Abstract
Adamts16 encodes a disintegrin-like and metalloproteinase with thrombospondin motifs, 16, a member of a family of multi-domain, zinc-binding proteinases. ADAMTS-16 is implicated in a number of pathological conditions, including hypertension, cancer and osteoarthritis. A large number of observations, including a recent report of human ADAMTS16 variants in cases of 46,XY disorders/differences of sex development (DSD), also implicate this gene in human testis determination. We used CRISPR/Cas9 genome editing to generate a loss-of-function allele in the mouse in order to examine whether ADAMTS-16 functions in mouse testis determination or testicular function. Male mice lacking Adamts16 on the C57BL/6N background undergo normal testis determination in the fetal period. However, adult homozygotes have an average testis weight that is around 10% lower than age-matched controls. Cohorts of mutant males tested at 3-months and 6-months of age were fertile. We conclude that ADAMTS-16 is not required for testis determination or male fertility in mice. We discuss these phenotypic data and their significance for our understanding of ADAMTS-16 function.
Subject terms: Germline development, Organogenesis, Animal breeding
Introduction
ADAMTS (a disintegrin-like and metalloproteinase with thrombospondin motifs) proteins comprise a 19-member family of extracellular metalloproteinases implicated in a number of molecular processes, including collagen processing, proteoglycan cleavage and inhibition of angiogenesis1. Indeed, their control of the structure and function of the extracellular matrix (ECM) is thought to underpin diverse roles in tissue morphogenesis and patho-physiological remodelling2. ADAMTS-16 is a family member expressed in chondrocytes3. A number of observations also implicate ADAMTS-16 in reproductive function. It is expressed at high levels in human ovary4, where it is detected in the granulosa cells of pre-ovulatory follicles5. Its expression in mature follicles can be stimulated by follicle-stimulating hormone5.
In XY males, ADAMTS-16 is co-expressed with the gene encoding Wilms’ tumour protein 1, Wt1, in both embryonic kidneys and gonads, and in adult testes, suggesting a role in genitourinary development6. Indeed, a recent report describes the identification of three human ADAMTS16 variants, resulting in amino acid substitutions predicted to be damaging, in cases of 46,XY disorders/differences of sex development (DSD)7. Two of these variants were identified in individuals with 46,XY complete gonadal dysgenesis i.e. XY female presentation. A third case had ambiguous genitalia at birth. No loss-of-function mouse model exists that allows the determination of ADAMTS-16’s physiological function, including any role in sex development. A rat model has been described; in addition to a role in blood pressure regulation8, disruption of ADAMTS16 in rats causes male infertility and failure of testicular descent (cryptorchidism)9. The testes of homozygous mutant rats were significantly smaller than controls and histological examination revealed that the seminiferous tubules could not support spermatogenesis and progressively lost germ cells. In contrast, female rats lacking ADAMTS-16 were fertile.
In order to determine whether ADAMTS-16 plays a role in XY mouse reproductive biology, in particular sex determination and testicular function, we used CRISPR/Cas9 genome editing to introduce a 906 bp deletion into the Adamts16 gene that removes exon 5. This deletion event removes part of the highly conserved protease domain of the enzyme and results in a predicted premature stop codon and nonsense-mediated decay. Homozygous XY mutants undergo normal primary sex determination and develop testes in the fetal period. However, adult mutant testes are smaller on average than those of controls. Nevertheless, all mutant males in two cohorts tested at 3 and 6 months of age had comparable fertility to controls. We discuss the significance of these data for our understanding of ADAMTS-16 function in reproduction.
Results
Analysis of Adamts16 gene expression in the developing gonads
We used wholemount in situ hybridisation (WMISH) and quantitative reverse transcriptase PCR (qRT-PCR) to examine Adamts16 expression in the developing gonads and reproductive tracts, between 11.5 and 14.5 dpc (Fig. 1). Expression was detected by WMISH in XY gonads at all stages from 11.5 dpc, and between 12.5 and 14.5 dpc this was prominent in the cords of the developing testis in a profile consistent with Sertoli cell expression (Fig. 1A–D). In contrast, significant signal in the XX gonads was not detected until 14.5 dpc (Fig. 1E–H). Sexually dimorphic expression was confirmed by qRT-PCR of gonadal tissue at 12.5 dpc (Fig. S1). These data, including the observed expression in the supporting cell lineage at the time of sex determination, are consistent with those generated previously by microarray-based expression profiling and single-cell RNA sequencing10,11.
Figure 1.
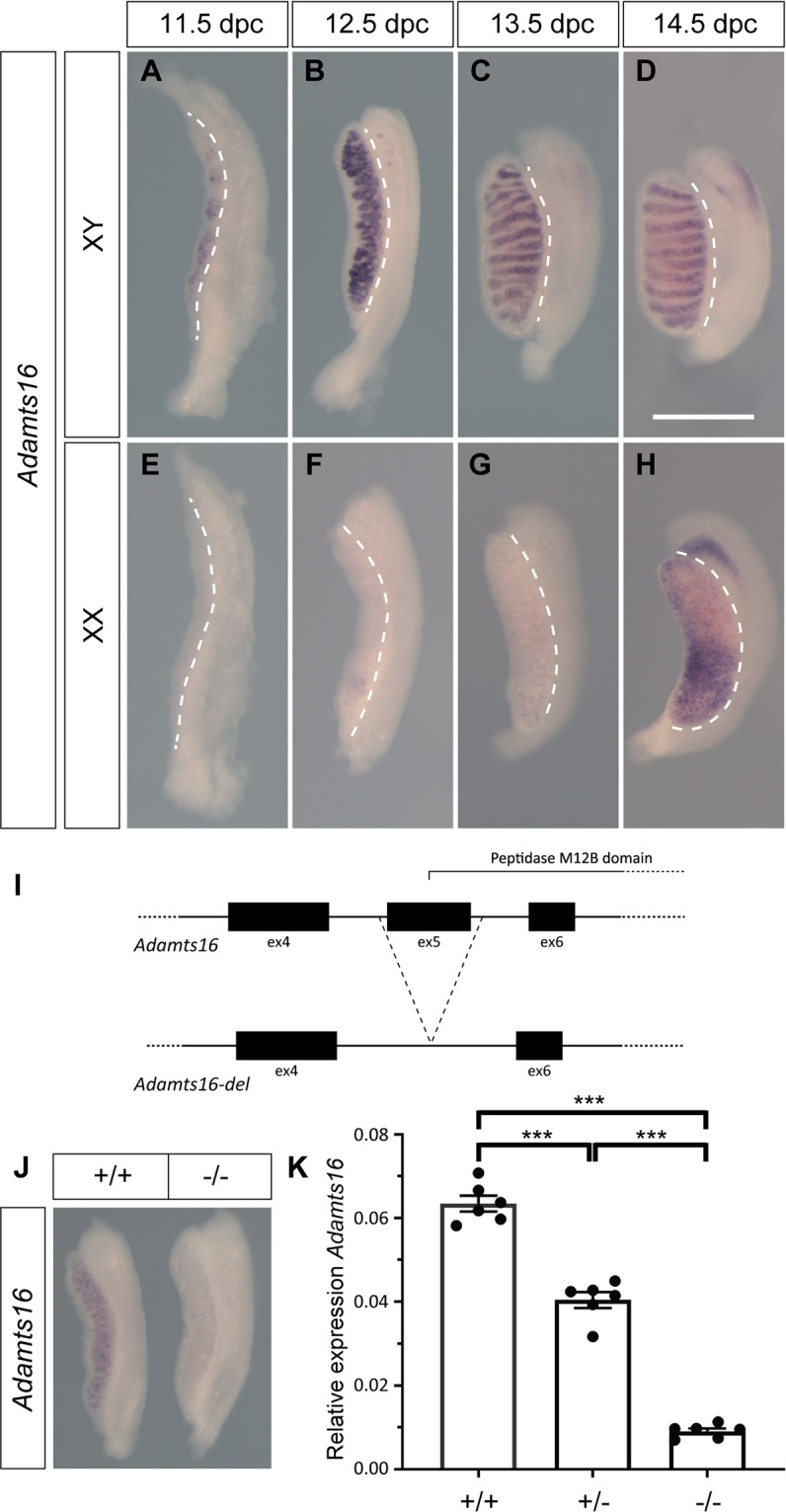
Adamts16 expression and impact of targeted exon deletion. (A–D) Wholemount in situ hybridisation (WMISH) with Adamts16 probe reveals expression at all developmental stages between 11.5 and 14.5 dpc in XY gonads. Expression is prominent in the testis cords (B–D); (E–H) Significant expression in XX gonads is detected only at 14.5 dpc (H); (I) Exon 5 of Adamts16, encoding part of the peptidase domain, was deleted using CRISPR/Cas9-mediated genome editing (not to scale). See Materials and Methods; (J) WMISH reveals Adamts16 in control (+/+) gonad at 12.5 dpc, but negligible expression in the homozygous mutant (−/−) gonad at the same stage; signal in the mutant gonad was visible upon microscopic examination but was insufficient for photographic reproduction. (K) qRT-PCR shows dramatic reduction in Adamts16 expression in homozygous mutant kidneys (−/−), and intermediate levels in heterozygotes (+/−). In all images in (A–H), the developing gonad is to the left of the white dotted line and the mesonephros is to the right. ***p ≤ 0.01 (student’s t-test).
Generation and characterisation of an Adamts16 loss-of-function allele using CRISPR/Cas9
We sought to use genome editing to generate a mouse Adamts16 loss-of-function model on a defined genetic background to permit careful analysis of the role of ADAMTS-16 in sex determination and testis development. We chose C57BL/6N as a strain background because, like the closely related C57BL/6J, it is sensitised to disruptions to testis determination12–15. The Adamts16 gene comprises 23 exons. In order to disrupt Adamts16 we used microinjection of Cas9 RNA/guide RNAs into 1-cell C57BL/6Ntac embryos to target and excise exon 5, which encodes part of the highly conserved, zinc-dependent metalloprotease domain (see Fig. 1I and Methods). We identified founders harbouring two deletions of this exon, one of 906 nucleotides (line 1) and one of 923 nucleotides (line 2). Both alleles result in a complete loss of exon 5; splicing from exon 4 to exon 6 results in a predicted frame-shift and early chain termination due to a premature stop codon. Both alleles were transmitted to the next generation following breeding. We focus here on line 1, but equivalent data were obtained for line 2.
Analysis of gonad development in XY embryos lacking functional ADAMTS16 reveals no overt abnormalities
Male and female Adamts16 heterozygotes were normal and fertile. We generated homozygous mutants by inter-crossing heterozygotes. Semi-quantitative analysis of Adamts16 expression in the homozygous XY mutant gonad at 12.5 dpc showed a dramatic reduction in signal when compared to controls (Fig. 1J). To confirm this loss of transcript in the mutant, we carefully quantified Adamts16 expression in control and homozygous mutant kidneys, a known site of expression, using qRT-PCR (Fig. 1K). This analysis revealed robust wild-type expression, negligible expression in homozygous mutants, and intermediate levels in heterozygotes, possibly due to nonsense-mediated decay of the mutated mRNA transcript. Given this significantly reduced expression of the mutant allele, and the predicted disruption to functionality of the ADAMTS-16 protein encoded by it, which lacks a portion of the critical protease domain, we conclude that the edited Adamts16 variant is a null allele or a severely hypomorphic allele.
Given the testicular phenotype of rats lacking ADAMTS-16, we examined testis morphology and marker gene expression in homozygous mutants and controls at 14.5 dpc, a fetal stage at which sex determination is essentially complete. Wholemount in situ hybridisation (WMISH) analysis with Sox9, a key testis-determining gene and marker of Sertoli cells16, revealed testis cord formation in both mutant and control gonads, which were of comparable size (Fig. 2). There was no evidence of any disruption to testis cord morphology. Similarly, WMISH with Insl3, a marker of fetal Leydig cells, also revealed no overt abnormalities in mutant gonads (Fig. 2). Finally, WMISH with Stra8, which marks germ cells entering meiosis that are normally only found in ovaries at this stage, revealed no evidence of any feminisation of mutant XY gonads. From these data showing that somatic and germ cell development proceeds normally during testis development in XY mutant gonads, we conclude that ADAMTS-16 is not required for testis determination in mice. An examination of marker gene expression in XX mutant gonads at the same developmental stage also revealed no overt abnormalities (Fig. S2).
Figure 2.
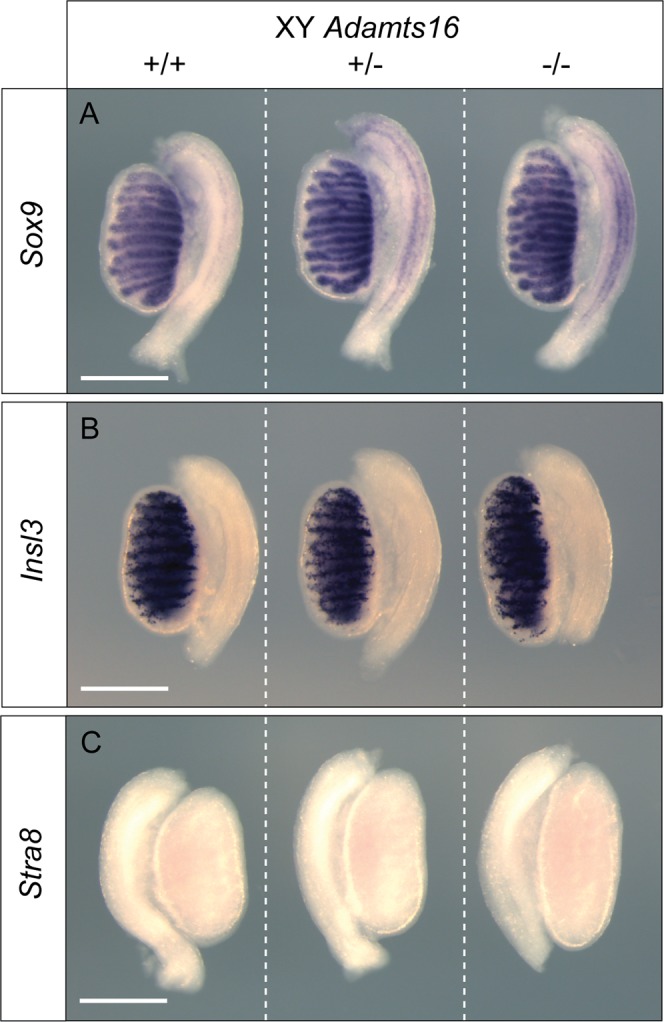
Marker gene analysis of XY gonad development in Adamts16 homozygous mutants. Expression of the Sertoli cell marker Sox9 (A), Leydig cell marker Insl3 (B) and the germ cell meiotic entry marker Stra8 are all unaffected in Adamts16 homozygous mutants (−/−) when compared to wild-type (+/+) and heterozygous (+/−) controls at 14.5 dpc. Scale bar = 0.5 mm.
Normal fertility in adult males lacking ADAMTS-16
Homozygous mutant animals were viable and appeared normal at weaning and later stages of adult life. Cohorts of homozygous mutant and control males were tested for fertility at 3 months and 6 months of age (see Materials and Methods). No significant differences were observed between mutant and control cohorts in respect of their ability to generate copulatory plugs in wild-type females and give rise to fetuses at 13.5 to 16.5 dpc (Fig. S3). Homozygous female mice were similarly fertile and exhibited no overt morphological abnormalities of the reproductive system (data not shown).
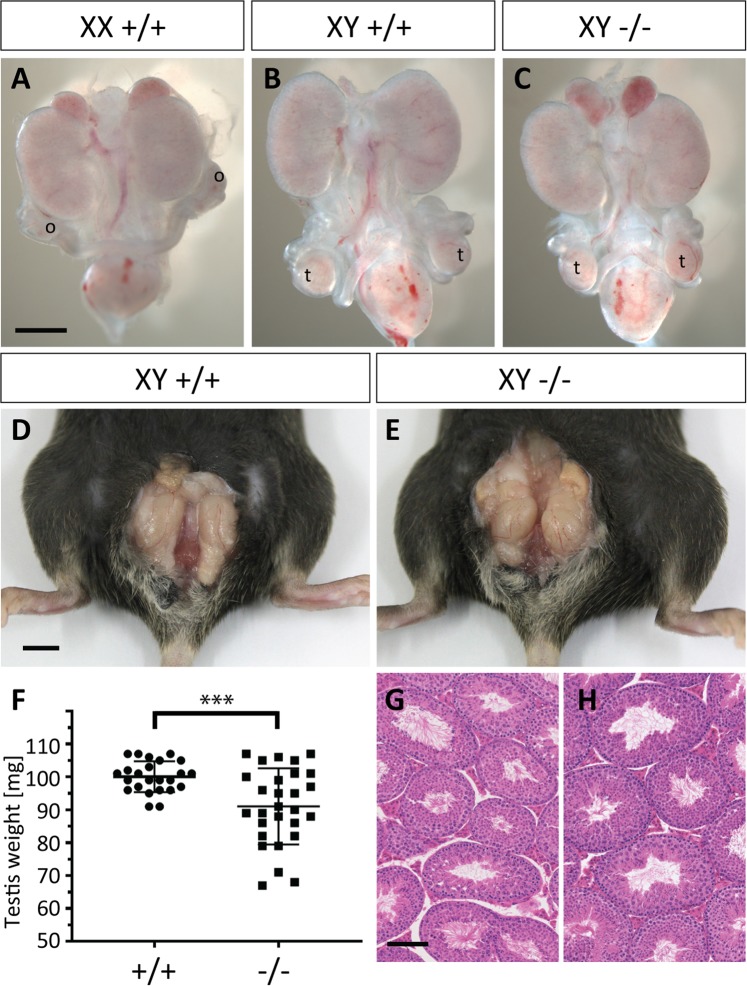
In rats lacking ADAMTS-16, testes appeared high in the abdomen, potentially indicating a defect in the first, trans-abdominal, phase of testicular descent9. We examined testicular position in mutants and controls at 16.5 dpc, by which time the transabdominal phase is essentially complete. Testes in both cohorts were positioned at the base of the abdomen (Fig. 3A–C), indicating that the trans-abdominal phase of testicular descent has occurred normally in ADAMTS-16 deficient mice. Consistent with this observation, Adamts16 expression was negligible in the gubernaculum at 14.5 dpc, when compared to its expression in testis and expression of the gubernacular marker gene, Lgr8 (Fig. S4). Following fertility testing, adult mutant males and controls were sacrificed and the testes examined. There were no overt differences between the positions of testes in mutants or controls (Fig. 3D,E); in both cases, the testes were commonly scrotal but occasionally found in a supra-scrotal position, as is common in mice. There was a small but significant reduction (9%) in average testis weight in mutants (Fig. 3F). We also examined the histology of control and mutant testes. Consistent with their normal fertility, the testes of mutant males also appeared overtly normal (Fig. 3G,H).
Figure 3.
Fetal and adult reproductive structures are overtly normal in Adamts16-deficient mice. (A–C) At 16.5 dpc, the fetal testes (t) have descended to a position adjacent to the bladder in wild-type (B) and Adamts16 −/− mutant males (C); this is in contrast to the para-renal position of the ovaries (o) in wild-type females at the same stage (A). Scale bar = 0.5 mm; (D,E) Testes in adult mice have descended in to the scrotum in wild-type (D) and Adamts16 −/− mutant males (E); scale bar = 0.5 cm; (F) The average weight of the adult testis is reduced in the Adamts16 −/− mutant males (n = 28) compared to wild-type controls (n = 24); ***p ≤ 0.01 (student’s t-test); (G,H) Testicular histology of H&E-stained section appears unaltered in mutant males (H) when compared to wild-type controls (G). Scale bar = 100 μm.
Discussion
Here, we describe the generation of an Adamts16 mutant allele through CRISPR/Cas9-mediated genome editing. Adamts16 transcript levels are negligible in homozygous mutant embryonic tissue, probably as a consequence of nonsense-mediated decay of the mutant transcript. Thus, the mutant allele described here is likely a null allele or a strongly hypomorphic allele. Homozygous XY male mice lacking functional ADAMTS-16 undergo normal testis determination. The testes of adult mutant males are somewhat smaller than controls but they exhibit normal histology. These observations are consistent with the fertility of homozygous male mutants, at 3 and 6 months of age. We conclude that ADAMTS-16 is not required for testis development in mice.
Several prior observations indicated a possible role for ADAMTS-16 in gonadal sex determination. First, Adamts16 transcripts are detected in the developing mouse gonad at the stage of sex determination and have been reported to be at higher levels in XY supporting cells (pre-Sertoli cells) than XX pre-granulosa cells10. Our WMISH data confirm male-enhanced expression of Adamts16 in the gonad at 11.5 dpc, the sex-determining stage of gonadogenesis; such sexual dimorphism in gene expression sometimes reflects a role in testis determination. Secondly, the Adamts16 locus is bound by SRY and SOX9 based on genome-wide ChIP-seq analyses of those transcription factors in the developing testis17, suggesting that it may be a regulatory target of these core testis-determining factors. Adamts16 has also been reported as a potential target of WT16, a transcription factor with known roles in sex determination. Finally, mis-sense variants of ADAMTS16, predicted to be damaging, are associated with human 46,XY DSD, including complete gonadal dysgenesis (female presentation), indicating a possible role in human testis determination7. However, such a role is not supported by the overtly normal sex determination in XY and XX mouse fetuses reported here. In this context, it is interesting to note that a large number of Adamts genes are detected in the developing gonad at the stage of sex determination10, suggesting the possibility of functional redundancy. Whilst it is not uncommon for a mouse knockout not to phenotypically model a human heterozygous variant associated with disease, we cannot exclude the possibility that ADAMTS16 variants associated with DSD are mere polymorphisms or, alternatively, not loss-of-function alleles, but rather gain-of-function or another class of disruptive allele.
Male rats lacking functional ADAMTS-16 exhibited significantly smaller testes than controls, with more pronounced differences as the rats aged9. Moreover, mutant males were infertile and bilaterally cryptorchid. One outstanding question concerns why the phenotype associated with loss of ADAMTS-16 in rats is much more severe than that detected in mice. Such questions can always be answered with reference to species-specific differences in particular biological processes: different genomes, different gene expression levels and temporal profiles, and different tissues and contexts. One other explanation might concern the alleles generated in rat and mice: are they both loss-of-function alleles? Expression analyses reported here indicate that in mouse mutants homozygous for the induced Adamts16 mutation there is significant loss of Adamts16 mRNA, this in addition to the loss of exon 5-encoded amino acids and predicted introduction of a premature stop codon. The rat mutation comprised a 17 base-pair deletion in exon 1, introducing a predicted premature stop codon and thus also constituting a likely null allele9. The modest alteration in testis weight observed in the homozygous mutant male mice is consistent with the absence of a cryptorchidism phenotype. The difference in phenotype between rat and mouse remains unexplained, but the species-specific compensatory action of other ADAMTS proteins may be a contributory factor. We did not observe significant expression of Adamts16 in the fetal mouse gubernaculum, a structure that controls testicular descent by undergoing significant remodelling during this period; it is unclear whether it is expressed in the developing rat gubernaculum.
The variability in testis weight of mutant males represents an example of phenotypic variation on a pure-bred genetic background. Whilst the mechanistic basis of such reduced penetrance, or variable expressivity, remains unclear it is not an uncommon observation even when little or no genetic variation exists between animals in a cohort. A recent report of a high-throughput study of mouse knockout phenotypes, performed on a cohort of isogenic mice, indicates that variable expressivity is widespread18. Stochastic influences on the severity of sex development phenotypes are also common19.
In summary, we report normal sexual development in male mice lacking functional ADAMTS-16. Since C57BL/6 comprises genetic backgrounds sensitised to disruptions to testis determination, it seems most unlikely that any sex development phenotypes would become apparent on any other mouse inbred strains. Therefore, we conclude that ADAMTS-16 is dispensable for testis determination and testicular function in mice, despite a wealth of data suggesting a potential role for this protein in sexual development.
Methods
Mouse strains
The Animal Welfare and Ethical Review Body (AWERB) at MRC Harwell approved the mouse experiments reported here. All mice were bred under license from the UK Home Office (PPL 70/8898) and experiments performed according to the Animals (Scientific Procedures) Act, 1986. Mice were kept in individually ventilated cages (IVCs) in an environment that was free of specific opportunistic pathogens (SPF). Further details of micro- and macro-environmental conditions are available upon request. Mice harbouring a 906 bp deletion of Adamts16 were generated using CRISPR/Cas9. Two pairs of single-guide RNAs (gRNAs) flanking ENSMUSE00000448614, along with Cas9 RNA, were injected into 1-cell mouse zygotes from C57BL6/NTac (B6/N) females, using methodologies as previously described20,21. gRNA pair sequences, including protospacer adjacent motifs (PAM), were: 5.1/5.2-TATATGATGCTCGTACAATC(TGG), CTCGTACAATCTGGATGTCT(TGG) and 3.1/3.2-AAGACTCAGCTACACAAATG(AGG), TCACACACTAGAAATCTCAT(TGG).
All lines were maintained on B6/N. Genotyping of Adamts16 mice was performed using a common forward primer (5′-GAGTCCTGCTCTGCAAGTCC-3′) and two reverse primers that distinguish between the wild-type and deletion alleles: 5′-AGGCATGTCTATGGAGGAACAT-3′ (wild-type) and 5′-TGTGTGAAGATATCAGTGAGCCC-3′ (deletion).
Generation of embryos
Noon on the day the copulatory plug was detected was counted as 0.5 days post coitum (dpc). Adult mice were sacrificed in humane fashion by dislocating the neck, and this was confirmed by cessation of circulation; embryos were decapitated in ice-cold, phosphate buffered saline (PBS). Accurate staging of embryos collected at 11.5 dpc was performed by carefully counting the number of tail somites (ts).
Wholemount in situ hybridization
Wholemount in situ hybridization (WMISH) of embryonic tissues, and the use of probes for the genes Sox9, Stra8, Insl3 and Lgr8, have been described previously22,23. A 566 bp Adamts16 probe was generated by PCR amplification of fetal kidney cDNA using primers 5′-GGCAACATTATGTCCCCAAC-3′ and 5′-TCCAGAGGGCATCTCTGACT-3′. At least three independent biological samples (per genotype) were analysed with a given marker.
Quantitative RT-PCR
Total gonadal RNA (following removal of the mesonephros) was extracted using the RNeasy plus micro kit (Qiagen). Reverse transcription (RT) was performed using 100 ng of total RNA, using the High capacity cDNA RT kit (Applied 24 Biosystem). Quantitative RT-PCR (qRT-PCR) was with Fast SYBR Green Master Mix (Life technologies), using a 7500 Fast Real-Time PCR system (Applied Biosystem). RNA expression levels were calculated following normalization to those of Hprt1 (endogenous control), employing the ΔΔCt method. At least five samples per genotype were included in the analysis. Sequences of primers are available on request.
Supplementary information
Acknowledgements
The authors would like to thank the husbandry team in MLC Ward 5 at MRC Harwell, especially Jackie Harrison, Michelle Sandell, Gemma Atkins, Rosie Hillier and Anju Paudyal. We also thank the FESA team and the Gene Delivery team in the MLC transgenic facility. This research was funded by the UK Medical Research Council through core funding to AG (MC_U142684167) at the Mammalian Genetics Unit, MRC Harwell Institute.
Author contributions
Experiments were conceived and designed by N.W., J.M., G.C., L.T., S.W. and A.G. C.L., N.W., N.C., P.S. and J.M. performed experiments. A.G. wrote the manuscript and it was approved by all authors.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-019-53900-0.
References
- 1.Porter S, Clark IM, Kevorkian L, Edwards DR. The ADAMTS metalloproteinases. Biochem J. 2005;386:15–27. doi: 10.1042/BJ20040424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kelwick R, Desanlis I, Wheeler GN, Edwards DR. The ADAMTS (A Disintegrin and Metalloproteinase with Thrombospondin motifs) family. Genome Biol. 2015;16:113. doi: 10.1186/s13059-015-0676-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Surridge AK, et al. Characterization and regulation of ADAMTS-16. Matrix Biol. 2009;28:416–424. doi: 10.1016/j.matbio.2009.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cal S, et al. Cloning, expression analysis, and structural characterization of seven novel human ADAMTSs, a family of metalloproteinases with disintegrin and thrombospondin-1 domains. Gene. 2002;283:49–62. doi: 10.1016/S0378-1119(01)00861-7. [DOI] [PubMed] [Google Scholar]
- 5.Gao S, De Geyter C, Kossowska K, Zhang H. FSH stimulates the expression of the ADAMTS-16 protease in mature human ovarian follicles. Mol. Hum. Reprod. 2007;13:465–471. doi: 10.1093/molehr/gam037. [DOI] [PubMed] [Google Scholar]
- 6.Jacobi, C. L., Rudigier, L. J., Scholz, H. & Kirschner, K. M. Transcriptional regulation by the Wilms tumor protein, Wt1, suggests a role of the metalloproteinase Adamts16 in murine genitourinary development. J Biol. Chem. 288 (2013). [DOI] [PMC free article] [PubMed]
- 7.Barseghyan H, et al. Identification of novel candidate genes for 46,XY disorders of sex development (DSD) using a C57BL/6J-Y (POS) mouse model. Biol. Sex. Differ. 2018;9:8. doi: 10.1186/s13293-018-0167-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gopalakrishnan K, et al. Targeted disruption of Adamts16 gene in a rat genetic model of hypertension. Proc Natl Acad Sci USA. 2012;109:20555–20559. doi: 10.1073/pnas.1211290109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abdul-Majeed S, Mell B, Nauli SM, Joe B. Cryptorchidism and infertility in rats with targeted disruption of the Adamts16 locus. PLoS One. 2014;9:e100967. doi: 10.1371/journal.pone.0100967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jameson SA, et al. Temporal transcriptional profiling of somatic and germ cells reveals biased lineage priming of sexual fate in the fetal mouse gonad. PLoS Genet. 2012;8:e1002575. doi: 10.1371/journal.pgen.1002575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stevant I, et al. Dissecting Cell Lineage Specification and Sex Fate Determination in Gonadal Somatic Cells Using Single-Cell Transcriptomics. Cell Rep. 2019;26:3272–3283 e3273. doi: 10.1016/j.celrep.2019.02.069. [DOI] [PubMed] [Google Scholar]
- 12.Munger SC, et al. Elucidation of the transcription network governing mammalian sex determination by exploiting strain-specific susceptibility to sex reversal. Genes Dev. 2009;23:2521–2536. doi: 10.1101/gad.1835809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Correa SM, et al. Sex reversal in C57BL/6J XY mice caused by increased expression of ovarian genes and insufficient activation of the testis determining pathway. PLoS Genet. 2012;8:e1002569. doi: 10.1371/journal.pgen.1002569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Warr N, et al. Gadd45gamma and Map3k4 interactions regulate mouse testis determination via p38 MAPK-mediated control of Sry expression. Dev Cell. 2012;23:1020–1031. doi: 10.1016/j.devcel.2012.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gierl MS, Gruhn WH, von Seggern A, Maltry N, Niehrs C. GADD45G functions in male sex determination by promoting p38 signaling and Sry expression. Dev Cell. 2012;23:1032–1042. doi: 10.1016/j.devcel.2012.09.014. [DOI] [PubMed] [Google Scholar]
- 16.Gonen N, Lovell-Badge R. The regulation of Sox9 expression in the gonad. Curr Top Dev Biol. 2019;134:223–252. doi: 10.1016/bs.ctdb.2019.01.004. [DOI] [PubMed] [Google Scholar]
- 17.Li Y, Zheng M, Lau YF. The sex-determining factors SRY and SOX9 regulate similar target genes and promote testis cord formation during testicular differentiation. Cell Reports. 2014;8:723–733. doi: 10.1016/j.celrep.2014.06.055. [DOI] [PubMed] [Google Scholar]
- 18.Dickinson ME, et al. High-throughput discovery of novel developmental phenotypes. Nature. 2016;537:508–514. doi: 10.1038/nature19356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harris A, et al. ZNRF3 functions in mammalian sex determination by inhibiting canonical WNT signaling. Proc Natl Acad Sci USA. 2018;115:5474–5479. doi: 10.1073/pnas.1801223115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mianne J, et al. Correction of the auditory phenotype in C57BL/6N mice via CRISPR/Cas9-mediated homology directed repair. Genome Med. 2016;8:16. doi: 10.1186/s13073-016-0273-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mianne J, et al. Analysing the outcome of CRISPR-aided genome editing in embryos: Screening, genotyping and quality control. Methods. 2017;121-122:68–76. doi: 10.1016/j.ymeth.2017.03.016. [DOI] [PubMed] [Google Scholar]
- 22.Bogani D, et al. Loss of mitogen-activated protein kinase kinase kinase 4 (MAP3K4) reveals a requirement for MAPK signalling in mouse sex determination. PLoS Biol. 2009;7:e1000196. doi: 10.1371/journal.pbio.1000196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Warr N, et al. Sfrp1 and Sfrp2 are required for normal male sexual development in mice. Dev Biol. 2009;326:273–284. doi: 10.1016/j.ydbio.2008.11.023. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.