Abstract
Expression of the vasa gene is associated with germline establishment. Therefore, identification of vasa activator(s) should provide insights into germline development. However, the genes sufficient for vasa activation remain unknown. Previously, we showed that the BTB/POZ-Zn-finger protein Mamo is necessary for vasa expression in Drosophila. Here, we show that the truncated Mamo lacking the BTB/POZ domain (MamoAF) is a potent vasa activator. Overexpression of MamoAF was sufficient to induce vasa expression in both primordial germ cells and brain. Indeed, Mamo mRNA encoding a truncated Mamo isoform, which is similar to MamoAF, was predominantly expressed in primordial germ cells. The results of our genetic and biochemical studies showed that MamoAF, together with CBP, epigenetically activates vasa expression. Furthermore, MamoAF and the germline transcriptional activator OvoB exhibited synergy in activating vasa transcription. We propose that a Mamo-mediated network of epigenetic and transcriptional regulators activates vasa expression.
Subject terms: Oogenesis, Cell biology
Shoichi Nakamura, Seiji Hira et al. demonstrate that a truncated form of Mamo lacking the BTB/POZ domain (MamoAF) is a potent transcriptional activator of a germline gene vasa. This study suggests that MamoAF cooperates with transcriptional activators CBP and OvoB to regulate the development of germline cells.
Introduction
The germline is the only cell lineage that contributes to the production of the next generation. Accordingly, germline cells exhibit properties, including stemness and longevity, that are distinct from those of somatic cells. In animals, the germline is specified by maternal determinants or inductive signals1. Irrespective of the mode of specification, primordial germ cells (PGCs) express conserved germline genes. vasa (vas), which encodes a DEAD-box RNA helicase2, is a highly conserved germline gene, and its expression is a marker of germline establishment in many species1. For example, Ddx4, the human ortholog of Vas, is used as a molecular biomarker for germ cells3. Moreover, genetic studies have demonstrated that Vas is required for gametogenesis in Drosophila, C. elegans, zebrafish and mouse4–7. Vas has essential roles that contribute to germline development, including translational regulation of specific mRNAs and production of Piwi-interacting RNAs8,9.
In Drosophila, maternal factors localised in the germ plasm are partitioned into PGCs to direct germline development. For example, the mitochondrial large ribosomal RNA and the Germ cell-less protein are both involved in PGC formation10. The maternal translational repressor Nanos is essential for repressing somatic differentiation of PGCs11. The Polar granule component protein has an essential role in PGC maintenance by inhibiting the positive transcription elongation factor b in newly formed PGCs12. The maternal Trapped in endoderm-1 protein is required for PGC migration through the midgut wall13. The germ plasm is necessary and sufficient for germ cell establishment14, suggesting that maternal transcription factors localised in the germ plasm activate vas expression in PGCs. Indeed, RNA interference-mediated knockdown of maternal mRNAs enriched in the germ plasm has revealed that six transcription factors are required for the expression of germline genes in PGCs15. Among them, the transcriptional activator OvoB, which is encoded by the ovo gene, is predominantly expressed in PGCs during embryogenesis, and its maternal function is essential for germline development16. However, the genes sufficient for vas activation remain unknown.
Previously, we showed that the maternal factor Mamo, which contains both a BTB/POZ domain and C2H2 Zn-finger domains and is enriched in PGCs, is necessary for vas expression in PGCs17. Mamo protein is detectable in the nuclei of PGCs at stage 15, in which vas is actively expressed17. Mamo can bind chromosomes when Mamo is expressed in salivary gland cells18. Biochemical analyses demonstrated that the C2H2 Zn-finger domains of Mamo directly bind specific DNA sequences18. C2H2 Zn-finger domains are among the most common domains in the transcription factors of metazoans19. The Zn-finger domains of Mamo are homologous to those of human Sp1;20 in vertebrates, Sp1-related transcription factors regulate gene expression in germ cells21–24. Accordingly, we focused on Mamo and investigated its role in vas activation in Drosophila embryos.
Biochemical analysis is an essential approach to understanding the molecular basis of transcriptional activation. To date, however, biochemical studies of vas activation have been difficult owing to the small number of PGCs in embryos. Accordingly, an experimental system capable of inducing forced expression of vas could be a powerful tool for analysing the mechanism of vas activation. In this study, we found that an N-terminal truncated Mamo isoform lacking the BTB/POZ domain but retaining the C2H2 Zn-finger domains was a potent vas activator. Maternal overexpression of the truncated Mamo (MamoAF) was sufficient to activate vas expression in both PGCs and brain in embryos. Indeed, Mamo mRNA encoding a truncated Mamo isoform, which is similar to MamoAF, was predominantly expressed in PGCs, in which vas is actively expressed. Moreover, MamoAF could partially rescue the mamo mutant phenotype. Therefore, through biochemical and genetic analyses in which vas expression was induced by MamoAF, we elucidated the molecular mechanisms of vas activation. MamoAF directly bound the vas locus and, together with CBP, epigenetically activated vas expression. In addition, we identified the germline transcriptional activator OvoB as a cofactor of MamoAF. MamoAF physically interacted with OvoB, and the two proteins exhibited synergy in activating vas transcription. Together, these findings demonstrate that Mamo-mediated transcriptional regulation directly activates vas expression.
Results
Mamo short isoform may be associated with vas activation
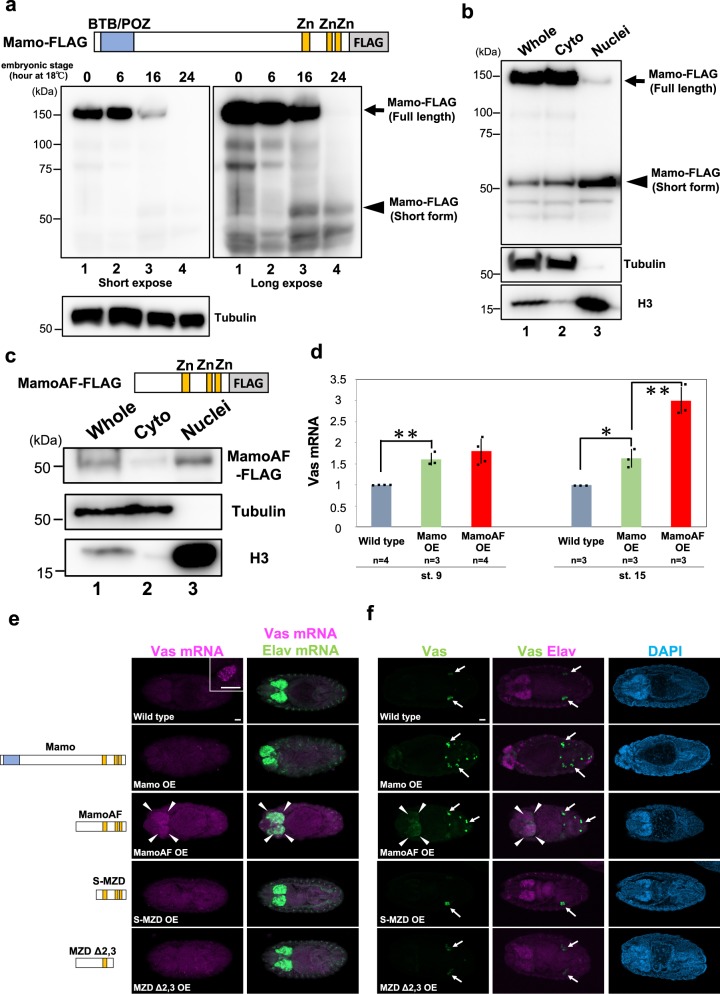
To evaluate the molecular function of Mamo protein, we expressed a C-terminally FLAG-tagged allele of Mamo that can rescue the mamo mutant phenotype17 in embryos, under the control of the maternal Gal4 driver (matα4-Gal-VP16). During embryogenesis, maternally expressed FLAG-tagged Mamo (~ 150 kDa) was processed into shorter proteins (~ 50 kDa) that lacked the N-terminal BTB/POZ domain but retained the C-terminal Zn-finger DNA-binding domains (Fig. 1a). Full-length Mamo, as well as its ~ 100-kDa and ~ 75-kDa derivatives, was detected in early embryos, in which zygotic vas expression is not yet activated (Fig. 1a, lanes 1 and 2). By contrast, in later embryos, in which zygotic vas expression is activated, the level of full-length Mamo was considerably reduced, and the ~ 50-kDa derivatives of Mamo became detectable (Fig. 1a, lanes 3 and 4). These observations imply that maternally provided Mamo is gradually processed into shorter derivatives through specific proteolytic processes. Moreover, in contrast to full-length Mamo, short derivatives of Mamo preferentially accumulated in the nuclear fraction (Fig. 1b). Based on these observations, we hypothesised that the short isoform of Mamo, which lacks the BTB/POZ domain but retains the Zn-finger domains, (hereafter, referred to as Mamo short isoform) is a potent vas activator.
Fig. 1.
MamoAF overexpression induces vas expression in embryos. a Developmental western blotting analysis of embryos expressing full-length Mamo-FLAG under the control of matα4-Gal-VP16 driver. Embryos were cultured at 18 °C for indicated hours and subjected to western blotting with anti-FLAG antibody. Embryos cultured at 18 °C for 0, 6, 16 and 24 h are corresponded to embryonic stage 4, 9, 12 and 15, respectively. b Analysis of fractionated samples extracted from embryos expressing full-length Mamo-FLAG by western blotting. Embryos expressing Mamo-FLAG under the control of matα4-Gal-VP16 driver were cultured at 25 °C for 0–16 h and fractionated. Normalised load of each extract (7 μg) were analysed by western blotting with anti-FLAG, anti-Tubulin and anti-Histone H3 antibodies. Whole extract (lane 1), cytoplasmic extract (lane 2) and nuclear extract (lane 3) were loaded. c Analysis of fractionated samples extracted from embryos expressing MamoAF-FLAG by western blotting. Embryos expressing MamoAF-FLAG under the control of mat4α-Gal-VP16 driver were cultured at 25 °C for 0–16 h and fractionated. Normalised load of each extract (11 μg) were analysed by western blotting with anti-FLAG, anti-Tubulin and anti-Histone H3 antibodies. Whole extract (lane 1), cytoplasmic extract (lane 2) and nuclear extract (lane 3) were loaded. Full images of the electrophoreses are shown in Supplementary Fig. 16. d qRT-PCR analyses of Vas mRNA in wild type, Mamo and MamoAF overexpressed embryos under the control of matα4-Gal-VP16 driver. Error bars are SD of the mean. **P < 0.01, *P < 0.05 (two-tailed Student’s t test). e Double fluorescence in situ hybridisation for Vas and Elav mRNAs of wild type, the embryos overexpressing Mamo, MamoAF and MamoAF fragments under the control of matα4-Gal-VP16 driver. Elav is a neural marker. Confocal sections of the brain of the embryos are shown. The inset shows close-up image of wild-type PGCs. f Double-staining for Vas and neural marker Elav of wild type, the embryos overexpressing Mamo, MamoAF and MamoAF fragments under the control of matα4-Gal-VP16 driver. Arrows indicate PGCs expressing Vas. Arrowheads indicate ectopic Vas expression. Scale bar: 30 μm
Mamo mRNA encoding Mamo short isoform is expressed in PGCs
Next, we investigated whether Mamo mRNA encoding the Mamo short isoform is expressed in PGCs. Expressed sequence tags (GM29022 and GM29051) have shown that the short Mamo mRNA is expressed in ovaries20. 5′-rapid amplification of cDNA ends (RACE) confirmed that short Mamo mRNA is expressed in ovaries. The sequences of 5′-RACE clones revealed that the short Mamo mRNA encodes the Mamo short isoform, which lacks the BTB/POZ domain, and arises owing to the use of alternative transcription initiation sites (Supplementary Fig. 1). To investigate the expression patterns of short Mamo mRNA, we performed in situ hybridisation using a Mamo Zn-finger probe, which detects both short and full-length Mamo mRNAs, and a Mamo BTB probe, which specifically detects the full-length Mamo mRNA (Supplementary Fig. 2a). Mamo mRNA signals were detected in germ cells in the ovaries when using a Mamo Zn-finger probe or Mamo BTB probe (Supplementary Fig. 2b), confirming that these probes are available. These results are consistent with previous studies showing that full-length Mamo mRNA is expressed in the ovary17,20. By contrast, during embryogenesis, Mamo mRNA signals were specifically detected in PGCs only when using a Mamo Zn-finger probe, but not a Mamo BTB probe (Supplementary Fig. 2b). In the early embryos, at stages 5–9, no Mamo mRNA signal was detected, whereas at stages 14–16, the Mamo mRNA signals were more intense in PGCs, suggesting that Mamo mRNA encoding the Mamo short isoform lacking the BTB/POZ domain is zygotically expressed in PGCs, in which vas is actively expressed. Antibodies specific for the Mamo short isoform are not yet available, because the amino-acid sequence of the Mamo short isoform is present within that of full-length Mamo. These observations indicate that Mamo short isoform is predominantly expressed in PGCs, and that this isoform may promote vas expression.
MamoAF can partially rescue the mamo mutant phenotype
Because a mamo mutant specific for the short isoform was not available, we used overexpression experiments to evaluate the function of the short isoform. Previously, we generated a UAS line (UAS-MZD-FLAG) to produce a truncated Mamo protein lacking the BTB/POZ domain18 (hereafter, referred to as MamoAF). When it was expressed in embryos under the control of matα4-Gal-VP16, MamoAF preferentially accumulated in the nuclear fraction (Fig. 1c), just like the short derivatives of Mamo (Fig. 1b). Next, we investigated whether MamoAF overexpression can rescue the mamo mutant phenotype. Egg chambers containing the germ cells derived from mamo mutant germline clones were rarely observed in ovarioles (1.4%, n = 74) (Supplementary Fig. 3, Supplementary Table 1). However, when full-length Mamo-FLAG or MamoAF-FLAG was expressed in germ cells under the control of the nanos-Gal4-VP16 (nos-Gal4) driver, differentiating egg chambers containing germ cells derived from mamo mutant germline clones after oogenic stage 6 were observed in 28.8% and 13.2% of ovarioles, respectively (Supplementary Fig. 3, Supplementary Table 1). Mamo or MamoAF overexpression could rescue the mamo mutant phenotype (P < 0.01, Fisher’s exact test). We speculated that the low rescue activity of Mamo may be due to the expression level of Mamo driven by nos-Gal4, which is different from the endogenous mamo promoter. Next, we investigated whether MamoAF overexpression can rescue the differentiation of mamo mutant germ cells into mature eggs. Overexpression of full-length Mamo, but not MamoAF, rescued the differentiation of mamo mutant germ cells into mature eggs (Supplementary Table 2). To assess the difference in rescue ability between full-length Mamo and MamoAF, we examined the expression patterns of full-length Mamo-FLAG or MamoAF-FLAG in ovaries when these proteins were expressed under the control of nos-Gal4. Full-length Mamo was enriched both in the nuclei of germline cysts and in the nuclei of the nurse cells in egg chambers after oogenic stage 6 (Supplementary Fig. 4). In 65.3% of egg chambers at stage 6, full-length Mamo signals were enriched in the nuclei of the nurse cells (n = 121): 84.3% at stage 7 (n = 115). By contrast, MamoAF was detected in the nuclei of germline cysts, whereas MamoAF signals were weak in the nuclei of nurse cells in egg chambers (Supplementary Fig. 4). In 25.3% of egg chambers at stage 6, MamoAF signals were enriched in the nuclei of the nurse cells (n = 87): 18.1% at stage 7 (n = 72). Together, these findings show that MamoAF could partially rescue the differentiation of mamo mutant germline clones. Accordingly, we used overexpression of MamoAF to investigate the effect of the Mamo short isoform on vas activation.
MamoAF is sufficient to activate vas expression
To determine whether MamoAF can induce vas expression, we monitored vas expression in embryos overexpressing full-length Mamo or MamoAF under the control of matα4-Gal-VP16 driver. Quantitative reverse-transcription PCR (qRT-PCR) revealed that maternal overexpression of full-length Mamo only moderately increased vas expression at stages 9 and 15, whereas MamoAF markedly increased expression of vas at stage 15 (Fig. 1d). In addition, we observed somatic vas expression, especially in brain of embryos overexpressing MamoAF (hereafter, referred to as MamoAF-OE embryos), but not in embryos overexpressing full-length Mamo (Fig. 1e, f). In MamoAF-OE embryos, PGCs formed normally at the posterior poles, and brain cells expressing Vas appeared at stage 14 (Supplementary Fig. 5). These results show that MamoAF overexpression is sufficient to induce vas expression in brain.
Next, we investigated whether MamoAF can promote vas expression in PGCs. Specifically, we monitored vas expression in PGCs when MamoAF was overexpressed under the control of the maternal nos-Gal4 driver16. MamoAF overexpression increased vas expression in PGCs at stages 14 and 15 (Supplementary Fig. 6). We also found that overexpression of full-length Mamo increased vas expression in PGCs at stage 15 (Supplementary Fig. 6). These results indicate that the Mamo short isoform is a potent inducer of vas expression in PGCs.
The mamo gene produces two types of vas activators
Full-length Mamo could promote vas expression in PGCs, but its activity was weaker than that MamoAF (Supplementary Fig. 6). In embryos, full-length Mamo preferentially accumulated in the cytoplasmic fraction relative to MamoAF (Fig. 1b), which may suppress its transcriptional activity. Thus, the BTB/POZ domain may regulate the nuclear localisation of full-length Mamo. Consistent with this notion, overexpression of full-length Mamo did not strongly induce vas expression in brain (Fig. 1e, f). These results suggest that full-length Mamo is a weak but specific activator of vas whose activity is restricted in PGCs. By contrast, MamoAF lacking the BTB/POZ domain preferentially accumulated in the nuclear fraction (Fig. 1c), and potently induced vas in PGCs and brain. Vas mRNA becomes detectable in PGCs at stage 9, when short Mamo mRNAs are not yet zygotically expressed in PGCs. Thus, we propose that maternal full-length Mamo establishes specific vas expression in early PGCs at stage 9. In later PGCs, after stage 14, zygotic Mamo short isoform contributes to the maintenance of robust vas expression. We found that, during embryogenesis, maternally expressed full-length Mamo was processed into shorter proteins (Fig. 1a). Thus, in addition to zygotic Mamo short isoform, the conversion from maternal full-length Mamo into short derivatives may promote vas expression in late PGCs.
Zn-finger domain of MamoAF is required for vas activation
To investigate the mechanisms underlying vas activation, we focused subsequent experiments on MamoAF owing to its potency as a vas activator. To evaluate the functional domains of MamoAF, we examined vas expression in embryos expressing fragments of the protein. Fragments of MamoAF proteins that lacked the N-terminal region (S-MZD) or the C-terminal Zn-finger DNA-binding domains (MZDΔ2, 3) failed to induce vas expression in brain (Fig. 1e, f), indicating that both of these domains are required to induce somatic vas expression. Thus, the Zn-finger domains of MamoAF may directly bind to vas.
MamoAF directly binds and epigenetically activates vas
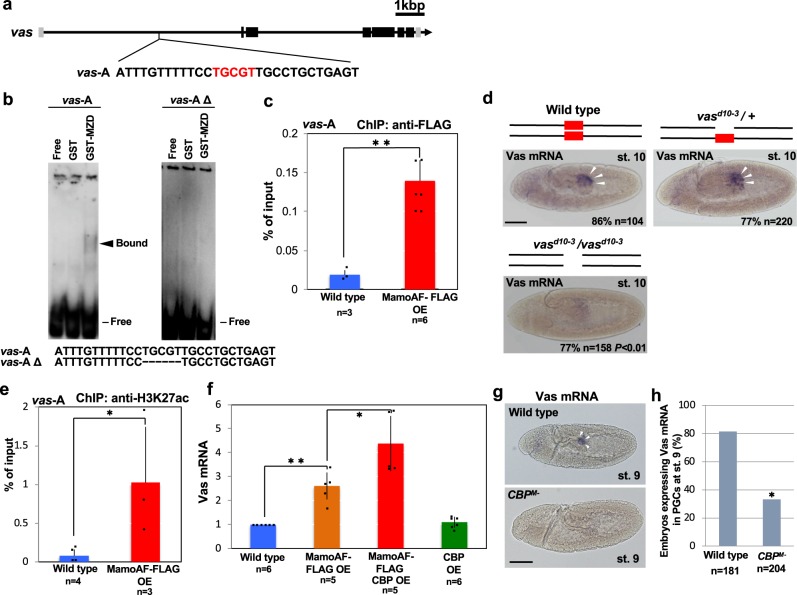
To determine whether MamoAF directly binds vas, we performed electrophoretic mobility shift assays (EMSA) and chromatin immunoprecipitation assays (ChIP) on MamoAF-OE embryos. Although the upstream region (~ 2 kbp) of the vas gene does not contain cis-elements containing a Mamo-binding consensus sequence18, the first intron of vas contains several possible cis-elements, suggesting that MamoAF activates vas through previously unidentified cis-elements in the first intron. One such cis-element, hereafter referred to as the vas-A element, is conserved among several Drosophila species (Supplementary Fig. 7). Hence, we focused subsequent experiments on the vas-A element.
EMSA revealed that the Zn-finger domains of MamoAF directly bound the vas-A element in the first intron of vas (Fig. 2a, b), and ChIP-qPCR assays confirmed that MamoAF bound to this sequence in vivo (Fig. 2c). Next, we investigated whether the vas-A element is required for vas expression. After removal of the Mamo-binding sequence by gene editing, vas expression in PGCs was considerably reduced (Fig. 2d; Supplementary Fig. 8). Vas mRNA signals were detected in PGCs of 86% of wild-type embryos (n = 104) and 77% of embryos heterozygous for vasd10-3 (n = 220), but only 23% of embryos homozygous for vasd10-3, which lacks the vas-A element (n = 158, P < 0.01). Together, these results demonstrate that MamoAF directly activates vas through the vas-A element.
Fig. 2.
MamoAF directly binds vas and epigenetically activates vas together with CBP. a vas-A element, which contains a Mamo-binding consensus sequence (red), in the first intron of vas. b EMSA analyses of Zn-finger domains of Mamo protein (MZD) using vas-A and vas-AΔ probes. vas-AΔ probe lacks a Mamo-binding consensus sequence. c ChIP-qPCR assays were performed with MamoAF-OE embryos and FLAG antibody. The precipitated DNA was detected by qPCR with primers targeting to vas-A. Error bars are SD of the mean. **P < 0.01 (two-tailed Student’s t test). d Vas mRNA in situ hybridisation of wild type, vasd10-3 / + and vasd10-3 / vasd10-3 embryos at stage 10. The Mamo-binding consensus sequence (red box) is deleted from vas-A element in vasd10-3 / vasd10-3embryo. Scale bar: 50 μm. e ChIP-qPCR assays were performed with MamoAF-OE embryos and H3K27ac antibody. The precipitated DNA was detected by qPCR with primers targeting to vas-A. Error bars are SD of the mean. *P < 0.05 (one-tailed Student’s t test). f qRT-PCR analyses of Vas mRNA in wild type, MamoAF, CBP and both CBP and MamoAF overexpressed embryos. Error bars are SD of the mean. *P < 0.05, **P < 0.01 (two-tailed Student’s t test). g Vas mRNA in situ hybridisation of wild type and CBPM- embryo derived from nej0.3 mutant germline clones at stage 9. White arrowheads indicate PGCs expressing Vas mRNA. Scale bar: 50 μm. h Percentages of embryos expressing Vas mRNA in wild type and CBPM- embryos at stage 9. *P < 0.01 (Fisher’s exact test)
MamoAF works with CBP to epigenetically activate vas
We hypothesised that the vas-A element acts as an enhancer. If so, acetylated H3K27 (H3K27ac), which marks active enhancers25–27, should accumulate in this region. ChIP assays revealed greater enrichment of H3K27ac at the vas-A element in MamoAF-OE than in wild-type embryos, suggesting that MamoAF activates vas-A through H3K27ac (Fig. 2e). Because MamoAF lacks a histone acetyltransferase (HAT) domain, MamoAF must collaborate with CBP, which is responsible for acetylation of H3K27 in Drosophila25. Accordingly, we investigated the genetic interaction between MamoAF and CBP. Because CBP is essential for embryogenesis, we examined the outcome of CBP overexpression. Overexpressing both CBP and MamoAF synergistically increased vas expression, whereas CBP overexpression alone did not (Fig. 2f). These results demonstrate that MamoAF collaborates with CBP to epigenetically activate vas.
Next, we investigated whether CBP is required for vas expression in PGCs. In Drosophila, CBP is encoded by nejire (nej)25. In embryos derived from nej germline clones, maternal CBP activity was required for both H3K27ac accumulation (Supplementary Fig. 9) and vas expression in PGCs (Fig. 2g, h). These results show that vas expression in PGCs is epigenetically activated through H3K27ac. These findings are consistent with our idea that Mamo collaborates with CBP to epigenetically activate vas in PGCs.
MamoAF collaborates with OvoB
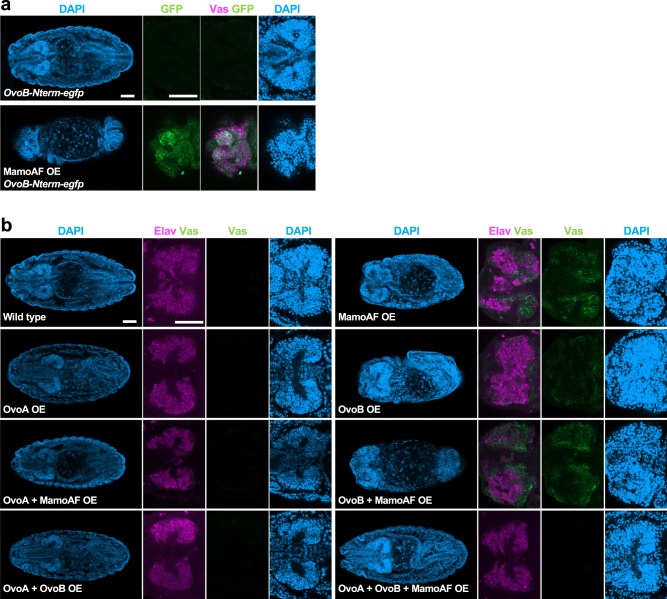
In addition to CBP, we assumed that a transcriptional activator collaborates with MamoAF for vas activation, because MamoAF lacks a transcriptional activation domain. The transcriptional activator OvoB, which is maternally expressed and maintained predominantly in PGCs, is required for vas expression15,16, making it a candidate cofactor of Mamo. However, because OvoB activity is essential for oogenesis28, loss-of-function alleles of ovo are not available for investigating the role of maternal OvoB in vas activation. To overcome this problem, we used MamoAF-induced vas expression to determine whether OvoB cooperates with MamoAF to promote vas expression. If OvoB is essential for vas expression, then MamoAF should induce OvoB expression in brain. We examined the effects of MamoAF on OvoB expression using the ovoB-Nterm-egfp knock-in allele, which enabled us to detect OvoB as GFP fluorescence16 (Supplementary Fig. 10a). As expected, MamoAF overexpression induced GFP expression from the ovoB-Nterm- egfp allele, especially in brain expressing vas (Fig. 3a). The ovo locus produces three proteins: OvoA and OvoB, which serve, respectively, as a transcriptional repressor and activator in germ cells, and Shavenbaby (Svb), which regulates epidermal differentiation29. In situ hybridisation of MamoAF-OE embryos detected Ovo mRNA signals in brain when using an Ovo common probe, but not an Svb-specific probe (Supplementary Fig. 10b–q). Because OvoA is a transcriptional repressor, MamoAF probably induces OvoB in brain expressing vas. Although the mechanism remains elusive, these observations show that MamoAF induces OvoB expression in brain. Therefore, MamoAF may collaborate with OvoB transcriptional activator to promote vas expression in brain.
Fig. 3.
MamoAF collaborates with OvoB to induced Vas expression in brain. a Immunostaining of embryos carrying OvoB-Nterm-egfp allele and MamoAF OE carrying OvoB-Nterm-egfp allele showing MamoAF induces ectopic GFP expression from the OvoB-Nterm-egfp allele. MamoAF is overexpressed under the control of maternal matα4-Gal-VP16 driver. Scale bar: 50 μm. b Wild type, MamoAF OE, OvoA OE, OvoB OE, OvoA + MamoAF OE, OvoB + MamoAF OE, OvoA + OvoB OE and OvoA + OvoB + MamoAF OE are stained for Vas, Elav and DAPI. The transgenes are overexpressed under the control of maternal matα4-Gal-VP16 driver. Whole embryos are shown in left, close-up views of brain of the embryos are in right panels. Scale bar: 50 μm
The OvoA transcriptional repressor can antagonise OvoB-induced transcription;16,28 therefore, if MamoAF collaborates with OvoB, then OvoA should suppress MamoAF-induced vas expression. Indeed, OvoA overexpression suppressed MamoAF-induced vas expression in brain. No somatic Vas expression was observed in embryos overexpressing both MamoAF and OvoA (n = 147) (Fig. 3b). We also confirmed the absence of somatic Vas expression in wild-type embryos (n = 175), embryos expressing OvoA (n = 116), embryos expressing both OvoA and OvoB (n = 82), and embryos expressing OvoA, OvoB and MamoAF (n = 35) (Fig. 3b). Taken together, these results show that MamoAF collaborates with OvoB to induce Vas expression in brain.
Because MamoAF induces OvoB expression in brain (Fig. 3a, Supplementary Fig. 10b–q), OvoB may act downstream of MamoAF. Thus, we tested whether OvoB is sufficient to induce Vas expression in brain. We evaluated vas expression in embryos overexpressing OvoB. Strong Vas signals in brain were detected in MamoAF-OE embryos (58.3%, n = 127), but not in OvoB-OE embryos (n = 71). OvoB overexpression induced much weaker somatic Vas expression (35.2%, n = 71) than MamoAF-OE embryos (Fig. 3b). This observation indicates that OvoB alone is not sufficient to induce strong Vas expression in brain. Vas expression in brain was increased in embryos expressing both MamoAF and OvoB compared with embryos expressing OvoB alone. The increased Vas expression in brain appears to be owing to the enhancement of OvoB expression induced by MamoAF. However, Vas expression in brain of embryos expressing both MamoAF and OvoB (66.7%, n = 63) was not significantly increased relative to embryos expressing MamoAF alone (58.3%, n = 127) (Fig. 3b). This indicates that the promotion of vas expression in brain cannot be explained by the increased expression of OvoB alone. These results suggest that MamoAF collaborates with OvoB to promote vas expression in brain.
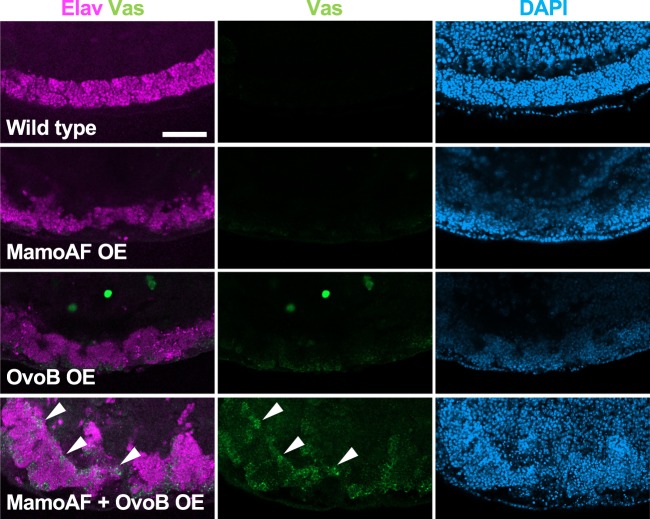
Given that the collaboration between MamoAF and OvoB are essential for vas activation, overexpression of both MamoAF and OvoB should activate vas expression in somatic cells, where MamoAF cannot induce OvoB. Indeed, overexpression of both MamoAF and OvoB synergistically induced Vas expression in somatic cells in the ventral nerve cord (VNC) (55.6%, n = 63), a region where no expression was observed in embryos expressing MamoAF or OvoB alone (n = 127, n = 71, respectively) (Fig. 4). Overexpression of both MamoAF and OvoB is sufficient for activating vas expression in VNC. However, other somatic cells exhibited little response to MamoAF and OvoB. Thus, vas expression in somatic cells is dependent on their cellular contexts. Although the pertinent differences in cellular context remain elusive, these observations show that both MamoAF and OvoB are essential for vas activation both in brain and VNC.
Fig. 4.
Overexpression of both MamoAF and OvoB is sufficient to induce Vas expression in VNC. Wild type, MamoAF OE, OvoB OE and MamoAF + OvoB OE are stained for Vas, Elav and DAPI. The transgenes are overexpressed under the control of maternal matα4-Gal-VP16 driver. Close-up views of ventral region of embryos are shown. White arrowheads indicate ectopic Vas expression in the VNC. Scale bar: 50 μm
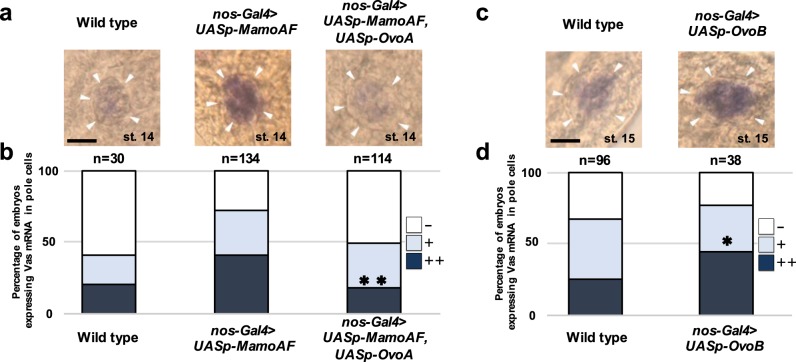
Next, we investigated whether Mamo collaborates with OvoB to promote vas expression in PGCs. Because OvoB activity is essential for oogenesis28, genetic approaches using loss-of-function alleles of ovo are not available for investigating the role of maternal OvoB in PGCs16 of progeny. To circumvent this obstacle, we paternally introduced the UASp-ovoA transgene into embryos derived from female adults carrying both UASp-MamoAF and the nos-Gal4 driver. We found that OvoA suppressed MamoAF-induced vas expression in PGCs (Fig. 5a, b), and that OvoB overexpression was sufficient to promote vas expression (Fig. 5c, d). These results show that Mamo collaborates with OvoB to promote vas in PGCs.
Fig. 5.
MamoAF collaborates with OvoB to induce Vas expression in PGCs. a Vas mRNA in situ hybridisation of wild type, MamoAF overexpressed and MamoAF and OvoA overexpressed embryos under the control of maternal nos-Gal4 driver at stage 14. White arrowheads indicate embryonic gonads. b Embryos were classified into three groups depending on their strong (++), middle (+) and low (−) signal intensities of Vas mRNA in PGCs. Percentages of the embryos carrying PGCs with strong, middle and low signals in wild type, MamoAF-OE and MamoAF + OvoA OE embryos. **P < 0.01 (Significance is calculated with MamoAF OE by Fisher’s exact test). c Vas mRNA in situ hybridisation of wild type and OvoB overexpressed embryos under the control of maternal nos-Gal4 driver at stage 15. White arrowheads indicate embryonic gonads. d Embryos were classified into three groups depending on their strong (++), middle (+) and low (−) signal intensities of Vas mRNA in PGCs. Percentages of the embryos carrying PGCs with strong, middle and low signals in wild type and OvoB-OE embryos. *P < 0.05 (Significance is calculated with wild type by Fisher’s exact test). Scale bar: 20 μm
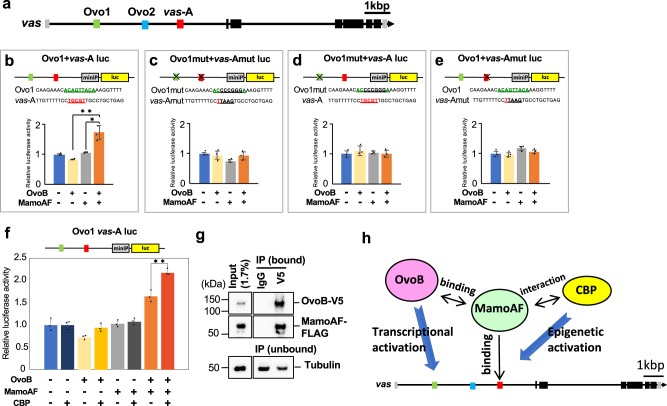
If MamoAF collaborates with OvoB to activate vas, Ovo enhancers30 should promote vas expression. In addition to the vas-A element required for Mamo binding (Fig. 2b), the first intron of vas contains Ovo-binding consensus sequences (Ovo1 and Ovo2)30 (Fig. 6a), which are conserved in several Drosophila species (Supplementary Fig. 7). To investigate whether these cis-elements have enhancer activity, we performed luciferase reporter assays in Drosophila Schneider cells (S2 cells). We evaluated the effects of fragments of the first intron containing each cis-element (Ovo1, Ovo2 and vas-A) on luciferase expression, and found that none of the fragments were sufficient to promote luciferase expression in response to MamoAF and OvoB (Supplementary Figs 11, 12). These results suggested that, individually, none of the cis-elements has enhancer activity. Next, we examined combinations of these fragments containing the cis-elements. Although a reporter construct containing both Ovo2 and vas-A (Ovo2 + vas-A luc) did not exhibit enhancer activity (Supplementary Fig. 12d), a reporter construct containing both Ovo1 and vas-A (Ovo1 + vas-A luc) promoted luciferase expression in response to both MamoAF and OvoB (Fig. 6b). Furthermore, mutation analyses of Ovo1 and vas-A demonstrated that both Ovo1 and the vas-A element were required to stimulate transcription (Fig. 6c–e). Consistent with this, both MamoAF and OvoB were required to promote luciferase expression mediated by Ovo1 and vas-A (Fig. 6b). We concluded that MamoAF collaborates with OvoB to activate vas through Ovo1 and the vas-A element. Furthermore, CBP enhanced luciferase expression mediated by MamoAF and OvoB, suggesting that these factors worked together to stimulate transcription (Fig. 6f).
Fig. 6.
MamoAF and OvoB exhibit transcriptional synergy. a vas-A element and the Ovo-binding consensus sequences (Ovo1 and Ovo2) in the first intron of vas. b–e Luciferase reporter assays in S2 cells, showing that both Ovo1 and vas-A element are required for transcriptional activation of luciferase. All experiments were conducted as triplicate and the mean of relative luciferase activities and SD are shown. *P < 0.05, **P < 0.01 (two-tailed Student’s t test). f Luciferase reporter assays in S2 cells, showing that MamoAF, OvoB and CBP collectively stimulates luciferase expression. **P < 0.01 (two-tailed Student’s t test). All experiments were conducted as triplicate and the mean of relative luciferase activities and SD are shown. g Coimmunoprecipitation assays with nuclear extracts of S2 cells that had been transfected with both FLAG-tagged MamoAF and V5-tagged OvoB. The precipitate with a V5 antibody contains not only OvoB but also MamoAF-FLAG, showing MamoAF physically interacts with OvoB. Full images of the electrophoreses are shown in Supplementary Fig. 16. h A model of interaction between MamoAF, CBP and OvoB in the activation of vas expression
MamoAF physically interacts with OvoB
Next, we investigated the physical interaction between MamoAF and OvoB. Coimmunoprecipitation assays with nuclear extracts of S2 cells transfected with both FLAG-tagged MamoAF and V5-tagged OvoB revealed a physical interaction between MamoAF and OvoB (Fig. 6g; Supplementary Figs 13–15). We found that the N-terminal region encompassing residues 251–277 of MamoAF is necessary for the physical interaction with OvoB (Supplementary Fig. 14). Conversely, the region encompassing residues 470–757 of OvoB is required for the interaction with MamoAF (Supplementary Fig. 15). These results, together with the data from the luciferase reporter assay, demonstrate that MamoAF and OvoB exhibit transcriptional synergy in activating vas expression.
Discussion
Although it is well recognised that maternal translational and transcriptional repressors play essential roles in establishing PGCs in Drosophila11,12, the genes sufficient for vas activation in PGCs remain unknown. In this study, we identified two types of vas activators: full-length Mamo, a weak but specific inducer of vas, and Mamo short isoform, a potent inducer of vas activation. To clarify the molecular mechanisms of vas activation, we conducted biochemical and genetic analyses using MamoAF-induced vas expression, and revealed that two cofactors of MamoAF, CBP and OvoB, are both involved in activation of vas in PGCs. Thus, MamoAF-induced vas expression is useful for identifying cofactors of vas activation in PGCs. MamoAF can induce vas expression in both PGCs and brain. In both cellular contexts, the transcriptional activator OvoB is necessary for MamoAF-induced vas expression. Moreover, overexpression of both MamoAF and OvoB is sufficient to induce vas expression in VNC. Thus, the Mamo–OvoB axis is essential for directing vas activation. Our data revealed that MamoAF functions as a molecular hub: it collaborates with CBP to epigenetically activate the vas locus, and physically interacts with OvoB to stimulate vas transcription (Fig. 6h). Consistent with this notion, a reporter assay demonstrated that these factors worked together to stimulate transcription. We conclude that the Mamo-mediated network of epigenetic and transcriptional regulators directs vas activation in Drosophila embryos.
We successfully demonstrated that MamoAF directly activates vas expression through the vas-A element in the first intron, which is essential for endogenous vas expression in PGCs. However, MamoAF could also activate vas transcription through other cis-elements that remain to be identified. We previously reported that Mamo has a role in the regulation of chromatin structure18. Therefore, Mamo may regulate chromatin structure, in addition to transcriptional activation, to promote vas expression.
A previous study using reporter assays showed that a 40-bp element in the 5′ flanking region of vas, the up-40 element, is sufficient to recapitulate germline-specific expression during oogenesis and embryogenesis31. The up-40 element does not contain consensus sequences for either Mamo18 or Ovo30. This implies that different transcriptional factors must control vas expression through the up-40 element. Because removal of the vas-A element decreases endogenous vas expression in PGCs, vas-A and up-40 elements may act in a partially redundant manner to upregulate vas expression in PGCs. Therefore, multiple enhancers may act in parallel to activate vas expression in germ cells. However, it remains unclear whether the up-40 element is necessary for endogenous vas expression.
In this study, we focused on MamoAF to investigate the mechanisms of vas activation due to its potent activity for vas activation. Thus, it remains unclear how full-length Mamo activates vas expression. Understanding the mechanism requires the identification of factors regulating the nuclear localisation of full-length Mamo. Maternal full-length Mamo in the nuclei of early PGCs may interact with OvoB, which is maternally provided and enriched in PGC nuclei16. However, it will be necessary to investigate the interaction between full-length Mamo and OvoB under conditions in which full-length Mamo is not converted into short derivatives.
Both full-length and short form Mamo mRNAs are expressed in germ cells in ovaries. However, full-length Mamo rescues the differentiation of mamo mutant germline clones more efficiently than MamoAF. We found that full-length Mamo can be converted to truncated derivatives. Thus, full-length Mamo has the potential to complement the short isoform. By contrast, the short form of Mamo does not appear to complement the function of the full-length protein owing to the lack of the BTB/POZ domain. During oogenesis, the transcriptional activity of full-length Mamo may be regulated through interactions with epigenetic regulators via the BTB/POZ domain, as reported for other BTB/Zn-finger transcription factors32. Full-length Mamo is enriched in the nuclei of the nurse cells in egg chambers after oogenic stage 6, but MamoAF is only weakly enriched. Thus, full-length Mamo may play a role in regulating gene expression in nurse cells and promoting the differentiation of oocytes into mature eggs.
We propose that Mamo short isoform is a potent vas activator. The C2H2 Zn-finger domains of Mamo short isoform are homologous to those of human Sp1;20 Sp1-related transcription factors regulate gene expression in germ cells in vertebrates21–23. For example, chicken Sp1 promotes vas expression in PGCs24. Moreover, Ovo has conserved roles in germline development in mouse and Drosophila16,33. Accordingly, Sp1-related Zn-finger proteins and Ovo may also be key transcriptional activators of vas in other animals, including mouse and human. We anticipate that our results will facilitate understanding of the molecular mechanisms that regulate germline development in animals.
Methods
Fly stains
Oregon-R served as a wild-type control. UASp-Mamo-FLAG has been described previously17. UASp-MZD-FLAG18 was used to overexpress MamoAF. nos-Gal4 was a gift from Dr. R. Lehmann. UAS-CBP was a gift from Dr. J. Kumar. w ovoD1 v24 P{FRT}101;P{hsFLP}38 (BL1813), matα4-Gal-VP16 (BL7063), nej0.3(BL34040), nej3(BL3729) were obtained from the Bloomington Stock Center. ovoB-Nterm-egfp, UASp-OvoB and UASp-OvoA has been described previously16. All stocks were maintained at 25 °C or at room temperature in standard Drosophila medium unless otherwise noted.
Expression vectors for Mamo fragments in embryos
The fragment of s-MZD-FLAG was amplified from the pBluescript-mamo-FLAG plasmid17 by PCR. The fragment of MZDΔ2,3-FLAG was amplified from the pUASp-MZD-FLAG plasmid18 by PCR. For s-MZD-FLAG, p-UASp-d2,3 F (5′-CGCGGATCCATG CCACTGCACATGTTTCCG-3′) and T3 (5′-AATTAACCCTCACTAAAGGG-3′) primers were used. For MZDΔ2,3-FLAG, UASP-FA1 (5′-GTCCTGTTCATTGGTACCCG-3′) and 54 MZD d2,3 R (5′-GCTCTAGATTATTTATCATCATCATCTTTATAATCTGGTCGATCACACATGAC-3′) primers were used. The amplified fragments were cloned into the pUASp vector to construct pUASp-s-MZD-FLAG and pUASp-MZDΔ2,3-FLAG, respectively. These vectors were used to generate transgenic flies by standard injection protocols.
Overexpression of Mamo and Mamo fragments
matα4-Gal-VP16 flies were crossed with the flies carrying pUASp-Mamo-FLAG, pUASp-MZD-FLAG, pUASp-s-MZD-FLAG and pUASp-MZDΔ2,3-FLAG, respectively. The embryos (Mamo OE) derived from matα4-Gal-VP16 > UASp-Mamo-FLAG, the embryos (MamoAF OE) from matα4-Gal-VP16 > UASp-MZD-FLAG, the embryos (s-MZD OE) from matα4-Gal-VP16 > UASp-s-MZD-FLAG, the embryos (MZDΔ2,3 OE) from matα4-Gal-VP16 > UASp-MZDΔ2,3-FLAG were analysed by in situ hybridisation and immunostaining. To investigate the effects of MamoAF and Mamo overexpression on vas expression in PGCs, the PGCs in the embryos derived from nos-Gal4 > UASp-MZD-FLAG and nos-Gal4 > UASp-Mamo-FLAG females were analysed by in situ hybridisation and immunostaining.
Developmental western blotting of Mamo OE embryos
Mamo OE embryos were collected at 25 °C for 3 h, then the embryos were incubated at 18 °C for 0, 6, 16 and 24 h, respectively. The embryos were dechorionated and homogenised in the sample buffer, and then the samples (40 embryos/lane) were fractionated using a 5–20% gradient sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) gel (c520L, Atto) and analysed by western blotting with anti-FLAG antibody.
Analyses of fractionated Mamo OE and MamoAF OE embryos
Mamo OE and MamoAF OE embryos were collected at 25 °C for 0–16 h. Approximately 0.1 g of the embryos were dechorionated and homogenised using a Dounce tissue grinder in the sucrose solution (0.5 m sucrose, 5 mm HEPES, pH7.9, 5 mm KCl, 1.5 mm MgCl2, 0.25 mm DTT, 0.25 mm phenylmethylsulfonyl fluoride (PMSF) supplemented with Protease inhibitor complete EDTA-free protease inhibitor cocktail (Roche)). Debris was removed by centrifugation at 390 × g for 10 min at 4 °C and the supernatant was recovered. The supernatant was centrifuged at 6300 × g for 10 min at 4 °C. The resulting supernatant was used for cytoplasmic extract, the pellet was suspended in the sucrose solution and used for nuclear extract. Normalised load of each extract was fractionated using a 5–20% gradient SDS-PAGE gel, and then analysed by western blotting with anti-FLAG, anti-Tubulin and anti-Histone H3 antibodies.
Western blot assays
Samples were denatured in sample buffer and fractionated using SDS-PAGE gel. Then blots were transferred to polyvinylidene difluoride membranes (Immobilon-P, Merck Millipore), and reacted with mouse anti-FLAG M2 antibody (1:2000; Sigma, F1804), anti-V5 antibody (1:5000; abcam, ab27671), anti-tubulin antibody (1:2000; Sigma, T6199) or rabbit anti-histone H3 (1:2000; abcam, ab1791) for 1 h, subsequently with a horseradish peroxidase (HRP)-conjugated mouse IgG antibody (1:2000; Bio-Rad, immune-star anti-mouse HRP) or horseradish peroxidase (HRP)-conjugated rabbit IgG antibody (1:2000; Bio-Rad, immune-star anti-rabbit HRP) for 1 h. Clarity western ECL substrate (Bio-Rad) was used according to the manufacturer’s instructions to detect chemiluminescence. Fluorescent images were obtained using an ImageQuant LAS 4000 system (GE healthcare).
5′-RACE and cloning of RACE fragments
Total RNAs were prepared using the Qiagen RNeasy mini kit, according to the manufacturer’s instruction from wild type ovaries. Approximately 1 μg of total RNA was subjected to RNA ligase-mediated 5′-RACE reaction (5′-Full-RACE Core Set) (Takara), according to the manufacturer’s instructions. In brief, the first-strand DNA was synthesised by AMV reverse transcriptase XL using 5′-RACE Short Mamo RT (5′-CCATGTTGAGATCGCC-3′) primer. Inverse PCR was performed with following primer sets. For 1st PCR, 5′-RACE Short Mamo S1 (5′-GCATCAGCAGCAACAGCAG-3′) and 5′-RACE Short Mamo A1 (5′-GATTCGGACGTTTCCGACTC-3′) primer pairs were used. For 2nd PCR, 5′-RACE Short Mamo S2 (5′-GCACATGTTTCCGTGCATC-3′) and 5′-RACE Short Mamo A2 (5′-GATGCCGAACGCTGTTGCATG-3′) primer pairs were used. PCR products were separated on 1% agarose gel, extracted, subcloned into pBSK and sequenced.
Analyses of germline clones
To investigate the maternal function of CBP, we generated germline clones. We introduced nej3 or nej0.3 mutations into the chromosome carrying FRT (BL1844) by meiotic recombination. The nej3/Binsinscy and nej0.3/Binsinscy females were mated with w ovoD1 v24 P{FRT}101;P{hsFLP}38 males, respectively. Their progenies were then subjected to heat shock at 37 °C for 1 h in a water bath. Because the nej3 germline clones were degenerated during mid-oogenesis, we used CBPM- embryos derived from nej0.3 germline clones were analysed by in situ hybridisation and immunostaining.
In situ hybridisation to detect Vas mRNA
To detect Vas mRNA in CBPM- embryos and the embryos homozygous for vasd10-3 at stage 9–10, embryos were collected, dechorionated with sodium hypochlorite for 3 min, then fixed with PBS containing 4% formaldehyde/heptane for 25 min. PBS phase containing formaldehyde was discarded. The vitelline membranes were removed by adding methanol and vigorous shaking. Because vasd10-3was balanced with CyO, P{w[ + mC] = GAL4-Kr.C}DC3, P{w[ + mC] = UAS-GFP.S65T}DC7, the embryos homozygous for vasd10-3 were selected under the fluorescence stereoscopic microscope. The fixed embryos were hybridised with digoxigenin (DIG)-labelled anti-sense Vas RNA probe in hybridisation solution (50% formamide, 4× SSC, 5% dextransulfate, 0.01% Tween-20) at 55 °C for 16 h15, washed with wash solution 1 (50% formamide, 2× SSC, 0.01% Tween-20) for 10 min eight times, then washed with wash solution 1 at 55 °C overnight. Hybridised embryos were rinsed with PBS containing 0.1% Tween-20 (PBTw), then reacted with anti-DIG alkaline phosphatase-conjugated antibody (Roche) at 1:2000 in PBTw containing 5% goat serum for 2 h. The signals were visualised by incubating the embryos with colour developing solution (100 mm Tris-HCl, pH9.5, 100 mm NaCl, 50 mm MgCl2, 0.1% Tween-20) containing NBT-BCIP (Roche) at room temperature for 1 h in the dark. To detect Vas mRNA in PGCs of Mamo-OE and MamoAF-OE embryos under the control of maternal nos-Gal4 driver at stage 14–15, embryos were dechorionated with sodium hypochlorite for 15 s, then fixed with PBS containing 4% paraformaldehyde/heptane for 20 min. The fixed embryos were hybridised with DIG- labelled anti-sense Vas RNA probe in hybridisation solution (50% formamide, 5× SSC, 10% dextransulfate, 0.1% Tween-20, 0.1 mg ml−1 yeast RNA, 0.1 mg ml−1 heparin, 10 mm DTT) at 60 °C for 16 h15, washed with wash solution 2 (50% formamide, 2× SSC, 0.1% Tween-20) at 60 °C for 30 min three times, at room temperature for 10 min three times, and then with wash solution 3 (2× SSC, 0.1% Tween-20) at room temperature for 10 min three times. Hybridised embryos were rinsed with PBTw, then blocked with Blocking reagent (Roche) for 1 h, then reacted with anti-DIG alkaline phosphatase-conjugated antibody (Roche) at 1:3000 for 1 h. To detect Vas mRNA signals, hybridised embryos were incubated with colour developing solution containing NBT-BCIP at room temperature for 30 min in the dark.
In situ hybridisation to detect the short Mamo mRNA
To make a template for a Mamo BTB/POZ RNA probe, PCR was performed full-length Mamo cDNA as a template using Mamo FL probe Pst F (5′-CAGCTGCAGATGGGCAGTGAG-3′) and Mamo FL probe R (5′-CATGGTGGTGCGTGTGATGG-3′) primers. The resulted PCR product was subcloned and used as a template for producing a Mamo BTB/POZ RNA probe. To make a template for a Mamo Zn-finger RNA probe, PCR was performed MZD-FLAG cDNA18 as a template. The resulted PCR product was subcloned and used as a template for producing a Mamo Zn-finger RNA probe. Wild type embryos were collected, dechorionated with sodium hypochlorite for 15 s, then fixed with PBS containing 4% paraformaldehyde/heptane for 20 min. PBS phase containing formaldehyde was discarded. The vitelline membranes were removed by adding methanol and vigorous shaking. In situ hybridisation was performed as described above. To detect Mamo mRNAs encoding Mamo short-isoforms, hybridised embryos were incubated with colour developing solution containing NBT-BCIP at room temperature for 3 h in the dark.
Double fluorescence in situ hybridisation (double FISH)
To detect ectopic Vas and Ovo mRNA expression in MamoAF-OE embryos, we performed double FISH. To confirm whether ectopic vas and ovo were expressed in brains, the anti-sense Elav RNA probe, which can specifically detect neural cells34, was used. The template for Elav probe was amplified by PCR using Elav probe-F Eco (5′-CGGAATTCGCTAATGCAGAGTGCCGCTG-3′) and Elav probe-R Eco (5′-CGGAATTCCAGACCCTTGTTAACTGGCG-3′) primers from wild type genomic DNA and cloned into pBSK. Fluorescein isothiocyanate (FITC)-labelled anti-sense Elav RNA probe was synthesised from the template using the Fluorescein RNA Labeling Mix (Roche). We used both DIG- and FITC-labelled probes. The DIG-labelled probes for Vas and Ovo mRNAs were visualised by the anti-DIG alkaline phosphatase-conjugated antibody in combination with HNPPD/Fast Red TR (Roche). For FITC-labelled probe for Elav mRNA, the components and protocols of the TSA Plus Fluorescein kit (Perkin Elmer), including the anti-FITC horseradish peroxidase-conjugate antibody and FITC-tyramides as substrates, were used. For double FISH, the embryos were hybridised at the same time using DIG- and FITC-labelled anti-sense RNA probes in hybridisation solution (50% formamide, 5× SSC, 10% dextransulfate, 0.1% Tween-20, 0.1 mg ml−1 yeast RNA, 0.1 mg ml−1 heparin, 10 mm DTT) at 60 °C for 16 h, washed with solution 2 at 60 °C for 30 min three times, at room temperature for 10 min three times, and then with wash solution three at room temperature for 10 min three times. The embryos were washed with TNT Buffer (0.1 m Tris-HCl, pH7.5, 0.15 m NaCl, 0.05% Tween20) for 10 min three times and blocked with TNB buffer (Perkin Elmer). The embryos were reacted with anti-FITC horseradish peroxidase-conjugate antibody at 1:4000 at 4 °C overnight. The signals of FITC-labelled probe were visualised by incubating the embryos first for 30 min with the amplification solution of TSA kit, and then washed with TNT Buffer (0.1 m Tris-HCl, pH7.5, 0.15 m NaCl, 0.05% Tween20) for 10 min three times. The embryos were incubated with anti-DIG alkaline phosphatase-conjugated antibody at 1:3000 for 1 h. The signals of DIG-labelled probe were visualised by incubating HNPPD/Fast Red TR for 30 min twice. The embryos were mounted with CC/Mount (Diagnostic BioSystems) and imaged with a confocal microscope (LSM 800, Zeiss) within 2 days.
Rescue experiments
mamoSVA53 and UASp-Mamo-FLAG has been described previously17. To investigate whether MamoAF can rescue mamo mutation, we generated germline clones of mamoSVA53. Unfortunately, the w ovoD1 v24 P{FRT}101;P{hsFLP}38 strains (BL1813) in both the Bloomington Stock Center and the Genetic Resource Center in Kyoto Institute of Technology lost the hsFLP transgene, which is essential to produce germline clones. We crossed the strain with other hsFLP strain (Kyoto 108127) to obtain the strain transiently carrying the hsFLP transgene. Then, we crossed the strain with nos-Gal4 to obtain w ovoD1 v24 P{FRT}101;P{hsFLP}; nos-Gal4 strain. We crossed w ovoD1 v24 P{FRT}101/Y;P{hsFLP}; nos-Gal4 males with mamoSVA53/Binsinscy, mamoSVA53/Binsinscy carrying UASp-Mamo-FLAG and mamoSVA53/Binsinscy carrying UASp-MamoAF-FLAG females, respectively. Their progenies were then subjected to heat shock at 37 °C for 1 h in a water bath. The resulted females without Binsinscy balancer chromosome were crossed with y w males to investigate their progenies. The eggs of the females were collected for 3 h. The number of the eggs and their hatching rate were scored. We found that mamo phenotype was rescued by expressing Mamo-FLAG. But the phenotype of germline clones was different from that previously reported17. We could not obtain the mature eggs derived from germline clones homozygous for mamoSVA53 allele in this experimental condition. This difference is probably owing to w ovoD1 v24 P{FRT}101; P{hsFLP}38 strain described above. FLP activity may be weaker than original BL1813. Moreover, genetic background of w ovoD1 v24 P{FRT}101; P{hsFLP}38 strains may affect the efficiency of germline clone production.
Immunostaining
The embryos were collected and fixed as described above. The fixed embryos were washed with PBS containing 0.1% Triton X-100 (PBTx), blocked PBTx containing 5% goat serum for 1 h and then reacted with primary and secondary antibodies. For primary antibodies, a rabbit anti-Vas antibody at 1:500 from Dr. S. Kobayashi, a mouse anti-Elav monoclonal antibody (Developmental Studies Hybridoma Bank) at 1:100, and a mouse anti-GFP monoclonal antibody (3E6, Thermo Fisher) at 1:300 were used. A mouse anti-H3K27ac monoclonal antibody at 1:500, a mouse anti-H3K9ac monoclonal antibody at 1:500, a mouse anti-H4K16ac monoclonal antibody at 1:500, from Dr. H. Kimura35 were used. A mouse anti-Piwi antibody at 1:200 from Dr. M. Shiomi was used to detect PGCs36 in Supplementary Fig. 5. AlexaFlour-conjugated antibodies (Molecular Probes) at 1:1000 were used for secondary detection. DNA was stained with DAPI. Stained embryos were mounted with Vectashield (Vector laboratories) and imaged with a confocal microscope (FV2000, Olympus or LSM 800, Zeiss). Fluorescent imaging was acquired with Zen 2.1 Software (Zeiss) or the software for the FV1200 (Olympus).
Immunostaining of ovaries with mamo mutant germline clone
Rescue experiments were performed as described above. To investigate the rescue ability of Mamo and MamoAF, we stained the ovaries containing mamo mutant germline clone expressing full-length Mamo-FLAG or MamoAF-FLAG. Germline clones were generated as described above. Females carrying mamo mutant germline clones were dissected. The ovaries were fixed with PBS containing 4% paraformaldehyde for 20 min. The ovaries were washed with PBS containing 0.1% Triton X-100 (PBTx), blocked PBTx containing 5% goat serum for 1 h and then reacted with primary and secondary antibodies. For primary antibodies, a rabbit anti-Vas antibody at 1:500. AlexaFlour-conjugated antibodies (Molecular Probes) at 1:1000 were used for secondary detection. DNA was stained with DAPI. Stained ovaries were mounted with Vectashield (Vector laboratories) and imaged with a confocal microscope (FV2000, Olympus). Fluorescent imaging was acquired with the software for the FV1200 (Olympus).
Immunostaining of ovaries overexpressing Mamo
Ovaries were dissected from females of nos-Gal4, UASp-Mamo-FLAG and nos-Gal4, UASp-MamoAF-FLAG. The ovaries were fixed with PBS containing 4% paraformaldehyde for 20 min. The ovaries were washed with PBS containing 0.5% Triton X-100 (PBTx0.5), blocked PBTx0.5 containing 5% goat serum for 1 h and then reacted with primary and secondary antibodies. For primary antibodies, a rabbit anti-Vas antibody at 1:500, a mouse anti-FLAG monoclonal antibody M2 (Sigma) at 1:200 were used. AlexaFlour-conjugated antibodies (Molecular Probes) at 1:1000 were used for secondary detection. DNA was stained with DAPI. Stained ovaries were mounted with Vectashield (Vector laboratories) and imaged with a confocal microscope (LSM 800, Zeiss). Fluorescent imaging was acquired with Zen 2.1 Software (Zeiss).
Detection of ectopic Vas expression
Ectopic Vas expression was detected in heterogenous cell population in embryonic brain and VNC. To gain the image, a single section image was used. Ectopic Vas signals were normalised by DAPI signals.
RNA preparation and qRT-PCR
Total RNAs were prepared using the Qiagen RNeasy mini kit according to the manufacturer’s instructions from wild-type embryos, Mamo OE, MamoAF OE, CBP OE, and the embryos overexpressed both CBP and MamoAF. RNAs were reverse-transcribed using SuperScript III. Expression levels of vas were quantified using the SYBR Premix Ex TaqTM II (Tli RNaseH Plus) (Takara) using standard curve method. Expression levels of vas were normalised to Rp49 expression. Following primers were used: the RT-rp49F (5′-AGCGCACCAAGCACTTCATC-3′) and Rt-rp49 R (5′-GACGCACTCTGTTGTCGATACC-3′) primers for Rp49, the RT-vas sense (5′-TTGCGTTGCGCGAAGTGAT-3′) and RT-vas anti-sense (5′-CGCCGCGAGTATCAACAAT-3′) primers for vas. Expression levels of vas were quantified using the software for Thermal Cycler Dice TP700 (Takara).
EMSA
Bacterially expressed GST-MZD and GST proteins were purified18. Escherichia coli BL21 transformed with the GST-MZD bacterial expression vector, pGEX GST-MZD18, was grown at 37 °C to OD600 = ~ 0.5 and then induced by the addition of IPTG to a final concentration of 0.5 mm. The cells were grown for an additional 120 min at 37 °C. The cells were collected by centrifugation, sonicated in the sonication buffer (2 mm Na2HPO4, 0.2 mm KH2PO4, 13.7 mm NaCl containing 1 mm DTT, 0.5 mm PMSF, 0.1% Triton X-100) and affinity-purified by Glutathione-Sepharose 4B (GE Healthcare). Eluted proteins were dialysed against the dialysis buffer (20 mm HEPES, pH 7.9, 50 mm KCl, 2 mm MgCl2, 10% glycerol, 1 mm DTT, 1 mm PMSF) three times for 2 h at 4 °C. The purified proteins were concentrated by Microcon (Millipore). Sequences of the DNA probes were as follows: vas-A (5′-ATTTGTTTTTCCTGCGTTGCCTGCTGAGT-3′); vas-AΔ, (5′-ATTTGTTTTTCCTGCCTGCTGAGT-3′). Sense and anti-sense oligonucleotides for each probe were annealed, DIG-labelled and subjected to EMSA assay using the DIG Gel shift Kit 2nd Generation (Roche). Purified GST-MZD or GST protein (60 ng) and DIG-labelled probe (30 fmol) were incubated in the binding buffer (20 mm HEPES, pH 7.6, 1 mm EDTA, 10 mm (NH4)2SO4, 1 mm DTT, 0.2% Tween20, 30 mm KCl, 1 μg poly[d(I-C)], 0.1 μg poly l-lysine) for 15 min at 25 °C. Binding reactions were electrophoresed in 7% polyacrylamide gels, and transferred to a positively charged nylon membrane (Roche). The membrane was then UV cross-linked, blocked and probed with an anti-DIG Fab fragment (Roche). DIG-labelled DNA–protein complexes were detected by CSPD (Roche). Chemiluminescence images were obtained using an ImageQuant LAS 4000 system (GE healthcare).
Mutagenesis using CRISPR/Cas9 system
Guide RNAs (gRNAs) were designed (Supplementary Fig. 8), synthesised and purified using the SmartNuclease TM T7 gRNA vector (System Biosciences) according to the manufacturer’s instructions. gRNA was injected into the embryos derived from y2 cho2 v1; attP40{nos-Cas9}/CyO females by standard injection protocols. Target sequences were amplified by PCR, annealed and the resulted heteroduplexes were digested with T7 endonuclease I (NEB) to detect Cas9-induced mutations. Following primers were used: CRISPR vas Fv2 (5′-CAAACACCGAACGAATTCC-3′) and CRISPR vasA R (5′-TTCCGATAGAACTGGTGAG-3′). Deletions were confirmed by sequencing (accession No. LC503775 at DNA Data Bank of Japan).
ChIP analyses
Dechorionized wild type embryos and MamoAF-OE (~ 0.15 g) were homogenised with Dounce tissue grinder in the buffer (15 mm HEPES, pH 7.5, 15 mm NaCl, 60 mm KCl, 4 mm MgCl2, 0.5% Triton X-100, 0.5 mm DTT, complete EDTA-free protease inhibitor cocktail (Roche)) containing 0.8% formaldehyde on ice and then were fixed for 15 min at room temperature. The fixed embryos were recovered by centrifugation at 1430 x g for 5 min at 4 °C, and then washed with the buffer (15 mm HEPES, pH 7.5, 140 mm NaCl, 1 mm EDTA, 0.5 mm EGTA, 1% Triton-X-100, 0.1% sodium deoxycholate, 0.1% SDS, 0.5% N-lauroylsarcosine, supplemented with complete EDTA-free protease inhibitor cocktail (Roche)). The fixed embryos were sonicated with a Branson Model Sonifier 250 sonicator using a microtip, and then centrifuged at 15,880 x g for 30 min at 4 °C to recover the supernatant containing fragmented chromatin. Dynabeads M-280 sheep anti-mouse IgG (Thermo Fisher), mouse anti-FLAG monoclonal antibody M2 (Sigma) and mouse anti-H3K27ac monoclonal antibody35 were used for chromatin immunoprecipitation. Input DNA, mock-precipitated DNA with control mouse IgG and DNA from the immunoprecipitation were analysed by PCR. Quantitative PCR (qPCR) analyses were performed using SYBR Premix Ex TaqTM II (Tli RNaseH Plus) (Takara). Following primers were used: vasa A chip F (5′- GTAATTGGTTACGCGTAAAC-3′) and vasa A chip R (5′-AACGAGCGCGCCCATAC-3′).
S2 cell culture
Drosophila S2 cells were cultured in Schneider’s Drosophila medium (Gibco) supplemented with 10% heat-inactivated fetal bovine serum (Biosera).
Expression vectors for coimmunoprecipitation assays
The expression vectors of MamoAF and OvoB used in coimmunoprecipitation assays was constructed as follows. MamoAF fragment was amplified from the pUASp-MZD-FLAG plasmid18 by PCR. Eco-MZD-puastV2 (5′-CGGAATTCATGGATGCCATGCCCGTGATT-3′) and puast-MZD-R-EcoR1 (5′-GGAATTCTTATTTATCATCATCATCTTTAT-3′) primers were used. The amplified fragment was cloned into the pUASt vector to construct pUASt-MamoAF-FLAG. The coding sequence of OvoB was amplified from pAC-OvoB by PCR and cloned into the pUASt vector to construct pUASt-OvoB-V5. The expression vectors of MamoAF fragments used in coimmunoprecipitation assays (Supplementary Fig. 14) was constructed as follows. MamoAF fragments were amplified from the pUASt-MamoAF-FLAG plasmid by PCR. For s-MamoAF(251–550)-FLAG, s-MamoAF-1-SacIIF (5′-CTACCGCGGATGCCGCTGCACATGTTTCCG-3′) and MamoAF-R-Xba (5′- CGCGTCTAGATTATTTATCATCATCATCTTT-3′) primers were used. For s-MamoAF(278–550)-FLAG, s-MamoAF-2-SacIIF (5′- CTACCGCGGATGTCGCTGAAGACGCTCGAG-3′) and MamoAF-R-Xba (5′-CGCGTCTAGATTATTTATCATCATCATCTTT-3′) primers were used. For s-MamoAF(454–550)-FLAG, s-MamoAF-3-SacIIF (5′-CTACCGCGGATGGCCGCGTCCAGTGATCC-3′) and MamoAF-R-Xba (5′- CGCGTCTAGATTATTTATCATCATCATCTTT-3′) primers were used. The amplified fragments were cloned into the pUASt vector, respectively. The expression vectors of OvoB fragments used in coimmunoprecipitation assays (Supplementary Fig. 15) was constructed as follows. pUASt-OvoB-V5 plasmid was digested with SacII and the resulted fragment was removed and self-ligated to construct pUASt- OvoB(338–982)-V5. pUASt-OvoB-V5 plasmid was digested with NotI and the resulted fragment was removed and self-ligated to construct pUASt- OvoB(470–982)-V5. pUASt-OvoB-V5 plasmid was digested with EcoRI and the resulted fragment was removed and self-ligated to construct pUASt- OvoB(758–982)-V5.
Coimmunoprecipitation assays
In the coimmunoprecipitation assays for MamoAF and OvoB, both proteins were overexpressed in S2 cells. Approximately 1.4 μg of pWAGal4 (a gift from Dr. Y. Hiromi), 4 μg of pUASt-MamoAF-FLAG and 4 μg of pUASt-OvoB-V5 plasmid vectors were transfected into S2 cells (3.6 × 107) using Effectene transfection reagent (Qiagen) and then cultured in the culture medium supplemented with Penicillin–Streptomycin–Amphotericin B Suspension (Wako) for 3 days. The transfected S2 cells were incubated in the buffer (10 mm HEPES pH7.5, 10 mm KCl, 0.1 mm EDTA, 0.4% NP-40, supplemented with Protease inhibitor complete EDTA-free protease inhibitor cocktail) for 10 min on ice, and then were centrifuged to obtain the nuclear fraction. Nuclear extracts were prepared from the nuclear fraction by incubating in the nuclear lysis buffer (50 mm HEPES pH7.5, 3 mm MgCl2, 300 mm NaCl, 1 mm DTT, 0.1 mm PMSF, Protease inhibitor complete EDTA-free) for 2 h at 4 °C with shaking, then incubated with 8 μg ml−1 DNase, 8 μg ml−1 RNase and 40 U ml−1 Benzonase (Merck) at 4 °C for 30 min. The nuclear extracts were incubated with 2 μg of rabbit anti-V5 antibody (Abcam, ab9116) or 2 μg of rabbit anti-FLAG antibody (Sigma, F7425) or 2 μg of rabbit control IgG (Cell signalling technology, 2729 S) in the nuclear lysis buffer for 2 h at 4 °C. The immunocomplexes were precipitated with Dynabeads M-280 sheep anti-rabbit IgG (Thermo Fisher) and washed with the ice-cold buffer (50 mm Tris-HCl, pH7.5, 0.3 m NaCl, 10% glycerol, 0.1% NP-40) using DynaMagTM-2. Finally, the complexes were fractionated using 10% PAGE gel, and analysed by western blotting.
Expression vectors for reporter assays
MamoAF fragment was amplified from the pUASp-MZD-FLAG18 plasmids by PCR. The amplified fragment was cloned into the pAC vector. The coding sequences of OvoB16 and CBP, a gift from Dr. M. Mannervik, were cloned into the pAC vector to construct pAC-OvoB and pAC-CBP, respectively.
Luciferase reporter constructs
The genomic fragment of the first intron of vas (2 L: 15,061,805–15,068,333) was amplified by PCR and cloned into pBSK to obtain pBSK-vas 1st intron plasmid. Following the primers, vas 1st intron F2 H3 (5′-CCCAAGCTTAGTGATATGCAATTAGTTTT-3′) and vas 1st intron R2 H3 (5′-CCCAAGCTTAATACGTTCATTTCTAAAGT-3′) were used for PCR. pBSK-vas 1st intron plasmid was digested with restriction enzymes to produce the genomic fragments containing the cis-elements in the first intron of vas, Ovo1, Ovo2, vas-A and Ovo2 + vas-A fragments (Supplementary Fig. 11). These fragments were cloned into a luciferase reporter enhancer assay vector (Promega, pGL4.23), respectively. vas-A and Ovo1 Sac I fragments were amplified by PCR. For Ovo1 Sac I fragment, F Ovo1 up250-SacI(5′-GGATGAGCTCGAAAGTTTCCCTCCAAC-3′) and R Ovo1 down250-SacI(5′- CTTTGAGCTCAAGAAAAATACACTATTAC-3′) primers were used. For vas-A fragment, PstI-vas intron A up (5′-TGCACTGCAGATGTTCTGTCCGACCATTAG-3′) and vas intron A down500 (5′-TGCACTGCAGGTCGGAGCACCCGATGG-3′) primers were used. These fragments were ligated with linkers and cloned into pGL4.23 to construct Ovo1 + vas-A luc reporter construct. The PCR-based, site-directed mutagenesis procedure was used to produce Ovo1mut and vas-A mut fragments (Fig. 6 and Supplementary Fig. 11). For Ovo1mut, vas 1st intron ovo bind seq1 (5′-CAGGGATCTTCAAATTAGCG-3′) and 1R-Ovo1-del-sma1 (5′-CCTTTCCCGGGGTGTTTCTTGGATGCAAA-3′) primers were used for first round of PCR. 2F-ovo1-del-sma1 (5′-AACACCCCGGGAAGGTTTTAACACTTAC-3′) and 2R-ovo1-down700nde1 (5′-AAATACCAACCAACAAGCTAAGTAA-3′) primers were used second round of PCR reaction. For vas-A mut, 1 F StuIvasA 750up (5′-CAATGATTATGCCTGCAAAGGCCTC-3′) and 1R3 vasA MZD bind-afl2 (5′-CAGGCACTTAAGGAAAAACAAATTCCATC-3′) primers were used for first round of PCR. 2F3 vasA MZD bind-afl2 (5′-TTTTCCTTAAGTGCCTGCTGAGTTTGATT-3′) and 2 R vasA900down PstI (5′-CCATTAGATAATCTTGTGGCGCCGAC-3′) primers were used for second round of PCR reaction.
Luciferase reporter assays
The luciferase reporter constructs containing the cis-elements in the first intron of vas described above were used for luciferase reporter assays. The hRluc/TK (Promega, pGL4.74) served as an internal control. About 50 ng of a luc reporter construct and 1 ng of an internal control were co-transfected into S2 cells (3 × 104 cells) with either 10 ng of pAC-MamoAF-FLAG and/or 1 ng of pAC-OvoB and/or 1 ng of pAC-CBP and/or pBSK (for mocked transfected cells) using Effectene transfection reagent (Qiagen). The transfected cells were cultured in the culture medium supplemented with Penicillin–Streptomycin–Amphotericin B Suspension (Wako). Cells were lysed after 1 day of transfection with a passive lysis buffer (Promega) and luciferase activity was measured using a Dual-Luciferase reporter assay system (Promega). All experiments were carried out in triplicate.
Statistics and reproducibility
Statistical tests were employed for each experiment are indicated in the figure legends. Microsoft Excel was used for statistical analyses. For all data in these analyses, P values of < 0.05 were considered to be significant. Replicate experiments were successful.
Reporting summary
Further information on experiments and research design are available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Acknowledgements
We are grateful to R. Lehmann, J. Kumar, Y. Hiromi, M. Shiomi, the Bloomington Stock Center, the Genetic Resource Center in Kyoto Institute of Technology, National Institute of Genetics Fly Stock Center for fly stocks and reagents. We also thank N. Soga for technical assistance. This work was supported in part by Grants-in-Aid for Scientific Research from the Japan Society for the Promotion of Science (JSPS) (KAKENHI Grant Numbers: 21570233, 16K07429, 25114002 and 18H05552).
Author contributions
S.N., M.M., S.H., H.K., M.H., Y.S. and S.K. designed the experiments, S. N., M.M., S.H., M.F., N.M., T.T., Y.S. and A.K. performed experiments, M.M., S.H. and S.N. analysed data, M.M., S.H., M.H., S.N., H.K. and S.K. wrote the paper.
Data availability
The data sets generated during and/or analysed during the current study are available from the corresponding author on reasonable request. The source data underlying plots are provided in Supplementary Data 1. Full blots are shown in Supplementary Information. The sequencing data confirming vas mutants were deposited into the DNA Data Bank of Japan under the accession number LC503775.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Shoichi Nakamura, Seiji Hira.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s42003-019-0663-4.
References
- 1.Extavour CG, Akam M. Mechanisms of germ cell specification across the metazoans: epigenesis and preformation. Development. 2003;130:5869–5884. doi: 10.1242/dev.00804. [DOI] [PubMed] [Google Scholar]
- 2.Lasko P. The DEAD-box helicase Vasa: Evidence for a multiplicity of function in RNA processes and developmental biology. Biochim. Biophys. Acta. 2013;1829:810–816. doi: 10.1016/j.bbagrm.2013.04.005. [DOI] [PubMed] [Google Scholar]
- 3.Medrano JV, et al. Comparative analysis of the germ cell markers c-KIT, SSEA-1 and VASA in testicular biopsies from secretory and obstructive azoospermias. Mol. Hum. Reprod. 2010;16:811–817. doi: 10.1093/molehr/gaq044. [DOI] [PubMed] [Google Scholar]
- 4.Roussell DL, Bennett KL. glh-1, a germ-line putative RNA helicase from Caenorhabditis, has four zinc fingers. Proc. Natl. Acad. Sci. USA. 1993;90:9300–9304. doi: 10.1073/pnas.90.20.9300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Styhler S, Nakamura A, Swan A, Suter B, Lasko P. vasa is required for GURKEN accumulation in the oocyte, and is involved in oocyte differentiation and germline cyst development. Development. 1998;125:1569–1578. doi: 10.1242/dev.125.9.1569. [DOI] [PubMed] [Google Scholar]
- 6.Tanaka SS, et al. The mouse homolog of Drosophila Vasa is required for the development of male germ cells. Genes Dev. 2000;14:841–853. [PMC free article] [PubMed] [Google Scholar]
- 7.Hartung O, Forbes MM, Marlow FL. Zebrafish vasa is required for germ cell differentiation and maintenance. Mol. Reprod. Dev. 2014;81:946–961. doi: 10.1002/mrd.22414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu N, Han H, Lasko P. Vasa promotes Drosophila germline stem cell differentiation by activating mei-P26 translation by directly interacting with a (U)-rich motif in its 3′ UTR. Genes Dev. 2009;23:2742–2752. doi: 10.1101/gad.1820709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xiol J, et al. RNA clamping by Vasa assembles a piRNA amplifier complex on transposon transcripts. Cell. 2014;157:1698–1711. doi: 10.1016/j.cell.2014.05.018. [DOI] [PubMed] [Google Scholar]
- 10.Starz-Gaiano M, Lehmann R. Moving towards the next generation. Mech. Dev. 2001;105:5–18. doi: 10.1016/S0925-4773(01)00392-6. [DOI] [PubMed] [Google Scholar]
- 11.Hayashi Y, Hayashi M, Kobayashi S. Nanos suppresses somatic cell fate in Drosophila germ line. Proc. Natl. Acad. Sci. USA. 2004;101:10338–10342. doi: 10.1073/pnas.0401647101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hanyu-Nakamura K, Sonobe-Nojima H, Tanigawa A, Lasko P, Nakamura A. Drosophila Pgc protein inhibits P-TEFb recruitment to chromatin in primordial germ cells. Nature. 2008;451:730–733. doi: 10.1038/nature06498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kunwar PS, Starz-Gaiano M, Bainton RJ, Heberlein U, Lehmann R. Tre1, a G protein-coupled receptor, directs transepithelial migration of Drosophila germ cells. PLoS Biol. 2003;1:80. doi: 10.1371/journal.pbio.0000080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Illmensee K, Mahowald AP. Transplantation of posterior polar plasm in Drosophila. Induction of germ cell at the anterior pole of the egg. Proc. Natl. Acad. Sci. USA. 1974;71:1016–1020. doi: 10.1073/pnas.71.4.1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yatsu J, et al. Maternal RNAs encoding transcription factors for germline-specific gene expression in Drosophila embryos. Int. J. Dev. Biol. 2008;52:913–923. doi: 10.1387/ijdb.082576jy. [DOI] [PubMed] [Google Scholar]
- 16.Hayashi M, et al. Conserved role of Ovo in germline development in mouse and Drosophila. Sci. Rep. 2017;7:40056. doi: 10.1038/srep40056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mukai M, et al. MAMO, a maternal BTB/POZ-Zn-finger protein enriched in germline progenitors is required for the production of functional eggs in Drosophila. Mech. Dev. 2007;124:570–583. doi: 10.1016/j.mod.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 18.Hira S, et al. Binding of Drosophila maternal Mamo protein to chromatin and specific DNA sequences. Biochem. Biophys. Res. Commun. 2013;438:156–160. doi: 10.1016/j.bbrc.2013.07.045. [DOI] [PubMed] [Google Scholar]
- 19.Fedotova AA, Bonchuk AN, Mogila VA, Georgiev PG. C2H2 Zinc finger proteins: the largest but poorly explored family of higher eukaryotic transcription factors. Acta Nat. 2017;9:47–58. doi: 10.32607/20758251-2017-9-2-47-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thurmond J, et al. FlyBase 2.0: the next generation. Nucleic Acids Res. 2019;47:D759–D765. doi: 10.1093/nar/gky1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Worrad DM, Ram PT, Schultz RM. Regulation of gene expression in the mouse oocyte and early preimplantation embryo: developmental changes in Sp1 and TATA box-binding protein, TBP. Development. 1994;120:2347–2357. doi: 10.1242/dev.120.8.2347. [DOI] [PubMed] [Google Scholar]
- 22.Kim M, et al. Regulatory factor interactions and somatic silencing of the germ cell-specific ALF gene. J. Biol. Chem. 2006;281:34288–34298. doi: 10.1074/jbc.M607168200. [DOI] [PubMed] [Google Scholar]
- 23.Ma W, Horvath GC, Kistler MK, Kistler WS. Expression patterns of SP1 and SP3 during mouse spermatogenesis: SP1 down-regulation correlates with two successive promoter changes and translationally compromised transcripts. Biol. Reprod. 2008;79:289–300. doi: 10.1095/biolreprod.107.067082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sohn YA, et al. The CCAAT element in the CIWI promoter regulates transcriptional initiation in chicken primordial germ cells. Mol. Reprod. Dev. 2014;81:871–882. doi: 10.1002/mrd.22356. [DOI] [PubMed] [Google Scholar]
- 25.Tie F, et al. CBP-mediated acetylation of histone H3 lysine 27 antagonizes Drosophila Polycomp silencing. Development. 2009;136:3131–3141. doi: 10.1242/dev.037127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Creyghton MP, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. USA. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stasevich TJ, et al. Regulation of RNA polymerase II activation by histone acetylation in single living cells. Nature. 2014;516:272–275. doi: 10.1038/nature13714. [DOI] [PubMed] [Google Scholar]
- 28.Andrews J, et al. OVO transcription factors function antagonistically in the Drosophila female germline. Development. 2000;127:881–892. doi: 10.1242/dev.127.4.881. [DOI] [PubMed] [Google Scholar]
- 29.Kondo T, et al. Small peptides switch the transcriptional activity of Shavenbaby during Drosophila embryogenesis. Science. 2010;329:336–339. doi: 10.1126/science.1188158. [DOI] [PubMed] [Google Scholar]
- 30.Bielinska B, Lü J, Sturgill D, Oliver B. Core promoter sequences contribute to ovo-B regulation in the Drosophila melanogaster germline. Genetics. 2005;169:161–172. doi: 10.1534/genetics.104.033118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sano H, Nakamura A, Kobayashi S. Identification of a transcriptional regulatory region for germline-specific expression of vasa gene in Drosophila melanogaster. Mech. Dev. 2002;112:129–139. doi: 10.1016/S0925-4773(01)00654-2. [DOI] [PubMed] [Google Scholar]
- 32.Chaharbakhshi E, Jemc JC. Broad-complex, tramtrack, and bric-a-brac (BTB) proteins: critical regulators of development. Genesis. 2016;54:505–518. doi: 10.1002/dvg.22964. [DOI] [PubMed] [Google Scholar]
- 33.Unezaki S, et al. Characterization of the isoforms of MOVO zinc finger protein, a mouse homologue of Drosophila Ovo, as transcription factors. Gene. 2004;336:47–58. doi: 10.1016/j.gene.2004.03.013. [DOI] [PubMed] [Google Scholar]
- 34.Egger B, et al. Gliogenesis in Drosophila: genome-wide analysis of downstream genes of glial cells missing in the embryonic nervous system. Development. 2002;129:3295–3309. doi: 10.1242/dev.129.14.3295. [DOI] [PubMed] [Google Scholar]
- 35.Kimura H, Hayashi-Takanaka Y, Goto Y, Takizawa N, Nozaki N. The organization of histone H3 modifications as revealed by a panel of specific monoclonal antibodies. Cell Struct. Funct. 2008;33:61–73. doi: 10.1247/csf.07035. [DOI] [PubMed] [Google Scholar]
- 36.Saito K, et al. Specific association of Piwi with rasiRNAs derived from retrotransposon and heterochromatic regions in the Drosophila genome. Genes Dev. 2006;20:2214–2222. doi: 10.1101/gad.1454806. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
The data sets generated during and/or analysed during the current study are available from the corresponding author on reasonable request. The source data underlying plots are provided in Supplementary Data 1. Full blots are shown in Supplementary Information. The sequencing data confirming vas mutants were deposited into the DNA Data Bank of Japan under the accession number LC503775.