Abstract
Liver cancer is one of the most common malignant tumors, with the death rate ranking fourth among all types of cancer. Over the past few decades, several studies have reported that liver tumorigenesis is associated with dysfunction in autophagy. However, the detailed mechanism remains unclear. In this paper, we used tissue micro-array (TMA) of liver cancer to detect proteins associated with the regulation of autophagic signaling in non-cancerous and cancerous regions by immunohistochemical staining. Those proteins contained 4-HNE, p-AMPK, Erk1/2, p-Erk1/2, CARM1, TFEB, LAMP1, and p62. According to the degrees of tumor differentiation in patients (well differentiated group vs. moderately and poorly differentiated group), we analyzed each protein's expression in the ratio of the “cancerous region/non-cancerous region” in two groups. Current data showed that there were AMPK-ERK/CARM1 autophagic signaling pathways during the formation of liver cancer. The above-mentioned changes in signals indicated an upregulation of autophagy in cancerous regions, which means overactivated autophagy plays an important role in liver cancer.
Keywords: AMPK, autophagy, 4-HNE, liver cancer, poorly differentiated
Introduction
According to the statistics of the World Health Organization (WHO)1, cancer is a main factor to cause the death globally, estimating 9.6 million deaths in 2018. As the most common malignant tumors, liver cancer shows a death rate ranking fourth (782,000 deaths) among all types of cancer. Recently, more and more researches have focused on the involvement of autophagy in liver cancer. However, the detailed mechanism still requires further clarification.
Autophagy is an evolutionarily conserved pathway via lysosomes for the degradation of organelles and cytoplasmic proteins, and plays important roles in many diseases, such as clearance of dysfunctional mitochondria and protein aggregates (1), and damaged proteins and organelles (2). Particularly, changes in autophagic activity during the process of oxidative stress can regulate the effects of hydroxynonenal (HNE) on cellular bioenergetics and an impact on cell death was then occurred (3). The downstream effects of 4-HNE, a typical lipid aldehyde, are both multifactorial and cell-specific. Moreover, 4-HNE is also a major indicator of inflammation and oxidative stress, and is considered to initiate cellular damage and promote cellular survival, simultaneously (4). The inhibitory effect of 4-HNE on AMP-activated protein kinase (AMPK) has been reported previously (5).
AMPK is a crucial energy sensor and regulates cellular metabolism to maintain energy homeostasis, and can further promote the autophagy (6). The activity of autophagy is evidenced by elevating AMPK phosphorylation. It has been known to activate autophagy through inhibiting the activity of rapamycin complex 1 (mTORC1), a mammalian target, by directly phosphorylating its receptor and activating phosphorylation of Unc-51-like autophagy activating kinase 1 (ULK1) at early times of starvation (7). Wang et al. (8) has demonstrated that AMPK is an upstream regulator of extracellular signal-regulated kinase (ERK) in autophagy. The ERK (also known as Erk1/2) signaling pathway is indispensable in controlling diverse cellular processes (9). Erk1/2 also participates in the regulation of autophagy. It influences autophagosome formation and adjusts gene expression for cellular turnover as well as autophagy protein synthesis (10), and promote the hepatic autophagy both in vivo and in vitro (11, 12).
In another downstream of AMPK, co-activated factor-related arginine methyltransferase 1 (CARM1) Shin et al. (13) found that CARM1 expression in various tested cell lines could be increased by the starvation of amino acid or the treatment of rapamycin. An AMPK-SKP2-CARM1 signaling axis was also identified for the first time, in which CARM1 expression in the nucleus could be increased by the nutrient starvation-induced AMPK. Moreover, CARM1-dependent histone arginine methylation, functioning as a key nuclear event of autophagy for epigenetic and transcriptional regulation was further confirmed. Other autophagic proteins, such as the transcription factor EB (TFEB), lysosome-associated membrane protein 1 (LAMP1), and p62/SQSTM (sequestosome) was also detected in our present study. We hypothesize that the AMPK-ERK/CARM1 signaling pathways play important roles in the progression of liver cancer.
Materials and Methods
Reagents
The used reagents were summarized as follows: SQSTM1/p62 (Abcam, UK), LAMP1-lysosome marker (Abcam, UK), p44/42 MAPK (Erk1/2; Cell Signaling, US), p-AMPKα1/2 Thr172 (Santa Cruz Biotechnology, US), phospho-p44/42 MAPK (Erk1/2; Thr202/Tyr204; Cell Signaling, US), anti-4 hydroxynonenal antibody (Abcam, UK), rabbit monoclonal to PRMT4/CARM1 (C31G9), rabbit mAb (Cell Signaling, US), goat polyclonal to TFEB-ChIP grade (Abcam, UK), and horseradish peroxidase with a substrate solution of 3,3′-diaminobenzidine tetrahydrochloride (DAB) (Nichirei, Japan).
Human Samples and Their Tissue Micro-Array (TMA) Analysis
Liver tissue samples of 30 liver cancer patients were obtained from the Key Laboratory of Chinese Internal Medicine of MOE of the Beijing Dongzhimen Hospital of Beijing University of Chinese Medicine. The cancerous region was obtained in carcinoma tissues and reviewed by professional pathologists. The non-cancerous region was obtained in normal tissues over 3 cm apart from the borderline of the cancerous region in the same fixed slice. The collection and follow-up manipulation of all pathological tissue samples were approved by the Committee on the Ethics of Experiments of the South-Central University for Nationalities in China (Permit Number: 2017-SCUEC-MEC-007).
The tissues were extracted using a tissue chip handle (2 μm), then put into a 96-hole paraffin mold (2 μm). After that, the mold was heated to keep the tissue flat. The remaining paraffin was added to the mold to fill the gaps between holes. The TMA was made after cooling was over.
Immunohistochemical Analysis
After deparaffinization, the activity of endogenous peroxidase was blocked on slides by using 3% H2O2 in methanol, then pretreated in sodium citrate buffer, and heated in a microwave oven (except 4-HNE), incubated with serum in BSA. Subsequently, TMA sections were incubated with corresponding primary and secondary antibody, and the reaction products were visualized with DAB. Hematoxylin was used for the counterstain of all slides. As a negative control, 1% bovine serum albumin (BSA) was used to replace the primary antibody on sections that were proven to be positive for 4-HNE, p-AMPK, Erk1/2, p-Erk1/2, CARM1, p62, LAMP1, and TFEB in the present experiments.
Multispectral Imaging
Two colors of DAB and hematoxylin on single-stained slides or dual-stained slides were imaged by using the Nuance Multispectral Imaging System (Cambridge Research and Instrumentation Inc., Woburn, MA), and detailed procedures can be found in our previous study (14). Images at 200× magnification were obtained at 20-nm intervals across a range of 420-720 nm, and further used to create image cubes, which were then resolved into two individual monochrome images by Nuance software (v3.0.2) for subsequent image analyses. The total signal optical density (OD) values in red, green, and blue (RGB) and mass spectroscopy (MS) images were measured.
Multispectral Imaging Statistical Analysis
Excel (Microsoft Corporation) and Prism 5.01 (GraphPad Software) were used to process the data, before-after graphs and bar charts were then obtained, and the statistical analyses were further performed. Paired t-test was performed for each dependent variable. According to the degrees of tumor differentiation in patients (well-differentiated group vs. moderately and poorly differentiated group), we analyzed each protein's expression in the ratio of “cancerous region/non-cancerous region” in two groups. An unpaired t-test was conducted for each dependent variable. *p < 0.05, **p < 0.01, and ***p < 0.001 were considered statistically significant; very significant; and extremely significant, respectively, and p > 0.05 was considered not significant.
Result
4-HNE, Which Could Inhibit the Activity of AMPK, Was Downregulated in Cancerous Regions
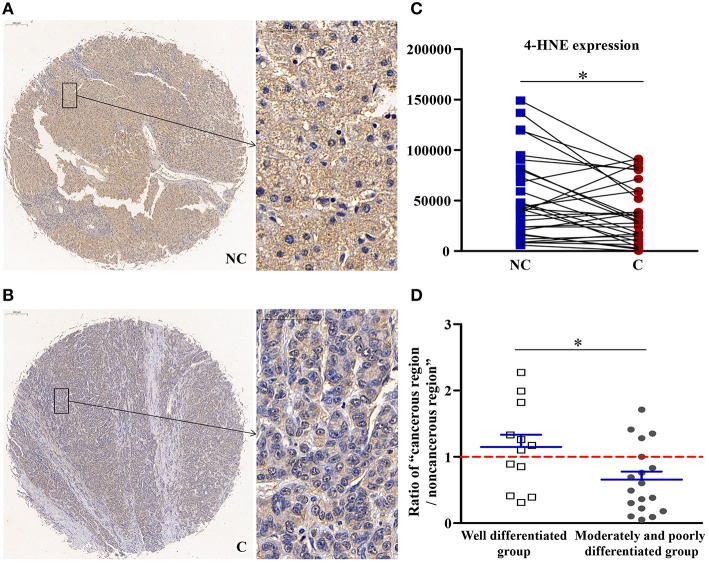
As shown in Figure 1, compared with non-cancerous regions, the expression of 4-HNE, an oxidative stress product, was significantly lower (p < 0.05) in cancerous regions. Compared with well-differentiated patients, the expression of 4-HNE in patients with moderately and poorly differentiated tumors was more significantly downregulated (p < 0.05). That is, as the degree of differentiation decreased, the extent of downregulation in the expression of 4-HNE increased.
Figure 1.
The detailed expression of 4-HNE. Representative images of 4-HNE protein expression in liver cancer's corresponding (A) non-cancerous (NC) and (B) cancerous (C) regions. (C) Quantification of 4-HNE expression level using multispectral image analysis in the liver cancer samples. (D) The ratio of “cancerous region/non-cancerous region” of 4-HNE expression in two groups (well-differentiated group vs. moderately and poorly differentiated group). The red line indicates that the ratio is equal to 1. Scale bar: 200 or 50 μm. *Denotes significant differences from the noncancerous region, or well-differentiated group with P < 0.05.
p-AMPK Upregulated in Cancerous Regions
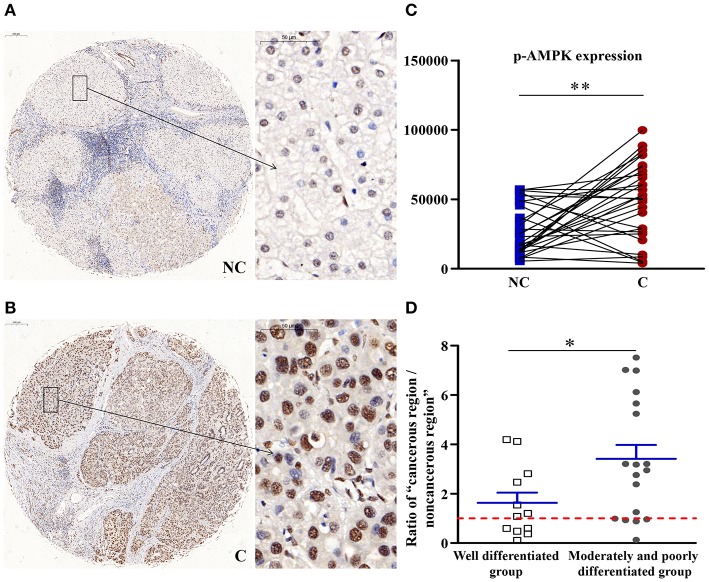
As shown in Figure 2, compared with the non-cancerous region, the phosphorylation of the energy regulator AMPK (p-AMPK) was significantly higher in the cancerous tissues (p < 0.01). Compared with well-differentiated patients, the expression of p-AMPKα in patients with moderately and poorly differentiated tumors was upregulated more significantly (p < 0.05). As the degree of differentiation decreased, the extent of upregulation in the expression of p-AMPKα increased.
Figure 2.
The detailed expression of p-AMPK. Representative images of p-AMPK protein expression in liver cancer's corresponding (A) non-cancerous (NC) and (B) cancerous (C) regions. (C) Quantification of p-AMPK expression level using multispectral image analysis in the liver cancer samples. (D) The ratio of “cancerous region/non-cancerous region” of p-AMPK expression in two groups (well-differentiated group vs. moderately and poorly differentiated group). The red line indicates that the ratio is equal to 1. Scale bar: 200 or 50 μm. *Denotes significant differences from the well-differentiated group with P < 0.05; **Denotes significant differences from the noncancerous region with P < 0.01.
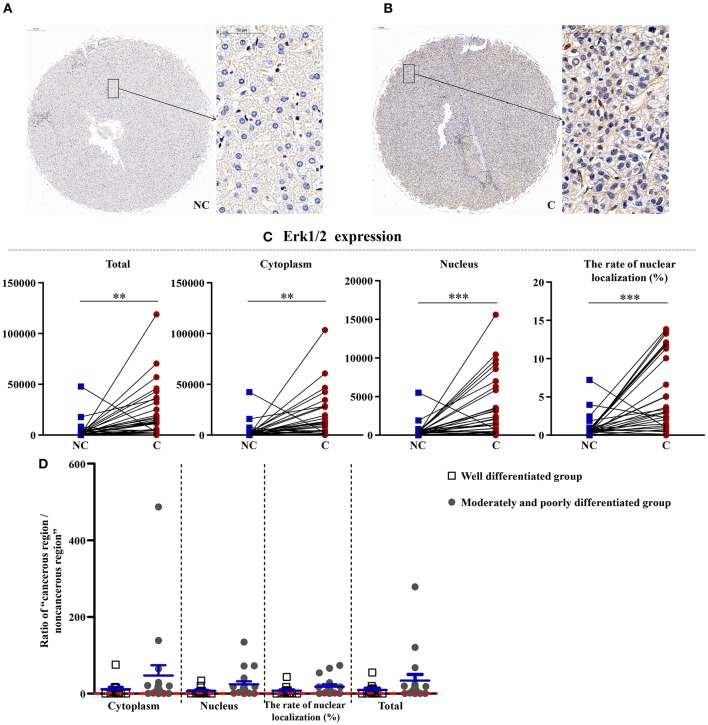
Upregulation of Erk1/2 in Cancerous Regions
As shown in Figures 3, 4, the total (p < 0.01), cytoplasmic (p < 0.01), and nuclear expressions (p < 0.001) and the rate of nuclear localization (p < 0.001) of Erk1/2 were significantly higher in the cancerous regions than that in non-cancerous regions. No significant differences in the total and cytoplasmic expressions of the p-Erk1/2 were found between the cancerous and non-cancerous regions. However, its nuclear expression (p < 0.05) and the rate of nuclear localization (p < 0.01) were significantly upregulated in the cancerous regions compared to that in non-cancerous regions. The up-regulation of Erk1/2 may be caused by the increase of p-Erk1/2 in the nucleus. Although no significant difference was occurred in the expressions of Erk1/2 in patients between the two groups, the nuclear expression (p < 0.05) and rate of nuclear localization (p < 0.05) of p-Erk1/2 showed a significant difference in the two groups. That is, as the degree of differentiation became decreased, the extent of upregulation in the nuclear expression of p-Erk1/2 increased.
Figure 3.
The detailed expression of Erk1/2. Representative images of Erk1/2 protein expression in liver cancer's corresponding (A) non-cancerous (NC) and (B) cancerous (C) regions. (C) Quantification of Erk1/2 expression (total, cytoplasmic, and nuclear expressions and the rate of nuclear localization) levels using multispectral image analysis in the liver cancer samples. (D) The ratio of “cancerous region/non-cancerous region” of Erk1/2 expression in two groups (well-differentiated group vs. moderately and poorly differentiated group). The red line indicates that the ratio is equal to 1. Scale bar: 200 or 50 μm. **, ***Denote significant differences from the well-differentiated group with P < 0.01, 0.001, respectively.
Figure 4.
The detailed expression of p-Erk1/2. Representative images of p-Erk1/2 protein expression in liver cancer's corresponding (A) non-cancerous (NC) and (B) cancerous (C) regions. (C) Quantification of p-Erk1/2 expression (total, cytoplasmic, and nuclear expressions and the rate of nuclear localization) levels using multispectral image analysis in the liver cancer samples. (D) The ratio of “cancerous region/non-cancerous region” of p-Erk1/2 expression in two groups (well-differentiated group vs. moderately and poorly differentiated group). The red line indicates that the ratio is equal to 1. Scale bar: 200 or 50 μm. *, **Denote significant differences from the noncancerous region, or well-differentiated group with P < 0.05, 0.01, respectively.
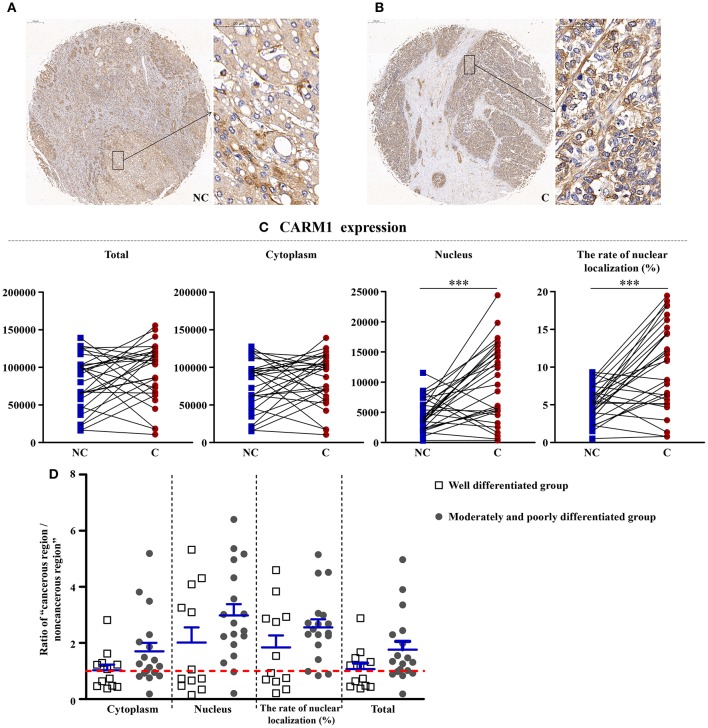
CARM1, a Downstream of AMPK, Was Higher in Cancerous Regions
As shown in Figure 5, CARM1 is the downstream factor of AMPK, although the total and cytoplasm expressions of it have no significant difference, the nucleus expression (p < 0.001) and the rate of nucleus localization (p < 0.001) of CARM1 were significantly higher in cancerous region than that in non-cancerous region.
Figure 5.
The detailed expression of CARM1. Representative images of CARM1 protein expression in liver cancer's corresponding (A) non-cancerous (NC) and (B) cancerous (C) regions. (C) Quantification of CARM1 expression (total, cytoplasmic, and nuclear expressions and the rate of nuclear localization) levels using multispectral image analysis in the liver cancer samples. (D) The ratio of “cancerous region/non-cancerous region” of CARM1 expression in two groups (well-differentiated group vs. moderately and poorly differentiated group). The red line indicates that the ratio is equal to 1. Scale bar: 200 or 50 μm. ***Denotes significant differences from the well-differentiated group with P < 0.001.
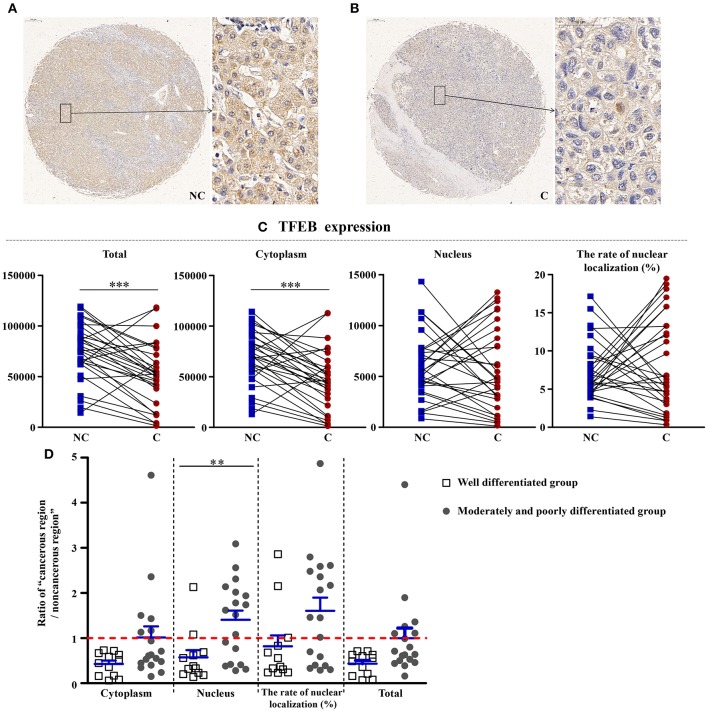
Reduction of TFEB in Cancerous Regions
As shown in Figure 6, compared with non-cancerous regions, the total (p < 0.001) and cytoplasmic expressions (p < 0.001) of TFEB, a main gene that controls the biosynthesis of lysosomes, were significantly reduced in cancerous regions. However, the nuclear expression and the rate of nuclear localization had no significant difference in two regions, showing that the synthesis of lysosomes was affected in the course of liver cancer. Interestingly, compared with well-differentiated liver cancer patients, the nuclear expression of TFEB in patients with moderately and poorly differentiated cancer was upregulated more significantly (p < 0.01).
Figure 6.
The detailed expression of TFEB. Representative images of TFEB protein expression in liver cancer's corresponding (A) non-cancerous (NC) and (B) cancerous (C) regions. (C) Quantification of TFEB expression (total, cytoplasmic, and nuclear expressions and the rate of nuclear localization) levels using multispectral image analysis in the liver cancer samples. (D) The ratio of “cancerous region/non-cancerous region” of TFEB expression in two groups (well-differentiated group vs. moderately and poorly differentiated group). The red line indicates that the ratio is equal to 1. Scale bar: 200 or 50 μm. **Denotes significant differences from the well-differentiated group with P < 0.01; ***Denotes significant differences from the noncancerous region with P < 0.001.
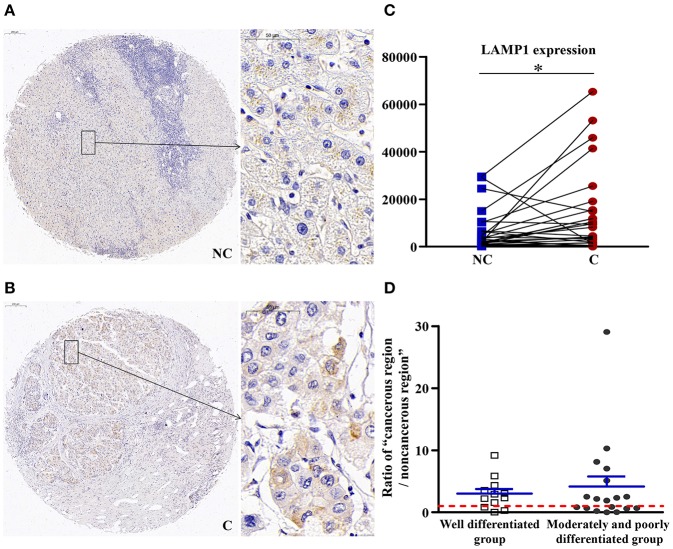
The Number of Lysosomes Was Increased in Cancerous Regions
As shown in Figure 7, compared with non-cancerous regions, the expression of LAMP1 was significantly higher in cancerous regions (p < 0.05), which suggested an increase in the number of lysosomes in cancerous regions.
Figure 7.
The detailed expression LAMP1. Representative images of LAMP1 protein expression in liver cancer's corresponding (A) non-cancerous (NC) and (B) cancerous (C) regions. (C) Quantification of LAMP1 expression level using multispectral image analysis in the liver cancer samples. (D) The ratio of “cancerous region/non-cancerous region” of LAMP1 expression in two groups (well-differentiated group vs. moderately and poorly differentiated group). The red line indicates that the ratio is equal to 1. Scale bar: 200 or 50 μm. *Denotes significant differences from the noncancerous region with P < 0.05.
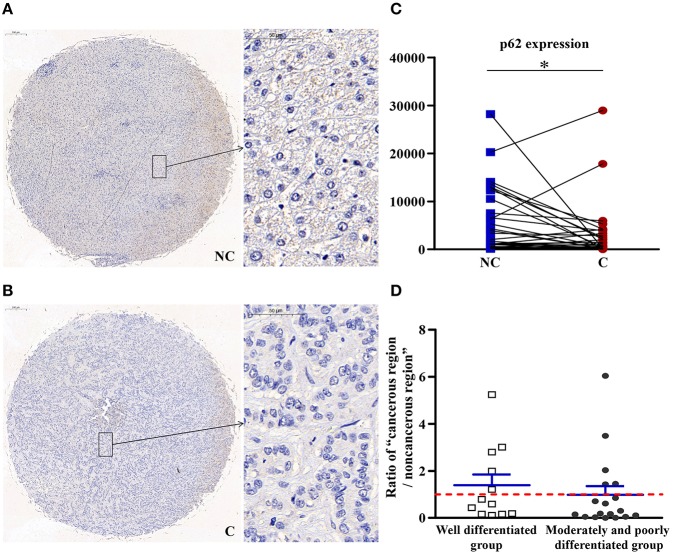
Autophagy in Cancerous Regions Was More Active Than in Non-cancerous Regions
As shown in Figure 8, compared with non-cancerous regions, p62/SQSTM1, a substrate specifically degraded in autophagy, was negatively correlated with autophagy and significantly downregulated (p < 0.05) in cancerous regions, suggesting that autophagy in cancerous regions was more active than in non-cancerous regions.
Figure 8.
The detailed expression of p62. Representative images of p62 protein expression in liver cancer's corresponding (A) non-cancerous (NC) and (B) cancerous (C) regions. (C) Quantification of p62 expression level using multispectral image analysis in the liver cancer samples. (D) The ratio of “cancerous region/non-cancerous region” of p62 expression in two groups (well-differentiated group vs. moderately and poorly differentiated group). The red line indicates that the ratio is equal to 1. Scale bar: 200 or 50 μm. *Denotes significant differences from the noncancerous region with P < 0.05.
Discussion
Liver cancer is the sixth most commonly diagnosed cancer worldwide, resulting in hundreds of thousands of deaths every year. The Global Cancer Observatory (GCO) estimated that it would have 1.36 million (+61.9%) incident cases and 1.28 million (+64.3%) deaths of liver cancer from 2018 to 2040 worldwide2. Recently, more and more researches have focused on the involvement of autophagy in liver cancer. Researchers believe that clarifying the molecular mechanisms underlying the autophagy modulation and liver cancer development can promote the translational studies, and new therapeutic strategies will be ultimately developed for liver cancer.
An Oxidative Product, 4-HNE, Was Downregulated in Cancerous Regions
Autophagy is essential for clearing nonfunctional organelles and proteins during the process of oxidative stress, and in this role, it serves an essential antioxidant function (2). Electrophiles, such as 4-HNE can lead to the dysregulation of cell signaling, which plays a main role in the pathogenesis of chronic inflammatory liver disease. It has also been effective in decreasing the cellular proliferation in several cell types, such as breast cancer cells and prostate (15, 16), and it is involved in the pathogenesis of several degenerative diseases and cancer types (17). Some studies (18, 19) have shown that 4-HNE-induced aggregates can inhibit autophagy. However, the mechanism underlying 4-HNE regulation of autophagy remains unknown. Recently, Zhong et al. (20) found that 4-HNE-protein adducts in tumor tissues of hepatocellular carcinoma decreased gradually. In our study, 4-HNE was significantly downregulated in cancerous regions, which suggests that its inhibitory effect on the AMPK had diminished. Additionally, compared with well-differentiated patients, the expression of 4-HNE in patients with moderately and poorly differentiated tumors was more significantly downregulated. That is, as the degree of differentiation decreased, the extent of downregulation in the expression of 4-HNE increased, which suggests that 4-HNE may be an indicator of the degree of differentiation of liver cancer.
When AMPK, a Direct Target of 4-HNE, Was Upregulated in Cancerous Tissues, Energy Consumption Increased
In response to environmental stress or nutritional factors, which will deplete intracellular adenosine triphosphate (ATP), AMPK is activated by the allosteric binding of AMP and the phosphorylation via upstream AMPK kinases (21). Activation of AMPK plays an important role in maintaining homeostasis of cellular energy under stress conditions at both cellular and physiological level (22–24). An anti-cancer function after the activation of AMPK has been proposed (25, 26). Activation of AMPK upregulates the autophagic pathway. We found that the expression of p-AMPK in cancerous regions was much more higher than that in non-cancerous regions, which means that autophagy is upregulated in liver cancer. Compared with well-differentiated patients, the expression of p-AMPKα in patients with moderately and poorly differentiated tumors was upregulated more significantly. As the degree of differentiation decreased, the extent of upregulation in the expression of p-AMPKα increased. 4-HNE can directly action on AMPK. In HepG2 cells, 4-HNE works in two ways. Firstly, 4-NHE can inhibit the activation of AMPK by H2O2. Secondly, 4-NHE inhibits the activity of AMPK via the direct modification of recombinant AMPK (27). Our results showed that the 4-HNE'capacity of inhibiting the activity of AMPK decreased, the expression of p-AMPK was upregulated, and the energy consumption of tumor cells increased, which affected subsequent changes.
The Downstream of AMPK, Nuclear Expression of Erk1/2 and p-Erk1/2 U-Regulated in Cancerous Tissues, and p-Erk1/2 Is Associated With Liver Differentiation
Aberrant activation of the ERK pathway has been confirmed to be an essential feature for various human tumors (9). The activated Erk1/2 can phosphorylate various downstream substrates involved in the cellular responses of cell survival, cell motility, cell proliferation, and cell differentiation. Recently, Xiao et al. (11) reported that Erk1/2 can promote the ATG7-dependent autophagy, which is a novel beneficial effect for Erk1/2 in liver steatosis. The MEK/ERK pathway was upstream of TSC and mTOR and downstream of AMPK (8). The MEK/ERK module can regulate the Beclin 1 level via the AMPK-MEK/ERK-TSC-mTOR pathway, thus regulating the autophagy. In our study, the total, cytoplasmic, and nuclear expressions and the rate of nuclear localization of Erk1/2 were significantly higher in the cancerous regions than that in non-cancerous regions, which was in accordance with other scientists' studies (28). Total and cytoplasmic expressions of p-Erk1/2 showed no significant difference between the cancerous and non-cancerous regions. However, its nuclear expression and the rate of nuclear localization were significantly upregulated in the cancerous regions compared to that in non-cancerous regions. The upregulation of Erk1/2 may be caused by the increase of p-Erk1/2 in the nucleus. Although no significant difference was occurred in the expressions of Erk1/2 in patients between the two groups, the nuclear expression and rate of nuclear localization of p-Erk1/2 showed significant difference in the two groups. That is, as the degree of differentiation decreased, the extent of upregulation in the nuclear expression of p-Erk1/2 increased. Combined with the downregulation of p-AMPK, this further confirmed the up-regulation of autophagy in cancerous regions.
Another Downstream of AMPK, Nuclear Expression of CARM1, Increased in Cancerous Tissues
Biochemically, as a cofactor for several transcription factors and nuclear hormone receptors, the histone methyltransferase of CARM1 belongs to a family of arginine methyltransferases, and is involved in the tumorigenesis of various cancers, such as ovarian cancer (29). Osada et al. (30) found that the expression of CARM1 during hepatocellular carcinogenesis increased in adenomas and was aberrant in carcinomas. Shin et al. (13) recently found that CARM1 was a key component among the AMPK-SKP2-CARM1 signaling axis in autophagy in mammals. In the current study, the nucleus expression and the rate of nucleus localization of CARM1 were significantly higher in cancerous regions than that in non-cancerous regions, which is in accordance with the results mentioned above. Combined with the downregulation of p-AMPK, this further confirmed the upregulation of autophagy in cancerous regions.
Reduction of Total Expression of TFEB in Cancerous Tissues, Indicating Impaired Lysosomal Synthesis
Autophagy has been found to selectively degrade specific cellular constituents in the lysosome (31). TFEB controls the process of lysosomal biogenesis and autophagy. It always localizes to the cytosol, and will mobilize into the nucleus when lysosomal function is compromised or starvation conditions was reached (32). Study has shown that TFEB is regulated by Erk1/2 (33). TFEB overexpression can lead to an increase of autophagosomes and autophagic flux, generation of new lysosomes, and clearance of storage material in several disordered lysosomal storage through the promotion of lysosomal secretion (34). Overexpression of TFEB was occurred for the patients with renal cell carcinoma (35), and it was confirmed to accelerate tumorigenesis by inducing various oncogenic signals. Recently, a study (36) showed that TFEB in the liver of mice reduced the liver damage caused by alcohol. In our study, compared with noncancerous regions, the total and cytoplasmic expressions of TFEB were significantly reduced in cancerous regions. However, the nuclear expression and the rate of nuclear localization had no significant difference in two regions, showing that the synthesis of lysosomes was affected in the course of liver cancer. Interestingly, compared with well-differentiated liver cancer patients, the nuclear expression of TFEB in patients with moderately and poorly differentiated tumors was upregulated significantly.
The Downstream of TFEB, Expression of LAMP1 Was Upregulated in Cancerous Tissues, Indicating an Increase in the Number of Lysosomes in Cancerous Tissues
LAMP1, as a downstream effector of TFEB (37), is a key protein in the biogenesis of lysosomal. It has assumed importance as a marker of lysosomes in various biological studies, with the most abundant lysosomal membrane proteins being LAMP-1, which represents membrane proteins of the lysosomal membrane. The expression of LAMP-1 occurred on the surface of various cancer cells, such as colon cancer (38). LAMP1 can significantly promote the development, proliferation, and metastasis of cancer, and the downregulation of LAMP1 can inhibit the metastasis of cancer (39). In our study, LAMP1 was upregulated significantly in cancerous regions, which suggested an increase in the number of lysosomes in cancerous regions. Combined with the results of TFEB mentioned above, we hypothesized that the function of lysosomes was not seriously damaged in the well-differentiated stage of liver cancer. In moderately and poorly differentiated tumors, the damage to the lysosomes was aggravated. The nuclear expression of TFEB in cancerous tissues was significantly increased, which led to upregulation of LAMP1 expression, but the factors that affect changes in LAMP1 in well-differentiated tumors require further research.
Specific Autophagy Degradation Substrate p62/SQSTM1 Was Downregulated in Cancerous Tissues, Indicating That Autophagy May Be More Active in Cancerous Areas
The expression of many autophagy-lysosomal genes, including p62/SQSTM1, was controlled by TFEB (40). p62 can achieve their degradation in the lysosome by linking ubiquitinated proteins to the autophagic machinery (41). The amount of p62/SQSTM1 will markedly accumulates when autophagy is suppressed, while decreases when autophagy is activated. Therefore, p62 is a common marker for autophagic flux (42). The expression of p62 was downregulated in cancerous regions, which suggested that autophagy in cancerous regions was more active than in non-cancerous regions.
Conclusion
Based on these facts, we hypothesized that, during the progression of liver tumors, there were AMPK-ERK/CARM1 autophagic signaling pathways. Followed by oxidative stress, the expression of 4-HNE was downregulated, and hence its ability to inhibit AMPK decreased. This subsequently led to increase in phosphorylation and alteration in the nuclear localization of AMPK, which further enhanced the rate of nuclear localization of p-Erk1/2 and elevated the expression of Erk1/2. In addition, the rate of nuclear localization of CARM1, another downstream of AMPK, was also enlarged. Other changes, including the reduction of TFEB and p62 and the up-regulation of LAMP1, were also observed. The above-mentioned changes in signals indicated an upregulation of autophagy in cancerous regions, which means that overactivated autophagy plays an important role in liver cancer.
Data Availability Statement
All datasets generated for this study are included in the article/supplementary material.
Ethics Statement
The studies involving human participants were reviewed and approved by the Committee on the Ethics of Experiments of the South-Central University for Nationalities in China (Permit Number: 2017-SCUEC-MEC-007). The patients/participants provided their written informed consent to participate in this study.
Author Contributions
Q-FQ, Y-SL, and H-BT performed the experiments. Q-FQ, X-JL, Y-SL, WZ, G-HT, H-CS, and H-BT analyzed the data. Y-SL, WZ, G-HT, H-CS, and H-BT contributed reagents, materials, and analysis tools. Q-FQ, Y-SL, WZ, G-HT, H-CS, and H-BT wrote the paper.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding. This work was supported by the National Natural Science Foundation of China (81573887, 81673711, and 81674050).
References
- 1.Choi AM, Ryter SW, Levine B. Autophagy in human health and disease. N Engl J Med. (2013) 368:1845–6. 10.1056/NEJMc1303158 [DOI] [PubMed] [Google Scholar]
- 2.Sarkar S, Ravikumar B, Floto RA, Rubinsztein DC. Rapamycin and mTOR-independent autophagy inducers ameliorate toxicity of polyglutamine-expanded huntingtin and related proteinopathies. Cell Death Differ. (2009) 16:46–56. 10.1038/cdd.2008.110 [DOI] [PubMed] [Google Scholar]
- 3.Dodson M, Liang Q, Johnson MS, Redmann M, Fineberg N, Darley-Usmar VM, et al. Inhibition of glycolysis attenuates 4-hydroxynonenal-dependent autophagy and exacerbates apoptosis in differentiated SH-SY5Y neuroblastoma cells. Autophagy. (2013) 9: 1996–2008. 10.4161/auto.26094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chaudhary P, Sharma R, Sharma A, Vatsyayan R, Yadav S, Singhal SS, et al. Mechanisms of 4-hydroxy-2-nonenal induced pro- and anti-apoptotic signaling. Biochemistry. (2010) 49:6263–75. 10.1021/bi100517x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang Z, Wang S, Zhou S, Yan X, Wang Y, Chen J, et al. Sulforaphane prevents the development of cardiomyopathy in type 2 diabetic mice probably by reversing oxidative stress-induced inhibition of LKB1/AMPK pathway. J Mol Cell Cardiol. (2014) 77:42–52. 10.1016/j.yjmcc.2014.09.022 [DOI] [PubMed] [Google Scholar]
- 6.Hardie DG. AMP-activated/SNF1 protein kinases: conserved guardians of cellular energy. Nat Rev Mol Cell Biol. (2007) 8:774–85. 10.1038/nrm2249 [DOI] [PubMed] [Google Scholar]
- 7.Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. (2011) 13:132–41. 10.1038/ncb2152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang J, Whiteman MW, Lian H, Wang G, Singh A, Huang D, et al. A non-canonical MEK/ERK signaling pathway regulates autophagy via regulating Beclin 1. J Biol Chem. (2009) 284:21412–24. 10.1074/jbc.M109.026013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kohno M, Pouyssegur J. Targeting the ERK signaling pathway in cancer therapy. Ann Med. (2006) 38:200–11. 10.1080/07853890600551037 [DOI] [PubMed] [Google Scholar]
- 10.Thapalia BA, Zhou Z, Lin X. Sauchinone augments cardiomyocyte viability by enhancing autophagy proteins -PI3K, ERK(1/2), AMPK and Beclin-1 during early ischemia-reperfusion injury in vitro. Am J Transl Res. (2016) 8:3251–65. [PMC free article] [PubMed] [Google Scholar]
- 11.Xiao Y, Liu H, Yu J, Zhao Z, Xiao F, Xia T, et al. Activation of ERK1/2 ameliorates liver steatosis in leptin receptor-deficient (db/db) mice via stimulating ATG7-dependent autophagy. Diabetes. (2016) 65:393–405. 10.2337/db15-1024 [DOI] [PubMed] [Google Scholar]
- 12.Yin Y, Zhao Y, Han S, Zhang N, Chen H, Wang X. Autophagy-ERK1/2-involved disinhibition of hippocampal neurons contributes to the pre-synaptic toxicity induced by a β42 exposure. J Alzheimers Dis. (2017) 59: 851–69. 10.3233/JAD-170246 [DOI] [PubMed] [Google Scholar]
- 13.Shin HJ, Kim H, Oh S, Lee JG, Kee M, Ko HJ, et al. AMPK-SKP2-CARM1 signalling cascade in transcriptional regulation of autophagy. Nature. (2016) 534:553–7. 10.1038/nature18014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li YS, Wang JX, Jia MM, Liu M, Li XJ, Tang HB. Dragon's blood inhibits chronic inflammatory and neuropathic pain responses by blocking the synthesis and release of substance P in rats. J Pharmacol Sci. (2012) 118:43–54. 10.1254/jphs.11160FP [DOI] [PubMed] [Google Scholar]
- 15.Covey TM, Edes K, Coombs GS, Virshup DM, Fitzpatrick FA. Alkylation of the tumor suppressor PTEN activates Akt and β-catenin signaling: a mechanism linking inflammation and oxidative stress with cancer. PLoS ONE. (2010) 5:e13545. 10.1371/journal.pone.0013545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pettazzoni P, Pizzimenti S, Toaldo C, Sotomayor P, Tagliavacca L, Liu S, et al. Induction of cell cycle arrest and DNA damage by the HDAC inhibitor panobinostat (LBH589) and the lipid peroxidation end product 4-hydroxynonenal in prostate cancer cells. Free Radic Biol Med. (2011) 50:313–22. 10.1016/j.freeradbiomed.2010.11.011 [DOI] [PubMed] [Google Scholar]
- 17.Uchida K. 4-Hydroxy-2-nonenal: a product and mediator of oxidative stress. Prog Lipid Res. (2003) 42:318–43. 10.1016/S0163-7827(03)00014-6 [DOI] [PubMed] [Google Scholar]
- 18.Dodson M, Wani WY, Redmann M, Benavides GA, Johnson MS, Ouyang X, et al. Regulation of autophagy, mitochondrial dynamics, and cellular bioenergetics by 4-hydroxynonenal in primary neurons. Autophagy. (2017) 13:1828–40. 10.1080/15548627.2017.1356948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bonet-Ponce L, Saez-Atienzar S, da Casa C, Sancho-Pelluz J, Barcia JM, Martinez-Gil N, et al. Rotenone induces the formation of 4-hydroxynonenal aggresomes. Role of ROS-mediated tubulin hyperacetylation and autophagic flux disruption. Mol Neurobiol. (2016) 53:6194–208. 10.1007/s12035-015-9509-3 [DOI] [PubMed] [Google Scholar]
- 20.Zhong H, Xiao M, Zarkovic K, Zhu M, Sa R, Lu J, et al. Mitochondrial control of apoptosis through modulation of cardiolipin oxidation in hepatocellular carcinoma: a novel link between oxidative stress and cancer. Free Radic Biol Med. (2017) 102:67–76. 10.1016/j.freeradbiomed.2016.10.494 [DOI] [PubMed] [Google Scholar]
- 21.Jung SN, Park IJ, Kim MJ, Kang I, Choe W, Kim SS, et al. Down-regulation of AMP-activated protein kinase sensitizes DU145 carcinoma to Fas-induced apoptosis via c-FLIP degradation. Exp Cell Res. (2009) 315:2433–41. 10.1016/j.yexcr.2009.05.018 [DOI] [PubMed] [Google Scholar]
- 22.Shackelford DB, Shaw RJ. The LKB1-AMPK pathway: metabolism and growth control in tumour suppression. Nat Rev Cancer. (2009) 9:563–75. 10.1038/nrc2676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mihaylova MM, Shaw RJ. The AMPK signalling pathway coordinates cell growth, autophagy and metabolism. Nat Cell Biol. (2011) 13:1016–23. 10.1038/ncb2329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vakana E, Altman JK, Platanias LC. Targeting AMPK in the treatment of malignancies. J Cell Biochem. (2012) 113:404–9. 10.1002/jcb.23369 [DOI] [PubMed] [Google Scholar]
- 25.Zhao Z, Feng L, Wang J, Cheng D, Liu M, Ling M, et al. NPC-26 kills human colorectal cancer cells via activating AMPK signaling. Oncotarget. (2017) 8 18312–21. 10.18632/oncotarget.15436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Law BY, Mok SW, Chan WK, Xu SW, Wu AG, Yao XJ, et al. Hernandezine, a novel AMPK activator induces autophagic cell death in drug-resistant cancers. Oncotarget. (2016) 7:8090–104. 10.18632/oncotarget.6980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shearn CT, Backos DS, Orlicky DJ, Smathers-McCullough RL, Petersen DR. Identification of 5' AMP-activated kinase as a target of reactive aldehydes during chronic ingestion of high concentrations of ethanol. J Biol Chem. (2014) 289:15449–62. 10.1074/jbc.M113.543942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang X, Yang H, Yue S, He G, Qu S, Zhang Z, et al. The mTOR inhibition in concurrence with ERK1/2 activation is involved in excessive autophagy induced by glycyrrhizin in hepatocellular carcinoma. Cancer Med. (2017) 6:1941–51. 10.1002/cam4.1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakayama N, Sakashita G, Nariai Y, Kato H, Sinmyozu K, Nakayama JI, et al. Cancer-related transcription regulator protein NAC1 forms a protein complex with CARM1 for ovarian cancer progression. Oncotarget. (2018) 9:28408–20. 10.18632/oncotarget.25400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Osada S, Suzuki S, Yoshimi C, Matsumoto M, Shirai T, Takahashi S, et al. Elevated expression of coactivator-associated arginine methyltransferase 1 is associated with early hepatocarcinogenesis. Oncol Rep. (2013) 30:1669–74. 10.3892/or.2013.2651 [DOI] [PubMed] [Google Scholar]
- 31.Chourasia AH, Boland ML, Macleod KF. Mitophagy and cancer. Cancer Metab. (2015) 3:4. 10.1186/s40170-015-0130-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sardiello M, Palmieri M, di Ronza A, Medina DL, Valenza M, Gennarino VA, et al. A gene network regulating lysosomal biogenesis and function. Science. (2009) 325:473–7. 10.1126/science.1174447 [DOI] [PubMed] [Google Scholar]
- 33.Shen HM, Mizushima N. At the end of the autophagic road: an emerging understanding of lysosomal functions in autophagy. Trends Biochem Sci. (2014) 39:61–71. 10.1016/j.tibs.2013.12.001 [DOI] [PubMed] [Google Scholar]
- 34.Medina DL, Fraldi A, Bouche V, Annunziata F, Mansueto G, Spampanato C, et al. Transcriptional activation of lysosomal exocytosis promotes cellular clearance. Dev Cell. (2011) 21:421–30. 10.1016/j.devcel.2011.07.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Calcagni A, Kors L, Verschuren E, De Cegli R, Zampelli N, Nusco E, et al. Modelling TFE renal cell carcinoma in mice reveals a critical role of WNT signaling. Elife. (2016) 5:e17047. 10.7554/eLife.17047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chao XJ, Wang SG, Zhao K, Li Y, Williams JA, Li TG, et al. Impaired TFEB-mediated lysosome biogenesis and autophagy promote chronic ethanol-induced liver injury and steatosis in mice. Gastroenterology. (2018) 155:865–6. 10.1053/j.gastro.2018.05.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li F, Lang F, Zhang H, Xu L, Wang Y, Hao E. Role of TFEB Mediated autophagy, oxidative stress, inflammation, and cell death in endotoxin induced myocardial toxicity of young and aged mice. Oxid Med Cell Longev. (2016) 2016:5380319. 10.1155/2016/5380319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Piao S, Amaravadi RK. Targeting the lysosome in cancer. Ann N Y Acad Sci. (2016) 1371:45–54. 10.1111/nyas.12953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Agarwal AK, Srinivasan N, Godbole R, More SK, Budnar S, Gude RP, et al. Role of tumor cell surface lysosome-associated membrane protein-1 (LAMP1) and its associated carbohydrates in lung metastasis. J Cancer Res Clin Oncol. (2015) 141:1563–74. 10.1007/s00432-015-1917-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Palmieri M, Impey S, Kang H, di Ronza A, Pelz C, Sardiello M, et al. Characterization of the CLEAR network reveals an integrated control of cellular clearance pathways. Hum Mol Genet. (2011) 20:3852–66. 10.1093/hmg/ddr306 [DOI] [PubMed] [Google Scholar]
- 41.Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. (2007) 282:24131–45. 10.1074/jbc.M702824200 [DOI] [PubMed] [Google Scholar]
- 42.Cheng Y, Ren X, Hait WN, Yang JM. Therapeutic targeting of autophagy in disease: biology and pharmacology. Pharmacol Rev. (2013) 65:1162–97. 10.1124/pr.112.007120 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All datasets generated for this study are included in the article/supplementary material.