Antibiotic resistance is a growing clinical threat. One of the most serious areas of concern is the ability of some bacteria to degrade carbapenems, drugs that are often reserved as last-resort antibiotics. Resistance to carbapenems can be conferred by a large group of related enzymes called metallo-β-lactamases that rely on zinc ions for function and for overall stability. Here, we studied an extensive panel of 45 different metallo-β-lactamases from a subfamily called VIM to discover what changes are emerging as resistance evolves in clinical settings. Enhanced resistance to some antibiotics was observed. We also found that at least one VIM variant developed a new ability to remain more stable under conditions where zinc availability is limited, and we determined the origin of this stability in atomic detail. These results suggest that zinc scarcity helps drive the evolution of this resistance determinant.
KEYWORDS: VIM, VIM-20, carbapenem resistance, metallo-β-lactamase, protein stability, salt bridge, zinc
ABSTRACT
To understand the evolution of Verona integron-encoded metallo-β-lactamase (VIM) genes (blaVIM) and their clinical impact, microbiological, biochemical, and structural studies were conducted. Forty-five clinically derived VIM variants engineered in a uniform background and expressed in Escherichia coli afforded increased resistance toward all tested antibiotics; the variants belonging to the VIM-1-like and VIM-4-like families exhibited higher MICs toward five out of six antibiotics than did variants belonging to the widely distributed and clinically important VIM-2-like family. Generally, maximal MIC increases were observed when cephalothin and imipenem were tested. Additionally, MIC determinations under conditions with low zinc availability suggested that some VIM variants are also evolving to overcome zinc deprivation. The most profound increase in resistance was observed in VIM-2-like variants (e.g., VIM-20 H229R) at low zinc availability. Biochemical analyses reveal that VIM-2 and VIM-20 exhibited similar metal binding properties and steady-state kinetic parameters under the conditions tested. Crystal structures of VIM-20 in the reduced and oxidized forms at 1.25 Å and 1.37 Å resolution, respectively, show that Arg229 forms an additional salt bridge with Glu171. Differential scanning fluorimetry of purified proteins and immunoblots of periplasmic extracts revealed that this difference increases thermostability and resistance to proteolytic degradation when zinc availability is low. Therefore, zinc scarcity appears to be a selective pressure driving the evolution of multiple metallo-β-lactamase families, although compensating mutations use different mechanisms to enhance resistance.
INTRODUCTION
By inactivation of bacterial transpeptidases, β-lactam antibiotics (penicillins, cephalosporins, and carbapenems) have been long used to treat bacterial infections (1). The emergence of new and catalytically versatile β-lactamases, which are the most common mechanism of resistance to β-lactams (2), is an increasingly important public health threat. An estimated 2 million people are infected with antibiotic-resistant bacteria in the United States, causing >23,000 deaths each year (3). The five β-lactamases currently of primary public health concern are Klebsiella pneumoniae carbapenemase (KPC), New Delhi metallo-β-lactamase (NDM), Verona integron-encoded metallo-β-lactamase (VIM), imipenemase (IMP), and oxacillinase-48-like carbapenemase (OXA-48) (4). These periplasmic enzymes appear in bacterial strains that are linked to increased mortality rates (4). NDM, VIM, and IMP belong to the B1 subclass of β-lactamases and are called metallo-β-lactamases (MBLs) because these enzymes require Zn(II) for catalytic activity (5). To date, inhibitors for the MBLs have not been commercially developed (6).
The IMP-type MBLs were first found in Japan in the late 1980s (7), while the VIM-type MBLs were first discovered in Europe in the latter half of the 1990s (8). NDM-1 was first detected in a strain of Klebsiella pneumoniae isolated in 2008 from a patient returning to Sweden after elective surgery in India (9). The bla genes encoding NDM, VIM, and IMP continue to evolve, with the recent report of 27 NDM variants, 62 VIM variants, and 80 IMP variants currently listed in the β-lactamase database (10). Some of the variants have been biochemically and microbiologically characterized, and even a few of the variants have been structurally characterized (11). While the heavy use of β-lactams is obviously a leading reason for the increasing number of MBLs and other β-lactamases, it is not always clear what selective pressures are driving the evolution of these enzymes. Recently, our labs reported a comprehensive study on NDM variants, and it appears that NDM variants are evolving to overcome the selective pressure of zinc deprivation imposed by host defenses (12, 13). This result was also confirmed by another group (14).
Recently, Galán and coworkers analyzed the phylogenetic relationship within the VIM family (from VIM-1 to VIM-54) (15). BEAST phylogenetic analysis suggested that VIM-2 and VIM-4 arose separately between 1987 and 1989 (15). Then, VIM-1 (R205S) diversified from VIM-4 around 1994 (15). Among the VIM variants, VIM-2 is the most commonly found VIM variant in the clinic setting, and bacteria harboring VIM-2 have been found in many countries (16). VIM-2 has been extensively characterized using microbiological, biochemical, crystallographic, and spectroscopic studies by several groups, including ours (16–18). In contrast, there are relatively few studies on any of the other VIM variants (11).
In the present study, antibiotic susceptibility studies, under standard conditions and under conditions with low zinc availability, were used to evaluate the significance of amino acid differences in 45 clinically derived VIM variants. Several variants were shown to exhibit increased MIC values specifically when zinc availability is low, and one variant in the VIM-2-like family, VIM-20, was further characterized using biochemical and structural studies. Metal analyses and steady-state kinetics were used to compare properties of VIM-2 and VIM-20, and crystallographic studies were conducted to determine whether structural features could help explain the different MIC values. Differential scanning fluorimetry and immunoblotting studies were used to compare the thermal and proteolytic stabilities of VIM-2 and VIM-20 at different zinc concentrations. These studies reveal that variants of VIM MBLs use several strategies to overcome selective pressures and that VIM-20 in particular employs a single amino acid difference to form a strategically located salt bridge that increases protein stability that results in increased antibiotic resistance during zinc deprivation.
RESULTS
MIC studies under standard conditions.
Based on previous phylogenetic analyses (15), we divided the VIM variants into 3 distinct subgroups, (i) VIM-2-like, (ii) VIM-4-like, and (iii) VIM-1-like. Presently, 60 known variants of VIM are known, and we chose to examine only those variants that had 1 or 2 amino acid differences from VIM-2 (25 variants), VIM-4 (9 variants), or VIM-1 (12 variants) (Fig. 1). Escherichia coli DH10B strains harboring genes encoding VIM-2-like, VIM-4-like, and VIM-1-like variants were used to assess the impact of substitutions on phenotype in antibiotic susceptibility studies. Initially, we determined the MIC values of these variants under standard conditions (19) against a panel of three structural classes of β-lactams, including a penicillin (ampicillin), two cephalosporins (ceftazidime and cephalothin), and three carbapenems (imipenem, meropenem, and ertapenem) (Table 1; see also Table S1 in the supplemental material). Not surprisingly and confirming a previous report (15), all strains containing VIM-like variants exhibited increased resistance to all tested antibiotics compared to strains harboring the empty vector (Tables 1 and S1). A comparison of our MIC data to those of a previous study demonstrates similar MIC values for ampicillin and ceftazidime but larger differences when carbapenems were used in the antibiotic susceptibility studies (Table S2) (15). The observed differences may be due to the different E. coli cell lines and VIM expression plasmids used in the two studies.
FIG 1.
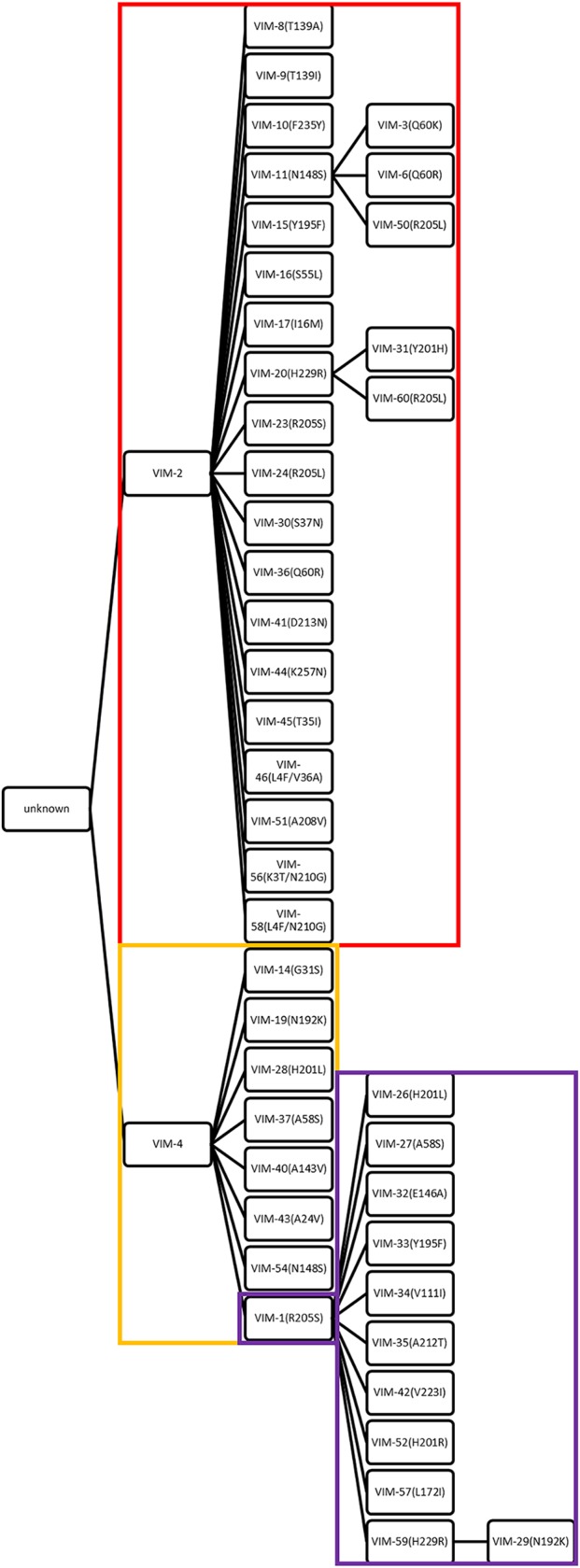
Mutated residues in clinically derived VIM gene family. In total, 25 members of VIM-2-like variants (boxed in red), 9 members of VIM-4-like variants (boxed in orange), and 12 members of VIM-1-like variants (boxed in purple) were selected, from VIM-1 to VIM-60. The amino acid changes acquired with respect to the sequence of the ancestor are indicated in the branches. VIM-1 is the node and was analyzed in both VIM-4-like variants and VIM-1-like variants.
TABLE 1.
Summary of MIC values for select VIM variantsa
| Drug(s)b | MIC (μg/ml) for E. coli DH10B/pBC variant (mutation): |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| SK(-) | SK(-) blaVIM-2 | SK(-) blaVIM-20 (H229R) | SK(-) blaVIM-4 | SK(-) blaVIM-28 (H201L) | SK(-) blaVIM-37 (A58S) | SK(-) blaVIM-1 | SK(-) blaVIM-59 (H229R) | All others (range) | |
| AMP | 4 | 1,024 | 1,024 | 8,192 | 8,192 | 8,192 | 8,192 | 8,192 | 32–8,192 |
| AMP + EDTA | 4 | 16 | 256 | 64 | 1,024 | 512 | 128 | 512 | 4–512 |
| CAZ | 0.5 | 32 | 32 | 64 | 128 | 32 | 1,024 | 1,024 | 2–1,024 |
| CAZ + EDTA | 0.5 | 1 | 8 | 1 | 16 | 2 | 32 | 128 | 0.25–128 |
| CF | 4 | 256 | 512 | 1,024 | 1,024 | 1,024 | 2,048 | 2,048 | 32–2,048 |
| CF + EDTA | 4 | 16 | 128 | 32 | 256 | 128 | 64 | 256 | 4–256 |
| IMI | 0.25 | 4 | 8 | 8 | 16 | 16 | 8 | 8 | 0.5–16 |
| IMI + EDTA | 0.25 | 0.25 | 0.5 | 0.25 | 1 | 1 | 0.5 | 1 | 0.25–1 |
| MER | 0.03 | 0.5 | 0.5 | 1 | 2 | 4 | 1 | 1 | 0.125–4 |
| MER + EDTA | 0.015 | 0.03 | 0.125 | 0.03 | 0.25 | 0.125 | 0.06 | 0.25 | 0.03–0.25 |
| ERT | 0.015 | 0.25 | 0.5 | 1 | 4 | 4 | 1 | 1 | 0.015–8 |
| ERT + EDTA | 0.0075 | 0.015 | 0.06 | 0.015 | 0.5 | 0.25 | 0.015 | 0.125 | 0.015–0.25 |
See Tables S1 to S3 for more details.
AMP, ampicillin; CAZ, ceftazidime; CF, cephalothin; IMI, imipenem; MER, meropenem; ERT, ertapenem.
MIC values (μg/ml) for VIM-2-like (A strain), VIM-4 (B strain), and VIM-1-like (C strain) variants under standard conditions. Download Table S1, DOCX file, 0.1 MB (21KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Comparison of MIC values (μg/ml) between our work and Martinez-Garcia et al. (15). Download Table S2, DOCX file, 0.1 MB (17.6KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
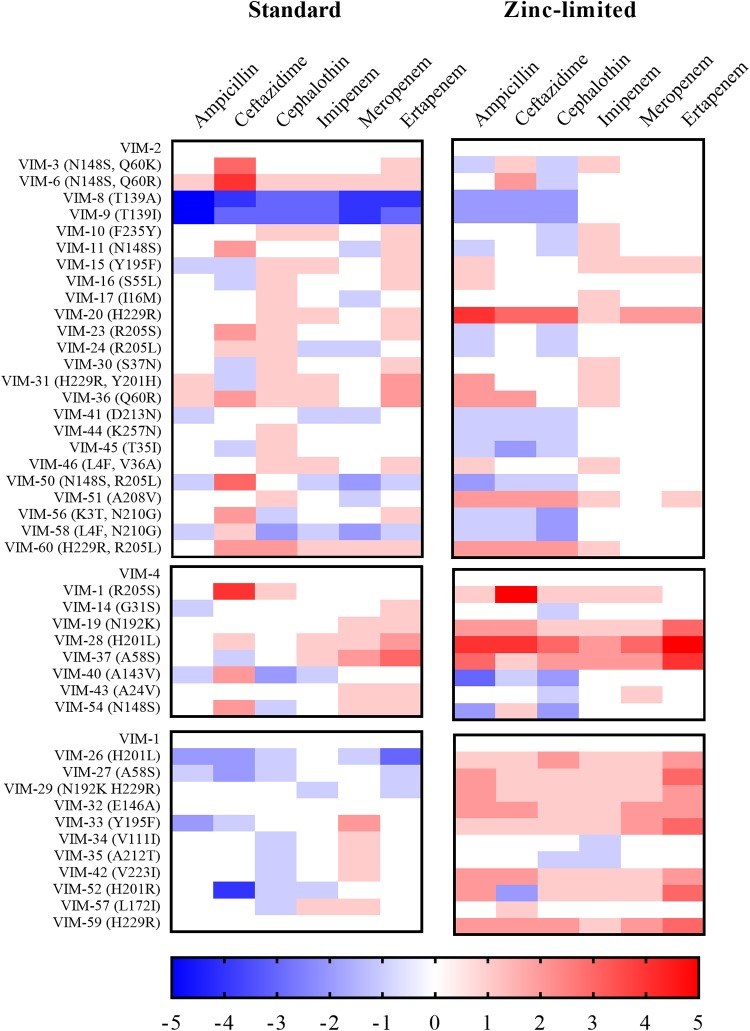
To facilitate visualization of the MIC data, a heat map was generated (Fig. 2). In this map, the fold changes in MIC values for the strains containing VIM-2-like, VIM-4-like, and VIM-1-like variants were determined by normalizing to the MIC values of strains containing the parent VIM-2, VIM-4, and VIM-1 genes, respectively. What is striking about this representation is that there are generally small differences in MIC values of the strains containing the variants compared to strains containing the parent enzymes, with the exception of strains expressing VIM-8 and VIM-9, which exhibited significantly reduced resistance toward four of the antibiotics compared to the strain with the parent VIM-2 MBL (Fig. 2, left). Therefore, VIM-8 and VIM-9 were excluded from subsequent analyses.
FIG 2.
Heat maps of fold changes in MIC values against a panel of antibiotics representing three structural classes of β-lactams. Heat map intensities for each family represent the fold change in MIC (see Tables S1 and S3) compared to that of the parent family member (VIM-2, VIM-4, or VIM-1) under standard or zinc-limited conditions.
MIC values (μg/ml) for VIM-2-like (A strain), VIM-4 (B strain), and VIM-1-like (C strain) variants under zinc-limited conditions (50 μM EDTA). Download Table S3, DOCX file, 0.1 MB (20.8KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
MIC studies under Zn(II)-limiting conditions.
Previously, we found the M154L substitution in NDM variants increases antibiotic resistance when zinc availability is low (12, 13). Therefore, we tested for a similar effect by determining MIC values for strains expressing VIM-2-like, VIM-4-like, and VIM-1-like variants in the presence of EDTA against the same panel of antibiotics (Table S3).
Generally, the MIC values of strains harboring the VIM variants determined in the presence of EDTA were substantially lower than those determined under standard conditions. A heat map of the factor change in MIC values was generated from raw data as described above for the MIC values determined under standard conditions (Fig. 2, right). Interestingly, strains with several of the VIM-1-like variants exhibited increased resistance toward ertapenem, compared to the strain containing the VIM-1 parent (Fig. 2, right). The zinc-limited conditions yielded the greatest enhancements in strains with VIM-1- and VIM-4-like variants. Unlike the heat map generated with data collected under standard conditions, the heat map from data collected at zinc-limited conditions (Fig. 2, right) shows consistent increases in resistance in strains with most VIM variants when imipenem was used as the antibiotic. Significantly, strains with VIM-20 and VIM-28 exhibited larger MIC values when data were collected under zinc-limited conditions than when collected under standard conditions, compared to their corresponding parent family member. This result strongly suggests that strains containing these VIM variants are better able to overcome zinc scarcity (Fig. 2).
Overexpression, purification, and characterization of VIM variants.
Antibiotic susceptibility studies revealed that a number of variants that deserve further study due to their increased resistance under conditions where zinc availability was low, as follows: VIM-20 in the VIM-2-like variants, VIM-28 and VIM-37 in the VIM-4-like variants, and VIM-59 in the VIM-1-like variants. Small-scale overexpression cultures revealed that the VIM-2-like variants are produced at much higher levels than are the VIM-4- or VIM-1-like variants and that the VIM-4 and VIM-1 variants could not easily be obtained at concentrations necessary for spectroscopic or crystallographic studies. Therefore, we focused on the VIM-2-like variants, and in particular on VIM-20, which contains the H229R substitution and exhibits the largest and broadest spectrum of increased MIC values of any VIM-2 family variants, compared to VIM-2, under low zinc conditions. It is important to note that the H229R substitution is also found in VIM-31, VIM-60, VIM-29, and VIM-59.
VIM-2 and VIM-20 were overexpressed and purified. The yields for tag-free VIM-2 and VIM-20 were 47 and 39 mg of protein/liter of LB medium, respectively. The predicted molecular weights for tag-free VIM-2 and VIM-20 are 25,840.80 Da and 25,859.85 Da, respectively, as determined by the ExPASy-Compute pI/MW tool (20). The purities of the proteins were shown to be >95% by SDS-PAGE. Inductively coupled plasma-atomic emission spectroscopy (ICP-AES) revealed that VIM-2 and VIM-20 bind 2.6 ± 0.1 and 2.8 ± 0.1 molar equivalents of Zn(II), respectively. Metal analyses indicate that VIM-2 and VIM-20 have a third Zn(II) binding site in addition to the two Zn(II) ions at the active site (21). To confirm the metal content, electrospray mass spectrometry (ESI-MS) was used to probe metal binding. After buffer exchange into 100 mM ammonium acetate (pH 7.5), native MS showed that VIM-2 and VIM-20 have molecular weights of 26,032.6 and 26,048.2 Da, respectively, corresponding to their tag-free MW plus three Zn(II) ions (Fig. S1).
Native mass spectrometry data of VIM-2 (left) and VIM-20 (right). Both protein samples were in 50 μM ammonium acetate (pH 7.5). The 10+ and 9+ charged peaks were observed from both samples in positive mode. Download FIG S1, PDF file, 0.1 MB (57.8KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Steady-state kinetics.
Steady-state kinetic studies were conducted on VIM-2 and VIM-20 using a penam (ampicillin), two cephems (chromacef and ceftazidime), and a carbapenem (meropenem). A comparison of steady-state kinetic constants of VIM-2 and VIM-20 with 10 μM added Zn(II) revealed <2-fold differences in kcat, KM, and kcat/KM values for all substrates tested (Table 2); a similar result was observed in studies conducted at 0.1 nM added Zn(II). Interestingly, there are only small (<4-fold) differences in steady-state kinetic constants between VIM-2 and VIM-20 at 0.1 nM and 10 μM of added Zn(II). Among clinical NDM variants, we previously observed significant differences in kinetic parameters that strongly depended upon zinc concentration in this range and found that NDM variants that conferred more resistance also bound Zn2 more tightly (12, 13), Notably, VIM appears to have used a different mechanism to improve resistance when zinc availability is low. The steady-state kinetic parameters of both VIM-2 and VIM-20 do not change significantly at low zinc concentrations [0.1 nM Zn(II)], indicating that the dinuclear zinc site likely stays intact under these conditions, and that the improvement of MIC values in VIM-20 is not due to a direct improvement of catalytic parameters at low zinc concentrations.
TABLE 2.
Steady-state kinetic parameters for VIM-2 and VIM-20
| Substrate by Zn(II) concn | Parameter by metallo-β-lactamase |
|||||
|---|---|---|---|---|---|---|
| VIM-2 |
VIM-20 |
|||||
| KM (μM) | kcat (s−1) | kcat/KM (s−1 μM−1) | KM (μM) | kcat (s−1) | kcat/KM (s−1 μM−1) | |
| 10 μM | ||||||
| Meropenem | 5.0 ± 0.8 | 2.0 ± 0.1 | 0.4 | 9.9 ± 0.5 | 2.7 ± 0.1 | 0.27 |
| Ceftazidime | 90 ± 11 | 1.0 ± 0.1 | 0.01 | 105 ± 8 | 1.0 ± 0.1 | 0.01 |
| Ampicillin | 84 ± 7 | 125 ± 3 | 1.5 | 120 ± 9 | 176 ± 5 | 1.5 |
| Chromacef | 11 ± 2 | 27 ± 2 | 2.5 | 14 ± 1 | 49 ± 2 | 3.5 |
| 0.1 nM | ||||||
| Meropenem | 8 ± 1 | 1.6 ± 0.1 | 0.2 | 7.0 ± 0.5 | 2.0 ± 0.1 | 0.28 |
| Ceftazidime | 100 ± 10 | 0.76 ± 0.04 | 0.008 | 130 ± 10 | 0.68 ± 0.03 | 0.005 |
| Ampicillin | 82 ± 7 | 37.7 ± 0.9 | 0.46 | 49 ± 4 | 45.4 ± 0.8 | 0.93 |
| Chromacef | NDa | ND | ND | ND | ND | ND |
ND, not determined.
Metal coordination of Co(II)-substituted VIM-2 and VIM-20.
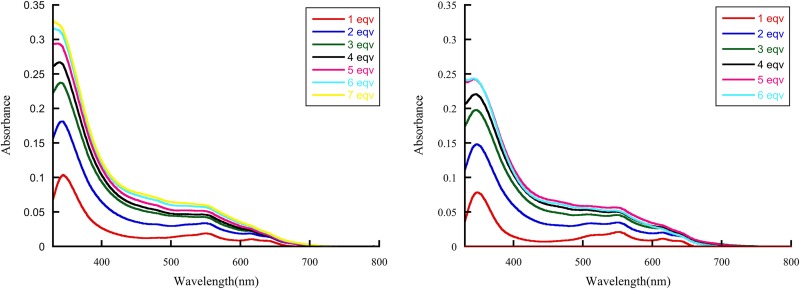
Conversion of purified VIM proteins into dicobalt (II) metalloforms provides a spectroscopic window to probe the structure at the active site. The UV-visible spectra of VIM-2 and VIM-20 revealed broad absorption peaks at 342 nm that are readily assigned to a Cys-S-to-Co(II) ligand-to metal charge-transfer (LMCT) transitions, requiring Co(II) binding at the Zn2 site (Fig. 3) (16). The ligand field (d-d) transitions between 500 and 650 nm are similar in shape to those of other B1 MBLs (22). The LMCT and d-d transitions increase proportionally as CoCl2 is added, demonstrating that Co(II) binds to the Zn1 and Zn2 sites in both variants. A similar result was reported previously for VIM-2 (16). Even though there are only 3 Zn(II) binding sites in VIM-2 or VIM-20, it took >5 equivalents (equiv.) of Co(II) to yield spectra that no longer changed upon the addition of more Co(II); for VIM-2 with 6 equiv. of Co(II), the ε342 value was 1,100 M−1 cm−1 and the ε550 value was 200 M−1 cm−1; and for VIM-20 with 5 equiv. of Co(II), the ε342 value was 833 M−1 cm−1 and the ε550 value was 200 M−1 cm−1. The ε550 values suggest a mixture of 5/6 coordinate Co(II) sites, and the difference in ε342 values suggests changes in orientation of Cys198 in VIM-2 (23). These spectra show little difference in metal binding to VIM-2 compared to VIM-20.
FIG 3.
Difference UV-Vis spectra of apo-VIM-2 (left) and apo-VIM-20 (right) titrated with CoCl2. eqv, equivalents.
Structural characterization of VIM-20.
Our biochemical studies demonstrated that the increased resistance afforded by VIM-20 in our antibiotic susceptibility studies under zinc-limited conditions is not obviously caused by different metal binding properties of VIM-20 compared to VIM-2. Therefore, we conducted structural studies in an effort to reveal differences in the two VIM variants. Our structures of VIM-20 in the reduced and oxidized states overlay with 0.274 Å root mean square deviation (RMSD) over all protein heavy atoms (Fig. 4). The structures of VIM-20 in the reduced and oxidized states overlay with VIM-2 (PDB identifier [ID] 4NQ2) with 0.217 Å and 0.228 Å RMSD for all heavy atoms. Differences between the two VIM-20 structures are primarily restricted to the active site resulting from the presence of Cys198 as the reduced sulfhydryl (PDB ID 6OP6) or the oxidized sulfonic acid (PDB ID 6OP7). In the reduced form, Cys198, Asp118, and His240 coordinate Zn2, while His114, His116, and His179 coordinate Zn1. In comparison, oxidation of Cys198 to the sulfonic acid prevents metalation at the Zn2 site. Apart from the absence of Zn2, small shifts to the position of Zn1 and residues His114, His116, Asp118, and His179 are observed, relative to the reduced structure, that serve to shift the position of Zn1 by 0.5 Å and expand the distance between active-site atoms His116ND1 and His240NE2 by 1.5 Å. Outside the active site, the structures are highly similar. A third Zn observed in the reduced structure is coordinated to the side chains of His251 and Glu266 and makes contact to His153 of another VIM-20 monomer in the unit cell. Similarly, the third Zn is observed in the oxidized VIM-20 structure; however, this zinc ion is only coordinated with the side chains of His251 and the His153 of another VIM-20 monomer in the unit cell. Since electron density for residues 262 to 266 was not available for the oxidized structure, we did not observe an interaction with Glu266 and the third Zn for oxidized VIM-20. However, because this zinc ion forms crystal contacts between different copies of VIM-20 within the unit cell, it is likely that binding to this fortuitous site only occurs when the crystal is formed and not in monomeric VIM-20 where only two coordinating residues would be available.
FIG 4.
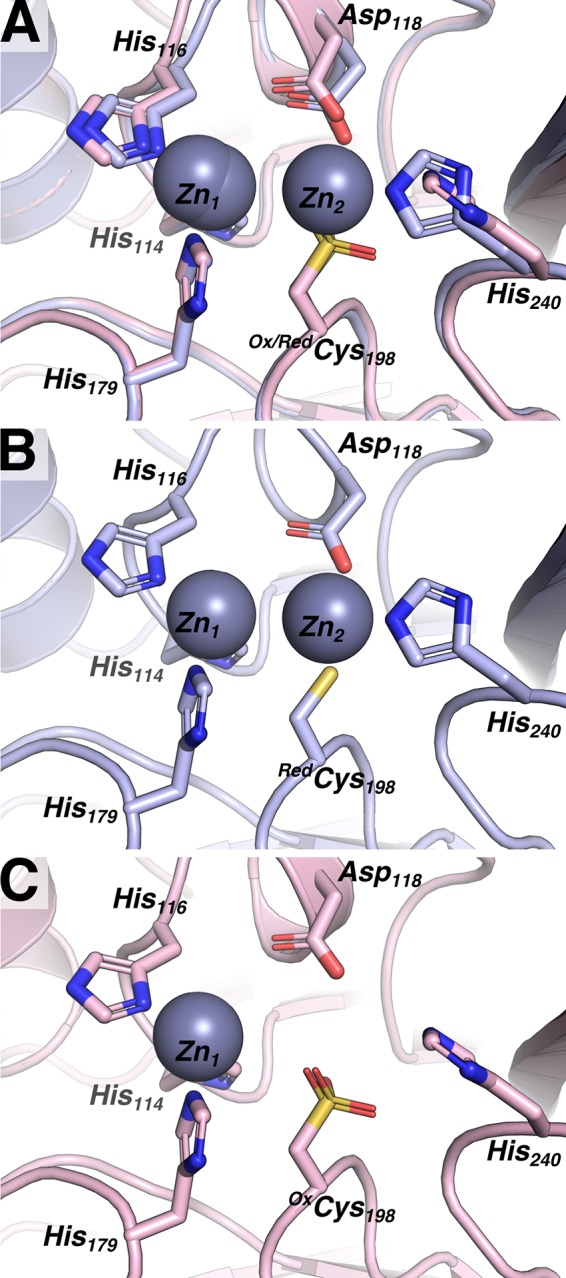
Reduced VIM20 and oxidized VIM20 feature overall similar structures, with differences localized to the active site. (A) An overlay of oxidized VIM-20 (OxVIM-20, pink) and reduced VIM-20 (RedVIM-20, light blue) highlights key differences. (B) The structure of RedVIM-20 features two zinc ions and a reduced Cys198 sulfhydryl. (C) The structure of OxVIM-20 features a single zinc ion and Cys198 oxidized to a sulfonic acid.
Beyond the oxidation/reduction state of Cys198, the most intriguing feature we identified was a structural role of Arg229. Relative to VIM-2 (PDB ID 4NQ2), the H229R mutation present in both the reduced and oxidized VIM-20 structures results in the introduction of a salt bridge between the Glu171 side-chain carboxylate and the Arg229 side-chain guanidino group (Fig. 5A). For clarity and due to the high similarity of the reduced and oxidized structures, in the subsequent discussion and in Fig. 5, we only provide information for the structure of the physiologically relevant reduced form of VIM-20. The distances between the VIM-20 Glu171 carboxylate oxygens to the guanidine NH2 nitrogen and the Nε of the Arg229 guanidino group are 3.0 Å and 3.2 Å, respectively. The distances and geometries between the Glu171 and ArgR229 side-chain functional groups are suggestive of a salt bridge (Fig. 5A). In comparison, the position of His229 in VIM-2 places the side-chain Nε 3.3 Å from the nearest carboxylate oxygen of Glu171; however, the geometry is not optimal for hydrogen bonding or the formation of a salt bridge. Thus, while a weak electrostatic interaction may be possible between VIM-2 His229 and Glu171, clear potential exists for stronger electrostatic interactions to be formed between Arg229 and Glu171 of VIM-20.
FIG 5.
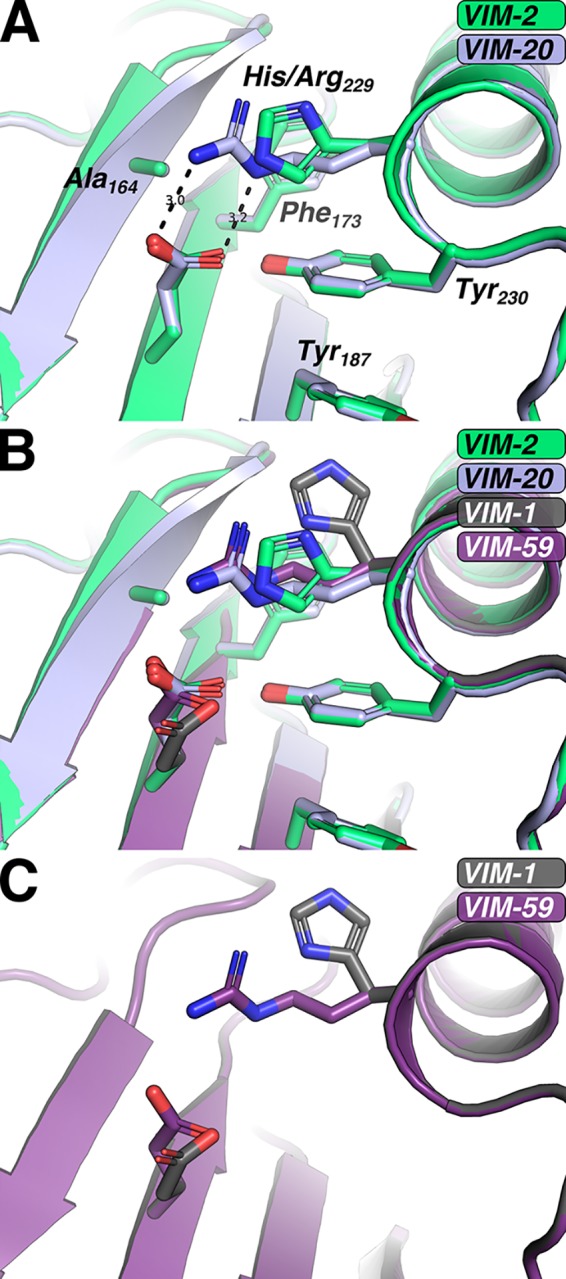
Role of VIM-20 H229R mutation and accommodation in VIM-59. (A) Compared to VIM-2 (green), VIM-20 (blue) features a salt bridge between residues 229 and 171. (B) An overlay of VIM-2 (green, 4NQ2), VIM-20 (blue), VIM-59 (purple), and VIM-1 (gray) indicate similarities between VIM-20 and VIM-59. (C) Mutation in silico indicates that the H229R mutation is accommodated in the VIM-1 background.
Building on our VIM-20 structure, we sought to examine whether the H229R substitution could enable the formation of salt bridges or additional hydrogen bonds within VIM-31 and VIM-59, two additional VIM variants harboring the H229R mutation. VIM-31, like VIM-20, is a VIM-2-like variant with one additional mutation, Y201H. The tyrosine-to-histidine substitution is located 18 Å, a Cα-Cα distance, from the H229R change, and in silico mutation of VIM-20 Tyr201 to histidine does not suggest any perturbation to the position of Arg229. In comparison, VIM-59 is a VIM-1-like variant; thus, modeling of the H229R substitution required using VIM-1 as the starting point (PDB ID 5N5G) (24). After introducing the H229R change in silico in the VIM-1 background, we found that H229R was easily accommodated (Fig. 5B and C). While our introduction of H229R incorporated only favored side-chain rotamers for all altered residues, it did require small adjustments to the side-chain rotamers of Phe173 and Tyr230. Compared to VIM-20, our model of VIM-59 allows for a single hydrogen bond to form between Arg229 and Glu171 (Fig. 5B and C). However, it is possible that with minor adjustment of the VIM-59 backbone that Arg229 and Glu171 could undergo slight (<0.2 Å) shifts that would enable formation of the salt bridge observed in VIM-20 between Arg229 and Glu171 while still maintaining allowed rotamers for each side chain. It is important to note that the MIC data showed increased resistance in strains with VIM-59 under zinc-limited conditions, but not to the extent observed with VIM-20 or VIM-28 (Fig. 2).
Thermal stability of VIM-20.
Previously, we utilized differential scanning fluorimetry (DSF) to evaluate the thermal stability of NDM variants, and our data showed that blaNDM was evolving to encode a more thermally stable protein (13). DSF was used to evaluate the relative temperature stabilities of VIM-2 and VIM-20 as a function of Zn(II) (13). A sigmoidal fit to the melting temperature data identified an apparent affinity for zinc of 17.5 ± 0.3 μM for VIM-2 and 19.8 ± 0.3 μM for VIM-20 (Fig. 6). These similar binding affinities are consistent with the metal analyses and UV-Vis studies presented above. VIM-2 and VIM-20 exhibited melting temperatures (Tm) of 56.8 ± 0.4°C and 60.9 ± 0.4°C, respectively, at high Zn(II) concentrations (Fig. 6). At lower Zn(II) concentrations, the Tm of VIM-2 is 36.6 ± 0.3°C, and the Tm of VIM-20 is 44.3 ± 0.2°C. In addition to characterizing the fraction of unfolded protein, we capitalized on our prior published approach (25) to determine the Gibbs free energy of unfolding (ΔuG) for VIM-2 and VIM-20. This analysis indicates that at 310 K, VIM-20 is more stable than VIM-2, with approximate changes in the Gibbs free energy of unfolding (ΔΔuG) of 2.3 kJ mol−1 and 1.8 kJ mol−1 under high Zn(II) and low Zn(II) concentrations, respectively.
FIG 6.
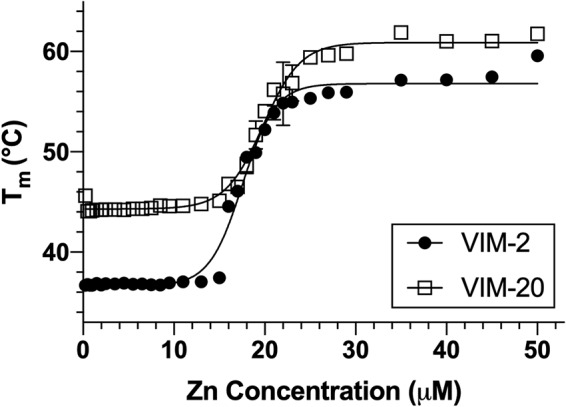
Differential scanning fluorimetry of VIM-2 and VIM-20 variants with various Zn(II) concentrations. Melting temperatures (Tm) for VIM-2 (black) and VIM-20 (red) were determined by differential scanning fluorimetry at a range of ZnCl2 concentrations from 0 to 50 μM. Error bars are shown for all data points; some error bars are smaller than the symbol used.
This result suggests that the H229R substitution in VIM-20 not only stabilizes the protein under high Zn(II) conditions but also under low-Zn(II) conditions. Applying Boltzmann sigmoidal fits to the raw DSF data for VIM-2 and VIM-20 in high zinc and low zinc concentrations allows for extraction of the folded protein fraction (Pf) and unfolded protein fraction (Pu) at each temperature along the DSF unfolding curves (25). Using this process (Fig. S2), we determined that at both high and low Zn(II) concentrations, VIM-2 and VIM-20 each exhibited fractions of unfolded protein of less than 0.1% at 25°C, the temperature used to determine steady-state kinetic parameters (Table 1). In contrast, at 37°C, VIM-2 exhibits fractions of unfolded protein of 2.6% under high Zn(II) concentrations and 51.9% under low Zn(II) concentrations. In comparison, VIM-20 exhibits an unfolded protein fraction of less than 0.1% at high Zn(II) concentrations and 9.2% under low Zn(II) concentrations. Thus, under high zinc conditions at physiological temperature, we propose that VIM-2 and VIM-20 are both properly folded. However, at physiological temperature under low zinc conditions, VIM-20 is likely to have a high percentage of folded protein, but VIM-2 will be approximately 50% unfolded, raising the possibility of zinc loss, and increased proteolysis as has been noted before for apo VIM-2 (26). This result is different from those reported for NDM-1 and NDM-4 (13). NDM-1 exhibited a lower Tm (57.2 ± 0.3°C) than did NDM-4 (59.5 ± 0.1°C) at high Zn(II) concentrations (13). In contrast, Tm values of NDM-1 (39.1 ± 0.3°C) and NDM-4 (40.6 ± 0.1°C) are similar at low Zn(II) concentrations, indicating that the amino acid difference does not confer overall NDM thermostability.
Fitting of DSF data to the equation ΔuG = −RT ln K to enable extrapolation of ΔuG from the unfolding temperature (Tm) back to 310 K. (A and B) Calculated ΔuG values at temperatures immediately preceding the Tm for VIM-20 (open squares) and VIM-2 (black circles) were plotted for high Zn(II) (A) and low Zn(II) (B) concentrations. (C) Predicted fraction unfolded as a function of temperature at low and higher concentrations of Zn(II). Download FIG S2, TIF file, 0.3 MB (365.3KB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Periplasmic stability of VIM-20.
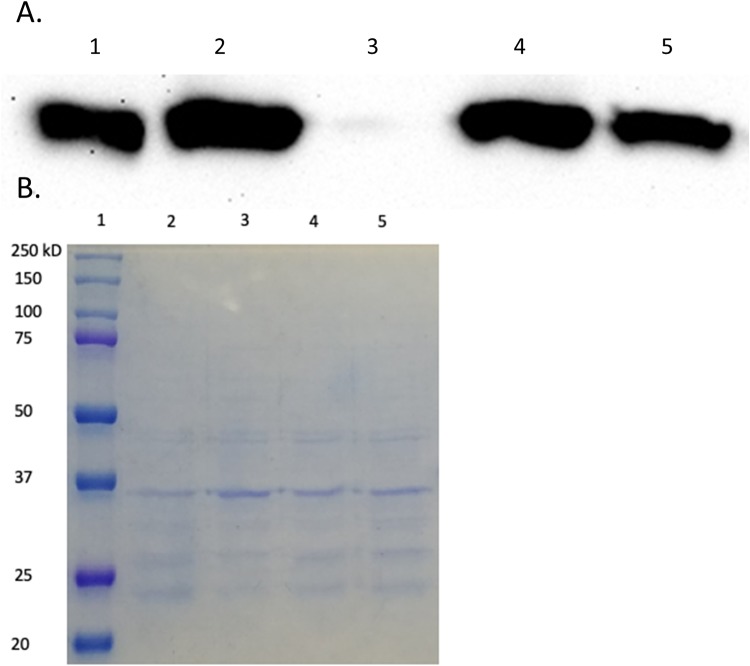
In an effort to probe the in vivo stability of VIM-20 compared to VIM-2, immunoblotting was used to detect periplasmic protein levels of VIM-2 and VIM-20 under normal and low zinc conditions (Fig. 7). The protein levels of VIM-2 and VIM-20 are identical under normal conditions (Fig. 7A, lanes 2 and 4). However, VIM-2 could not be detected in the extracts of cells treated with EDTA and grown to the same optical density, while levels of VIM-20, albeit lower than under standard conditions, are clearly detectable, with all other conditions being equal (Fig. 7A and B). This result is consistent with our DSF results indicating stability of both VIM-20 and VIM-2 at high zinc concentrations and a markedly lower stability for VIM-2 than for VIM-20 at low zinc concentrations. Similarly, previous studies by Vila and coworkers reported that the metal-free analog of VIM-2, which is likely also less structured, is quickly degraded under zinc-limited conditions (27).
FIG 7.
Results of Western blotting. (A) Protein levels of VIM-2 and VIM-20 in periplasmic extracts of E. coli DH10B containing pBCSK-/VIM-2 or pBCSK-/VIM-20 grown with or without added 50 μM EDTA. Anti-VIM-2 polyclonal antibodies were used for detection. Lane 1, 100 ng pure VIM-2; lane 2, VIM-2 (30 μl) periplasmic extract; lane 3, VIM-2 plus 50 μM EDTA (30 μl) periplasmic extract; lane 4, VIM-20 (30 μl) periplasmic extract; lane 5, VIM-20 plus 50 μM EDTA (30 μl) periplasmic extract. (B) Coomassie-stained gel with the same samples. Lane 1, protein ladder; lane 2, VIM-2 (30 μl) periplasmic extract; lane 3, VIM-2 plus 50 μM EDTA (30 μl) periplasmic extract; lane 4, VIM-20 (30 μl) periplasmic extract; lane 5, VIM-20 plus 50 μM EDTA (30 μl) periplasmic extract. The expected molecular weight of VIM-2 and VIM-20 is approximately 26.0 kDa.
DISCUSSION
Recently, Galán and coworkers reported MIC determinations of 20 clinically derived VIM variants, and their major conclusion was that the clinical use (overuse) of ceftazidime is driving the evolution of blaVIM (15). This report identified five VIM-2-like variants (VIM-6, VIM-11, VIM-23, VIM-24, VIM-36, and VIM-50), four VIM-4-like variants (VIM-1, VIM-19, VIM-28, and VIM-54), and three VIM-1 variants (VIM-26, VIM-27, and VIM-33) that exhibited ≥4× increased resistance for ceftazidime (15). In our studies under standard conditions, all of these variants, except VIM-24, VIM-19, and VIM-28, also exhibited ≥4× increased resistance toward ceftazidime. In addition, we examined 25 additional VIM variants and identified nine variants that also exhibit increased resistance toward ceftazidime, including VIM-3, VIM-56, VIM-60, VIM-32, VIM-34, VIM-40, VIM-42, VIM-57, and VIM-59. Our study demonstrates that 46% of the tested clinically derived VIM variants exhibit increased resistance toward ceftazidime. Therefore, our data confirm and support the previous conclusion that at least some blaVIM evolution may be driven by the selective pressure of ceftazidime use. However, our data also show that this effect is not particularly selective for ceftazidime. Our data also predict that VIM-4- and VIM-1-like variants may emerge as more of a potential clinical threat than VIM-2 variants, due to the larger MIC values for all antibiotics tested, although they are currently less common in clinical settings (28, 29).
We and others previously discovered that the gene for the homologous enzyme NDM is evolving in response to selective pressures imparted by the limited availability of zinc at infection sites (12–14).We suspected that zinc scarcity may also be driving the evolution of other MBL genes, so we repeated antibiotic susceptibility studies of VIM variants in the presence of EDTA to mimic conditions at infection sites where zinc availability is expected to be low. Under these conditions, MIC values for all tested VIM variants decreased compared to those obtained under standard conditions, indicating that zinc scarcity in conjunction with antibiotic treatment can provide a selective pressure for VIM-harboring bacteria. Notably, several variants stood out for their ability to better confer resistance when zinc availability was low. For the VIM-2 family, one variant, VIM-20, conferred greater resistance to all six tested antibiotics. Two of the nine VIM-4 family variants, VIM-19 and VIM-28, conferred greater resistance to the entire panel of antibiotics. Finally, seven of 12 VIM-1 family variants exhibited greater resistance to the panel of antibiotics. In whole, 22% of the VIM variants tested conferred greater resistance to the entire panel of six antibiotics compared to the original parent enzymes (VIM-1, VIM-2, and VIM-4) under conditions of low zinc availability. These MIC results indicate that blaVIM may be evolving to enhance resistance in low-zinc environments, similar to our findings for blaNDM (12–14), although we discovered that this effect is achieved through a different mechanism (see below).
In the zinc-limited MIC studies, one of the most significant variants was VIM-20, a VIM-2-like variant that showed a prominent improvement in resistance to multiple antibiotics. Therefore, we overexpressed, purified, and characterized VIM-20 in an effort to understand how the H229R substitution affords the highest resistance of all VIM-2-like variants under conditions of low zinc availability. Steady-state kinetics (at low and high zinc concentrations), metal analyses, and UV-Vis spectroscopic studies on Co(II)-substituted analogs demonstrated very little difference between VIM-2 and VIM-20, suggesting similar metal binding and catalytic properties of these two enzymes under the tested conditions. This finding differs significantly from the earlier studies of NDM variants. NDM has a dinuclear zinc ion cluster at the active site in which Zn2 is bound more weakly than Zn1 (23). Most of the clinical NDM variants showed an increased affinity for Zn2 that resulted in enhanced catalytic activity at low zinc concentrations compared to NDM-1. In contrast, VIM-2 is known to bind both active-site zinc ions tightly in a cooperative manner (16). Here, we find no evidence that steady-state kinetic parameters are perturbed for VIM variants under conditions of zinc scarcity. Under the conditions tested, we also find no evidence that VIM variants exhibit increased activity against β-lactams or have altered Zn binding. Therefore, although each of these MBL genes is evolving to provide enhanced resistance when zinc availability is low, they appear to be using different mechanisms to overcome the same selective pressure.
Previous studies have revealed that some VIM variants have improved thermostability (30), so we also investigated heat-induced unfolding of VIM variants over a wide range of zinc concentrations by using DSF. Somewhat unexpectedly, we observed a zinc-dependent transition to a more stable form for both VIM-2 and VIM-20 at high zinc concentrations. The apparent Kd (dissociation constant) values derived from this transition for each variant were similar (approximately 18 and 20 μM, respectively) and of a magnitude much higher than expected for dissociation of either of the two active-site zinc ions. Since activity is not significantly perturbed at low zinc concentrations and formation of the dizinc site is cooperative, we interpret that the activity at low zinc concentrations to be due to dizinc VIM metalloforms. We therefore assign the zinc-dependent transition in DSF instead to the weaker binding of a third, noncatalytic Zn(II) ion, likely the same Zn(II) observed when we copurify VIM-2 and VIM-20 along with three equivalents of Zn(II) ions, and this was verified by ESI-MS experiments. Although this third Zn(II) ion provides considerable thermostability (inducing a Tm increase of approximately 18°C), this stabilization is not likely to be physiologically relevant due to the high concentration of zinc required. We also note that the binding site for Zn3 studied in our DSF and ESI-MS experiments is not yet defined and is not likely represented by the third zinc site visualized in our X-ray crystal structures of VIM-20, which were determined without excess added zinc and consist of ligands from neighboring symmetry mates that are serendipitously positioned by the crystal lattice and would thus require crystallization to orient.
However, the thermal denaturation at low zinc concentrations does represent conditions more similar to those expected to be encountered in vivo. Surprisingly, at 37°C and low Zn(II) concentrations, approximately 50% of VIM-2 is unfolded, indicating poor stability under conditions expected at the sites of infection. In contrast, the VIM-20 variant exhibits greater thermostability (Tm is increased by approximately 8°C) and could thereby afford a significant survival advantage to bacteria challenged by β-lactams under these conditions by maintaining a larger proportion of folded VIM, all other things being equal. Again, both VIM and NDM variants show enhanced resistance under low zinc concentrations but appear to use different mechanisms to overcome this challenge since all of the characterized NDM variants have similar Tm values at low Zn(II) concentrations (13).
A previous report by Vila and coworkers offers an additional explanation of how increased stability, particularly under low zinc conditions, could result in increased antibiotic resistance (27). All MBLs are thought to be synthesized intracellularly, transported into the periplasm via the secretory (Sec) transport system, and folded and metallated in the periplasm (31). The concentration of Zn(II) in the periplasm is not as tightly controlled as intracellular Zn(II) concentrations and is largely controlled by the presence or absence of host proteins that bind zinc in the extracellular environment (32). The host’s innate immune response can take advantage of this scenario to fight infections from bacteria that produce MBLs (33). Specifically, calprotectin and psoriasin are secreted by neutrophils at sites of infection and reduce effective extracellular zinc availability by sequestering Zn(II) and thereby preventing metalation of bacterial Zn(II)-metalloenzymes (34). Metal-free Zn(II)-metalloenzymes, and specifically apo-VIM-2, are readily degraded in the periplasm (27), presumably due to a larger proportion of unfolded protein that would be more susceptible to proteolysis.
Vila and coworkers proposed that NDM overcomes this proteolytic degradation through N-terminal lipidation and subsequent anchoring to outer membrane vesicles (27). VIM does not undergo N-terminal lipidation, but the increased Tm of VIM-20 suggests that this variant may be more resistant to proteolysis. Indeed, our immunoblotting studies clearly demonstrate that VIM-2 protein levels are significantly decreased in the periplasm when zinc availability is restricted, likely due to an increased susceptibility to proteolytic degradation. In contrast, VIM-20 concentrations remain constant regardless of zinc availability. This effect is not likely due to an increase in overall proteolysis stimulated by EDTA because proteases are not observed to be upregulated during zinc deprivation (35).
Our X-ray crystal structures of reduced and oxidized VIM-20, in comparison with that of VIM-2, provide structural insights into how thermostability of this variant is increased. Although the H229R substitution seems to be a conservative change, the longer side chain of Arg229 enables the formation of a direct salt bridge with Glu171, linking more tightly a peripheral α-helix with the β-sheet sandwich that serves as the core of all MBL structures. Salt bridge formation has been shown to stabilize some protein structures up to 12 to 21 kJ/mol−1 (36), but the degree of stabilization is strongly dependent on whether the salt bridge is mobile and surface accessible (weaker) or rigid and buried (stronger) (37, 38). Our analyses of the DSF data to extract ΔuG values indicate that, relative to VIM-2, stabilization of VIM-20 by the H229R substitution yields a ΔΔuG of approximately 2 kJ/mol−1, which is similar in magnitude to the 0 to 2 kJ/mol−1 range reported for protein stabilization due to salt bridges found at the surface of a protein. In VIM-20, the Arg229/Glu171 salt bridge is solvent accessible but appears to be held in place by close packing of the Arg side chain with surrounding residues. The resulting ΔTm (+8°C) is similar to that assigned to stabilization by a similarly placed salt bridge in T4 lysozyme (+11°C). Taken together, these comparisons indicate that the magnitude of stabilization assigned to this newly formed salt bridge is reasonable (36).
This report demonstrates that VIM variants are emerging due to numerous selective pressures placed on blaVIM, including antibiotic treatment and low zinc availability at infection sites. Previously, it was established that clinical NDM variants use increased Zn2 affinity and monozinc activity to overcome this pressure. Here, we discovered that the VIM-20 variant instead uses a different mechanism. The introduction of a new salt bridge increases thermostability and stabilizes the folded and metallated enzyme at low zinc concentrations, reducing susceptibility to proteolysis by disfavoring the formation of the unfolded apo protein. Based on our results with these two MBL families, we anticipate that zinc scarcity will likely continue driving the evolution of many MBL variants in clinical settings and that different strategies will emerge to overcome these selective pressures. The benefits conferred by amino acid changes in VIM and NDM variants would not have been detected without studying these systems at low zinc availability, indicating the importance of assessing the impact of zinc deprivation in future studies of clinical MBL variants.
MATERIALS AND METHODS
Cloning of blaVIM-2-like variants for cell viability assays.
The blaVIM-2 gene, including the region encoding the leader sequence, was cloned from Pseudomonas aeruginosa PA3 into the pET-24a(+) vector (Novagen, Darmstadt, Germany) between the NdeI and BamHI restriction sites. After verification by DNA sequencing, the blaVIM-2 gene, including the pET24a(+) ribosomal binding site, was subcloned into phagemid pBC SK(-) (Agilent, Santa Clara, CA) using SacI and BamHI sites, as described previously (39). Therefore, the blaVIM-2 open reading frame was placed under the control of a lac promoter. Coding sequences for VIM-2-like variants were generated by site-directed mutagenesis using nonoverlapping primers and the pBC SK(-) blaVIM-2 plasmid as the template. After PCR, the amplified genes were added directly to a kinase-ligase-DpnI enzyme mix (NEB) for rapid, room-temperature circularization and template removal. A 2-μl aliquot of the ligation mixture was used to transform 30 μl of Escherichia coli DH5α chemically competent cells (Lucigen), and the transformation mixtures were spread on lysogeny broth (LB) plates containing 50 μg/ml chloramphenicol. Plasmid DNA, purified from a single colony, was used to sequence and confirm the insert DNAs (Eurofins Genomics). Plasmids encoding VIM-2-like variants were transformed into E. coli DH10B for subsequent MIC measurements.
Cloning of blaVIM-4/1-like variants for cell viability assays.
The bla gene encoding VIM-1 (GenBank accession no. AJ278514), including the signal peptide sequence, was synthesized by GenScript Biotech Corp. and inserted into a pUC57 vector. The synthesized fragment was subsequently subcloned into the pBC SK(-) vector, which contains the lac promoter and operator but not the lacI repressor. Similar constructs encoding VIM-1-like variants (12 in total) and VIM-4-like variants (9 in total) were generated using the same procedure, and DNA sequencing was used to confirm the expected sequence of each gene. Plasmids encoding VIM-4- and VIM-1-like variants were transformed into E. coli DH10B, which does not produce the lac repressor, for subsequent MIC measurements.
MIC measurements.
MIC measurements were performed in triplicate using the Mueller-Hinton (MH) agar dilution method according to the Clinical Laboratory and Standards Institute (CLSI) protocol (19). All cell viability assays were conducted in E. coli DH10B in order to provide a uniform genetic background in which to evaluate the variants. Briefly, bacterial cultures containing blaVIM variants cloned into a uniform vector were grown overnight at 37°C in cation-adjusted Mueller-Hinton broth (CAMHB). Since the pBC SK(-) vector constitutively expresses the bla gene, no isopropyl-β-d-thiogalactopyranoside (IPTG) was added for these MIC studies. The cultures were diluted, and a Steers replicator was used to deliver 10 μl of a diluted overnight culture containing approximately 104 CFU. MICs were determined for the following: meropenem (Fresnius Kabi), imipenem, ertapenem (Merck), ampicillin, cephalothin, and ceftazidime. MICs were also determined (as described above) with the addition of 50 μM EDTA (Promega Corp.) to the Mueller-Hinton agar in order to evaluate antibiotic susceptibility under conditions where Zn(II) availability was low. The MIC values are reported in Tables S1 and S3.
To facilitate interpretation of the MIC data, we generated a heat map. In Fig. 2, heat map intensities were determined by determining the fold change of MIC for each VIM variant compared to the parent variant; therefore, all VIM-2-like variants were compared to VIM-2 only, and VIM-4-like and VIM-1-like variants were compared to their corresponding parent.
Cloning, overexpression, and purification of VIM-2-like variants.
A codon-optimized VIM-2 encoding DNA sequence (corresponding to residues 27 to 266) was synthesized and inserted into a pUC57 vector by GenScript Biotech Corp. The insert was subsequently subcloned into a pET-28a(+)-TEV plasmid between NdeI/XhoI restriction sites to generate the expression plasmid pET-28a(+)-TEV-VIM-2. An expression plasmid for VIM-20, which harbors the H229R mutation, was generated by site-directed mutagenesis using a forward primer (5′-TATTCAGCAACGTTACCCGGAAGCGCAATTC-3′), a reverse primer (5′-CGTTCGATGCTGGTCGGC-3′), and the pET-28a(+)-TEV-VIM-2 plasmid as the template.
After the PCR, the amplified sequences were added directly to a kinase-ligase-DpnI enzyme mix (NEB) for rapid, room-temperature circularization and template removal. A 2-μl aliquot of the ligation mixture was used to transform 30 μl of E. coli DH5α chemically competent cells (Lucigen), and the transformation mixture was spread onto a lysogeny broth (LB) plate containing 50 μg/ml kanamycin. Plasmid DNA, purified from a single colony, was used to sequence and confirm the inserted sequence (Eurofins Genomics) in the final expression vector, pET-28a(+)-TEV-VIM-20.
Identical overexpression and purification procedures were utilized for VIM-2 and VIM-20 constructs. The expression plasmid [either pET-28a(+)-TEV-VIM-2 or pET-28a(+)-TEV-VIM-20] was transformed into E. coli Express BL21(DE3) chemically competent cells (Lucigen), and the transformation mixture was plated on an LB agar plate containing 50 μg/ml kanamycin. A single colony was transferred into 50 ml of LB broth, containing 50 μg/ml kanamycin, and the culture was shaken overnight at 30°C. The overnight culture (10 ml) was transferred into 2 flasks with each containing 1 liter of LB with 50 μg/ml kanamycin. The resulting culture was grown at 37°C with a shaking speed of 220 rpm until an optical density at 600 nm (OD600) of 0.6 was reached. Protein production was induced by adding ZnCl2 (final concentration, 100 μM) and IPTG (final concentration, 0.5 mM) to each culture. The cell cultures were shaken overnight at 18°C, and cells were harvested by centrifugation for 10 min at 8,000 × g. The resulting pellets were resuspended in 40 ml of 50 mM HEPES (pH 7.5) containing 500 mM NaCl. Cells were lysed by passing the mixture two times through a French press at a pressure between 15,000 and 20,000 lb/in2. The insoluble components were removed after pelleting in a centrifuge for 1 h at 32,000 × g. The retained supernatant was dialyzed overnight versus 1 liter of 50 mM HEPES (pH 7.5) containing 500 mM NaCl and 100 μM ZnCl2. The resulting solution was mixed with imidazole (final concentration, 50 mM) and loaded onto a HisTrap HP (5 ml) column (GE). The column was washed with 10 column volumes of 50 mM HEPES (pH 7.5) containing 500 mM NaCl. Bound proteins were eluted using 50 mM HEPES (pH 7.5) containing 500 mM NaCl and 500 mM imidazole. The imidazole in the eluted protein mixture was removed by dialysis against 2 liters of 50 mM HEPES (pH 7.5) containing 500 mM NaCl. TEV protease was added at a protease-to-target protein ratio of 1:20 (wt/wt), and the mixture was incubated overnight in the presence of 1 mM dithiothreitol (DTT). The resulting truncated VIM variants were then repurified by passage through a HisTrap column, which removed the both the His6-TEV enzyme and the His6 tag peptide cleaved from the VIM constructs.
Purified recombinant VIM-2 and VIM-20 (before and after TEV digestion) were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) using Fisher EZ-Gel solution (12.5%), a Mini-PROTEAN system (Bio-Rad Laboratories), and Coomassie blue stain. Protein concentrations were determined by UV-Vis absorbance at 280 nm using the calculated molar absorptivities (ε280 nm = 31,400 M−1 cm−1 for VIM-2 and VIM-20 containing the poly-His tag and ε280 nm = 29,910 M−1 cm−1 for the VIM-2 and VIM-20 without the poly-His tag).
Metal analyses.
The zinc contents of purified VIM-2 and VIM-20 samples were determined by using a PerkinElmer Optima 7,300 V inductively coupled plasma-optical emission spectroscopy (ICP-OES) detector (40). Protein samples, used without modification after purification, were diluted to 4 μM with 100 mM ammonium acetate (pH 7.5). Calibration curves were generated using serial dilutions of Fisherbrand zinc metal standard, and the emission line at 202.548 nm was used for zinc.
Nano-ESI-MS-based analysis.
To confirm the third Zn(II) binding site in VIM-2 and VIM-20, nano-ESI-MS-analysis was used. Tris(2-carboxyethyl) phosphine hydrochloride (TCEP) (final concentration, 1 mM) and ZnCI2 (final concentration, 100 μM) were added to purified VIM-2 and VIM-20 (50 μM), and the mixtures were incubated for 1 h. Adventitiously bound Zn(II) was removed by dialysis against 100 mM ammonium acetate (pH 7.5) overnight. A Thermo Scientific LTQ Orbitrap XL hybrid ion trap-orbitrap mass spectrometer equipped with a nanoelectrospray ionization (nano-ESI) probe (Thermo Fisher Scientific, San Jose, CA, USA) with positive-mode protein detection was used to analyze the samples. The major parameters were capillary temperature, 180°C; sheath gas, 0; auxiliary gas, 0; sweep gas, 0; spray voltage, 1.1 to 1.9 kV; tube-lens, 150 V; and capillary voltage, 35 V. A full scan ranging from m/z 1,000 to 4,000 was used. The resolution was set to 30,000. Automated gain control was set at 3 × 104 in full scan, 1 × 104 in structured illumination microscopy (SIM), 1 × 104 in sequential mass spectrometry (MSn), and 3,000 in zoom for ion trap, 3 × 106 in full scan, 1 × 105 in SIM and 1 × 105 in MSn for Fourier transform. The nano-ESI source was equipped with an offline unit ES260 (ThermoFisherScientific), and the source was constructed based on published work, with modifications (41). Briefly, a platinum wire (0.25-mm diameter) was inserted in the center of the offline unit and in a pulled-glass capillary with a tip inner diameter (i.d.) of about 1 μm (produced in-house from a glass capillary (i.d., 0.8 mm; outer diameter [o.d.], 1.5 mm) using a micropipette puller (model P-87 Flaming/Brown micropipette puller; Sutter Instrument, Inc., USA). The sample solution (5 μl) was loaded into the pulled-glass capillary by an infusion syringe (Thermo Scientific, USA). The platinum wire was inserted in the center of the capillary. The position of capillary tip was then adjusted to approximately 3 mm away from the MS inlet.
Steady-state kinetics.
Initial rates for substrate hydrolysis were determined by following linear changes in absorbance over time, and changes in substrate concentration were calculated using difference extinction coefficients for ampicillin (Δε235 nm = −755 M−1 cm−1), meropenem (Δε300 nm = −6,929 M−1 cm−1), and ceftazidime (Δε260 nm = −9,114 M−1 cm−1) (Sigma-Aldrich; Δε values were determined experimentally under assay conditions similar to a previously described method [23]). The total volume used in quartz cuvettes was 300 μl. The rates of product formation upon hydrolysis of chromacef (a generous gift from L. D. Sutton, Benedictine College, Atchison, KS) were determined using the difference extinction coefficient Δε442 nm of 14,500 M−1 cm−1 and disposable polystyrene cuvettes (42). For all substrates, assay buffer contained 50 mM HEPES (pH 7.0) containing 0.1 nM or 10 μM added ZnSO4. Extensive washing of quartz cuvettes was completed before assaying with 0.1 nM ZnSO4, as described previously (12). The concentration of enzyme used for each substrate was adjusted to maintain linear initial rate conditions for the first 0.3 min of the reaction at 25°C, and the final enzyme concentration was always significantly less than the lowest substrate concentration tested. Initial rates were plotted versus substrate concentrations and directly fitted to the Michaelis-Menten equation using KaleidaGraph (Synergy Software) to determine KM and Vmax values, and the kcat values were calculated using the respective enzyme concentrations. Fitting errors are reported for each parameter.
UV-visible spectroscopy.
VIM-2 and VIM-20 samples were dialyzed twice against 50 mM HEPES (pH 6.8) containing 500 mM NaCl and 2 mM EDTA, followed by three separate dialysis steps against 50 mM HEPES (pH 6.8) containing 500 mM NaCl and Chelex resin (final concentration, 0.5 g/liter). Buffers were exchanged at approximately 12-h intervals. Zinc-depleted VIM-2 and VIM-20 samples were then diluted to a final concentration of 300 μM with 50 mM HEPES (pH 6.8) containing 500 mM NaCl, 10% glycerol, and 2 mM TCEP. Aliquots of CoCl2 (15 to 105 mM) were added to 500-μl samples of metal-free VIM-2 and VIM-20. After the samples had been gently mixed and incubated at 25°C for 1 min, UV-Vis spectra were collected on a PerkinElmer Lambda 750 UV-Vis-near-infrared (UV-Vis-NIR) spectrometer between 300 to 800 nm at 25°C. Blank spectra of 300 μM metal-free VIM-2 or VIM-20 in 50 mM HEPES (pH 6.8) containing 500 mM NaCl, 10% glycerol, and 2 mM TCEP were used to generate difference spectra. All data were normalized to the absorbance at 800 nm.
Crystallization, diffraction data collection, and structural refinement.
VIM-20 crystals were obtained by sitting drop vapor diffusion in 96-well IntelliPlates (Art Robbins) set up with a Phoenix crystallization robot (Art Robbins). Protein (10 mg/ml) was mixed with crystallization conditions from MCSG1-4 (Microlytic) sparse matrix crystallization screens at volumes of 400 nl VIM-20 and 400 nl crystallization condition. Crystals of reduced VIM-20 were harvested and frozen in liquid nitrogen within 7 days of setting up sitting drop vapor diffusion trials. Crystals of oxidized VIM-20 were frozen in liquid nitrogen 60 to 90 days after setting up sitting drop vapor diffusion trials. Holo VIM-20 in the reduced form was crystallized in 0.2 M magnesium chloride and 20% (wt/vol) polyethylene glycol 3350 (PEG 3350). Oxidized VIM-20 was crystallized in 0.17 M ammonium acetate, 0.085 M sodium acetate-HCl (pH 4.6), 25.5% (wt/vol) PEG 4000, and 15% (vol/vol) glycerol. Harvested crystals were swished through LV CryoOil (MiTeGen) for cryoprotection immediately prior to freezing in liquid nitrogen. X-ray diffraction data were collected on beamline 4.2.2 at a 1.00-Å wavelength at the Advanced Light Source at Lawrence Berkeley National Laboratory. X-ray diffraction data were processed in XDS (43), followed by molecular replacement with PHASER (44) using our prior VIM-2 structure (PDB ID 4NQ2) (16) as the molecular replacement search model. Iterative rounds of model building and refinement were conducted using Coot (45) and PHENIX (46). Stereochemical and geometrical validations of each structure were conducted using MolProbity (47). All figures were prepared using PyMOL (version 1.8; Schrödinger).
In silico substitutions.
Comparative models for VIM-1 and VIM-59 began using the holo structure of reduced VIM-1 (PDB ID 5N5G) (24). A model for VIM-59 was prepared by introducing the H229R mutation in silico using Coot (45). The initial side-chain rotamer for arginine 229, ttt90°, was selected from a group of most preferred rotamers (48). The R229 side-chain rotamer, and the side-chain rotamers for E171 and F173, underwent minor adjustments to accommodate the increased bulk of the R229 side chain compared to histidine. The resulting structure, analyzed by MolProbity (47), features favored rotamers for E171 (tt0°), F173 (t80°), and R229 (ttt90°) and no steric clashes of >0.5 Å.
Differential scanning fluorimetry studies.
VIM-2 and VIM-20 samples were injected into Slide-A-Lyzer dialysis cassettes and dialyzed twice against 20 mM HEPES (pH 7.5) containing 150 mM NaCl and 2 mM EDTA, followed by three dialysis steps against 20 mM HEPES (pH 7.5) containing 0.5 mM TCEP and Chelex resin (final concentration, 0.5 g/liter). Dialysis buffers were exchanged approximately every 12 h. Zinc-depleted VIM-2 and VIM-20 samples were then diluted to a final concentration of 5 μM with 20 mM HEPES (pH 7.5) containing 150 mM NaCl, 5× Sypro Orange (Pierce Thermo, Inc.), and 0.25 to 50 μM ZnCl2. Protein samples were dispensed into a 96-well FrameStar PCR plate sealed with a clear thermal-seal film to prevent evaporation. DSF data were collected in triplicate on a CFX96 real-time PCR (RT-PCR) system (Bio-Rad) via the preset “HEX” channel for fluorescence excitation and emission. The temperature was ramped from 25 to 95°C using 0.5°C increments with a 5-s equilibration at each step. Fluorescence intensity data were fitted to a Boltzmann sigmoidal curve using Prism 8 (GraphPad, Inc.) to determine the melting temperature (Tm), maximal fluorescence value (Fmax), and minimal fluorescence value (Fmin) for all proteins (25). As described by Wright et al., the fraction of folded protein, Pf, at a specific temperature is extracted from Boltzmann sigmoidal curve fits using Equation 1, where F is the fluorescence at the temperature of interest (25).
| (1) |
The fraction unfolded protein (Pu) at a given temperature is then determined using Equation 2.
| (2) |
Apparent affinities for the third equivalent of Zn(II) were obtained by fitting Tm values across the range of Zn(II) concentrations to sigmoidal curves using Prism. The Gibbs free energy of unfolding (Δμg) was calculated for zinc-loaded and zinc-depleted VIM-2 and VIM-20 samples, as previously described (25).
Periplasmic extraction.
Overnight bacterial cultures (in CAMHB) of E. coli DH10B, transformed with either pBC SK(-)-VIM-2 or pBC SK(-)-VIM-20, were diluted (1/100) in 60 ml of CAMHB (with or without added 50 μM EDTA) and grown to an OD600 of 1. An aliquot (50 ml) of each culture was centrifuged for 10 min at 6,000 × g, and the supernatant was discarded. Pellets were washed by resuspending in 10 ml of cold 10 mM Tris-HCl (pH 7.3), containing 30 mM MgCl2, and the mixtures were centrifuged 10 min at 6,000 × g. The supernatants were discarded, and an additional pellet wash was conducted. The pellets were then resuspended in 900 μl of cold 10 mM Tris-HCl (pH 7.3) containing 30 mM MgCl2 and 20 μl/ml chloroform, and mixtures were incubated on ice for 15 min. Following centrifugation for 15 min at 4,500 × g, the supernatants (periplasmic extracts) were removed.
Immunoblotting.
Immunoblotting was used to assess the expression levels of VIM-2 and VIM-20 present under standard conditions or conditions under which zinc availability is limited. Periplasmic extracts were combined with loading dye in a 1:1 volume, separated by SDS-PAGE, and transferred to a polyvinylidene difluoride membrane (Novex, Life Technologies, Carlsbad, CA) by electroblotting. After blocking for 1 h with 5% nonfat dry milk, the presence of VIM was detected by incubation in 5% nonfat dry milk with anti-VIM-2 polyclonal antibodies overnight at 4°C. The membrane was washed four times, 15 min each, in Tris-buffered saline (pH 7.4) containing 0.1% Tween 20 and subsequently incubated in 5% nonfat dry milk with a 1/10,000 dilution of horseradish peroxidase (HRP)-protein G conjugate (Bio-Rad). After four additional washes, the membrane was processed for exposure using the ECL kit (GE Healthcare) and FOTO/AnalystVR FX (Fotodyne).
Data availability.
The coordinates and experimental data for reduced VIM-20 and oxidized VIM-20 were deposited in the PDB with accession codes 6OP6 and 6OP7, respectively.
Data collection and refinement statistics for VIM-20 crystal structures. Download Table S4, DOCX file, 0.1 MB (17.2KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
ACKNOWLEDGMENTS
We acknowledge support from National Institutes of Health under grants R01 GM111926 (to W.F., M.W.C., R.A.B., D.L.T., and R.C.P.), R35 GM128595 (to R.C.P.), R01 AI072219 (to R.A.B.), R01 AI100560 (to R.A.B.), R21 AI114508 (to R.A.B.), and R01 AI063517 (to R.A.B. and R.C.P.), R15 GM134454 (to M.W.C.), NSF grant CHE-1509285 (to M.W.C. and D.L.T.), and a grant from the Robert A. Welch foundation (F-1572 to W.F.). R.C.P. also acknowledges institutional support from Miami University through the Robert H. and Nancy J. Blayney Professorship. This research was also supported by funds and/or facilities provided by the Louis Stokes Cleveland VA Medical Center to R.A.B., and the Veterans Affairs Merit Review Program Award 1I01BX001974 to R.A.B. from the Biomedical Laboratory Research & Development Service of the VA Office of Research and Development and the Geriatric Research Education and Clinical Center VISN 10.
The content of this paper is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health, the U.S. Department of Veterans Affairs, or the U.S. Government.
We declare no competing financial interest.
Footnotes
This article is a direct contribution from Robert A. Bonomo, a Fellow of the American Academy of Microbiology, who arranged for and secured reviews by Patricia Bradford, Antimicrobial Development Specialists, LLC, and Timothy Palzkill, Baylor College of Medicine.
Citation Cheng Z, Shurina BA, Bethel CR, Thomas PW, Marshall SH, Thomas CA, Yang K, Kimble RL, Montgomery JS, Orischak MG, Miller CM, Tennenbaum JL, Nix JC, Tierney DL, Fast W, Bonomo RA, Page RC, Crowder MW. 2019. A single salt bridge in VIM-20 increases protein stability and antibiotic resistance under low-zinc conditions. mBio 10:e02412-19. https://doi.org/10.1128/mBio.02412-19.
REFERENCES
- 1.Fernandes R, Amador P, Prudêncio C. 2013. β-Lactams: chemical structure, mode of action and mechanisms of resistance. Rev Med Microbiol 24:7–17. doi: 10.1097/MRM.0b013e3283587727. [DOI] [Google Scholar]
- 2.Worthington RJ, Melander C. 2013. Overcoming resistance to β-lactam antibiotics. J Org Chem 78:4207–4213. doi: 10.1021/jo400236f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lim C, Takahashi E, Hongsuwan M, Wuthiekanun V, Thamlikitkul V, Hinjoy S, Day NP, Peacock SJ, Limmathurotsakul D. 2016. Epidemiology and burden of multidrug-resistant bacterial infection in a developing country. Elife 5:e18082. doi: 10.7554/eLife.18082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nordmann P, Naas T, Poirel L. 2011. Global spread of carbapenemase-producing Enterobacteriaceae. Emerg Infect Dis 17:1791–1798. doi: 10.3201/eid1710.110655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Palzkill T. 2013. Metallo-β-lactamase structure and function. Ann N Y Acad Sci 1277:91–104. doi: 10.1111/j.1749-6632.2012.06796.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ju LC, Cheng Z, Fast W, Bonomo RA, Crowder MW. 2018. The continuing challenge of metallo-β-lactamase inhibition: mechanism matters. Trends Pharmacol Sci 39:635–647. doi: 10.1016/j.tips.2018.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Watanabe M, Iyobe S, Inoue M, Mitsuhashi S. 1991. Transferable imipenem resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother 35:147–151. doi: 10.1128/aac.35.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lauretti L, Riccio ML, Mazzariol A, Cornaglia G, Amicosante G, Fontana R, Rossolini GM. 1999. Cloning and characterization of blaVIM, a new integron-borne metallo-β-lactamase gene from a Pseudomonas aeruginosa clinical isolate. Antimicrob Agents Chemother 43:1584–1590. doi: 10.1128/AAC.43.7.1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yong D, Toleman MA, Giske CG, Cho HS, Sundman K, Lee K, Walsh TR. 2009. Characterization of a new metallo-β-lactamase gene, blaNDM-1, and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob Agents Chemother 53:5046–5054. doi: 10.1128/AAC.00774-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Naas T, Oueslati S, Bonnin RA, Dabos ML, Zavala A, Dortet L, Retailleau P, Iorga BI. 2017. Beta-Lactamase Database (BLDB)–structure and function. J Enzyme Inhib Med Chem 32:917–919. doi: 10.1080/14756366.2017.1344235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mojica MF, Bonomo RA, Fast W. 2016. B1-metallo-β-lactamases: where do we stand? Curr Drug Targets 17:1029–1050. doi: 10.2174/1389450116666151001105622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheng Z, Thomas PW, Ju L, Bergstrom A, Mason K, Clayton D, Miller C, Bethel CR, VanPelt J, Tierney DL, Page RC, Bonomo RA, Fast W, Crowder MW. 2018. Evolution of New Delhi metallo-β-lactamase (NDM) in the clinic: effects of NDM mutations on stability, zinc affinity, and mono-zinc activity. J Biol Chem 293:12606–12618. doi: 10.1074/jbc.RA118.003835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stewart AC, Bethel CR, VanPelt J, Bergstrom A, Cheng ZS, Miller CG, Williams C, Poth R, Morris M, Lahey O, Nix JC, Tierney DL, Page RC, Crowder MW, Bonomo RA, Fast W. 2017. Clinical variants of New Delhi metallo-β-lactamase are evolving to overcome zinc scarcity. ACS Infect Dis 3:927–940. doi: 10.1021/acsinfecdis.7b00128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bahr G, Vitor-Horen L, Bethel CR, Bonomo RA, González LJ, Vila AJ. 2018. Clinical evolution of New Delhi metallo-β-lactamase (NDM) optimizes resistance under Zn(II) deprivation. Antimicrob Agents Chemo 62:e01849-17. doi: 10.1128/AAC.01849-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martínez-García L, González-Alba JM, Baquero F, Cantón R, Galán JC. 2018. Ceftazidime is the key diversification and selection driver of VIM-type carbapenemases. mBio 9:e02109-17. doi: 10.1128/mBio.02109-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aitha M, Marts AR, Bergstrom A, Moller AJ, Moritz L, Turner L, Nix JC, Bonomo RA, Page RC, Tierney DL, Crowder MW. 2014. Biochemical, mechanistic, and spectroscopic characterization of metallo-β-lactamase VIM-2. Biochemistry 53:7321–7331. doi: 10.1021/bi500916y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Poirel L, Naas T, Nicolas D, Collet L, Bellais S, Cavallo JD, Nordmann P. 2000. Characterization of VIM-2, a carbapenem-hydrolyzing metallo-β-lactamase and its plasmid- and integron-borne gene from a Pseudomonas aeruginosa clinical isolate in France. Antimicrob Agents Chemother 44:891–897. doi: 10.1128/aac.44.4.891-897.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garcia-Saez I, Docquier JD, Rossolini GM, Dideberg O. 2008. The three-dimensional structure of VIM-2, a Zn-β-lactamase from Pseudomonas aeruginosa in its reduced and oxidised form. J Mol Biol 375:604–611. doi: 10.1016/j.jmb.2007.11.012. [DOI] [PubMed] [Google Scholar]
- 19.Clinical and Laboratory Standards Institute. 2017. Performance standards for antimicrobial susceptibility testing, 27th ed. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 20.Wilkins MR, Gasteiger E, Bairoch A, Sanchez JC, Williams KL, Appel RD, Hochstrasser DF. 1999. Protein identification and analysis tools in the ExPASy server. Methods Mol Biol 112:531–552. [DOI] [PubMed] [Google Scholar]
- 21.Kupper MB, Herzog K, Bennink S, Schlomer P, Bogaerts P, Glupczynski Y, Fischer R, Bebrone C, Hoffmann KM. 2015. The three-dimensional structure of VIM-31–a metallo-β-lactamase from Enterobacter cloacae in its native and oxidized form. FEBS J 282:2352–2360. doi: 10.1111/febs.13283. [DOI] [PubMed] [Google Scholar]
- 22.Yang H, Aitha M, Marts AR, Hetrick A, Bennett B, Crowder MW, Tierney DL. 2014. Spectroscopic and mechanistic studies of heterodimetallic forms of metallo-β-lactamase NDM-1. J Am Chem Soc 136:7273–7285. doi: 10.1021/ja410376s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thomas PW, Zheng M, Wu S, Guo H, Liu D, Xu D, Fast W. 2011. Characterization of purified New Delhi metallo-β-lactamase-1. Biochemistry 50:10102–10113. doi: 10.1021/bi201449r. [DOI] [PubMed] [Google Scholar]
- 24.Salimraj R, Hinchliffe P, Kosmopoulou M, Tyrrell JM, Brem J, van Berkel SS, Verma A, Owens RJ, McDonough MA, Walsh TR, Schofield CJ, Spencer J. 2019. Crystal structures of VIM-1 complexes explain active site heterogeneity in VIM-class metallo-β-lactamase. FEBS J 286:169–183. doi: 10.1111/febs.14695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wright TA, Stewart JM, Page RC, Konkolewicz D. 2017. Extraction of thermodynamic parameters of protein unfolding using parallelized differential scanning fluorimetry. J Phys Chem Lett 8:553–558. doi: 10.1021/acs.jpclett.6b02894. [DOI] [PubMed] [Google Scholar]
- 26.Ahmad S, Kumar V, Ramanand KB, Rao NM. 2012. Probing protein stability and proteolytic resistance by loop scanning: a comprehensive mutational analysis. Protein Sci 21:433–446. doi: 10.1002/pro.2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.González LJ, Bahr G, Nakashige TG, Nolan EM, Bonomo RA, Vila AJ. 2016. Membrane anchoring stabilizes and favors secretion of New Delhi metallo-β-lactamase. Nat Chem Biol 12:516–522. doi: 10.1038/nchembio.2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pournaras S, Tsakris A, Maniati M, Tzouvelekis LS, Maniatis AN. 2002. Novel variant (blaVIM-4) of the metallo-β-lactamase gene blaVIM-1 in a clinical strain of Pseudomonas aeruginosa. Antimicrob Agents Chemother 46:4026–4028. doi: 10.1128/aac.46.12.4026-4028.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Franceschini N, Caravelli B, Docquier JD, Galleni M, Frere JM, Amicosante G, Rossolini GM. 2000. Purification and biochemical characterization of the VIM-1 metallo-β-lactamase. Antimicrob Agents Chemother 44:3003–3007. doi: 10.1128/aac.44.11.3003-3007.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Makena A, Duzgun AO, Brem J, McDonough MA, Rydzik AM, Abboud MI, Saral A, Cicek AC, Sandalli C, Schofield CJ. 2015. Comparison of Verona integron-borne metallo-β-lactamase (VIM) variants reveals differences in stability and inhibition profiles. Antimicrob Agents Chemother 60:1377–1384. doi: 10.1128/AAC.01768-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morán-Barrio J, Limansky AS, Viale AM. 2009. Secretion of GOB metallo-β-lactamase in Escherichia coli depends strictly on the cooperation between the cytoplasmic DnaK chaperone system and the Sec machinery: completion of folding and Zn(II) ion acquisition occur in the bacterial periplasm. Antimicrob Agents Chemother 53:2908–2917. doi: 10.1128/AAC.01637-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cerasi M, Ammendola S, Battistoni A. 2013. Competition for zinc binding in the host-pathogen interaction. Front Cell Infect Microbiol 3:108. doi: 10.3389/fcimb.2013.00108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hood MI, Skaar EP. 2012. Nutritional immunity: transition metals at the pathogen-host interface. Nat Rev Microbiol 10:525–537. doi: 10.1038/nrmicro2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brophy MB, Hayden JA, Nolan EM. 2012. Calcium ion gradients modulate the zinc affinity and antibacterial activity of human calprotectin. J Am Chem Soc 134:18089–18100. doi: 10.1021/ja307974e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sigdel TK, Easton JA, Crowder MW. 2006. Transcriptional response of Escherichia coli to TPEN. J Bacteriol 188:6709–6713. doi: 10.1128/JB.00680-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Anderson DE, Becktel WJ, Dahlquist FW. 1990. pH-induced denaturation of proteins: a single salt bridge contributes 3–5 kcal/mol to the free energy of folding of T4 lysozyme. Biochemistry 29:2403–2408. doi: 10.1021/bi00461a025. [DOI] [PubMed] [Google Scholar]
- 37.Sun DP, Sauer U, Nicholson H, Matthews BW. 1991. Contributions of engineered surface salt bridges to the stability of T4 lysozyme determined by directed mutagenesis. Biochemistry 30:7142–7153. doi: 10.1021/bi00243a015. [DOI] [PubMed] [Google Scholar]
- 38.Takano K, Tsuchimori K, Yamagata Y, Yutani K. 2000. Contribution of salt bridges near the surface of a protein to the conformational stability. Biochemistry 39:12375–12381. doi: 10.1021/bi000849s. [DOI] [PubMed] [Google Scholar]
- 39.Mojica MF, Mahler SG, Bethel CR, Taracila MA, Kosmopoulou M, Papp-Wallace KM, Llarrull LI, Wilson BM, Marshall SH, Wallace CJ, Villegas MV, Harris ME, Vila AJ, Spencer J, Bonomo RA. 2015. Exploring the role of residue 228 in substrate and inhibitor recognition by VIM metallo-β-lactamases. Biochemistry 54:3183–3196. doi: 10.1021/acs.biochem.5b00106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cheng Z, VanPelt J, Bergstrom A, Bethel C, Katko A, Miller C, Mason K, Cumming E, Zhang H, Kimble RL, Fullington S, Bretz SL, Nix JC, Bonomo RA, Tierney DL, Page RC, Crowder MW. 2018. A noncanonical metal center drives the activity of the Sediminispirochaeta smaragdinae metallo-β-lactamase SPS-1. Biochemistry 57:5218–5229. doi: 10.1021/acs.biochem.8b00728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Harvey SR, Porrini M, Stachl C, MacMillan D, Zinzalla G, Barran PE. 2012. Small-molecule inhibition of c-MYC:MAX leucine zipper formation is revealed by ion mobility mass spectrometry. J Am Chem Soc 134:19384–19392. doi: 10.1021/ja306519h. [DOI] [PubMed] [Google Scholar]
- 42.Yu S, Vosbeek A, Corbella K, Severson J, Schesser J, Sutton LD. 2012. A chromogenic cephalosporin for β-lactamase inhibitor screening assays. Anal Biochem 428:96–98. doi: 10.1016/j.ab.2012.06.006. [DOI] [PubMed] [Google Scholar]
- 43.Kabsch W. 2010. Integration, scaling, space-group assignment and post-refinement. Acta Crystallogr D Biol Crystallogr 66:133–144. doi: 10.1107/S0907444909047374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. 2007. Phaser crystallographic software. J Appl Crystallogr 40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Emsley P, Lohkamp B, Scott WG, Cowtan K. 2010. Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen VB, Arendall WB III, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. 2010. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr 66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lovell SC, Word JM, Richardson JS, Richardson DC. 2000. The penultimate rotamer library. Proteins 40:389–408. doi:. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
MIC values (μg/ml) for VIM-2-like (A strain), VIM-4 (B strain), and VIM-1-like (C strain) variants under standard conditions. Download Table S1, DOCX file, 0.1 MB (21KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Comparison of MIC values (μg/ml) between our work and Martinez-Garcia et al. (15). Download Table S2, DOCX file, 0.1 MB (17.6KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
MIC values (μg/ml) for VIM-2-like (A strain), VIM-4 (B strain), and VIM-1-like (C strain) variants under zinc-limited conditions (50 μM EDTA). Download Table S3, DOCX file, 0.1 MB (20.8KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Native mass spectrometry data of VIM-2 (left) and VIM-20 (right). Both protein samples were in 50 μM ammonium acetate (pH 7.5). The 10+ and 9+ charged peaks were observed from both samples in positive mode. Download FIG S1, PDF file, 0.1 MB (57.8KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Fitting of DSF data to the equation ΔuG = −RT ln K to enable extrapolation of ΔuG from the unfolding temperature (Tm) back to 310 K. (A and B) Calculated ΔuG values at temperatures immediately preceding the Tm for VIM-20 (open squares) and VIM-2 (black circles) were plotted for high Zn(II) (A) and low Zn(II) (B) concentrations. (C) Predicted fraction unfolded as a function of temperature at low and higher concentrations of Zn(II). Download FIG S2, TIF file, 0.3 MB (365.3KB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Data collection and refinement statistics for VIM-20 crystal structures. Download Table S4, DOCX file, 0.1 MB (17.2KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Data Availability Statement
The coordinates and experimental data for reduced VIM-20 and oxidized VIM-20 were deposited in the PDB with accession codes 6OP6 and 6OP7, respectively.
Data collection and refinement statistics for VIM-20 crystal structures. Download Table S4, DOCX file, 0.1 MB (17.2KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.