Abstract
Thymocyte development requires a complex orchestration of multiple factors. Ablating either Tcf-1 or HEB in CD4+CD8+-thymocytes elicits similar developmental outcomes including increased proliferation, decreased survival, and reduced late Tcra rearrangements. Here, we provide a mechanistic explanation for these similarities by showing that Tcf-1 and HEB share ~7000 DNA binding-sites genome- wide and promote chromatin accessibility. The binding of both Tcf-1 and HEB is required at these shared sites for epigenetic and transcriptional gene regulation. Binding of Tcf-1 and HEB to their conserved motifs in enhancer regions of T-cell differentiation and survival genes promotes their expression. Binding to sites that lack conserved motifs in promoter regions of cell-cycle genes limits proliferation. Tcf-1 alone displaces nucleosomes to allow for chromatin accessibility. Importantly, Tcf-1 inhibits Notch-signaling to protect HEB from Notch mediated proteasomal degradation. Thus, Tcf-1 shifts nucleosomes and safeguards HEB to enable their cooperation in establishing the epigenetic and transcription profile of CD4+CD8+-thymocytes.
Introduction
The CD4+CD8+ double-positive (DP) stage of T-cell development encompasses critical developmental checkpoints, including rearrangement of the T-cell receptor alpha (Tcrα) locus, assembly of the αβ-T-cell receptor (αβTCR), and passage through positive and negative-selection1. DP thymocytes arise from the proliferative double-negative-4 (DN4) stage, which is controlled by cooperative pre-TCR and Notch signals2. Coordination between transcriptional regulators has been established for early stages of T-cell development, where Notch signals activate Tcf-1 (Tcf7)3, 4 and Gata35, 6 transcription, while Pu.1 regulates the binding site choice of Runx1 to initiate the T-cell program7, 8, 9. Together, Notch, Tcf-1, Gata3, and Runx1 activate Bcl11b and seal the T-cell fate7, 10. T-cell commitment in DN2 cells coincides with the upregulation of HEB (Tcf12), HEBAlt (HEB isoform), Runx1, Gfi1, and Ets1, which control subsequent T-cell developmental stages7, 11, 12, 13. Despite intense investigation, the orchestration of factors that regulate entry to and differentiation through the DP stages remains poorly understood.
Regulatory proteins implicated in the transition to and progression through the DP stage include Tcf-114, 15 and HEB16. Tcf-1 is a member of the HMG domain-containing Tcf/Lef family of transcriptional regulators and participates in complex transcriptional and epigenetic processes throughout T-cell development. It interacts with β-catenin to activate Wnt target genes, and with Groucho to silence genes17, 18, 19 Although Tcf-1 has intrinsic HDAC activity20, it has also been shown to promote chromatin accessibility and displace nucleosomes at its binding-sites21. The specific transcription and epigenetic functions of Tcf-1 at the DP stage and potential cooperation with other regulators remain unclear. HEB is a member of the E-protein family of transcription regulators, which are essential for the development of both B and T cells22. Through its helix-loop-helix domain, HEB can form homodimers as well as heterodimers with other E proteins to mediate positive and negative regulation of gene expression. Although HEB has been shown to bind the acetyltransferase p30023, its genome-wide chromatin remodeling functions remain unclear.
In germline Tcf-1-deficient thymocytes the transition to the DP stage is impaired 14, 15 Tcf-1 also controls the lifespan of DP thymocytes by upregulating the anti- apoptotic protein Bcl-xL24, 25,26. These shorter-lived DPs have less time to undergo distal Tcrα gene rearrangements, and therefore generate fewer NKT-cells, which are dependent on these rearrangements26. Like Tcf-1, HEB, regulates the transition to the DP stage as well as the survival of DP thymocytes and its deletion limits distal Tcrα rearrangements and NKT-cell development27. Following the DP stage, Tcf-1 promotes the CD4+ versus CD8+ T-cell fate28. HEB is also required for CD4+ lineage commitment29. Thus, Tcf-1 and HEB are fundamental in guiding thymocytes into and beyond the DP stages of development, it is however unclear whether they collaborate directly in DP thymocytes.
Here we provide a mechanistic understanding for the parallel phenotypes observed in Tcf-1- and HEB-deficient DP-thymocytes. We show that the majority of HEB-bound DNA sites throughout the genome were also bound by Tcf-1. Tcf-1 and HEB co-binding promoted chromatin accessibility, however, Tcf-1 had a predominant role in limiting nucleosome occupancy. Importantly, Tcf-1 and HEB co-binding to their conserved motifs correlated with positive transcriptional regulation of the associated genes. Positively regulated genes were involved in processes of αβ-T-cell development and TCR signaling and were predominantly bound by Tcf-1 and HEB in their enhancer regions. By contrast, Tcf-1 and HEB bound to sites that were not enriched for their conserved motifs in promoters of cell-cycle genes, and ablation of either Tcf-1 or HEB increased DP-thymocyte proliferation. Importantly we show that Tcf-1 limited Notch signaling, which targets HEB for proteasomal degradation. As a result, Tcf-1-deficient DP-thymocytes also had reduced HEB protein levels and severely limited HEB binding to DNA. Therefore, Tcf-1 both enhances HEB protein stability and co-operates functionally with HEB on DNA to define the epigenetic and transcriptional status of DP-thymocytes.
Results
Tcf-1 binds accessible regulatory regions of actively transcribed genes
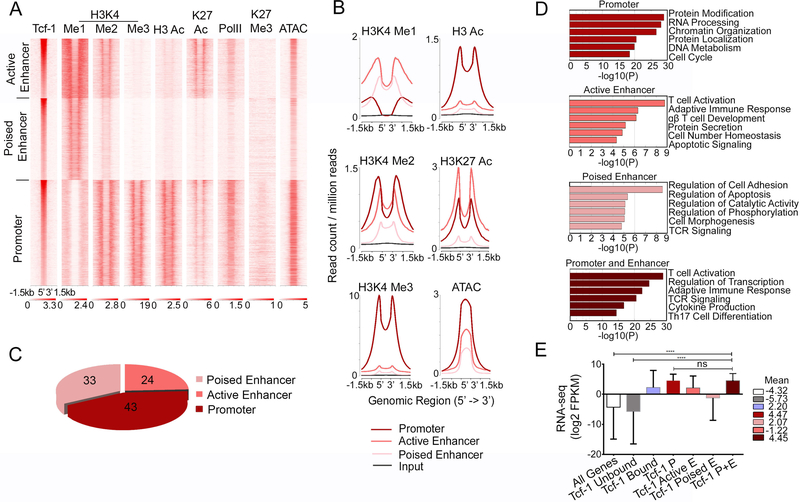
We previously established through chromatin immunoprecipitation followed by deep sequencing (ChIP-seq)30, 31 that in WT-thymocytes Tcf-1 bound to over 16,000 sites genome wide (p=10e-5). We performed ATAC-seq (Assay for Transposase-Accessible Chromatin followed by deep sequencing) to identify regions of accessible chromatin in WT DP-thymocytes. We then aligned the ATAC-seq results to our previous Tcf-1 ChIP-seq data and we established that the chromatin surrounding Tcf-1 peaks in DP-thymocytes was highly accessible (Figure 1A). We also mapped the landscape of histone-marks indicating poised/active (H3K4me1, H3K4me2), active (H3k4me3, H3Ac, H3K27Ac) and repressed chromatin (H3K27me3) proximal to Tcf-1 sites. Tcf-1 bound to gene promoters (43% Figure 1C) that were enriched for H3K4me2/me3, as well as H3Ac and H3K27Ac histone-marks, indicating that they were active promoters (Figure 1A and B). These sites were also enriched for RNA PolII (Figure 1A), indicating that they were transcriptionally active. Enhancer-bound Tcf-1 sites included active enhancers (24% Figure 1C), enriched for H3K4me1/me2, H3K27Ac, and RNA PolII, as well as poised enhancers (33% Figure 1C) marked by H3K4me1 but displaying reduced levels of H3K4me2, H3K27Ac and lacking RNA PolII (Figure 1A, & B). In contrast, Tcf-1 binding rarely overlapped with the repressive H3K27me3 mark (Figure 1A). Thus, Tcf-1 occupies gene regulatory regions enriched for marks of poised or active chromatin, indicating that it may directly regulate gene expression.
Figure 1: Tcf-1 binds Accessible Chromatin of Highly Expressed Genes.
A. K-means clustered heatmap centered on Tcf-1 binding sites (±1.5 kb) showing enrichment of the indicated histone modifications and RNA PolII in WT thymocytes, and ATAC-seq in DP thymocytes. B. Comparative enrichment histograms of permissive histone modifications (H3Ac, H3K4me1, H3K4me2, H3K4me3 and H3K27ac) and chromatin accessibility (ATAC-seq) at Tcf-1 binding sites (±1.5 kb) in K-means clusters shown in A. C. Genomic distribution of Tcf-1 peaks in wildtype thymocytes. D. Functional pathways enriched in genes bound by Tcf-1 in the indicated regions, identified by Metascape. E. Average expression in DP thymocytes of genes in the indicated groups E=enhancer, P=promoter. Values are log2 FPKM, shown as mean with s.d. (****=p≤0.0001) P-values determined by Kruskal-Wallis statistical test.
Tcf-1 binding at sites of open chromatin, enriched for RNA PolII, suggested that its gene targets are actively transcribed. Indeed, RNA-seq analysis of sorted WT DP-thymocytes showed that the average expression of Tcf-1 bound genes was higher than the average expression of all genes expressed in DPs (Figure 1E). In particular, genes bound by Tcf-1 either at promoter or at both promoter and enhancer regions had significantly higher expression levels than genes bound at either active or poised enhancer regions only. Thus, Tcf-1 binds accessible regulatory regions of actively transcribed genes. Interestingly, pathway-enrichment analysis (http://metascape.org) revealed that Tcf-1 binding to promoters alone versus both promoters and enhancers marks genes involved in distinct processes (Figure 1D). Genes bound by Tcf-1 only at promoter regions were involved in cell-cycle regulation. By contrast, Tcf-1 binding at enhancer or both promoter and enhancer regions marked genes involved in T-cell developmental processes. These findings suggest that Tcf-1 may differentially regulate these distinct processes through specific chromatin binding patterns, potentially in coordination with other factors.
Tcf-1 shares binding sites with other lymphoid factors critical for T-cell development
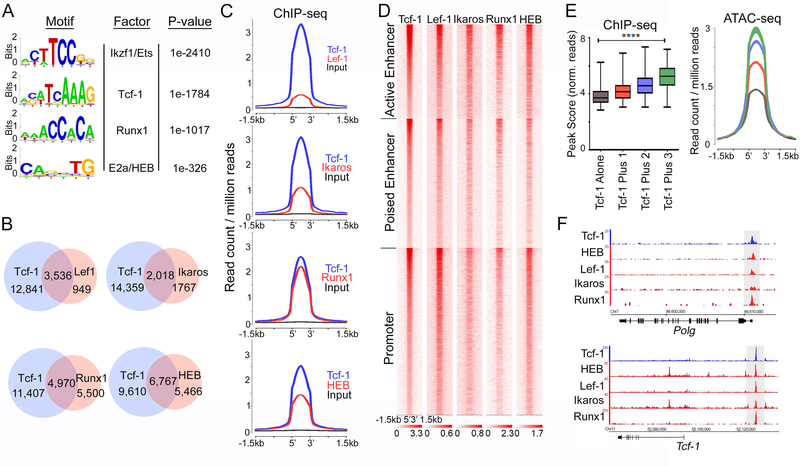
To identify additional factors that may cooperate with Tcf-1, we first analyzed Tcf-1 binding sites for common motifs. Tcf-1-bound sites were highly enriched for the conserved Tcf/Lef motif (p=1e-1784). Additionally, motifs for Ikaros/Ets proteins, Runx and basic helix-loop-helix (bHLH) domain containing proteins were also significantly enriched at Tcf-1-bound sites (Figure 2A). These factors are essential during the transitions from the DN to DP and SP stages of thymocyte development. Ikaros regulates differentiation from the CD4+CD8lo post-selected DP to the SP subsets and its deletion predisposes mice to Notch dependent thymic lymphomas32, 33 Runx1 is involved in the progression from the DN to DP and SP stages and, with Runx3, is essential for CD8+ lineage commitment34. Likewise, the bHLH domain containing E-proteins E2A and HEB regulate thymocyte progression into, and exit from, the DP stage. Interestingly, HEB-deficiency impairs thymic development in a manner akin to the deletion of Tcf-116, 27.
Figure 2: Tcf-1 Binding Overlaps with Lef1, HEB, Runx1, and Ikaros Binding.
A. De novo transcription factor-binding motif analysis (HOMER) within Tcf-1 binding sites in WT thymocytes. The most significantly enriched motifs and associated P-values are shown. B. Venn diagrams of the number of overlapping peaks between Tcf-1 and the indicated factors. C. Comparative enrichment histograms at overlapping DNA binding sites of Tcf-1 and indicated factors (±1.5 kb). D. Heatmap of ChIP-seq of indicated factors centered on K-means clustered Tcf-1 binding sites (±1.5 kb). E. Box plots (left panel) and comparative accessibility histograms (right panel) depicting average peak score of Tcf-1 binding at sites where it binds alone or with 1, 2 or three factors as indicated. Values are mean log2 peak scores. (****=p≤0.0001). P-value was determined by Kruskal-Wallis statistical test. F. Representative multifactor binding regions at the indicated loci (Integrated Genome Browser). ChIP-seq enrichment tracks are shown for Tcf-1, HEB, Lef-1, Ikaros and Runx1.
The involvement of these regulators in the DP stage of thymocyte development, and the enrichment of their motifs at Tcf-1-bound sites, prompted us to examine whether they share overlapping binding sites with Tcf-1. Hence, we performed ChlP-seq for HEB and analyzed previously published ChlP-seq for Ikaros35, and Runx136 in WT thymocytes. We also performed ChIP-seq for Lef-1, which recognizes the same motif as Tcf-1 and is thought to have redundant functions37, 38. As expected, even though Lef-1 occupied significantly fewer sites than Tcf-1 (4476/16377), 79% of these sites (3536) overlapped with Tcf-1 (Figure 2B). Tcf-1 also overlapped with 53% of Ikaros (2018) and 47% of Runx1 (4790) binding sites. HEB shared the largest number of overlapping sites with Tcf-1 (6767), representing nearly 55% of all HEB peaks (Figure 2B). The peak summits of Ikaros, Runx1, and HEB in common sites with Tcf-1 completely overlapped with Tcf-1 peak summits, indicating that binding of these factors centered on the same sequences (Figure 2C, D, and F).
We compared chromatin accessibility at sites bound by Tcf-1 alone (6883) with sites where Tcf-1 overlapped with one other factor (5,482 sites; 63% HEB, 28% Runx1, 9% Ikaros), two factors (3280, 76% HEB/Runx1, 24% HEB/Ikaros), or sites at which all four factors overlapped (732). Tcf-1 enrichment (peak-score) and chromatin accessibility (ATAC-seq) progressively increased at sites where Tcf-1 overlapped with additional factors (Figure 2E). It is unclear whether the increased accessibility facilitates the binding of multiple factors or the binding of multiple factors enhances accessibility at these sites. Among the co-bound factors, HEB was the preferential binding partner of Tcf-1. HEB shared an extensive number of sites with Tcf-1 and also co-bound all multifactor Tcf-1 sites, which led us to further investigate the functions that Tcf-1 and HEB regulate in DP-thymocytes.
Tcf-1 and HEB guide similar developmental processes
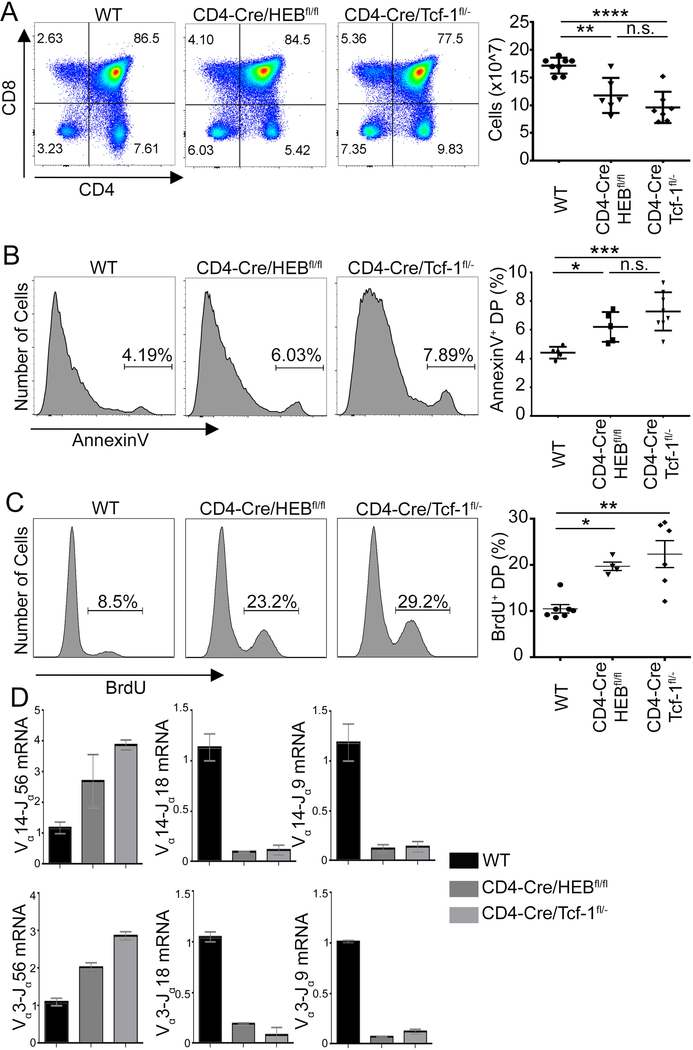
To understand the potential cooperative functions of Tcf-1 and HEB, we compared the phenotypes that result from DP-specific deletion of either Tcf-1 or HEB on thymocyte development. To accomplish this, we conditionally ablated Tcf-1 or HEB using a CD4-Cre transgene. While crossing CD4-Cre/HEBfl/fl mice exhibit sufficiently reduced HEB protein levels in preselected DPs, effective reduction of Tcf-1 protein in preselection DPs required crossing CD4-Cre to Tcf-1fl/- mice, as previously described28. Multiple studies have established that heterozygous deletion of Tcf-1 has no effect on DP-thymocyte development14, 15, 39. CD4-Cre mediated ablation of Tcf-1 (CD4-Cre/Tcf-1fl/-) or HEB (CD4-Cre/HEBfl/fl) mildly reduced thymic cellularity (Figure 3A). In accordance with published observations on HEB and Tcf-1 deficiencies26, 27, both Tcf-1 and HEB-deficient DP-thymocytes increased apoptosis (Annexin V+), than WT (CD4-Cre) littermate controls (Figure 3B). Additionally, qPCR showed that Tcrα gene rearrangements were strongly biased for proximal and against distal Jα genes, in both CD4-Cre/Tcf-1fl/- and CD4-Cre/HEBfl/fl DPs (Figure 3D). This impairment is likely due to the reduced lifespan of the mutant DPs, preventing cells from rearranging distal Ja genes. We further found that DP-thymocytes conditionally lacking either Tcf-1 or HEB were significantly more proliferative (22.33%, s.d.=7.137 and 19.7%, s.d.=1.824) than WT (CD4-Cre) littermate controls (10.49%, s.d.=2.404) (Figure 3C). Higher proliferation rates did not result from increased DP-blast thymocytes in CD4-Cre/Tcf-1fl/- and CD4-Cre/HEBfl/fl thymi compared to WT, as the fraction of CD71+FSChi blast DPs was not significantly altered (Supplemental-Figure 1). Thus, in addition to overlapping genome-wide binding, Tcf-1 and HEB regulate the same critical properties of DP-thymocytes. These observations suggest that Tcf-1 and HEB may cooperate to regulate the DP stage of thymocyte development.
Figure 3: Tcf-1 or HEB Deficiency Elicits Similar Developmental Impairments.
A. FACS plots of CD4 versus CD8 staining in WT (CD4Cre), CD4-Cre/HEBfl/fl and CD4-Cre/Tcf-1fl/- thymocytes from 6–8 week old mice. (Right) Thymocyte numbers from the indicated mouse strains (n=6–8) (**=p≤0.01, ****= p≤0.0001). B. Histogram plots of Annexin V staining of gated DPs in the indicated mouse strains. (Right) Cumulative plots of the fraction of Annexin V+ DPs in the indicated mouse strains (n=5–8). (*=p≤0.05, ****= p≤0.0001). C. Histogram plots of BrdU staining in gated DPs in the indicated mouse strains. Cumulative plots of the fraction of BrdU+ DPs. (*=p≤0.05, **=p≤0.01). D. Quantitative PCR analysis of transcripts of Vα14 and Vα3 rearrangements to Jα56, Jα18 or Jα9 segments in sorted DPs relative to the expression of Cα transcripts. Data are representative of two experiments, shown as mean with s.e.m. All P-values were determined by Ordinary one-way Anova.
Tcf-1/HEB share binding sites in regulatory regions of genes involved in T-cell developmental processes
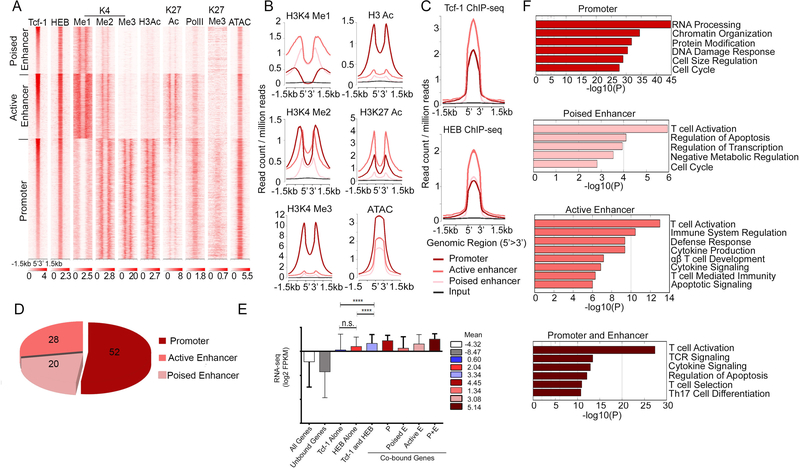
Our HEB ChIP-seq showed that, like Tcf-1, HEB bound to accessible chromatin sites. The mean expression of HEB-bound genes was higher than that of all genes expressed in DP thymocytes. Genes bound by HEB at promoter or both promoter and enhancer sites had the highest expression (Supplemental-Figure 2). To assess the potential cooperation between Tcf-1 and HEB in DP thymocytes we analyzed their overlapping binding-sites (Figure 4). Sites bound by both Tcf-1 and HEB were distributed to promoters as well as poised and active enhancers of the associated genes (Figure 4A, B and D). The average expression of HEB and Tcf-1 co-bound genes was significantly higher than the average expression of all genes in DP- thymocytes or genes uniquely bound by either Tcf-1 or HEB (Figure 4E). In particular, genes with overlapping Tcf-1 and HEB binding at both promoter and enhancer regions showed the highest expression, followed by promoter-only bound genes.
Figure 4: Tcf-1 and HEB Co-bind Accessible Chromatin of Highly Expressed Genes.
A. K-means clustered heatmap centered on Tcf-1 and HEB overlapping sites (±1.5 kb) showing enrichment of the indicated histone modifications and RNA PolII in WT thymocytes, and ATAC-seq in DP thymocytes. (±1.5 kb). B. Comparative enrichment histograms of permissive histone modifications (H3K9/14ac, H3K4me1, H3K4me2, H3K4me3 and H3K27ac) and chromatin accessibility (ATAC-seq) at overlapping Tcf-1 and HEB binding sites (±1.5 kb) in clusters identified in A. C. ChIP-seq enrichment of Tcf-1 (top) and HEB (bottom) at regions of overlapping Tcf-1/HEB binding identified in A (±1.5 kb). D. Genomic distribution of overlapping Tcf-1/HEB binding sites in wildtype thymocytes. E. Average expression in DP thymocytes of genes in the indicated groups E=enhancer, P=promoter. Numbers are mean log2 FPKM. (****=p≤0.0001). P-values determined by Kruskal-Wallis statistical test. F. Functional pathways enriched in genes bound by both Tcf-1 and HEB in promoter or promoters and enhancers.
Shared Tcf-1 and HEB binding at promoter versus enhancer sites differed by several parameters. First, Tcf-1 and HEB enrichment was highest at active enhancers (Figure 4C). Second, overlapping Tcf-1 and HEB binding at promoters versus enhancers marked distinct groups of genes. Binding to only promoter regions marked genes involved in general cellular processes, such as cell division and DNA damage response (Figure 4G, Supplemental-Figure 3A). However, binding to enhancers or enhancers as well as promoters marked genes involved in T-cell- specific processes, such as T-cell activation, and TCR signaling (Figure 4F). This binding pattern is similar to that observed for all Tcf-1 bound sites (Figure 1D, Supplemental Figure 3B, C, D). Finally, we identified regions with extensive Tcf-1 and HEB binding using a rank order super-cluster algorithm that stitches together adjacent transcription factor enrichment peaks within 12.5 kb regions, similar to the strategy described by Whyte et al 40. Of the 271 Tcf-1 and 213 HEB super-clusters identified (Supplemental Figure 4), 126 overlapped and the majority were in enhancer regions, with only 11 located in promoter regions. Overlapping super-clusters mostly occurred at enhancers of genes involved in TCR signaling, recombination, and apoptosis, highlighting DP thymocyte processes impacted by ablation of Tcf-1 or HEB (Supplemental Figure 3C). Thus, Tcf-1 and HEB shared binding to gene promoters versus enhancers identifies distinct processes. Independently deleting either protein functionally impairs these processes in DP thymocytes (Figure 3), suggesting that Tcf-1 and HEB cooperate to regulate the transcriptional and/or the epigenetic state of the associated genes.
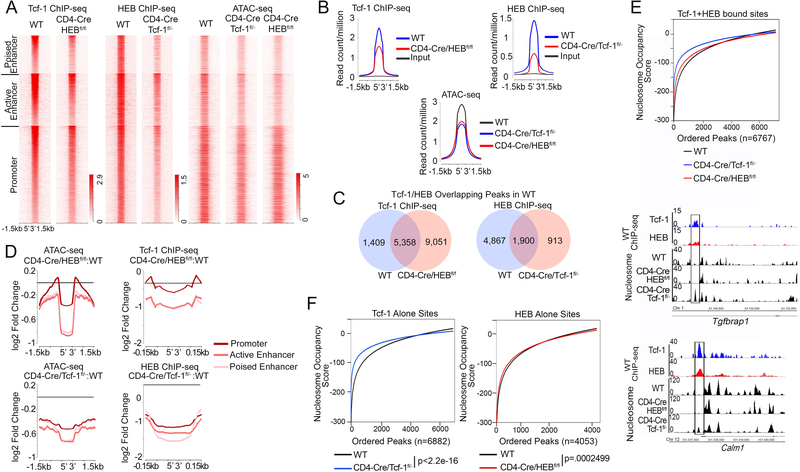
Tcf-1/HEB promote chromatin accessibility at co-bound sites
To determine the significance of the extensive sharing of binding sites between Tcf-1 and HEB, we first assessed the binding of Tcf-1 in CD4-Cre/HEBfl/fl and of HEB in CD4-Cre/Tcf-1fl/- thymocytes by ChIP-seq. Surprisingly HEB binding was reduced by more than 73% in the absence of Tcf-1; only 2,813 high-confidence (p=10e-5) HEB binding sites were identified in Tcf-1-deficient thymocytes, compared to 12,233 sites in WT thymocytes (Figure 5A, C). 84% of these sites were also bound by HEB in WT thymocytes while 441 were new. 68% of the HEB sites in CD4-Cre/Tcf-1fl/- thymocytes (1900/2813 Figure 5C) were co-bound by Tcf-1 and HEB in WT thymocytes, indicating that HEB binding at these sites does not require the presence of Tcf-1. In addition to the reduced number of HEB binding sites in CD4-Cre/Tcf-1fl/−. the average enrichment of HEB at the remaining sites was also greatly reduced compared to WT thymocytes (Figure 5A and B). HEB enrichment in CD4-Cre/Tcf-1fl/- DP-thymocytes was most reduced at poised-enhancer sites. followed by active- enhancer and then promoter sites (Figure 5D). Compared to HEB. the number of Tcf-1 binding sites was only moderately reduced in CD4-Cre/HEBfl/fl thymocytes from 16.377 to 14.409 sites. 10.007 of these remaining sites (70%) overlapped with Tcf-1 binding sites detected in WT thymocytes. while 4.402 were new (Figure 5 A, C). Although the majority of Tcf-1-bound sites were maintained in the absence of HEB. Tcf-1 enrichment in the remaining sites was reduced (Figure 5B). Similar to HEB. Tcf-1 binding was also primarily reduced at active and poised enhancers (Figure 5D). Thus. both Tcf-1 and HEB impact the enrichment of the other on DNA. however the drastic reduction in the number of HEB binding sites in CD4-Cre/Tcf-1fl/- cells may be an indirect effect of the absence of Tcf-1 (Figure 5B).
Figure 5: Tcf-1 and HEB Promote Chromatin Accessibility at Co-bound Sites.
A. Heatmap of Tcf-1 and HEB ChIP-seq enrichment and chromatin accessibility (ATAC-seq) in thymocytes from WT, CD4-Cre/Tcf-1fl/-, and CD4-Cre/HEBfl/fl mice (±1.5 kb around overlapping WT Tcf-1 and HEB sites). ATAC-seq was performed in sorted DPs. B. Histogram of Tcf-1 enrichment in WT and CD4-Cre/HEBfl/fl thymocytes (top left), HEB enrichment in WT and CD4-Cre/Tcf-1fl/- thymocytes (top right) and chromatin accessibility (bottom, ATAC-seq) in WT, CD4-Cre/Tcf-1fl/- and CD4-Cre/HEBfl/fl DPs. Plots are centered on overlapping WT Tcf-1 and HEB sites (±1.5 kb). C. Venn diagrams comparing the number of Tcf-1 bound sites in CD4-Cre/HEBfl/fl thymocytes that overlap with WT Tcf-1/HEB co-bound sites (left), and HEB bound sites in CD4-Cre/Tcf-1fl/- thymocytes that overlap with WT Tcf-1/HEB co-bound sites (right). D. Histograms of log2 fold change in accessibility (ATAC-seq) and transcription factor enrichment in the indicated mutants compared to WT thymocytes, at genomic regions centered on Tcf-1/HEB overlapping sites in WT thymocytes (±1.5 kb). E. (Upper) Nucleosome occupancy scores at Tcf-1/HEB overlapping sites in WT thymocytes. Negative values indicate nucleosome absence and positive values indicate the presence of nucleosomes (WT= −50.1028, HEBfl/fl=−46.2251, CD4-Cre/Tcf-1fl/-=−30.5216). (Lower) Representative tracks of nucleosome appearance in the absence of Tcf-1 within Tcf-1 and HEB overlapping sites at the Tgfbrap1 and Calm1 loci. F. (Left) Nucleosome occupancy scores in WT and CD4-Cre/Tcf-1fl/- DP thymocytes at WT Tcf-1 binding sites without HEB. (Right) Nucleosome occupancy scores in WT and CD4-Cre/HEBfl/fl DP thymocytes at WT HEB binding sites without Tcf-1. P-values were determined by the Mann-Whitney-Wilcoxon Test.
Tcf-1 has multiple epigenetic as well as chromatin conformation functions 18 and a recent study suggested that Tcf-1 plays an active role in defining chromatin accessibility throughout T-cell development21. HEB has been shown to bind acetyltransferases. including p300. but its direct function in shaping the chromatin landscape is unknown23. Therefore. we examined the effect of Tcf-1 and HEB on the chromatin landscape. We compared chromatin accessibility in WT and Tcf-1- or HEB-deficient DP-thymocytes using ATAC-seq. We found that 51.452 (p<1.0e-5) sites were accessible in WT cells. which was reduced by deletion of either Tcf-1 (48.479 sites) or HEB (42.237 sites). In WT accessible regions. two of the top five enriched motifs were Tcf-1 (p=1e-198) and HEB (p=1e-274) motifs (Supplemental Figure 5A). Tcf-1 (p=1e-82) and HEB (p=1e-112) motifs were also among the five most enriched motifs (Supplemental-Figure 4D) in the 7,241 sites that were accessible in WT but lost accessibility in Tcf-1- or HEB-deficient thymocytes. However, accessible regions in CD4-Cre/Tcf-1fl/-, and CD4-Cre/HEBfl/fl Ps were relatively depleted for Tcf-1 and HEB conserved motifs compared to WT (Supplemental-Figure 5B, C). Importantly, regions that gained accessibility in Tcf-1- or HEB-deficient DP-thymocytes were not enriched for Tcf-1 or HEB motifs WT (Supplemental-Figure 5F, G). Additionally, novel HEB and Tcf-1 peaks detected in Tcf-1 and HEB-deficient thymocytes respectively were not at these newly accessible regions that lack Tcf-1 and HEB motifs. These findings demonstrate that Tcf-1 and HEB promote accessibility at Tcf-1/HEB motif-containing sites. Moreover, they indicate that E2A, the partner of HEB, does not significantly compensate for the loss of accessibility at HEB motif containing sites.
Genomic sites bound by Tcf-1 or HEB in WT thymocytes were less accessible in HEB- or Tcf-1-deficient DP-thymocytes respectively and binding enrichment was greatly reduced (Supplemental-Figure 6). Importantly, Tcf-1 and HEB co-bound sites also had reduced accessibility in the absence of either Tcf-1 or HEB (Figure 5A, B). The degree of accessibility-loss was more pronounced at co-bound enhancers, particularly active enhancers (Figure 5D), where Tcf-1 and HEB binding enrichments are highest (Figure 4C). The novel finding that HEB deletion reduces chromatin accessibility is consistent with its known interaction with p30023 and establishes that HEB has genome-wide epigenetic functions.
We next examined whether reduced chromatin accessibility in the absence of Tcf-1 or HEB also reflects changes in the nucleosome landscape. Nucleosome tracks were generated from paired-end sequenced ATAC-seq of WT, CD4-Cre/Tcf-1fl/-, and CD4-Cre/HEBfl/fl DP-thymocytes. The nucleosome occupancy at Tcf-1/HEB co-bound sites was calculated using the UCSC tool bigWigAverageOverBed. This tool assigns a probability score for the presence of a nucleosome at each site. Positive values indicate high probability and negative values indicate low probability. Consistent with the reduced chromatin accessibility, the probability that nucleosomes occupied Tcf-1/HEB co-bound sites increased in Tcf-1- and HEB-deficient DP-thymocytes compared to WT (Figure 5E). However, while the probability of nucleosome occupancy compared to WT was dramatically increased in Tcf-1-deficient DP-thymocytes (p<2.2e-16) the increase in HEB-deficient DP thymocytes was only marginally significant (p=0.0012) (Figure 5E). De novo nucleosome occupancy at co-bound Tcf-1 and HEB sites in Tcf-1 deficient thymocytes is also shown for the Tgfbrap1 and Calm1 genes (Figure 5E). This finding is in line with a recent report demonstrating that Tcf-1 can shift nucleosomes21. The dominant role of Tcf-1 in controlling nucleosome occupancy was also confirmed for sites uniquely bound by Tcf-1 or by HEB in WT thymocytes (Figure 5F). Nucleosome occupancy at sites bound by Tcf-1-alone increased dramatically in Tcf-1-deficient DP thymocytes compared to WT (p<2.2e-16). However, at sites bound by HEB-alone, this increase in HEB-deficient thymocytes was only marginally significant (p=0.00025) (Figure 5F). Overall, our findings show that Tcf-1 and HEB coordinately regulate chromatin accessibility, while Tcf-1 has a dominant role in controlling the presence of nucleosomes at co-bound sites in DP-thymocytes.
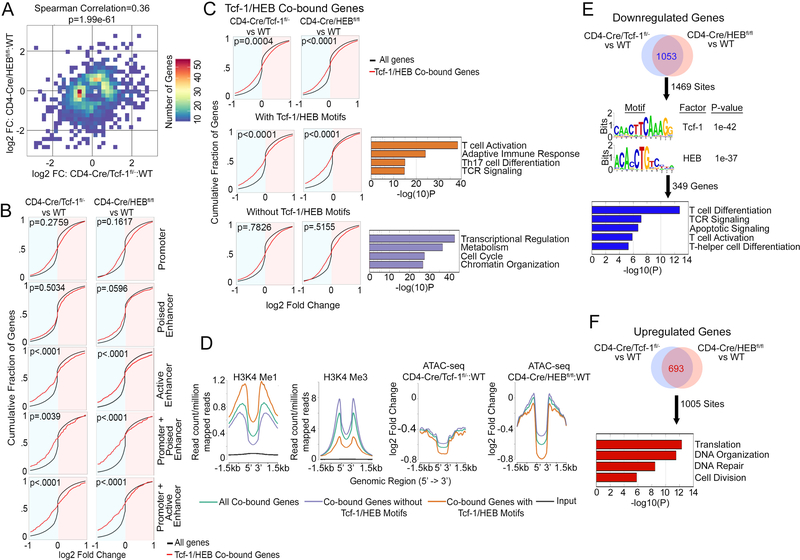
Co-binding of Tcf-1 and HEB to their cognate sites promotes gene expression
Several DP-thymocyte processes depend on the presence of Tcf-1 and HEB (Figure 3). Therefore, we investigated whether the overlapping binding of Tcf-1 and HEB in DP-thymocytes (Figure 4) reflects their ability to regulate overlapping transcriptional programs. Expression changes resulting from the deletion of Tcf-1 were compared to changes resulting from HEB-deletion. We identified significant (p<0.05) gene-expression changes in both Tcf-1- (n=1,269) and HEB- (n=838) deficient DP-thymocytes compared to WT. Spearman’s Rank Correlation comparison of the two sets established that the transcriptional changes associated with Tcf-1-deletion mirrored the transcriptional changes associated with loss of HEB (Spearman Correlation=0.36, p-value=1.99e-61, Figure 6A). This establishes that through their extensive sharing of binding sites and epigenetic functions, Tcf-1 and HEB cooperatively regulate the transcriptional profile of DP-thymocytes.
Figure 6: Tcf-1 and HEB Cooperatively Influence DP-Thymocyte Gene Expression.
A. Spearman’s Correlation analysis performed on genes that significantly changed expression between WT vs CD4-Cre/HEBfl/fl DPs (y-axis), and WT vs CD4-Cre/Tcf-1fl/- DPs (x-axis). Positive Spearman’s correlation, 0.36, indicates changes in the same direction. FC= fold change. B. Cumulative Distribution Function (CDF) plot of expression changes in CD4-Cre/Tcf-1fl/- versus WT (left column) or CD4-Cre/HEBfl/fl versus WT (right column) of genes bound by Tcf-1/HEB in WT. Black curves are expression changes of all genes expressed in DPs, red curves indicate genes bound by Tcf-1/HEB in indicated genomic regions. log2 expression changes are shown, P-values indicate significance of difference between red and black curves. P-values were determined using the Kolmogorov-Smirnov statistical test. C. CDF plot of expression changes as in B of all genes bound by Tcf-1/HEB in WT (top), genes bound by Tcf-1/HEB at sites that contain their conserved motifs (middle) and genes bound by Tcf-1/HEB at sites that do not contain their conserved motifs (lower). Pathways enriched within genes containing or lacking motifs are shown next to the relevant CDF plot. P-values were determined using the Mann-Whitney statistical test. D. Comparative enrichment histogram plots of the indicated histone marks and changes in chromatin accessibility centered on Tcf-1/ HEB binding sites (±1.5 kb) at the indicated sites. E. Number of overlapping downregulated genes (top) in Tcf-1 or HEB-deficient DP thymocytes (RNA-seq) within 20kb of Tcf-1/HEB co-bound sites identified using Binding and Expression Target Analysis (BETA). (Middle) Enriched motifs in the Tcf-1/HEB co-bound sites (1469) within 20kb of downregulated genes. (Bottom) Pathways enriched within downregulated genes with Tcf-1/HEB motifs identified by BETA (349 genes). F. Number of overlapping upregulated genes (top) identified by BETA (693 within 20kb of Tcf-1/HEB co-bound sites (1005 sites). (Bottom) Pathways enriched in upregulated genes.
Ablation of either Tcf-1 or HEB limited binding of the other factor to DNA and reduced chromatin accessibility particularly at active enhancer sites (Figure 5). To determine whether Tcf-1 and HEB binding at distinct regulatory regions differentially modulates gene transcription, we compared expression changes in WT versus Tcf-1- or HEB-deficient DP-thymocytes according to the region bound. Genes co-bound by Tcf-1/HEB in WT thymocytes were divided into clusters exhibiting promoter-only, enhancer-only (poised, or active), or both promoter and enhancer (poised, or active) binding. Expression changes in WT versus CD4-Cre/Tcf-1fl/- and WT versus CD4-Cre/HEBfl/fl DPs within these clusters were analyzed by cumulative distribution function (CDF) plots and compared to expression changes of all genes (Figure 6B). Tcf-1/HEB binding at promoter-only or poised enhancer-only was equally likely to confer up- or down-regulation of genes. Notably, Tcf-1 and HEB binding at active enhancer or both enhancer and promoter sites was significantly more likely to promote upregulation of the associated gene. Thus, DNA binding, epigenetic, and transcriptional analyses cumulatively establish that Tcf-1/HEB binding to active enhancers or enhancers and promoters increases chromatin accessibility and promotes expression of the associated genes.
We found that Tcf-1 and HEB promote accessibility at Tcf-1/HEB motif-containing sites (Supplemental-Figure 4). However, not all Tcf-1 and HEB co-bound sites contain Tcf-1 and HEB motifs. Therefore we tested whether transcription of gene targets was dependent on Tcf-1 and HEB binding to their motifs. Co-bound regions were subdivided into two clusters based on whether or not they contained Tcf-1 and HEB motifs. Expression changes of genes associated with each cluster in WT versus CD4-Cre/Tcf-1fl/- and WT versus CD4-Cre/HEBfl/fl were compared to expression changes of all genes using CDF analyses (Figure 6C). Genes associated with motif containing Tcf-1 and HEB peaks were significantly more likely to become downregulated upon ablation of either Tcf-1 or HEB and were enriched for T-cell development pathways (Supplemental-Table 1). However, genes associated with Tcf-1 and HEB peaks that lacked conserved Tcf-1/HEB motifs did not show consistent changes and were enriched for cell cycle and chromatin processes (Supplemental Table 2). Our two independent CDF analyses showed that Tcf-1 and HEB co-binding to active enhancers or to their conserved motifs promotes gene expression. To link these findings, we compared the chromatin landscape of co-bound regions containing Tcf-1/HEB motifs to regions lacking such motifs. Enrichment of the enhancer histone mark H3Kme1 was higher while enrichment of the promoter mark H3Kme3 was lower in sites containing Tcf-1 and HEB motifs (Figure 6D). Thus, binding of Tcf-1/HEB to sites that contain their conserved motif located predominantly at enhancer regions promotes the expression of the corresponding genes.
Our unsupervised analyses linked Tcf-1 and HEB co-binding to their conserved motif with gene upregulation. To obtain independent evidence for this finding we performed Binding and Expression Target Analysis (BETA) 41. Using BETA, we integrated Tcf-1 and HEB ChIP-seq data in WT with RNA-seq in WT versus Tcf-1- or HEB-deficient DP-thymocytes and identified shared peaks within 20kb of the TSS for genes that changed expression in the absence of Tcf-1 or HEB. The 1053 genes commonly downregulated in the absence of Tcf-1 or HEB represented 1,469 of the 6,767 Tcf-1/HEB co-bound sites (Figure 6E, top). Additionally, the 693 commonly upregulated genes represented 1005 of the Tcf-1/HEB co-bound sites (Figure 6F, top). Consistent with the unsupervised CDF analyses, motif enrichment at shared sites differentiated downregulated from upregulated genes. Only downregulated genes showed enrichment for Tcf-1 (p=1e-42) and HEB (p=1e-37) motifs (Figure 6E, middle). The conserved motifs were found in 349 genes that were involved in T-cell differentiation processes (Figure 6E, bottom). By contrast the 693 upregulated genes that lacked Tcf-1 and HEB motif enrichment were involved in proliferation and DNA repair processes. Overall, our analyses indicate that co-binding of Tcf-1/HEB to their conserved motifs predominantly at enhancer regions promotes expression of genes that are essential for DP-thymocyte development.
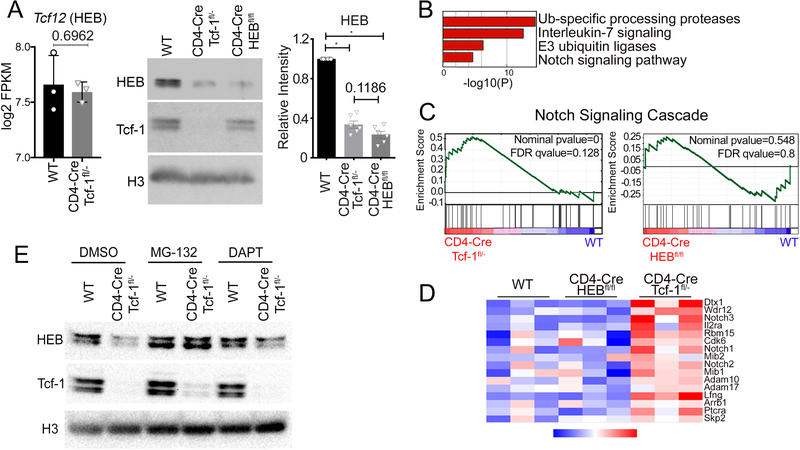
Tcf-1 regulates HEB stability by limiting Notch signaling
We showed that Tcf-1 and HEB coordinately regulate the chromatin landscape and transcription profiles of DP-thymocytes. However, it’s unclear why HEB binding was dramatically reduced in Tcf-1-deficient thymocytes while Tcf-1 binding was not as severely affected by deletion of HEB. HEB mRNA levels were similar between WT and Tcf-1-deficient T-cells (Figure 7A). Surprisingly western blot analyses showed that HEB protein was dramatically reduced (Figure 7A) to levels statistically comparable (p=0.1186) to the HEB protein levels in CD4-Cre/HEBfl/fl DP-thymocytes (Figure 7A). Notch signaling was previously reported to induce the ubiquitination of E2A, resulting in its degradation42, 43, 44. We investigated if a similar process reduced HEB protein levels in the absence of Tcf-1. Indeed, Notch and ubiquitination pathways were upregulated in Tcf-1-deficient DP-thymocytes (Figure 7B). Gene set enrichment analysis (GSEA) showed that the Notch Signaling cascade (Hallmark_Notch_signaling) was specifically upregulated in Tcf-1-deficient but not in HEB-deficient DP-thymocytes compared to WT (Figure 7C). Core components of the Notch cascade were significantly upregulated in the absence of Tcf-1 (Figure 7D). In particular our ChIP-Seq analyses showed that Tcf-1 bound alone without HEB to 103 regions of Notch pathway genes. Two thirds of these regions were at enhancer sites and their histone mark enrichment as well as accessibility patterns most closely corresponded to poised enhancers. However, Tcf-1 ablation did not limit accessibility at these sites to the same extend as co-bound Tcf-1 and HEB poised/active enhancer sites potentially allowing for transcription of the corresponding Notch pathway genes (Supplemental-Figure-7D).
Figure 7: Tcf-1 Promotes HEB Stability by Inhibiting Notch Signaling.
A. (Left) log2 FPKM of HEB transcripts in RNA-seq from sorted WT and Tcf-1-deficient DPs (n=3) P-value determined using a two-tailed unpaired t test. (Middle) Immunoblot analysis of HEB and Tcf-1 protein expression in WT, CD4-Cre/Tcf-1fl/-, and CD4-Cre/HEBfl/fl sorted DPs; Histone 3 (H3) as loading control. (Right) HEB protein quantification (Image Lab) in FACS sorted WT, CD4-Cre/Tcf-1fl/-, and CD4-Cre/HEBfl/fl DP thymocytes, normalized to WT quantities. N=6. P-values were determined using the Wilcoxon Signed Rank statistical test. B. Metascape pathway enrichment of genes only upregulated in CD4-Cre/Tcf-1fl/- DPs compared to WT DPs. C. Geneset Enrichment Analysis (GSEA) of the Notch Signaling Cascade (Hallmark_Notch_Signaling) in Tcf-1-deficient and HEB-deficient DPs. X-axis, genes are ranked from most upregulated (left end) to most downregulated (right end) between mutant and WT DPs. Y-axis indicates enrichment score of genes in pathway. D. Heatmap of the expression (RNA-seq) of Notch Signaling Cascade genes, in triplicate samples of WT, CD4-Cre/HEBfl/fl, and CD4-Cre/Tcf-1fl/- DPs. E. Immunoblot analysis of HEB and Tcf-1 protein levels in sorted DPs from WT and CD4-Cre/Tcf-1fl/- mice cultured with DMSO, MG132 (5μm), or DAPT (10μm) for 6 hours; Histone 3 (H3) as loading control.
To determine whether the upregulation of Notch and ubiquitin ligase pathways were responsible for the reduced HEB protein levels in Tcf-1-deficient thymocytes, we sorted WT and Tcf-1-deficient DP-thymocytes and assessed HEB protein levels following treatment with the proteasome inhibitor MG132 (5μM) or the Notch inhibitor DAPT (10μM) by immunoblot analysis. Both, proteasome inhibition by MG132 and Notch inhibition by DAPT, restored HEB protein levels in Tcf-1-deficient DPs (Figure 7E). These findings indicate that increased Notch Signaling in the absence of Tcf-1 promotes proteasomal degradation of HEB. Therefore, by inhibiting Notch signaling, Tcf-1 functions to stabilize HEB, allowing for their coordinated functions at the DP stage of thymocyte development.
Discussion
Thymocytes transitioning to the DP stage must cease proliferating to allow Rag- mediated rearrangement of the Tcrα gene and surface assembly of the αβ T-cell receptor45. The coordination of these events requires precise transcriptional and epigenetic reprograming of the incoming cells. Here, we identify Tcf-1 and HEB as cooperating partners in DP-thymocytes, regulating protein stability, chromatin accessibility, and gene expression. We show that Tcf-1 and HEB cooperate to halt the proliferation of early DP-thymocytes and promote survival and T-cell development. This cooperation involves extensive overlap of Tcf-1- and HEB-DNA binding. Tcf-1 and HEB promote chromatin accessibility while predominantly Tcf-1 minimizes nucleosome occupancy. Importantly, Tcf-1 stabilizes HEB levels by limiting the Notch-mediated proteasomal degradation of HEB protein.
Developmental programs depend on the coordinated actions of regulators and epigenetic organizers3, 4, 7, 8, 10, 46, 47. Here we show that Tcf-1, which reaches its highest expression levels in DP-thymocytes (Immgen), binds to the promoter and enhancer regions of highly expressed genes where it shares binding sites with other factors, including Runx1, Ikaros, and HEB. The essential functions of Tcf-14, 14, 15, 39, HEB16, 29, 48, 49, Runx134, and Ikaros50 in thymocyte development, and the differential abilities of these factors to modulate chromatin landscapes, suggest that they regulate this developmental process through a complex interplay. In early thymic development, Tcf-1 has been shown to coordinate with Notch1, Gata3, and Runx1, culminating in Bcl11b activation and T-cell commitment7. Furthermore, a recent report demonstrated that Tcf-1 can promote de novo chromatin opening 21.
Our study focused on the cooperation between Tcf-1 and HEB at the DP stage, where the two factors share an extensive number of binding sites as well as common developmental functions. Particularly, ablation of either Tcf-1 or HEB reduces DP- thymocyte survival, impacts NKT-cell development26, 27, and increases DP- thymocyte proliferation (Figure 3C). Tcf-1 and HEB share binding to sites that contain their conserved motifs predominantly at enhancer sequences of genes involved in T-cell development and positively co-regulate their expression. By contrast, Tcf-1 and HEB share binding sites that do not contain their conserved motifs and negatively co-regulate the expression of genes involved in proliferation.
Although HEB binds the histone acetyltransferase p30023, 51, it has not been directly shown to modulate the chromatin landscape. Here, we demonstrate that HEB promotes chromatin accessibility genome wide in DP-thymocytes. This epigenetic function of HEB is distinct from the epigenetic functions of Tcf-1 since HEB-deficient DP-thymocytes have reduced chromatin accessibility despite expressing normal levels of Tcf-1. Additionally, the accessibility-promoting functions of HEB and Tcf-1 are not complementary, since chromatin closing is comparable in HEB-deficient cells, which maintain Tcf-1 protein, and in Tcf-1-deficient cells, which also lose HEB protein expression. Moreover, E2A, the interacting partner of HEB22, 29, does not compensate for the epigenetic functions of HEB, since HEB-deficient DP-thymocytes specifically lose accessibility in regions containing the common HEB/E2A binding motif (Supplemental-Figure 4B). Consistent with promoting chromatin accessibility, predominantly Tcf-1, and marginally HEB, limit nucleosome occupancy in their co-bound sites. This is supported by the finding that, while genomic sites bound only by Tcf-1 have dramatically increased nucleosome presence after Tcf-1 deletion, HEB-only-bound sites show only a marginally significant increase upon loss of HEB. Altogether, our findings suggest that Tcf-1 and HEB coordinately shape the chromatin landscape of DP-thymocytes; they are both needed for promoting chromatin accessibility but they don’t have complementary effects. By contrast Tcf-1 has a dominant role over HEB in regulating the nucleosomal landscape.
Unexpectedly, we discovered that beyond Tcf-1 and HEB binding the same genomic locations and mediating epigenetic and transcriptional regulation, Tcf-1 directly regulates HEB protein stability. Tcf-1 accomplishes this by inhibiting Notch signaling, which targets HEB for ubiquitination and proteasomal degradation. This implicates a higher-level coordination whereby Tcf-1 controls the levels of Notch signaling and, in turn, Notch signaling controls HEB stability. Previous studies showed that Notch signaling targets E2A, a possible heterodimerization partner of HEB, for ubiquitination and proteasomal degradation42, 43, 44. Our finding that HEB levels are diminished in Tcf-1-deficient DPs and restored after inhibiting Notch activity or proteasomal degradation shows that Notch regulates HEB stability in a similar fashion as it regulates E2A stability. Notch signaling is progressively downregulated as cells advance from the DN to the DP stage, while Tcf-1 and HEB activities are essential for the transition into the DP stage. Our data suggest that Tcf-1 mediates the downregulation of Notch to facilitate stabilization of the HEB protein, which is essential for their coordinated actions promoting DP-thymocyte development.
Our studies offer a novel understanding of the complex regulatory network that controls the DP stage of thymocyte development. We demonstrate that Tcf-1/HEB binding to specific regulatory regions, promoters versus enhancers, identifies genes involved in distinct processes and differentially impacts their transcription and epigenetic status. Tcf-1 has an epistatic role in regulating common Tcf-1 and HEB functions through its ability to stabilize HEB levels by modulating Notch signaling. Tcf-1/HEB co-bound sites are enriched for different transcription factor motifs. In particular co-bound genes that are negatively impacted by the presence of Tcf-1 and HEB do not show enrichment of the conserved Tcf-1 and HEB motifs. This suggests that Tcf-1 and HEB may organize and/or participate in a potentially complex network of regulators that may change as cells enter and progress within the DP stage. Future studies will decipher the complex orchestration between Tcf-1 and Notch signaling, in regulating HEB. Additionally future studies will address the coordination of Tcf-1 and HEB with other lymphoid factors such as Runx1, Ikaros, and their combined impact on chromatin organization, gene expression and DP-thymocyte development.
Materials and Methods
Mice
CD4-Cre+xTcf-1fl/- (CD4-Cre/Tcf-1fl/-), CD4-Cre+xHEBfl/fl (CD4-Cre/HEBfl/fl) and littermate control WT (CD4-Cre+ or CD4Cre-), mice were used in all experiments described. Mice were maintained on the C57BL/6 background and experiments were performed with 6 to 8 week old mice. Mice were housed in the animal facilities at the University of Chicago in accordance with protocol #71880, approved by the University of Chicago Institutional Animal Care and Use Committee.
BrdU uptake experiments
Mice were injected intraperitoneally with 1.8mg of BrdU. 3 hours later, mice were euthanized and thymocytes were surface stained, followed by intracellular detection of BrdU incorporation using (8817–6600-42, eBioscience), following manufacturer’s protocol.
Cell culture and inhibitor treatment
Thymocytes from WT, CD4-Cre/Tcf-1fl/-, and CD4-Cre/HEBfl/fl mice and were labeled with fluorochrome-conjugated antibodies to CD4 (anti-CD4; 11–0042-82 eBiosciences) and CD8 (anti-CD8; 17–0081-82 eBiosciences) in FACS buffer (2% fetal bovine serum in PBS). Double positive thymocytes were sorted and cultured for 6 hours with DMSO, 5μg/ml MG-132 (m7449, Sigma), or 10μg/ml DAPT (D5942, Sigma).
Flow cytometry and antibodies
Thymocytes from mice were surface stained in FACS Buffer (2% FBS in PBS) for 30 minutes on ice. Samples were washed with FACS Buffer and acquired on an LSRII flow cytometer (Becton Dickinson). Data were analyzed with FlowJo software (Becton Dickinson). Antibodies were from CD4 (clone GK1.5, BD Biosciences), CD8 (clone 53–6.7, eBiosciences), Tcr-β (clone H57–597, eBiosciences), CD71 (clone R17217, BioLegend), Live/dead (L34957, Molecular Probes), BrdU (8811–6600-42, eBiosciences). An Annexin-V PE kit (556422, BD Biosciences) was used to measure apoptosis according to the manufacturer’s instructions.
Western Blot
Nuclear extracts from CD4+CD8+ T-cells were prepared with the Subcellular Protein Fractionation Kit (78840, Life Technologies) following the manufacturer’s instructions. Extracts were electrophoresed on a NuPAGE 4–12% Bis-Tris gel (NP0323, Invitrogen) and blotted. Membranes were probed with antibodies against Tcf-1 (2203, Cell Signaling), HEB/Tcf12 (SAB3500566, Sigma), and Histone 3 (2650, Cell Signaling).
mRNA isolation
1×107 CD4+CD8+ T-cells were sorted from 3 WT, Tcf-1fl/-, and CD4-Cre/HEBfl/fl mice each and total RNA was isolated using Trizol (15596026, Invitrogen) following the protocol described by the Immunological Genome Project (http://Immgen.org). Libraries were generated and sequenced by the University of Chicago Genomics Facility.
Chromatin immunoprecipitation (ChIP)
2×107 total thymocytes from 6–8 week old mice were paraformaldehyde fixed at a final concentration of 1% (2106–01, J.T. Baker) for 15 minutes at room temperature, quenched with glycine (0.125M) and washed with ice cold PBS containing protease inhibitors. Cells were resuspended in lysis buffer (10mM Tris (pH7.4), 1mM EDTA, 1% TritonX-100, 0.1% NaDOC, 0.8M NaCl, 0.1% SDS) for 10 minutes at 4°C and sonicated to an average size of 300bp. Chromatin was incubated overnight with Protein G Dynabeads (10004D, Invitrogen) coupled to 5ĝ of antibodies against Tcf-1 (2203, Cell Signaling), HEB/Tcf12 (SAB3500566, Sigma), Lef-1(2230, Cell Signaling), H3Ac (06–599, Millipore), H3K27Ac (ab4729, Abcam). Protocols for Ikaros, H3K4me1, H3K4me2, H3K4me3, H3K27me3, and RNA PolII were previously described 35 as was Runx1 52. Beads were washed 5x with lysis buffer (10mM Tris (pH7.4), 1mM EDTA, 1% TritonX-100, 0.1% NaDOC, 0.5M NaCl, 0.1% SDS) and once with 1X TE. Chromatin was eluted with elution buffer (2% SDS, 20mM Tris-HCl (pH6.8)) and reverse-crosslinked overnight at 65°C. RNaseA was added (50 μg/ml) and incubated at 37°C for 1 hour. Proteinase K was added to a final concentration of 240 μg/ml and incubated at 56°C for 2 hours. DNA was ethanol precipitated and resuspended in elution buffer (Qiagen).
ChIP-qPCR primers
Quantitative real time PCR was performed to assess HEB enrichment in DMSO or DAPT treated DPs from WT and Tcf-1fl/- mice using primers corresponding to regions co-bound by Tcf-1 and HEB: CCR9-F: 5’-TGC AAA GGC AAA GAT GAA AG-3’, CCR9-R: 5’-GCA GGA CAT GAA GGT GGA GT-3’; CD8-F: 5’-CTG TGC CTG AGT TTG TGA-3’, CD8-R: 5’-GGG AGT CTA GGG CAC AAT GA-3’; RORC-F: 5’-GGG TCC TGT CAC CAT TCC TC-3’, RORC-R: 5’-TTG ACC TTG ACT GGG ACA CA-3’; β-globin-F: 5’-GCC ATC GTT AAA GGC AGT TAT CA-3’, β-globin-R: 5’-TGC TAT CAT GGG TAA TGC CAA A-3’.
Tcra rearrangements
DPs from mice were sorted and RNA was extracted using Trizol (15596026, Invitrogen) and DNAse treated. cDNA was synthesized using a SuperScript III kit (18080093, Invitrogen). Quantitative real-time PCR was performed to assess the abundance of Tcra rearrangements using primers described previously 27.
Chromatin immunoprecipitation sequencing (ChIP-Seq)
ChIP material was prepared for sequencing in accordance with the Illumina/Solexa Genomic DNA protocol. Approximately 20ng of immunoprecipitated DNA was end repaired, polyadenylated, ligated to Illumina Truseq indexed adaptors, and purified with AMPure XP Beads (A63880, Beckman Coulter). Adaptor-ligated DNA was PCR amplified using KAPA Hifi DNA Polymerase (KK2601, Kapa Biosystems). PCR products were separated on a 2% agarose gel and DNA fragments between 200 and 500bp were excised and purified (28706, Qiagen). Libraries were sequenced on a HiSeq 4000 sequencer at the University of Chicago Genomics Facility.
Assay for transposase accessible chromatin and sequencing (ATAC-seq)
1×105 CD4+CD8+ T-cells sorted from WT, Tcf-1fl/- and CD4-Cre/HEBfl/fl mice were used for ATAC-seq. Cells were centrifuged at 500g at 4°C for 5 minutes, washed with 1X PBS, and centrifuged again. Cells were resuspended in lysis buffer (10mM Tris-HCl pH7.4,10mM NaCl, 3mM MgCl2, 0.1% IGEPAL CA-630) and immediately centrifuged at 500g at 4°C for 10 minutes. Pellets were resuspended in transposition reaction buffer (25μl 2x Tagment Buffer (FC-121–1030. Illumina). 2.5μ1 Tagment DNA Enzyme. 22.5^l nuclease free H2O) for 30 minutes at 37°C. DNA was purified with a Qiagen MinElute Kit and amplified with Nextera PCR Primers (Illumina Nextera Index Kit) and NEBNext PCR Master Mix (M0541. New England BioLabs) for 11 cycles. Amplified DNA was purified with a Qiagen PCR cleanup kit. Libraries were sequenced on a HiSeq 4000 sequencer at the University of Chicago Genomics Facility.
Genome mapping and data analysis
Sequenced ChIP data sets were mapped using the Galaxy (http://usegalaxy.org) suite of tools. Data were groomed and aligned to the mouse mm9 genome using Bowtie. allowing up to 1 mismatch. retaining only uniquely mapped reads. and unmapped reads were filtered. For transcription factors (Tcf-1. HEB. Runx1. Ikaros. Lef-1) peak calling was performed with MACS via HOMER 53. Transcription factor peak calling was performed relative to input controls with the requirement that peaks be at a minimum 5-fold enriched over input and meet a p-value cutoff of 10e-5. Transcription factor super-clusters were identified using HOMER’s findPeaks command using the –style factor option. following the strategy described by Whyte et al. 2013 40. This method combines transcription factor peaks within a 12.5kb region into a single cluster then ranks these regions by their score.
Sequenced RNA data sets were aligned to the mouse mm9 genome similarly to the ChIP-seq datasets. Differential expression analysis was performed with Cuffdiff 2 54. Genes with transcript abundance differences below p<.05 were considered to be significantly differentially expressed. Heat maps of normalized reads for gene subsets in WT, Tcf-1fl/-, and CD4-Cre/HEBfl/fl DP T-cells were generated with the Cluster software.
Motif analysis was performed using HOMER’s motif discovery algorithm and transcription factor overlap analysis was conducted with HOMER’s mergePeaks command, only considering peaks that directly overlapped. Peaks were annotated to the mm9 genome with annotatePeaks.pl in HOMER. Histograms for transcription factors and histone modifications were generated with the NGS.PLOT software 55. K-means clustering of ChIP-seq datasets and heat maps were also generated with NGS.PLOT.
Transcription factor binding and nucleosome positioning were visualized with the Integrated Genome Browser software 56. Pathway enrichment analysis for genes identified by ChIP-seq and RNA-seq analysis was performed via Metascape (57. http://metascape.org).
ATAC-seq peak analysis
Peak calling
Read alignments were first adjusted to account for TAC transposon binding: +4bp for + strand alignments, −5bp for - strand alignments. The open chromatin enrichment track was generated by first creating a bedGraph from the raw reads using bedtools genomcov 58, then converted to bigWig using the UCSC tool bedGraphToBigWig 59; tracks were normalized by the sum of alignment lengths over 1 billion. The start position track was generated by taking just the first base of the alignment for + strand alignments or the last base of the alignment for - strand alignments, then creating bedGraph and bigWig tracks as for the open chromatin; tracks were normalized to the alignment count over 1 million. Open chromatin peaks were called using Macs2 60 with --nomodel set and no background provided; peaks with a score >5 were retained.
Nucleosome positioning
Properly paired read pairs were first put into nucleosome-free or nucleosome-containing bins, based on their fragment size. Fragments <=100bp were considered nucleosome free (background) and converted to a single read covering the length of the fragment. Fragments 180–247bp were considered single-nucleosome-containing and converted to a single read the length of the fragment; similarly, fragments 315–473bp or 558–615bp were considered to contain two- or three-nucleosomes, and split into 2 or 3 reads covering half or 1/3 of the total fragment length. Single-, two-, and three- nucleosome reads were combined into the nucleosome signal read set. Nucleosome positioning analysis was run using danpos 61 with command depose and parameters -m 1 -a 10 -jd 20 --clonalcut 0, and contrasting nucleosome signal to nucleosome background for each sample. Wig tracks from danpos were re-normalized to counts per billion bases using the sum of alignment lengths over 1 billion from the original BAM file, and converted to bigWig using UCSC tool wigToBigWig 59.
Nucleosome quantification
Nucleosome presence or absence was determined using the UCSC tool bigWigAverageOverBed (https://genome.ucsc.edu/). Nucleosome occupancy was scored over genomic regions representing Tcf-1 and HEB binding sites and reported as confidence scores. Negative values indicated the absence of nucleosomes, while positive values represented an increased likelihood of the presence of a nucleosome at each region.
Density plots (Spearman Correlation)
Density plots were created using the stat_bin2d function in the ggplot2 package in R, with 30 bins in each dimension. For visualization purposes, the axis ranges of some density plots were limited to highlight the high-probability regions of the plot. Spearman correlation coefficients and p-values were computed in R using the cor and cor.test functions.
Supplementary Material
A. Representative CD71 expression versus FSC on gated WT, Tcf-1fl/+, CD4-Cre/Tcf-1fl/- and CD4-Cre/HEBfl/fl DP thymocytes. Numbers indicate the percent of cells in each quadrant. B. Cumulative data of CD71+FSChi DP thymocytes in indicated mice (n=3–10 mice). P-values determined by Ordinary one-way Anova statistical test.
A. K-means clustered heatmap centered on HEB binding sites in WT thymocytes (±1.5 kb) showing enrichment of indicated histone modifications, RNA PollI, and accessibility (ATAC-seq) in DP thymocytes. B. Comparative enrichment histograms of permissive histone modifications (H3Ac, H3K4me1, H3K4me2, H3K4me3 and H3K27ac) and chromatin accessibility (ATAC-seq) at HEB binding sites (±1.5 kb) in K-means clusters shown in A. C. Genomic distribution of HEB peaks in wildtype thymocytes. D. Average expression in DP thymocytes of genes in the indicated groups E=enhancer, P=promoter. Numbers are mean log2 FPKM. (****= p<0.0001). P-values determined by Kruskal-Wallis statistical test.
A. ChIP-seq tracks of Tcf-1 and HEB co-binding to the promoter of E2f5. B. ChIP-seq tracks of Tcf-1 and HEB co-binding to the poised enhancer of Ifng. C. ChIP-seq tracks of Tcf-1 and HEB co-binding at the active enhancer of Runx3. D. ChIP-seq tracks of Tcf-1 and HEB co-binding at the promoter and enhancer of Rag2. Relative enrichments of H3K4 Me1 and H3K4 Me3, as well as distance from the transcriptional start site were used in all tracks to identify the promoter, poised enhancer, and active enhancer of the associated gene.
A. Tcf-1 superclusters representing adjacent Tcf-1 binding sites in WT thymocytes within 12.5kb regions (Homer). Regions were ranked (x-axis) based on number of sequenced tags (y-axis) per region. The top 271 regions and genes annotated to these regions are shown in red. B. HEB superclusters representing adjacent HEB binding sites in HEB thymocytes within 12.5kb regions (Homer). Regions were ranked (x-axis) based on number of sequenced tags (y-axis) per region. The top 213 regions and genes annotated to these regions are shown in red. C. Overlapping Tcf-1 and HEB superclusters and pathways enriched (Metascape) within these clusters.
A. Motif enrichment analysis (HOMER) indicating the top 5 enriched motifs (P- value) in chromatin accessible sites (ATAC-seq) in sorted WT DP thymocytes (51, 452 sites, p<10e-5). B. Motif enrichment analysis indicating the top 5 enriched motifs in sorted CD4-Cre/HEBfl/fl DP thymocytes (42,237 sites, p<10e-5). C. Motif enrichment analysis indicating the top 5 enriched motifs in chromatin accessible sites in sorted CD4-Cre/Tcf-1fl/- DP thymocytes (48,479 sites, p<10e-5). D. Venn diagram indicating commonly accessible (34,866) and uniquely accessible sites in WT, CD4-Cre/HEBfl/fl, and CD4-Cre/Tcf-1fl/- DP thymocytes. E. Motif enrichment analysis indicating the top 5 enriched motifs in 7,241 sites only accessible in WT DP thymocytes. F. Motif enrichment analysis indicating the top 5 enriched motifs in 3,485 sites only accessible in CD4-Cre/HEBfl/fl DP thymocytes. G. Motif enrichment analysis indicating the top 5 enriched motifs in 4,295 sites only accessible in CD4- Cre/Tcf-1fl/- DP thymocytes.
A. Heat map of Tcf-1 ChIP-seq enrichment (left) and chromatin accessibility (right) in WT and CD4-Cre/HEBfl/fl thymocytes (±1.5 kb) around indicated WT Tcf-1 binding sites. ATAC-seq was performed in sorted DPs. B. Histogram representation of Tcf-1 enrichment (top) and chromatin accessibility (bottom) shown in A. C. Comparison of total Tcf-1 bound sites in WT and CD4-Cre/HEBfl/fl thymocytes. D. Heat map of HEB ChIP-seq enrichment (left) and chromatin accessibility (right) in WT and CD4-Cre/Tcf-1fl/- thymocytes (±1.5 kb) around indicated WT HEB binding sites. ATAC-seq was performed in sorted DPs. E. Histogram representation of HEB enrichment (top) and chromatin accessibility (bottom) shown in D. F. Comparison of total HEB bound sites in WT and CD4-Cre/Tcf-1fl/- thymocytes.
Tcf-1 and HEB binding within core Notch Signaling Cascade genes Dtx1 (A), Lfng (B), and Notch1 (C), that are significantly upregulated in CD4-Cre/Tcf-1fl/- DP thymocytes in comparison to WT or CD4-Cre/HEBfl/fl DP thymocytes. D. ChIP-seq enrichments of Tcf-1, H3K27 Ac, H3K4 Me1, and chromatin accessibility (ATAC-seq) at Tcf-1 binding sites in Notch Signaling/Ubiquitination genes compared to Tcf- 1/HEB co-bound promoters, poised enhancers, and active enhancers. E. log2 fold change in chromatin accessibility between WT and CD4-Cre/Tcf-1fl/- DP thymocytes at genomic regions described in D.
Acknowledgments
We thank H. Kawamoto for reagents, M. Mandal for help with the ATAC seq, B. Kee, M. Clark A. Khan and A. Bendelac for advice. M. Krishnan, is thanked for helpful suggestions. This work was supported by National Institutes of Health Grants R21AI076720, R01AI108682 and ASH bridge grant (to F.G.), R01CA160436 (to K.K). Further support came from the Chicago Biomedical Consortium (to F.G.); UL1TR002003 (to M. M-C); A.O.E. was supported by an NIH minority supplement; P.S.M. was supported by T32 HL07605 Institutional NRSA and is currently an LLS Fellow. J.Q. was supported by an AAI Careers in Immunology Fellowship, M.O. is a T32HD007009 recipient.
Footnotes
Data availability. The source data that support the findings of this study are available from the corresponding author upon request. RNA-seq, ChIP-seq and ATAC-seq data sets have been deposited in the GEO database with accession code SRP142342.
References
- 1.Klein L, Kyewski B, Allen PM & Hogquist KA Positive and negative selection of the T cell repertoire: what thymocytes see (and don’t see). Nat Rev Immunol 14, 377–391 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maillard I, Fang T. & Pear WS Regulation of lymphoid development, differentiation, and function by the Notch pathway. Annu Rev Immunol 23, 945–974 (2005). [DOI] [PubMed] [Google Scholar]
- 3.Weber BN et al. A critical role for TCF-1 in T-lineage specification and differentiation. Nature 476, 63–68 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Germar K. et al. T-cell factor 1 is a gatekeeper for T-cell specification in response to Notch signaling. Proc Natl Acad Sci U S A 108, 20060–20065 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garcia-Ojeda ME et al. GATA-3 promotes T-cell specification by repressing B-cell potential in pro-T cells in mice. Blood 121, 1749–1759 (2013). [DOI] [PubMed] [Google Scholar]
- 6.Scripture-Adams DD et al. GATA-3 dose-dependent checkpoints in early T cell commitment. J Immunol 193, 3470–3491 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kueh HY et al. Asynchronous combinatorial action of four regulatory factors activates Bcl11b for T cell commitment. Nat Immunol 17, 956–965 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kumata K. et al. Development of [(11)C]MFTC for PET imaging of fatty acid amide hydrolase in rat and monkey brains. ACS Chem Neurosci 6, 339–346 (2015). [DOI] [PubMed] [Google Scholar]
- 9.Hosokawa H. et al. Transcription Factor PU.1 Represses and Activates Gene Expression in Early T Cells by Redirecting Partner Transcription Factor Binding. Immunity 48, 1119–1134 e1117 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Longabaugh WJR et al. Bcl11b and combinatorial resolution of cell fate in the T-cell gene regulatory network. Proc Natl Acad Sci U S A 114, 5800–5807 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang D. et al. The basic helix-loop-helix transcription factor HEBAlt is expressed in pro-T cells and enhances the generation of T cell precursors. J Immunol 177, 109–119 (2006). [DOI] [PubMed] [Google Scholar]
- 12.Yucel R, Kosan C, Heyd F. & Moroy T. Gfi1:green fluorescent protein knock-in mutant reveals differential expression and autoregulation of the growth factor independence 1 (Gfi1) gene during lymphocyte development. J Biol Chem 279, 40906–40917 (2004). [DOI] [PubMed] [Google Scholar]
- 13.David-Fung ES et al. Transcription factor expression dynamics of early T- lymphocyte specification and commitment. Developmental biology 325, 444–467 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Verbeek S. et al. An HMG-box-containing T-cell factor required for thymocyte differentiation. Nature 374, 70–74 (1995). [DOI] [PubMed] [Google Scholar]
- 15.Schilham MW et al. Critical involvement of Tcf-1 in expansion of thymocytes. J Immunol 161, 3984–3991 (1998). [PubMed] [Google Scholar]
- 16.Barndt R, Dai MF & Zhuang Y. A novel role for HEB downstream or parallel to the pre-TCR signaling pathway during alpha beta thymopoiesis. J Immunol 163, 3331–3343 (1999). [PubMed] [Google Scholar]
- 17.Clevers H. Wnt/beta-catenin signaling in development and disease. Cell 127, 469–480 (2006). [DOI] [PubMed] [Google Scholar]
- 18.Mosimann C, Hausmann G. & Basler K. Beta-catenin hits chromatin: regulation of Wnt target gene activation. Nat Rev Mol Cell Biol 10, 276–286 (2009). [DOI] [PubMed] [Google Scholar]
- 19.Clevers H. & Nusse R. Wnt/beta-catenin signaling and disease. Cell 149, 1192–1205 (2012). [DOI] [PubMed] [Google Scholar]
- 20.Xing S. et al. Tcf1 and Lef1 transcription factors establish CD8(+) T cell identity through intrinsic HDAC activity. Nat Immunol 17, 695–703 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson JL et al. Lineage-Determining Transcription Factor TCF-1 Initiates the Epigenetic Identity of T Cells. Immunity 48, 243–257 e210 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kee BLE and ID proteins branch out. Nat Rev Immunol 9, 175–184 (2009). [DOI] [PubMed] [Google Scholar]
- 23.Williams CJ et al. The chromatin remodeler Mi-2beta is required for CD4 expression and T cell development. Immunity 20, 719–733 (2004). [DOI] [PubMed] [Google Scholar]
- 24.Huang Z. et al. Transcriptional regulation of CD4 gene expression by T cell factor-1/beta-catenin pathway. J Immunol 176, 4880–4887 (2006). [DOI] [PubMed] [Google Scholar]
- 25.Ioannidis V, Beermann F, Clevers H. & Held W. The beta-catenin--TCF-1 pathway ensures CD4(+)CD8(+) thymocyte survival. Nat Immunol 2, 691–697 (2001). [DOI] [PubMed] [Google Scholar]
- 26.Sharma A, Berga-Bolanos R. & Sen JM T cell factor-1 controls the lifetime of CD4+ CD8+ thymocytes in vivo and distal T cell receptor alpha-chain rearrangement required for NKT cell development. PLoS One 9, e115803 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.D’Cruz LM, Knell J, Fujimoto JK & Goldrath AW An essential role for the transcription factor HEB in thymocyte survival, Tcra rearrangement and the development of natural killer T cells. Nat Immunol 11, 240–249 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Steinke FC et al. TCF-1 and LEF-1 act upstream of Th-POK to promote the CD4(+) T cell fate and interact with Runx3 to silence Cd4 in CD8(+) T cells. Nat Immunol 15, 646–656 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jones-Mason ME et al. E protein transcription factors are required for the development of CD4(+) lineage T cells. Immunity 36, 348–361 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dose M. et al. beta-Catenin induces T-cell transformation by promoting genomic instability. Proc Natl Acad Sci U S A 111, 391–396 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li L. et al. A far downstream enhancer for murine Bcl11b controls its T-cell specific expression. Blood 122, 902–911 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Winandy S, Wu P. & Georgopoulos K. A dominant mutation in the Ikaros gene leads to rapid development of leukemia and lymphoma. Cell 83, 289–299 (1995). [DOI] [PubMed] [Google Scholar]
- 33.Tinsley KW et al. Ikaros is required to survive positive selection and to maintain clonal diversity during T-cell development in the thymus. Blood 122, 2358–2368 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Egawa T, Tillman RE, Naoe Y, Taniuchi I. & Littman DR The role of the Runx transcription factors in thymocyte differentiation and in homeostasis of naive T cells. J Exp Med 204, 1945–1957 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang J. et al. Harnessing of the nucleosome-remodeling-deacetylase complex controls lymphocyte development and prevents leukemogenesis. Nat Immunol 13, 86–94 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu M. et al. Direct recruitment of polycomb repressive complex 1 to chromatin by core binding transcription factors. Molecular cell 45, 330–343 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Okamura RM et al. Redundant regulation of T cell differentiation and TCRalpha gene expression by the transcription factors LEF-1 and TCF-1. Immunity 8, 11–20 (1998). [DOI] [PubMed] [Google Scholar]
- 38.Held W, Clevers H. & Grosschedl R. Redundant functions of TCF-1 and LEF-1 during T and NK cell development, but unique role of TCF-1 for Ly49 NK cell receptor acquisition. Eur J Immunol 33, 1393–1398 (2003). [DOI] [PubMed] [Google Scholar]
- 39.Yu S. et al. The TCF-1 and LEF-1 Transcription Factors Have Cooperative and Opposing Roles in T Cell Development and Malignancy. Immunity 37, 813–826 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Whyte WA et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 153, 307–319 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang S. et al. Target analysis by integration of transcriptome and ChIP-seq data with BETA. Nature protocols 8, 2502–2515 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nie L, Xu M, Vladimirova A. & Sun XH Notch-induced E2A ubiquitination and degradation are controlled by MAP kinase activities. The EMBO journal 22, 5780–5792 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nie L. et al. Notch-induced Asb2 expression promotes protein ubiquitination by forming non-canonical E3 ligase complexes. Cell research 21, 754–769 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li X, Gounari F, Protopopov A, Khazaie K. & von Boehmer H. Oncogenesis of T-ALL and nonmalignant consequences of overexpressing intracellular NOTCH1. J Exp Med 205, 2851–2861 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stritesky GL, Jameson SC & Hogquist KA Selection of self-reactive T cells in the thymus. Annu Rev Immunol 30, 95–114 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Baruch-Morgenstern NB et al. Paired immunoglobulin-like receptor A is an intrinsic, self-limiting suppressor of IL-5-induced eosinophil development. Nat Immunol 15, 36–44 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Adey A. et al. Rapid, low-input, low-bias construction of shotgun fragment libraries by high-density in vitro transposition. Genome Biol 11, R119 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jones ME & Zhuang Y. Acquisition of a functional T cell receptor during T lymphocyte development is enforced by HEB and E2A transcription factors. Immunity 27, 860–870 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wojciechowski J, Lai A, Kondo M. & Zhuang Y. E2A and HEB are required to block thymocyte proliferation prior to pre-TCR expression. J Immunol 178, 5717–5726 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Winandy S, Wu L, Wang JH & Georgopoulos K. Pre-T cell receptor (TCR) and TCR-controlled checkpoints in T cell differentiation are set by Ikaros. J Exp Med 190, 1039–1048 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Qiu Y, Sharma A. & Stein R. p300 mediates transcriptional stimulation by the basic helix-loop-helix activators of the insulin gene. Molecular and cellular biology 18, 2957–2964 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huang H. et al. Differentiation-dependent interactions between RUNX-1 and FLI-1 during megakaryocyte development. Molecular and cellular biology 29, 4103–4115 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Heinz S. et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Molecular cell 38, 576–589 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Trapnell C. et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nature protocols 7, 562–578 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shen L, Shao N, Liu X. & Nestler E. ngs.plot: Quick mining and visualization of next-generation sequencing data by integrating genomic databases. BMC genomics 15, 284 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Freese NH, Norris DC & Loraine AE Integrated genome browser: visual analytics platform for genomics. Bioinformatics 32, 2089–2095 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tripathi P, Guragain RP, Bhusal CL, Karna SL & Borgstein J. Response to the Letter to the Editor regarding “A comparison of two myringoplasty techniques in Nepalese children: A prospective randomized trial” by Tripathi et al. [Int. J. Pediatr. Otorhinolaryngol. 2015 (79) (9) 1556–1560]. Int J Pediatr Otorhinolaryngol 79 1967 (2015). [DOI] [PubMed] [Google Scholar]
- 58.Quinlan AR & Hall IM BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26 841–842 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kent WJ, Zweig AS, Barber G, Hinrichs AS & Karolchik D. BigWig and BigBed: enabling browsing of large distributed datasets. Bioinformatics 26 2204–2207 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang Y. et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol 9 R137 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen K. et al. DANPOS: dynamic analysis of nucleosome position and occupancy by sequencing. Genome research 23 341–351 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A. Representative CD71 expression versus FSC on gated WT, Tcf-1fl/+, CD4-Cre/Tcf-1fl/- and CD4-Cre/HEBfl/fl DP thymocytes. Numbers indicate the percent of cells in each quadrant. B. Cumulative data of CD71+FSChi DP thymocytes in indicated mice (n=3–10 mice). P-values determined by Ordinary one-way Anova statistical test.
A. K-means clustered heatmap centered on HEB binding sites in WT thymocytes (±1.5 kb) showing enrichment of indicated histone modifications, RNA PollI, and accessibility (ATAC-seq) in DP thymocytes. B. Comparative enrichment histograms of permissive histone modifications (H3Ac, H3K4me1, H3K4me2, H3K4me3 and H3K27ac) and chromatin accessibility (ATAC-seq) at HEB binding sites (±1.5 kb) in K-means clusters shown in A. C. Genomic distribution of HEB peaks in wildtype thymocytes. D. Average expression in DP thymocytes of genes in the indicated groups E=enhancer, P=promoter. Numbers are mean log2 FPKM. (****= p<0.0001). P-values determined by Kruskal-Wallis statistical test.
A. ChIP-seq tracks of Tcf-1 and HEB co-binding to the promoter of E2f5. B. ChIP-seq tracks of Tcf-1 and HEB co-binding to the poised enhancer of Ifng. C. ChIP-seq tracks of Tcf-1 and HEB co-binding at the active enhancer of Runx3. D. ChIP-seq tracks of Tcf-1 and HEB co-binding at the promoter and enhancer of Rag2. Relative enrichments of H3K4 Me1 and H3K4 Me3, as well as distance from the transcriptional start site were used in all tracks to identify the promoter, poised enhancer, and active enhancer of the associated gene.
A. Tcf-1 superclusters representing adjacent Tcf-1 binding sites in WT thymocytes within 12.5kb regions (Homer). Regions were ranked (x-axis) based on number of sequenced tags (y-axis) per region. The top 271 regions and genes annotated to these regions are shown in red. B. HEB superclusters representing adjacent HEB binding sites in HEB thymocytes within 12.5kb regions (Homer). Regions were ranked (x-axis) based on number of sequenced tags (y-axis) per region. The top 213 regions and genes annotated to these regions are shown in red. C. Overlapping Tcf-1 and HEB superclusters and pathways enriched (Metascape) within these clusters.
A. Motif enrichment analysis (HOMER) indicating the top 5 enriched motifs (P- value) in chromatin accessible sites (ATAC-seq) in sorted WT DP thymocytes (51, 452 sites, p<10e-5). B. Motif enrichment analysis indicating the top 5 enriched motifs in sorted CD4-Cre/HEBfl/fl DP thymocytes (42,237 sites, p<10e-5). C. Motif enrichment analysis indicating the top 5 enriched motifs in chromatin accessible sites in sorted CD4-Cre/Tcf-1fl/- DP thymocytes (48,479 sites, p<10e-5). D. Venn diagram indicating commonly accessible (34,866) and uniquely accessible sites in WT, CD4-Cre/HEBfl/fl, and CD4-Cre/Tcf-1fl/- DP thymocytes. E. Motif enrichment analysis indicating the top 5 enriched motifs in 7,241 sites only accessible in WT DP thymocytes. F. Motif enrichment analysis indicating the top 5 enriched motifs in 3,485 sites only accessible in CD4-Cre/HEBfl/fl DP thymocytes. G. Motif enrichment analysis indicating the top 5 enriched motifs in 4,295 sites only accessible in CD4- Cre/Tcf-1fl/- DP thymocytes.
A. Heat map of Tcf-1 ChIP-seq enrichment (left) and chromatin accessibility (right) in WT and CD4-Cre/HEBfl/fl thymocytes (±1.5 kb) around indicated WT Tcf-1 binding sites. ATAC-seq was performed in sorted DPs. B. Histogram representation of Tcf-1 enrichment (top) and chromatin accessibility (bottom) shown in A. C. Comparison of total Tcf-1 bound sites in WT and CD4-Cre/HEBfl/fl thymocytes. D. Heat map of HEB ChIP-seq enrichment (left) and chromatin accessibility (right) in WT and CD4-Cre/Tcf-1fl/- thymocytes (±1.5 kb) around indicated WT HEB binding sites. ATAC-seq was performed in sorted DPs. E. Histogram representation of HEB enrichment (top) and chromatin accessibility (bottom) shown in D. F. Comparison of total HEB bound sites in WT and CD4-Cre/Tcf-1fl/- thymocytes.
Tcf-1 and HEB binding within core Notch Signaling Cascade genes Dtx1 (A), Lfng (B), and Notch1 (C), that are significantly upregulated in CD4-Cre/Tcf-1fl/- DP thymocytes in comparison to WT or CD4-Cre/HEBfl/fl DP thymocytes. D. ChIP-seq enrichments of Tcf-1, H3K27 Ac, H3K4 Me1, and chromatin accessibility (ATAC-seq) at Tcf-1 binding sites in Notch Signaling/Ubiquitination genes compared to Tcf- 1/HEB co-bound promoters, poised enhancers, and active enhancers. E. log2 fold change in chromatin accessibility between WT and CD4-Cre/Tcf-1fl/- DP thymocytes at genomic regions described in D.