Abstract
目的
探讨lncRNA在胎盘植入中的表达特征及潜在的ceRNA调控机制。
方法
随机选取2017年12月~2018年6月于我院行剖宫产合并胎盘植入的植入部分胎盘组织与邻近正常胎盘组织各5例,采用基因芯片技术检测组织lncRNA的表达水平;植入组与对照组比较,筛选差异表达lncRNAs;随机选取5个差异表达lncRNAs进行实时荧光定量PCR检测,验证基因芯片结果的准确性与可靠性,对差异表达基因进行GO功能聚类分析及KEGG通路分析;选取芯片扫描与qRT-PCR结果均显示显著差异的ENST00000511361(RP5-875H18.4)、NR_027457(LINC00221)和NR_126415(FOXP4-AS1)3条lncRNAs构建ceRNA调控网络。
结果
植入组与对照组比较,筛选出329个差异表达lncRNAs与179个差异表达mRNAs,实时荧光定量PCR变化趋势与芯片结果相一致;差异表达mRNAs主要参与TGF-β通路的调控;ceRNA调控网络的构建提示RP5-875H18.4--miRNA-218-- SLIT2在胎盘植入的发生中存在潜在ceRNA调节机制。
结论
差异表达lncRNAs可能通过调控TGF-β通路参与胎盘植入的发生发展过程,RP5-875H18.4--miRNA-218--SLIT2在胎盘植入的发生中存在潜在ceRNA调节机制,但该结果还需进一步验证。
Keywords: 胎盘植入, lncRNA, ceRNA, 调控网络
Abstract
Objective
To investigate the expression profile of long non-coding RNAs (lncRNA) and identify potential lncRNA-related competing endogenous RNAs (ceRNA) in placenta accrete spectrum disorders (PAS).
Methods
Five tissue specimens of placental implantation and 5 adjacent normal placental tissues were collected from cesarean section deliveries complicated by PAS in our hospital between December, 2017 and June, 2018. Human microarrays were used to identify the lncRNAs that were differentially expressed in PAS, and 5 of the identified lncRNAs were further validated using qRT-PCR. GO and KEGG pathway analyses were performed to indentify the most significant enrichment functions. A ceRNA network was constructed based on ENST00000511361 (RP5-875H18.4), NR_027457 (LINC00221) and NR_126415 (FOXP4-AS1) to pinpoint the potential lncRNAs-related ceRNA.
Results
A total of 329 lncRNAs and 179 mRNAs were identified to have differential expression in PAS. The results of qRT-PCR were consistent with the human microarrays results. Transforming growth factor-β (TGF-β) signaling pathway was the most significantly enriched pathway. The constructed ceRNA network suggested that RP5-875H18.4--miRNA-218--SLIT2 had a potential ceRNA regulatory mechanism in PAS.
Conclusion
The differentially expressed lncRNAs are involved in the occurrence and progression of PAS possibly by regulating the TGF-β signaling pathway. The ceRNA network of RP5-875H18.4--miRNA-218--SLIT2 may play a role in the occurrence of PAS.
Keywords: long non-coding RNAs, placenta accreta spectrum, expression profile, competing endogenous RNA network
胎盘植入是胎盘绒毛异常侵入子宫肌层的一种产科并发症,根据胎盘绒毛侵入子宫肌层深度分为:胎盘粘连(胎盘绒毛黏附于子宫肌层表面),胎盘植入(胎盘绒毛深入子宫肌壁间未到达子宫浆膜层)以及穿透性胎盘植入(胎盘绒毛穿过子宫肌层到达或超过子宫浆膜层甚至侵入膀胱或者其他器官)[1]。胎盘植入是产科最严重的并发症之一,由于胎盘绒毛异常侵袭,胎盘难以从子宫分离,导致在尝试分离胎盘的过程中常引起大出血,大出血又会带来继发感染、多器官功能衰竭、DIC、失血性休克甚至产妇死亡[2]。胎盘植入的早期诊断有利于将此类型的危重孕产妇往高级别医院分流,对降低孕产妇不良结局意义重大。然而,目前胎盘植入的分娩前诊断主要依靠超声及影像学检查,到目前为止仍然有1/2~2/3的孕妇被漏诊[3]。为了提高孕产妇预后,寻找胎盘植入新的生物标志物,了解胎盘植入发生的分子机制将有助于合并胎盘植入孕产妇的早期诊断与治疗。
近年来的研究发现,lncRNA能在表达遗传调控、转录及转录后调控等多层面调控基因表达,并与靶基因构成复杂的调控网络,参与机体的病理生理过程[4-5],人类胎盘中发现的具有调节作用的lncRNA数量也急剧上升[6-7]。研究表明,许多胎盘lncRNA通过调节滋养细胞分化、增殖与迁移参与了子痫前期[8]、胎儿生长受限[9]、妊娠期糖尿病[10]、早产[11]、未足月胎膜早破[11]等疾病,有学者发现lncRNA MALAT1参与了胎盘植入[12],但是lncRNA在胎盘植入中的表达谱特征及调控网络尚未有完整研究报道。因此,本研究采用lncRNA芯片技术筛选胎盘植入患者差异表达lncRNA,并对相应的lncRNA进行基因调控网络分析,从而以lncRNA为切入点揭示胎盘植入发生的分子机制,为寻找胎盘植入的早期诊断生物标志物提供新的依据。
1. 资料和方法
1.1. 研究对象
随机选取2017年12月~2018年6月于我院行剖宫产合并胎盘植入的患者的植入胎盘组织与邻近的正常胎盘组织各5例,所有组织标本取样后用PBS缓冲液洗涤5遍,并于10 min内放入液氮中冷冻,液氮冻存3 h后转入-80 ℃冰箱中待测。纳入标准:①分娩方式为剖宫产;②手术诊断为胎盘植入且病理确诊为胎盘植入;③知情同意。排除标准:①病理诊断不确定胎盘植入;②胎儿畸形;③患者拒绝。本研究方案通过了本院的伦理委员会伦理审批,所有患者均签署了知情同意书。
1.2. 试剂和仪器
TRIzol试剂(Invitrogen),Rneasy Mini kit RNA提取试剂盒(Qiagen),人lncRNA芯片V4.0(Arraystar),杂交盒(Agilent),单色标记试剂盒(Agilent),基因表达杂交试剂盒(Agilent),Baseline-ZERO DNase(Epicentre),引物(英骏生物),-80 ℃超低温冰箱(ThermoFisher),液氮罐(ThermoFisher),低温离心机(Eppnedoff),G2565BA微阵列芯片扫描仪(Agilent),NanoDrop ND-1000紫外可见分光光度计(NanoDrop),ABI7900荧光定量PCR仪(ABI)。
1.3. RNA的提取和质量检测
将胎盘组织从-80 ℃超低温冰箱中取出,立即放入装有适量液氮的研钵中迅速研磨成粉末,然后加入TRIzol采用一步法提取细胞中的总RNA,使用NanoDrop ND-1000评估RNA量和质量,RNA完整性通过Agilent 2100生物分析仪进行评估,随后使用RNeasyMicro Kit和RNase-Free DNase Set纯化总RNA。
1.4. 基因芯片杂交和扫描
本研究采用的是Arraystar公司的人lncRNA芯片V4.0,委托上海康成生物工程有限公司进行。芯片能够检测40173个lncRNAs和20730个蛋白质编码转录本。将10个标本制作完成芯片10张,使用Arraystar RNA Flash Labeling Kit对样本进行标记,使用Agilent SureHyb进行杂交实验,然后洗涤芯片,洗涤后的芯片使用Agilent DNAMicroarray Scanner进行扫描。
1.5. 芯片数据的生物信息学分析
应用Agilent Feature Extraction V11.0.1.1软件采集芯片探针信号值,使用Agilent GeneSpring GXv12.1软件进行芯片数据均一化处理和差异分析,经过折叠倍率(fold change,FC)筛选出所有差异表达的lncRNAs和mRNAs,筛选标准为倍数变化>1.5,P < 0.05。对差异表达基因进行基因本体论(gene ontology,GO)分析,描述差异表达基因的功能属性;KEGG(Kyoto Encyclopedia of Genes and Genomes)分析差异表达基因的生物学途径;构建编码基因和非编码基因的共表达网络。
1.6. ceRNA network的构建
根据筛选出的差异表达lncRNAs,应用预测数据库miRcode匹配出与其相互作用的miRNA,应用miRNA靶基因预测数据库(miRDB、miRTarBase和TargetScan),筛选出靶基因。基于上述筛选出的lncRNA,miRNA与mRNA,采用cytoscape构架并绘制ceRNA调控网络。
1.7. 实时荧光定量PCR验证
采用TRIzol试剂提取lncRNA芯片组织剩余部分的RNA,进行实时定量聚合酶链反应(qRT-PCR)验证,然后使用SuperScript<sup>TM</sup> III逆转录酶(Invitrogen公司)生成第一链cDNAs。qRT-PCR采用ViiA 7 Real-time PCR系统(Applied Biosystems公司)和2*PCR master mix(Arraystar)进行相对表达水平测定,lncRNAs量化使用2ΔΔCt β-action方法计算。qRT-PCR引物的设计基于来自NONCODE(<a href="http://www.noncode.org" target="_blank">http://www.noncode.org</a>)的lncRNA序列,并在上海英骏生物技术有限公司合成与纯化,引物序列见<xref ref-type="table" rid="Table1">表 1</xref>。
1.
qRT-PCR验证的引物序列
Primers designed for qRT-PCR validation of the candidate lncRNAs
| Gene name | Primers (5' to 3') | Tm° | Lenth (bp) |
| Tm: Temperature; Bp: Base pair. | |||
| β | F: 5' GGGAAACTGTGGCGTGAT3' | 60 | 299 |
| NR_027457 | R: 5' GAGTGGGTGTCGCTGTTGA3' | 60 | 126 |
| NR_126415 | F: 5' CAGTAACTGTTGGTTGGGATGG 3' | 60 | 150 |
| NR_023387 | R: 5' GCCACTGAGATTGACAGGCTTA 3' | 60 | 83 |
| ENST00000442026 | F: 5' CTTCTGGGTTCGACAGTGGG 3' | 60 | 131 |
| ENST00000511361 | R: 5' TTCGGGATTGAGGGTTAGGG 3' | 60 | 97 |
2. 结果
2.1. 差异表达的lncRNA筛选结果
芯片一共检测了40 173 lncRNAs和20 730 mRNAs。其中在植入胎盘组织中差异表达的lncRNAs共有329个,其中101个lncRNAs表达上调,228个lncRNAs表达下调,差异表达倍率最大的各10个表达上调及表达下调的lncRNAs在表 2中列出。同时,我们筛选出差异表达的mRNAs共有179个,其中95个表达上调,84个表达下调,差异表达倍率最大的各10个表达上调及表达下调的mRNAs在表 3中列出。
2.
差异表达倍率最大的10组lncRNAs
Top 10 significantly dysregulated lncRNAs in PAS
| Seq name | Gene Symbol | Fold Chang | Lenth | Chr | P | Regulation |
| ENST00000602313 | AF003625.3 | 3.34 | 470 | chrX | 0.032 | Up |
| NR_026801 | FAM74A3 | 2.47 | 1883 | chr9 | 0.023 | Up |
| GSE61474_TCONS_00008727 | GSE61474_XLOC_001661 | 2.27 | 442 | chr1 | 0.039 | Up |
| T190418 | G043839 | 2.26 | 1114 | chr2 | 0.034 | Up |
| NR_038890 | LOC100132781 | 2.17 | 2138 | chr9 | 0.049 | Up |
| NR_027457 | LINC00221 | 2.08 | 1752 | chr14 | 0.02 | Up |
| T131415 | G030770 | 2.05 | 443 | chr16 | 0.001 | Up |
| ENST00000521991 | RP11-136O12.2 | 2.04 | 785 | chr8 | 0.011 | Up |
| ENST00000607665 | RP11-531A24.7 | 2.04 | 437 | chr8 | 0.027 | Up |
| NR_026803 | FAM74A1 | 2.03 | 623 | chr9 | 0.003 | Up |
| T160626 | G037111 | 3.87 | 4635 | chr18 | 0.004 | Down |
| ENST00000549951 | CTD-2311B13.2 | 3.69 | 381 | chr14 | 0.008 | Down |
| T145284 | G033813 | 3.68 | 1244 | chr17 | 0.001 | Down |
| T138116 | G032345 | 3.49 | 4337 | chr16 | 0.006 | Down |
| T255516 | G058891 | 3.4 | 603 | chr3 | 0.001 | Down |
| T292865 | G068537 | 3.33 | 8328 | chr5 | 0.02 | Down |
| T239943 | G055244 | 3.31 | 1192 | chr3 | 0.008 | Down |
| ENST00000442026 | AC009499.1 | 3.14 | 677 | chr2 | 0.022 | Down |
| T207150 | G047970 | 3.13 | 2245 | chr2 | 0.041 | Down |
| T238076 | G054648 | 2.85 | 1461 | chr3 | 0.01 | Down |
3.
差异表达倍率最大的10组mRNAs
Top 10 significantly dysregulated mRNAs in PAS
| Seq name | Gene symbol | Fold chang | Lenth | Chr | P | Regulation |
| NM_001010847 | LRRC38 | 3.14 | 1926 | chr1 | 0.003 | Up |
| NM_207328 | GPAT2 | 2.69 | 2840 | chr2 | 0.044 | Up |
| NM_005708 | GPC6 | 2.52 | 7114 | chr13 | 0.024 | Up |
| NM_003802 | MYH13 | 2.35 | 5992 | chr17 | 0.001 | Up |
| NM_153362 | PRSS35 | 2.26 | 2527 | chr6 | 0.048 | Up |
| NM_001031702 | SEMA5B | 2.13 | 4793 | chr3 | 0.045 | Up |
| NM_003283 | TNNT1 | 1.98 | 1259 | chr19 | 0.021 | Up |
| NM_000358 | TGFBI | 1.98 | 2805 | chr5 | 0.047 | Up |
| NM_001362 | DIO3 | 1.98 | 2120 | chr14 | 0.008 | Up |
| NM_021223 | MYL7 | 1.98 | 619 | chr7 | 0.039 | Up |
| NM_018712 | ELMOD1 | 3.04 | 3006 | chr11 | 0.002 | Down |
| NM_033132 | ZIC5 | 2.80 | 4607 | chr13 | 0.048 | Down |
| NM_007053 | CD160 | 2.43 | 1633 | chr1 | 0.029 | Down |
| NM_014465 | SULT1B1 | 2.15 | 1320 | chr4 | 0.003 | Down |
| NM_170776 | ADGRG3 | 2.12 | 2676 | chr16 | 0.001 | Down |
| NM_001199867 | MARK4 | 2.10 | 5163 | chr19 | 0.045 | Down |
| NM_153038 | CCDC140 | 2.08 | 1699 | chr2 | 0.004 | Down |
| NM_017413 | APLN | 2.08 | 3238 | chrX | 0.018 | Down |
| NM_173493 | PASD1 | 2.03 | 3250 | chrX | 0.050 | Down |
| NM_001114598 | ASPDH | 2.02 | 1061 | chr19 | 0.012 | Down |
2.2. PCR验证结果
随机选取5个差异表达的lncRNAs用定量PCR对基因芯片的结果进行验证。结果显示,5条用于PCR验证的lncRNAs中NR-027457表达上调(P < 0.01),NR-023387表达上调(P < 0.05),NR-126415、ENST00000442026和ENST00000511361均表达下调(P < 0.01,图 1)。qRTPCR结果与芯片扫描结果具有很好的一致性,表明芯片扫描具有很好的可靠性和可重复性。
1.
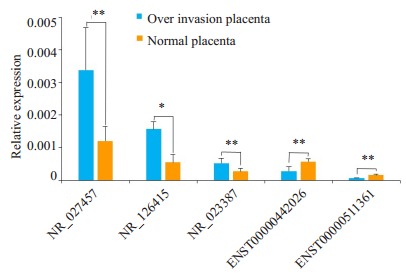
实时定量PCR验证基因芯片结果
Validation of the microarray data by quantitative RT-PCR. The relative expression levels of 5 dysregulated lncRNAs in invasive placenta are compared with those in adjacent normal placental specimens. NR_027457 and NR_023387 are up-regulated lncRNAs, and NR_126415, ENST00000442026 and ENST00000511361 are down-regulated lncRNAs. Data are presented as Mean±SD. *P < 0.05, **P < 0.01 by paired Mann-Whitney U test
2.3. 聚类分析
对差异表达基因进行GO功能聚类分析,可以了解相关基因主要参与的生命过程、细胞成分与分子功能。表达上调基因主要聚集的生物过程为multicellular organismal process(GO:0032501);主要聚集的细胞成分为proteinaceous extracellular matrix(GO:0005578);主要聚集的分子功能为receptor binding(GO:0005102)。前10功能聚类在表 4中列出,并且我们发现,所有聚类靠前的基因均为上调基因。
4.
前10功能聚类
The top 10 enriched functions
| GO.ID | Term | Ontology | Enrichment.Score | P |
| BP: Biological process; CC: Cellular component; MF: Molecular function. | ||||
| GO:0005578 | Proteinaceous extracellular matrix | CC | 8.88 | < 0.001 |
| GO:0031012 | Extracellular matrix | CC | 8.28 | < 0.001 |
| GO:0044420 | Extracellular matrix component | CC | 5.85 | < 0.001 |
| GO:0032501 | Multicellular organismal process | BP | 5.74 | < 0.001 |
| GO:0005604 | Basement membrane | CC | 5.33 | < 0.001 |
| GO:0044421 | Extracellular region part | CC | 5.13 | < 0.001 |
| GO:0019932 | Second-messenger-mediated signaling | BP | 4.93 | < 0.001 |
| GO:0007275 | Multicellular organism development | BP | 4.81 | < 0.001 |
| GO:0005102 | Receptor binding | MF | 4.51 | < 0.001 |
| GO:0006936 | Muscle contraction | BP | 4.36 | 0.00 |
此外,为了解差异表达基因聚集的分子调节通路,我们还进行了KEGG通路分析。结果显示,上调基因主要聚集的通路为TGF-beta signaling pathway(ko04350);下调基因主要聚集在cAMP signaling pathway(ko04024)。
2.4. ceRNA调控网络构建
为更深入了解差异表达lncRNA是否通过ceRNA机制参与了胎盘植入的形成,我们挑选了芯片扫描与qRT-PCR结果均显示显著差异的ENST00000511361(RP5-875H18.4)、NR_027457(LINC00221)和NR_ 126415(FOXP4-AS1)3条lncRNAs进行构建,最终获得的ceRNA network包含了3条lncRNAs、47条mRNAs与325个miRNAs(图 2)。
2.
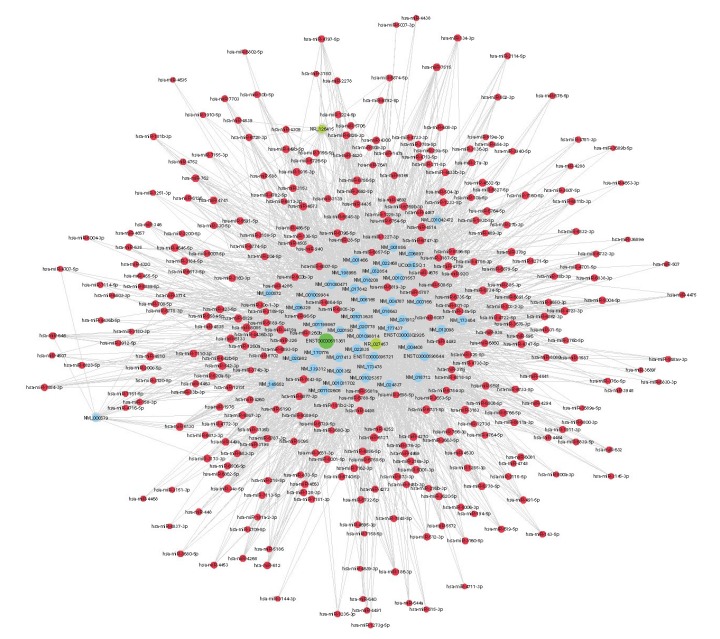
基于lncRNA/miRNA/mRNA间的相互作用的ceRNA调控网络
The ceRNA network constructed based on lncRNA/miRNA/mRNA interactions. The lncRNAs are indicated by green circles, miRNAs by red circles, and mRNAs by blue circles
3. 讨论
lncRNA参与细胞的生命活动,其表达异常与多种疾病的发生发展相关。既往研究曾报道lncRNA MALAT1参与胎盘植入的发生[12],lncRNA通过与miRNA竞争mRNAs结合位点调节mRNA表达[13],可能影响胎盘植入的发生发展,但目前仍未有对胎盘植入lncRNA表达特征全面深入的研究。在本研究中,我们发现lncRNA和mRNA在植入胎盘组织和正常胎盘组织之间的表达存在明显差异。在植入部分胎盘组织中,共筛选出329个差异表达lncRNAs(101个上调,228个下调,倍数变化>1.5,P < 0.05)和179个差异表达mRNA(95个上调,84个下调,倍数变化>1.5,P < 0.05),表明lncRNAs可能在胎盘植入的形成过程中发挥了重要作用。为了验证我们的基因芯片结果的可靠性和可重复性,随机选取了2个上调的和3个下调的lncRNA在5对标本中进行实时荧光定量PCR检测,qRT-PCR的结果与芯片扫描结果是一致的,表明芯片扫描结果具有很好的可靠性。
我们对差异表达基因进行GO功能聚类分析显示,所有失调基因的前三个功能都富集在蛋白质细胞外基质、细胞外基质(ECM)和细胞外基质成分。在生物学中,ECM是由细胞合成并分泌到胞外、分布在细胞表面或细胞之间的大分子,其支持并连接组织结构、调节组织的发生和细胞的生理活动,其主要组成包括胶原蛋白、弹性蛋白、蛋白多糖和糖蛋白等[14]。在既往的研究中已发现ECM与胎盘和滋养细胞的侵袭相关,如Haimov-Kochman等[15]发现一种名叫酸乙酰肝素蛋白多糖的ECM成分,在胎盘和蜕膜中富集,它们的降解可以促进胎盘和滋养细胞的侵袭。Borbely等[16]研究表明,一些ECM组分如decorin和biglycan是富含亮氨酸的蛋白多糖家族的成员,它们与胎盘侵袭性疾病相关。因此,ECM可能在胎盘植入过程中发挥重要作用。然而,既往研究中尚未发现多细胞生物体过程和胎盘基底板与胎盘植入、滋养细胞侵袭的关系,本研究中GO功能分析结果表明,多细胞生物体过程和胎盘基底板与胎盘植入的发生密切相关。Kuo等[17]研究报道,胎盘基底板中超过80%为ECM蛋白组成的基底膜蛋白,而对于有效的细胞滋养层侵袭是必不可少的,我们推测胎盘基底板通过基底膜蛋白影响胎盘植入的发生。而关于多细胞生物体过程与胎盘和滋养细胞侵袭的研究国内外尚未有相关报道,其作用机制有待于我们进一步研究。
本研究中,KEGG通路分析的结果与GO功能聚类分析的结果是一致的,DEmRNA的KEGG通路分析显示,6种上调的通路和11种下调的通路共同参与胎盘植入的发生发展,对于上调的mRNA,最主要聚集的通路为TGF-β通路。目前的研究发现,TGF-β通路调节许多细胞过程,包括上皮间质转化(EMT)[18-20]、ECM再生和胎盘形成[21]。虽然尚缺乏关于lncRNA调控胎盘植入发生发展的研究报道,但基于本研究提供的线索,差异表达lncRNA可能通过调控TGF-β通路,调节ECM再生和胎盘形成,从而促进胎盘植入的发生发展。
越来越多的研究发现,ceRNA在许多生物过程中起着关键作用,尤其是癌细胞的增殖、迁移和侵袭[22-24],至今已经发现超过10 000个lncRNA作为潜在的ceRNA起作用[25]。本研究挑选了芯片扫描与qRT-PCR结果均显示显著差异的ENST00000511361(RP5- 875H18.4)、NR_027457(LINC00221)和NR_126415(FOXP4-AS1)3条lncRNAs进行构建,成功构建了基于lncRNA/ miRNA/mRNA间的相互作用的ceRNA调控网络,与正常胎盘组织相比,通过基因芯片检测和qRT-PCR证实RP5-875H18.4(lncRNA-ENST00000511361)在胎盘植入中表达降低,RP5-875H18.4(lncRNA-ENST00000511361)作为表达下调的第top26的lncRNA,起到上调SLIT2的作用。既往的研究已表明,SLIT2受miRNA-218调节,miRNA-218是RP5-875H18.4和SLIT2共有的miRNA之一[26],SLIT2/ROBO信号调节滋养细胞分化[27]、血管再生[28-29]和侵袭过程[27-28],并在糖尿病[30]、子痫前期[29]、异位妊娠[28]和先兆流产[27]中起着重要作用。因此我们推测RP5-875H18.4-miRNA-218-SLIT2在胎盘植入的发生中存在潜在ceRNA调节机制,该ceRNA网络中的所有RNA都有可能成为胎盘植入诊断的生物标志物,其在胎盘植入过程中的潜在作用需要在将来研究中进一步验证。
综上所述,本研究找到的差异表达lncRNAs和mRNAs可能参与了胎盘植入的发生,差异表达lncRNAs可能通过调控TGF-β通路,调节ECM的生成与胎盘的形成,进而参与胎盘植入的发生。此外,我们还构建的差异表达lncRNA相关的ceRNA调控网络,其中RP5-875H18.4--miRNA-218--SLIT2可能在胎盘植入的发生中存在潜在ceRNA调节机制,它们可以作为分娩前诊断胎盘植入潜在的生物标志物。
Biography
吴淑贞,硕士研究生,主治医师,E-mail: szwu041@126.com
Funding Statement
广东省医学科研基金(B2017030);佛山市医学类科技攻关项目(2018AB000271);佛山市科技创新项目(2016AB001761)
Contributor Information
吴 淑贞 (Shuzhen WU), Email: szwu041@126.com.
刘 正平 (Zhengping LIU), Email: liuzphlk81@outlook.com.
References
- 1.Cahill AG, Beigi R, Heine RP, et al. Placenta accreta spectrum. Am J Obstet Gynecol. 2018;219(6):B2–B16. doi: 10.1016/j.ajog.2018.09.042. [Cahill AG, Beigi R, Heine RP, et al. Placenta accreta spectrum[J]. Am J Obstet Gynecol, 2018, 219(6): B2-B16.] [DOI] [PubMed] [Google Scholar]
- 2.Silver RM, Barbour KD. Placenta accreta spectrum: accreta, increta, and percreta. Obstet Gynecol Clin North Am. 2015;42(2):381–402. doi: 10.1016/j.ogc.2015.01.014. [Silver RM, Barbour KD. Placenta accreta spectrum: accreta, increta, and percreta[J]. Obstet Gynecol Clin North Am, 2015, 42(2): 381- 402.] [DOI] [PubMed] [Google Scholar]
- 3.Jauniaux E, Bhide A, Kennedy A, et al. FIGO consensus guidelines on placenta accreta spectrum disorders: Prenatal diagnosis and screening. Int J Gynaecol Obstet. 2018;140(3):274–80. doi: 10.1002/ijgo.12408. [Jauniaux E, Bhide A, Kennedy A, et al. FIGO consensus guidelines on placenta accreta spectrum disorders: Prenatal diagnosis and screening[J]. Int J Gynaecol Obstet, 2018, 140(3): 274-80.] [DOI] [PubMed] [Google Scholar]
- 4.Grozhik AV, Jaffrey SR. Distinguishing RNA modifications from noise in epitranscriptome maps. Nat Chem Biol. 2018;14(3):215–25. doi: 10.1038/nchembio.2546. [Grozhik AV, Jaffrey SR. Distinguishing RNA modifications from noise in epitranscriptome maps[J]. Nat Chem Biol, 2018, 14(3): 215-25.] [DOI] [PubMed] [Google Scholar]
- 5.Kopp F, Mendell JT. Functional classification and experimental dissection of long noncoding RNAs. Cell. 2018;172(3):393–407. doi: 10.1016/j.cell.2018.01.011. [Kopp F, Mendell JT. Functional classification and experimental dissection of long noncoding RNAs[J]. Cell, 2018, 172(3): 393- 407.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tao H, Liu X, Liu X, et al. LncRNA MEG3 inhibits trophoblast invasion and trophoblast-mediated VSMC loss in uterine spiral artery remodeling. Mol Reprod Dev. 2019;86(6):686–95. doi: 10.1002/mrd.23147. [Tao H, Liu X, Liu X, et al. LncRNA MEG3 inhibits trophoblast invasion and trophoblast-mediated VSMC loss in uterine spiral artery remodeling[J]. Mol Reprod Dev, 2019, 86(6): 686-95.] [DOI] [PubMed] [Google Scholar]
- 7.Azari I, Ghafouri-Fard S, Omrani MD, et al. Expression of Long Non-Coding RNAs in Placentas of Intrauterine Growth Restriction (IUGR) Pregnancies. Rep Biochem Mol Biol. 2019;8(1):25–31. [Azari I, Ghafouri-Fard S, Omrani MD, et al. Expression of Long Non-Coding RNAs in Placentas of Intrauterine Growth Restriction (IUGR) Pregnancies[J]. Rep Biochem Mol Biol, 2019, 8(1): 25-31.] [PMC free article] [PubMed] [Google Scholar]
- 8.Moradi MT, Rahimi Z, Vaisi-Raygani A. New insight into the role of long non-coding RNAs in the pathogenesis of preeclampsia. Hypertens Pregnancy. 2019;38(1):41–51. doi: 10.1080/10641955.2019.1573252. [Moradi MT, Rahimi Z, Vaisi-Raygani A. New insight into the role of long non-coding RNAs in the pathogenesis of preeclampsia[J]. Hypertens Pregnancy, 2019, 38(1): 41-51.] [DOI] [PubMed] [Google Scholar]
- 9.Zhang L, Deng X, Shi X, et al. Silencing H19 regulated proliferation, invasion, and autophagy in the placenta by targeting miR-18a-5p. J Cell Biochem. 2019;120(6):9006–15. doi: 10.1002/jcb.28172. [Zhang L, Deng X, Shi X, et al. Silencing H19 regulated proliferation, invasion, and autophagy in the placenta by targeting miR-18a-5p[J]. J Cell Biochem, 2019, 120(6): 9006-15.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leng L, Zhang C, Ren L, et al. Construction of a long noncoding RNA-mediated competitive endogenous RNA network reveals global patterns and regulatory markers in gestational diabetes. http://med.wanfangdata.com.cn/Paper/Detail/PeriodicalPaper_PM12377295. Int J Mol Med. 2019;43(2):927–35. doi: 10.3892/ijmm.2018.4026. [Leng L, Zhang C, Ren L, et al. Construction of a long noncoding RNA-mediated competitive endogenous RNA network reveals global patterns and regulatory markers in gestational diabetes[J]. Int J Mol Med, 2019, 43(2): 927-35.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burris HH, Just AC, Haviland MJ, et al. Long noncoding RNA expression in the cervix mid-pregnancy is associated with the length of gestation at delivery. Epigenetics. 2018;13(7):742–50. doi: 10.1080/15592294.2018.1503490. [Burris HH, Just AC, Haviland MJ, et al. Long noncoding RNA expression in the cervix mid-pregnancy is associated with the length of gestation at delivery[J]. Epigenetics, 2018, 13(7): 742-50.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pan J, Mor G, Ju W, et al. Viral infection-induced differential expression of LncRNAs associated with collagen in mouse placentas and amniotic sacs. Am J Reprod Immunol. 2015;74(3):237–57. doi: 10.1111/aji.12406. [Pan J, Mor G, Ju W, et al. Viral infection-induced differential expression of LncRNAs associated with collagen in mouse placentas and amniotic sacs[J]. Am J Reprod Immunol, 2015, 74 (3): 237-57.] [DOI] [PubMed] [Google Scholar]
- 13.Tseng JJ, Hsieh YT, Hsu SL, et al. Metastasis associated lung adenocarcinoma transcript 1 is up-regulated in placenta previa increta/percreta and strongly associated with trophoblast-like cell invasion in vitro. Mol Hum Reprod. 2009;15(11):725–31. doi: 10.1093/molehr/gap071. [Tseng JJ, Hsieh YT, Hsu SL, et al. Metastasis associated lung adenocarcinoma transcript 1 is up-regulated in placenta previa increta/percreta and strongly associated with trophoblast-like cell invasion in vitro[J]. Mol Hum Reprod, 2009, 15(11): 725-31.] [DOI] [PubMed] [Google Scholar]
- 14.Michel G, Tonon T, Scornet D, et al. The cell wall polysaccharide metabolism of the brown alga Ectocarpus siliculosus. Insights into the evolution of extracellular matrix polysaccharides in Eukaryotes. New Phytol. 2010;188(1):82–97. doi: 10.1111/j.1469-8137.2010.03374.x. [Michel G, Tonon T, Scornet D, et al. The cell wall polysaccharide metabolism of the brown alga Ectocarpus siliculosus. Insights into the evolution of extracellular matrix polysaccharides in Eukaryotes [J]. New Phytol, 2010, 188(1): 82-97.] [DOI] [PubMed] [Google Scholar]
- 15.Haimov-Kochman R, Friedmann Y, Prus D, et al. Localization of heparanase in normal and pathological human placenta. Mol Hum Reprod. 2002;8(6):566–73. doi: 10.1093/molehr/8.6.566. [Haimov-Kochman R, Friedmann Y, Prus D, et al. Localization of heparanase in normal and pathological human placenta[J]. Mol Hum Reprod, 2002, 8(6): 566-73.] [DOI] [PubMed] [Google Scholar]
- 16.Borbely AU, Daher S, Ishigai MM, et al. Decorin and biglycan immunolocalization in non-villous structures of healthy and pathological human placentas. Histopathology. 2014;64(5):616–25. doi: 10.1111/his.12304. [Borbely AU, Daher S, Ishigai MM, et al. Decorin and biglycan immunolocalization in non-villous structures of healthy and pathological human placentas[J]. Histopathology, 2014, 64(5): 616- 25.] [DOI] [PubMed] [Google Scholar]
- 17.Kuo CY, Guo T, Cabrera-Luque J, et al. Placental basement membrane proteins are required for effective cytotrophoblast invasion in a three-dimensional bioprinted placenta model. J Biomed Mater Res A. 2018;106(6):1476–87. doi: 10.1002/jbm.a.36350. [Kuo CY, Guo T, Cabrera-Luque J, et al. Placental basement membrane proteins are required for effective cytotrophoblast invasion in a three-dimensional bioprinted placenta model[J]. J Biomed Mater Res A, 2018, 106(6): 1476-87.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miyazono K, Katsuno Y, Koinuma D, et al. Intracellular and extracellular TGF-beta signaling in cancer: some recent topics. Front Med. 2018;12(4):387–411. doi: 10.1007/s11684-018-0646-8. [Miyazono K, Katsuno Y, Koinuma D, et al. Intracellular and extracellular TGF-beta signaling in cancer: some recent topics[J]. Front Med, 2018, 12(4): 387-411.] [DOI] [PubMed] [Google Scholar]
- 19.Huang TW, Li ST, Fang KM, et al. Hyaluronan antagonizes the differentiation effect of TGF-beta1 on nasal epithelial cells through down-regulation of TGF-beta type I receptor. Artif Cells Nanomed Biotechnol. 2018;46(sup3):S254–S63. doi: 10.1080/21691401.2018.1491477. [Huang TW, Li ST, Fang KM, et al. Hyaluronan antagonizes the differentiation effect of TGF-beta1 on nasal epithelial cells through down-regulation of TGF-beta type I receptor[J]. Artif Cells Nanomed Biotechnol, 2018, 46(sup3): S254-S63.] [DOI] [PubMed] [Google Scholar]
- 20.Qu H, Sun H, Wang X. Neogenin-1 promotes cell proliferation, motility, and adhesion by Up-regulation of zinc finger E-Box binding homeobox 1 via activating the Rac1/PI3K/AKT pathway in gastric cancer cells. http://med.wanfangdata.com.cn/Paper/Detail/PeriodicalPaper_PM16378853. Cell Physiol Biochem. 2018;48(4):1457–67. doi: 10.1159/000492255. [Qu H, Sun H, Wang X. Neogenin-1 promotes cell proliferation, motility, and adhesion by Up-regulation of zinc finger E-Box binding homeobox 1 via activating the Rac1/PI3K/AKT pathway in gastric cancer cells[J]. Cell Physiol Biochem, 2018, 48(4): 1457-67.] [DOI] [PubMed] [Google Scholar]
- 21.Prossler J, Chen Q, Chamley L, et al. The relationship between TGFbeta, low oxygen and the outgrowth of extravillous trophoblasts from anchoring villi during the first trimester of pregnancy. Cytokine. 2014;68(1):9–15. doi: 10.1016/j.cyto.2014.03.001. [Prossler J, Chen Q, Chamley L, et al. The relationship between TGFbeta, low oxygen and the outgrowth of extravillous trophoblasts from anchoring villi during the first trimester of pregnancy[J]. Cytokine, 2014, 68(1): 9-15.] [DOI] [PubMed] [Google Scholar]
- 22.Liu J, Wang D, Long Z, et al. CircRNA8924 promotes cervical cancer cell proliferation, migration and invasion by competitively binding to MiR-518d-5p /519-5p family and modulating the expression of CBX8. https://www.researchgate.net/publication/8047062_Recovery_of_Focal_Brain_Ischemia-Induced_Behavioral_Dysfunction_by_Intracerebroventricular_Injection_of_Microglia. Cell Physiol Biochem. 2018;48(1):173–84. doi: 10.1159/000491716. [Liu J, Wang D, Long Z, et al. CircRNA8924 promotes cervical cancer cell proliferation, migration and invasion by competitively binding to MiR-518d-5p /519-5p family and modulating the expression of CBX8[J]. Cell Physiol Biochem, 2018, 48(1): 173-84.] [DOI] [PubMed] [Google Scholar]
- 23.Hu CE, Du PZ, Zhang HD, et al. Long noncoding RNA CRNDE promotes proliferation of gastric cancer cells by targeting miR-145. https://www.researchgate.net/publication/5961277_DJ-1_protects_against_neurodegeneration_caused_by_focal_cerebral_ischemia_and_reperfusion_in_rats?ev=prf_cit. Cell Physiol Biochem. 2017;42(1):13–21. doi: 10.1159/000477107. [Hu CE, Du PZ, Zhang HD, et al. Long noncoding RNA CRNDE promotes proliferation of gastric cancer cells by targeting miR-145 [J]. Cell Physiol Biochem, 2017, 42(1): 13-21.] [DOI] [PubMed] [Google Scholar]
- 24.Xiong DD, Li ZY, Liang L, et al. The LncRNA NEAT1 accelerates lung adenocarcinoma deterioration and binds to Mir-193a-3p as a competitive endogenous RNA. https://www.researchgate.net/publication/19816740_Hargreaves_A_Hargreaves_K_Dubner_R_Brown_F_Flores_C_Joris_JA_new_and_sensitive_method_for_measuring_thermal_nociception_in_cutaneous_hyperalgesia_Pain_3277-88. Cell Physiol Biochem. 2018;48(3):905–18. doi: 10.1159/000491958. [Xiong DD, Li ZY, Liang L, et al. The LncRNA NEAT1 accelerates lung adenocarcinoma deterioration and binds to Mir-193a-3p as a competitive endogenous RNA[J]. Cell Physiol Biochem, 2018, 48 (3): 905-18.] [DOI] [PubMed] [Google Scholar]
- 25.Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505(7483):344–52. doi: 10.1038/nature12986. [Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition[J]. Nature, 2014, 505(7483): 344-52.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou X, Yao Q, Sun X, et al. Slit2 ameliorates renal inflammation and fibrosis after hypoxia-and lipopolysaccharide-induced epithelial cells injury in vitro. Exp Cell Res. 2017;352(1):123–29. doi: 10.1016/j.yexcr.2017.02.001. [Zhou X, Yao Q, Sun X, et al. Slit2 ameliorates renal inflammation and fibrosis after hypoxia-and lipopolysaccharide-induced epithelial cells injury in vitro[J]. Exp Cell Res, 2017, 352(1): 123-29.] [DOI] [PubMed] [Google Scholar]
- 27.Li P, Shi Y, Shuai H, et al. Alterted SLIT2/ROBO1 signalling is linked to impaired placentation of missed and threatened miscarriage in early pregnancy. Histopathology. 2017;71(4):543–52. doi: 10.1111/his.13250. [Li P, Shi Y, Shuai H, et al. Alterted SLIT2/ROBO1 signalling is linked to impaired placentation of missed and threatened miscarriage in early pregnancy[J]. Histopathology, 2017, 71(4): 543-52.] [DOI] [PubMed] [Google Scholar]
- 28.Li P, Peng H, Lu WH, et al. Role of Slit2/Robo1 in trophoblast invasion and vascular remodeling during ectopic tubal pregnancy. Placenta. 2015;36(10):1087–94. doi: 10.1016/j.placenta.2015.08.002. [Li P, Peng H, Lu WH, et al. Role of Slit2/Robo1 in trophoblast invasion and vascular remodeling during ectopic tubal pregnancy [J]. Placenta, 2015, 36(10): 1087-94.] [DOI] [PubMed] [Google Scholar]
- 29.Liao WX, Laurent LC, Agent S, et al. Human placental expression of SLIT/ROBO signaling cues: effects of preeclampsia and hypoxia. http://med.wanfangdata.com.cn/Paper/Detail/PeriodicalPaper_PM10319789. Biol Reprod. 2012;86(4):1–7. doi: 10.1095/biolreprod.110.088138. [Liao WX, Laurent LC, Agent S, et al. Human placental expression of SLIT/ROBO signaling cues: effects of preeclampsia and hypoxia [J]. Biol Reprod, 2012, 86(4): 1-7.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pan Y, Jiang S, Hou Q, et al. Dissection of glomerular transcriptional profile in patients with diabetic nephropathy: SRGAP2a protects podocyte structure and function. Diabetes. 2018;67(4):717–30. doi: 10.2337/db17-0755. [Pan Y, Jiang S, Hou Q, et al. Dissection of glomerular transcriptional profile in patients with diabetic nephropathy: SRGAP2a protects podocyte structure and function[J]. Diabetes, 2018, 67(4): 717-30.] [DOI] [PubMed] [Google Scholar]


