (see Fig. S3 in the supplemental material). A peptide displaying an increased molecular weight compared to the full-length LL-37 peptide was also observed in both 33277 and 381 strain reactions (Fig. 6, asterisk), but it appears to be a structural variant of LL-37 that is present in the synthetic peptide preparation (data not shown). Taken together the above data in Fig. 5 and 6 provide novel evidence that the differential expression of gingipain proteases may influence the immune-modulatory phenotypes of strains 33277 and 381.
FIG 5.
Strains 33277 and 381 display distinct cell-surface-associated gingipain activities. (A) Arginine (Rgp)-dependent and (B) lysine (Kgp)-dependent gingipain substrates were coincubated with strains 33277 and 381 (108 bacteria) and the OD450 was measured. Fold change in gingipain activity is reported relative to the PBS control. Data are presented as means ± SD from triplicate sample determinations and are representative of three independent experiments. Asterisks indicate statistical significance as determined by Student’s unpaired t test (**, P < 0.001; ***, P < 0.0001).
FIG 6.
Strains 33277 and 381 display pronounced differences in cell surface gingipain activity toward a defined substrate. Western blot analysis of degradation reactions of the gingipain substrate LL-37. For degradation reactions, synthetic LL-37 was exposed to 33277, 381, or the gingipain-deficient strain 33.k.rA.rB TKO for the indicated times. All samples loaded were from equivalent reaction volumes, except for the 33.k.rA.rB TKO sample, which was loaded at a 1/10 reaction volume relative to the other samples. The asterisk indicates the position of a minor variant form of LL-37 peptide present in the synthetic peptide preparation. The data shown are representative of three independent experiments.
Gingipains contribute to the distinct immune-stimulatory capacities of strain 33277 and strain 381.
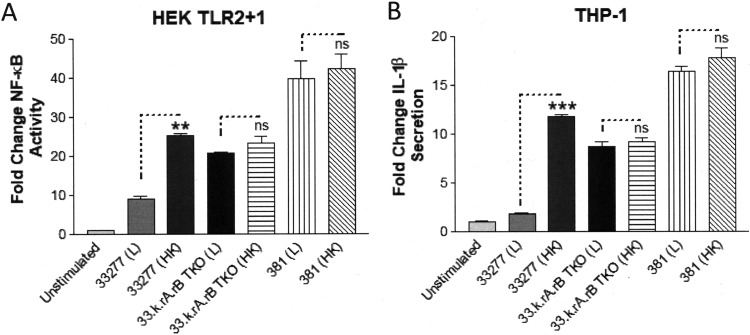
We next tested the 33277 isogenic gingipain-deficient strain (33.k.rA.rB) for its capacity to activate TLR2 relative to the wild-type 33277 and 381 strains (Fig. 7). The results in Fig. 7 demonstrate that the 33277 isogenic gingpain-deficient mutant strain is significantly more potent in its capacity to stimulate TLR2 activity than the parental 33277 strain. As an alternative approach to validate the role of gingipains, we compared the immune-stimulatory capacities of live (L) and heat-inactivated (HK) strains, based upon evidence that heat-inactivation eliminates the ability of strain 33277 to degrade the gingipain substrate, LL-37. Heat-inactivation of the 33277 strain significantly increased its ability to activate TLR2 relative to the live 33277 strain (Fig. 8A), consistent with a reduction of gingipain activity. In contrast, heat inactivation of the 33.k.rA.rB TKO isogenic mutant did not normally increase its immune-stimulatory capacity relative to the parental 33277 strain. Furthermore, the heat-inactivated 381 strain did not always exhibit statistically significant increases in immune-stimulatory activity relative to the live 381 strain, consistent with a more limited immune-suppressive role for the gingipains in determining the immune-modulatory characteristics of strain 381. Similar results were obtained when the samples were tested on THP-1 cells, confirming that higher levels of gingipains contribute to the reduced ability of strain 33277 to stimulate inflammasome-dependent IL-1β production (Fig. 8B).
FIG 7.
The 33277 isogenic mutant strain deficient for gingipains exhibits increased immune-stimulatory capacity. HEK cells expressing TLR2 were stimulated with wild-type strains 33277 and 381 and the gingipain-deficient strain, 33.k.rA.rB TKO (105 bacteria). The fold change in NF-κB activation from bacterial stimulations is shown relative to the unstimulated control. Data are presented as means ± SD from triplicate sample determinations and are representative of three independent experiments. Asterisks indicate statistical significance as determined by Student’s unpaired t test (***, P < 0.0001).
FIG 8.
Heat inactivation increases the immune-stimulatory activity of strain 33277. (A) HEK cells expressing TLR2 were stimulated with either live (L) or heat-inactivated (HK) strains 33277, 33.k.rA.rB TKO, and 381 (105 bacteria). Fold change in NF-κB activation from bacterial stimulations is shown relative to the unstimulated control. (B) Differentiated THP-1 cells were stimulated with either live (L) or heat-inactivated (HK) strains 33277, 33.k.rA.rB TKO, or 381 (105 bacteria). Fold change in IL-1β production from bacterial stimulations is shown relative to the unstimulated control. Data are presented as means ± SD from triplicate sample determinations and are representative of two independent experiments. Asterisks indicate statistical significance as determined by Student’s unpaired t test (**, P < 0.001; ***, P < 0.0001). ns, not statistically significant.
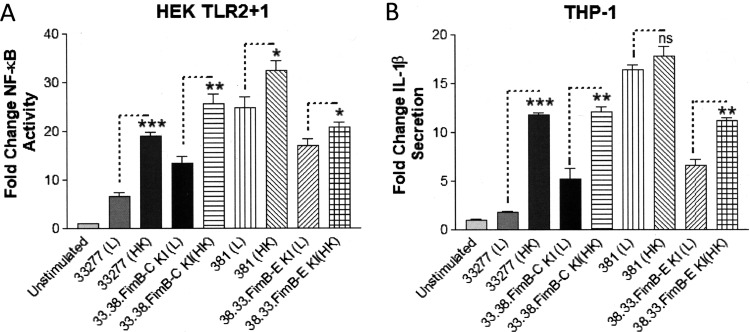
We also examined the combined influence of gingipain activity and the fimB allele in both strains 33277 and 381 on TLR2 activity by comparing the live parental strains and isogenic fimB mutant strains with their corresponding heat-inactivated strains (Fig. 9A). As expected, the live 33277 isogenic mutant strain bearing the 381 fimB allele (33.38.fimB-C KI) displayed a partially increased capacity to activate TLR2 compared to the live parental 33277 strain. The potencies of the 33277 and 33.38.fimB-C KI mutant, which contains the 381-type fimB allele, were further increased by heat inactivation. However, neither of these strains displayed maximal immune-stimulatory characteristics of the 381 strain. In contrast to strain 33277, heat inactivation of the parental 381 strain typically induced a more modest increase in its capacity to activate TLR2, and this outcome was not always statistically significant. These results are consistent with gingipain activity not contributing as substantially to immune repression by strain 381, due to a lower total cell surface expression of gingipains in strain 381 relative to strain 33277. In support of this, the strains having a 33277 background displayed a more marked increase in immune-stimulatory activity following heat inactivation compared to the strains having a 381 background. Similar results were obtained with respect to the abilities of these strains to stimulate IL-1β production in THP-1 cells (Fig. 9B). Based upon the cumulative evidence, we conclude that strain-specific differences in both gingipain activity and the fimB allele confer the distinct immune-stimulatory phenotypes characteristic for strains 33277 and 381. In other words, increased cell surface gingipain expression and a defective fimB variant render strain 33277 significantly less immune stimulatory than the 381 strain.
FIG 9.
Influence of heat inactivation on the immune-stimulatory activity of strains 33277 and 381 and the isogenic fimB mutant strains. (A) HEK cells expressing TLR2 were stimulated with live (L) or heat-inactivated (HK) wild-type strains 33277 and 381 and the indicated isogenic fimB mutant strains (105 bacteria). Fold change in NF-κB activation from bacterial stimulations is shown relative to the unstimulated control. (B) Differentiated THP-1 cells were stimulated with live (L) or heat-inactivated (HK) wild-type strains 33277 and 381 and the indicated isogenic fimB mutant strains (105 bacteria). Fold change in IL-1β production from bacterial stimulations is shown relative to the unstimulated control. Data are presented as means ± SD from triplicate sample determinations and are representative of two independent experiments. Asterisks indicate statistical significance as determined by Student’s unpaired t test (*, P < 0.05; **, P < 0.001; ***, P < 0.0001). ns, not significant.
Cell surface CTD proteins limit the immune-stimulatory capacity of P. gingivalis strains 33277 and 381.
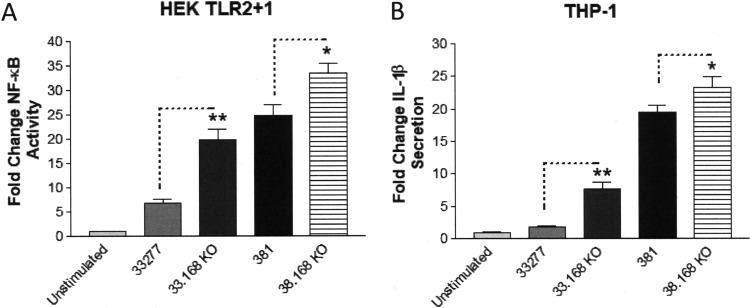
As mentioned above, the gingipains are outer-cell-surface CTD protein family members (44). Consequently, we expected that 33277 and 381 P. gingivalis strains devoid of CTD proteins would exhibit enhanced immune-stimulatory capacities similar to the phenotype observed for the 33277 gingipain-deficient strain. We decided to assess the potential immune-suppressive roles of CTD proteins, including the gingipains, by eliminating all of the CTD-containing proteins from the cell surface of both strains 33277 and 381. For this purpose, we used the isogenic 332277 and 381 mutant strains, which are deficient for the PGN_0168 gene, designated 33.168 KO and 38.168 KO, respectively. PGN_0168 encodes an enzyme that is essential for A-LPS synthesis, and A-LPS constitutes the membrane anchor to which CTD proteins are attached (44, 51–53). Therefore, these mutant strains are expected to express reduced levels of CTD family members, including gingipains, on their cell surface, resulting in a nonpigmented phenotype. We verified both the pigmentation-negative phenotypes (see Fig. S4 in the supplemental material), and the significantly reduced gingipain activities (Fig. S3) of these mutant strains. As shown in Fig. 10A, both the 33.168 KO strain and the 38.168 KO strain displayed enhanced capacities to stimulate TLR2 compared to their respective parental 33277 and 381 strains, consistent with reduced levels of gingipains on the cell surface. Likewise, both the 33.168 KO and 38.168 KO strains potently stimulated IL-1β production in THP-1 cells compared to their respective parental 33277 and 381 strains (Fig. 10B). These data provide additional novel evidence that both strains 33277 and 381 express A-LPS-linked cell surface CTD proteins, including the gingipains, which are capable of exerting immune-suppressive effects on host cells.
FIG 10.
Isogenic mutant strains of 33277 and 381 that are devoid of CTD proteins display increased immune-stimulatory activity. (A) HEK cells expressing TLR2 were stimulated with wild-type strains 33277 and 381 and the indicated isogenic mutant strains (105 bacteria). Fold change in NF-κB activation from bacterial stimulations is shown relative to the unstimulated control. (B) Differentiated THP-1 cells were stimulated with wild-type strains 33277 and 381 and the indicated isogenic mutant strains (105 bacteria). Fold change in IL-1β production from bacterial stimulations is shown relative to the unstimulated control. Data are presented as means ± SD from triplicate sample determinations and are representative of two independent experiments. Asterisks indicate statistical significance as determined by Student’s unpaired t test (*, P < 0.05; **, P < 0.001).
DISCUSSION
Previous studies by Rodrigues et al. provided compelling evidence that the highly related P. gingivalis strains 33277 and 381 display remarkably divergent capacities to interact with vascular endothelial cells and induce proinflammatory cytokine profiles, although the genetic basis for these strain-specific activities was not determined (33). In the present study, we investigated genetic differences between P. gingivalis strains 33277 and 381 in order to more fully understand the basis of their distinct capacities to activate TLR2-dependent signaling and stimulate IL-1β production in human cells. Our data indicate that the distinct immune-stimulatory capacities of these strains is determined in part by functional contributions of the fimB allele present within each genome of these strains. This finding reveals a previously unappreciated function of the FimB subunit of major fimbriae as an important cell surface activator of host TLR2-dependent innate immune responses for strain 381. However, we note that the 381 strain reported by Nagano et al. (38) differed from the 381 strain used in the present study (54). Specifically, the 381 strain used in our study contains an intact fimB allele, whereas the 381 strain used by Nagano et al. contains a 33277-type fimB allele bearing the premature stop codon in the coding region. The fimB gene is part of a gene cluster containing the major fimbrial subunit genes fimB to fimE (38), whereas the fimA gene, encoding the major structural fimbrial subunit, is expressed by an independent promoter (39). The major fimbria in P. gingivalis represents a prominent virulence factor in this organism that regulates a number of functions associated with pathogenicity, including its capacity to invade host cells and promote inflammatory disorders such as periodontitis and atherosclerosis (6, 35, 55). Early studies established the FimA component of major fimbria as a specific ligand for TLR2 in epithelial cells (41). Further peptide mapping studies demonstrated that FimA was capable of eliciting CD14-dependent TLR2/CD11b/CD18 activation in macrophages (56). In addition, the accessory structural subunits FimC, FimD, and FimE, which are associated with FimA, were reported to confer virulence characteristics to the 33277 strain by activating TLR2/CR3-dependent signaling (40, 57). However, these earlier studies did not examine the contribution of FimB, which may be required for the full biological activity of the major fimbria in the context of live P. gingivalis. Notably, during fimbria assembly and maturation, the N-terminal lipoprotein regions of FimA and FimC to FimE are removed by gingipain processing during fimbrial maturation, suggesting that these proteins do not function as conventional triacylated lipoprotein agonists for TLR2 signaling (37, 40). In line with these data, our previous evidence indicated that both 33277 and 381 isogenic mutant strains deficient in the fimA gene were not appreciably altered in their capacities to stimulate TLR2/TLR1/CD14 activation (15), supporting the idea that FimA is not required for live P. gingivalis to activate TLR2 signaling. In contrast to the fimA and fimC to -E genes, others have demonstrated that the fimB gene produces a mature fimbrial lipoprotein subunit that both controls the length of the fimbria and serves to anchor the major fimbrial structural proteins (FimA, FimC, FimD, and FimE) to the cell surface via its lipoprotein moiety (38, 40). Our data support the concept that the specific immune-stimulatory capacity of the major fimbria in strain 381 is partly due to the full-length FimB protein functioning as a direct ligand for TLR2 activation, which can also partially account for this strain’s robust ability to promote THP-1 cell-dependent inflammasome priming and IL-1β production via TLR2 activation (22). Conversely, our studies indicate that part of the reduced capacity of the 33277 strain to elicit TLR2 signaling and IL-1β production is conferred by aberrant FimB protein expression on the outer surface of the bacterium, presumably due to the premature stop codon in the 33277 fimB variant (38). Formal validation of this model will require examining 381 and 33277 isogenic mutant strains that contain the functional fimB allele but are deficient for the fimA and fimC to -E alleles. These types of experiments will clarify the respective functional immune-modulatory roles of FimB and the other Fim proteins.
Another important finding in the present study is that strains 381 and 33277 display remarkably distinct gene expression patterns. The transcriptomic profiling of these strains validated RNA seq as a powerful approach to identify genetic and functional differences between two closely related strains. For example, we discovered that the position of an insertion sequence (PGN_1319) is unique to the 33277 genome, and it potently interferes with the expression of a putative operon containing three genes (PGN_1320, PGN_1318, and PGN_1317) in strain 33277. However, despite the pronounced differential regulation of these genes, they do not appear to be involved in conferring the immune-modulatory phenotypes examined in the present study. Nevertheless, it is possible that they mediate another strain-specific phenotype or phenotypes between strains 381 and 33277.
The transcriptome comparison also demonstrated that select genes encoding CTD family proteins, as well as genes encoding factors that are involved in regulating CTD family protein expression, are upregulated in strain 33277 relative to strain 381. The CTD protein family consists of approximately 30 or more distinct proteins, including the gingipains, and these proteins are transported to the cell surface via the type IX secretory system (T9SS) (44). Subsequently, CTD family proteins are either attached to the cell surface via an A-LPS anchor or secreted (45). We observed upregulated genes in 33277, including those genes encoding proteins required for CTD protein transport (e.g., porL to porN and porT-porW), major virulence-associated CTD family proteins (e.g., kgp, rgpA, rgpB, and PPAD), and A-LPS synthesis (e.g., wbaP and waaL) (7, 44, 52, 53, 58). The mechanism underlying the strain-specific upregulation of genes involved in CTD protein expression is currently unknown. However, there is currently intense interest in understanding the regulation of genes governing CTD protein expression given the potential pathological role of CTD proteins such as gingipains and PPAD in human inflammatory diseases, including, chronic periodontitis, Alzheimer’s disease, and cardiovascular disease (5, 7). It is noteworthy that there was a global increase in genes encoding RNA polymerase and ribosomal subunit proteins in 33277 relative to 381, suggesting that the 33277 strain is metabolically more active. However, this distinction does not account for the specificity of differential gene expression patterns between the two strains. It is possible that specific response regulators may be differentially expressed in these two strains. We did not observe appreciable strain-specific differences in the expression of the two-component system genes (porX and porY), which have been implicated in regulating the gene expression of T9SS components (44, 59). On the other hand, the gene (sigP) encoding the extracytoplasmic function sigma factor, SigP, was upregulated approximately 1.8-fold in strain 33277. Notably, the SigP factor functions as a response regulator for the PorXY complex to promote expression of the por genes (44, 59). These types of observations suggest that further comparison of these two strains may be a useful tool for future studies aimed at elucidating the strain-specific regulation of the T9SS system and CTD protein expression revealed in this study.
Collectively, the observations regarding strain differences in T9SS and gingipain gene expression patterns led to the finding that cell surface gingipain activity in combination with the fimB allele significantly contributes to the strain-specific differences in HEK cell TLR2 activation and THP-1 cell IL-1β production between the two strains. Specifically, our data demonstrate that the elevated level of gingipain expression in strain 33277, compared to strain 381, serves to dampen its capacity to elicit robust innate immune responses. Our experiments did not elucidate the mechanism regarding how the gingipains diminish HEK cell TLR2-dependent NF-κB activation or THP-1 cell-dependent IL-1β production. However, evidence from other investigators has shown that P. gingivalis gingipains can directly degrade secreted IL-1β following inflammasome activation of THP-1 cells (60). This could also potentially contribute to diminished NF-κB activity in the macrophage via a reduction of autocrine IL-1R-dependent signaling. Other studies have indicated that gingipains directly target membrane-bound CD14 to accomplish P. gingivalis immune-dampening activity of TLR2-dependent responses in macrophages (24, 61), and both HEK cell assays and differentiated THP-1 cell assays in the present study employed CD14-mediated TLR2 signaling. In addition, the gingipains have been shown to modulate TLR2 signaling via complement 5a receptor (C5aR) cross talk in neutrophils and macrophages (8, 62). A direct comparison of strains 33277 and 381 and the isogenic gingipain mutant strains’ abilities to modulate CD14 binding function or C5aR-dependent signaling should address these possibilities.
It is also appropriate to consider how our findings regarding the fimB gene and gingipain activities in the present study may provide mechanistic explanations for a previous strain comparison by Rodrigues et al. examining the capacities of 33277 and 381 to interact with vascular endothelial cells (33). In that study, the authors examined multiple strain-specific characteristics, including the relative capacities of these strains to adhere to, traffic, and survive within human coronary artery endothelial (HCAE) cells, as well as the strains’ capacities to elicit proinflammatory host cell responses. The authors observed that the 381 strain exhibited significantly increased adherence to HCAE cells, and 381 trafficked intracellularly through the autophagic pathway, whereas 33277 trafficked via the endosomal pathway. In addition, the two strains exhibited distinct intracellular survival patterns. It is possible that differential gingipain expression contributed to the strain-specific adherence, trafficking, and survival phenotypes observed in that study, given the purported roles of gingipains in mediating strain 33277 adherence to and release from epithelial cells (63) and their roles in mediating 381 and 33277 intracellular invasion, trafficking, and survival in aortic endothelial cells (64, 65). Similarly, it is plausible that FimB-dependent fimbrial structure contributes to the strain-specific modes of adherence and intracellular entry in HCAE cells based upon previous evidence indicating that 381-type fimbriae mediate invasion into aortic endothelial cells (64, 66), as well as bacterial adherence and intracellular uptake by macrophages (67). In this regard, FimB might be a virulence determinant conferring the capacity of the invasive strain, 381, to promote periodontal disease or atherosclerosis, in contrast to the avirulent, noninvasive strain, 33277 (32, 35, 68). Perhaps both the cell surface gingipains and FimB influence the distinct capacities of strains 33277 and 381 to elicit TLR2-dependent cytokine responses (e.g., IL-6, MCP-1, VCAM, and E-selectin) observed in HCAE cells, since these cells express functional TLR2 (69). Formal investigation into the roles of FimB and gingipains in determining the strain-specific interactions of P. gingivalis with vascular endothelial cells and disease models will be required to test these possibilities.
We also note that it is likely that one or more factors in addition to fimB and gingipains contribute to the reduced immune-inflammatory potential to the 33277 strain, since the 33277 mutant strains (both single- and multiple-gene knockouts) did not typically achieve the maximal immune-stimulatory capacity displayed by strain 381. Additional candidate factors could be CTD family proteins other than the gingipains since there are more than 30 members of the CTD protein family (44). In line with this idea, we identified additional upregulated CTD family genes, PGN_0898 and PGN_0900, which encode a peptidyl arginine deiminase (PPAD) and a PrtT-related thiol protease, respectively. It is possible that the gingipains and PPAD play a cooperative role in promoting some aspects of dampened innate immune responses to strain 33277, given the dependence that PPAD activity has for Rgp protease activity and the potential for PPAD-dependent citrullination to modulate host cell responses (7). Interestingly, an early study found a single-nucleotide polymorphism creates a stop codon in the kgp genes of 381 and 33277 produces a separation of the Kgp catalytic domain from the adhesin domain relative to strains such as W12 (70). These types of strain-specific variations in the lysine gingipain locus emphasize the need to further explore the roles of these proteases in determining strain-specific virulence. These observations encourage future investigations into the role that these and other CTD proteins play in mediating the immune-modulatory activity of P. gingivalis.
In summary, our findings demonstrate that both the fimB allele and levels of gingipain expression are key determinants responsible for conferring the distinct immune-modulatory capacities of the highly related P. gingivalis strains 381 and 33277. These findings support the current focus on the fimbriae and CTD family proteins such as the gingipains as key virulence factors mediating the pathogenic role of P. gingivalis in chronic human inflammatory diseases (5–8, 25, 55, 71). The present study also validates further mining of the genomic and transcriptomic results obtained from this strain comparison as an efficient experimental approach to elucidate additional factors and regulatory pathways that determine virulence characteristics for P. gingivalis.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
P. gingivalis strains ATCC 33277 and 381 were from our culture collection, and the identity of each strain was verified by whole-genomic sequencing. All bacterial stains used in this study were grown on blood agar plates containing 5% sheep’s blood. Liquid cultures were grown in TYHK broth (30 g/liter Trypticase soy broth, 5 g/liter yeast extract, and 1 mg/liter vitamin K3). Following sterilization by autoclaving, filter-sterilized hemin was added to TYHK broth, just prior to inoculation, to a final concentration of 1 μg/ml. All strains were grown in an anaerobic growth chamber (5% H2, 5% CO2, 90% N2) at 37°C.
Generation of isogenic mutant strains.
The 33277 and 381 isogenic mutant strains used in this study were constructed using antibiotic resistance cassettes (ermF, tetQ, and cepA) in gene-targeting plasmids designed to either delete specific coding regions or insert desired replacement sequences by homologous recombination. The gene-targeting plasmids were introduced into strains by natural transformation and selected on TYHK agar plates containing the appropriate selection antibiotic (erythromycin [5 μg/ml], tetracycline [1 μg/ml], or ampicillin 2 μg/ml]). The genotype for each mutant strain was verified by PCR analyses. For construction of the isogenic mutant strain 33.381.fimB-C KI, the 33277 fimB to -E gene cluster was initially replaced with an ermF cassette. Subsequently, a gene-targeting plasmid bearing the 381 fimB-fimC gene cluster positioned between the upstream fimA gene and a downstream tetQ cassette joined to the fimE 3′-flanking region was introduced into this strain, replacing the ermF cassette and leaving an intact 381-type fimB-fimC gene cluster, but lacking the fimD and fimE genes in the 33277 background. For construction of the isogenic mutant strain 38.33.fimB-E KI, the 381 fimB to -E gene cluster was initially replaced with an ermF cassette. Subsequently, a targeting plasmid bearing the 33277 fimB to -E gene region positioned between the upstream fimA gene and downstream tetQ cassette and fimE flanking sequence was then introduced into this strain, replacing the ermF cassette and leaving an intact 33277-type fimB to -E gene cluster in the 381 background. For construction of the 381 isogenic mutant strains, 38.1320 KO, 38.1318 KO, and 38.1317 KO, deletions were generated in either PGN_1320, PGN_1318, or PGN_1317 coding regions by targeting plasmids containing an ermF cassette located between the upstream and downstream flanking regions for the gene to be deleted. The same three targeting plasmids were separately introduced into strain 38.33.fimB-E to construct the double mutant strains 38.1320 KO.33.fimB-E KI, 38.1318 KO.33.fimB-E KI, and 38.1317 KO.33.fimB-E KI, all in the 381 background. The isogenic mutant strain 38.mfa1-5 KO was generated by using a gene-targeting plasmid containing an ermF cassette located between the upstream and downstream flanking regions for mfa1 to mfa5, resulting in deletion of the mfa1 to -5 gene cluster from the 381 background. The 33277 isogenic mutant strain bearing deletions in all three gingipain alleles was created using three sequential gene-targeting plasmids designed to replace the kgp coding sequence with cepA, the rgpA coding sequence with ermF, and the rgpB coding sequence with tetQ. The 33277 and 381 isogenic mutant strains 33.168 KO and 38.168 KO are deficient for expression of cell surface CTD family proteins and were created by replacing the coding sequence for PGN_0168 with ermF. The black pigmentation-negative phenotype characteristic of gingipain-deficient strains used in this study was verified on blood agar plates (Fig. S4).
Whole-genome resequencing and variant analyses.
Duplicate bacterial genomic DNA preparations from 1-ml stationary-phase cultures were generated for both 33277 and 381 strains using the DNeasy Blood and Tissue kit (Qiagen). The samples were then purified further using the ZymoDNA Clean and Concentrator-5 kit (Zymo Research).
Genomic sequencing was performed as previously described (72). Briefly, DNA libraries were prepared using the Nextera XT kit (Illumina, San Diego, CA, USA), according to the manufacturer’s specifications. Libraries were the then sequenced using an Illumina MiSeq platform (2 × 300 bp). The BBDuk (73) tool for Geneious software version 9.1.8 (74) was used to quality trim and filter Illumina adapters, artifacts, and PhiX from reads. Paired reads with quality scores averaging <6 before trimming or with a length of <20 bp after trimming were discarded. The remaining reads were mapped using Geneious mapper to the published reference genomes of 33277 (accession no. AP009380.1) (75) and 381 (accession no. CP012889.1) (54), resulting in 40 to 70× mean coverage per duplicate sample. Polymorphisms were then identified using a minimum variant frequency of 0.25 and minimum coverage of 10 reads relative to the reference. Polymorphism frequencies in each culture were determined and gated at a >10% threshold. We summarize the data in Table 1. Only unique mutations found in both replicate cultures (frequency of >80%) were summarized.
RNA-seq and transcriptome analyses of strains 33277 and 381.
RNA was purified from triplicate P. gingivalis cultures of each strain that were treated with TRIzol (Invitrogen) to facilitate cell lysis. Briefly, a fully grown 3-ml culture was spun, the pellet was brought up in 900 μl TRIzol, mixed with 200 μl chloroform, and the sample was spun for 15 min. The upper aqueous layer was transferred to a new tube, an equal volume of isopropanol added, mixed well and frozen in a –80°C freezer. The next day, the samples were thawed, pelleted, and washed with 70% ethanol, and then resuspended in H2O. Twenty micrograms of this sample, comprising crude RNA, was subjected to DNase treatment using RQ1 DNase (Promega). The sample was again subjected to TRIzol and chloroform treatment and centrifuged, and the upper aqueous layer was concentrated using the Zymo RNA Clean and Concentrator-5 kit (Zymo Research), according to the manufacturer’s instructions.
RNA samples were then processed for sequencing (Center for Precision Diagnostics, University of Washington). rRNA was removed from the total RNA using the Ribo-Zero (Bacteria) kit (Epicentre, Madison, WI). mRNA was purified using the Zymo RNA Clean and Concentrator kit (Zymo Research, Irvine, CA). The RNA concentration and integrity were monitored before and after rRNA removal using a Qubit 3.0 fluorometer (Life Technologies, Waltham, MA) and Agilent RNA ScreenTape Assay for the 4200 TapeStation system (Agilent Technologies, Santa Clara, CA). cDNA libraries from rRNA-depleted RNA were generated using random-primed cDNA synthesis methods according to the ScriptSeq v2 RNASeq Library preparation protocol (Epicentre, Madison, WI). Prior to second-strand cDNA synthesis, the di-tagged cDNA was purified using the Agencourt AMPure XP system (Beckman Coulter, Brea, CA). ScriptSeq index primers were added to the libraries, which were then PCR amplified for 15 cycles. The clustering of the index-coded samples was performed on a cBot Cluster Generation system using TruSeq PE Cluster kit v3-cBot-HS (Illumina, San Diego, CA) according to the manufacturer’s instructions. After cluster generation, the library preparations were sequenced on an Illumina Hiseq 2500 platform, and paired-end reads were generated (2× 100 bp).
Barcodes were trimmed, and low-quality and short sequences (<100 bp) were filtered using BBDuk (https://sourceforge.net/projects/bbmap) (76), implemented in Geneious version 11 (74). Expression values for each mRNA sample were generated by mapping of paired reads onto the annotated reference genome of P. gingivalis ATCC 33277, resulting in >500× coverage per sample (range of 17 to 20 million reads mapped per sample). Expression-level counts and differential expression calculations in triplicate between strains were performed using Geneious version 11 using DESeq2 normalization (77). DESeq2 uses a model based on the negative binomial distribution with variance and mean linked by local regression (78).
HEK cell TLR2 and TLR4 reporter assays.
HEK293 cells were cultured in high-glucose Dulbecco’s modified Eagle’s medium (DMEM), containing 10% heat-inactivated fetal bovine serum (FBS), and supplemented with penicillin and streptomycin. HEK293 cells were plated in 96-well plates (4 × 104 cells per well) and transfected the following day with plasmids encoding NF-κB-dependent firefly luciferase reporter, β-actin promoter-dependent Renilla luciferase reporter, in combination with either plasmids containing human TLR2, human TLR1, and human CD14, or plasmids containing human TLR4, human MD-2, and human CD14 as described previously (15). At 18 to 20 h posttransfection, test wells were stimulated in triplicate with 105 bacteria in stimulation medium (DMEM containing 10% human serum) for 4 h at 37°C. For initial preparation of bacterial stimulation samples, strains 33277 and 381 were grown to the stationary phase (24 to 48 h) pelleted, and washed once in TYHK, and their concentration was determined by measuring the optical density at 600 nm (OD600). For experiments using heat-inactivated bacteria, the bacteria were heated at 100°C for 5 to 10 min prior to suspension in stimulation medium. After the stimulation reactions were terminated, luciferase activity of the plate samples was assayed using a dual-luciferase assay reporter system (Promega, Madison, WI). NF-κB activity was measured as the ratio of NF-κB-dependent firefly luciferase activity to β-actin promoter-dependent Renilla luciferase activity, which served as an internal standard. The data were plotted as the fold difference of NF-κB activity of the test samples relative to the unstimulated control.
THP-1 cell assay for IL-1β production.
Human THP-1 monocytic cells were cultured in growth medium (RPMI 1640 medium; Life Technologies, Carlsbad, CA) containing 10% heat-inactivated FBS and supplemented with penicillin (100 U/ml) and streptomycin (100 μg/ml). The cells were plated in 96-well plates (4 × 104 cells per well) in growth medium containing phorbol-12 myristate-13 acetate (100 nM) to promote differentiation for 20 to 24 h. The next day, the cells were rinsed once with growth medium and exposed to RPMI 1640 medium containing 10% heat-inactivated FBS supplemented with 100 nM vitamin D3 (Sigma-Aldrich, St. Louis, MO) for an additional 24 to 48 h. For preparation of bacterial stimulation samples, bacterial strains were grown to the stationary phase (24 to 48 h), pelleted, and washed once in TYHK medium, and their concentration was determined by measuring the OD600, and suspended in stimulation medium. Subsequently, the differentiated THP-1 cells were stimulated in triplicate with 105 bacteria suspended in RPMI 1640 medium containing 10% heat-inactivated FBS for 5 h at 37°C. For experiments using heat-inactivated bacteria, the bacteria were heated at 100°C for 5 to 10 min prior to suspension in stimulation medium. Following the 5-h stimulation, THP-1 cell supernatants (100 μl) analyzed by enzyme-linked immunosorbent assay (ELISA) to quantify human IL-1β production according to the manufacturer’s instructions (BD Biosciences). IL-1β production was determined by measuring the absorbance at OD450 on a microplate reader. The data were plotted as the fold change in IL-1β production of the test samples relative to the unstimulated control.
Gingipain activity assays.
Lysine (Kgp)-dependent and arginine (Rgp)-dependent gingipain activities were measured for the wild-type, gingipain-deficient, and A-LPS-deficient P. gingivalis strains using chromogenic substrates N-α-acetyl-l-lysine-p-nitroanilide and N-α-benzoyl-l-Arg-p-nitroanilide as previously described with slight modifications (48, 49). For each test sample, 108 bacteria were washed, pelleted, and suspended in 100 μl of phosphate-buffered saline (PBS) and combined with 100 μl of the appropriate substrate solution (either 0.75 mM N-α-acetyl-l-lysine-p-nitroanilide, 10 mM l-cysteine, 10 mM CaCl2, 100 mM Tris-HCl (pH 8.0), or 0.75 mM N-α-benzoyl-l-Arg-p-nitroanilide, 10 mM l-cysteine, 10 mM CaCl2, 100 mM Tris-HCl [pH 8.0]). Reactions proceeded for 5 to 15 min at 30°C. Subsequently, OD450 measurements were recorded to quantify the Rgp-dependent and Kgp-dependent gingipain activities of the wild-type and mutant P. gingivalis strains.
Strain-dependent gingipain activity toward a defined biological substrate was measured as the capacity of live bacterial strains to degrade a synthetic LL-37 peptide (AnaSpec). For each reaction, 5 μl of LL-37 (5 μg) suspended in H2O was combined with 45 μl of P. gingivalis strain 33277 or 381 suspended in TYHK (OD600 = 0.1). The reaction mixtures were incubated at 37°C for the times specified in Fig. 6. At the appropriate time points, 10 μl of each reaction mixture was removed, and the reaction was terminated by addition of 140 μl of LDS buffer (Invitrogen) and the sample heated for 3 min at 100°C. For Western blotting of the reactions, terminated samples (20 μl) were resolved in nonreducing, denaturing 4 to 20% Tris-glycine polyacrylamide gels (Invitrogen, Carlsbad, CA) and then transferred to a 0.2-μm-pore nitrocellulose membrane (Invitrogen, Carlsbad, CA). Membranes were probed overnight at 4°C using a monoclonal anti-LL37 antibody (Santa Cruz). The blot was washed and probed with goat anti-mouse IgG antibody-horseradish peroxidase conjugate (Jackson ImmunoResearch Laboratories, West Grove, PA) for 1 h at room temperature. Detection of proteins was achieved by treating the blots with SuperSignal West Pico chemiluminescent substrate (Pierce, Rockford, IL) followed by exposure to BioMax MS film (Kodak).
Accession number(s).
RNA-seq data have been deposited in the GEO database under accession no. GSE128899.
Supplementary Material
ACKNOWLEDGMENTS
We thank Erik L. Hendrickson for assisting with the RNA-seq data processing. We thank Lil M. Pabón for critical reading of the manuscript.
Research reported in this publication was supported by the National Institute of Dental and Craniofacial Research of the National Institutes of Health (NIH) under awards R21DE026338 (to S.R.C. and R.P.D.) and 1R01DE026186 (to J.S.M.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/IAI.00319-19.
REFERENCES
- 1.Darveau RP. 2009. The oral microbial consortium’s interaction with the periodontal innate defense system. DNA Cell Biol 28:389–395. doi: 10.1089/dna.2009.0864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Darveau RP. 2010. Periodontitis: a polymicrobial disruption of host homeostasis. Nat Rev Microbiol 8:481–490. doi: 10.1038/nrmicro2337. [DOI] [PubMed] [Google Scholar]
- 3.Hajishengallis G, Darveau RP, Curtis MA. 2012. The keystone-pathogen hypothesis. Nat Rev Microbiol 10:717–725. doi: 10.1038/nrmicro2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hajishengallis G, Liang S, Payne MA, Hashim A, Jotwani R, Eskan MA, McIntosh ML, Alsam A, Kirkwood KL, Lambris JD, Darveau RP, Curtis MA. 2011. Low-abundance biofilm species orchestrates inflammatory periodontal disease through the commensal microbiota and complement. Cell Host Microbe 10:497–506. doi: 10.1016/j.chom.2011.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dominy SS, Lynch C, Ermini F, Benedyk M, Marczyk A, Konradi A, Nguyen M, Haditsch U, Raha D, Griffin C, Holsinger LJ, Arastu-Kapur S, Kaba S, Lee A, Ryder MI, Potempa B, Mydel P, Hellvard A, Adamowicz K, Hasturk H, Walker GD, Reynolds EC, Faull RLM, Curtis MA, Dragunow M, Potempa J. 2019. Porphyromonas gingivalis in Alzheimer’s disease brains: evidence for disease causation and treatment with small-molecule inhibitors. Sci Adv 5:eaau3333. doi: 10.1126/sciadv.aau3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hayashi C, Gudino CV, Gibson FC III, Genco CA. 2010. Review: pathogen-induced inflammation at sites distant from oral infection: bacterial persistence and induction of cell-specific innate immune inflammatory pathways. Mol Oral Microbiol 25:305–316. doi: 10.1111/j.2041-1014.2010.00582.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Olsen I, Singhrao SK, Potempa J. 2018. Citrullination as a plausible link to periodontitis, rheumatoid arthritis, atherosclerosis and Alzheimer’s disease. J Oral Microbiol 10:1487742. doi: 10.1080/20002297.2018.1487742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hajishengallis G. 2015. Periodontitis: from microbial immune subversion to systemic inflammation. Nat Rev Immunol 15:30–44. doi: 10.1038/nri3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jain S, Darveau RP. 2010. Contribution of Porphyromonas gingivalis lipopolysaccharide to periodontitis. Periodontol 2000 54:53–70. doi: 10.1111/j.1600-0757.2009.00333.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coats SR, Jones JW, Do CT, Braham PH, Bainbridge BW, To TT, Goodlett DR, Ernst RK, Darveau RP. 2009. Human Toll-like receptor 4 responses to P. gingivalis are regulated by lipid A 1- and 4′-phosphatase activities. Cell Microbiol 11:1587–1599. doi: 10.1111/j.1462-5822.2009.01349.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jain S, Chang AM, Singh M, McLean JS, Coats SR, Kramer RW, Darveau RP. 2019. Identification of PGN_1123 as the gene encoding lipid A deacylase, an enzyme required for Toll-like receptor 4 evasion, in Porphyromonas gingivalis. J Bacteriol 201:e00683-18. doi: 10.1128/JB.00683-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Slocum C, Coats SR, Hua N, Kramer C, Papadopoulos G, Weinberg EO, Gudino CV, Hamilton JA, Darveau RP, Genco CA. 2014. Distinct lipid A moieties contribute to pathogen-induced site-specific vascular inflammation. PLoS Pathog 10:e1004215. doi: 10.1371/journal.ppat.1004215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burns E, Eliyahu T, Uematsu S, Akira S, Nussbaum G. 2010. TLR2-dependent inflammatory response to Porphyromonas gingivalis is MyD88 independent, whereas MyD88 is required to clear infection. J Immunol 184:1455–1462. doi: 10.4049/jimmunol.0900378. [DOI] [PubMed] [Google Scholar]
- 14.Hayashi C, Madrigal AG, Liu X, Ukai T, Goswami S, Gudino CV, Gibson FC III, Genco CA. 2010. Pathogen-mediated inflammatory atherosclerosis is mediated in part via Toll-like receptor 2-induced inflammatory responses. J Innate Immun 2:334–343. doi: 10.1159/000314686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jain S, Coats SR, Chang AM, Darveau RP. 2013. A novel class of lipoprotein lipase-sensitive molecules mediates Toll-like receptor 2 activation by Porphyromonas gingivalis. Infect Immun 81:1277–1286. doi: 10.1128/IAI.01036-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Papadopoulos G, Weinberg EO, Massari P, Gibson FC III, Wetzler LM, Morgan EF, Genco CA. 2013. Macrophage-specific TLR2 signaling mediates pathogen-induced TNF-dependent inflammatory oral bone loss. J Immunol 190:1148–1157. doi: 10.4049/jimmunol.1202511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zenobia C, Hajishengallis G. 2015. Porphyromonas gingivalis virulence factors involved in subversion of leukocytes and microbial dysbiosis. Virulence 6:236–243. doi: 10.1080/21505594.2014.999567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cunha LD, Zamboni DS. 2013. Subversion of inflammasome activation and pyroptosis by pathogenic bacteria. Front Cell Infect Microbiol 3:76. doi: 10.3389/fcimb.2013.00076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lamkanfi M, Dixit VM. 2011. Modulation of inflammasome pathways by bacterial and viral pathogens. J Immunol 187:597–602. doi: 10.4049/jimmunol.1100229. [DOI] [PubMed] [Google Scholar]
- 20.Latz E, Xiao TS, Stutz A. 2013. Activation and regulation of the inflammasomes. Nat Rev Immunol 13:397–411. doi: 10.1038/nri3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sahoo M, Ceballos-Olvera I, del Barrio L, Re F. 2011. Role of the inflammasome, IL-1beta, and IL-18 in bacterial infections. ScientificWorldJournal 11:2037–2050. doi: 10.1100/2011/212680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Park E, Na HS, Song YR, Shin SY, Kim YM, Chung J. 2014. Activation of NLRP3 and AIM2 inflammasomes by Porphyromonas gingivalis infection. Infect Immun 82:112–123. doi: 10.1128/IAI.00862-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shi X, Xie WL, Kong WW, Chen D, Qu P. 2015. Expression of the NLRP3 Inflammasome in Carotid Atherosclerosis. J Stroke Cerebrovasc Dis 24:2455–2466. doi: 10.1016/j.jstrokecerebrovasdis.2015.03.024. [DOI] [PubMed] [Google Scholar]
- 24.Hajishengallis G. 2011. Immune evasion strategies of Porphyromonas gingivalis. J Oral Biosci 53:233–240. doi: 10.2330/joralbiosci.53.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dashper SG, Mitchell HL, Seers CA, Gladman SL, Seemann T, Bulach DM, Chandry PS, Cross KJ, Cleal SM, Reynolds EC. 2017. Porphyromonas gingivalis uses specific domain rearrangements and allelic exchange to generate diversity in surface virulence factors. Front Microbiol 8:48. doi: 10.3389/fmicb.2017.00048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen T, Hosogi Y, Nishikawa K, Abbey K, Fleischmann RD, Walling J, Duncan MJ. 2004. Comparative whole-genome analysis of virulent and avirulent strains of Porphyromonas gingivalis. J Bacteriol 186:5473–5479. doi: 10.1128/JB.186.16.5473-5479.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hall LM, Fawell SC, Shi X, Faray-Kele MC, Aduse-Opoku J, Whiley RA, Curtis MA. 2005. Sequence diversity and antigenic variation at the rag locus of Porphyromonas gingivalis. Infect Immun 73:4253–4262. doi: 10.1128/IAI.73.7.4253-4262.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Igboin CO, Griffen AL, Leys EJ. 2009. Porphyromonas gingivalis strain diversity. J Clin Microbiol 47:3073–3081. doi: 10.1128/JCM.00569-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aduse-Opoku J, Slaney JM, Hashim A, Gallagher A, Gallagher RP, Rangarajan M, Boutaga K, Laine ML, Van Winkelhoff AJ, Curtis MA. 2006. Identification and characterization of the capsular polysaccharide (K-antigen) locus of Porphyromonas gingivalis. Infect Immun 74:449–460. doi: 10.1128/IAI.74.1.449-460.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Amano A. 2010. Bacterial adhesins to host components in periodontitis. Periodontol 2000 52:12–37. doi: 10.1111/j.1600-0757.2009.00307.x. [DOI] [PubMed] [Google Scholar]
- 31.Maekawa T, Takahashi N, Tabeta K, Aoki Y, Miyashita H, Miyauchi S, Miyazawa H, Nakajima T, Yamazaki K. 2011. Chronic oral infection with Porphyromonas gingivalis accelerates atheroma formation by shifting the lipid profile. PLoS One 6:e20240. doi: 10.1371/journal.pone.0020240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pereira RB, Vasquez EC, Stefanon I, Meyrelles SS. 2011. Oral P gingivalis infection alters the vascular reactivity in healthy and spontaneously atherosclerotic mice. Lipids Health Dis 10:80. doi: 10.1186/1476-511X-10-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rodrigues PH, Reyes L, Chadda AS, Belanger M, Wallet SM, Akin D, Dunn W Jr, Progulske-Fox A. 2012. Porphyromonas gingivalis strain specific interactions with human coronary artery endothelial cells: a comparative study. PLoS One 7:e52606. doi: 10.1371/journal.pone.0052606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen T, Siddiqui H, Olsen I. 2017. In silico comparison of 19 Porphyromonas gingivalis strains in genomics, phylogenetics, phylogenomics and functional genomics. Front Cell Infect Microbiol 7:28. doi: 10.3389/fcimb.2017.00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gibson FC III, Hong C, Chou HH, Yumoto H, Chen J, Lien E, Wong J, Genco CA. 2004. Innate immune recognition of invasive bacteria accelerates atherosclerosis in apolipoprotein E-deficient mice. Circulation 109:2801–2806. doi: 10.1161/01.CIR.0000129769.17895.F0. [DOI] [PubMed] [Google Scholar]
- 36.Maekawa T, Krauss JL, Abe T, Jotwani R, Triantafilou M, Triantafilou K, Hashim A, Hoch S, Curtis MA, Nussbaum G, Lambris JD, Hajishengallis G. 2014. Porphyromonas gingivalis manipulates complement and TLR signaling to uncouple bacterial clearance from inflammation and promote dysbiosis. Cell Host Microbe 15:768–778. doi: 10.1016/j.chom.2014.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nakayama H, Kurokawa K, Lee BL. 2012. Lipoproteins in bacteria: structures and biosynthetic pathways. FEBS J 279:4247–4268. doi: 10.1111/febs.12041. [DOI] [PubMed] [Google Scholar]
- 38.Nagano K, Hasegawa Y, Murakami Y, Nishiyama S, Yoshimura F. 2010. FimB regulates FimA fimbriation in Porphyromonas gingivalis. J Dent Res 89:903–908. doi: 10.1177/0022034510370089. [DOI] [PubMed] [Google Scholar]
- 39.Park Y, Xie H, Lamont RJ. 2007. Transcriptional organization of the Porphyromonas gingivalis fimA locus. FEMS Microbiol Lett 273:103–108. doi: 10.1111/j.1574-6968.2007.00782.x. [DOI] [PubMed] [Google Scholar]
- 40.Xu Q, Shoji M, Shibata S, Naito M, Sato K, Elsliger MA, Grant JC, Axelrod HL, Chiu HJ, Farr CL, Jaroszewski L, Knuth MW, Deacon AM, Godzik A, Lesley SA, Curtis MA, Nakayama K, Wilson IA. 2016. A distinct type of pilus from the human microbiome. Cell 165:690–703. doi: 10.1016/j.cell.2016.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Asai Y, Ohyama Y, Gen K, Ogawa T. 2001. Bacterial fimbriae and their peptides activate human gingival epithelial cells through Toll-like receptor 2. Infect Immun 69:7387–7395. doi: 10.1128/IAI.69.12.7387-7395.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hasegawa Y, Iwami J, Sato K, Park Y, Nishikawa K, Atsumi T, Moriguchi K, Murakami Y, Lamont RJ, Nakamura H, Ohno N, Yoshimura F. 2009. Anchoring and length regulation of Porphyromonas gingivalis Mfa1 fimbriae by the downstream gene product Mfa2. Microbiology 155:3333–3347. doi: 10.1099/mic.0.028928-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hiramine H, Watanabe K, Hamada N, Umemoto T. 2003. Porphyromonas gingivalis 67-kDa fimbriae induced cytokine production and osteoclast differentiation utilizing TLR2. FEMS Microbiol Lett 229:49–55. doi: 10.1016/S0378-1097(03)00788-2. [DOI] [PubMed] [Google Scholar]
- 44.Lasica AM, Ksiazek M, Madej M, Potempa J. 2017. The type IX secretion system (T9SS): highlights and recent insights into its structure and function. Front Cell Infect Microbiol 7:215. doi: 10.3389/fcimb.2017.00215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gorasia DG, Veith PD, Chen D, Seers CA, Mitchell HA, Chen YY, Glew MD, Dashper SG, Reynolds EC. 2015. Porphyromonas gingivalis type IX secretion substrates are cleaved and modified by a sortase-like mechanism. PLoS Pathog 11:e1005152. doi: 10.1371/journal.ppat.1005152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li N, Collyer CA. 2011. Gingipains from Porphyromonas gingivalis—complex domain structures confer diverse functions. Eur J Microbiol Immunol 1:41–58. doi: 10.1556/EuJMI.1.2011.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sugawara S, Nemoto E, Tada H, Miyake K, Imamura T, Takada H. 2000. Proteolysis of human monocyte CD14 by cysteine proteinases (gingipains) from Porphyromonas gingivalis leading to lipopolysaccharide hyporesponsiveness. J Immunol 165:411–418. doi: 10.4049/jimmunol.165.1.411. [DOI] [PubMed] [Google Scholar]
- 48.Aduse-Opoku J, Davies NN, Gallagher A, Hashim A, Evans HE, Rangarajan M, Slaney JM, Curtis MA. 2000. Generation of Lys-gingipain protease activity in Porphyromonas gingivalis W50 is independent of Arg-gingipain protease activities. Microbiology 146:1933–1940. doi: 10.1099/00221287-146-8-1933. [DOI] [PubMed] [Google Scholar]
- 49.Rangarajan M, Smith SJ, U S, Curtis MA. 1997. Biochemical characterization of the arginine-specific proteases of Porphyromonas gingivalis W50 suggests a common precursor. Biochem J 323:701–709. doi: 10.1042/bj3230701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gutner M, Chaushu S, Balter D, Bachrach G. 2009. Saliva enables the antimicrobial activity of LL-37 in the presence of proteases of Porphyromonas gingivalis. Infect Immun 77:5558–5563. doi: 10.1128/IAI.00648-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Paramonov N, Rangarajan M, Hashim A, Gallagher A, Aduse-Opoku J, Slaney JM, Hounsell E, Curtis MA. 2005. Structural analysis of a novel anionic polysaccharide from Porphyromonas gingivalis strain W50 related to Arg-gingipain glycans. Mol Microbiol 58:847–863. doi: 10.1111/j.1365-2958.2005.04871.x. [DOI] [PubMed] [Google Scholar]
- 52.Shoji M, Sato K, Yukitake H, Kondo Y, Narita Y, Kadowaki T, Naito M, Nakayama K. 2011. Por secretion system-dependent secretion and glycosylation of Porphyromonas gingivalis hemin-binding protein 35. PLoS One 6:e21372. doi: 10.1371/journal.pone.0021372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shoji M, Sato K, Yukitake H, Naito M, Nakayama K. 2014. Involvement of the Wbp pathway in the biosynthesis of Porphyromonas gingivalis lipopolysaccharide with anionic polysaccharide. Sci Rep 4:5056. doi: 10.1038/srep05056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chastain-Gross RP, Xie G, Belanger M, Kumar D, Whitlock JA, Liu L, Raines SM, Farmerie WG, Daligault HE, Han CS, Brettin TS, Progulske-Fox A. 2017. Genome sequence of Porphyromonas gingivalis strain 381. Genome Announc 5. doi: 10.1128/genomeA.01467-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Enersen M, Nakano K, Amano A. 2013. Porphyromonas gingivalis fimbriae. J Oral Microbiol 5. doi: 10.3402/jom.v5i0.20265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hajishengallis G, Ratti P, Harokopakis E. 2005. Peptide mapping of bacterial fimbrial epitopes interacting with pattern recognition receptors. J Biol Chem 280:38902–38913. doi: 10.1074/jbc.M507326200. [DOI] [PubMed] [Google Scholar]
- 57.Wang M, Shakhatreh MA, James D, Liang S, Nishiyama S, Yoshimura F, Demuth DR, Hajishengallis G. 2007. Fimbrial proteins of porphyromonas gingivalis mediate in vivo virulence and exploit TLR2 and complement receptor 3 to persist in macrophages. J Immunol 179:2349–2358. doi: 10.4049/jimmunol.179.4.2349. [DOI] [PubMed] [Google Scholar]
- 58.Vanterpool E, Roy F, Fletcher HM. 2005. Inactivation of vimF, a putative glycosyltransferase gene downstream of vimE, alters glycosylation and activation of the gingipains in Porphyromonas gingivalis W83. Infect Immun 73:3971–3982. doi: 10.1128/IAI.73.7.3971-3982.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kadowaki T, Yukitake H, Naito M, Sato K, Kikuchi Y, Kondo Y, Shoji M, Nakayama K. 2016. A two-component system regulates gene expression of the type IX secretion component proteins via an ECF sigma factor. Sci Rep 6:23288. doi: 10.1038/srep23288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jung YJ, Jun HK, Choi BK. 2015. Contradictory roles of Porphyromonas gingivalis gingipains in caspase-1 activation. Cell Microbiol 17:1304–1319. doi: 10.1111/cmi.12435. [DOI] [PubMed] [Google Scholar]
- 61.Wilensky A, Tzach-Nahman R, Potempa J, Shapira L, Nussbaum G. 2015. Porphyromonas gingivalis gingipains selectively reduce CD14 expression, leading to macrophage hyporesponsiveness to bacterial infection. J Innate Immun 7:127–135. doi: 10.1159/000365970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Olsen I, Hajishengallis G. 2016. Major neutrophil functions subverted by Porphyromonas gingivalis. J Oral Microbiol 8:30936. doi: 10.3402/jom.v8.30936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen T, Nakayama K, Belliveau L, Duncan MJ. 2001. Porphyromonas gingivalis gingipains and adhesion to epithelial cells. Infect Immun 69:3048–3056. doi: 10.1128/IAI.69.5.3048-3056.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Deshpande RG, Khan MB, Genco CA. 1998. Invasion of aortic and heart endothelial cells by Porphyromonas gingivalis. Infect Immun 66:5337–5343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yamatake K, Maeda M, Kadowaki T, Takii R, Tsukuba T, Ueno T, Kominami E, Yokota S, Yamamoto K. 2007. Role for gingipains in Porphyromonas gingivalis traffic to phagolysosomes and survival in human aortic endothelial cells. Infect Immun 75:2090–2100. doi: 10.1128/IAI.01013-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chou HH, Yumoto H, Davey M, Takahashi Y, Miyamoto T, Gibson FC III, Genco CA. 2005. Porphyromonas gingivalis fimbria-dependent activation of inflammatory genes in human aortic endothelial cells. Infect Immun 73:5367–5378. doi: 10.1128/IAI.73.9.5367-5378.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hajishengallis G, Wang M, Harokopakis E, Triantafilou M, Triantafilou K. 2006. Porphyromonas gingivalis fimbriae proactively modulate beta2 integrin adhesive activity and promote binding to and internalization by macrophages. Infect Immun 74:5658–5666. doi: 10.1128/IAI.00784-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Baek KJ, Ji S, Kim YC, Choi Y. 2015. Association of the invasion ability of Porphyromonas gingivalis with the severity of periodontitis. Virulence 6:274–281. doi: 10.1080/21505594.2014.1000764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Erridge C, Spickett CM, Webb DJ. 2007. Non-enterobacterial endotoxins stimulate human coronary artery but not venous endothelial cell activation via Toll-like receptor 2. Cardiovasc Res 73:181–189. doi: 10.1016/j.cardiores.2006.11.004. [DOI] [PubMed] [Google Scholar]
- 70.Han N, Lepine G, Whitlock J, Wojciechowski L, Progulske-Fox A. 1998. The porphyromonas gingivalis prtP/kgp homologue exists as two open reading frames in strain 381. Oral Dis 4:170–179. [DOI] [PubMed] [Google Scholar]
- 71.How KY, Song KP, Chan KG. 2016. Porphyromonas gingivalis: an overview of periodontopathic pathogen below the gum line. Front Microbiol 7:53. doi: 10.3389/fmicb.2016.00053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.McLean JS, Liu Q, Bor B, Bedree JK, Cen L, Watling M, To TT, Bumgarner RE, He X, Shi W. 2016. Draft genome sequence of Actinomyces odontolyticus subsp. actinosynbacter strain XH001, the basibiont of an oral TM7 epibiont. Genome Announc 4:e01685-15. doi: 10.1128/genomeA.01685-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bushnell B. 2015. BBMap (version 35.14). https://sourceforge.net/projects/bbmap/.
- 74.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Naito M, Hirakawa H, Yamashita A, Ohara N, Shoji M, Yukitake H, Nakayama K, Toh H, Yoshimura F, Kuhara S, Hattori M, Hayashi T, Nakayama K. 2008. Determination of the genome sequence of Porphyromonas gingivalis strain ATCC 33277 and genomic comparison with strain W83 revealed extensive genome rearrangements in P. gingivalis. DNA Res 15:215–225. doi: 10.1093/dnares/dsn013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bushnell B., Jr 2016. BBMap (version 36.30). https://sourceforge.net/projects/bbmap/.
- 77.Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Anders S, Huber W. 2010. Differential expression analysis for sequence count data. Genome Biol 11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.