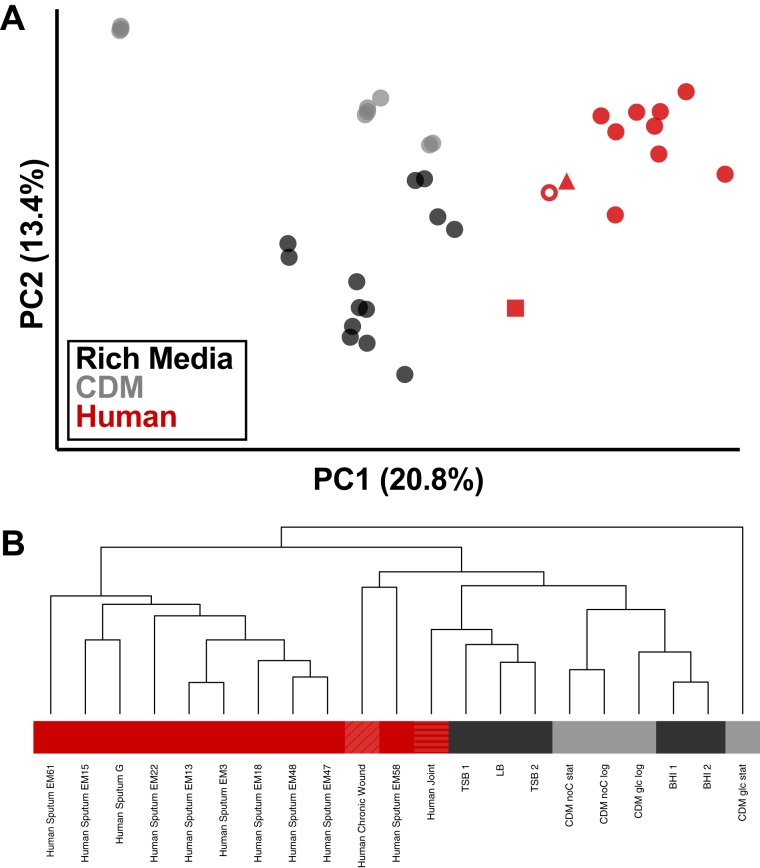
FIG 1.
S. aureus transcriptomes from human infection cluster independently from in vitro samples. (A) Principal-component analysis of variance stabilizing transformation (VST)-normalized reads from S. aureus RNA-seq. Transcriptomes from human infection are shown in red, with closed circles representing human CF sputum from Emory (n = 9), open circle representing human CF sputum from Denmark (n = 1), square representing human joint infection (n = 1), and triangle representing human chronic wound infection (n = 1). Transcriptomes from in vitro conditions are shown indicating the type of medium in which they were grown, with black representing rich medium (LB, TSB, or BHI; n = 13) and gray representing chemically defined medium with two separate carbon sources (CDM; n = 9). (B) Hierarchal clustering of sample types using normalized reads per gene for each condition. Under conditions with replicates (in vitro), replicates were averaged and the mean counts for each gene were used. Black indicates samples from rich medium, gray indicates samples from CDM, and red indicates samples from human infection (solid, sputum; diagonally striped, chronic wound; horizontally striped, joint). For in vitro samples, “CDM glc log” is samples 1 and 2, “CDM glc stat” is samples 3 to 5, “CDM noC log” is samples 6 and 7, “CDM noC stat” is samples 8 and 9, “BHI 1” is samples 10 and 11, “BHI 2” is samples 12 and 13, “LB” is samples 14 to 16, “TSB 1” is samples 17 to 19, and “TSB 2” is samples 20 to 22 listed in Table 2.