Abstract
Background and Aims
At the rear edge of the distribution of species, extreme isolation and small population size influence the genetic diversity and differentiation of plant populations. This may be particularly true for Arctic-alpine species in mid-latitude mountains, but exactly how peripherality has shaped their genetic and reproductive characteristics is poorly investigated. The present study, focused on Salix herbacea, aims at providing new insights into the causes behind ongoing demographic dynamics and their consequences for peripheral populations of Arctic-alpine species.
Methods
We performed a whole-population, highly detailed sampling of the only two S. herbacea populations in the northern Apennines, comparing their clonal and genetic diversity, sex ratio and spatial genetic structure with a reference population from the Alps. After inspecting ~1800 grid intersections in the three populations, 563 ramets were genotyped at 11 nuclear microsatellite markers (nSSRs). Past demography and mating patterns of Apennine populations were investigated to elucidate the possible causes of altered reproductive dynamics.
Key Results
Apennine populations, which experienced a Holocene bottleneck and are highly differentiated (FST = 0.15), had lower clonal and genetic diversity compared with the alpine population (RMLG = 1 and HE = 0.71), with the smaller population exhibiting the lowest diversity (RMLG = 0.03 and HE = 0.24). An unbalanced sex ratio was found in the larger (63 F:37 M) and the smaller (99 F:1 M) Apennine population. Both were characterized by the presence of extremely large clones (up to 2500 m2), which, however, did not play a dominant role in local reproductive dynamics.
Conclusions
Under conditions of extreme isolation and progressive size reduction, S. herbacea has experienced an alteration of genetic characteristics produced by the prevalence of clonal growth over sexual reproduction. However, our results showed that the larger Apennine population has maintained levels of sexual reproduction enough to counteract a dramatic loss of genetic and clonal diversity.
Keywords: Fragmentation, snowbed, peripherality, clonal growth, genetic diversity, sex ratio, spatial genetic structure, Salix herbacea, gene flow, isolation, marginality, microsatellite markers
INTRODUCTION
Genetic drift and gene flow interact with natural selection in shaping allele frequencies of populations over time. Demographic processes and spatial distribution of populations influence the strength of evolutionary forces modulating the outcome of their interplay. In particular, towards the species distribution edges, population size is expected to progressively decrease and genetic isolation to increase until the disappearance of the species. Peripheral populations are therefore assumed to exhibit lower genetic diversity and greater genetic differentiation compared with populations located in the core area of species distribution, as predicted by the centre–periphery hypothesis (Vucetich and Waite, 2003). In the survey by Eckert et al. (2008), it was shown that such signals, although often weak, were confirmed in approximately two-thirds of the reviewed studies. The centre–periphery hypothesis has, however, been challenged by several studies showing that, at least on a broad geographical scale, the distribution of genetic diversity might have been markedly affected by past climate-driven range dynamics (Hewitt, 2000, 2004) rather than by demo-genetic stochasticity per se (Hampe and Petit, 2005). In particular, due to the location of refugial areas, peripheral meta-populations of Northern Hemisphere species at the rear edge of their distribution might represent long-term stores of genetic diversity that could be of crucial importance to species survival in the face of climate change (Hampe and Petit, 2005; Rehm et al., 2015).
At the rear edge of their distribution, Arctic-alpine plant species (i.e. species with a predominantly arctic distribution, but growing in mid-latitude mountain ranges as well; Hultén and Fries, 1986) exhibit a paradigmatic case of isolated, peripheral populations. For instance, the central and southern European mountain systems are home to a number of disjunct populations of Arctic-alpine species (Schönswetter et al., 2005). Moving southwards from their core distribution area, Arctic-alpine plants still occur on mountain ranges, but at progressively higher elevation sites, restricted to habitat patches with micro-environmentally favourable conditions (Birks and Willis, 2008). Studies on population genetics of Arctic-alpine species in central and southern Europe have proliferated in recent years, most of them being carried out in the Alps (e.g. Schönswetter et al., 2003, 2005, 2008; Skrede et al., 2006; Kropf et al., 2006, 2008; Alvarez et al., 2012). On the other hand, the levels of genetic diversity in peripheral populations of Arctic-alpine plants located in the Apennines (i.e. the main mountain system of the Italian Peninsula) have been less well studied and understood (Ansell et al., 2008). This is somewhat surprising in view of the fact that the Apennines were one of the main refugial areas where cold-adapted taxa potentially survived over consecutive Quaternary glacial cycles (Ansell et al., 2008; Gentili et al., 2015a).
This lack of knowledge stimulated us to focus on the genetic consequences of extreme peripherality for Arctic-alpine species in the northern Apennines, one of the southernmost outposts where these species are well represented (Foggi, 1990; Tomaselli and Agostini, 1994; Abeli et al., 2018). We thus focused our attention on the Apennine populations of the dwarf willow, Salix herbacea, one of the most common vascular plants living in late-exposed snowbeds (Björk and Molau, 2007), habitats that host a unique set of species sharing adaptations to temperature-, nutrient- and, frequently, wind-limited environmental conditions. Snowbeds represent a key component of alpine landscape diversity, acting as warm-stage microrefugia for high-mountain plants (Gentili et al., 2015b) and providing important ecosystem services to neighbouring plant communities and herbivores (e.g. Billings and Bliss, 1959; Fox, 1991). Nowadays, snowbeds are considered to be one of the most vulnerable mountain habitats threatened by climate change (e.g. Björk and Molau, 2007; Matteodo et al., 2016).
In the highly heterogeneous alpine landscape, the spatial pattern of snowmelt determines an often strong small-scale differentiation in environmental conditions influencing the genetic differentiation of alpine plant populations (e.g. Shimono et al., 2009). In this context, snowbed habitats can act as reservoirs of genetic diversity (Cortés et al., 2014). However, under future climatic conditions, characterized by warmer and longer snow-free periods, it is likely that snowbed habitats will shrink or even disappear at southern latitudes (Theurillat and Guisan, 2001). In the Apennines, the populations of snowbed species, such as the forbs Veronica alpina, Omalotheca supina and Leucanthemopsis alpina, the graminoid Luzula alpinopilosa and the moss Polytrichastrum sexangulare, as well as populations of several other snowbed bryophytes and lichens, are indeed largely isolated both from one another and from the alpine ones, highly fragmented and small in size (Tomaselli, 1991; Petraglia and Tomaselli, 2007). These conditions allow an intensive genetic characterization at the whole-population level to be performed, with such an approach providing new insights into the effects of extreme isolation on the genetic characteristics of snowbed species.
Salix herbacea is regarded as an ideal species for studying the effect of climate change in Arctic and alpine tundra due to its ecological characteristics and the amphi-Atlantic, Arctic-alpine distribution range (Alsos et al., 2009; Wheeler et al., 2016; Abeli et al., 2018). In Europe, it spans from Svalbard to the Mediterranean mountains with the southernmost outposts in central Spain, the central Apennines and Macedonia (Rechinger and Akeroyd, 1993). This species has a number of life-history traits that may interact with isolation and demography in shaping the genetic characteristics of populations. Salix herbacea is a dioecious species that, being self-incompatible, should be more vulnerable to habitat fragmentation and small population size than self-compatible species (Leimu et al., 2006; Kramer et al., 2008). Moreover, as a primarily wind-pollinated species it should be less prone to pollen limitation than insect-pollinated species (Beerling, 1998), although seed set of wind-pollinated species may depend more on population density than on population size and isolation, as in insect-pollinated plants (Steven and Waller, 2007). Salix herbacea has the potential for clonal as well as sexual reproduction (Beerling, 1998). The ecological advantages of clonal growth include the ability to effectively acquire resources such as light, water and nutrients, and share them through clonal integration, reducing the mortality of genets (e.g. Klimeš et al., 1997). Clonal growth also avoids the costs and the risks associated with sexual reproduction (Maynard Smith, 1978), allowing populations to persist in habitats in which ecological conditions can be frequently or periodically unfavourable for sexual reproduction. On the other hand, clonal growth can also affect sexual reproduction, reducing fitness due to a strong unbalance of mating groups required for outcrossing (Vallejo-Marin et al., 2010). When sexual reproduction is very limited, the accumulation of somatic mutations could also reduce fertility, potentially leading to the loss of sex (Eckert, 2001).
In continuous populations, S. herbacea can maintain both frequent sexual reproduction and high genetic diversity (Reisch et al., 2007). However, it is well known that peripheral populations of species characterized by a mixed reproductive capability may lose genetic diversity and even become monoclonal as a result of restricted gene flow, genetic drift and prevailing asexual reproduction (e.g. Beatty et al., 2008). Single genets of S. herbacea have been shown to reach crown sizes as large as 10 m (Stamati et al., 2007) and survive up to 450 years (de Witte et al., 2012), suggesting that populations and even individuals might have persisted locally despite considerable climatic oscillations. Long-lived species are expected to cope better with habitat fragmentation than short-lived ones because the former may experience smaller fluctuations in abundance (Marini et al., 2012), with a positive effect on meta-population persistence (Eriksson, 1996; Bossuyt and Honnay, 2006). As a consequence, long-lived species may exhibit extensive time delays in extinctions due to their longevity (Eriksson, 1996; Honnay and Bossuyt, 2005; Lindborg, 2007; Saar et al., 2012).
The primary objective of this study was to assess the effects of extreme isolation in the only two remnant populations of S. herbacea in the northern Apennines, along the rear edge of the species distribution, with reference to a population located in the Alps, where the species is widespread in high-elevation snowbed habitats. Our investigation was not limited to a comparison of summary parameters among populations but rather consisted of a highly detailed sampling at the whole-population level for the two Apennine populations. This approach is the only one that can reveal the hitherto unknown spatial complexity behind the transition from sexual to clonal reproduction expected in peripheral populations. The two northern Apennine populations of S. herbacea are extremely small (~1.50 and ~0.16 ha) and highly isolated (~23 km apart and ~160 km from the nearest population growing in the Alps). At such a degree of isolation, and considering the biological and ecological characteristics of the species, we expected to find trends among populations going from low peripherality and larger size to high peripherality and smaller size in terms of (1) progressively lower occurrence of sexual reproduction and higher occurrence of clonal propagation, (2) more skewed distribution of clone sizes, (3) higher impact of genetic drift (reduction of genetic diversity and high genetic differentiation), and (4) increase in both the intensity and heterogeneity of the spatial genetic structure. Once the genetic consequences of isolation were assessed, we further investigated past demography and mating patterns of Apennine populations to elucidate the possible causes of altered reproductive dynamics.
MATERIALS AND METHODS
Study area
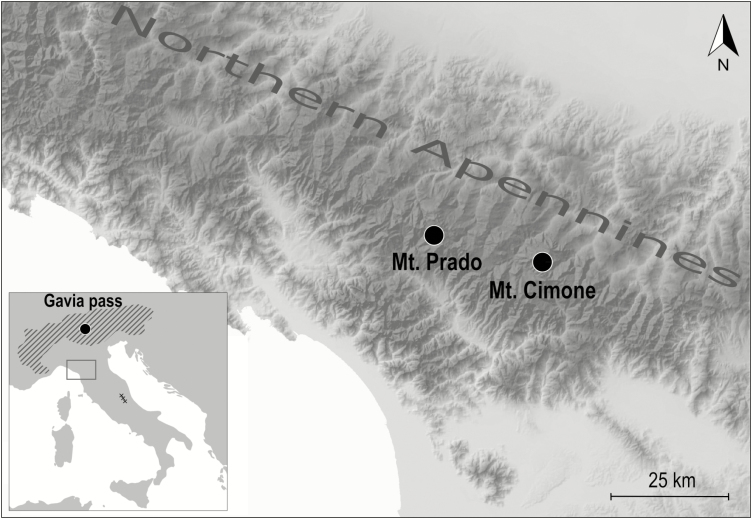
The northern Apennines harbour two highly isolated dwarf willow populations. These are ~160 km from the geographically closest populations occurring in the south-eastern Alps to the north, ~220 km from those occurring in the Maritime Alps to the west and ~260 km from those occurring in the central Apennines to the south-east. The present study is focused on these two Salix herbacea populations, lying on the northern slope of the Tuscan–Emilian Apennines 23 km apart (Fig. 1, Table 1). At both sites the dwarf willow is fairly abundant, growing as a loose mat of prostrate twigs strictly limited to snowbeds, which are surrounded by sub-shrub vegetation dominated by Vaccinium species. The easternmost population (hereafter referred to as Cimone) occurs not far from the summit of Mt Cimone (2165 m a.s.l.), the highest peak in the northern Apennines. The second population (hereafter referred to as Prado) is located on the northern slope of Mt Prado (2054 m a.s.l.). The area covered by these two populations differs considerably: Cimone covers ~0.16 ha, whereas Prado is almost ten times larger (~1.50 ha).
Fig. 1.
Location of the two remnant S. herbacea populations in the northern Apennines at Mt Prado and Mt Cimone. The insert at bottom left shows the Alpine distribution of the species (hatched area) according to Jalas and Suominen (1976) and the location of the Alpine population at Gavia pass (black circle). The box in the insert represents the area of the northern Apennines included in the main figure and × symbols show the location of S. herbacea remnants in the central Apennines.
Table 1.
Number of samples, numbers of MLGs and MLLs, genetic parameters (inbreeding coefficient was calculated at both ramet and MLG levels), clonality index (R) for MLGs and MLLs, and Shannon–Wiener (S-W) diversity index for MLGs and MLLs computed for the three sampled populations. Clonality index R was computed as (G − 1)/(N − 1), where G is the number of genotypes (either MLGs or MLLs) and N is sample size (Dorken and Eckert, 2001)
| Population | Latitude | Longitude | Altitude (m a.s.l.) |
Samples | MLG | MLL | N a | A r | H O | H E |
F
IS
ramets |
F
IS
MLGs |
R index MLGs | R index MLLs |
S-W index MLGs |
S-W index MLLs |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gavia | 46°21′ N | 10°29′ E | 2700 | 30 | 30 | 30 | 8.64 | 8.64 | 0.55 | 0.71 | 0.19 | 0.19 | 1.00 | 1.00 | 3.40 | 3.40 |
| Prado | 44°14′ N | 10°23′ E | 2000 | 347 | 108 | 89 | 5.18 | 4.09 | 0.55 | 0.51 | −0.07 | 0.06 | 0.32 | 0.26 | 3.18 | 2.95 |
| Cimone | 44°11′ N | 10°42′ E | 1750 | 196 | 7 | 5 | 2.46 | 1.87 | 0.46 | 0.24 | −0.32 | -0.21 | 0.03 | 0.02 | 0.26 | 0.17 |
N a, number of alleles; Ar, allelic richness; HO, observed heterozygosity; HE, expected heterozygosity; FIS, inbreeding coefficient.
We compared the genetic and reproductive characteristics of northern Apennine populations with a ‘geographically central’ population, i.e. lying at the distributional core of S. herbacea in the Alps. This alpine population is located close to the Gavia Pass (2700 m a.s.l.) within the Stelvio National Park. This population (hereafter referred to as Gavia), situated at the bottom of a glacial cirque, was selected because snowbed communities are widespread in this area (Carbognani et al., 2014a), where S. herbacea is currently undergoing a phase of expansion (Carbognani et al., 2014b). The Gavia population is ~230 km from those in the northern Apennines (Fig. 1, Table 1).
Sampling procedure
The areas occupied by the two Apennine populations were fully inspected using a removable square grid consisting of measuring tapes fixed on the soil surface with permanent metal sticks. Mesh size differed between the two sites according to the area occupied by each population, being 2 and 5 m for Cimone and Prado, respectively. Each grid intersection was used as the centre of a square in which the occurrence of male and/or female individuals was recorded (Supplementary Data Fig. S1).
During successive surveys from June to August 2014, 638 squares for Cimone (i.e. 29 × 22 grid intersections) and 1150 squares for Prado (i.e. 46 × 25 grid intersections) were carefully inspected by a team of eight operators searching for all Salix ramets with reproductive structures. Around each grid, a 50-m-wide buffer area was delineated and checked for the presence of disjunct ramets. When a disjunct ramet was found, a buffer area with a radius of 50 m around the ramet was further inspected. This approach allowed us to sample the entire area within the 3 weeks following snowmelt, when the flowering peak of S. herbacea usually takes place (Carbognani et al., 2016). At each grid intersection, the nearest generative ramet was marked with a coloured wool thread and a plastic tag and its position relative to the grid intersection was recorded. In areas where no generative ramets were found, a vegetative ramet was tagged and subsequently sexed in 2015 whenever possible. In the Apennine populations, the sex of 722 ramets was determined out of a total of 818.
For Cimone and Prado, sex ratio was calculated at the whole-population level on the basis of the occurrence of female and/or male individuals within all the square areas of the grid (Supplementary Data Fig. S1). For Gavia, sex ratio was calculated based on the occurrence of female and/or male individuals in 38 plots of ~1 m2, randomly located in stands dominated by S. herbacea within an area of ~2 ha.
Genetic analyses were performed on 563 ramets, from which five to ten fresh leaves were collected for DNA extraction. At the northern Apennine sites, 196 samples for Cimone and 337 for Prado were randomly selected from the samples collected according to the sampling strategy described above (Fig. 2). At the alpine site, a square grid with a mesh size of 20 m was positioned on the previously described area. Then, an S. herbacea ramet was sampled within 1 m from 30 randomly selected grid intersections, as described above. All samples were dried with silica gel and stored at −20 °C until DNA extraction was performed.
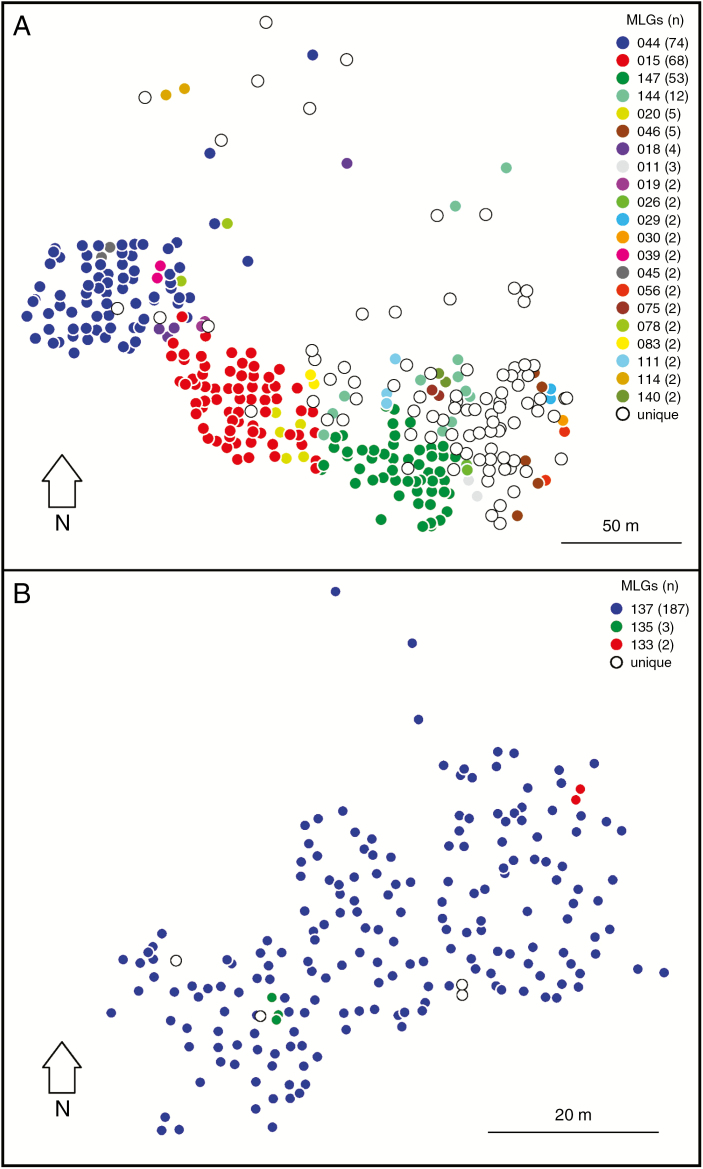
Fig. 2.
Location of sampled ramets in the Prado (A) and Cimone (B) populations. Open circles represent ramets that, after genetic analyses, were classified as unique MLGs. Coloured dots represent >1 ramets MLGs. The key shows the number of ramets associated with each MLG identity code.
DNA extraction and genotyping
DNA extraction was carried out using the DNeasy 96 Plant Kit (Qiagen) following the manufacturer’s instructions. Thirty milligrams of dried tissue for each sample was put in each well of 96-well plates together with a 3-mm diameter tungsten bead. Plates were frozen in liquid nitrogen for 30 s before two cycles of 30 s of disruption at 28 Hz using a Mixer Mill MM300 (Retsch, Germany). DNA quality was checked on a 1 % agarose gel stained with GelRed (Biotium, USA). DNA concentration was measured with a NanoDrop ND-1000 (Thermo Scientific, Wilmington, USA).
Sixteen nuclear microsatellite markers (nSSRs) were initially tested on a subset of samples: seven nSSRs already used on S. herbacea (Cortés et al., 2014) and nine nSSRs transferred from other Salix spp. (Stamati et al., 2003; Barker et al., 2005; Steltzer et al., 2008). After excluding low-quality and multi-banding loci as well as loci that were found to be monomorphic in all populations, 11 nSSRs were retained for subsequent analyses. The final marker set was arranged in two PCR multiplexes (Supplementary Data Table S1). Multiple PCR reactions were carried out using the Type-it Microsatellite PCR kit (Qiagen, Germany); the final PCR volume was optimized to 6 μL and the PCR mix for both multiplexes consisted of 3 μL of Qiagen Multiplex PCR Master Mix, 1 μL of DNA (20 ng μL−1) and 0.2 μm primers. Multiplex composition is presented in Supplementary Data Table S1. For both multiplexes, the PCR thermal profile was as follows: an initial step at 95 °C for 5 min followed by 32 cycles at 95 °C for 30 s, 57 °C for 90 s and 72 °C for 30 s, with a final 30-min extension step at 60 °C. PCR products were run on an AB 3500 automatic sequencer (Applied Biosystems, USA) using LIZ-500 as internal size standard. Chromatograms were analysed using GeneMapper v4.1 (Applied Biosystems, USA). For two nSSRs (SB38 and ORPM_312) single PCRs were performed, multiplexing the products with the other nSSRs before loading the sequencer. A touchdown PCR with the following thermal profile was used: initial denaturation at 94 °C for 3 min, followed by ten cycles at 94 °C for 30 s, 60 °C (decreasing by 1 °C per cycle) for 30 s and 72 °C for 40 s, followed by 25 cycles at 94 °C for 30 s, 50 °C for 30 s and 72 °C for 40 s, with a final 10-min extension step at 72 °C.
Data analysis
The power of the marker set to distinguish all detectable multilocus genotypes (MLGs) within sampled ramets was evaluated through a Monte Carlo procedure (Arnaud-Haond et al., 2007). The probability of finding identical MLGs generated by sexual reproduction (Psex) in Cimone and Prado was estimated following Arnaud-Haond et al. (2007). On the other hand, the occurrence of different MLGs characterized by slight genetic differences due to few somatic mutations (i.e. derived from asexual reproduction) or scoring errors was assessed through the inspection of pairwise genetic distances among MLGs (Rozenfeld et al., 2007) within each population. In this case, MLGs differentiated by few somatic mutations can be collapsed into the same multilocus lineage (MLL) (Arnaud-Haond et al., 2007). All subsequent analyses were carried out at the MLG and MLL levels. For all the computations of clonal variation we used the RClone R package (Bailleul et al., 2016; R Core Team, 2017).
Allelic frequencies and standard genetic parameters [i.e. FIS (inbreeding coefficient), HO (observed heterozygosity) and HE (expected heterozygosity)] were computed with GenAlEx v 6.5 (Peakall and Smouse, 2012). Since there is no standardized way to compute genetic variability indexes for clonal populations, these indexes were computed on datasets built relying on two extreme and opposite approaches. The first dataset comprised all sampled ramets (N = 563), whereas the second dataset included only one sample for each MLG (N = 145).
To assess the potential influence of the different mesh sizes used on diversity estimates, we performed a spatial rarefaction of samples by means of computer simulations. We subsampled the Prado dataset with the same mesh size used for Gavia (20 m) and the Cimone dataset with the same mesh size used for Prado (5 m). We started from a single random sample from each population and added further samples randomly, one at a time, exclusively when each new sample was >20 m apart for Prado (or 5 m apart for Cimone) from all other samples already present in the subsampled population. This procedure was repeated 30 times. We did not subsample Cimone with the mesh size used for Gavia because in this case only a handful of samples would have been included.
Fine-scale spatial genetic structure (SGS) of populations was investigated by spatial autocorrelation analysis performed using RClone and the pairwise kinship coefficient (Fij) of Loiselle et al. (1995). Tests for statistical significance of mean Fij values in each distance class were carried out by random shuffling of individual geographical locations (1000 times) to define the upper and lower bounds of the 95 % confidence interval around the null hypothesis that Fij and pairwise distances were uncorrelated. Analyses were performed using both the even distance classes option (using 20-m-wide distance classes) and the even sample size option (distributing all possible pairs in ten distance classes with similar numbers of pairs per class). The intensity of SGS was also measured by the Sp statistic (Vekemans and Hardy, 2004), computed as Sp = bF/(F1 – 1), where F1 is the average kinship coefficient between individuals of the first distance class (0–20 m) and bF is the regression slope of the kinship estimator Fij computed among all pairs of individuals against their geographical distances. The statistical significance of F1 and bF was assessed through 1000 permutations of spatial coordinates among individuals. For Prado, spatial autocorrelation analysis was run both on all sampled ramets and only considering one sample for each MLG (located at the barycentre of each MLG complex). Given the extremely low number of MLGs detected in Cimone (see the Results section), in this population spatial autocorrelation analysis was solely run on all samples.
To investigate how past demography and current mating patterns have shaped the extant genetic and clonal structure of Apennine populations we assessed (1) the occurrence, timing and intensity of population size changes, and (2) parentage relationships among MLGs.
Demographic inference was conducted on the MLG datasets of Apennine populations with the approach developed in the Migraine v.0.5.4 software (Leblois et al., 2014). Model parameters (duration of the population size change in number of generation Dg/2N; current population size 2Nμ; ancestral population size 2Nancμ), scaled by mutation rate (μ) and population size (2N), were estimated under a maximum likelihood framework using the single-population model with a single continuous past variation in population size (OnePopVarSize). The generalized stepwise mutation model (GSM) was selected to describe nSSR variation. An iterative procedure based on five subsequent steps was used to gradually increase the focus on the high-likelihood region of the parameter space. At each iteration, the total number of points considered in the parameter range was 500 and the number of runs per point was 2000.
Parentage relationships were assessed through parentage analysis considering MLGs as offspring and candidate parents, as in Macaya-Sanz et al. (2016). Parentage analysis was carried out based on a maximum likelihood approach using the software FaMoz (Gerber et al., 2003). Parentage was categorically assigned to compatible parent pairs or single parents (when it was not possible to identify a parent pair) with the highest LOD score above the threshold. The LOD score threshold was estimated following the simulation method implemented in FaMoz, by randomly generating 5 × 104 seedlings both from MLG genotypes and from MLG allele frequencies. Once parentage relationships were assessed, we calculated the immigration rate as well as the individual reproductive success of MLGs. Possible dispersal distances were also estimated connecting the spatial centroids of MLGs considered as offspring and their assigned parents.
Due to the extremely low number of MLGs detected at Cimone (see the Results section), demographic inference and parentage analysis were performed solely on the Prado dataset.
RESULTS
Clonal diversity
The set of 11 SSRs used in this study allowed an accurate determination of the number of genotypes present in the samples (Supplementary Data Table S2). Overall, 145 MLGs were identified using the entire marker set. Monte Carlo simulations showed that values approaching this plateau would have been reached even using fewer markers: 142 (98 %) MLGs were detected on average using ten markers, 140 (97 %) using nine markers and 136 (94 %) using eight markers (Supplementary Data Fig. S2). The probability of a second encounter of an identical MLG via sexual reproduction was always very low (Psex < 0.001) for Prado. In three MLGs of Cimone Psex was >0.05, but it should be noted that this computation has limitations when almost monoclonal populations are considered (Arnaud-Haond et al., 2007).
The number of MLGs varied considerably among the sampled populations and clonal diversity was markedly lower in the two Apennine populations compared with Gavia (Table 1). The Shannon–Wiener diversity index for the three populations indicated the same pattern, with the Apennine populations being less diverse than the alpine reference population and, in particular, Cimone being by far the least diverse. The general pattern of clonal diversity did not change if we considered MLLs (i.e. distinct MLGs with a genetic distance below a threshold of six base pairs) instead of MLGs (Table 1). Therefore, hereafter we decided to present results based exclusively on MLGs.
The three populations did not share any MLG. In the Apennine populations, the distribution of clone abundances was highly skewed (Fig. 2, Supplementary Data Fig. S3). However, while in Prado there were three distinct abundant clones with >50 ramets (Fig. 2A), in Cimone only a single clone was abundant (Fig. 2B), accounting for >95 % of the sampled ramets. The distribution of clone abundances in Gavia was flat, since every ramet was a different MLG (Fig. 2, Supplementary Data Fig. S3).
Spatial rarefaction had the expected effect in terms of reduction of samples. In rarefied populations there are far fewer samples than in the original populations: over the 30 simulated rarefaction replicates, the number of samples at Prado (rarefied to 20 m) was reduced from 347 to an average of 40.53 (s.e. ±0.27) and the number of samples at Cimone (rarefied to 5 m) was reduced from 196 to an average of 41.3 (±0.32). Consequently, in rarefied populations there were also fewer MLGs and fewer ramets per MLG. However, such reduction did not change the overall picture emerging from the comparison of clonal diversity among populations as calculated on the original dataset. The number of MLGs was reduced, on average, from 108 to 24.2 (s.e. ±0.29) at Prado and from 7 to 2.27 (±0.16) at Cimone, while the clonal index increased, on average, from 0.32 to 0.61 (s.e. ±0.01) for the rarefied Prado population and did not change for Cimone [from 0.031 to 0.032 (±0.004)]. On the other hand, the effect of rarefaction on the Shannon–Wiener diversity index was negligible.
Sex ratio
Sex ratio varied considerably among populations. Indeed, Gavia showed a balanced sex ratio of 51 F:49 M, Prado had a slightly unbalanced ratio of 63 F:37 M and Cimone was characterized by an extremely unbalanced ratio of 99 F:1 M. In addition, the spatial distribution of sexes in Prado was not random, but it was highly clumped, reflecting the spatial distribution of clones (see below).
Genetic diversity
The overall pattern of genetic variability did not change between the two approaches used to compute genetic variability at the population level. The two Apennine populations had a reduced genetic variability compared with the alpine reference population (Table 1), which had more alleles (Na) and exhibited higher allelic richness (Ar) and expected heterozygosity (HE). Again, Prado showed intermediate values and Cimone was by far the most genetically impoverished. The only difference between the two methods employed to compute variability indexes was the FIS value for Prado, which was negative when using only one ramet per genet (i.e. with MLGs), showing an excess of heterozygotes, whereas it was positive when using all the ramets of genets, showing an excess of homozygotes.
Values of genetic differentiation indices did not change between the two approaches employed for computing allelic frequencies. The FST values were generally high and were not related to the geographical distance among populations (Table 2). The lowest value recorded was between Gavia and Prado, while the highest value was found for the two nearby Apennine populations. These patterns remained consistent using other methods for assessing genetic distances among population, such as Hedrick’s GST and Jost’s D (results not shown).
Table 2.
F ST values computed between pairs of populations. Allelic frequencies were computed considering all sampled ramets and only one sample for each MLG
| Ramets | MLGs | |||||
|---|---|---|---|---|---|---|
| Gavia | Prado | Cimone | Gavia | Prado | Cimone | |
| Gavia | – | *** | *** | – | *** | *** |
| Prado | 0.09 | – | *** | 0.05 | – | *** |
| Cimone | 0.18 | 0.24 | – | 0.10 | 0.15 | – |
Significance levels: *P < 0.05, **P < 0.01, ***P < 0.001.
Spatial genetic structure
Given the clonal structure presented above, it was possible to produce spatial autocorrelograms only when all ramets in Gavia and Cimone were considered. In these two populations, an absence of SGS was detected (Fig. 3). A significant SGS up to ~50 m was found in Prado, considering both all ramets and MLGs only. The intensity of SGS was high in Prado, with a steep and negative regression slope (b = −0.1427) and Sp up to 0.1842 when calculated on all samples (b = −0.0316 and Sp = 0.0333 on MLGs).
Fig. 3.
Correlograms from spatial autocorrelation analysis based on the correlation coefficient Fij by Loiselle et al. (1995) and even sample size classes for the three S. herbacea populations. Filled dots represents distance classes for which significant spatial structuring of individuals was observed. Black and grey lines indicated autocorrelograms calculated at the ramet and MLG level, respectively.
Besides SGS indices and the spatial autocorrelogram, which provide a global evaluation of SGS in Prado, it is worth noting that the spatial distribution of MLGs was highly heterogeneous, with three large MLGs (i.e. mlg044, mlg015 and mlg147 in Fig. 2A) in the western part of the population and a high number of small MLGs in the eastern part.
Demographic inference
A bottleneck of moderate intensity was detected for Prado. The duration of the population size change in terms of number of generations (Dg/2N) was 0.121 (CI95%: 0.0134–0.552), the current (2Nμ) and ancestral (2Nancμ) population sizes were 0.474 (CI95%: 9.36 × 10−5–1.159) and 2.022 (CI95%: 1.192–3.995) and the N/Nanc ratio was 0.234 with no overlap between the confidence intervals of 2Nμ and 2Nancμ. Considering that 2N stands for the number of gene copies (i.e. 4 times the number of diploid individuals) and assuming a mutation rate equal to 1 × 10−3, these estimates indicate a bottleneck that occurred 58 (CI95%: 6–262) generations ago, reducing the effective population size from 505 (CI95%: 298–999) to 119 (CI95%: 1–290) individuals. Although there are no estimates available for clonal, dwarf willows, we can consider a reasonable generation time varying between 20 and 200 years (de Witte et al., 2012), so that the median estimate of 58 generations would indicate a period ranging from 1160 to 11 600 years BP.
Potential parentage relationships among MLGs
Potential parentage relationships among MLGs were investigated through parentage analysis. Single-parent and parent-pair exclusion probabilities calculated from MLG allele frequencies were 0.9541 for the single parent and 0.9999 for the parent pair. LOD score thresholds chosen after simulations guaranteed 0.83 and 0.94 confidence levels for the assignment of single parents and parent pairs, respectively. Using such thresholds, 51 out of the 108 MLGs (47.2 %) had a compatible parent pair inside the population, 44 MLGs (40.7 %) had a single compatible parent (22 male and 22 female parents) and 13 MLGs (12 %) had no compatible local parents. At the gamete level, this translates into 70 (i.e. 13 × 2 + 44) out of 216 gametes (32.4 %) originated from external, not sampled or extirpated MLGs. Individual MLG reproductive success was not correlated to MLG size expressed in terms of number of ramets, either globally (r = −0.07; t-test, t = −0.74, d.f. = 106, P = 0.46) or as male parents (r = −0.04; t-test, t = −0.44, d.f. = 106, P = 0.66) or female parents (r = −0.04; t-test, t = −0.46, d.f. = 106, P = 0.64).
The distributions of effective seed and pollen dispersal distances were inferred from parentage assignments using MLG geographical centroids as spatial data. The mean seed and pollen dispersal distances were 61.27 and 67.54 m, respectively. The longest detected dissemination and pollination events were 188.79 and 197.8 m, respectively.
DISCUSSION
Plant populations at the rear edge of species distributions have a remarkable evolutionary and conservation importance as an inheritance from glacial–interglacial dynamics (Hampe and Petit, 2005). Such peripheral populations may harbour rare genetic variants potentially advantageous for both species persistence and migration, which is particularly important considering that climate warming is expected to shift their climatic optima (Davis and Shaw, 2001; Jump and Peñuelas, 2005; Rabasa et al., 2013). Due to ecological and biogeographical marginality, peripheral plant populations are often small and isolated (Hampe and Petit, 2005), which makes them more prone to the loss of genetic variability and to stochastic demographic events (Eckert et al., 2008). The present study was primarily aimed at quantifying the effects of extreme isolation in the only two remnant populations of the dwarf willow S. herbacea in the northern Apennines through a whole-population genetic characterization. Overall, results regarding clonal structure, levels of genetic diversity and within-population SGS are all consistent in showing a marked effect of isolation on these two populations (Prado and Cimone). This effect is more marked for the extremely small Cimone population, which is almost monoclonal with a highly unbalanced production of reproductive structures between sexes (i.e. only two ramets with male flowers). Overall, our results suggest the dramatically decreased role of sexual reproduction for S. herbacea at such a degree of isolation.
In general, the clonality index ranges from 0.02 to 1 in plant species (Ellstrand and Roose, 1987), with an average value of 0.42 as calculated on 195 taxa by Vallejo-Marin et al. (2010). A large difference in clonal diversity was found in the three populations included in our study, with extremely low values for Cimone and Prado populations (0.03 and 0.32, respectively) when compared with the Alpine population (1.00) and what was found in previous studies carried out in Scotland (0.54) and Switzerland (0.78) at a much lower spatial scale (Stamati et al., 2007; Cortés et al., 2014). Our simulations of spatial rarefaction demonstrated that the number of MLGs detected is sensitive to the mesh size used for sampling the populations. However, simulations also showed that the mesh size has a limited impact on estimates of clonal diversity and, in particular, on the general picture concerning differences among populations in terms of clonality indexes. The only marked difference found after rarefaction is the increase in the clonality index at Prado. Although this difference is obviously due to less intensive sampling of extremely large clones when the mesh size is larger, it highlights the importance of accounting for the sampling design when comparing results from different populations and/or studies. However, in our study the advantages of a whole-population sampling approach outweighed the possible limitations related to the need to apply different mesh sizes, as can be clearly shown by the effectiveness of such an approach in revealing the highly complex fine-scale SGS in the Prado population.
Sexual reproduction is highly limited at the degree of isolation and population size that characterizes the Cimone population. A low clonal diversity has often been found in studies exploring the effects of isolation on clonal plants (Eckert, 2001). Low clonal diversity has been especially recorded in populations at the leading edge of species distribution. This is generally due to the combination of a limited number of founders together with the prevalence of vegetative propagation during the colonization process, plus the effects of stochastic events related to peripherality itself (e.g. Dorken and Eckert, 2001; Sugai et al., 2016). Eckert (2001) noted that, among peripheral populations, low clonal diversity is more common near the northern limit of the species’ range in comparison with the southern limit, and such a pattern was confirmed by more recent investigations (Silvertown, 2008). Considering that the Apennine populations were not included in the only large-scale genetic study on S. herbacea by Alsos et al. (2009), it is difficult to understand whether these populations are at the trailing edge of what the authors call the ‘alpine genetic cluster’ or at the leading edge of an Apennine genetic cluster. However, considering the latitudinal pattern found by previous literature reviews (Eckert, 2001; Silvertown, 2008), our study reveals a strong impact of peripherality on clonal diversity even at southern latitudes.
Low clonal diversity combines with other signals pointing to the prevalence of vegetative propagation over sexual reproduction in Apennine peripheral populations. A skewed sex ratio was found in both populations and an excess of heterozygotes was detected at the MLG level at Cimone. In S. herbacea, a sex ratio of 60 F:40 M (Crawford and Balfour, 1983) and a slight excess of homozygote individuals up to FIS = 0.09 (Cortés et al., 2014) were recorded in populations located in the Alps. Overall, the deviation detected in the mating system of Apennine populations, especially in the smallest one, is consistent with what is to be expected in plant populations when vegetative propagation prevails over sexual reproduction. In fact, although high isolation and low density of populations are expected to increase genetic drift and positively correlate with inbreeding levels (Wright, 1946), an excess of heterozygote individuals could occur in species with mixed reproductive systems, such as apomictic, clonal or self-fertilizing species (Peck et al., 1998; Dorken and Eckert, 2001). In particular, high rates of clonal reproduction might determine a heterozygote excess due to the accumulation of somatic mutations over time (Balloux et al., 2003). Indeed, geographical and subsequent genetic isolation are thought to outweigh the advantages of sex in marginal populations and to induce a shift towards asexual reproduction in clonal plants (Eckert, 2001). Such a transition towards clonality can also result in profound consequences for the genetic variation within small populations (Raabová et al., 2015). In fact, we found that both expected heterozygosity and allelic richness significantly decreased from the alpine to the smallest Apennine population. In Cimone, allelic richness and expected heterozygosity are only 46 and 59 %, respectively, of those found in Gavia. Such a dramatic decrease in genetic diversity is linked with the strong effect of genetic drift due to the small population size of the Apennine populations and the likely historical absence of gene flow. The genetic differentiation between Prado and Cimone, besides being high compared with what was detected among fragmented populations of congeneric species (Sochor et al., 2013; Berlin et al., 2014; Perderau et al., 2014), is higher than alpine–Apennine differentiation. Moreover, demographic inference on the Prado dataset confirms the scenario depicted by Alsos et al. (2009) for the species in the northern Apennines. Based on modelling climatically suitable areas, these authors hypothesized a widespread distribution of S. herbacea across the Last Glacial Maximum (~21 000 BP) followed by a demographic contraction and a migration towards mountain summits in the Holocene. The northern Apennines were, in fact, characterized by an expansion of arboreal species from 10 400 BP (Watson, 1996) and a concurrent strong warming phase from 11 000 to 5000 BP (Samartin et al., 2017). Our data indicate the occurrence of a bottleneck that reduced the effective population size of S. herbacea by ~80 % during the Holocene in this area.
Genetic variation can be highly and heterogeneously structured even at the microgeographical scale in trees, as shown by recent findings based on neutral and potentially adaptive markers (reviewed by Scotti et al., 2016). The closer a genetic survey is to a whole-stand genetic characterization, the higher the chance for such complexity to come to light (Aleksić et al., 2017) and, in turn, to successfully show possible differences in SGS between contrasting populations. Indeed, our intensive sampling of northern Apennine populations provides a comprehensive picture of the modification on the fine-scale SGS of S. herbacea induced by peripherality, confirming that the prevalence of clonal reproduction alters the spatial distribution of genotypes in plants with mixed reproductive capabilities (Binks et al., 2015; Raabová et al., 2015; Jankowska-Wroblewska et al., 2016). A recent review has shown the importance of clonality as a driver of spatial structuring of genetic variation, with clonal tree species characterized by a spatial clustering of genotypes at the genet level up to approximately three times higher than non-clonal species (Dering et al., 2015). A formal assessment of SGS in Cimone is of little relevance because of its almost monoclonal structure, but spatial structuring in Prado is indeed much higher than that in Gavia. Although such a comparison should be viewed with caution due to different sampling strategies employed in Prado and Gavia, it is worth noting that the Sp values estimated for Prado are high even compared with what is generally found in wind-dispersed species (Vekemans and Hardy, 2004). Moreover, a significant SGS up to 50 m is much larger than that which had previously been found in S. herbacea core populations (i.e. 7 m; Stamati et al., 2007). Among the demographic factors and life history traits that can produce patches of genetically related individuals within natural plant populations, limited seed and pollen dispersal and small effective density are particularly relevant (Vekemans and Hardy, 2004; Sagnard et al., 2011). Parentage analysis shows that, at Prado, it is unlikely that seed and pollen dispersal have been spatially limited. On the other hand, the presence of large clones has the effect of lowering the effective population density (van Loo et al., 2007) and this might contribute to an increase in the intensity of SGS. As a result, the presence of extremely large clones at Prado is the most likely cause of the extent of the spatial clustering of genotypes detected.
Spatial statistics inferred from the relationship between pairwise genetic similarity and spatial distance (i.e. Sp, Fij) show a strong spatial impact of prevailing clonal reproduction in peripheral populations, but the main differences in SGS among the three populations emerge when the spatial distribution and extent of MLGs are considered. From previous research on core populations, it emerged that the mean size of clones varies between 0.70 and 0.96 m2 (Reisch et al., 2007; Stamati et al., 2007). A highly heterogeneous spatial distribution of clone size, with MLGs ranging from a few square metres to ~2000 m2, can be found in both Apennine populations. A similar heterogeneity in the spatial distribution of different-sized clones was recorded in Populus tremula populations as a consequence of the prevalence of vegetative propagation over sexual reproduction in rear-edge populations (Cristóbal et al., 2014). In P. alba, Macaya-Sanz et al. (2016) found large clones with peculiar genomic characteristics that may have promoted their spread under stressful environmental conditions. The conclusion that isolation exacerbates the effect of clonality, increasing the strength and complexity of SGS, is corroborated by the study of two rare species of sedges with different levels of geographical isolation (Binks et al., 2015). Besides confirming that clonality is one of the life-history traits that can have a profound impact on spatial processes in plants with mixed reproductive capabilities (Dering et al., 2015), our results show how far the complexity of fine-scale SGS can be increased by prolonged conditions of peripherality (i.e. a highly skewed distribution of genet sizes with a few dominant and probably more resilient individuals covering most of the population area).
An interesting aspect that has emerged from the spatial distribution of MLGs in Prado is that the western part of the population is dominated by three large clones (mlg044, 72 × 53 m; mlg015: 60 × 42 m; mlg147: 56 × 40 m), whereas the eastern part is mainly composed of unique or small MLGs. The particular spatial distribution of clones in Prado leads to an interesting hypothesis: whether the three large clones are the parents of the remaining 105 MLGs. To test this hypothesis, we ran a parentage analysis to check how many of the remaining individuals are compatible with being the offspring of parent pairs comprising mlg044 (male), mlg015 and mlg147 (females). Based on genetic compatibility, only two out of 105 MLGs were likely to have originated from these three large clones and, in general, there was no correlation between reproductive success and MLG size. This rules out a possible recent origin from a limited number of individuals of the eastern part of the population, as confirmed by the current effective population size estimated by demographic inference, which is similar to the number of MLGs detected. These results indicate that reproductive dynamics are not extremely simplified in this isolated population. Contrary to what has been found in other clonal tree species (Macaya-Sanz et al., 2016), large clones do not dominate sexual reproduction in isolated S. herbacea populations and therefore the causes of heterogeneous mating patterns are not dependent on the potential reproductive output of single individuals. Considering that the large dissemination and pollination distances detected would seem to exclude possible dispersal limitations, our results suggest that micro-environmental variation might have an important role in determining the spatial distribution of MLGs, as already demonstrated for S. herbacea by Cortés et al. (2014).
To conclude, our comprehensive survey at the whole-population level has helped to provide a quantitative assessment of the effects of and the demographic dynamics induced by extreme isolation in the only two remnant populations of S. herbacea in the northern Apennines. Clonal and genetic diversity has been shown to be extremely impoverished and the sex ratio highly distorted in the smallest population on Mt Cimone. Conversely, it seems that the larger population occurring on Mt Prado has at least buffered the major impact of a marked reduction in its effective population size during the Holocene. These results contribute to the knowledge concerning the reproductive and genetic characteristics of cold-adapted species at the rear (warm) edge of their distribution, and are of maximum importance in addressing conservation efforts for highly endangered marginal populations of Arctic-alpine species (Abeli et al., 2018). The occurrence of large-sized genets suggests that S. herbacea has very long-lived individuals, which have persisted in the northern Apennines for a long time (centuries or even millennia), experiencing climatic oscillations as well as land-use changes. However, while clonal growth could be essential in postponing the extinction of these populations, the extremely low level of sexual reproduction exhibited by the smallest population will likely prevent the opportunity for both local adaptation and further resilience in the long run. The topography of the northern Apennines (low-elevation summits and few, small areas with long-lasting snow cover) substantially impedes the migration of alpine tundra species to more favourable locations (Petraglia and Tomaselli, 2007). Further studies are therefore needed to assess the levels of local adaptation of remnant populations of S. herbacea and to forecast their fate under global climate change scenarios. Indeed, the current and unprecedented warming trend calls into question whether these S. herbacea populations can keep up with fast and dramatic environmental changes, including prolonged snow-free periods, a higher frequency of summer heatwaves and increasing competition with taller sub-alpine shrubs.
SUPPLEMENTARY DATA
Supplementary data are available online at https://academic.oup.com/aob and consist of the following. Figure S1: schematic representation of the approach used for sampling at Mt Prado. Figure S2: results of the Monte Carlo procedure to assess the power of the marker set to distinguish all detectable multilocus genotypes within sampled ramets. Figure S3: MLG abundances in the three studied populations. Table S1: description of the 11 nuclear SSRs used in this study and composition of the two multiplexes. Table S2: genotypes at the 11 microsatellite loci of all individuals from the three populations.
ACKNOWLEDGEMENTS
We thank the Ente Parco Nazionale Appennino Tosco-Emiliano, Ente di gestione per i Parchi e la Biodiversità Emilia Centrale, and Parco Nazionale dello Stelvio for logistic support. Sincere thanks are due to F. Perucco, M. Petit Bon, N. Delnevo, C. Zanni, P. Bertolini Zaccara, G. Chiari, T. Vanini, E. Bianchi, S. Chiussi, E. Vajana and L. Inserviente for field-work support, and to F. Degola, M. Labriola and C. Boggi for technical assistance during laboratory work. We are also grateful to three anonymous reviewers for their useful comments on the manuscript and to T. G. W. Forte for revising the English text.
FUNDING
This work was supported by the Ente Parco Nazionale Appennino Tosco-Emiliano project ex cap. 1551–2014 ‘Conservazione di Salix herbacea L. nel Parco Nazionale dell’Appennino Tosco-Emiliano’ and the COST Action FP1202 ‘Strengthening conservation: a key issue for adaptation of marginal/peripheral populations of forest trees to climate change in Europe (MaP-FGR)’.
LITERATURE CITED
- Abeli T, Vamosi JC, Orsenigo S. 2018. The importance of marginal population hotspots of cold-adapted species for research on climate change and conservation. Journal of Biogeography 45: 977–985. [Google Scholar]
- Aleksić JM, Piotti A, Geburek T, Vendramin GG. 2017. Exploring and conserving a “microcosm”: whole-population genetic characterization within a refugial area of the endemic, relict conifer Picea omorika. Conservation Genetics 18: 777–788. [Google Scholar]
- Alsos IG, Alm T, Normand S, Brochmann C. 2009. Past and future range shifts and loss of diversity in dwarf willow (Salix herbacea L.) inferred from genetics, fossils and modelling. Global Ecology and Biogeography 18: 223–239. [Google Scholar]
- Alvarez N, Manel S, Schmitt T, the IntraBioDiv Consortium. 2012. Contrasting diffusion of Quaternary gene pools across Europe: the case of the arctic–alpine Gentiana nivalis L. (Gentianaceae). Flora 207: 408–413. [Google Scholar]
- Ansell SW, Grunmann M, Russel SJ, Schneider H, Vogel JC. 2008. Genetic discontinuity, breeding-system change and population history of Arabis alpina in the Italian Peninsula and adjacent Alps. Molecular Ecology 17: 2245–2257. [DOI] [PubMed] [Google Scholar]
- Arnaud-Haond S, Duarte CM, Alberto F, Serrão EA. 2007. Standardizing methods to address clonality in population studies. Molecular Ecology 16: 5115–5139. [DOI] [PubMed] [Google Scholar]
- Bailleul D, Stoeckel S, Arnaud-Haond S. 2016. RClone: a package to identify multilocus clonal lineages and handle clonal data sets in R. Methods in Ecology and Evolution 7: 966–970. [Google Scholar]
- Balloux F, Lehmann L, de Meeûse T. 2003. The population genetics of clonal and partially clonal diploids. Genetics 164: 1635–1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker CL, Donald T, Pauquet J, et al. . 2005. Genetic and physical mapping of the grapevine powdery mildew resistance gene, Run1, using a bacterial artificial chromosome library. Theoretical and Applied Genetics 111: 370–377. [DOI] [PubMed] [Google Scholar]
- Beatty GE, McEvoy PM, Sweeney O, Provan J. 2008. Range-edge effects promote clonal growth in peripheral populations of the one-sided wintergreen Orthilia secunda. Diversity and Distributions 14: 546–555. [Google Scholar]
- Beerling DJ. 1998. Biological flora of the British Isles: Salix herbacea L. Journal of Ecology 86: 872–895. [Google Scholar]
- Berlin S, Ghelardini L, Bonosi L, Weih M, Rönnberb-Wästljung AC. 2014. QTL mapping of biomass and nitrogen economy traits in willows (Salix spp.) grown under contrasting water and nutrient conditions. Molecular Breeding 34: 1987–2003. [Google Scholar]
- Billings WD, Bliss LC. 1959. An alpine snowbank environment and its effects on vegetation, plant development, and productivity. Ecology 40: 388–397. [Google Scholar]
- Binks RM, Millar MA, Byrne M. 2015. Contrasting patterns of clonality and fine-scale genetic structure in two rare sedges with differing geographic distributions. Heredity 115: 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birks HJB, Willis KJ. 2008. Alpines, trees and refugia in Europe. Plant Ecology & Diversity 1: 147–160. [Google Scholar]
- Björk RG, Molau U. 2007. Ecology of alpine snowbeds and the impact of global change. Arctic, Antarctic and Alpine Research 39: 34–43. [Google Scholar]
- Bossuyt B, Honnay O. 2006. Interactions between plant life span, seed dispersal capacity and fecundity determine metapopulation viability in a dynamic landscape. Landscape Ecology 21: 1195–1205. [Google Scholar]
- Carbognani M, Petraglia A, Tomaselli M. 2014a Warming effects and plant trait control on the early-decomposition in alpine snowbeds. Plant and Soil 376: 277–290. [Google Scholar]
- Carbognani M, Tomaselli M, Petraglia A. 2014b Current vegetation changes in an alpine late snowbed community in the south-eastern Alps (N-Italy). Alpine Botany 124: 105–113. [Google Scholar]
- Carbognani M, Bernareggi G, Perucco F, Tomaselli M, Petraglia A. 2016. Micro-climatic controls and warming effects on flowering time in alpine snowbeds. Oecologia 182: 573–585. [DOI] [PubMed] [Google Scholar]
- Cortés AJ, Waeber S, Lexer C, et al. . 2014. Small-scale patterns in snowmelt timing affect gene flow and the distribution of genetic diversity in the alpine dwarf shrub Salix herbacea. Heredity 113: 233–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristóbal D, Martínez-Zurimendi P, Villamediana I, et al. . 2014. Clonal structure and dynamics of peripheral Populus tremula L. populations. iForest 7: 140–149. [Google Scholar]
- Crawford RMM, Balfour J. 1983. Female predominant sex ratios and physiological differentiation in arctic willows. Journal of Ecology 71: 149–160. [Google Scholar]
- Davis MB, Shaw RG. 2001. Range shifts and adaptive responses to Quaternary climate change. Science 292: 673–679. [DOI] [PubMed] [Google Scholar]
- Dering M, Chybicki IJ, Rączka G. 2015. Clonality as a driver of spatial genetic structure in populations of clonal tree species. Journal of Plant Research 128: 731–745. [DOI] [PubMed] [Google Scholar]
- Dorken ME, Eckert CG. 2001. Severely reduced sexual reproduction in northern populations of a clonal plant, Decodon verticillatus (Lythraceae). Journal of Ecology 89: 339–350. [Google Scholar]
- Eckert CG. 2001. The loss of sex in clonal plants. Evolutionary Ecology 15: 501–520. [Google Scholar]
- Eckert CG, Samis KE, Lougheed SC. 2008. Genetic variation across species’ geographical ranges: the central-marginal hypothesis and beyond. Molecular Ecology 17: 1170–1188. [DOI] [PubMed] [Google Scholar]
- Ellstrand NC, Roose M. 1987. Patterns of genotypic diversity in clonal plant species. American Journal of Botany 74: 123–131. [Google Scholar]
- Eriksson O. 1996. Regional dynamics of plants: a review of evidence for remnant, source-sink and metapopulations. Oikos 77: 248–258. [Google Scholar]
- Foggi B. 1990. Analisi fitogeografica del distretto appenninico tosco-emiliano. Webbia 44: 169–196. [Google Scholar]
- Fox JL. 1991. Forage quality of Carex macrochaeta emerging from Alaskan alpine snowbanks through the summer. American Midland Naturalist 126: 287–293. [Google Scholar]
- Gentili R, Bacchetta G, Fenu G, et al. . 2015. a From cold to warm-stage refugia for boreo-alpine plants in southern European and Mediterranean mountains: the last chance to survive or an opportunity for speciation? Biodiversity 16: 247–261. [Google Scholar]
- Gentili R, Baroni C, Caccianiga M, Armiraglio S, Ghiani A, Citterio S. 2015b Potential warm-stage microrefugia for alpine plants: feedback between geomorphological and biological processes. Ecological Complexity 21: 87–99. [Google Scholar]
- Gerber S, Chabrier P, Kremer A. 2003. FAMOZ: a software for parentage analysis using dominant, codominant and uniparentally inherited markers. Molecular Ecology Notes 3: 479–481. [Google Scholar]
- Hampe A, Petit RJ. 2005. Conserving biodiversity under climate change: the rear edge matters. Ecology Letters 8: 461–467. [DOI] [PubMed] [Google Scholar]
- Hewitt GM. 2000. The genetic legacy of the ice ages. Nature 405: 907–913. [DOI] [PubMed] [Google Scholar]
- Hewitt GM. 2004. Genetic consequences of climatic changes in the Quaternary. Philosophical Transactions of the Royal Society B 359: 183–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honnay O, Bossuyt B. 2005. Prolonged clonal growth: escape route or route to extinction? Oikos 108: 427–432. [Google Scholar]
- Hultén E, Fries M. 1986. Atlas of North European vascular plants. North of the Tropic of Cancer. Köningstein: Koeltz Scientific Books. [Google Scholar]
- Jalas J, Suominen J. 1976. Atlas Florae Europaea 3. Salicaceae to Balanophoraceae. Helsinki: The Committee for Mapping the Flora of Europe & Societas Biologica Fennica Vanamo. [Google Scholar]
- Jankowska-Wroblewska S, Meyza K, Sztupecka E, Kubera L, Burczyk J. 2016. Clonal structure and high genetic diversity at peripheral populations of Sorbus torminalis (L.) Crantz. iForest 9: 892–900. [Google Scholar]
- Jump AS, Peñuelas J. 2005. Running to stand still: adaptation and the response of plants to rapid climate change. Ecology Letters 8: 1010–1020. [DOI] [PubMed] [Google Scholar]
- Klimeš L, Klimesová J, Hendriks R, van Groenendael JM. 1997. Clonal plant architecture: a comparative analysis of form and function. In: de Kroon H, van Groenendael J, eds. The ecology and evolution of clonal plants. Leiden: Backhuys Publishers, 1–29. [Google Scholar]
- Kramer AT, Ison JL, Ashley MV, Howe HF. 2008. The paradox of forest fragmentation genetics. Conservation Biology 22: 878–885. [DOI] [PubMed] [Google Scholar]
- Kropf M, Comes HP, Kadereit JW. 2006. Long-distance dispersal vs vicariance: the origin and genetic diversity of alpine plants in the Spanish Sierra Nevada. New Phytologist 172: 169–184. [DOI] [PubMed] [Google Scholar]
- Kropf M, Comes HP, Kadereit JW. 2008. Causes of the genetic architecture of south-west European high mountain disjuncts. Plant Ecology & Diversity 1: 217–228. [Google Scholar]
- Leblois R, Pudlo P, Néron J, et al. . 2014. Maximum-likelihood inference of population size contractions from microsatellite data. Molecular Biology and Evolution 31: 2805–2823. [DOI] [PubMed] [Google Scholar]
- Leimu R, Mutikainen P, Koricheva J, Fischer M. 2006. How general are positive relationships between plant population size, fitness and genetic variation? Journal of Ecology 94: 942–952. [Google Scholar]
- Lindborg R. 2007. Evaluating the distribution of plant life-history traits in relation to current and historical landscape configurations. Journal of Ecology 95: 555–564. [Google Scholar]
- Loiselle BA, Sork VL, Nason J, Graham C. 1995. Spatial genetic structure of a tropical understory shrub, Psychotria officinalis (Rubiaceae). American Journal of Botany 82: 1420–1425. [Google Scholar]
- van Loo M, Joseph JA, Heinze B, Fay MF, Lexer C. 2007. Clonality and spatial genetic structure in Populus × canescens and its sympatric backcross parent P. alba in a Central European hybrid zone. New Phytologist 177: 506–516. [DOI] [PubMed] [Google Scholar]
- Macaya-Sanz D, Heuertz M, Lindtke D, Vendramin GG, Lexer C, Gonzalez-Martinez SC. 2016. Causes and consequences of large clonal assemblies in a poplar hybrid zone. Molecular Ecology 25: 5330–5344. [DOI] [PubMed] [Google Scholar]
- Marini L, Bruun HH, Heikkinen RK, et al. . 2012. Traits related to species persistence and dispersal explain changes in plant communities subjected to habitat loss. Diversity and Distribution 18: 898–908. [Google Scholar]
- Matteodo M, Ammann K, Verecchia EP, Vittoz P. 2016. Snowbeds are more affected than other subalpine-alpine plant communities by climate change in the Swiss Alps. Ecology and Evolution 6: 6969–6982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maynard Smith J. 1978. The evolution of sex. Cambridge: Cambridge University Press. [Google Scholar]
- Peakall R, Smouse PE. 2012. GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research – an update. Bioinformatics 28: 2537–2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peck JR, Yearsley JM, Waxman D. 1998. Explaining the geographic distributions of sexual and asexual populations. Nature 391: 889–892. [Google Scholar]
- Perderau AC, Kelleher CT, Douglas GC, Hodkinson TR. 2014. High levels of gene flow and genetic diversity in Irish populations of Salix caprea L. inferred from chloroplast and nuclear SSR markers. BMC Plant Biology 14: 202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petraglia A, Tomaselli M. 2007. Phytosociological study of the snowbed vegetation in the Northern Apennines (Northern Italy). Phytocoenologia 37: 67–98. [Google Scholar]
- R Core Team. 2017. R: a language and environment for statistical computing. Vienna: R Foundation for Statistical Computing. [Google Scholar]
- Rabasa SG, Granda E, Benavides R, et al. . 2013. Disparity in elevational shifts of European trees in response to recent climate warming. Global Change Biology 19: 2490–2499. [DOI] [PubMed] [Google Scholar]
- Raabová J, Van Rossum F, Jacquemart A-L, Raspé O. 2015. Population size affects genetic diversity and fine-scale spatial genetic structure in the clonal distylous herb Menyanthes trifoliata. Perspectives in Plant Ecology, Evolution and Systematics 17: 193–200. [Google Scholar]
- Rechinger KH, Akeroyd JR. 1993. Salix L. In: Tutin TG, Burges NA, Chater AO, et al. eds. Flora Europaea. Vol. 1. Psilotaceae to Platanaceae. Cambridge: Cambridge University Press, 53–64. [Google Scholar]
- Rehm EM, Olivas P, Stroud J, Feeley KJ. 2015. Losing your edge: climate change and the conservation value of range-edge populations. Ecology and Evolution 5: 4315–4326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reisch C, Schurm S, Poschlod P. 2007. Spatial genetic structure and clonal diversity in an alpine population of Salix herbacea (Salicaceae). Annals of Botany 99: 647–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozenfeld AF, Arnaud-Haond S, Hernández-García E, et al. . 2007. Spectrum of genetic diversity and networks of clonal organisms. Journal of the Royal Society Interface 4: 1093–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saar L, Takkis K, Pärtel M, Helm A. 2012. Which plant traits predict species loss in calcareous grassland with extinction debt? Diversity and Distribution 18: 808–817. [Google Scholar]
- Sagnard F, Oddou-Muratorio S, Pichot C, Vendramin GG, Fady B. 2011. Effects of seed dispersal, adult tree and seedling density on the spatial genetic structure of regeneration at fine temporal and spatial scales. Tree Genetics & Genomes 7: 37–48. [Google Scholar]
- Samartin S, Heiri O, Joos F, et al. . 2017. Warm Mediterranean mid-Holocene summers inferred from fossil midge assemblages. Nature Geoscience 10: 207–212. [Google Scholar]
- Schönswetter P, Paun O, Tribsch A, Niklfeld H. 2003. Out of the Alps: colonization of the Arctic by East Alpine populations of the glacier buttercup Ranunculus glacialis L. (Ranunculaceae). Molecular Ecology 12: 3371–3381. [DOI] [PubMed] [Google Scholar]
- Schönswetter P, Stehlik I, Holderegger R, Tribsch A. 2005. Molecular evidence for glacial refugia of mountain plants in the European Alps. Molecular Ecology 14: 3547–3555. [DOI] [PubMed] [Google Scholar]
- Schönswetter P, Elven R, Brochmann C. 2008. Trans-Atlantic dispersal and large-scale lack of genetic structure in the circumpolar, arctic-alpine sedge Carex bigelowii s. l. (Cyperaceae). American Journal of Botany 95: 1006–1014. [DOI] [PubMed] [Google Scholar]
- Scotti I, Gonzalez-Martinez SC, Budde KB, Lalagüe H. 2016. Fifty years of genetic studies: what to make of large amounts of variation found within populations? Annals of Forest Science 73: 69–75. [Google Scholar]
- Shimono Y, Watanabe M, Hirao AS, Wada N, Kudo G. 2009. Morphological and genetic variations of Potentilla matsumurae (Rosaceae) between fellfield and snowbed populations. American Journal of Botany 96: 728–737. [DOI] [PubMed] [Google Scholar]
- Silvertown J. 2008. The evolutionary maintenance of sexual reproduction: evidence from the ecological distribution of asexual reproduction in clonal plants. International Journal of Plant Sciences 169: 157–168. [Google Scholar]
- Skrede I, Eidesen PB, Portela RP, Brochmann C. 2006. Refugia, differentiation and postglacial migration in arctic-alpine Eurasia, exemplified by the mountain avens (Dryas octopetala L.). Molecular Ecology 15: 827–1840. [DOI] [PubMed] [Google Scholar]
- Sochor M, Vašut RJ, Bártová E, Majeský Ľ, Mráček J. 2013. Can gene flow among populations counteract the habitat loss of extremely fragile biotopes? An example from the population genetic structure in Salix daphnoides. Tree Genetics & Genomes 9: 1193–1205. [Google Scholar]
- Stamati K, Blackie S, Brown WS, Russel J. 2003. A set of polymorphic SSR loci for subarctic willow (Salix lanata, S. lapponum and S. herbacea). Molecular Ecology Notes 3: 280–282. [Google Scholar]
- Stamati K, Hollingsworth PM, Russell J. 2007. Patterns of clonal diversity in three species of sub-arctic willow (Salix lanata, Salix lapponum and Salix herbacea). Plant Systematics and Evolution 269: 75–88. [Google Scholar]
- Steltzer H, Hufbauer RA, Welker JM, Casalis M, Sullivan PF, Chimner R. 2008. Frequent sexual reproduction and high intraspecific variation in Salix arctica: implications for a terrestrial feedback to climate change in the High Arctic. Journal of Geophysical Research 113: G03S10. [Google Scholar]
- Steven JC, Waller DM. 2007. Isolation affects reproductive success in low-density but not high-density populations of two wind-pollinated Thalictrum species. Plant Ecology 190: 131–141. [Google Scholar]
- Sugai K, Watanabe S, Kuishi T, et al. . 2016. Extremely low genetic diversity of the northern limit populations of Nypa fruticans (Arecaceae) on Iriomote Island, Japan. Conservation Genetics 17: 221–228. [Google Scholar]
- Theurillat JP, Guisan A. 2001. Potential impact of climate change on vegetation in the European Alps: a review. Climatic Change 50: 529–530. [Google Scholar]
- Tomaselli M. 1991. The snow-bed vegetation in the Northern Apennines. Vegetatio 94: 177–189. [Google Scholar]
- Tomaselli M, Agostini N. 1994. A comparative phytogeographic analysis of the summit flora of the Tuscan-Emilian Apennines and of the Apuan Alps (Northern Apennines). Fitosociologia 26: 99–109. [Google Scholar]
- Vallejo-Marín M, Dorken ME, Barrett SCH. 2010. The ecological and evolutionary consequences of clonality for plant mating. Annual Review of Ecology, Evolution, and Systematics 41: 193–213. [Google Scholar]
- Vekemans X, Hardy OJ. 2004. New insights from fine-scale spatial genetic structure analyses in plant populations. Molecular Ecology 13: 921–935. [DOI] [PubMed] [Google Scholar]
- Vucetich JA, Waite TA. 2003. Spatial patterns of demography and genetic processes across the species’ range: null hypotheses for landscape conservation genetics. Conservation Genetics 4: 639–645. [Google Scholar]
- Watson CS. 1996. The vegetational history of the northern Apennines, Italy: information from three new sequences and a review of regional vegetational change. Journal of Biogeography 23: 805–841. [Google Scholar]
- Wheeler JA, Cortés AJ, Sedlacek J, et al. . 2016. The snow and the willows: earlier spring snowmelt reduces performance in the low-lying alpine shrub Salix herbacea. Journal of Ecology 104: 1041–1050. [Google Scholar]
- de Witte LC, Armbruster GFJ, Gielly L, Taberlet P, Stöcklin J. 2012. AFLP markers reveal high clonal diversity and extreme longevity in four key arctic-alpine species. Molecular Ecology 21: 1081–1097. [DOI] [PubMed] [Google Scholar]
- Wright S. 1946. Isolation by distance under diverse systems of mating. Genetics 31: 39–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.