Abstract
Background:
Pseudomonas aeruginosa is a pit of an enormous group of free-living bacteria that are able to live everywhere and suggested to be the causative agent of great scope of acute and chronic animal infections.
Aim:
The current study was carried out to illustrate the prevalence of P. aeruginosa in small ruminants and existence of some virulence operons as well as its antimicrobial resistance.
Materials and Methods:
A total of 155 samples from sheep and 105 samples from goats (mouth abscesses, fecal swabs, nasal, tracheal swabs, and lung tissue) were collected for bacteriological study, existence of some virulence expression operons with the study of their sensitivity to the antimicrobials using disc diffusion and presence of mexR operon which is responsible for multidrug resistance (MDR).
Results:
The bacteriological examination revealed that P. aeruginosa was isolated from nine out of 155 samples from sheep (5.8%) and four isolates out of 105 samples from goat (3.8%). It is found that 12 (92.3%), 10 (76.9 %), and 8 (61.5%) of P. aeruginosa isolates harbored hemolysin phospholipase gene (pcl H), gene (exo S), and enterotoxin gene (tox A), respectively. The results of antibiotic sensitivity test showed that all tested isolates were resistant to ampicillin, bacitracin, erythromycin, streptomycin, tetracycline, trimethoprim-sulfamethoxazole, and tobramycin but sensitive to ciprofloxacin and norfloxacin. The MDR (mex R) operon was existed in all isolates.
Conclusion:
There is a growing risk for isolation of virulent MDR P. aeruginosa from sheep and goat illness cases, and this should be regarded in the efficient control programs.
Keywords: drug resistance, goat, Pseudomonas aeruginosa, sheep, virulence
Introduction
Pseudomonas aeruginosa is a Gram-negative, encapsulated, nonsporulated, and strict aerobic motile rod. It is an opportunistic pathogen, widely exists in various ecosystems and believed to be implemented in several serious human and animal diseases [1,2]. P. aeruginosa causes numerous diseases in sheep and goats; respiratory illness, which is one of the major issues particularly pneumonia, associated with physical and physiological stress, leading to significant mortality rates, and increased economic loss [3]. A number of mastitis cases and the pathogen can reside in the udder for many years [4]. Moreover, P. aeruginosa infection may lead to urogenital disorders, gastrointestinal illness sinusitis, and osteomyelitis [5-8].
The organism declares plenty of virulence agents, which share in its pathogenicity. These comprise enterotoxins, exocytotoxins, and toxins produced by protein secretion systems, as a result of expression of certain virulence operons. Consequently, many of these have been implemented in infection, septicemia, and fatal condition [9,10]. The mortality rate is usually higher than bacteremia sourced with other Gram-negative pathogens due to its ability to secrete these several products that after successive colonization can induce extensive tissue damage, bloodstream invasion, and dissemination [11]. Through all resistant pathogens, the condition is most significant for P. aeruginosa as its incidence of resistance to antimicrobial agents in continuous increasing and has been accounted worldwide [12]. This critical ability of the pathogen could be attributed to the existence of the unusually restricted outer membrane permeability which acts as a safeguard barrier for antibiotics to overcome. Besides that, there were other secondary intrinsic factors as energy-dependent multidrug efflux and chromosomally encoded periplasmic beta-lactamase [13].
This study was conducted to address its isolation and identification from ovine and caprine population, with stressing on its toxigenic expressed operons as well as the associated multidrug resistance (MDR) property.
Materials and Methods
Ethical approval
As per CPCSEA guidelines, a study involving clinical samples does not require the approval of the Institute Animal Ethics Committee.
Samples
A total of 260 different samples were collected from sheep (155) and from goat (105) selectively suffering from respiratory manifestation (40 from sheep, and 22 from goats) in triplicate as nasal, tracheal swabs, and lung tissues. Also, fecal samples were collected from diarrheic (14 sheep and 23 goats) and gathered swabs from skin lesion with abscesses (21 sheep and 16 goats). The samples were gathered from scattered local farms or owners in Great Cairo and Delta rural areas. The samples were gathered from September 2017 to April 2018. The samples were bacteriologically examined and the selected colonies were biochemically identified [14].
Antimicrobial sensitivity assay
The susceptibility of the isolates to various antibiotics was achieved using the diffusion technique [15], the following antibiotic discs were used; amikacin (30 mg), ampicillin (10 µg), bacitracin (10 µg), chloramphenicol (30 µg), erythromycin (15 µg), gentamycin (10 µg), streptomycin (10 µg), tetracycline (30 µg), trimethoprim-sulfamethoxazole (2.25/7.75 µg), tobramycin (30 µg), ciprofloxacin (5 µg), and norfloxacin (10 µg) [16].
DNA extraction
DNA extraction from the samples was performed using the QIAamp DNA Mini kit (Qiagen, Germany, GmbH) with modifications from the manufacturer’s recommendations. Briefly, 200 µl of the sample suspension was incubated with 10 µl of proteinase K and 200 µl of lysis buffer at 56°C for 10 min. After incubation, 200 µl of 100% ethanol was added to the lysate. The sample was then washed and centrifuged following the manufacturer’s recommendations. Nucleic acid was eluted with 100 µl of elution buffer provided in the kit.
Polymerase chain reaction (PCR) amplification using oligonucleotide primers
Primers used were supplied from Metabion (Germany), as shown in Tables-1 and 2 [16,17]. Primers were utilized in a 25 µl reaction containing 12.5 µl of Emerald Amp Max PCR Master Mix (Takara, Japan), 1 µl of each primer of 20 pmol concentrations, 4.5 µl of water, and 6 µl of DNA template. The reactions were performed either uniplex (mex R) or multiplex (exo S, pcl H, and tox A), as described in Tables-1 and 2 [17,18], in a T3 Biometra thermal cycler.
Table 1.
Uniplex PCR: Primers sequences, target operons, amplicon sizes, and cycling conditions.
| Target operon | Primers sequences | Amplified segment (bp) | Primary denaturation | Amplification (35 cycles) | Final extension | Reference | ||
|---|---|---|---|---|---|---|---|---|
| Secondary denaturation | Annealing | Extension | ||||||
| mex R | GCGCCATGGC CCATAT TCAG | 637 | 94°C 5 min | 94°C 45 s | 57°C 45 s | 72°C 45 s | 72°C 10 min | [16] |
| GGCATTC GCC AGTAAGCGG | ||||||||
PCR=Polymerase chain reaction
Table 2.
Multiplex PCR: Primers sequences, target genes, amplicon sizes, and cycling conditions.
| Target gene | Primers sequences | Amplified segment (bp) | Primary denaturation | Amplification (30 cycles) | Final extension | Reference | ||
|---|---|---|---|---|---|---|---|---|
| Secondary denaturation | Annealing | Extension | ||||||
| exo S | CCTTCCCT CCTTCCCCC CGGCGATCTGGA AAAG AAATG | 270 | 95°C 5 min | 95°C 30 s | 58°C 30 s | 72°C 50 s | 72°C 10 min | [17] |
| CATCCTCA GGCG TACATCCT | ||||||||
| pcl H | GAAGCCAT GGGCT ACTTCAA | 307 | ||||||
| AGAGTGA CGAGG AGCGGTAG | ||||||||
| tox A | ATGGTGTA GATC GGCGACAT | 433 | ||||||
| AAGCCTTC GACC TCTGGAAC | ||||||||
PCR=Polymerase chain reaction
Analysis of the PCR products
The products of PCR were electrophoresed on 1.5% agarose gel (Applichem, Germany, GmbH) in 1× TBE buffer at room temperature using gradients of 5V/cm. For gel analysis, 15 µl of the products were loaded in each gel slot. A 100 bp DNA Ladder (Qiagen, Germany, GmbH) was used to determine the fragment sizes. The gel was photographed by a gel documentation system (Alpha Innotech, Biometra) and the data were analyzed through computer software. P. aeruginosa (ATCC 27853) was used as positive controls and distilled water (Merck, Germany) was used as a negative control in all PCR reactions.
P. aeruginosa (ATCC 15442) was used as a positive control while Staphylococcus aureus (ATCC 25923) was used as a negative one.
Results and Discussion
P. aeruginosa is an environmental ubiquitous nonpathogenic saprophyte present in animate and inanimate media. Otherwise, in many statuses, this microorganism becomes a primary pathogen and is encountered in severe localized or generalized infections in human, animals, and birds particularly among immune-compromised hosts [19]. The organism was implicated in multiple diseases in sheep and goat; respiratory manifestation, diarrhea, skin abscess, and the severity of the condition may result in death cases [20,21].
In this study, as shown in Table-3, it was found that P. aeruginosa was successfully isolated from (6/40) sheep and (2/22) goat with respiratory manifestation with an incidence of 15% and 9.1%, respectively. The isolates were recovered from 3 (7.5%) sheep and 2 (9.1%) goat nasal swabs. Furthermore, the organism was recovered from 2 (5%) tracheal swabs and 1 (2.5%) lung tissue belonged only to sheep samples.
Table 3.
Prevalence of Pseudomonas aeruginosa recovered from living and slaughtered sheep and goat.
| Groups | Type of samples | Sheep | Goat | ||||
|---|---|---|---|---|---|---|---|
| No. of samples | Positive samples number | Positive samples % | No. of samples | Positive samples number | Positive samples % | ||
| I–animals with respiratory manifestation | Nasal swabs | 40 | 3 | (7.5) | 22 | 2 | (9.1) |
| Tracheal swabs | 40 | 2 | (5) | 22 | 0 | 0 | |
| Lung tissues | 40 | 1 | (2.5) | 22 | 0 | 0 | |
| II–animals with skin lesions | Wound and abscesses | 21 | 2 | (9.5) | 16 | 1 | (6.25) |
| III–diarrheic animals | Fecal | 14 | 1 | (7.1) | 23 | 1 | (4.3) |
| Total | 155 | 9 | (5.8) | 105 | 4 | (3.8) | |
These average percentage results agreed with that formerly reported [22,23] while the lower incidence was 3.6% [24] and one necropsied goat [25]. In contrast, higher isolations were determined [26]. Two P. aeruginosa isolates were recovered from diarrheic sheep 1/14 (7.1%) and goat 1/23 (4.3%), respectively. On the other hand, the animals infected with wound and abscesses, the percentage of isolated P. aeruginosa from sheep was 2/21 (9.5%) and goat 1/16 (6.25%), respectively. These obtained data close to that stated by Hears et al. [27], who isolated three strains P. aeruginosa from one of the infected sheep flocks. Abd El-Rahman [28] deduced that the incidence of P. aeruginosa recovered from diarrheic sheep was higher than that obtained among specimens of other infected animals. Furthermore, current results less than that obtained by Alkeshan [29], who isolated P. aeruginosa from abscesses with an incidence (6.18%).
The unselective use of antibiotics is potentially leading to a higher incidence of infections with resistant microorganisms such as P. aeruginosa, worse which may be transmitted from animal to human complicating the treatment of human diseases [30]. Regarding, the antimicrobial sensitivity agar test shown in (Table-4); it was noticed that the isolates were lack of susceptibility to many tested antimicrobial agents. The primary mechanism of the microorganism’s resistance relies on its ability to shut out various agents rather than the production of antibiotic inactivating enzymes. Consequently, most of antibiotics are of limited value in the treatment of P. aeruginosa infection in animals [31].
Table 4.
Antibiotic sensitivity test of (13) Pseudomonas aeruginosa strains isolated from sheep and goat.
| Antimicrobial agents | Sensitive | Resistant |
|---|---|---|
| n (%) | n (%) | |
| Amikacin | 3 (23.1) | 10 (76.9) |
| Ampicillin | 0 (0) | 13 (100) |
| Bacitracin | 0 (0) | 13 (100) |
| Chloramphenicol | 2 (15.4) | 11 (84.6) |
| Erythromycin | 0 (0) | 13 (100) |
| Gentamicin | 4 (30.8) | 9 (69.2) |
| Streptomycin | 0 (0) | 13 (100) |
| Tetracycline | 0 (0) | 13 (100) |
| Trimethoprirn/sulfamethoxazole | 0 (0) | 13 (100) |
| Tobramycin | 0 (0) | 13 (100) |
| Ciprofloxacin | 13 (100) | 0 (0) |
| Norfloxacin | 12 (92.3) | 1 (4.5) |
The results indicated that the isolates were susceptible to both ciprofloxacin 13 (100%) and norfloxacin 12 (92.3%), while completely resistant to ampicillin, bacitracin, erythromycin, streptomycin, tetracycline, trimethoprim-sulfamethoxazole, and tobramycin, most of the isolates were resistant to amikacin, chloramphenicol, and gentamycin. These findings agreed with the other previous studies which illustrated that the organism is resistant to all used antibiotics except quinolones [32-35].
Assorted chromosomally encoded efflux systems and outer membrane porins have been distinguished as essential contributors to MDR phenotype resistance. The MexAB-OprM is the only pump which is expressed at a level enough to allow intrinsic MDR in wild type P. aeruginosa strains. Mutations in mex R cause over-expression of MexAB-OprM efflux pump [36]. It is obvious in our study as shown in Figure-1; that all isolates exhibited the amplification of 637 bp which represent the mex R operon. The emergence of MDR P. aeruginosa is turning out a challenging issue in infection control schemes.
Figure-1.
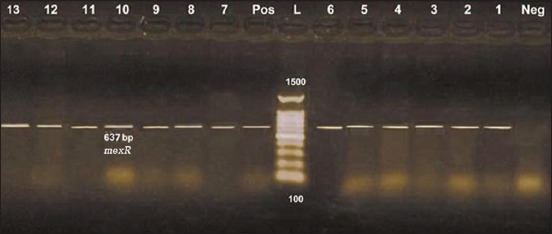
Uniplex polymerase chain reaction detection of antibiotic-resistant mexR operon in Pseudomonas aeruginosa isolates showing: L: 100 bp DNA ladder. Lanes 1-13: P. aeruginosa isolates. Lane Pos.: Positive control; amplification of 637 bp represented mexR. Lane Neg.: Negative control.
Figure-2 shows multiplex PCR detection of virulence genes in P. aeruginosa isolates P. aeruginosa is considered a potent opportunistic pathogen and this is contributed to its ability to produce arrays of virulence agents which aid the organism to attach, penetrate, disseminate, and inhibit phagocytosis, and chemotaxis [37]. The data obtained from Figure-2 revealed the multiplex PCR reaction of three genes amplification; plc H (307bp), exo S (270bp), and tox A (433 bp) which responsible for expression of cytotoxins, phospholipase, and enterotoxins as 12 (92.3%), 10 (76.9%), and 8 (61.5%), respectively. These results drew near the other previous investigations [38-42].
Figure-2.
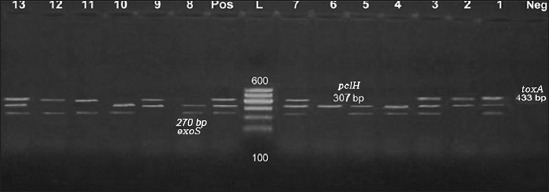
Multiplex polymerase chain reaction detection of virulence genes in Pseudomonas aeruginosa isolates showing: L: 100 bp DNA ladder. Lanes 1-13: P. aeruginosa isolates. Lane Pos.: Positive control; amplification of 270 bp represented exoS, 307 bp represented plcH, and 433 bp represented toxA. Lane Neg.: Negative control.
Conclusion
P. aeruginosa could be implicated in sheep and goat infections, and the isolates showed high resistance to commonly used antibiotics as well as having numerous agents of virulence. A strict antibiotic policy and establishment of infection control programs will help to lower the incidence of resistance in P. aeruginosa.
Authors’ Contributions
ASH and AND supervised the experiment. AMA and EAE shared in the collection of the samples, AND and HAA did the isolation and identification of microorganism. EAE performed the antibiotic susceptibility assay. HAA and AMA performed PCR. ASH and AND prepared and reviewed the manuscript. All authors read and approved the final manuscript.
Acknowledgments
The authors are thankful to National Research Centre, Egypt for providing necessary facilities for this study. The authors would like to thank Editage (www.editage.com) for English language editing. The authors did not receive any fund for this study.
Competing Interests
The authors declare that they have no competing interests.
Publisher’s Note
Veterinary World remains neutral with regard to jurisdictional claims in published institutional affiliation.
References
- 1.Yang J, Zhao H.L, Ran L.Y, Li C.Y, Zhang X.Y, Su H.N, Shi M, Zhou B.C, Chen X.L., Zhang Y.Z. Mechanistic insights into elastin degradation by pseudolysin, the major virulence factor of the opportunistic pathogen Pseudomonas aeruginosa. Sci. Rep. 2015;23(5):9936. doi: 10.1038/srep09936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bird V.Y, Chastain-Gross R, Sutkowski R, Bird V.G, Vyas P, Joseph R. Pseudomonas aeruginosa as an etiologic agent of nephrolithiasis in deep water divers. J. Endourol. Case Rep. 2017;3(1):4–6. doi: 10.1089/cren.2016.0117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bangar Y.C, Pachpute S.T, Nimase R.G. The survival analysis of the potential risk factors affecting lamb mortality in Deccani sheep. J. Dairy Vet. Anim. Res. 2016;4(2):266–270. [Google Scholar]
- 4.Wright E.A, Di Lorenzo V, Trappetti C, Liciardi M, Orru G, Viti C, Bronowski C, Hall A.J, Darby A.C, Oggioni M.R, Winstanley C. Divergence of a strain of Pseudomonas aeruginosa during an outbreak of ovine mastitis. Vet. Microbiol. 2015;175(1):105–113. doi: 10.1016/j.vetmic.2014.11.011. [DOI] [PubMed] [Google Scholar]
- 5.Chigerwe M, Mavangira V, Byrne B.A, Angelos J.A. Antibiotic resistance patterns of bacteria isolated from indwelling foley catheters following tube cystostomy in goats with obstructive urolithiasis. J. Vet. Diagn. Invest. 2017;29(3):316–320. doi: 10.1177/1040638717695607. [DOI] [PubMed] [Google Scholar]
- 6.Hwang I.Y, Koh E, Wong A, March J.C, Bentley W.E, Lee Y.S, Chang M.W. Engineered probiotic Escherichia coli can eliminate and prevent Pseudomonas aeruginosa gut infection in animal models. Nat. Commun. 2017;11(8):15028. doi: 10.1038/ncomms15028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fong S.A, Drilling A.J, Ooi M.L, Paramasivan S, Finnie J.W, Morales S, Psaltis A.J, Vreugde S, Wormald P.J. Safety and efficacy of a bacteriophage cocktail in an in vivo model of Pseudomonas aeruginosa sinusitis. Transl. Res. 2018;206(4):41–56. doi: 10.1016/j.trsl.2018.12.002. [DOI] [PubMed] [Google Scholar]
- 8.Rasooli A, Nouri M, Esmaeilzadeh S, Ghadiri A, Gharibi D, Koupaei M.J, Moazeni M. Occurrence of purulent mandibular and maxillary osteomyelitis associated with Pseudomonas aeruginosa in a sheep flock in south-west of Iran. Iran J. Vet. Res. 2018;19(2):133–136. [PMC free article] [PubMed] [Google Scholar]
- 9.Mccarty S.M, Cochrane C.A, Clegg P.D, Percival S.L. The role of endogenous and exogenous enzymes in chronic wounds:A focus on the implications of aberrant levels of both host and bacterial proteases in wound healing. Wound Repair Regen. 2012;20(2):125. doi: 10.1111/j.1524-475X.2012.00763.x. [DOI] [PubMed] [Google Scholar]
- 10.Tayabali A.F, Gordon C, Kathy C.N. Virulence attributes and host response assays for determining pathogenic potential of Pseudomonas strains used in biotechnology. PLoS One. 2015;10(11):e0143604. doi: 10.1371/journal.pone.0143604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hu P, Chen J, Chen Y, Zhou T, Xu X, Pei X. Molecular epidemiology, resistance, and virulence properties of Pseudomonas aeruginosa cross-colonization clonal isolates in the non-outbreak setting. Infect Genet. Evol. 2017;55(11):288–296. doi: 10.1016/j.meegid.2017.09.010. [DOI] [PubMed] [Google Scholar]
- 12.Briers Y, Walmagh M, Grymonprez B, Biebl M, Pirnay J.P, Defraine V, Michiels J, Cenens W, Aertsen A, Miller S, Lavigne R. Art-175 is a highly efficient antibacterial against multidrug-resistant strains and persisters of Pseudomonas aeruginosa Antimicrob. Agents Chemother. 2014;58(7):3774–3784. doi: 10.1128/AAC.02668-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu Z.Q, Flavin M.T, Flavin J. Combating multidrug-resistant Gram-negative bacterial infections. Expert Opin. Invest. Drugs. 2014;23(2):163–182. doi: 10.1517/13543784.2014.848853. [DOI] [PubMed] [Google Scholar]
- 14.Quinn P.J, Markyl B.K, Carter M.E, Donnelly W.J, Leonard F.C. Veterinary Microbiology anal Microbial Disease. United Kingdom: Blackwell; 2002. [Google Scholar]
- 15.Clinical Laboratory Standards Institute. Performance Standards for Antimicrobial Disk Susceptibility Tests;Approved Standards. 12th ed. USA: Clinical Laboratory Standards Institute; 2015. pp. M02–A12. [Google Scholar]
- 16.Magiorakos A.P, Srinivasan A, Carey R.B, Carmeli Y, Harbarth S, Hindler J.F, Kahlmeter G. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria:An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012;18(3):268–281. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 17.Odumosu B.T, Adeniyi B.A, Chandra R. First detection of OXA-10 extended-spectrum beta-lactamases and the occurrence of mex R and nfx B in clinical isolates of Pseudomonas aeruginosa from Nigeria. Chemotherapy. 2016;61(2):87–92. doi: 10.1159/000441712. [DOI] [PubMed] [Google Scholar]
- 18.Hui S, Quoclinh T, Wentao X, Baiqiang Z.H, Yunbo L, Kunlun H. A universal primer multiplex PCR method for typing of toxinogenic Pseudomonas aeruginosa. Appl. Microbiol. Biotechnol. 2012;95(6):1579–1587. doi: 10.1007/s00253-012-4277-8. [DOI] [PubMed] [Google Scholar]
- 19.Gellatly H, Gellatly S.L, Hancock R.E.W. Pseudomonas aeruginosa:New insights into pathogenesis and host defenses. Pathog. Dis. 2013;67(3):159–173. doi: 10.1111/2049-632X.12033. [DOI] [PubMed] [Google Scholar]
- 20.Cassirer E.F, Sinclair A.R.E. Dynamics of pneumonia in a bighorn sheep meta population. J. Wildl. Manage. 2007;71(4):1080–1088. [Google Scholar]
- 21.Khaled B.H. Prevalence and etiology of abscess disease of sheep and goats at Qassim Region, Saudi Arabia. Vet. World. 2011;4(11):495–499. [Google Scholar]
- 22.Almeida P.F, Alves F.S.F, Santos L.F, Rosa J.S. Survey on bacterial agents associated with respiratory diseases of goats in North-Eastern Brazil. Rev. Microbiol. 1986;17(3):213–215. [Google Scholar]
- 23.Pasic S, Popvic M. Study of ovine Mycoplasma in Bosnia and Herzegovina. Mycoplasma and bacteria in the pneumonic lungs of sheep Vet. Yugoslavia. 1988;37(1):35–39. [Google Scholar]
- 24.Baysal T, Guler L. Isolation of bacteria from enzootic pneumonia in lambs and kids in the Kenya region. Veterinarium. 1992;3(1):1–5. [Google Scholar]
- 25.Shiferaw G, Tariku S, Ayelet G, Abebe Z. Contagious caprine pleuropneumonia and Mannheimia haemolytica-associated acute respiratory disease of goats and sheep in Afar Region, Ethiopia. Rev. Sci. Tech. 2006;25(3):1153–1163. [PubMed] [Google Scholar]
- 26.Mishra K.C. Antimicrobial sensitivity of bacteria isolated from respiratory tract infection of goats in Sikkim. Ind. J. Anim. Sci. 1992;26(7):635–636. [Google Scholar]
- 27.Hears A, Dominguez L, Lopez J, Hears A.L, Garayzabal J. Outbreak of ovine mastitis associated with P. aeruginosa infection Vet. Rec. 1999;145(4):111–112. doi: 10.1136/vr.145.4.111. [DOI] [PubMed] [Google Scholar]
- 28.Abd El-Rahman W.M. Thesis (Microbiology) Egypt: Faculty of Veterinary Medicine, Cairo University; 2004. Further Studies on Strains Isolated from Broiler Chickens. Ph.D. [Google Scholar]
- 29.Alkeshan Y.M, Alharbi S, Alrehaili F, Almutiri J, Althobity M, Alotaibi N. Antimicrobial resistance pattern of Pseudomonas aeruginosa in regional tertiary care hospitals of Saudi Arabia. IOSR J. Nurs. Health Sci. 2015;5(2):54–62. [Google Scholar]
- 30.Wiedmann M, Weilmeier D, Dineen S.S, Ralyea R, Boor K.J. Molecular and phenotypic characterization of Pseudomonas spp. isolated from milk Appl. Environ. Microbiol. 2000;66(5):2085–2095. doi: 10.1128/aem.66.5.2085-2095.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bhullar K, Waglechner N, Pawlowski A, Koteva K, Banks E.D, Johnston M.D, Barton H.A, Wright G.D. Antibiotic resistance is prevalent in an isolated cave microbiome. PLoS One. 2012;7(4):e34953. doi: 10.1371/journal.pone.0034953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Siew M.L, Ganeswrei R, Ampalam P, Kek H.C. Antimicrobial susceptibility and virulence genes of clinical and environmental isolates of Pseudomonas aeruginosa. PeerJ. 2019;7:e6217. doi: 10.7717/peerj.6217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Enas G, Samah A, Magdy K. Prevalence of multi-drug resistant Pseudomonas spp. and Acinetobacter spp. causing nosocomial infection in intensive care unit (ICU) of National Liver Institute Egypt. J. Med. Microbiol. 2010;19(1):1. [Google Scholar]
- 34.Sahar M.H. Proceeding Veterinary Science Conference. 11th. 2012. Comparison of Microbial Isolates from External Ear Canal of Sheep and their Susceptibility to Antibiotics; pp. 41–48. [Google Scholar]
- 35.Hannah J, Kathryn S.L, Martin W. Ciprofloxacin binding to GyrA causes global changes in the proteome of Pseudomonas aeruginosa. FEMS Microbiol. Lett. 2018;365(13):134. doi: 10.1093/femsle/fny134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thanyaluck S, Potjanee S, Sasitorn C, Boonek Y, Channarong S, Varomyalin T, Supayang P.V. Conessine as a novel inhibitor of multidrug efflux pump systems in Pseudomonas aeruginosa. BMC Complement. Altern. Med. 2017;17(1):405. doi: 10.1186/s12906-017-1913-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Habibi A., Honarmand R. Profile of virulence factors in the multi-drug resistant Pseudomonas aeruginosa strains of human urinary tract infections (UTI) Iran Red. Crescent Med. J. 2015;17(12):e26095. doi: 10.5812/ircmj.26095. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 38.Mitov I, Strateva T, Markova B. Prevalence of virulence genes among Bulgarian nosocomial and cystic fibrosis isolates of Pseudomonas aeruginosa. Braz. J. Microbiol. 2010;41(3):588–595. doi: 10.1590/S1517-83822010000300008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Heidary Z, Bandani E, Eftekhary M, Jafari A.A. Virulence genes profile of multidrug-resistant Pseudomonas aeruginosa isolated from Iranian children with UTIs. Acta Med. Iran. 2016;54(3):201–210. [PubMed] [Google Scholar]
- 40.Khosravi A.D, Shafie F, Montazeri E.A, Rostami S. The frequency of genes encoding exotoxin A and exoenzyme S in Pseudomonas aeruginosa strains isolated from burn patients. Burns. 2016;42(5):1116–1120. doi: 10.1016/j.burns.2016.02.012. [DOI] [PubMed] [Google Scholar]
- 41.Pobiega M, Maciag J, Pomorska-Wesolowska M, Chmielarczyk A, Romaniszyn D, Ziolkowski G, Heczko P.B, Wojkowska-Mach J, Bulanda M. Urinary tract infections caused by Pseudomonas aeruginosa among children in Southern Poland:Virulence factors and antibiotic resistance . J. Pediatr. Urol. 2016;12(1):36.e1–6. doi: 10.1016/j.jpurol.2015.05.034. [DOI] [PubMed] [Google Scholar]
- 42.Badamchi A, Masoumi H, Javadinia S, Asgarian R, Tabatabaee A. Molecular detection of six virulence genes in Pseudomonas aeruginosa isolates detected in children with urinary tract infection. Microb. Pathog. 2017;107(6):44–47. doi: 10.1016/j.micpath.2017.03.009. [DOI] [PubMed] [Google Scholar]


