Acinetobacter baumannii is an opportunistic bacterial pathogen capable of causing a variety of infections, including pneumonia, sepsis, wound, and burn infections. A. baumannii is an increasing threat to public health due to the prevalence of multidrug-resistant strains, leading the World Health Organization to declare A. baumannii a “Priority 1: Critical” pathogen, for which the development of novel antimicrobials is desperately needed.
KEYWORDS: Acinetobacter, metal transporters, zinc
ABSTRACT
Acinetobacter baumannii is an opportunistic bacterial pathogen capable of causing a variety of infections, including pneumonia, sepsis, wound, and burn infections. A. baumannii is an increasing threat to public health due to the prevalence of multidrug-resistant strains, leading the World Health Organization to declare A. baumannii a “Priority 1: Critical” pathogen, for which the development of novel antimicrobials is desperately needed. Zinc (Zn) is an essential nutrient that pathogenic bacteria, including A. baumannii, must acquire from their hosts in order to survive. Consequently, vertebrate hosts have defense mechanisms to sequester Zn from invading bacteria through a process known as nutritional immunity. Here, we describe a Zn uptake (Znu) system that enables A. baumannii to overcome this host-imposed Zn limitation. The Znu system consists of an inner membrane ABC transporter and an outer membrane TonB-dependent receptor. Strains of A. baumannii lacking any individual Znu component are unable to grow in Zn-starved conditions, including in the presence of the host nutritional immunity protein calprotectin. The Znu system contributes to Zn-limited growth by aiding directly in the uptake of Zn into A. baumannii cells and is important for pathogenesis in murine models of A. baumannii infection. These results demonstrate that the Znu system allows A. baumannii to subvert host nutritional immunity and acquire Zn during infection.
INTRODUCTION
Acinetobacter baumannii is a Gram-negative pathogen that causes pneumonia, sepsis, wound, and burn infections (1). A. baumannii has many intrinsic mechanisms of resistance to antibiotics and is capable of surviving inhospitable environments through resistance to both desiccation (2) and reactive oxygen species (3). These factors, coupled with the increasing prevalence of A. baumannii infections, both in hospitals and in the community, have led to the classification of A. baumannii by the CDC as a threat level “Serious” (4) and by the World Health Organization as a “Priority 1: Critical” pathogen (5). Taken together, these facts highlight the importance of understanding A. baumannii physiological pathways that contribute to virulence.
A promising area for the development of new therapeutics targeting A. baumannii is metal acquisition, as metals are required for a variety of functions that are essential for life (6). Zinc (Zn) is a first-row d-block metal that serves as a structural and catalytic cofactor for a multitude of proteins (7). Zn-requiring proteins are needed for numerous essential functions within bacterial cells, including transcription (8), amino acid biosynthesis (9), carbon metabolism (10), DNA repair (11), and pH balance (12).
Maintaining homeostatic Zn balance during infection is a key contributor to the virulence of A. baumannii. During Zn starvation, A. baumannii upregulates expression of a Zn-binding GTPase, ZigA (13). Deletion of zigA reduces flavin pools within the cell and disrupts overall metal homeostasis (14). Additionally, a Zn-requiring peptidase, ZrlA, maintains cell wall integrity during Zn limitation, contributing to virulence and resistance to antibiotics in a mouse model of pneumonia (15). A putative Zn acquisition system has been identified in A. baumannii (16), but only one gene out of this multistep pathway was characterized, leaving questions regarding the mechanism of Zn uptake through the inner and outer membrane.
Vertebrates have evolved to exploit the microbial requirement for Zn in a process known as nutritional immunity, in which the availability of nutrient metals, such as Zn, is restricted from the pathogen during the innate immune response (17). Mechanisms used by vertebrates to reduce free extracellular Zn include sequestration by secreted proteins and import into host cells. One secreted Zn-binding protein is calprotectin, a heterodimer of S100A8/A9 proteins that binds metal at two sites. One site has high affinity for Zn, while the other is promiscuous, binding multiple metals (18). Calprotectin is released in response to bacterial pathogens, including Staphylococcus aureus (19), Clostridium difficile (20), and A. baumannii, and calprotectin-deficient mice have higher bacterial burdens in their lungs during A. baumannii pneumonia (16). Additionally, Zn transporters on host cells, such as ZIP8 on airway epithelial cells and ZIP14 on hepatocytes, are induced during inflammation and remove Zn from the extracellular space (21, 22). Therefore, in order to colonize the vertebrate host, pathogens like A. baumannii must have mechanisms to subvert nutritional immunity and acquire the Zn needed for survival.
A primary microbial strategy for overcoming host-mediated Zn restriction is the expression of high-affinity Zn transporters; however, the transporters used by A. baumannii for Zn acquisition are not well characterized. A transport system responsible for Zn uptake in many Gram-negative bacteria is the Znu system. These systems consist of a periplasmic binding protein, ZnuA, which is thought to confer specificity of the transporter for Zn by binding Zn with a triad of conserved histidine residues. The membrane-spanning portion of the transporter, ZnuB, allows Zn ions to enter the cell, while ZnuC, the ATPase, provides the energy for this import to occur against a concentration gradient. ZnuABC systems have been described in Gram-negative bacteria, including Escherichia coli (23), Pseudomonas aeruginosa (24), Salmonella enterica (25), Vibrio cholerae (26), Moraxella catarrhalis (27), Brucella abortus (28), Treponema pallidum (29), Yersinia pestis (30), Francisella tularensis (31), and Neisseria gonorrhoeae (32). However, whether A. baumannii employs a similar system to acquire nutrient Zn has not been uncovered.
Because Gram-negative bacteria have an inner and outer membrane, Zn must cross the outer membrane before it can be bound by ZnuA and transported across the inner membrane. Despite the prevalence of identified ZnuABC transporters, few outer membrane Zn receptors have been described. The most extensively characterized outer membrane Zn transporter is the Neisseria meningitidis TonB-dependent receptor, ZnuD, which binds Zn with high affinity (33, 34). ZnuD has been implicated as a promising vaccine candidate for N. meningitidis (35), but studies interrogating the functional conservation of the outer membrane ZnuD for Zn transport in other Gram-negative pathogens, including A. baumannii, have not been reported.
As the vertebrate host is a Zn-deplete environment, bacteria must have mechanisms to sense metal limitation and express Zn acquisition systems. However, excess Zn is toxic to cells, so these systems must be tightly regulated to ensure proper expression. Bacterial systems for Zn acquisition are commonly under the control of the Zn uptake repressor (Zur). When Zn is abundant, Zur binds Zn and Zur-Zn complexes undergo high-affinity engagement with promoters of genes within the Zur regulon (36). However, when Zn becomes scarce, Zur no longer binds Zn, and the Zur regulon is derepressed, allowing for the expression of genes aiding in Zn acquisition, including the znu genes. A. baumannii strains with zur deleted exhibit reduced bacterial burdens in mice (37), further suggesting that proper regulation of Zn acquisition systems is essential for the pathogenesis of this organism.
In this work, we report the identification and functional characterization of the Acinetobacter baumannii inner membrane (ZnuABC) and outer membrane (ZnuD) Zn acquisition system. Both outer and inner membrane systems are required for growth in low Zn conditions as well as for efficient uptake of Zn. Additionally, we demonstrate the requirement of the Znu system in overcoming nutrient metal limitation imposed by calprotectin and show the impact of impaired Zn acquisition on virulence in multiple murine models of A. baumannii infection.
RESULTS
A. baumannii encodes a candidate Znu system.
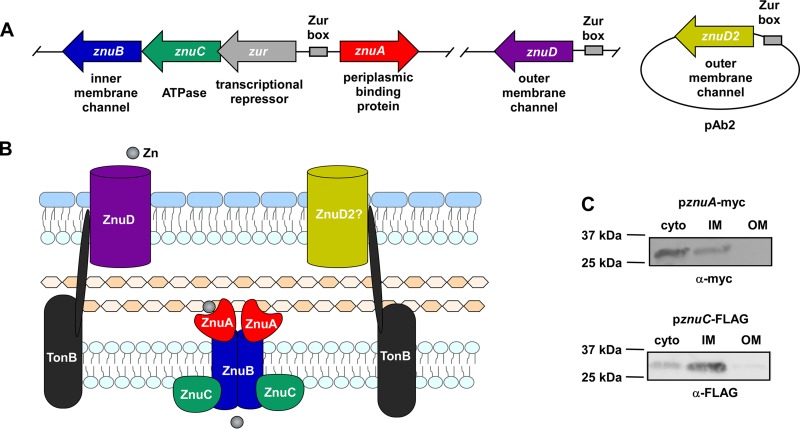
Previous work has identified an A. baumannii gene with homology to znuB as being required for growth in calprotectin (16). Bioinformatic analysis of the genomic context surrounding znuB revealed that znuB is in a candidate operon with znuC and zur. A znuA ortholog is encoded upstream of the candidate operon in the reverse orientation (Fig. 1A). These proteins are homologous to the ZnuABC system in E. coli with 47% (ZnuA), 36% (ZnuB), and 27% (ZnuC) amino acid identity. Based on the presence of a candidate Znu inner membrane Zn transport system, we searched the genome for potential znuD orthologs. Two genes in A. baumannii strain ATCC 17978 encode proteins with 42% and 40% identity to N. meningitidis ZnuD and are annotated as encoding putative outer membrane TonB-dependent receptor proteins. One gene, designated znuD, is part of the A. baumannii chromosome and conserved among all sequenced strains in the NCBI database with greater than 90% sequence identity, while the other gene, znuD2, is present in less than half of A. baumannii strains. znuD2 is found on pAb2 in strain ATCC 17978 and on a similar plasmid in all strains that have the gene. To validate the predicted subcellular localization of the Znu system proteins (Fig. 1B), strains of A. baumannii harboring tagged versions of ZnuA and ZnuC were grown in Zn-limited conditions, and differential centrifugation was used to fractionate the cells. Immunoblotting of the cellular fractions revealed that ZnuA and ZnuC are inner membrane proteins (Fig. 1C; see also Fig. S1 in the supplemental material). These observations suggest that the A. baumannii genome harbors genes involved in Zn uptake across the inner and outer membranes.
FIG 1.
The A. baumannii genome contains genes for an inner and outer membrane Zn acquisition system. (A) Schematic of the genomic context in A. baumannii ATCC 17978 of genes predicted to be involved in Zn acquisition and the putative functions of the encoded proteins. (B) Model with the predicted locations of each Znu system component. (C) ΔznuA pznuA-myc and ΔznuC pznuC-FLAG strains were grown in 30 μM TPEN, and localization of ZnuA-myc and ZnuC-FLAG was determined by immunoblot on cellular fractions. Blots shown are representative of at least three independent experiments, and a fractionation control is shown in Fig. S1 in the supplemental material.
The A. baumannii ZnuABC system is required for growth in low Zn conditions.
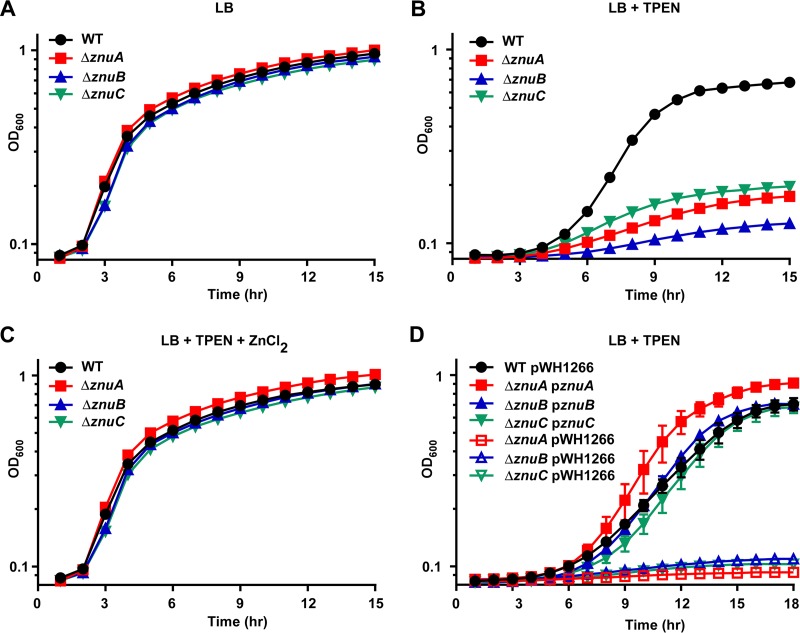
To investigate the contribution of the A. baumannii ZnuABC system to growth in low Zn conditions, deletion mutants in the corresponding genes were constructed in A. baumannii 17978 using allelic exchange. None of the deletion mutants, ΔznuA, ΔznuB, or ΔznuC mutants, exhibited a growth defect compared to wild-type (WT) A. baumannii when grown in lysogeny broth (LB), a rich medium (Fig. 2A). However, the growth of all three deletion mutants was severely reduced in the presence of the Zn chelator N,N,N′,N′-tetrakis(2-pyridinylmethyl)-1,2-ethanediamine (TPEN) (Fig. 2B). This growth defect was rescued by addition of ZnCl2 to the medium (Fig. 2C). In addition, providing an intact copy of the corresponding gene in trans fully restored the growth of each mutant during Zn limitation (Fig. 2D). These results establish the assignment of the ZnuABC system in A. baumannii and suggest that ZnuABC is required to overcome Zn starvation.
FIG 2.
The ZnuABC system is required for growth in low Zn conditions. WT, ΔznuA, ΔznuB, and ΔznuC strains were grown in LB (A), LB + 40 μM TPEN (B), and LB + 40 μM TPEN + 40 μM ZnCl2 (C), and OD600 was monitored over time. (D) WT, ΔznuA, ΔznuB, and ΔznuC strains harboring either an empty pWH1266 expression vector or pWH1266 providing a copy of the listed gene in trans were grown in LB + 40 μM TPEN and 75 μg/ml carbenicillin, and OD600 was monitored over time. Curves are shown in biological triplicate as mean ± standard deviation (SD) and are representative of at least three independent experiments.
ZnuA is a conserved Zn-binding protein induced during Zn starvation.
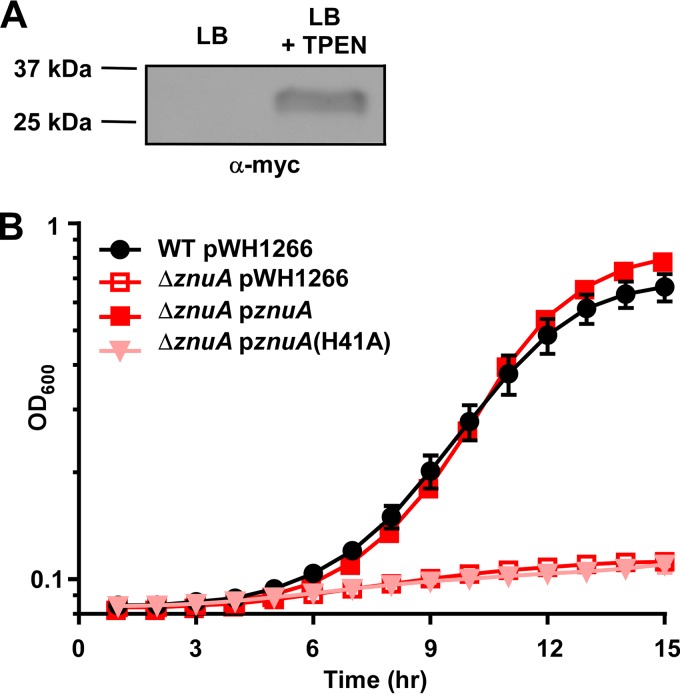
To determine the conditions that induce expression of the ZnuABC system, a ΔznuA strain that expresses a myc-tagged version of ZnuA under the control of its native promoter on the pWH1266 plasmid was created. When grown in LB, the ZnuA-myc protein was not detected by immunoblot; however, expression was induced upon TPEN treatment (Fig. 3A; see also Fig. S2 in the supplemental material). An alignment of A. baumannii ZnuA with the proteins from five other pathogens that use the ZnuABC system for Zn import (23–25, 29, 30) revealed a high degree of similarity, including conservation of the three His residues predicted to be involved in Zn binding (38) (see Fig. S3 in the supplemental material). Therefore, a construct was created where His41, one of the conserved residues, was mutated to Ala, and growth in low Zn medium was assessed. The ΔznuA strains harboring an empty vector, a vector with a WT copy of znuA under the control of its native promoter, or a vector with His41Ala mutated in znuA were grown in TPEN-chelated medium. The single His41Ala point mutation completely abolished growth in the Zn-deplete medium (Fig. 3B), demonstrating that His41 is required for ZnuA-mediated Zn acquisition and implicating this residue in Zn binding.
FIG 3.
ZnuA is produced during, and required for, growth in low Zn conditions. (A) ΔznuA strains harboring pWH1266 alone or pWH1266 expressing znuA under the control of its native promoter were grown in either LB or LB + 30 μM TPEN, and 10 μg of cell lysate was added to each lane for immunoblotting. Equal loading of lanes is shown in Fig. S2 in the supplemental material, and the blot shown is representative of four independent experiments. (B) WT cells harboring pWH1266 and ΔznuA strains harboring an empty vector, a vector containing a WT copy of znuA, or a vector containing znuA with a substitution of A for H at residue 41 were grown in 40 μM TPEN and 75 μg/ml carbenicillin, and OD600 was monitored over time. Expression of the ZnuA(H41A)-myc protein is demonstrated with an α-myc immunoblot in Fig. S2 in the supplemental material. Data are shown in biological triplicate ± SD and are representative of three independent experiments.
A. baumannii ZnuD is required for growth in low Zn conditions.
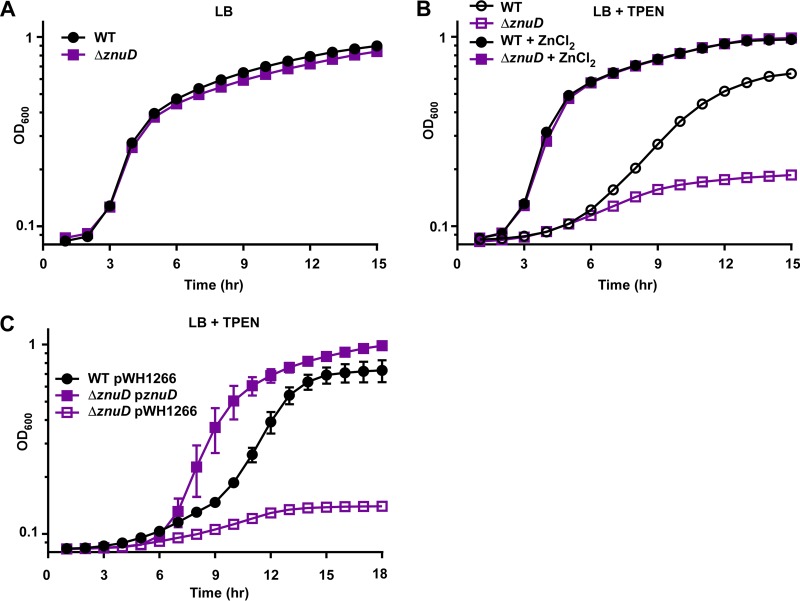
In addition to the inner membrane ZnuABC system, A. baumannii also encodes a candidate Zur-regulated TonB-dependent receptor, ZnuD. A. baumannii ZnuD is homologous with ZnuD from N. meningitidis, including conservation of the Zn-binding His residues (see Fig. S4 in the supplemental material). A strain lacking znuD was generated by allelic exchange and grown in Zn-replete and -deplete conditions. The ΔznuD strain grew comparably to the wild-type in LB (Fig. 4A) but had a growth defect in TPEN rescued by the addition of ZnCl2 (Fig. 4B). The defect of the ΔznuD strain in low Zn conditions was complemented by expression of znuD in trans from the pWH1266 expression vector (Fig. 4C). These data show that A. baumannii requires ZnuD for optimal growth during Zn starvation.
FIG 4.
ZnuD is required for growth in low Zn conditions. WT and ΔznuD strains were grown in LB (A) and LB + 40 μM TPEN or LB + 40 μM TPEN + 40 μM ZnCl2 (B), and OD600 was monitored over time. (C) WT and ΔznuD strains harboring empty pWH1266 expression vector or pWH1266 containing znuD were grown in LB + 20 μM TPEN, and OD600 was monitored over time. Data are shown in biological triplicate as mean ± SD and are representative of three independent experiments.
The Znu system contributes to growth during nutrient limitation imposed by the host protein calprotectin.
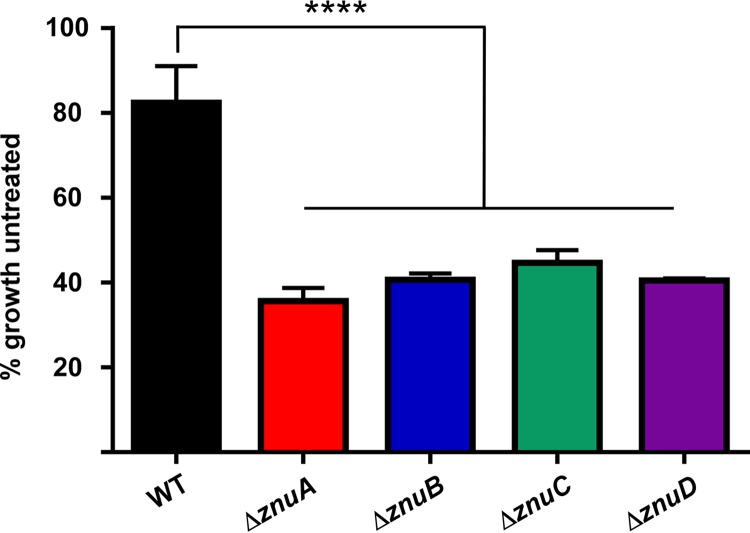
Calprotectin chelates Zn in vitro and is abundant in the lungs of mice infected with A. baumannii (16). Based on the observed defect of znu mutants in the presence of the synthetic Zn chelator TPEN, calprotectin was used to determine if the Znu system is required to overcome host-mediated Zn sequestration. WT, ΔznuA, ΔznuB, ΔznuC, and ΔznuD strains of A. baumannii were grown in the presence of 150 μg/ml calprotectin for 6 h, then growth was assessed by measuring optical density at 600 nm (OD600). This concentration of calprotectin modestly reduced growth of WT A. baumannii, whereas all four strains defective for a Znu system component exhibited severe growth restriction in the presence of calprotectin (Fig. 5). These data suggest that the Znu system aids A. baumannii in combating nutrient limitation imposed by host calprotectin.
FIG 5.
The Znu system contributes to growth during nutrient limitation imposed by the host protein calprotectin. WT, ΔznuA, ΔznuB, ΔznuC, and ΔznuD strains were grown for 6 h in 150 μg/ml recombinant human calprotectin, and OD600 was measured to determine growth compared to untreated. ****, P < 0.0001 by one-way analysis of variance (ANOVA) followed by Dunnett’s multiple-comparison test. Data shown in biological triplicate as mean ± SD.
The Znu system contributes to Zn uptake.
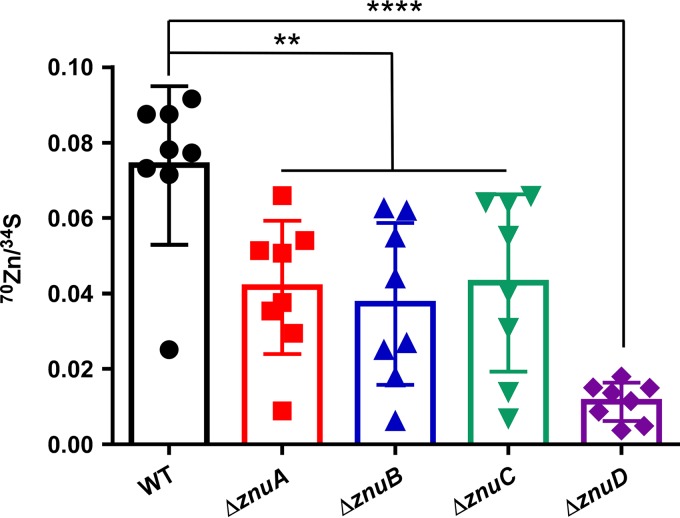
The requirement for the Znu system for growth during conditions of Zn restriction suggests that this system contributes to Zn uptake. To test the mechanism by which the Znu system contributes to growth during Zn starvation, a Zn uptake assay using a heavy isotope of Zn was performed. Cells were grown to mid-exponential phase, and then 70Zn was added for 5 min before the cells were processed for inductively coupled plasma mass spectrometry (ICP-MS) analysis. All Znu system mutants, ΔznuA, ΔznuB, ΔznuC, and ΔznuD mutants, were significantly impaired for 70Zn uptake, with the ΔznuD strain containing the lowest internal 70Zn levels (Fig. 6). This suggests that the Znu system aids in growth during Zn starvation by directly contributing to Zn uptake, with ZnuD being critical for Zn to enter the cell in vitro.
FIG 6.
The Znu system is required for efficient Zn uptake. WT, ΔznuA, ΔznuB, ΔznuC, and ΔznuD strains were grown in 30 μM TPEN to mid-exponential phase when 5 μM 70Zn was spiked into each culture and incubated 5 additional minutes. Cells were then pelleted, washed, and digested for quantification of metal levels by ICP-MS. 70Zn uptake is normalized to sulfur content of each cell pellet. **, P < 0.01; ****, P < 0.0001 by one-way ANOVA followed by Dunnett’s multiple-comparison test. Each symbol represents an independent biological replicate, error bars ± SD.
ZnuA and ZnuD contribute differentially to A. baumannii pathogenesis during sepsis.
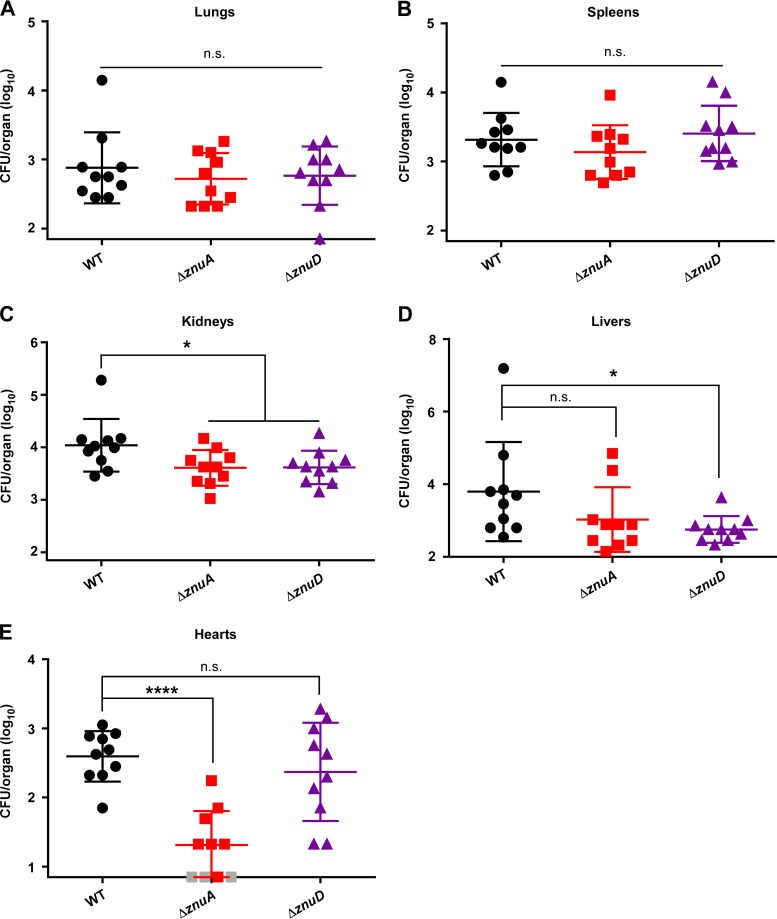
To test the hypothesis that the Znu system is required for A. baumannii pathogenesis, a sepsis model of A. baumannii infection was employed. The ΔznuA strain was selected to test the requirement for inner membrane components, while the ΔznuD strain was used to determine the role of the outer membrane system. Eight-week-old female C57BL/6J mice were infected retro-orbitally with the WT, ΔznuA, or ΔznuD strain and monitored as the infection proceeded. At 24 h postinfection, mice were humanely euthanized and lungs, spleens, kidneys, livers, and hearts collected for enumeration of bacterial burdens on LB agar. WT, ΔznuA, and ΔznuD strains were found at equal numbers in the lungs (Fig. 7A) and spleens (Fig. 7B) of infected mice; however, both mutants were reduced for bacterial burdens in the kidneys (Fig. 7C). In the livers, the ΔznuD strain was attenuated while the ΔznuA strain was not (Fig. 7D). This phenotype was reversed in the heart, where levels of the ΔznuA strain were lower than those of the WT or ΔznuD strain (Fig. 7E). These findings suggest that Zn acquisition is important during A. baumannii sepsis but that the inner membrane ZnuA and outer membrane ZnuD contribute differentially to pathogenesis in particular tissues.
FIG 7.
ZnuA and ZnuD contribute differentially to A. baumannii survival in specific tissues during sepsis. Mice were infected retro-orbitally with the WT, ΔznuA, or ΔznuD strain and bacterial burdens in organs assessed at 24 h postinfection in the lungs (A), spleens (B), kidneys (C), livers (D), and hearts (E). Data shown as mean ± SD with each symbol representing bacterial burdens in the specified organ from an individual mouse. The y axis begins at the limit of detection in each organ, and gray symbols represent mice with bacterial burdens below the limit of detection. *, P < 0.05 by one-way ANOVA followed by Dunnett’s multiple comparison test; ****, P < 0.0001 by one-way ANOVA followed by Dunnett’s multiple comparison test.
ZnuA is required for survival in the spleen during a murine model of pneumonia.
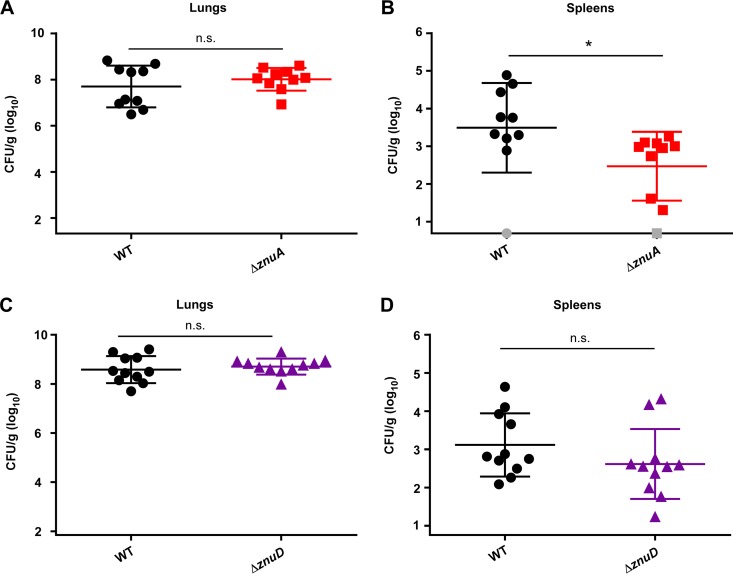
In addition to sepsis, a common presentation of A. baumannii infection is pneumonia. To test the contribution of znuA and znuD to infection by a different route, a murine pneumonia model was used. Competitive infections were performed between WT A. baumannii and each mutant strain using 9-week-old female C57BL/6J mice. Mice were infected intranasally and monitored as the infection proceeded. At 36 h postinfection, mice were humanely euthanized. Lungs and spleens were homogenized and plated to LB agar and LB agar plus kanamycin for enumeration of each bacterial strain. While the ΔznuA mutant was present in the lungs of infected mice at levels comparable to those of the WT strain (Fig. 8A), the ΔznuA strain had reduced bacterial burdens compared to those of the WT in the spleen (Fig. 8B). Surprisingly, the bacterial burdens of the WT and ΔznuD strains were similar in both the lungs (Fig. 8C) and the spleens (Fig. 8D) after 36 h of infection. These data support the conclusion that Zn acquisition is important during infection but suggest that ZnuD does not play a major role in A. baumannii survival in this model of pneumonia.
FIG 8.
ZnuA contributes to A. baumannii survival in the spleen during pneumonia. Mice were intranasally infected with a 1:1 mixture of either WT:ΔznuA strain (A, B) or WT:ΔznuD strain (C, D), and bacterial burdens were assessed at 36 h postinfection in the lungs (A, C) and the spleen (B, D). Data shown as mean ± SD with each symbol representing bacterial burdens in the specified organ from an individual mouse. The y axis begins at the limit of detection, and gray symbols represent mice with bacterial burdens below the limit of detection. *, P < 0.05 by unpaired t test.
DISCUSSION
In this study, we have identified an A. baumannii inner and outer membrane Zn uptake system and ascribed a role for this system during pathogenesis. Each of the components of the inner membrane ZnuABC system is required for growth in low Zn conditions and for efficient Zn uptake. ZnuA, the periplasmic binding portion of the ABC transporter, is induced in low Zn conditions and localizes to the cytoplasm and inner membrane. Mutation of a conserved His residue involved in Zn binding completely abrogates its function, suggesting that Zn binding is the primary function of this protein. The ΔznuA mutant is attenuated compared to the WT strain in both sepsis and pneumonia models of infection. The outer membrane receptor ZnuD is essential for overcoming in vitro Zn limitation by the synthetic chelator TPEN and by the host protein calprotectin. A strain lacking znuD is also severely impaired for Zn uptake in vitro. The livers and kidneys of mice infected with the ΔznuD strain had lower bacterial burdens than mice infected with WT A. baumannii during sepsis, but the ΔznuD strain was not defective for survival following coinfection with WT A. baumannii in a murine model of pneumonia.
The role of the putative outer membrane transporter ZnuD2 in Zn acquisition is still unclear, as we were unable to make a ΔznuD2 strain. We hypothesize that the difficulty in generating this strain is due to challenges associated with inactivating a gene present on a multicopy plasmid. Efforts to delete the Zur-regulated tonB (A1S_0452) (37), which we hypothesize provides the energy for ZnuD- and ZnuD2-mediated Zn uptake, were also unsuccessful.
A 2014 study by Wang et al. (39) used insertion sequencing to determine genes important for A. baumannii survival within murine lungs. Insertions in all three genes that make up the inner membrane transporter, znuA, znuB, and znuC, as well as the outer membrane transporter, znuD, were found at significantly lower frequencies after passage through murine lungs. While we did not observe a difference in bacterial burdens between the WT strain and the ΔznuA or ΔznuD mutants in the lungs in either our sepsis or pneumonia model, we did observe decreased burdens in other tissues, supporting the overall conclusion that Zn acquisition by ZnuABCD is important to A. baumannii survival within the vertebrate host. Of note, although Wang et al. (39) did use A. baumannii strain ATCC 17978, insertions were not mapped to plasmid genes, so no conclusions can be drawn about the role of znuD2. While most genes in the library had thousands of transposon insertions, the Zn-regulated tonB had less than five reads that mapped to that gene during growth in rich medium and none in murine lungs, which is in agreement with our work suggesting that this gene is essential.
Several studies interrogating the impact of Zn acquisition on bacterial pathogenesis suggest a varied role for the Znu system during infection. In a mouse model of invasive neisserial disease, a ΔznuD mutant does not kill mice to the same extent as WT N. meningitidis, although there was no difference between the ΔznuD and WT strains in an asymptomatic nasopharynx carriage model (34). The Yersinia pestis ZnuABC system is essential for in vitro Zn-limited growth but not for pathogenesis in a murine plague model (30) similar to what we report here for ZnuD in the A. baumannii pneumonia model. Additionally, Vibrio cholerae encodes two Zn acquisition systems, ZnuABC as well as ZrgABCDE (26). In vitro, the Znu system is the primary transporter for Zn, but both systems contribute to the colonization of mice. Overall, these data, along with our tissue-specific phenotypes for the A. baumannii ΔznuA and ΔznuD mutants, support a model where Zn availability is not uniform across body sites and infection pathologies, and consequently, Zn uptake by the Znu system is more important in certain niches than others. Bacteria encounter many stressors other than Zn limitation during infection and may have mechanisms for activating other Zn transporters that are not required during in vitro studies of Zn limitation.
TonB-dependent receptors are responsible for the transport of many substrates across the Gram-negative outer membrane, including metals such as Fe, in both free and siderophore-bound forms (40). In N. meningitidis, ZnuD binds free Zn(II) through two critical His residues (34), and A. baumannii ZnuD contains His residues at similar positions. However, other mechanisms for Zn acquisition by pathogens during infection have been described. The fungal pathogen Candida albicans secretes a protein “zincophore,” Pra1, which binds Zn in the environment before being imported back into the cell for use (41). Additionally, N. meningitidis steals Zn from calprotectin using the TonB-dependent receptor CbpA (42). S. aureus is also capable of using calprotectin as a Zn source and secretes staphylopine, a broad-spectrum metallophore that binds Zn (43). A. baumannii may possess such alternative mechanisms for Zn acquisition during infection, and ZnuD2 may be the protein in certain strains of A. baumannii that carries out these functions. Zn piracy from host proteins and other Zn acquisition mechanisms may not be important for growth in vitro but may be essential in the highly metal-restricted host environment. Our data suggest that the functions of ZnuD and ZnuD2 are not purely redundant, as the ΔznuD mutant is more sensitive to Zn starvation than the WT strain, even with an intact znuD2. ZnuD2 could serve a distinct function in nutrient acquisition or ZnuD and ZnuD2 could be playing similar but not overlapping roles. Future studies that probe the role of ZnuD in strains of A. baumannii that lack ZnuD2 and investigation into other putative Zn transporters, including ZnuD2 or currently unidentified systems, could help shed light on the Zn uptake mechanisms used by A. baumannii during infection.
In vitro, the Znu system contributes directly to the uptake of Zn. Zn must first enter the cell through the outer membrane transporter ZnuD, where it is bound in the periplasm by ZnuA to enter the cytoplasm through ZnuB. However, once Zn arrives in the cytoplasm, little is known about downstream processing and allocation mechanisms. The Zn metallochaperone ZigA (13) may be important for directing Zn to metalloproteins essential for survival during Zn restriction. What these critical Zn-requiring processes are and how ZigA is loaded with Zn are outstanding questions in the field of A. baumannii physiology.
Overall, this work describes the role of the ZnuABCD system in the nosocomial pathogen A. baumannii. Each individual component of the Znu system contributes directly to Zn uptake and is required for overcoming in vitro Zn limitation, both by the synthetic chelator TPEN and by the host protein calprotectin. ZnuA, the periplasmic binding protein, and ZnuD, the outer membrane transporter, contribute to virulence in mouse models of A. baumannii infection. Taken together, these studies reveal a mechanism employed by A. baumannii to acquire the essential nutrient Zn within the hostile environment of the host.
MATERIALS AND METHODS
Bacterial strains and reagents.
All experiments were performed using Acinetobacter baumannii ATCC 17978 or its derivatives. Plasmids for allelic exchange and expression of genes in trans were constructed in Escherichia coli DH5α. Bacteria were routinely cultured in lysogeny broth (LB) at 37°C with shaking. Kanamycin was used at 40 μg/ml for selection in A. baumannii, and carbenicillin was used at 50 μg/ml for E. coli selection and 75 μg/ml for A. baumannii selection and maintenance of the pWH1266 vector.
Generation of znuA, znuC, and znuD deletion mutants.
znuA, znuC, and znuD were replaced with a kanamycin resistance cassette using allelic exchange. Constructs were generated with 1 kb of the genomic region immediately upstream and downstream of each gene flanking the kanamycin resistance gene aph from pUCK18-K1. These constructs were cloned into the pFlp2 vector using either overlap extension PCR (znuA, znuD) or NEBuilder HiFi assembly (New England Biolabs) (znuC). pFlp2 constructs were electroporated into wild-type A. baumannii and resolved to kanamycin-resistant deletion mutants using sucrose selection.
Construction of complementation vectors.
The plasmid pWH1266 was digested with BamHI and SalI and used as the vector backbone for each construct. Primers were designed using the NEBuilder assembly tool (http://nebuilder.neb.com/), and genes were amplified from wild-type genomic DNA. znuA and znuC complementation constructs included the complete open reading frame and the intergenic region preceding it, while znuB and znuD constructs included the r01 promoter directly upstream of the beginning of the open reading frame. The NEBuilder HiFi assembly kit (New England BioLabs) was used to make each complementation construct, following the manufacturer’s instructions.
Bacterial growth assays in TPEN.
Overnight cultures of A. baumannii WT and deletion mutants were subcultured at 1:100 in LB with shaking at 37°C for 1 h and then inoculated at 1:50 into fresh medium ± 40 μM TPEN in a 96-well plate. Plates were placed in a BioTek Epoch plate reader with shaking at 37°C, and optical density at 600 nm was measured every hour for 15 h. For growth of strains harboring a pWH1266 expression vector, 75 μg/ml carbenicillin was present in all steps. Optical density was measured for 18 h, and concentrations of TPEN used are indicated in the figure legends.
Immunoblotting for protein abundance.
A. baumannii cells were grown to mid-exponential phase and then pelleted and resuspended in lysis buffer (150 mM NaCl, 20 mM Tris-HCl, pH 7.5). Cell suspensions were then transferred to Lysing Matrix B tubes (MP Biologicals) and lysed on a FastPrep-24 (MP Biologicals) bead beater. IGEPAL was added to 0.1% and the cells run on the bead beater again. Supernatant was transferred to a fresh 1.5-ml tube and protein quantified by BCA (Thermo Fisher Scientific). Equal volumes normalized to total protein were loaded into wells of 4 to 20% gradient Mini-Protean SDS-PAGE gels (Bio-Rad) and run in the Mini-Protean electrophoresis system (Bio-Rad). Proteins were transferred to a polyvinylidene difluoride (PVDF) membrane, stained using Ponceau S stain (Sigma), and imaged on a ChemiDoc MP imaging system (Bio-Rad). Immunoblotting was performed using mouse anti-c-Myc 9E10 and Alexa Fluor conjugated anti-mouse secondary. Protein bands were visualized on a ChemiDoc MP imaging system (Bio-Rad).
Subcellular fractionation.
A. baumannii was grown to mid-exponential phase and then pelleted and resuspended in lysis buffer as above. Cells were lysed by sonication, and then lysate was ultracentrifuged at 100,000 × g for 90 min. The soluble cytoplasmic fraction was removed, and the insoluble membrane fraction was resuspended in lysis buffer plus 0.5% sarkosyl. The inner and outer membrane fractions were separated by an additional 60 min of ultracentrifugation at 100,000 × g. Protein concentrations in each fraction were determined by BCA (Thermo Fisher Scientific).
Site-directed mutagenesis.
Mutation of His41 to Ala in znuA on the pWH1266 expression vector was performed using NEB’s Q5 site-directed mutagenesis kit, according to the manufacturer’s instructions. Primers were designed using the online tool NEBaseChanger (http://nebasechanger.neb.com/).
Zn uptake assay.
WT A. baumannii and each znu deletion mutant were grown in LB plus 30 μM TPEN to late exponential phase. A stock of 70ZnO (Cambridge Isotope Laboratories Inc.) was made in aqueous solution with HCl (Thermo Fisher Scientific) added until all 70ZnO was dissolved. After addition of 5 μM 70ZnO, cells were incubated with shaking for 5 min at 37°C and then pelleted and washed 2× with phosphate-buffered saline (PBS) (Corning). Cell pellets were then resuspended in 70% nitric acid (Thermo Fisher Scientific), transferred to metal-free tubes (VWR), and incubated at 60°C overnight. Digested samples were centrifuged again to remove any insoluble debris, and supernatant was transferred to fresh metal-free tubes. Elemental quantification was performed using an Agilent 7700 inductively coupled plasma mass spectrometer attached to an ASX-560 autosampler (Teledyne CETAC Technologies). The following settings were fixed for the analysis: cell entrance = −40 V, cell exit = −60 V, plate bias = −60 V, OctP bias = −18 V, and collision cell helium flow = 4.5 ml/min. Optimal voltages for extract 2, omega bias, omega lens, OctP RF, and deflect were determined empirically before each sample set was analyzed. Element calibration curves were generated using ARISTAR ICP standard mix (VWR). Samples were introduced by peristaltic pump with 0.5-mm-internal-diameter tubing through a MicroMist borosilicate glass nebulizer (Agilent). Samples were initially up taken at 0.5 rps for 30 s followed by 30 s at 0.1 rps to stabilize the signal. Samples were analyzed in Spectrum mode at 0.1 rps collecting three points across each peak and performing three replicates of 100 sweeps for each element analyzed. Sampling probe and tubing were rinsed for 20 s at 0.5 rps with 2% nitric acid between every sample. Data were acquired and analyzed using the Agilent MassHunter workstation software version A.01.02.
Mouse model of sepsis.
Female C57BL/6J mice were purchased from Jackson Laboratories. Mice were housed in a Vanderbilt University Medical Center (VUMC) facility with a 12-h light-dark cycle and fed the standard VUMC chow. At 8 weeks of age, mice were infected retro-orbitally with approximately 1 × 108 CFU of the WT, ΔznuA, or ΔznuD strain. Mice were monitored for 24 h as the infection proceeded and then were humanely euthanized. Lungs, spleens, kidneys, livers, and hearts were harvested, homogenized, and plated to LB agar to determine the bacterial burdens in each organ. All animal experiments were approved by the VUMC Institutional Care and Use Committee and conform to policies and guidelines established by VUMC, the Animal Welfare Act, the National Institutes of Health, and the American Veterinary Medical Association.
Mouse model of pneumonia.
Female C57BL/6J mice were purchased from Jackson Laboratories at 4 weeks of age. Mice were housed in the VUMC facility with a 12-h light-dark cycle. For all pneumonia infections, mice were fed a control Zn diet from Dyets Inc. (Dyet no. 515260) for 5 weeks, and then mice were infected at 9 weeks of age. Approximately 4 × 108 CFU of total bacteria (WT and mutant A. baumannii) were administered to each mouse intranasally. Mice were monitored as the infection proceeded for 36 h and then were humanely euthanized. Lungs and spleens were homogenized and plated to LB agar and LB agar plus kanamycin for enumeration of each bacterial strain.
Supplementary Material
ACKNOWLEDGMENTS
We thank members of the Skaar laboratory for review of this manuscript.
The work presented here was supported by NIH R01 AI101171 (to E.P.S.). L.E.H. was supported by NIH T32 GM065086, Z.R.L. was supported by NIH F31 AI136255, and W.N.B. was supported by AHA 18POST34030426.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/IAI.00746-19.
REFERENCES
- 1.Weinstein RA, Gaynes R, Edwards JR. 2005. Overview of nosocomial infections caused by Gram-negative bacilli. Clin Infect Dis 41:848–854. doi: 10.1086/432803. [DOI] [PubMed] [Google Scholar]
- 2.Antunes LCS, Imperi F, Carattoli A, Visca P. 2011. Deciphering the multifactorial nature of Acinetobacter baumannii pathogenicity. PLoS One 6:e22674. doi: 10.1371/journal.pone.0022674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Juttukonda LJ, Green ER, Lonergan ZR, Heffern MC, Chang CJ, Skaar P. 2019. Acinetobacter baumannii OxyR regulates the transcriptional response to hydrogen peroxide. Infect Immun 87:e00413-18. doi: 10.1128/IAI.00413-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention. 2013. Antibiotic resistance threats. Centers for Disease Control and Prevention, Atlanta, GA. [Google Scholar]
- 5.Tacconelli E, Magrini N. 2017. Global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 6.Maret W. 2016. The metals in the biological periodic system of the elements: concepts and conjectures. Int J Mol Sci 17:E66. doi: 10.3390/ijms17010066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Capdevila DA, Wang J, Giedroc DP. 2016. Bacterial strategies to maintain zinc metallostasis at the host-pathogen interface. J Biol Chem 291:20858–20868. doi: 10.1074/jbc.R116.742023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scrutton MC, Wu CW, Goldthwait DA. 1971. The presence and possible role of zinc in RNA polymerase obtained from Escherichia coli. Proc Natl Acad Sci U S A 68:2497–2501. doi: 10.1073/pnas.68.10.2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peariso K, Goulding CW, Huang S, Matthews RG, Penner-Hahn JE. 1998. Characterization of the zinc binding site in methionine synthase enzymes of Escherichia coli: the role of zinc in the methylation of homocysteine. J Am Chem Soc 120:8410–8416. doi: 10.1021/ja980581g. [DOI] [Google Scholar]
- 10.Sun HW, Plapp BV. 1992. Progressive sequence alignment and molecular evolution of the Zn-containing alcohol dehydrogenase family. J Mol Evol 34:522–535. doi: 10.1007/bf00160465. [DOI] [PubMed] [Google Scholar]
- 11.Myers LC, Verdine GL, Wagner G. 1993. Solution structure of the DNA methyl phosphotriester repair domain of Escherichia coli Ada. Biochemistry 32:14089–14094. doi: 10.1021/bi00214a003. [DOI] [PubMed] [Google Scholar]
- 12.Merlin C, Masters M, McAteer S, Coulson A. 2003. Why is carbonic anhydrase essential to Escherichia coli? J Bacteriol 185:6415–6424. doi: 10.1128/jb.185.21.6415-6424.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nairn BL, Lonergan ZR, Wang J, Braymer JJ, Zhang Y, Calcutt MW, Lisher JP, Gilston BA, Chazin WJ, De Crécy-Lagard V, Giedroc DP, Skaar EP. 2016. The response of Acinetobacter baumannii to zinc starvation. Cell Host Microbe 19:826–836. doi: 10.1016/j.chom.2016.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang J, Lonergan ZR, Gonzalez-Gutierrez G, Nairn BL, Maxwell CN, Zhang Y, Andreini C, Karty JA, Chazin WJ, Trinidad JC, Skaar EP, Giedroc DP. 2019. Multi-metal restriction by calprotectin impacts de novo flavin biosynthesis in Acinetobacter baumannii. Cell Chem Biol 26:745–755. doi: 10.1016/j.chembiol.2019.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lonergan ZR, Nairn BL, Wang J, Hsu YP, Hesse LE, Beavers WN, Chazin WJ, Trinidad JC, VanNieuwenhze MS, Giedroc DP, Skaar EP. 2019. An Acinetobacter baumannii, zinc-regulated peptidase maintains cell wall integrity during immune-mediated nutrient sequestration. Cell Rep 26:2009–2018. doi: 10.1016/j.celrep.2019.01.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hood MI, Mortensen BL, Moore JL, Zhang Y, Kehl-Fie TE, Sugitani N, Chazin WJ, Caprioli RM, Skaar EP. 2012. Identification of an Acinetobacter baumannii zinc acquisition system that facilitates resistance to calprotectin-mediated zinc sequestration. PLoS Pathog 8:e1003068. doi: 10.1371/journal.ppat.1003068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hood MI, Skaar EP. 2012. Nutritional immunity: transition metals at the pathogen-host interface. Nat Rev Microbiol 10:525–537. doi: 10.1038/nrmicro2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Damo SM, Kehl-Fie TE, Sugitani N, Holt ME, Rathi S, Murphy WJ, Zhang Y, Betz C, Hench L, Fritz G, Skaar EP, Chazin WJ. 2013. Molecular basis for manganese sequestration by calprotectin and roles in the innate immune response to invading bacterial pathogens. Proc Natl Acad Sci U S A 110:3841–3846. doi: 10.1073/pnas.1220341110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Corbin B, Seeley E, Raab A, Feldmann J, Miller M, Torres V, Anderson K, Dattilo B, Dunman P, Gerads R, Caprioli R, Nacken W, Chazin W, Skaar EP. 2008. Metal chelation and inhibition of bacterial growth in tissue abscesses. Science 319:962–965. doi: 10.1126/science.1152449. [DOI] [PubMed] [Google Scholar]
- 20.Zackular JP, Moore JL, Jordan AT, Juttukonda LJ, Noto MJ, Nicholson MR, Crews JD, Semler MW, Zhang Y, Ware LB, Washington MK, Chazin WJ, Caprioli RM, Skaar EP. 2016. Dietary zinc alters the microbiota and decreases resistance to Clostridium difficile infection. Nat Med 22:1330–1334. doi: 10.1038/nm.4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liuzzi JP, Lichten LA, Rivera S, Blanchard RK, Aydemir TB, Knutson MD, Ganz T, Cousins RJ. 2005. Interleukin-6 regulates the zinc transporter Zip14 in liver and contributes to the hypozincemia of the acute-phase response. Proc Natl Acad Sci U S A 102:6843–6848. doi: 10.1073/pnas.0502257102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu MJ, Bao S, Gálvez-Peralta M, Pyle CJ, Rudawsky AC, Pavlovicz RE, Killilea DW, Li C, Nebert DW, Wewers MD, Knoell DL. 2013. ZIP8 regulates host defense through zinc-mediated inhibition of NF-κB. Cell Rep 3:386–400. doi: 10.1016/j.celrep.2013.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Patzer SI, Hantke K. 1998. The ZnuABC high-affinity zinc uptake system and its regulator Zur in Escherichia coli. Mol Microbiol 28:1199–1210. doi: 10.1046/j.1365-2958.1998.00883.x. [DOI] [PubMed] [Google Scholar]
- 24.D’Orazio M, Mastropasqua MC, Cerasi M, Pacello F, Consalvo A, Chirullo B, Mortensen B, Skaar EP, Ciavardelli D, Pasquali P, Battistoni A. 2015. The capability of Pseudomonas aeruginosa to recruit zinc under conditions of limited metal availability is affected by inactivation of the ZnuABC transporter. Metallomics 7:1023–1035. doi: 10.1039/C5MT00017C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ammendola S, Pasquali P, Pistoia C, Petrucci P, Petrarca P, Rotilio G, Battistoni A. 2007. High-affinity Zn2+ uptake system ZnuABC is required for bacterial zinc homeostasis in intracellular environments and contributes to the virulence of Salmonella enterica. Infect Immun 75:5867–5876. doi: 10.1128/IAI.00559-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sheng Y, Fan F, Jensen O, Zhong Z, Kan B, Wang H, Zhu J. 2015. Dual zinc transporter systems in Vibrio cholerae promote competitive advantages over gut microbiome. Infect Immun 83:3902–3908. doi: 10.1128/IAI.00447-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Murphy TF, Brauer AL, Kirkham C, Johnson A, Koszelak-Rosenblum M, Malkowski MG. 2013. Role of the zinc uptake ABC transporter of Moraxella catarrhalis in persistence in the respiratory tract. Infect Immun 81:3406–3413. doi: 10.1128/IAI.00589-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim S, Watanabe K, Shirahata T, Watarai M. 2004. Zinc uptake system (znuA locus) of Brucella abortus is essential for intracellular survival and virulence in mice. J Vet Med Sci 66:1059–1063. doi: 10.1292/jvms.66.1059. [DOI] [PubMed] [Google Scholar]
- 29.Desrosiers DC, Sun YC, Zaidi AA, Eggers CH, Cox DL, Radolf JD. 2007. The general transition metal (Tro) and Zn2+ (Znu) transporters in Treponema pallidum: analysis of metal specificities and expression profiles. Mol Microbiol 65:137–152. doi: 10.1111/j.1365-2958.2007.05771.x. [DOI] [PubMed] [Google Scholar]
- 30.Desrosiers DC, Bearden SW, Mier I, Abney J, Paulley JT, Fetherston JD, Salazar JC, Radolf JD, Perry RD. 2010. Znu is the predominant zinc importer in Yersinia pestis during in vitro growth but is not essential for virulence. Infect Immun 78:5163–5177. doi: 10.1128/IAI.00732-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moreau GB, Qin A, Mann BJ. 2018. Zinc acquisition mechanisms differ between environmental and virulent Francisella species. J Bacteriol 200:e00587-17. doi: 10.1128/JB.00587-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen C, Morse SA. 2001. Identification and characterization of a high-affinity zinc uptake system in Neisseria gonorrhoeae. FEMS Microbiol Lett 202:67–71. doi: 10.1111/j.1574-6968.2001.tb10781.x. [DOI] [PubMed] [Google Scholar]
- 33.Stork M, Bos MP, Jongerius I, de Kok N, Schilders I, Weynants VE, Poolman JT, Tommassen J. 2010. An outer membrane receptor of Neisseria meningitidis involved in zinc acquisition with vaccine potential. PLoS Pathog 6:e1000969. doi: 10.1371/journal.ppat.1000969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Calmettes C, Ing C, Buckwalter CM, El Bakkouri M, Chieh-Lin Lai C, Pogoutse A, Gray-Owen SD, Pomès R, Moraes TF. 2015. The molecular mechanism of Zinc acquisition by the neisserial outer-membrane transporter ZnuD. Nat Commun 6:7996. doi: 10.1038/ncomms8996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vogel U, Mordhorst I, Weynants V, Hubert K, Baudoux G, Tommassen J, Poolman JT, Devos N, Feron C, Tans C, Goraj K. 2013. ZnuD, a potential candidate for a simple and universal Neisseria meningitidis vaccine. Infect Immun 81:1915–1927. doi: 10.1128/IAI.01312-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Outten CE, Tobin DA, Penner-Hahn JE, O'Halloran TV. 2001. Characterization of the metal receptor sites in Escherichia coli Zur, an ultrasensitive zinc(II) metalloregulatory protein. Biochemistry 40:10417–10423. doi: 10.1021/bi0155448. [DOI] [PubMed] [Google Scholar]
- 37.Mortensen BL, Rathi S, Chazin WJ, Skaar EP. 2014. Acinetobacter baumannii response to host-mediated zinc limitation requires the transcriptional regulator Zur. J Bacteriol 196:2616–2626. doi: 10.1128/JB.01650-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ilari A, Alaleona F, Petrarca P, Battistoni A, Chiancone E. 2011. The X-ray structure of the zinc transporter ZnuA from Salmonella enterica discloses a unique triad of zinc-coordinating histidines. J Mol Biol 409:630–641. doi: 10.1016/j.jmb.2011.04.036. [DOI] [PubMed] [Google Scholar]
- 39.Wang N, Ozer EA, Mandel MJ, Hauser AR. 2014. Genome-wide identification of Acinetobacter baumannii genes necessary for persistence in the lung. mBio 5:e01163-14. doi: 10.1128/mBio.01163-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zimbler DL, Arivett BA, Beckett AC, Menke SM, Actis LA. 2013. Functional features of TonB energy transduction systems of Acinetobacter baumannii. Infect Immun 81:3382–3394. doi: 10.1128/IAI.00540-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Citiulo F, Jacobsen ID, Miramón P, Schild L, Brunke S, Zipfel P, Brock M, Hube B, Wilson D. 2012. Candida albicans scavenges host zinc via Pra1 during endothelial invasion. PLoS Pathog 8:e1002777. doi: 10.1371/journal.ppat.1002777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stork M, Grijpstra J, Bos MP, Mañas Torres C, Devos N, Poolman JT, Chazin WJ, Tommassen J. 2013. Zinc piracy as a mechanism of Neisseria meningitidis for evasion of nutritional immunity. PLoS Pathog 9:e1003733. doi: 10.1371/journal.ppat.1003733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Grim KP, San Francisco B, Radin JN, Brazel EB, Kelliher JL, Párraga Solórzano PK, Kim PC, McDevitt CA, Kehl-Fie TE. 2017. The metallophore staphylopine enables Staphylococcus aureus to compete with the host for zinc and overcome nutritional immunity. mBio 8:e01281-17. doi: 10.1128/mBio.01281-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.