Abstract
Telomere maintenance is critical for chromosome stability. Here we report that periodic tryptophan protein 1 (PWP1) is involved in regulating telomere length homeostasis. Pwp1 appears to be essential for mouse development and embryonic stem cell (ESC) survival, as homozygous Pwp1-knockout mice and ESCs have never been obtained. Heterozygous Pwp1-knockout mice had shorter telomeres and decreased reproductive capacity. Pwp1 depletion induced rapid telomere shortening accompanied by reduced shelterin complex and increased DNA damage in telomeric regions. Mechanistically, PWP1 bound and stabilized the shelterin complex via its WD40 domains and regulated the overall level of H4K20me3. The rescue of telomere length in Pwp1-deficient cells by PWP1 overexpression depended on SUV4-20H2 co-expression and increased H4K20me3. Therefore, our study revealed a novel protein involved in telomere homeostasis in both mouse and human cells. This knowledge will improve our understanding of how chromatin structure and histone modifications are involved in maintaining telomere integrity.
Subject terms: Telomeres, Embryonic stem cells
Introduction
Telomeres are special structures located at chromosomal ends. They are composed of variable numbers of the simple DNA repeat TTAGGG, which is associated with large shelterin complex proteins1. This type of telomeric nucleoprotein structure is conserved in the majority of eukaryotic species. The variable numbers of telomeric DNA repeats result in differences in telomere length2. Telomeres are indispensable for chromosome stability in all species, including humans, mice, and ciliates3. Abnormal telomere shortening causes the development of telomere-related diseases, such as Dyskeratosis Congenital4. Therefore, elucidating the molecular mechanisms underlying the regulation of telomere length is particularly important. Currently, the known mechanisms of telomere length regulation5,6 are as follows: (1) telomerase-dependent telomere regulation, involving telomerase reverse transcriptase (TERT), RNA template (Terc), or other telomerase-associated factors7; (2) alternative lengthening of telomeres (ALT), involving proteins such as ZSCAN48,9; (3) telomere protection by the shelterin complex, involving TRF1 (telomeric repeat-binding factor 1), TRF2 (telomeric repeat-binding factor 2), RAP1 (repressor and activator protein 1), POT1 (protection of telomeres 1), TPP1 (TIN2-interacting protein 1), and TIN2 (TRF1-interacting nuclear protein 2)10; (4) telomeric chromatin structure maintenance by the covalent modification of histones, such as H4K20me3 and H3K9me311; and (5) telomere length regulation by non-coding RNA, such as TERRA12,13.
Previous studies have shown that telomeres have important functions in embryonic stem cells (ESCs)14. In Tert- or Terc-deficient ESCs, telomere length is decreased, genomic DNA is abnormally methylated, and H3K27me3 exhibits abnormal enrichment. These changes decrease the differentiation potential of ESCs and their ability to form teratomas and chimeras. Telomeres rapidly shorten upon ESC differentiation and gradually lengthen during the reprogramming of somatic cells into induced pluripotent stem cells15. Zscan4, identified as a specific marker for two-cell embryos and ESCs, is involved in telomere maintenance and long-term genomic stability in ESCs16. Overexpression of ZSCAN4 in ESCs rapidly increases telomere length through telomeric sister chromatid exchange (T-SCE). Decreasing Zscan4 expression through RNA interference decreases telomere length and proliferation of ESCs after passaging for 7–8 generations16–18.
The shelterin complex plays an important role in remodeling telomeric structures19. TRF1 and TRF2, two telomeric repeat-binding proteins in the shelterin complex, are abundant in telomeric regions and interact with telomeric repeats and other proteins, facilitating the formation of telomeric loops and the synthesis of short telomere-like small fragments20–23. POT1 proteins bind primarily to the single-stranded region of telomeric DNA24. In mice, POT1a inhibits ATR kinase-mediated telomere signals, and POT1b regulates the protruding end of the single-stranded telomeric DNA. Heterozygous Pot1b-knockout mice develop normally, but homozygous Pot1a-knockout mice is embryonic lethal25,26. Therefore, the shelterin complex is a critical regulator for maintaining telomere integrity.
The histone marks H3K9me3 and H4K20me3 are highly enriched in subtelomeric and telomeric regions11. Mouse embryonic fibroblasts from telomerase-deficient mice have shortened telomeres and reduced H3K9me3 and H4K20me3 in subtelomeric and telomeric regions27. However, it has also been shown that after Suv4-20h2 knockout, the H4K20me3 mark is significantly decreased in telomeric and subtelomeric regions, but the telomere length increases28. Abnormal DNA methylation and reduced H3K9me3 and H4K20me3 have been found in telomeric and subtelomeric regions in cancer cells. These changes might help to activate the telomere elongation mechanism and maintain the proliferative capacity of cells that have lost telomerase activity. These data have exposed the complexity of telomere length regulation by histone modifications in subtelomeric and telomeric regions28,29. Moreover, the molecular mechanism underlying the regulation of telomere length in ESCs also awaits further investigation. In particular, few studies have been performed to understand how histone modifications collaborate with the shelterin complex in telomere length regulation.
Proteins with the WD40 domain have a wide variety of biological functions. They are involved in the stabilization of protein complexes, RNA processing, DNA replication, transcriptional regulation, the maintenance of genome stability, histone modifications, cell cycle regulation, and tumor development30,31. For example, WD repeat domain 5 (WDR5), a core component of the TrxG complex, acts as an effector molecule of H3K4 methylation to regulate the self-renewal of ESCs32. Periodic tryptophan protein 1 (PWP1) is a typical WD40 repeat protein33. Our previous studies indicated that this protein affected the multipotent differentiation capacity of ESCs by influencing the level of H4K20me334. Here, we report that Pwp1 is present in mouse testicular tissues, where telomere lengthening mainly occurs. Mice with heterozygous Pwp1-knockout exhibited significant telomere shortening accompanied by a reduced reproductive capacity. RNA interference-mediated Pwp1 silencing resulted in a decrease in the accumulation of shelterin and H4K20me3 in telomeric regions and induced rapid telomere shortening.
Results
Pwp1 depletion shortened telomere length
Our previous studies showed that Pwp1 regulated the differentiation of mouse ESCs by inhibiting the LIF/Stat3 signaling pathway34. In addition, we detected high levels of Pwp1 mRNA expression in the 2-cell stage of mouse embryonic development (Supplementary Fig. S1a, b). To better understand the function of Pwp1 in mouse embryonic development, we construct Pwp1-knockout mice using the CRISPR/Cas9 method. Of the 180 embryos that received the plasmid containing a Pwp1 gRNA, only six mice were born. In comparison, of the 120 embryos that received control plasmids, 48 mice were successfully born. As determined by PCR-DNA sequencing, among these six mice, two mice had the Pwp1+/− genotype, and no mice were Pwp1−/− (Supplementary Fig. S1c). One of these Pwp1+/− mice contained a 5-bp deletion, resulting in a frameshift in the Pwp1 coding region (Supplementary Fig. S1d). When Pwp1+/− mice were used for crossbreeding, there were only offspring with wild-type and Pwp1+/− phenotypes, and no mice with the Pwp1−/− phenotype were obtained (Fig. 1a). The offspring birth rate of Pwp1+/− mouse breeding was also significantly lower than that of wild-type mice (Fig. 1b), indicating that the Pwp1 mutation affected embryonic development and embryo survival. Thus, we used mouse embryos to establish ES cell lines. Only 6 Pwp1+/− cell lines were obtained from a total of 20 Pwp1+/− embryos, and no Pwp1−/− cell line was found (Supplementary Fig. S1e). In contrast, four cell lines were obtained from five wild-type embryos. In addition, we attempted to generate Pwp1-knockout ESCs using the CRISPR/Cas9 method. From the 56 ES clones screened, 10 Pwp1+/− cell lines were obtained, but no Pwp1−/− cell lines were found (Supplementary Fig. S1f). Our previous studies showed that Pwp1 was required for the exit of ESCs from the pluripotent state into all lineages34. Together, our data suggested that Pwp1 was essential for mouse embryonic development and ESC survival.
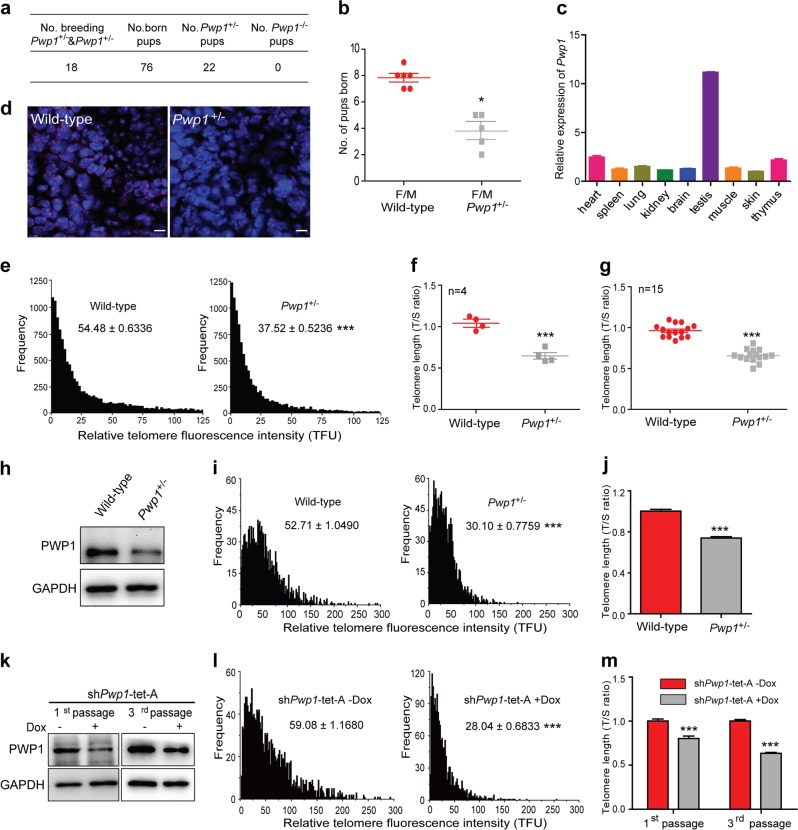
Fig. 1. Pwp1 is required to maintain telomere length. See also Supplementary Fig. S1.
a Numbers of pups born by breeding Pwp1+/− male and female mice. b Numbers of pups obtained by breeding wild-type and Pwp1+/− mice. c Pwp1 mRNA levels in mouse tissues. d Telomere Q-FISH images of wild-type and Pwp1+/− mouse testes. Mouse testes were stained for telomeres (TTAGGG; red) and nuclei (DAPI; blue). The scale bar represents 10 μm. e Q-FISH analysis of telomere length in mouse testicular tissues. Telomere fluorescence intensity (TFU) determined by telomere Q-FISH. Shown are diagrams of the relative telomere length distribution in wild-type and Pwp1+/− mouse testes. f qPCR analysis of telomere length in mouse testicular tissues. Assays were performed using four pairs of mice. g qPCR analysis of telomere length in mouse tails. Assays were performed using 15 pairs of mice. h PWP1 protein levels in wild-type and Pwp1+/− mouse ESCs. i Q-FISH analysis of telomere length in ESCs. The data are shown as TFU determined by telomere Q-FISH in wild-type and Pwp1+/− ESCs. j qPCR analysis of telomere length in ESCs. The data are shown as the T/S ratio determined by qPCR in wild-type and Pwp1+/− ESCs. k Knockdown of PWP1 protein expression in ESCs. ESCs expressing shPwp1 (shPwp1-tet-A) were passaged in the absence or presence of doxycycline (Dox). l Effect of Pwp1 knockdown on telomere length in ESCs as measured by Q-FISH. Telomere lengths in shPwp1-tet-A ESCs at passage 1 were determined by telomere Q-FISH. m Effect of Pwp1 knockdown on telomere length in ESCs as measured by qPCR. The data are shown as the T/S ratio determined by qPCR in shPwp1-tet-A ESCs at passages 1 and 3. The data are presented as the mean ± SEM of three independent experiments. *P < 0.05, ***P < 0.001
To determine the role of Pwp1 in mouse development, we examined its mRNA levels in several mouse tissues by RT-qPCR. As shown in Fig. 1c, Pwp1 was expressed at the highest level in the testes (Fig. 1c), suggesting that it played a role in spermatogenesis and reproduction. Interestingly, ~15% of Pwp1+/− mice exhibited a congenital absence of facial hair (Supplementary Fig. S1g) similar to mouse models of Dyskeratosis Congenita with mutations in genes of telomerase or the shelterin component Tin235,36. PWP1, as a chromatin-binding protein but not telomere specific, was widely expressed in the nucleus and also indeed localized at the telomere region (Supplementary Fig. S1h, i). Together with the telomere-specific ChIP-dot blot assays, we indeed detected an association between PWP1 and telomeres which determined the presence of PWP1 at telomeres (Supplementary Fig. S1j, k). Furthermore, PWP1 bound with the subtelomeric regions of chromosomes in mouse ESCs (Supplementary Fig. S1l). These results suggested that PWP1 could have an ability to regulate telomere homeostasis. To determine whether Pwp1 is involved in telomere homeostasis, we measured the telomere length in testicular tissues and in the tails of wild-type and Pwp1+/− mice using Q-FISH (Fig. 1d, e) and quantitative PCR (qPCR) methods (Fig. 1f, g). The results showed that the telomeres in the testes and tails of Pwp1+/− mice were significantly shorter than those in wild-type mice, suggesting that Pwp1 regulated telomere homeostasis.
In addition, telomeres were also shorter in Pwp1+/− ESCs than in wild-type ESCs (Fig. 1h–j and Supplementary Fig. S1m). We therefore constructed ESC lines with an inducible Pwp1 small hairpin RNA. Upon addition of doxycycline (Dox), both Pwp1 mRNA and protein levels were down-regulated in ESCs of the first (2 days) and third (6 days) passages (Fig. 1k and Supplementary Fig. S1n). Accordingly, telomere length was markedly reduced within 48 h after Pwp1 knockdown (Fig. 1l, m). Therefore, decreased Pwp1 expression resulted in telomere shortening both in the mouse testis in vivo and in ESCs in vitro.
Telomere shortening induced by Pwp1 deficiency was independent of the expression of telomerase and Zscan4, and accompanied by reduced telomerase-telomere association
Telomere length homeostasis can be mediated by activating telomerase or other telomere regulatory factors6. Dox-induced Pwp1 knockdown in ESCs did not cause changes in the expression of Tert, Terc or telomerase activity (Supplementary Fig. S2a–c). No change of TERT-Terc association was observed following Pwp1 knockdown (Supplementary Fig. S2d). However, Pwp1 depletion had a weak effect on telomerase-telomere association (Supplementary Fig. S1e, f). To further verify the role of telomerase on telomere shortening in Pwp1 depletion ESC, we knocked down Pwp1 in Terc−/− ESCs. In the Terc−/−&shPwp1-tet-A ESCs, the reduction of Pwp1 did cause telomere length shortening (Supplementary Fig. S1g, h). These indicted that the telomere shortening induced by Pwp1 deficiency was not owing to telomerase. In addition, we also examined the changes in Zscan4, a key molecule in ALT16,17,37,38, in Pwp1-depleted cells. Pwp1 knockdown induced the downregulation of ZSCAN4 protein (Supplementary Fig. S2i). However, telomere shortening was not detectable after Zscan4 knockdown for 48 h (Supplementary Fig. S2j, k), and ZSCAN4 overexpression did not restore the shortened telomeres in Pwp1+/− ESCs (Supplementary Fig. S2l, m). Thus, regulation of neither telomerase nor Zscan4 was a major factor in the telomere shortening induced by Pwp1 deficiency.
Telomeric enrichment of shelterin complex was reduced in Pwp1-depleted ESCs
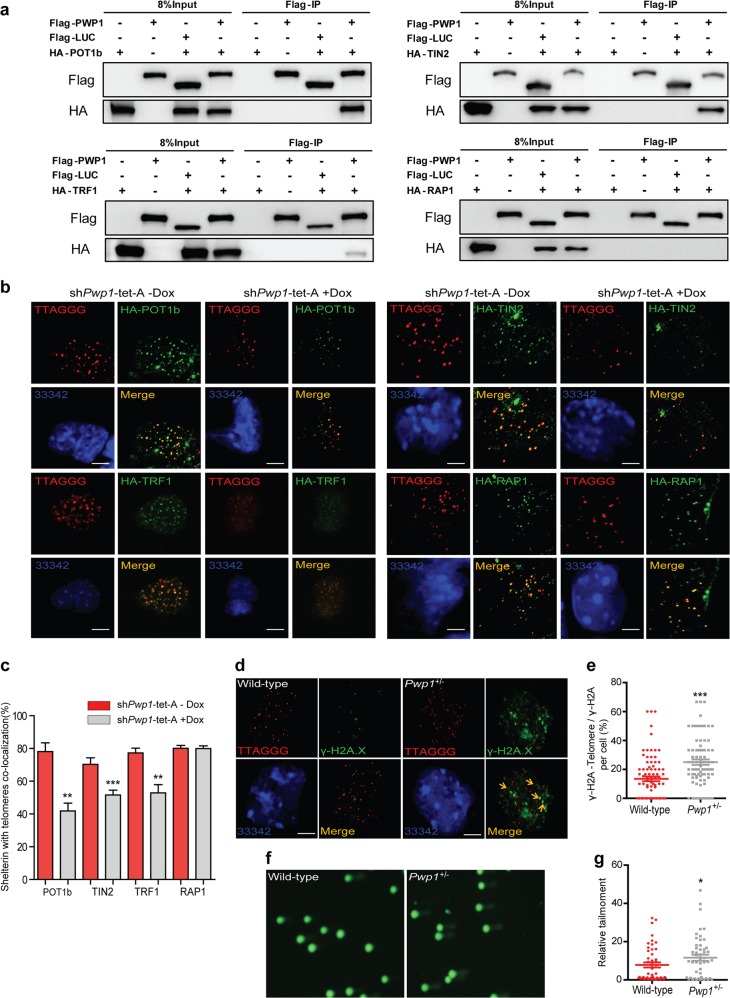
The shelterin complex consists of a group of proteins at the telomere ends and functions to protect telomeres from DNA damage19,39. To investigate its role in Pwp1 deficiency-induced telomere shortening, we performed the reciprocal co-immunoprecipitation (co-IP) to examine the association between PWP1 and six shelterin proteins (POT1 has two subunit structures, POT1a and POT1b, in mice25). PWP1 bound strongly to hemagglutinin (HA)-tagged POT1a, POT1b, TIN2, and TPP1, weakly to TRF1, but did not to TRF2 or RAP1 (Fig. 2a and Supplementary Fig. S3a, b). To further verify the shelterin binding with PWP1, we constructed two cell lines: (1) Pot1-knockdown with PWP1 and TIN2 over-expression ESCs; (2) Tin2-knockdown with PWP1 and POT1b over-expression ESCs. In our results, there were no significant binding changes in the both cell lines. These results showed that the binding of PWP1 and POT1b or PWP1 and TIN2 was not affected by Tin2 depletion or Pot1 depletion (Supplementary Fig. S3c, d). Next, plasmids expressing HA-tagged shelterin proteins (POT1b, TIN2, TRF1, and RAP1) were introduced into shPwp1-tet-A ESCs. After induction of shPwp1 by Dox for 48 h, the enrichment of shelterin proteins (POT1b, TIN2, and TRF1) at telomeres was markedly decreased, whereas the telomere localization of RAP1, which did not bind to PWP1, did not change significantly (Fig. 2b, c). To determine the effect of Pwp1 knockdown on endogenous shelterin proteins, an 3xHA tag was knocked into the carboxyl terminus of POT1b protein at the endogenous POT1b locus in the shPwp1-tet-A ESC line, allowing us to track the endogenous POT1b protein. In this cell line, the enrichment of endogenous POT1b protein at telomeres decreased in response to Dox treatment (Supplementary Fig. S4a, b). The reduction of shelterin enrichment at telomeres was corroborated by a telomere-specific ChIP-dot blot assay showing reduced association between POT1 (Supplementary Fig. S4c, d). In addition, we examined the expression of shelterin complex proteins in shPwp1-tet-A ESCs. Both mRNA and protein levels of shelterin complex proteins were reduced after Pwp1 depletion (Supplementary Fig. S4e, f). Next, the DNA damage signals at the telomeric regions, as measured by γ-H2A.X staining and telomere-specific ChIP-dot blot assays, the DNA damage signals at the subtelomeric regions, as measured by ChIP-qPCR, and the overall levels of DNA damage, as determined by comet electrophoresis, significantly increased in Pwp1+/− ESCs compared with those in wild-type cells (Fig. 2d–g and Supplementary Fig. S4g-i). Furthermore, we measured the telomere aberrations in our experiments. Among the four types of telomere aberrations, there were only slightly more terminal deletions after Pwp1 reduction (Supplementary Fig. S4j, k). Therefore, our data indicated that decreases in Pwp1 expression resulted in a reduction of shelterin complex assembly at telomeric regions, attenuating its telomere protective function.
Fig. 2. Pwp1 depletion reduced shelterin complex enrichment at telomeres. See also Supplementary Fig. S2–S4.
a Binding between PWP1 and the shelterin proteins. Plasmids expressing Flag-PWP1, Flag-Luciferase, and HA-shelterin were transfected into 293FT cells for 48 h. Cell extracts were immunoprecipitated using anti-Flag antibody, followed by Western blotting with an anti-HA antibody. b, c Effects of Pwp1 knockdown on telomere recruitment of shelterin proteins in ESCs. ESCs containing shPwp1-tet-A were treated with Dox or without for 48 h and stained for telomeres (TTAGGG; red), shelterin (POT1b, TIN2, TRF1, and RAP1; HA antibody; green), and nuclei (Hoechst 33342; blue). b Representative immunofluorescence (IF)-FISH images. c Quantifications of co-localizing foci between shelterin proteins (POT1b, TIN2, TRF1, and RAP1) and telomeres. d, e Localization of γ-H2A.X in telomric region. Wild-type and Pwp1+/− ESCs were subjected to IF-FISH analysis using a telomere probe (red) and an antibody against γ-H2A.X (green). d Representative images of γ-H2A.X-Telomere. e Quantification of co-localizing foci of γ-H2A.X and telomeres. The graph shows the percentage of γ-H2A.X-Telomere co-localizing foci among the total γ-H2A.X foci per cell. f, g DNA damage in wild-type and Pwp1+/− ESCs. Representative comet assay images (f). Quantification of DNA damage as measured by the comet assay (g). The scale bar represents 10 μm. The data are presented as the mean ± SEM of three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001
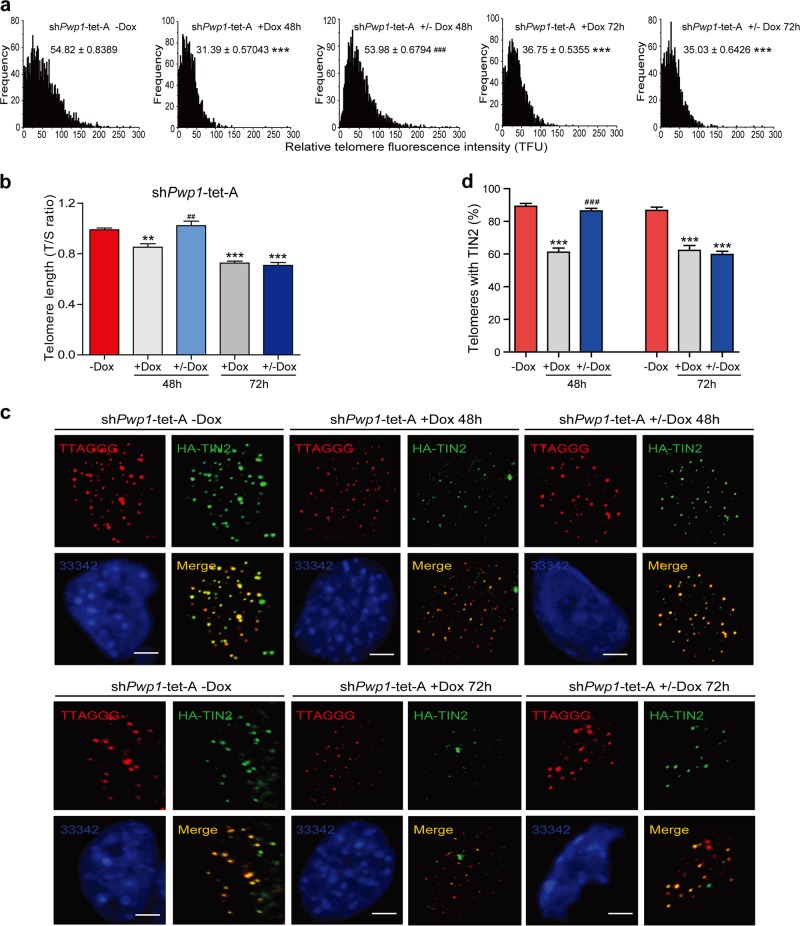
To further study the impact of Pwp1 depletion on telomere regulation, we assessed telomere length and the telomeric association of shelterin proteins in shPwp1-tet-A ESCs upon restoration of Pwp1 expression. First, telomere length was measured using Q-FISH and qPCR. As shown in Fig. 3a, b, telomere length shortened when Pwp1 levels were reduced for 48 h but was restored to the pretreatment level when Pwp1 was re-expressed in shPwp1-tet-A ESCs by removing Dox. However, after 72 h of Pwp1 depletion, telomere length shortening could not be reversed upon Pwp1 restoration. Second, the enrichment of exogenous shelterin protein (TIN2) at the telomeres began to show significant reductions after 48 h of Dox treatment. The reduced enrichment of the exogenous shelterin protein (TIN2) could be restored by withdrawing Dox after 48 h but not after 72 h (Fig. 3c, d), consistent with that of the endogenous shelterin protein (POT1b) in shPwp1-tet-A KI POT1b ESCs (Supplementary Fig. S5a, b). These data suggested that prolonged Pwp1 depletion resulted in more permanent changes in telomere length and structures.
Fig. 3. Only Pwp1 could not rescue telomere length in prolonged-Pwp1- depletion ESCs. See also Supplementary Fig. S5.
a, b Telomere length regulation by Pwp1 knockdown. shPwp1-tet-A ESCs were treated with 1 µg/ml Dox for 48 or 72 h, and telomere length was determined by telomere Q-FISH. -Dox, no Dox treatment; +Dox, treated with Dox for 48 h or 72 h; +/− Dox, treated with Dox for 48 h or 72 h and then without Dox for 48 h or 72 h. a The data are shown as TFU determined by telomere Q-FISH. b The data are shown as the T/S ratio determined by telomere qPCR. c, d Shelterin formation in Pwp1-knockdown cells. shPwp1-tet-A ESCs were treated with Dox for 48 h or 72 h. Cells were stained for telomeres (TTAGGG; red), shelterin (TIN2; green), and nuclei (Hoechst 33342; blue). c Representative images. −Dox, without Dox treatment; +Dox, treated with 1 µg/ml Dox for 48 h or 72 h; +/− Dox, treated with Dox for 48 h or 72 h and then without Dox for 48 h or 72 h. d Quantification of shelterin protein (TIN2) and telomere co-localizing foci. The scale bar represents 10 μm. The data are presented as the mean ± SEM of three independent experiments. **P < 0.01, ***P < 0.001 compared with shPwp1-tet-A ESCs without Dox; ##P < 0.01, ###P < 0.001 compared with shPwp1-tet-A ESCs +Dox for 48 h
Chromosomal association of Pwp1 correlated with H4K20me3
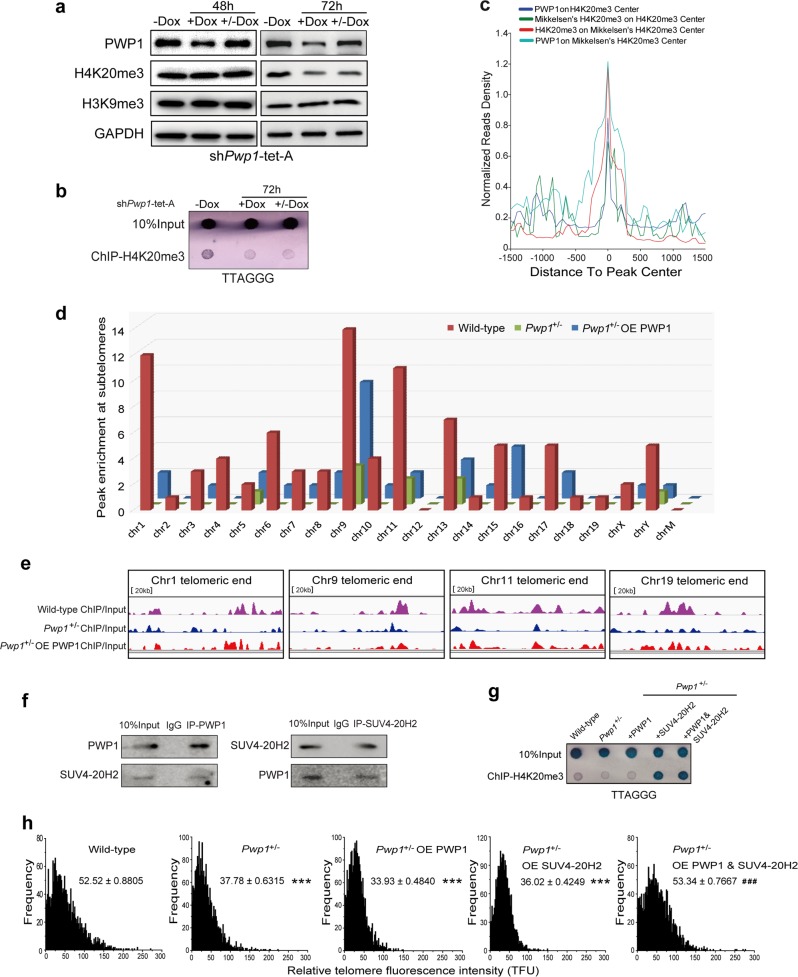
Trimethylations at H3K9 and H4K20 (H3K9me3 and H4K20me3, respectively) are the most important histone modifications in regulating telomere length11. To study the effects of Pwp1 expression on H3K9me3 and H4K20me3 levels, shPwp1-tet-A ESCs were first treated with Dox for 48 or 72 h and then cultured without Dox for an additional 48 or 72 h. Neither H4K20me3 nor H3K9me3 levels changed significantly after Dox treatment for 48 h (Fig. 4a), despite the fact that telomere length had already decreased at this time. These data suggested that telomere shortening was not a direct result of H4K20me3 reduction. Further, H4K20me3, but not H3K9me3, decreased after Dox treatment for 72 h. Upon Dox withdrawal, the PWP1 protein level recovered to the pretreatment level, but the H4K20me3 level was not restored. ChIP-dot blot assays showed that decreasing Pwp1 expression also reduced H4K20me3 enrichment in telomeric regions (Fig. 4b). To further explore the changes related to H4K20me3 modifications in the telomeric and subtelomeric regions of chromosomes after Pwp1 downregulation, ChIP-seq experiments using the H4K20me3 antibody were performed using wild-type, Pwp1+/−, and Pwp1+/−/PWP1-overexpression (OE) ESCs. As shown in Fig. 4c, H4K20me3 genome enrichment profiles were similar to those of PWP1. This result was consistent with the data reported by Mikkelsen et al.40 regarding H4K20me3 in ESCs. Compared with wild-type ESCs, H4K20me3 enrichment was significantly decreased in the subtelomeric regions of Pwp1+/− cells and mostly maintained in those of Pwp1+/−/PWP1-OE cells (Fig. 4d). Specifically, H4K20me3 levels were substantially reduced at subtelomeric regions of Chr1, Chr9, Chr11, and Chr19 in Pwp1+/− cells. These changes were not reversed upon subsequent PWP1 overexpression (Fig. 4e). Thus, Pwp1 depletion-induced changes in chromosomal H4K20me3 modification, especially in the telomeric and subtelomeric regions.
Fig. 4. The role of H4K20me3 in the rescue of telomere length in Pwp1-depleted ESCs. See also Supplementary Fig. S6.
a H4K20me3 and H3K9me3 levels in shPwp1-tet-A ESCs. Cells were treated with 1 µg/ml Dox for 48 h or 72 h. Cell extracts were analyzed by Western blot analysis using the antibodies indicated on the left. −Dox, without Dox treatment; +Dox, treated with Dox for 48 h or 72 h; +/−Dox, treated with Dox for 48 h or 72 h and then without Dox for 48 h or 72 h. b H4K20me3 at telomeres. shPwp1-tet-A ESCs were treated with Dox for 72 h. Chromatin fragments were immunoprecipitated using H4K20me3 antibody, and telomere sequences were detected by dot blot analysis. c Distributions of PWP1 sequence reads relative to the H4K20me3 center. Read densities were normalized to reads per kilobases per million reads (RPKM). d H4K20me3-devoid islands at subtelomeric regions in wild-type, Pwp1+/−, and Pwp1+/−/PWP1-OE ESCs (P < 10−5 versus random genomic regions). The enrichment (versus genome random) of such islands on each subtelomeric region is shown. e H4K20me3 enrichment in subtelomeric regions of multiple chromosomes in mouse ESCs. f Interaction between PWP1 and SUV4-20H2 proteins. Extracts from ESCs were immunoprecipitated using antibodies against PWP1 or SUV4-20H2 proteins, followed by Western blot analysis using both antibodies. g H4K20me3 enrichment at telomeres. Chromatin fragments from wild-type, Pwp1+/−, PWP1- and SUV4-20H2-overexpressing Pwp1+/− ESCs were immunoprecipitated with an antibody against H4K20me3, and telomere sequences were detected on a dot blot. h Telomere length in Pwp1+/− ESCs. Pwp1+/− ESCs were infected with lentiviruses-overexpressing PWP1 or SUV4-20H2. Telomere length was determined by telomere Q-FISH, and the data are shown as TFU. The data are presented as the mean ± SEM of three independent experiments. ***P < 0.001 compared with wild-type ESCs; ###P < 0.001 compared with Pwp1+/− ESCs
Restoration of telomere length following Pwp1 depletion required H4K20me3 catalyzed by SUV4-20H2
Prolonged Pwp1 depletion led to decreases of both telomere length and H3K20me3 level that were not rescued by subsequent PWP1 overexpression, suggesting that H4K20me3 level was important for the rescue of telomere shortening following Pwp1 depletion. In mouse ESCs, PWP1 had nuclear localization pattern that was remarkably similar to that of SUV4-20H2, the trimethyltransferase for H4K20me3 (Supplementary Fig. S6a). As shown in Fig. 4f, PWP1 and SUV4-20H2 associated with each other in reciprocal co-IP assays (Fig. 4f and Supplementary Fig. S6b), and Pwp1 depletion resulted in reduced Suv4-20h2 expression (Supplementary Fig. S6e). Moreover, the depletion of both Pwp1 and Suv4-20h2 led to a shortened telomere length, same as the Pwp1 depletion (Supplementary Fig. S6c, d). We thought depletion of Pwp1 had played the leading role on telomere length in the Pwp1&Suv4-20h2 KD ESCs. Next, PWP1 and SUV4-20H2 were overexpressed either individually or in combination in Pwp1+/− cells (Supplementary Fig. S6e). H4K20me3 enrichment in telomeric regions was significantly increased in Pwp1+/−/PWP1 & SUV4-20H2-OE cells (Fig. 4g). The restoration of telomere length required the expression of both PWP1 and SUV4-20H2 proteins (Fig. 4h and Supplementary Fig. S6f). Moreover, an HA-tagged methyltransferase-deficient SUV4-20H2 fragment without the SET domain (HA-SUV4-20H2 DeSET) did not restore the telomere length (Supplementary Fig. S6g, h). Together, these results indicated that H4K20me3 was critical for the regulation of telomere homeostasis by Pwp1.
The second WD40 domain of PWP1 is important for its telomeric functions
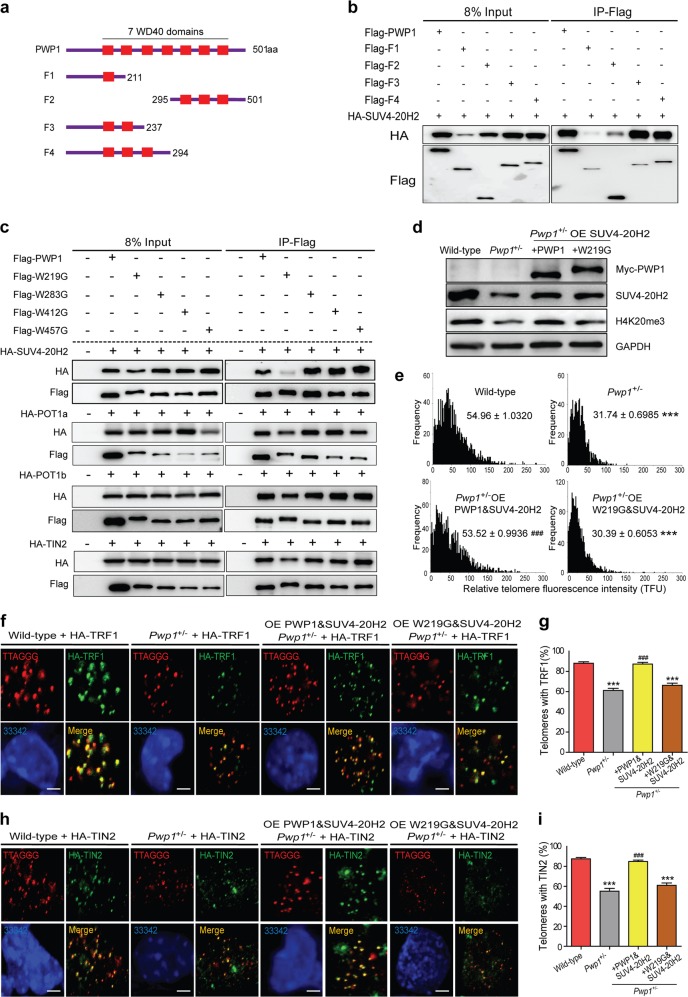
The PWP1 protein contains multiple WD40 repeats. To study the roles of these domains in regulating telomeric functions by PWP1, deletions and point mutations that specifically target its WD40 domains were created41–43 (Fig. 5a and Supplementary Fig. S7a–b). As shown in Fig. 5a, we generated four PWP1 deletions: F1 contained the first 211 amino acids containing with one WD40 domain; F2 was a C-terminal fragment containing the last three WD40 repeats; and F3 and F4 were two N-terminal fragments of 237 and 294 amino acids, containing with two and three WD40 domains, respectively. Co-IP of these overexpressed protein fragments in 293FT cells showed that SUV4-20H2 interacted more strongly with F3 and F4 fragments than with F1 and F2 fragments (Fig. 5b), indicating that the second and the third WD40 domains of PWP1, especially the second WD40 domain, played an important role in the interaction between PWP1 and SUV4-20H2. On the other hand, the fragments that contained multiple WD40 domains, F2 and F4, had strong bindings with shelterin (Supplementary Fig. S7c–e). To extend these findings, the interactions between SUV4-20H2 and full-length PWP1 proteins with point mutations at the conserved tryptophan residues (W to G) in WD40 domains were evaluated in 293FT cells. Some of these mutations caused a slight increase in the apparent protein sizes. As shown in Fig. 5c, the interaction with SUV4-20H2 was severely attenuated by the W219G mutation in the second WD40 domain of PWP1 protein, PWP1(W219G). On the other hand, the interaction between PWP1 and shelterin proteins was only marginally affected by the same mutation. Our results suggested that the second WD40 domain of PWP1 protein was critical for the specific association between PWP1 and SUV4-20H2.
Fig. 5. The second WD40 domain of PWP1 plays key roles in stabilizing H4K20 methylation. See also Supplementary Fig. S7.
a Schematic representation of constructs expressing PWP1 fragments. b Interaction between SUV4-20H2 and PWP1 fragments. 293FT cells were cotransfected with plasmids expressing SUV4-20H2 and fragments of PWP1 for 48 h. Cell extracts were immunoprecipitated using an anti-Flag antibody, followed by Western blot analysis using an HA antibody. c Binding of SUV4-20H2 and shelterin proteins, POT1a, POT1b, and TIN2, to full-length PWP1 with mutations in WD40 repeats. Plasmids expressing various proteins were co-transfected into 293FT cells for 48 h. Cell extracts were immunoprecipitated using an anti-Flag antibody, followed by Western blotting using an HA antibody. d Role of the second WD40 domain of PWP1 in the regulation of SUV4-20H2 expression and cellular H4K20me3 in ESCs. Western blot analysis was performed using ESCs of wild-type, Pwp1+/−, Pwp1+/−/SUV4-20H2-OE ESCs transfected with wild-type PWP1 or PWP1(W219G). e Relative telomere lengths in wild-type ESCs, Pwp1+/− ESCs, Pwp1+/− ESCs over-expressing SUV4-20H2 and PWP1 or PWP1(W219G). The telomere lengths were determined by telomere Q-FISH analysis, and the data are shown as TFU. f–i Telomeric recruitment of shelterin in wild-type ESCs, Pwp1+/− ESCs over-expressing SUV4-20H2 and PWP1 or PWP1(W219G). f, h Representative IF-FISH images. Cells were stained for telomeres (TTAGGG; red), shelterin (TRF1 (f) or TIN2 (h); green), and nuclei (Hoechst 33342; blue). g, i Quantifications of (f) and (h), respectively. The graph shows shelterin proteins (TRF1 or TIN2) and telomere co-localizing foci. The scale bar represents 10 μm. The data are presented as the mean ± SEM of three independent experiments. ***P < 0.001 compared with wild-type ESCs; ###P < 0.001 compared with Pwp1+/− ESCs
Next, we determined the role of the second WD domain of PWP1 protein in rescuing H4K20me3 and telomere defects following Pwp1 depletion. A plasmid expressing wild-type PWP1 protein or PWP1(W219G) was transfected into Pwp1+/− ESCs overexpressing SUV4-20H2. Whereas the H4K20me3 level was restored in cells co-expressing SUV4-20H2 and wild-type PWP1, its level remained lower in cells expressing PWP1(W219G) (Fig. 5d). In addition, overexpression of PWP1(W219G) together with SUV4-20H2 could not restore the telomere length to the same extent as wild-type PWP1 (Fig. 5e). Finally, enrichment of shelterin proteins in telomeric regions was evaluated. The enrichment of shelterin proteins (TRF1 and TIN2) in the telomeric regions of Pwp1+/−/PWP1(W219G) + SUV4-20H2-OE cells did not recover, compared with those in wild-type ESCs (Pwp1+/+) and Pwp1+/−/ PWP1 + SUV4-20H2-OE cells (Fig. 5f–i). Together, these results indicated that WD40 repeats of the PWP1 protein were involved in protein-protein interaction with SUV4-20H2 and shelterin, and the second WD40 domain was especially important for its interaction with SUV4-20H2. As a result, this domain was critical for the involvement of PWP1 in the regulation of telomere length and the enrichment of shelterin proteins in telomeric regions.
PWP1 is involved in telomere length regulation in human cells
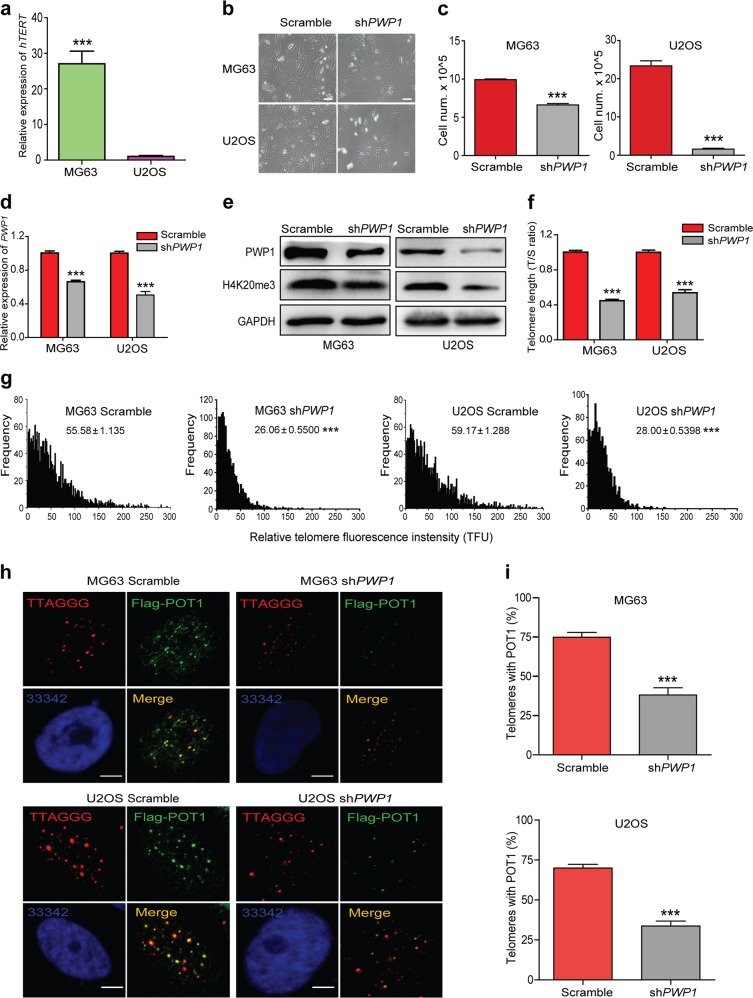
To determine whether PWP1 regulated telomere length/structure in human cells, we performed PWP1 knockdown in human osteosarcoma cells, MG63 and U2OS, using an shRNA targeting PWP1 (shPWP1). MG63 and U2OS cells were telomerase-positive and -negative cell lines, respectively (Fig. 6a). Cells were infected with lentiviruses, and puromycin-resistant cells were selected. As shown in Fig. 6b, c, following PWP1 depletion, both cell lines exhibited reduced cell proliferation capacity, with U2OS cells being affected more substantially. Overall, H4K20me3 levels were decreased in PWP1-knockdown cells compared with those in control cells (Fig. 6d, e). Because human cells have shorter telomeres (5–15 kb) than mouse cells (50-150 kb), we conducted TRF analysis to determine telomere length in human cells (Supplementary Fig. S8a). As shown in Fig. 6f, g, telomere shortening was apparent after PWP1 depletion, similar to that observed in mouse ESCs. We then investigated whether PWP1 depletion could affect shelterin binding at telomeres in these two cell lines. Upon PWP1 knockdown, the binding of exogenous shelterin protein (POT1) to telomeres was markedly reduced in MG63 and U2OS cells (Fig. 6h, i). Endogenous shelterin protein (TIN2) also decreased significantly in U2OS cells upon PWP1 depletion (Supplementary Fig. S8b, c). Together, these data showed that PWP1 depletion in human cells resulted in reduction in H4K20me3 levels and shelterin assembly in human cells, leading to shortened telomeres.
Fig. 6. PWP1 is involved in the regulation of telomere length in human cells. See also Supplementary Fig. S8.
a hTERT expression in human cell lines. hTERT mRNA levels in MG63 and U2OS cells were determined by RT-qPCR analysis. b, c Effects of PWP1 knockdown in human cell lines. b Representative images and (c) proliferation of 105 MG63 and U2OS cells. d, e Knockdown of PWP1 expression (mRNA (d) and protein (e) levels) in MG63 and U2OS cells. PWP1 expression was determined by RT-qPCR analysis, and Western blotting was performed using antibodies against PWP1 protein and H4K20me3. f, g Changes of telomere length upon PWP1 knockdown in MG63 and U2OS cells. f Relative telomere lengths are shown as the T/S ratio as determined by qPCR. g A distribution diagram of relative telomere length. The data are shown as TFU determined by telomere Q-FISH. h, i Effect of PWP1 knockdown on Pot1 binding to telomeres. h Representative IF-FISH images of MG63 and U2OS cells following PWP1 knockdown. Cells were stained for telomeres (TTAGGG; red), shelterin (POT1; green), and nuclei (Hoechst 33342; blue). i Quantification of POT1 and telomere co-localizing foci. The scale bar represents 10 μm. The data are presented as the mean ± SEM of three independent experiments. ***P < 0.001
Discussion
Compared with somatic cells, ESCs have longer telomeres, which are critical for maintaining their unlimited proliferation and multipotent differentiation capacities. However, mechanisms underlying telomere homeostasis in ESCs remains unclear. Telomeres gradually shorten as ESCs lose telomerase activity. Here, our results showed that Pwp1 knockdown resulted in a rapid decrease in telomere length, in contrast to what was observed after the loss of telomerase activity. Pwp1 downregulation did not cause the downregulation of telomerase gene expression or induce changes of telomerase activity or stability. Zscan4 is an important molecule in the ALT-dependent (telomerase-independent) telomere maintenance mechanism. It has been shown that Zscan4 silencing leads to shortened telomere length, increases T-SCE, and stalls cell proliferation after 7–8 divisions16. Our data showed that, although Zscan4 decreased upon Pwp1 depletion, Zscan4 knockdown did not result in rapid telomere shortening or inhibition of cell proliferation within 48 h. Moreover, ZSCAN4 overexpression did not restore the shortened telomere length in Pwp1+/− ESCs. Therefore, Pwp1 depletion-induced telomere shorten did not involve reduced telomerase activity or Zscan4-dependent ALT mechanism.
Studies have shown that the shelterin complex plays a very important role in telomere protection. Shelterin maintains telomeric 3′ overhang, forms and protects telomere t-loop structure44. Formation of the 3′ overhang at mouse telomeres involves an intricate set of steps that are controlled by POT1b and TRF245. In human cells, the TPP1-POT1 unit can cap telomere ends and regulate telomerase access to telomere ssDNA46,47. Abnormal expression, mutation or depletion of different shelterin components have different effects on telomere length and stability. For example, heterozygous mutation of TPP1 causes telomere shortening and the development of telomere-related diseases48. Removing TRF1 leads to very frequent replication fork stalling in the telomeric region49,50. Moreover, combinational depletion of several shelterin components has more dramatic effects on telomere structures than single depletion of one protein51. Our results showed that PWP1 associated with shelterin proteins (POT1a, POT1b, TPP1, and TIN2). Pwp1 depletion led to a simultaneous reduction of enrichment of most shelterin proteins at telomeres, which impaired shelterin protection and disturbed telomere homeostasis. Therefore, our results indicated that the telomere shortening in Pwp1-deficient cells was triggered by the loss of shelterin enrichment in the telomeric region.
Pwp1 depletion activated DNA damage signaling in both the telomeric region and the nucleus. PWP1 has the WD40 repeats structure, which endows it with an ability of protein binding and a possibility of participating in different regulatory networks52,53. In the telomeric region, depletion of Pwp1 reduced the shelterin protein enrichment and impaired the shelterin complex protection, in turn aggravating the DNA damage. In Pwp1-deficient ESCs, DNA damage was also aggravated in the nucleus, possibly because of other impaired PWP1 binding relationships. The DNA damage has many factors and the WD40 repeats protein could affect DNA damage in the nucleus54,55. PWP1 was widely expressed and present throughout the nucleus, suggesting that it played roles at and outside telomeres. Therefore, PWP1, which indeed had locations at the telomeric and subtelomeric regions and bindings with the shelterin complex, could prevent DNA damage at telomeres, and it might play roles in DNA damage of non-telomeric region.
Studies from de Lange laboratory have shown that Tin2 heterozygous deletion causes telomere shortening and that its homozygous deletion is embryonic lethal in mice. This telomere length shortening occurs despite the presence of sufficient telomerase activity35. This finding is similar to ours regarding Pwp1-knockout mice and ESCs. Furthermore, telomere lengthening occurs when germ cells develop into haploid sperm cells in mammals. Our previous studies showed that telomerase was highly expressed in spermatids of mouse testicular tissues56. In this study, we showed that the Pwp1 was highly expressed in testicular tissues and that Pwp1 heterozygous-knockout mice had significantly reduced reproductive capability, suggesting that Pwp1 played a role in telomere lengthening during spermatogenesis.
Histone modifications are critical for telomere maintenance and regulation. Liu et al. has shown that RIF1 interacted with the H3K9 methylation complex to stabilize the level of H3K9me3 modification and negatively regulate ZSCAN4 expression to achieve the dynamic regulation of telomere length57. Recently, Konishi et al. also found that the interaction between TRF2 and core histones was important for the stabilization of chromosome ends. Mutations in the GAR domain of TRF2 caused a loss of telomere protection and resulted in rapid telomere shortening58. In our current study, the co-IP data showed that PWP1 interacted with SUV4-20H2, a methyltransferase for H4K20 trimethylation. ChIP-seq data showed that genome-wide PWP1 binding sites matched H4K20me3 sites. Lastly, both H4K20me3 level and telomere length were reduced upon Pwp1 depletion. Consistently, it was previously reported that H4K20me3 enrichment in telomeric and subtelomeric regions was significantly decreased in Terc-knockout mice with shortened telomeres27. Our data also showed that PWP1 overexpression in ESCs with Pwp1 knockdown or heterozygous knockout could not restore telomere length or the level of H4K20me3. However, overexpression of both PWP1 and SUV4-20H2 restored the H4K20me3 level and telomere length and increased localization of shelterin complexes to telomeric regions, indicating that H4K20me3 is crucial in Pwp1-mediated telomere maintenance.
PWP1 has seven WD40 repeats41. These domains have important functions in protein-protein interactions. Our mutagenesis study showed that the second WD40 repeat of PWP1 was important for its interaction with SUV4-20H2. Overexpression of PWP1W219G and SUV4-20H2 in Pwp1+/− ESCs failed to restore H4K20me3 levels, telomere length, or the localization of the shelterin complex to telomeric regions. Hence, the interaction between the second WD40 repeat of PWP1 protein and SUV4-20H2 was likely important not only for restoring the H4K20me3 level, but also for shelterin complex stabilization and telomere maintenance. While shelterin instability might be the primary reasons for the rapid telomere shortening induced by Pwp1 deficiency, the restoration of telomere length by Pwp1 could not occur without an adequate level of H4K20me3 (Fig. 7). In conclusion, our study revealed a new telomere regulatory mechanism involving the chromatin-binding protein PWP1 as a bridge that facilitated both the recruitment of telomere end protective proteins and histone modifications at telomeric regions.
Fig. 7. A model of Pwp1’s roles in regulating telomere length and function.
Left: In wild-type cells, PWP1 binds with and stabilizes shelterin protein complexes and regulates the overall level of H4K20me3 to maintain telomere homeostasis. Right: Within 48 h, the absence of Pwp1 resulted in the reduction of shelterin enrichment in telomeres and rapid telomere shortening. The rescue of telomere length in Pwp1-deficient cells depended on PWP1 expression alone. After a prolonged Pwp1 depletion, the H4K20me3 enrichment was reduced in the subtelomeric and telomeric regions of chromosomes. The rescue of telomere length in Pwp1-deficient cells by PWP1 overexpression also depended on SUV4-20H2 co-expression and increased H4K20me3
Materials and methods
Cell culture
Mouse ESCs (E14.1) were cultured on plates coated with 0.1% gelatin (Millipore) in DMEM (Gibco) supplemented with 15% (v/v) fetal bovine serum (FBS, Gibco), 2 mM GlutaMAX (Gibco), 1 mM sodium pyruvate (Gibco), 0.1 mM non-essential amino acids (Gibco), 1000 units/ml leukemia inhibitory factor (LIF, Millipore), and 0.18% β-mercaptoethanol (Sigma). Medium was changed daily, and cells were routinely passaged every two days. Additionally, 293FT, MG63, and U2OS cells were cultured in DMEM containing 10% FBS.
Gene knockdown, knockout, knock in or overexpression
In mouse ESCs, for Pwp1 knockdown, an shRNA targeting Pwp1 mRNA was designed (shPwp1-tet-A, Table S1) and cloned into the lentivirus vector pLKO-Tet-On (a gift from Zhang Xiaoqing Lab, Tongji University, China). For Pwp1 knockout, a gRNA targeting the major open reading frame of Pwp1 was designed (Pwp1 gRNA, Table S1) and cloned as previously described59. For HA-POT1b knock in, a gRNA was designed (HA-Pot1b gRNA, Table S1) through the website Optimized CRISPR Design (http://crispr.mit.edu/). For protein overexpression, cDNAs were cloned into the vector FUGW (Addgene), and plasmids were electroporated into E14.1 mESCs using Gene Pulser X Cell System (Bio-Rad).
To knockdown PWP1 in human cells, an shRNA targeting PWP1 mRNA was designed (shPWP1-A, Table S1) and cloned into the lentivirus vector pLKO.1 (Addgene). Human shelterin plasmids (gifts from Dr. Mao Zhiyong Lab, Tongji University, China) were electroporated into cells using Gene Pulser X Cell System (Bio-Rad).
Reverse transcription and quantitative PCR (RT-qPCR)
Total RNAs were isolated using TRIzol (Invitrogen) and reverse transcribed to generate cDNAs using a Prime-Script RT reagent kit (TaKaRa). cDNAs were amplified with the SYBR Premix Ex Taq (TaKaRa). The primer sequences used for qPCR are listed in Table S1.
Telomere measurement by qPCR
Genomic DNA was prepared using the TIANamp Genomic DNA Kit (DP304) (TIANGEN). Average telomere length was measured from total genomic DNA using a modified qPCR assay60,61. Equal amounts of DNA were used for each reaction. The primers used in the PCR reactions were telomeric primers and primers for the reference control gene (Mouse 36B4 single copy gene and human hBg single copy gene). The primer sequences are listed in Table S1.
Western blot and co-IP
Cells were lysed in RIPA buffer containing a protease inhibitor cocktail (Roche). Total protein from cell extracts were resolved by 10–12% Bis-Tris SDS-PAGE and transferred to polyvinylidene difluoride (PVDF) membranes (Millipore). Blots were blocked by incubation in 3% BSA in TBST at RT for 1 h, probed with various primary antibodies overnight at 4 °C in TBST and with secondary antibodies at RT for 1 h. Signals were visualized using enhanced chemiluminescence (ECL Hakata, ImageQuant LAS 4000 mini).
Co-IP was performed as previously described62. For the endo-co-IP, cell lysates were incubated with antibodies or control normal IgG overnight at 4 °C. Then, a mixture of Ezview Red Protein A affinity gel (Sigma) and Ezview Red Protein G affinity gel (Sigma) (1:1) was added to the antibody-containing lysates for 2 h. For the exogenous co-IP, cell lysates were immunoprecipitated with anti-FLAG or anti-HA M2 magnetic beads (Sigma) for 4 h at 4 °C. Bead-protein mixtures were subjected to Western blotting after washing. The antibodies used for western blot and co-IP are listed in Table S2.
Telomere quantitative fluorescence in situ hybridization (Q-FISH)
Cells were incubated with 0.2 μg/ml colcemid (Sigma) for 4 h to enrich cells at metaphase. Metaphase-enriched cells were harvested by trypsinization, resuspended in 75 mM KCl at 37 °C for 5 min, fixed with methanol/glacial acetic acid (3:1) at room temperature (RT) for 1 h, and spread onto slides. Telomere FISH and quantification were performed as described previously63 except that a fluorescein isothiocyanate (Cy3)-labeled (TAAGGG)3 peptide nucleic acid (PNA) probe (Panagene) was used in this study. Telomeres were denatured at 80 °C for 10 min and hybridized with the telomere-specific PNA probe (0.5 mg/ml). Chromosomes were counterstained with 0.5 mg/ml DAPI.
Immunofluorescence-telomere FISH (IF-FISH)
The IF-FISH method was performed based on an established protocol64. Cells were grown on gelatin-treated coverslips, fixed with 2% paraformaldehyde for 10 min, and then blocked with blocking solution (1 mg/ml BSA, 3% goat serum, 0.1% Triton X-100, and 1 mM EDTA, pH 8.0) for 1 h at RT, followed by incubation with primary antibodies in blocking solution at 4 °C overnight. The coverslips were then incubated for 1 h at RT with secondary antibodies and again fixed with 2% paraformaldehyde for 5 min FISH was performed as described earlier. Cells were stained with 0.5 mg/ml Hoechst 33342 for 10 min at RT.
RNA immunoprecipitation (RIP)
RIP was performed as previously described65. Briefly, 5 × 106 cells were lysed in RIP buffer (10mM HEPES pH 7.0, 5mM MgCl2, 100mM KCl, 1mM Dithiothrectol, and 0.5% NP-40) for 30 min at 4 °C. And 4 μg of antibody was used in each RIP assay to pull down the RNAs associated with the corresponding proteins. Finally, the co-immunoprecipitated RNAs were extracted, reverse transcribed to generate cDNAs and then analyzed by RT-qPCR.
Chromatin immunoprecipitation (ChIP)
ChIP assays were performed using the Simple ChIP Plus Enzymatic Chromatin IP Kit (Magnetic Beads) (#9005, Cell Signaling Technology). Briefly, 107 cells were crosslinked with 1% formaldehyde for 10 min at RT, and formaldehyde was quenched with 0.125 M glycine. Samples were lysed and sonicated to generate fragments of 500–750 bp. The fragments were immunoprecipitated in ChIP buffer with ChIP-Grade Protein G magnetic beads coupled to specific antibodies and eluted in ChIP Elution Buffer, and the cross-linking was reversed overnight at 65 °C. Samples were treated with Proteinase K and RNase A and extracted with DNA purification columns.
ChIP sequencing (ChIP-seq)
H4K20me3 ChIP-seq reads were processed to filter out reads with low-quality using cutadapt (version 1.8.3) and trimmomatic (version 0.36)66,67. The remaining reads were mapped to mm10 (GRCm38) using Bowtie2 with default parameters68. Only uniquely aligned reads were used for subsequent analysis. Peaks were detected using the MACS2 with parameters -p and -broad-cutoff set to 10-5 (version 2.1.0)69. The R package-ChIPseeker was used to annotate the identified peaks and visualize differential H4K20me3 enrichment following Pwp1 depletion and restoration70. The R language (https://www.r-project.org/) was used to perform hypothesis testing. PWP1 and Mikkelsen’s H4K20me3 ChIP data were downloaded from the Gene Expression Omnibus (GEO) database (GSE59389, GSM307622). The ChIP-seq data have been deposited in the European Bioinformatics Institute Database Array Express (https://www.ebi.ac.uk/arrayexpress/) under accession number E-MTAB-5985.
Dot blot
DNA samples were denatured in 0.1 M Tris-HCl, pH 8.5, and 0.1 M NaCl for 10 min at 95 °C, mixed with cold 2 M NH4Ac (equal volume), and spotted on a nitrocellulose membrane. The membrane was dried at 65 °C for 5 min and crosslinked for 90 s. A DIG-labeled probe was added to the hybridization buffer and incubated with the membrane overnight at 37 °C. The probes were performed as described previously71. The membrane was developed using the DIG Nucleic Acid Detection Kit (Catalog No. 11175041910, Roche).
TRAP assay
TRAP assays were performed using the TRAPeze® Telomerase Detection Kit (S7700, Millipore). Cells were resuspended in 1× CHAPS lysis buffer freshly supplemented with RNase inhibitors, incubated on ice for 30 min, and spun in a microcentrifuge at 12,000 × g for 20 min at 4 °C, followed by PCR analysis. The PCR products were run on a 10% or 12.5% non-denaturing PAGE gel and stained with ethidium bromide.
Comet assay
Comet assays were performed using Trevigen Comet Assay® Kit. Cells were mixed in Comet LMAgarose. The mixture (50 µl) was spread on the surface of Comet Slides. The slides were incubated at 4 °C for 30 min, immersed in lysis solution for 30 min at 4 °C, soaked in alkaline unwinding solution for 30 min at RT, and then electrophoresed in the Comet Assay® ES system and stained with SYBR Green I.
Telomere restriction fragment (TRF) analysis
The TRF analysis was performed based on an established protocol72. Southern blot assays were performed using the North2South™ Chemiluminescent Hybridization and Detection Kit (17097, Thermo Fisher Scientific). Equal amounts of genomic DNA were used for each reaction.
Protein structural model
The web server Hhpred (https://toolkit.tuebingen.mpg.de/#/tools/hhpred) was employed to model the protein structures. PDB_mmCIF70 was used as the target database to generate a homology model. The best hits were selected as templates for modeling.
Animal study
All procedures involving animals were approved by the Laboratory Animal Care Committee of Tongji University under the Guide for the Care and Use of Laboratory Animals (NIH Guide). All mice were maintained in a pathogen-free environment throughout the experiments, and all efforts were made to minimize the number of animals used and their suffering. Production of Pwp1+/− mice was conducted as previously described73.
Statistical analysis
The values are presented as the mean ± SEM. Significance was determined using two-tailed Student’s t tests and GraphPad Prism software.
Supplementary information
Acknowledgements
We thank Xiaoqing Zhang for the pLKO-Tet-On vector and Zhiyong Mao for the shelterin plasmid. We thank Lin Liu for the Terc−/− ESCs. This work was supported by grants obtained from the Major Program of Development Fund for Shanghai Zhangjiang National Innovation Demonstration Zone (ZJ2018-ZD-004); the National Natural Science Foundation of China (31671533, 81530042, 31571529, 31721003,31771506); the Ministry of Science and Technology (2016YFA0101300); National Institutes of Health grant (R01GM071725); the Shanghai municipal medical and health discipline construction project (2017ZZ02015); and the Fundamental Research Funds for the Central Universities (KX0151520172571).
Author contributions
W.J., Y.Y., and J.K. conceived the experiments. W.J. and Y.Y. performed most of the experiments. Y.L., D.S., M.B., J.S., J.Q., T.H., W.L., J.C., W.C., D.Y., X.G., S.Z., J.X., R.Z., X.W., and S.G. contributed reagents and helped with experiments. W.J., Y.Y., J.Z., and J.K. analyzed data and wrote the paper.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Wenwen Jia, Email: wuj3@tongji.edu.cn.
Jiyue Zhu, Email: jiyue.zhu@wsu.edu.
Jiuhong Kang, Email: jhkang@tongji.edu.cn.
Supplementary information
Supplementary Information accompanies the paper at (10.1038/s41421-019-0116-8).
References
- 1.McElligott R, Wellinger RJ. The terminal DNA structure of mammalian chromosomes. EMBO J. 1997;16:3705–3714. doi: 10.1093/emboj/16.12.3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Armanios M, et al. Short telomeres are sufficient to cause the degenerative defects associated with aging. Am. J. Hum. Genet. 2009;85:823–832. doi: 10.1016/j.ajhg.2009.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Feldser DM, Hackett JA, Greider CW. Telomere dysfunction and the initiation of genome instability. Nat. Rev. Cancer. 2003;3:623–627. doi: 10.1038/nrc1142. [DOI] [PubMed] [Google Scholar]
- 4.Blasco MA. Telomeres and human disease: ageing, cancer and beyond. Nat. Rev. Genet. 2005;6:611–622. doi: 10.1038/nrg1656. [DOI] [PubMed] [Google Scholar]
- 5.Levy MZ, Allsopp RC, Futcher AB, Greider CW, Harley CB. Telomere end-replication problem and cell aging. J. Mol. Biol. 1992;225:951–960. doi: 10.1016/0022-2836(92)90096-3. [DOI] [PubMed] [Google Scholar]
- 6.Lewis KA, Wuttke DS. Telomerase and telomere-associated proteins: structural insights into mechanism and evolution. Structure. 2012;20:28–39. doi: 10.1016/j.str.2011.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gocha AR, Nuovo G, Iwenofu OH, Groden J. Human sarcomas are mosaic for telomerase-dependent and telomerase-independent telomere maintenance mechanisms: implications for telomere-based therapies. Am. J. Pathol. 2013;182:41–48. doi: 10.1016/j.ajpath.2012.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Conomos D, Pickett HA, Reddel RR. Alternative lengthening of telomeres: remodeling the telomere architecture. Front. Oncol. 2013;3:27. doi: 10.3389/fonc.2013.00027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Neumann AA, et al. Alternative lengthening of telomeres in normal mammalian somatic cells. Genes Dev. 2013;27:18–23. doi: 10.1101/gad.205062.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Lange T. Shelterin: the protein complex that shapes and safeguards human telomeres. Gene Dev. 2005;19:2100–2110. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- 11.Gonzalo S, et al. DNA methyltransferases control telomere length and telomere recombination in mammalian cells. Nat. Cell Biol. 2006;8:416–424. doi: 10.1038/ncb1386. [DOI] [PubMed] [Google Scholar]
- 12.Deng Z, Norseen J, Wiedmer A, Riethman H, Lieberman PM. TERRA RNA binding to TRF2 facilitates heterochromatin formation and ORC recruitment at telomeres. Mol. Cell. 2009;35:403–413. doi: 10.1016/j.molcel.2009.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roake CM, Artandi SE. Approaching TERRA Firma: genomic functions of telomeric noncoding RNA. Cell. 2017;170:8–9. doi: 10.1016/j.cell.2017.06.020. [DOI] [PubMed] [Google Scholar]
- 14.Zeng S, et al. Telomerase-mediated telomere elongation from human blastocysts to embryonic stem cells. J. Cell. Sci. 2014;127:752–762. doi: 10.1242/jcs.131433. [DOI] [PubMed] [Google Scholar]
- 15.Marion RM, et al. Telomeres acquire embryonic stem cell characteristics in induced pluripotent stem cells. Cell Stem Cell. 2009;4:141–154. doi: 10.1016/j.stem.2008.12.010. [DOI] [PubMed] [Google Scholar]
- 16.Zalzman M, et al. Zscan4 regulates telomere elongation and genomic stability in ES cells. Nature. 2010;464:858–863. doi: 10.1038/nature08882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiang J, et al. Zscan4 promotes genomic stability during reprogramming and dramatically improves the quality of iPS cells as demonstrated by tetraploid complementation. Cell Res. 2013;23:92–106. doi: 10.1038/cr.2012.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakai-Futatsugi Y, Niwa H. Zscan4 is activated after telomere shortening in mouse embryonic stem cells. Stem Cell Rep. 2016;6:483–495. doi: 10.1016/j.stemcr.2016.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Palm W, de Lange T. How shelterin protects mammalian telomeres. Annu. Rev. Genet. 2008;42:301–334. doi: 10.1146/annurev.genet.41.110306.130350. [DOI] [PubMed] [Google Scholar]
- 20.Doksani Y, Wu JY, de Lange T, Zhuang X. Super-resolution fluorescence imaging of telomeres reveals TRF2-dependent T-loop formation. Cell. 2013;155:345–356. doi: 10.1016/j.cell.2013.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Houghtaling BR, Cuttonaro L, Chang W, Smith S. A dynamic molecular link between the telomere length regulator TRF1 and the chromosome end protector TRF2. Curr. Biol. 2004;14:1621–1631. doi: 10.1016/j.cub.2004.08.052. [DOI] [PubMed] [Google Scholar]
- 22.Lin J, et al. TRF1 and TRF2 use different mechanisms to find telomeric DNA but share a novel mechanism to search for protein partners at telomeres. Nucleic Acids Res. 2014;42:2493–2504. doi: 10.1093/nar/gkt1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zimmermann M, Kibe T, Kabir S, de Lange T. TRF1 negotiates TTAGGG repeat-associated replication problems by recruiting the BLM helicase and the TPP1/POT1 repressor of ATR signaling. Gene Dev. 2014;28:2477–2491. doi: 10.1101/gad.251611.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ray S, Bandaria JN, Qureshi MH, Yildiz A, Balci H. G-quadruplex formation in telomeres enhances POT1/TPP1 protection against RPA binding. Proc. Natl Acad. Sci. USA. 2014;111:2990–2995. doi: 10.1073/pnas.1321436111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hockemeyer D, Daniels JP, Takai H, de Lange T. Recent expansion of the telomeric complex in rodents: two distinct POT1 proteins protect mouse telomeres. Cell. 2006;126:63–77. doi: 10.1016/j.cell.2006.04.044. [DOI] [PubMed] [Google Scholar]
- 26.Frescas D, de Lange T. Binding of TPP1 protein to TIN2 protein is required for POT1a,b protein-mediated telomere protection. J. Biol. Chem. 2014;289:24180–24187. doi: 10.1074/jbc.M114.592592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Benetti R, Garcia-Cao M, Blasco MA. Telomere length regulates the epigenetic status of mammalian telomeres and subtelomeres. Nat. Genet. 2007;39:243–250. doi: 10.1038/ng1952. [DOI] [PubMed] [Google Scholar]
- 28.Benetti R, et al. Suv4-20h deficiency results in telomere elongation and derepression of telomere recombination. J. Cell. Biol. 2007;178:925–936. doi: 10.1083/jcb.200703081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Garcia-Cao M, O'Sullivan R, Peters AH, Jenuwein T, Blasco MA. Epigenetic regulation of telomere length in mammalian cells by the Suv39h1 and Suv39h2 histone methyltransferases. Nat. Genet. 2004;36:94–99. doi: 10.1038/ng1278. [DOI] [PubMed] [Google Scholar]
- 30.Li D, Roberts R. WD-repeat proteins: structure characteristics, biological function, and their involvement in human diseases. Cell. Mol. Life Sci. 2001;58:2085–2097. doi: 10.1007/PL00000838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Migliori V, Mapelli M, Guccione E. On WD40 proteins: propelling our knowledge of transcriptional control? Epigenetics. 2012;7:815–822. doi: 10.4161/epi.21140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ang YS, et al. Wdr5 mediates self-renewal and reprogramming via the embryonic stem cell core transcriptional network. Cell. 2011;145:183–197. doi: 10.1016/j.cell.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Duronio RJ, Gordon JI, Boguski MS. Comparative analysis of the beta transducin family with identification of several new members including PWP1, a nonessential gene of Saccharomyces cerevisiae that is divergently transcribed from NMT1. Proteins. 1992;13:41–56. doi: 10.1002/prot.340130105. [DOI] [PubMed] [Google Scholar]
- 34.Shen J, et al. Pwp1 is required for the differentiation potential of mouse embryonic stem cells through regulating Stat3 signaling. Stem Cells. 2015;33:661–673. doi: 10.1002/stem.1876. [DOI] [PubMed] [Google Scholar]
- 35.Frescas D, de Lange T. A TIN2 dyskeratosis congenita mutation causes telomerase-independent telomere shortening in mice. Genes Dev. 2014;28:153–166. doi: 10.1101/gad.233395.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hockemeyer D, Palm W, Wang RC, Couto SS, de Lange T. Engineered telomere degradation models dyskeratosis congenita. Genes Dev. 2008;22:1773–1785. doi: 10.1101/gad.1679208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Falco G, et al. Zscan4: a novel gene expressed exclusively in late 2-cell embryos and embryonic stem cells. Dev. Biol. 2007;307:539–550. doi: 10.1016/j.ydbio.2007.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pickett HA, Reddel RR. Molecular mechanisms of activity and derepression of alternative lengthening of telomeres. Nat. Struct. Mol. Biol. 2015;22:875–880. doi: 10.1038/nsmb.3106. [DOI] [PubMed] [Google Scholar]
- 39.Sfeir A, de Lange T. Removal of shelterin reveals the telomere end-protection problem. Science. 2012;336:593–597. doi: 10.1126/science.1218498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mikkelsen TS, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ma, J. et al. WDSPdb: an updated resource for WD40 proteins. Bioinformatics (2019). [DOI] [PMC free article] [PubMed]
- 42.Wang C, et al. Identification of WD40 repeats by secondary structure-aided profile-profile alignment. J. Theor. Biol. 2016;398:122–129. doi: 10.1016/j.jtbi.2016.03.025. [DOI] [PubMed] [Google Scholar]
- 43.Wang Y, et al. WDSPdb: a database for WD40-repeat proteins. Nucleic Acids Res. 2015;43:D339–D344. doi: 10.1093/nar/gku1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.de Lange T. Shelterin-mediated telomere protection. Annu. Rev. Genet. 2018;52:223–247. doi: 10.1146/annurev-genet-032918-021921. [DOI] [PubMed] [Google Scholar]
- 45.Wu P, Takai H, de Lange T. Telomeric 3' overhangs derive from resection by Exo1 and Apollo and fill-in by POT1b-associated CST. Cell. 2012;150:39–52. doi: 10.1016/j.cell.2012.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xin H, et al. TPP1 is a homologue of ciliate TEBP-beta and interacts with POT1 to recruit telomerase. Nature. 2007;445:559–562. doi: 10.1038/nature05469. [DOI] [PubMed] [Google Scholar]
- 47.Sexton AN, et al. Genetic and molecular identification of three human TPP1 functions in telomerase action: recruitment, activation, and homeostasis set point regulation. Genes Dev. 2014;28:1885–1899. doi: 10.1101/gad.246819.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bisht K, Smith EM, Tesmer VM, Nandakumar J. Structural and functional consequences of a disease mutation in the telomere protein TPP1. Proc. Natl Acad. Sci. USA. 2016;113:13021–13026. doi: 10.1073/pnas.1605685113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Martinez P, et al. Increased telomere fragility and fusions resulting from TRF1 deficiency lead to degenerative pathologies and increased cancer in mice. Genes Dev. 2009;23:2060–2075. doi: 10.1101/gad.543509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sfeir A, et al. Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell. 2009;138:90–103. doi: 10.1016/j.cell.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bandaria JN, Qin P, Berk V, Chu S, Yildiz A. Shelterin protects chromosome ends by compacting telomeric chromatin. Cell. 2016;164:735–746. doi: 10.1016/j.cell.2016.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stirnimann CU, Petsalaki E, Russell RB, Muller CW. WD40 proteins propel cellular networks. Trends Biochem. Sci. 2010;35:565–574. doi: 10.1016/j.tibs.2010.04.003. [DOI] [PubMed] [Google Scholar]
- 53.Xu C, Min J. Structure and function of WD40 domain proteins. Protein Cell. 2011;2:202–214. doi: 10.1007/s13238-011-1018-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang Q, et al. The WD40 domain of FBXW7 is a poly(ADP-ribose)-binding domain that mediates the early DNA damage response. Nucleic Acids Res. 2019;47:4039–4053. doi: 10.1093/nar/gkz058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu S, et al. RING finger and WD repeat domain 3 (RFWD3) associates with replication protein A (RPA) and facilitates RPA-mediated DNA damage response. J. Biol. Chem. 2011;286:22314–22322. doi: 10.1074/jbc.M111.222802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jia W, et al. A BAC transgenic reporter recapitulates in vivo regulation of human telomerase reverse transcriptase in development and tumorigenesis. FASEB J. 2011;25:979–989. doi: 10.1096/fj.10-173989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dan JM, et al. Rif1 maintains telomere length homeostasis of ESCs by mediating heterochromatin silencing. Dev. Cell. 2014;29:7–19. doi: 10.1016/j.devcel.2014.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Konishi A, Izumi T, Shimizu S. TRF2 protein interacts with core histones to stabilize chromosome ends. J. Biol. Chem. 2016;291:20798–20810. doi: 10.1074/jbc.M116.719021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Aparicio-Prat E, et al. DECKO: J. Cell. Biol.ingle-oligo, dual-CRISPR deletion of genomic elements including long non-coding RNAs. BMC Genom. 2015;16:846. doi: 10.1186/s12864-015-2086-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Callicott RJ, Womack JE, Real-time PCR. assay for measurement of mouse telomeres. Comp. Med. 2006;56:17–22. [PubMed] [Google Scholar]
- 61.Cawthon RM. Telomere measurement by quantitative PCR. Nucleic Acids Res. 2002;30:e47. doi: 10.1093/nar/30.10.e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Song CL, Zhu SC, Wu CY, Kang JH. Histone deacetylase (HDAC) 10 suppresses cervical cancer metastasis through inhibition of matrix metalloproteinase (MMP) 2 and 9 expression. J. Biol. Chem. 2013;288:28021–28033. doi: 10.1074/jbc.M113.498758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Poon SSS, Martens UM, Ward RK, Lansdorp PM. Telomere length measurements using digital fluorescence microscopy. Cytometry. 1999;36:267–278. doi: 10.1002/(SICI)1097-0320(19990801)36:4<267::AID-CYTO1>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 64.Sfeir A, Kabir S, van Overbeek M, Celli GB, de Lange T. Loss of Rap1 Induces Telomere Recombination in the Absence of NHEJ or a DNA Damage Signal. Science. 2010;327:1657–1661. doi: 10.1126/science.1185100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ng SY, Johnson R, Stanton LW. Human long non-coding RNAs promote pluripotency and neuronal differentiation by association with chromatin modifiers and transcription factors. EMBO J. 2012;31:522–533. doi: 10.1038/emboj.2011.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Marcelino E, et al. Besnoitia besnoiti protein disulfide isomerase (BbPDI): molecular characterization, expression and in silico modelling. Exp. Parasitol. 2011;129:164–174. doi: 10.1016/j.exppara.2011.06.012. [DOI] [PubMed] [Google Scholar]
- 68.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yu G, Wang LG, He QY. ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics. 2015;31:2382–2383. doi: 10.1093/bioinformatics/btv145. [DOI] [PubMed] [Google Scholar]
- 71.Loayza D, de Lange T. POT1 as a terminal transducer of TRF1 telomere length control. Nature. 2003;423:1013–1018. doi: 10.1038/nature01688. [DOI] [PubMed] [Google Scholar]
- 72.Mender, I. & Shay, J. W. Telomere restriction fragment (TRF) analysis. Bio Protoc5, e1658 (2015). [DOI] [PMC free article] [PubMed]
- 73.Liu W, et al. Dosage effects of ZP2 and ZP3 heterozygous mutations cause human infertility. Hum. Genet. 2017;136:975–985. doi: 10.1007/s00439-017-1822-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.