Intercellular mitochondrial transfer mediated by the endoplasmic reticulum is essential for tissue homeostasis.
Abstract
Mitochondrial transfer plays a crucial role in the regulation of tissue homeostasis and resistance to cancer chemotherapy. Osteocytes have interconnecting dendritic networks and are a model to investigate its mechanism. We have demonstrated, in primary murine osteocytes with photoactivatable mitochondria (PhAM)floxed and in MLO-Y4 cells, mitochondrial transfer in the dendritic networks visualized by high-resolution confocal imaging. Normal osteocytes transferred mitochondria to adjacent metabolically stressed osteocytes and restored their metabolic function. The coordinated movement and transfer of mitochondria within the dendritic network rely on contact between the endoplasmic reticulum (ER) and mitochondria. Mitofusin 2 (Mfn2), a GTPase that tethers ER to mitochondria, predominantly mediates the transfer. A decline in Mfn2 expression with age occurs concomitantly with both impaired mitochondrial distribution and transfer in the osteocyte dendritic network. These data show a previously unknown function of ER-mitochondrial contact in mediating mitochondrial transfer and provide a mechanism to explain the homeostasis of osteocytes.
INTRODUCTION
Mitochondria play an essential role in cellular metabolism (1–4). A great deal of attention has been paid recently to the process of intercellular mitochondrial transfer, which enables recipient cells to regulate tissue damage, inflammation, and even resistance to cancer chemotherapy with the acquisition of functional mitochondria from donor cells (5–9). For example, endogenous neuroprotection during transient focal cerebral ischemia has been demonstrated when mouse astrocytes transfer their functional mitochondria to neighboring neurons (10). Conversely, damaged mitochondria in neurons have been shown to transfer into astrocytes for disposal and recycling (11).
While the biological impact of intercellular mitochondrial transfer among different cell types has been demonstrated over the past few years, the specific mechanism by which mitochondria are transferred from one cell to another remains unclear. Since contact between the endoplasmic reticulum (ER) and mitochondria appears to play a crucial role in coordinating communication between these two organelles (12, 13), we propose that the ER may also play a role in mediating intercellular mitochondrial transfer. We have used terminally differentiated osteocytes that lie within the lacunae of mineralized bone to investigate the role of mitochondrial transfer. Osteocytes are interconnected by a highly developed dendritic network that is known to influence the balance between bone resorption and bone formation via communication with osteoblasts and osteoclasts (14–17). Osteocytes therefore provide an ideal model for studying mitochondrial transfer between cells and its role in regulating cellular homeostasis.
RESULTS
Mitochondrial distribution in the osteocyte dendritic network
First, we examined whether a uniform and metabolically synchronized distribution of mitochondria existed within the dendritic networks of primary osteocytes. Using an established immunostaining and visualization protocol for primary osteocytes, we demonstrated in situ abundant mitochondria, labeled with the mitochondrial outer membrane protein TOM20 (translocase of outer mitochondrial membrane 20), along phalloidin-labeled calvarial osteocyte dendrites (Fig. 1, A and B). As mitochondrial metabolism is related to aging (18), we then examined mitochondrial mass in the primary osteocyte networks of cortical bones at 4, 12, and 18 months of age. Quantitative confocal images revealed that the distribution of TOM20-labeled mitochondria within the osteocyte dendritic processes decreased significantly with increasing age (Fig. 1, C and D).
Fig. 1. Mitochondria distributed within the osteocyte dendritic network.
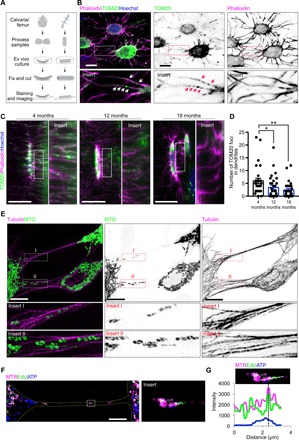
(A) Flowchart of imaging ex vivo cultured primary osteocytes. (B) Confocal analyses of primary osteocytes from ex vivo cultured calvariae show the distribution of TOM20-labeled mitochondria in dendrites of phalloidin-labeled primary osteocytes. (C) Confocal images of primary osteocytes from cortical bone of 4-, 12-, and 18-month-old mice femurs. (D) Quantitative analysis of mitochondrial distribution in the osteocyte dendritic network with age. Kruskal-Wallis test with Dunn’s post hoc. Three independent experiments were conducted. At least three fields and 30 cells in each group were analyzed. Data are presented as means ± SEM. (E) SIM images of the interconnected dendritic processes of MLO-Y4 cells show the association of mitochondria (MTO) in tubulin along dendrites of osteocytes. (F and G) Confocal images and fluorescence intensity analysis show the association of MitoTracker Red (MTR)–labeled mitochondria, EdU (5-ethynyl-2′-deoxyuridine)–labeled mtDNA, and Alexa Fluor 647–labeled ATP. Scale bars, 10 μm. *P < 0.05, **P < 0.01.
Previous studies have reported mitochondrial movement in the dendritic process of a single MLO-Y4 cell (19, 20). We therefore chose MLO-Y4 cells to further investigate the dynamic movement and distribution of mitochondria in the osteocyte dendritic network. Confocal microscope and structured illumination microscope (SIM) revealed similar characteristics between the mitochondrial distribution in the dendritic networks of primary calvarial osteocytes and cultured MLO-Y4 cells. Mitochondria, labeled with MitoTracker Orange (MTO), were distributed along with tubulin in the dendritic processes of MLO-Y4 cells (Fig. 1E). Mitochondria within the dendritic network of MLO-Y4 cells were functional, demonstrated by adenosine 5′-triphosphate (ATP) production and mitochondrial DNA (mtDNA) synthesis using fluorescence intensity analysis (Fig. 1, F and G). Moreover, confocal live-cell images also demonstrated the dynamic movement of MTO-labeled mitochondria along the track of tubulin in the osteocyte dendritic process (Fig. 2A).
Fig. 2. Mitochondrial transfer within the osteocyte dendritic process.
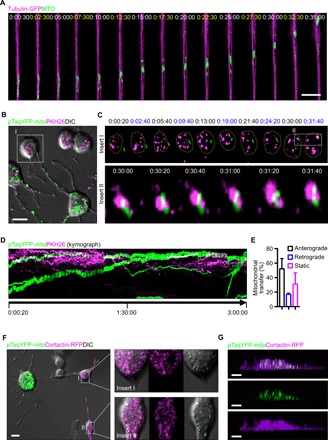
(A) Dynamic movement of MTO-labeled mitochondria with GFP-labeled dendritic tubulin in MLO-Y4 cells using confocal live-cell images. (B) PKH26-labeled recipient MLO-Y4 cells cocultured with pTaqYFP-mito–transfected donor MLO-Y4 cells. (C) Time-lapse confocal images show dendritic mitochondria labeled with pTaqYFP-mito moving dynamically toward the adjacent PKH26-labeled MLO-Y4 cell. Inserted images show the contacts between pTaqYFP-mito–labeled donor mitochondria and PKH26-labeled recipient cell membrane. (D) Kymograph analysis shows pTaqYFP-mito–labeled donor mitochondria moving toward PKH26-labeled recipient cell. (E) Quantitative assessment of mitochondrial movement in dendrites. Data are presented as means ± SEM. (F) Cocultures of MLO-Y4 cells transfected with pTaqYFP-mito or cortactin-RFP, respectively. Transferred mitochondria from donor cell (enlarged images in insert I) and transferred mitochondria from recipient cell (enlarged images in insert II) shown via differential interference contrast (DIC)–labeled dendrites. (G) 3D confocal images of replated cocultured MLO-Y4 cells show that the transferred pTaqYFP-mito–labeled mitochondria are present inside the cortactin-RFP–labeled recipient cell. Scale bars, 10 μm.
Mitochondrial transfer within the osteocyte dendritic network
We next investigated whether mitochondrial transfer occurs between osteocytes via the dendritic network. We cocultured MLO-Y4 cells expressing pTaqYFP-mito with PKH26-stained MLO-Y4 cells (Fig. 2B) to visualize mitochondrial motility and potential mitochondrial transfer between osteocytes within the dendritic network. Live-cell imaging showed that mitochondria (labeled green) in the dendrites dynamically migrated toward adjacent PKH26-labeled cells, as demonstrated by colocalization of mitochondria with the membrane of the recipient cells (Fig. 2, C and D, and movie S1). Quantitative confocal analysis and kymograph analysis showed anterograde movement of 52% of mitochondria toward adjacent cells and retrograde shuttling of 17% of mitochondria within the dendrites (Fig. 2E). To further validate the mitochondrial transfer between osteocytes, we cocultured pTaqYFP-mito–transfected donor cells with cortactin-RFP (red fluorescent protein)–transfected recipient cells. After 24 hours, we observed pTaqYFP-mito–labeled donor mitochondria in the cytoplasm of dendritically interconnected cortactin-RFP–labeled cells (Fig. 2F). Furthermore, three-dimensional (3D) confocal imaging on replated cortactin-RFP–labeled cells confirmed the transfer of donor mitochondria to the cytoplasm of recipient cells (Fig. 2G). To exclude the possibility that extracellular mitochondria released by cells may also enter other cocultured cells (10), we collected conditioned medium from cultured pTaqYFP-mito–transfected MLO-Y4 cells and added it to a culture of untransfected MLO-Y4 cells. After 24 hours, detection of transferred pTaqYFP-mito–labeled donor mitochondria within recipient cells was rare (fig. S1). These findings indicate that mitochondrial transfer occurs within the osteocyte dendritic network and that this process requires cell membrane contacts within the dendritic network of osteocytes.
To further demonstrate whether mitochondrial transfer occurs in the dendritic network of primary osteocytes in situ, we generated mice with osteocyte-specific photoactivatable mitochondria (PhAM). We used a Dendra2 green/red photoswitchable monomeric fluorescent protein fused to the mitochondrial targeting signal of subunit VIII of cytochrome c oxidase to generate mito-Dendra2. Mito-Dendra2 can produce fluorescence specifically in mitochondria without affecting function. Dmp1Cre excision of a floxed stop segment permits expression of green fluorescence in primary osteocyte mitochondria in situ (Fig. 3A). Upon exposure to a 405-nm laser in a designated area a (Fig. 3B) of mouse calvariae, mitochondria emitting green fluorescence can be irreversibly converted to mitochondria emitting red fluorescence (Fig. 3, B and C). The ex vivo mouse calvariae were cultured for 2 days to allow the transfer of mitochondria emitting red fluorescence, via the dendritic network, to other primary osteocytes with mitochondria emitting only green fluorescence in an area not activated by laser (area b; Fig. 3, B and C). We observed red fluorescence–emitting and green fluorescence–emitting mitochondria in primary calvarial osteocytes, indicating the transfer of mitochondria from osteocytes in area a into osteocytes in area b (Fig. 3D). To monitor and quantitate mitochondrial transfer, we isolated primary osteocytes from Dmp1CrePhAMfloxed mice (Fig. 3E). We then photoswitched primary osteocytes (Fig. 3, E and F) and cocultured them with non-photoswitched primary osteocytes for 12, 18, 24, and 48 hours (Fig. 3, E and G). Quantitative live-cell imaging further confirmed the transfer of mitochondria between primary osteocytes in a time-dependent manner (Fig. 3, H and I).
Fig. 3. Mitochondria transfer via the primary osteocyte dendritic network.
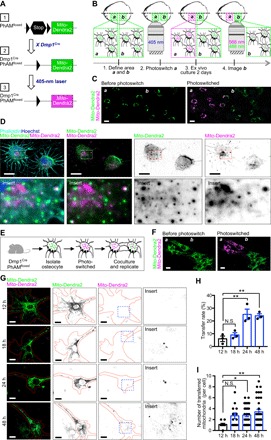
(A) A cre-dependent mito-Dendra2 cassette was inserted into the Rosa26 locus (schematic 1: black arrowheads, loxP sites; stop symbol, termination cassette). When crossed to Dmp1Cre mouse line, the termination signal is excised to produce the PhAM line with osteocyte-specific labeling of mitochondria (schematic 2). Irradiating mito-Dendra2 (green) with a 405-nm laser results in a photoswitch to mito-Dendra2 (red) (schematic 3). (B) Calvariae extracted from young Dmp1CrePhAMfloxed mice and irradiated by 405-nm laser in area a, but not in area b, are imaged after 2 days ex vivo culture. (C) Area a and area b before and after photoswitch. (D) Confocal images of photoswitched primary osteocytes in situ, after ex vivo culture, demonstrate the transfer of mito-Dendra2–labeled mitochondria (magenta) to the recipient osteocytes with mito-Dendra2–labeled mitochondria (green). (E) Schematic of photoswitching primary osteocytes and (F) confocal images of primary osteocytes isolated from Dmp1CrePhAMfloxed mice photoswitched in area a. (G) Photoswitched and non-photoswitched primary osteocytes were replated after coculture, and confocal images show the transfer of mitochondria between primary osteocytes. (H) Quantitative analysis shows the ratio of cells with transferred mito-Dendra2 (magenta) mitochondria at 12-, 18-, 24-, and 48-hour coculture. One-way analysis of variance (ANOVA) with Dunnett’s post hoc. Three independent experiments were conducted. Data are presented as means ± SEM. (I) Number of transferred Dendra2–labeled mitochondria (magenta) in each cell increases over time in culture. Kruskal-Wallis test with Dunn’s post hoc. Three independent experiments were conducted. At least 40 cells in each group were analyzed. Data are presented as means ± SEM. Scale bars, 10 μm. N.S., not significant; *P < 0.05, **P < 0.01.
Healthy osteocytes transfer mitochondria through the dendritic network to stressed osteocytes with nonfunctional mitochondria
Next, we investigated whether healthy osteocytes can rescue osteocytes under metabolic stress through the transfer of healthy mitochondria. We established a transwell culture system (Fig. 4A) to culture stressed MLO-Y4 with either healthy MLO-Y4 parental cells or primary osteocytes from Dmp1CrePhAMfloxed mice separated by a membrane. After 4 days, cells on either side of the membrane connect with each other using dendritic processes that pass through the pores of the membrane (Fig. 4B). No osteocytes passed through the membrane during coculture (fig. S2). To generate stressed osteocytes without mitochondrial function, we depleted mtDNA in MLO-Y4 cells (MLO-Y4 ρ° cells) by pretreating them with ethidium bromide (EtBr) in medium containing uridine and pyruvate (21, 22). Depletion of mtDNA in MLO-Y4 ρ° cells, as confirmed by confocal microscope and quantitative polymerase chain reaction (qPCR), resulted in reduced cellular ATP levels (Fig. 4C and fig. S3, A to C). Confocal live-cell imaging of the transwell culture system demonstrated the transfer of green fluorescence–labeled mito-Dendra2 mitochondria from primary osteocytes into MLO-Y4 ρ° cells through the pores of the membrane (Fig. 4, D and E). The confocal live-cell imaging revealed that both healthy parental MLO-Y4 cells and primary osteocytes can improve the mitochondrial function in MLO-Y4 ρ° cells, as indicated by the level of ATP 647 (Fig. 4, F and G), the fluorescence signal of an oxygen sensor (labeled by Mito ID) (fig. S4, A and B), and H2DCFDA (fig. S4, A and C), through the transfer of functional mitochondria via interconnected dendritic processes.
Fig. 4. Healthy osteocytes rescue MLO-Y4 ρ° cells via dendritic connection through the transwell membrane.
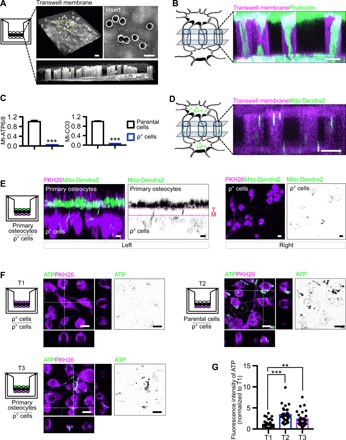
(A) Confocal images of the transwell membrane and its pores. (B) Dendritic processes of MLO-Y4 cells extend through the pores of the membrane and connect with cocultured MLO-Y4 cells. (C) qPCR results show the depletion of mitochondrial genes Mt-ATP6/8 and Mt-CO3 in MLO-Y4 ρ° cells. (D) Mito-Dendra2 green–labeled mitochondria from primary osteocytes transfer through the pores of the transwell membrane. (E) 3D confocal images of transwell membrane coculture show the transfer of mitochondria from mito-Dendra2 green–labeled primary osteocytes to MLO-Y4 ρ° cells (ρ° cells). Left: Transverse view of 3D cocultured primary osteocyte and ρ° cells (TM, transwell membrane). Right: View of ρ° cells from above. (F) T1: ρ° cells cocultured with ρ° cells in the transwell culture system; T2: ρ° cells cocultured with healthy MLO-Y4 cells (parental cells); T3: ρ° cells cocultured with primary osteocytes. (G) Quantitative analysis of 3D confocal images shows that the healthy osteocytes are able to rescue ATP production (labeled by Alexa Fluor 647) in ρ° cells. Kruskal-Wallis test with Dunn’s post hoc. Three independent experiments were conducted. At least 30 cells in each group were analyzed. Data are presented as means ± SEM. Scale bars, 10 μm. **P < 0.01, ***P < 0.001.
ER facilitates mitochondrial transfer between osteocytes
The close association between mitochondria and the ER (23–25) underpins the importance of structural and functional interactions required for cellular homeostasis (12, 13, 26–28). In MLO-Y4 cells expressing Mito-GFP (green fluorescent protein) and ER-RFP, we observed colocalization of mitochondria and ER within dendritic processes (fig. S5A) and the dynamic movement of RFP-labeled ER along with GFP-labeled dendritic tubulin in MLO-Y4 cells using live-cell imaging (fig. S5B). In ex vivo cultured calvarial primary osteocytes, TOM20-labeled mitochondria within the dendritic network also colocalized with calnexin-labeled ER (Fig. 5, A and B). Live-cell imaging showed that the dynamic movement of pTaqYFP-mito–labeled mitochondria from the cytoplasm to the dendritic process was associated with RFP-labeled ER (Fig. 5C and movie S2). Transfer of mitochondria within dendrites was dynamically colocalized with the ER (Fig. 5, D to I, and movie S3). Mitofusin 2 (Mfn2), a guanosine triphosphatase localized to the outer membrane of mitochondria, has been shown to tether mitochondria to the ER (29–31). We showed that Mfn2 was involved in the dynamic mitochondrial movement within the osteocyte dendritic process (Fig. 6, A to C, and movie S4).
Fig. 5. ER facilitates mitochondrial transfer between osteocytes.
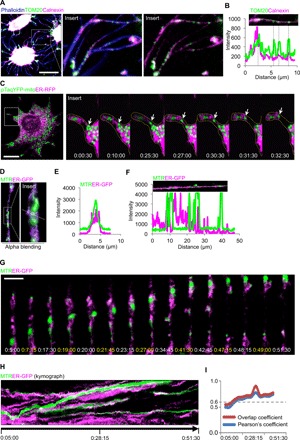
(A and B) 3D confocal analysis and fluorescence intensity analysis of primary osteocytes from ex vivo cultured calvariae. Images show the colocalization of TOM20-labeled mitochondria and calnexin-labeled ER in phalloidin-labeled dendrites. (C) Confocal time-lapse images of live MLO-Y4 cell show the colocalization of pTagYFP-mito–labeled mitochondria and RFP-labeled ER during transfer from the cytoplasm to the dendritic process. (D and E) 3D image and fluorescence intensity analysis show the colocalization between MTR-labeled mitochondria and GFP-labeled ER. (F) Fluorescence intensity analysis of dendritic processes shows the colocalization between GFP-labeled ER and MTR-labeled mitochondria. (G) Confocal time-lapse images show that GFP-labeled ER is dynamically colocalized with MTR-labeled mitochondria during movement toward the adjacent MLO-Y4 cell. (H and I) Kymograph and overlap and Pearson’s coefficient analyses show the dynamic association of GFP-labeled ER and MTR-labeled mitochondria. Scale bars, 10 μm.
Fig. 6. Mfn2 regulates mitochondrial transfer between osteocytes.
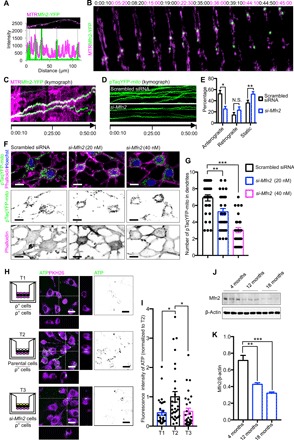
(A) YFP-labeled Mfn2 and MTR-labeled mitochondria are colocalized in the MLO-Y4 cell dendrite. (B and C) Colocalization of YFP-labeled Mfn2 and MTR-labeled mitochondria during movement toward adjacent MLO-Y4 cells. (D) Dynamic mitochondrial movement was impeded by knockdown of Mfn2. (E) Knockdown of Mfn2 attenuates movement of the pTaqYFP-mito–labeled mitochondria in dendrites. Two-tailed Student’s t test. Three independent experiments were conducted. At least 10 cells in each group were analyzed. Data presented as means ± SEM. (F and G) Mfn2 knockdown significantly reduces distribution of pTaqYFP-mito–labeled mitochondria within dendrites of MLO-Y4 cells. Kruskal-Wallis test with Dunn’s post hoc. Three independent experiments were conducted. At least 30 cells in each group were analyzed. Data presented as means ± SEM. (H and I) T1: MLO-Y4 ρ° cells (ρ° cells) cocultured with ρ° cells in the transwell culture system; T2: ρ° cells cocultured with healthy MLO-Y4 cells (parental cells); T3: ρ° cells cocultured with Mfn2 knockdown MLO-Y4 cells (si-Mfn2 cells). Healthy cells rescue ATP production (labeled by Alexa Fluor 647) in ρ° cells but not in cells lacking Mfn2. Kruskal-Wallis test with Dunn’s post hoc. Three independent experiments were conducted. At least 30 cells in each group were analyzed. Data presented as means ± SEM. (J and K) Protein level of Mfn2 in cortical bone decreases with age. One-way ANOVA with Dunnett’s multiple comparisons test. Data presented as means ± SEM. Scale bars, 10 μm. *P < 0.05, **P < 0.01.
To confirm the role of ER in mitochondrial transfer, we blocked the ER-mitochondria association by silencing Mfn2 in MLO-Y4 cells (fig. S6, A and B). Silencing Mfn2 had minimal effect on cellular activity and on mtDNA synthesis (fig. S6, C to E), but confocal imaging revealed significantly attenuated mitochondrial motility (Fig. 6, D and E) and reduced distribution of pTaqYFP-mito–labeled mitochondria within phalloidin-labeled dendrites in Mfn2 siRNA (small-interfering RNA) (si-Mfn2)–transfected MLO-Y4 cells (Fig. 6, F and G). Moreover, si-Mfn2–treated cells that lacked sufficient ER-mitochondria contacts were not able to restore metabolic function when cocultured with MLO-Y4 ρ° cells, as indicated by the levels of ATP 647 (Fig. 6, H and I), the fluorescence signal of an oxygen sensor (labeled by Mito ID) (fig. S7, A and B), and H2DCFDA (fig. S7, A and C). Immunoblotting revealed that the levels of Mfn2 in cortical bone protein lysates decreased with age (Fig. 6, J and K). This suggests that the decline of Mfn2 expression is associated with the decline in mitochondrial distribution, and possibly also mitochondrial transfer, that occurs in osteocyte dendritic networks with age (Fig. 1, C and D). Similarly, knockdown of vesicle-associated membrane protein–associated protein B (VAPB), another ER-mitochondria tethering protein located on the ER membrane (32), also impeded the mitochondrial transfer and the ability to rescue the metabolic function of MLO-Y4 ρ° cells (fig. S8, A to D).
Together, these results suggest that the ER mediates mitochondrial transfer between neighboring osteocytes to synchronize their energy metabolism. ER from the donor osteocyte may interact with the ER in the recipient osteocyte to initiate mitochondrial transfer. Our data demonstrate that ER regulates the distribution and transfer of mitochondria within the osteocyte dendritic processes via the tethering protein complexes including Mfn2 and VAPB.
DISCUSSION
Terminally differentiated osteocytes are trapped within the lacunae of the mineralized bone matrix, which renders them vulnerable to reduced mineral supply (33, 34). Osteocytes, however, are interconnected by their highly developed dendritic network (35–39). The osteocyte network is known to regulate the balance between bone resorption and bone formation via communication with osteoblasts and osteoclasts (14–17), primarily in response to mechanical stress (40). Although the dense osteocyte dendritic network is essential for regulating mineral homeostasis (40, 41), little is known about how osteocytes use the network to coordinate cellular function and communication.
We have used high-resolution live-cell imaging, in combination with an established ex vivo culture system, to demonstrate the dynamic transfer of mitochondria within the dendritic network in both primary osteocytes and MLO-Y4 cells. The transfer process is mediated by ER-mitochondria contacts. Regional differences in mitochondrial viability have been demonstrated between osteocytes located in endocortical surfaces and those embedded deeper in the bone matrix (42). This suggests that the synchronization of energy metabolism in different osteocyte networks may depend on the anatomical structure.
Donor mitochondrial transfer provides a potential mechanism for exogenous replacement of dysfunctional mitochondria in stressed recipient osteocytes, thereby rescuing the recipient osteocytes. Given the vulnerability of osteocytes to stresses such as hypoxia and mechanical overloading, as well as their long life span (33, 43–46), it is essential that osteocytes have survival mechanisms that are distinct from other cell types. Osteocyte mitochondrial transfer may provide protection against excessive cell damage and avoid subsequent permanent dysfunction of the “imprisoned” osteocytes in the bone matrix.
Our data suggest that stress-induced impairment of mitochondrial function in osteocytes may trigger donor mitochondrial transfer from neighboring healthy osteocytes. The intriguing questions regarding (i) the degree of cellular damage required for stressed cells to initiate mitochondria acquisition and (ii) the mechanisms by which healthy osteocytes identify stressed osteocytes and successfully restore their mitochondrial functionality remain to be investigated. Precedent for this type of cellular behavior has already been established in the activation of bystander osteocytes by apoptosing osteocytes (47). In addition, our study does not exclude the possibility that other organelles and metabolites may also be transferred between cells and affect osteocyte metabolism in other ways.
Aging is a chief determinant of fracture risk and osteoporosis (48) and is likely caused by the simultaneous deterioration of various inter-related cellular functions (49). Osteocytes, as the predominant regulator of bone mass, likely play a critical role in the progression of bone aging. We have shown that the distribution of mitochondria in dendritic processes and levels of Mfn2 are reduced in aging osteocytes. The age-associated decrease in Mfn2 levels may contribute to the impairment of mitochondrial transfer within the osteocyte dendritic network.
Using our 3D transwell coculture model, we observed that healthy MLO-Y4 parental cells can induce recovery of mitochondrial metabolic function of stressed MLO-Y4 ρ° cells. MLO-Y4 ρ° cells formed abundant dendritic interconnections with parental cells. Metabolic recovery in MLO-Y4 ρ° cells was assessed using semiquantitative measurements of the levels of ATP, oxygen consumption, and reactive oxygen species (ROS) accumulation. Direct quantitation was not feasible in our 3D coculture system, and further studies will be required to more accurately analyze these markers of metabolic function. Nevertheless, the semiquantitative results have shown the functional significance of mitochondrial transfer between osteocytes.
In conclusion, we provide evidence that mitochondria are shared between osteocytes through their dendritic network. Mitochondria are dynamically transferred into the network, and their movement is dependent on contact with the ER. Healthy osteocytes can transfer mitochondria into metabolically stressed osteocytes having nonfunctional mitochondria through the dendritic network. Such a mechanism enables recovery of ATP levels and oxygen consumption and attenuates ROS accumulation in the stressed osteocytes.
We propose a putative model of mitochondrial transfer between osteocytes in fig. S9. When mitochondria are transferred between osteocytes, a dendritic process from a donor osteocyte extends to the cytoplasmic membrane of a recipient osteocyte. The ER membrane of the donor cell makes contact with the recipient osteocyte, and donor mitochondria, closely apposed by the ER, are then transferred to the recipient osteocytes (fig. S9, A to E). The precise characteristics of the cytoplasmic extensions/dendrites that execute the process are yet to be defined.
MATERIALS AND METHODS
Reagents and animals
pTaqYFP-mito vector and cortactin-RFP vector were a gift from N. J. Pavlos (University of Western Australia), and Mfn2-YFP vector was a gift from R. Youle [University of Maryland Biotechnology Institute; Addgene plasmid no. 28010 (http://n2t.net/addgene:28010) and RRID: Addgene_28010]. PhAMfloxed mice [B6;129S-Gt(ROSA)26Sortm1(CAG-COX8A/Dendra2)Dcc/J] were purchased from the Jackson Laboratory. The 9.6-kb Dmp1Cre transgenic mice were provided by J. Q. (Jerry) Feng at Texas A&M College of Dentistry, USA. All animals were housed and maintained in accordance with the Institutional Animal Care and Use Committee guidelines.
Immunoblotting
Soft tissues and periosteum were removed from mice femora by microsurgery forceps under the dissecting microscope. Diaphyses were extracted, and bone marrow was flushed away. Femoral diaphyses were then crushed using a mortar and pestle in liquid nitrogen. Both bone and cellular proteins were extracted by incubating in radioimmunoprecipitation assay lysis buffer containing protease inhibitor (Roche) and a phosphatase inhibitor cocktail (Sigma-Aldrich) for 30 min at 4°C. Clarified bone and cell lysate (obtained by centrifugation at 16,000g for 20 min at 4°C) were diluted with 4× SDS sampling buffer and boiled for 5 min. For each sample, 4 μg of protein was fractionated on 10 to 17.5% SDS–polyacrylamide gel electrophoresis gel and transferred to a nitrocellulose membrane (Millipore). Membranes were incubated with primary antibodies, including β-actin JLA20 antibody (1:5000; Developmental Studies Hybridoma Bank), Mfn2 antibody (1:1000; 9482, Cell Signaling Technology), or VAPB antibody (1:1000; HPA013144, Sigma-Aldrich), and the corresponding horseradish peroxidase–conjugated secondary antibodies. Proteins were visualized by enhanced chemiluminescence and autoradiography (FujiFilm LAS-3000/4000 Gel Documentation System, Japan).
Cell culture and ex vivo calvarial culture
MLO-Y4 cells were plated in type I rat tail collagen-coated six-well plates (Biocoat, Becton Dickinson) or coverslips and maintained in α–minimal essential medium (α-MEM; Gibco) supplemented with 5% fetal bovine serum (FBS; Gibco, Thermo Fisher Scientific) and 1% penicillin/streptomycin (P/S) (Sigma-Aldrich). For transwell culture, MLO-Y4 ρ° cells were plated on the outer side of the transwell membrane. After 24 hours, MLO-Y4 ρ° cells, MLO-Y4 parental cells, primary osteocytes, or si-Mfn2 MLO-Y4 cells were plated on the inner side of the transwell membrane and cultured for up to 4 days. The cells were maintained at 37°C in a 5% CO2 humidified incubator.
For ex vivo culture, calvariae from 10-day-old mice were washed three times with phosphate-buffered solution (PBS) and three times with α-MEM containing 1% P/S. The periosteum was removed from the calvariae with scraping and extensive washing. Calvariae were maintained in α-MEM supplemented with 10% FBS and 1% P/S at 37°C and 5% CO2 in a humidified incubator. Ex vivo cultured calvariae were fixed in 4% paraformaldehyde (PFA) for immunostaining and maintained in culture medium in 35-mm glass-bottom petri dishes (P35g-1.5-14-C, MatTek) for subsequent confocal imaging. MLO-Y4 ρ° cells were generated by culturing parental MLO-Y4 for 15 weeks in α-MEM medium supplemented with EtBr (50 ng/ml), pyruvate (110 μg/ml), and uridine (50 μg/ml) and subsequently transferred to the medium lacking EtBr.
Cell transfections
MLO-Y4 cells were transfected with pTaqYFP-mito, cortactin-RFP, Mfn2 siRNA (172635, Thermo Fisher Scientific), Vapb siRNA (185633, Thermo Fisher Scientific), or negative control siRNA (AM4611, Thermo Fisher Scientific) using Lipofectamine LTX Plus reagents or Lipofectamine 3000 transfection reagents following the manufacturer’s protocol. All cells were routinely tested for mycoplasma contamination using a MycoTOOL kit (Roche). All experiments were performed at passages <10.
Isolation of primary osteocytes from long bone
The isolation of primary osteocytes was performed according to the published method (50). Primary osteocytes were isolated from mouse (4 months old) long bones according to the protocol (50). Long bones (femur and tibia) were aseptically dissected from skeletally mature mice and placed in α-MEM with 1% P/S. Following removal of epiphyses, bone marrow, and periosteum with scraping, extensive washing, and centrifugation, cortical bones were cut into 1 to 2 mm in length. Bone pieces were digested in collagenase solution [collagenase type IA (300 U/ml); Sigma-Aldrich] and dissolved in α-MEM for 25 min. The solution was aspirated and kept for cell plating, and the bone pieces were washed in Hanks’ balanced salt solution (HBSS). Subsequently, bone pieces were incubated with EDTA [5 mM (pH 7.4); Sigma-Aldrich] prepared in magnesium- and calcium-free Dulbecco’s PBS (Mediatech) with 1% bovine serum albumin (BSA; Sigma-Aldrich) for 25 min. The solution was aspirated, centrifuged (200g for 5 min), and kept for cell plating, and the bone pieces were washed again in HBSS. These steps were repeated for a total of three digestions. The remaining bone pieces were minced with a tissue homogenizer (Medimachine, BD Biosciences) in α-MEM, and the bone particles were directly plated. The digestions took place in a six-well petri dish on a rotating shaker set to 200 rpm at 37°C in a 5% CO2 humidified incubator. The collected cell suspensions were cultured on type I rat tail collagen-coated six-well plates at a seeding density of approximately 250,000 cells/9.5 cm2 in α-MEM supplemented with 10% FBS and 1% P/S. Cells were maintained at 37°C in a 5% CO2 humidified incubator for 7 days.
Immunofluorescence
Cortical bone and calvariae were cleaned of non-osseous tissue (soft tissues and periosteum) by microsurgery forceps under the dissecting microscope, washed three times with culture medium (α-MEM supplemented with 5% FBS and 1% P/S), and cultured on stainless steel grids (1210 T71, Thomas Scientific). Bone tissues were fixed in 4% PFA for 2 hours at room temperature and embedded in optimal cutting temperature compound (OCT compound) (Tissue-Tek). Bone tissues were cut using a cryostat microtome (Leica) to expose the embedded osteocytes in the central region of the cortical bone. After melting the OCT from the tissue, diaphyses were extracted and bone marrow was removed. The cortical or calvarial bones were washed three times in PBS, permeabilized in 0.1% Triton X-100 in PBS for 5 min at room temperature, washed again three times in PBS, and incubated with 3% BSA-PBS for 30 min at room temperature to block nonspecific antibody binding. Bone tissues were then incubated with TOM20 antibody (1:50; sc-17764, Santa Cruz Biotechnology) or calnexin antibody (1:50; sc-23954, Santa Cruz Biotechnology) in 0.2% BSA-PBS overnight at 4°C, with gentle shaking to thoroughly permeate antibodies into the osteocytes. Bone tissues were then washed three times in PBS with gentle shaking and incubated with goat anti-rabbit immunoglobulin G (IgG) (H + L) cross-adsorbed secondary antibody, Alexa Fluor 647 (1:1000; A-21244, Thermo Fisher Scientific), goat anti-mouse IgG (H + L) cross-adsorbed secondary antibody, Alexa Fluor 488 (1:1000; A-11001, Thermo Fisher Scientific), rhodamine phalloidin (1:500; R415, Thermo Fisher Scientific), or Alexa Fluor 647 phalloidin (1:500; A22287, Thermo Fisher Scientific) for 45 min with gentle shaking. Bone tissues were washed three times in 0.2% BSA-PBS and three times in PBS with gentle shaking and then incubated with Hoechst dye (1:5000; 33342, PerkinElmer) for 15 min at room temperature. After washing three times in 0.2% BSA-PBS, tissues were mounted onto 35-mm glass-bottom petri dishes (P35g-1.5-14-C, MatTek) by ProLong Diamond antifade medium (Invitrogen).
MLO-Y4 cells seeded in 5- or 8-mm coverslips (ProSciTech) were fixed in 4% PFA at room temperature for 20 min, washed three times in PBS, permeabilized in 0.1% Triton X-100 in PBS for 5 min at room temperature, and again washed three times in PBS. Nonspecific antibody binding was blocked by incubating in 3% BSA-PBS for 30 min at room temperature. MLO-Y4 cells were incubated with 50 nM MTO chloromethyltetramethylrosamine (M7510, Thermo Fisher Scientific) for 30 min, washed three times with culture medium, fixed, and immunostained with α-tubulin antibody (1:100; 2144, Cell Signaling Technology) for SIM.
For live-cell staining, MLO-Y4 cells were placed on 35-mm glass-bottom petri dishes (P35g-1.5-14-C, MatTek). Cells were transfected with Mito-GFP (C10600, CellLight) BacMam 2.0, ER-GFP BacMam 2.0 (C10590, CellLight), ER-RFP BacMam 2.0 (C10591, CellLight), or tubulin-GFP BacMam 2.0 (C10613, CellLight) by directly adding them to the culture medium, according to the online protocol.
Cells were stained with 5 μM PKH26 (MINI26, Sigma-Aldrich) and 50 nM MitoTracker Red CMXRos (M7512, Thermo Fisher Scientific) for 30 min and washed three times with culture medium for live-cell imaging or stained with 5 μM Alexa Fluor 647 ATP (Thermo Fisher Scientific), 5 μM H2DCFDA (general oxidative stress indicator; C6827, Thermo Fisher Scientific), MITO-ID Extracellular O2 Sensor kit (ENZ-51045-K500, Enzo), and 1 μM EdU (5-ethynyl-2′-deoxyuridine; E1018, Thermo Fisher Scientific), together with Click-iT EdU Alexa Fluor 488 Imaging Kit (C10337, Invitrogen), for 20 min and washed three times with culture medium for live-cell imaging.
Cell imaging and analysis
Fixed cell images were acquired on a Nikon A1 microscope using a 60× oil immersion objective, a Zeiss LSM 710 confocal microscope with EC-Plan-Neofluar 40× oil immersion objective, or a Nikon SIM (Ti2) using a 100× oil immersion objective with motorized correction collar. Live-cell images were acquired on a Nikon A1 microscope using a water objective in a temperature-controlled chamber (37°C, 5% CO2). To photoswitch Dendra2, a region was illuminated with the 405-nm line (4% laser power) for 90 bleaching iterations at a scan speed of 6.3 μs/pixel. No obvious cell death was observed during the photoswitch process.
Digital images were acquired using Nikon NIS Elements software or ZEISS ZEN microscope software. For 3D primary osteocyte imaging, 20- to 40-μm Z-stack images, examined in 1-μm steps, from the exposed surface to the bone matrix were processed with MaxIP or alpha blending using Nikon NIS Elements software. Cells were excluded from analysis if they displayed signs of phototoxicity such as blebbing or vacuolization. All images were assembled and analyzed using Fiji (National Institutes of Health).
RNA isolation and real-time qPCR
Total RNA was isolated using TRizol reagent (15596-026, Invitrogen). RNA (1 μg) was reverse-transcribed into complementary DNA using M-MLV (Moloney murine leukemia virus) reverse transcriptase (Promega). Real-time qPCR was performed using 2× SYBR Green Master Mix (Bio-Rad) in a CFX Connect Real-Time PCR Detection System (Bio-Rad). Each sample was run in triplicate, and gene expression levels were normalized to β-actin. The primer sequences are listed in table S1.
Statistical analysis
A two-tailed Student’s t test was used for means comparison between two groups. Comparisons of multiple individual datasets were performed using one-way analysis of variance (ANOVA) with Dunnett’s post hoc or Kruskal-Wallis test with Dunn’s post hoc in accordance with the normal or non-normal distribution of data. Statistical calculations were processed by GraphPad Prism software (version 8.0.0). Data are presented as the means ± SEM, and P values <0.05 were considered statistically significant and marked by asterisks (N.S., no significance; *P < 0.05, **P < 0.01, and ***P < 0.001).
Supplementary Material
Acknowledgments
We acknowledge P. Rigby and A. Buckley for the facilities and the scientific and technical assistance at the National Imaging Facility at the Centre for Microscope, Characterisation and Analysis, University of Western Australia. This facility is funded by the University, State, and Commonwealth Governments. We also thank D. Findlay from University of Adelaide and A. Carleton for syntax and grammar editing. Funding: This project was supported by the Australian National Health and Medical Research Council (NH&MRC) Project Grant (APP1049884) awarded to M.H.Z., by the Early Career Researcher Small Grant from Faculty of Health and Medical Sciences, University of Western Australia awarded to J.G., and by the National Natural Science Foundation of China awarded to C.Z. (grant no. 81820108020) and to A.Q. (grant nos. 81572167, 81201364, and 81772373). The content is solely the responsibility of the authors. Author contributions: J.G., H.T., C.Z., and M.H.Z. designed the research. J.G., A.Q., D.L., R.R., Q.W., J.Y., and T.S.C. performed the research. D.L., R.R., J.Y., X.G., and Q.J. analyzed the data. J.G., A.F., J.M.P., K.D., J.Q.F., and M.H.Z. wrote the paper. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/5/11/eaaw7215/DC1
Fig. S1. No mitochondrial transfer from conditioned medium to MLO-Y4 cells.
Fig. S2. Osteocytes do not pass through the pores of the transwell membrane.
Fig. S3. Depleted mtDNA and impeded ATP production in MLO-Y4 ρ° cells.
Fig. S4. Parental MLO-Y4 cells and primary osteocytes can transfer mitochondria to MLO-Y4 ρ° cells.
Fig. S5. Mitochondrial distribution and ER movement in osteocyte dendrites.
Fig. S6. Knockdown of Mfn2 has no effect on ATP production and the level of mtDNA.
Fig. S7. Mfn2 knockdown impedes mitochondrial transfer in MLO-Y4 cells.
Fig. S8. VAPB knockdown impedes mitochondrial transfer in MLO-Y4 cells.
Fig. S9. Hypothetical model of ER-mediated mitochondrial transfer.
Table S1. Primer sequences for qPCR.
Movie S1. pTaqYFP-mito–labeled mitochondria (green) in the dendrite dynamically migrate toward the PKH26-stained cells (magenta).
Movie S2. pTaqYFP-mito–labeled mitochondria (green) dynamically move from the cytoplasm to the dendritic process and are associated with RFP-labeled ER (magenta).
Movie S3. Transfer of MTR-labeled mitochondria (magenta) within dendrites is dynamically colocalized with GFP-labeled ER (green).
Movie S4. Transfer of MTR-labeled mitochondria (magenta) is dynamically colocalized with YFP-labeled Mfn2 (green) within dendrites.
REFERENCES AND NOTES
- 1.Kroemer G., Reed J. C., Mitochondrial control of cell death. Nat. Med. 6, 513–519 (2000). [DOI] [PubMed] [Google Scholar]
- 2.Balaban R. S., Nemoto S., Finkel T., Mitochondria, oxidants, and aging. Cell 120, 483–495 (2005). [DOI] [PubMed] [Google Scholar]
- 3.Schriner S. E., Linford N. J., Martin G. M., Treuting P., Ogburn C. E., Emond M., Coskun P. E., Ladiges W., Wolf N., van Remmen H., Wallace D. C., Rabinovitch P. S., Extension of murine life span by overexpression of catalase targeted to mitochondria. Science 308, 1909–1911 (2005). [DOI] [PubMed] [Google Scholar]
- 4.Chan D. C., Mitochondria: Dynamic organelles in disease, aging, and development. Cell 125, 1241–1252 (2006). [DOI] [PubMed] [Google Scholar]
- 5.Lee C. W., Peng H. B., The function of mitochondria in presynaptic development at the neuromuscular junction. Mol. Biol. Cell 19, 150–158 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Courchet J., Lewis T. L. Jr., Lee S., Courchet V., Liou D. Y., Aizawa S., Polleux F., Terminal axon branching is regulated by the LKB1-NUAK1 kinase pathway via presynaptic mitochondrial capture. Cell 153, 1510–1525 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cao X., Wang H., Wang Z., Wang Q., Zhang S., Deng Y., Fang Y., In vivo imaging reveals mitophagy independence in the maintenance of axonal mitochondria during normal aging. Aging Cell 16, 1180–1190 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yao Y., Fan X. L., Jiang D., Zhang Y., Li X., Xu Z. B., Fang S. B., Chiu S., Tse H. F., Lian Q., Fu Q. L., Connexin 43-mediated mitochondrial transfer of iPSC-MSCs alleviates asthma inflammation. Stem Cell Reports 11, 1120–1135 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Babenko V. A., Silachev D. N., Popkov V. A., Zorova L. D., Pevzner I. B., Plotnikov E. Y., Sukhikh G. T., Zorov D. B., Miro1 enhances mitochondria transfer from multipotent mesenchymal stem cells (MMSC) to neural cells and improves the efficacy of cell recovery. Molecules 23, E687 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hayakawa K., Esposito E., Wang X., Terasaki Y., Liu Y., Xing C., Ji X., Lo E. H., Transfer of mitochondria from astrocytes to neurons after stroke. Nature 535, 551–555 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davis C. H., Kim K. Y., Bushong E. A., Mills E. A., Boassa D., Shih T., Kinebuchi M., Phan S., Zhou Y., Bihlmeyer N. A., Nguyen J. V., Jin Y., Ellisman M. H., Marsh-Armstrong N., Transcellular degradation of axonal mitochondria. Proc. Natl. Acad. Sci. U.S.A. 111, 9633–9638 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rowland A. A., Voeltz G. K., Endoplasmic reticulum-mitochondria contacts: Function of the junction. Nat. Rev. Mol. Cell Biol. 13, 607–615 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Friedman J. R., Lackner L. L., West M., DiBenedetto J. R., Nunnari J., Voeltz G. K., ER tubules mark sites of mitochondrial division. Science 334, 358–362 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huiskes R., Ruimerman R., van Lenthe G. H., Janssen J. D., Effects of mechanical forces on maintenance and adaptation of form in trabecular bone. Nature 405, 704–706 (2000). [DOI] [PubMed] [Google Scholar]
- 15.Nakashima T., Hayashi M., Fukunaga T., Kurata K., Oh-hora M., Feng J. Q., Bonewald L. F., Kodama T., Wutz A., Wagner E. F., Penninger J. M., Takayanagi H., Evidence for osteocyte regulation of bone homeostasis through RANKL expression. Nat. Med. 17, 1231–1234 (2011). [DOI] [PubMed] [Google Scholar]
- 16.Tatsumi S., Ishii K., Amizuka N., Li M., Kobayashi T., Kohno K., Ito M., Takeshita S., Ikeda K., Targeted ablation of osteocytes induces osteoporosis with defective mechanotransduction. Cell Metab. 5, 464–475 (2007). [DOI] [PubMed] [Google Scholar]
- 17.Poole K. E., van Bezooijen R. L., Loveridge N., Hamersma H., Papapoulos S. E., Löwik C. W., Reeve J., Sclerostin is a delayed secreted product of osteocytes that inhibits bone formation. FASEB J. 19, 1842–1844 (2005). [DOI] [PubMed] [Google Scholar]
- 18.Giorgi C., Marchi S., Simoes I. C. M., Ren Z., Morciano G., Perrone M., Patalas-Krawczyk P., Borchard S., Jędrak P., Pierzynowska K., Szymański J., Wang D. Q., Portincasa P., Węgrzyn G., Zischka H., Dobrzyn P., Bonora M., Duszynski J., Rimessi A., Karkucinska-Wieckowska A., Dobrzyn A., Szabadkai G., Zavan B., Oliveira P. J., Sardao V. A., Pinton P., Wieckowski M. R., Mitochondria and reactive oxygen species in aging and age-related diseases. Int. Rev. Cell Mol. Biol. 340, 209–344 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bonewald L. F., Establishment and characterization of an osteocyte-like cell line, MLO-Y4. J. Bone Miner. Metab. 17, 61–65 (1999). [DOI] [PubMed] [Google Scholar]
- 20.Kitase Y., Vallejo J. A., Gutheil W., Vemula H., Jähn K., Yi J., Zhou J., Brotto M., Bonewald L. F., β-Aminoisobutyric acid, l-BAIBA, is a muscle-derived osteocyte survival factor. Cell Rep. 22, 1531–1544 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.King M. P., Attardi G., Human cells lacking mtDNA: Repopulation with exogenous mitochondria by complementation. Science 246, 500–503 (1989). [DOI] [PubMed] [Google Scholar]
- 22.Bodnar A. G., Cooper J. M., Holt I. J., Leonard J. V., Schapira A. H., Nuclear complementation restores mtDNA levels in cultured cells from a patient with mtDNA depletion. Am. J. Hum. Genet. 53, 663–669 (1993). [PMC free article] [PubMed] [Google Scholar]
- 23.Rizzuto R., Pinton P., Carrington W., Fay F. S., Fogarty K. E., Lifshitz L. M., Tuft R. A., Pozzan T., Close contacts with the endoplasmic reticulum as determinants of mitochondrial Ca2+ responses. Science 280, 1763–1766 (1998). [DOI] [PubMed] [Google Scholar]
- 24.de Brito O. M., Scorrano L., An intimate liaison: Spatial organization of the endoplasmic reticulum-mitochondria relationship. EMBO J. 29, 2715–2723 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rizzuto R., Marchi S., Bonora M., Aguiari P., Bononi A., de Stefani D., Giorgi C., Leo S., Rimessi A., Siviero R., Zecchini E., Pinton P., Ca2+ transfer from the ER to mitochondria: When, how and why. Biochim. Biophys. Acta 1787, 1342–1351 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Horner S. M., Liu H. M., Park H. S., Briley J., Gale M. Jr., Mitochondrial-associated endoplasmic reticulum membranes (MAM) form innate immune synapses and are targeted by hepatitis C virus. Proc. Natl. Acad. Sci. U.S.A. 108, 14590–14595 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marchi S., Patergnani S., Pinton P., The endoplasmic reticulum-mitochondria connection: One touch, multiple functions. Biochim. Biophys. Acta 1837, 461–469 (2014). [DOI] [PubMed] [Google Scholar]
- 28.Hoppins S., Nunnari J., Mitochondrial dynamics and apoptosis—The ER connection. Science 337, 1052–1054 (2012). [DOI] [PubMed] [Google Scholar]
- 29.de Brito O. M., Scorrano L., Mitofusin 2 tethers endoplasmic reticulum to mitochondria. Nature 456, 605–610 (2008). [DOI] [PubMed] [Google Scholar]
- 30.Merkwirth C., Langer T., Mitofusin 2 builds a bridge between ER and mitochondria. Cell 135, 1165–1167 (2008). [DOI] [PubMed] [Google Scholar]
- 31.de Brito O. M., Scorrano L., Mitofusin-2 regulates mitochondrial and endoplasmic reticulum morphology and tethering: The role of Ras. Mitochondrion 9, 222–226 (2009). [DOI] [PubMed] [Google Scholar]
- 32.Stoica R., de Vos K. J., Paillusson S., Mueller S., Sancho R. M., Lau K. F., Vizcay-Barrena G., Lin W. L., Xu Y. F., Lewis J., Dickson D. W., Petrucelli L., Mitchell J. C., Shaw C. E., Miller C. C. J., ER-mitochondria associations are regulated by the VAPB-PTPIP51 interaction and are disrupted by ALS/FTD-associated TDP-43. Nat. Commun. 5, 3996 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bonewald L. F., The amazing osteocyte. J. Bone Miner. Res. 26, 229–238 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Prideaux M., Findlay D. M., Atkins G. J., Osteocytes: The master cells in bone remodelling. Curr. Opin. Pharmacol. 28, 24–30 (2016). [DOI] [PubMed] [Google Scholar]
- 35.Plotkin L. I., Bellido T., Osteocytic signalling pathways as therapeutic targets for bone fragility. Nat. Rev. Endocrinol. 12, 593–605 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Klein-Nulend J., Bakker A. D., Bacabac R. G., Vatsa A., Weinbaum S., Mechanosensation and transduction in osteocytes. Bone 54, 182–190 (2013). [DOI] [PubMed] [Google Scholar]
- 37.Burra S., Nicolella D. P., Francis W. L., Freitas C. J., Mueschke N. J., Poole K., Jiang J. X., Dendritic processes of osteocytes are mechanotransducers that induce the opening of hemichannels. Proc. Natl. Acad. Sci. U.S.A. 107, 13648–13653 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bonewald L. F., Osteocytes as dynamic multifunctional cells. Ann. N. Y. Acad. Sci. 1116, 281–290 (2007). [DOI] [PubMed] [Google Scholar]
- 39.Dallas S. L., Prideaux M., Bonewald L. F., The osteocyte: An endocrine cell ... and more. Endocr. Rev. 34, 658–690 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Klein-Nulend J., van der Plas A., Semeins C. M., Ajubi N. E., Frangos J. A., Nijweide P. J., Burger E. H., Sensitivity of osteocytes to biomechanical stress in vitro. FASEB J. 9, 441–445 (1995). [DOI] [PubMed] [Google Scholar]
- 41.Aarden E. M., Burger E. H., Nijweide P. J., Function of osteocytes in bone. J. Cell. Biochem. 55, 287–299 (1994). [DOI] [PubMed] [Google Scholar]
- 42.Frikha-Benayed D., Basta-Pljakic J., Majeska R. J., Schaffler M. B., Regional differences in oxidative metabolism and mitochondrial activity among cortical bone osteocytes. Bone 90, 15–22 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Morinobu M., Ishijima M., Rittling S. R., Tsuji K., Yamamoto H., Nifuji A., Denhardt D. T., Noda M., Osteopontin expression in osteoblasts and osteocytes during bone formation under mechanical stress in the calvarial suture in vivo. J. Bone Miner. Res. 18, 1706–1715 (2003). [DOI] [PubMed] [Google Scholar]
- 44.You L., Temiyasathit S., Lee P., Kim C. H., Tummala P., Yao W., Kingery W., Malone A. M., Kwon R. Y., Jacobs C. R., Osteocytes as mechanosensors in the inhibition of bone resorption due to mechanical loading. Bone 42, 172–179 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brighton C. T., Krebs A. G., Oxygen tension of healing fractures in the rabbit. J. Bone Joint Surg. Am. 54, 323–332 (1972). [PubMed] [Google Scholar]
- 46.Kofoed H., Sjontoft E., Siemssen S. O., Olesen H. P., Bone marrow circulation after osteotomy. Blood flow, pO2, pCO2, and pressure studied in dogs. Acta Orthop. Scand. 56, 400–403 (1985). [DOI] [PubMed] [Google Scholar]
- 47.Kennedy O. D., Herman B. C., Laudier D. M., Majeska R. J., Sun H. B., Schaffler M. B., Activation of resorption in fatigue-loaded bone involves both apoptosis and active pro-osteoclastogenic signaling by distinct osteocyte populations. Bone 50, 1115–1122 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dominguez L. J., Di Bella G., Belvedere M., Barbagallo M., Physiology of the aging bone and mechanisms of action of bisphosphonates. Biogerontology 12, 397–408 (2011). [DOI] [PubMed] [Google Scholar]
- 49.Nussey D. H., Froy H., Lemaitre J. F., Gaillard J. M., Austad S. N., Senescence in natural populations of animals: Widespread evidence and its implications for bio-gerontology. Ageing Res. Rev. 12, 214–225 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stern A. R., Stern M. M., Van Dyke M. E., Jähn K., Prideaux M., Bonewald L. F., Isolation and culture of primary osteocytes from the long bones of skeletally mature and aged mice. Biotechniques 52, 361–373 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/5/11/eaaw7215/DC1
Fig. S1. No mitochondrial transfer from conditioned medium to MLO-Y4 cells.
Fig. S2. Osteocytes do not pass through the pores of the transwell membrane.
Fig. S3. Depleted mtDNA and impeded ATP production in MLO-Y4 ρ° cells.
Fig. S4. Parental MLO-Y4 cells and primary osteocytes can transfer mitochondria to MLO-Y4 ρ° cells.
Fig. S5. Mitochondrial distribution and ER movement in osteocyte dendrites.
Fig. S6. Knockdown of Mfn2 has no effect on ATP production and the level of mtDNA.
Fig. S7. Mfn2 knockdown impedes mitochondrial transfer in MLO-Y4 cells.
Fig. S8. VAPB knockdown impedes mitochondrial transfer in MLO-Y4 cells.
Fig. S9. Hypothetical model of ER-mediated mitochondrial transfer.
Table S1. Primer sequences for qPCR.
Movie S1. pTaqYFP-mito–labeled mitochondria (green) in the dendrite dynamically migrate toward the PKH26-stained cells (magenta).
Movie S2. pTaqYFP-mito–labeled mitochondria (green) dynamically move from the cytoplasm to the dendritic process and are associated with RFP-labeled ER (magenta).
Movie S3. Transfer of MTR-labeled mitochondria (magenta) within dendrites is dynamically colocalized with GFP-labeled ER (green).
Movie S4. Transfer of MTR-labeled mitochondria (magenta) is dynamically colocalized with YFP-labeled Mfn2 (green) within dendrites.


