Abstract
Establishment and subsequent maintenance of distinct chromatin domains during embryonic stem cell (ESC) differentiation are crucial for lineage specification and cell fate determination. Here we show that the histone chaperone Chromatin Assembly Factor 1 (CAF-1), which is recruited to DNA replication forks through its interaction with proliferating cell nuclear antigen (PCNA) for nucleosome assembly, participates in the establishment of H3K27me3-mediated silencing during differentiation. Deletion of CAF-1 p150 subunit impairs the silencing of many genes including Oct4, Sox2 and Nanog as well as the establishment of H3K27me3 at these gene promoters during ESC differentiation. Mutations of PCNA residues involved in recruiting CAF-1 to the chromatin also result in defects in differentiation in vitro and impair early embryonic development as p150 deletion. Together, these results reveal that the CAF-1-PCNA nucleosome assembly pathway plays an important role in the establishment of H3K27me3-mediated silencing during cell fate determination.
INTRODUCTION
Generation of a variety of specialized cell types from stem cells is fundamental for the development of multicellular organisms. Mouse embryonic stem cells (ESCs) possess self-renewal and pluripotency and are, therefore, a suitable model system to explore the molecular mechanisms governing cell identity transitions during development (1–4). The three core transcription factors (TFs) Oct4, Sox2 and Nanog are critical for the establishment and maintenance of the ESC pluripotent state (5,6). These three TFs form interconnected auto-regulatory loop to promote their own expression as well as the expression of genes critical for ESC stability (5). In addition, they also repress the expression of lineage-specific regulators to prevent differentiation (7–9). In response to differentiation stimuli or withdrawal of culture conditions favoring self-renewal, ESCs execute a differentiation program and generate cell types of all three germ layers, mimicking early embryonic development. During this process, the ESC-specific TF network collapses, which leads to silencing of many genes including pluripotency genes. Concurrently, commitment to a particular lineage is sealed by the induction of genes specific for that lineage. It is relatively underexplored how the ESC-specific TF network is turned off during pluripotency exit.
In addition to TFs, chromatin organization and modifications are also important for maintaining the pluripotent state and regulating lineage specification (10). Chromatin is a highly organized complex of DNA, RNA and proteins. The basic repeating unit of chromatin is the nucleosome, which consists of 147 bp of DNA wrapped around a histone octamer containing a H3-H4 tetramer and two H2A-H2B dimers (11). These histone proteins carry post-translationally modifications including acetylation and methylation, which are important in regulating gene expression in response to environmental and developmental stimuli (10). For instance, tri-methylation of histone H3 lysine 4 (H3K4me3) is found at the promoters of actively transcribed genes (12), whereas tri-methylation of histone H3 lysine 27 (H3K27me3), catalyzed by the Polycomb Repressive Complex 2 (PRC2) (13) is enriched at the promoters of silent genes and plays an important role in repressing gene transcription during development. In ESCs, H3K4me3 is detected at the promoters of pluripotency genes such as Oct4, Sox2 and Nanog (14), whereas acetylated H3K27 (H3K27ac), a mark associated with active (open) chromatin, is found at the enhancers and promoters of these genes (15). During differentiation, the silencing of the pluripotency gene network is associated with a dramatic reduction of the levels of H3K4me3 at the promoters of Oct4, Sox2 and Nanog and a concomitant increase of H3K27me3 (14). However, it remains unclear how the H3K27me3-mediated silencing is established during the transition from active to silent chromatin during differentiation.
During the process of chromatin replication, nucleosomes are first disassembled to allow the DNA replication machinery to access the DNA. Following DNA replication, newly synthesized DNA is assembled into nucleosomes in a process that is tightly coupled to ongoing DNA replication (16). This DNA replication-coupled nucleosome assembly is regulated by histone chaperones including Chromatin Assembly Factor 1 (CAF-1), the classical chaperone for histones H3-H4 (16). CAF-1 consists of three subunits, p150, p60 and p48 (17,18). CAF-1 is recruited to DNA replication forks through its interaction with proliferating cell nuclear antigen (PCNA), the ring-shaped homotrimer serving as the sliding clamp for DNA polymerases (19). CAF-1 interacts with PCNA mainly through the CAF-1 p150 subunit (19,20).
Experimental evidence accumulated thus far supports the idea that DNA replication-coupled nucleosome assembly plays an important role in the inheritance of chromatin states during mitotic cell divisions. For instance, mutations in CAF-1 and PCNA lead to defects in transcriptional silencing at heterochromatin in yeast (21–23). In mammalian cells, CAF-1 interacts with heterochromatin protein 1 (HP1) (24,25), and depletion of p150 and p60 subunits results in impairment of pericentromeric heterochromatin centers in mouse cells (24). In plants, CAF-1 interacts with PRC2, the H3K27 methyltransferase; and it has been proposed that the PRC2–CAF-1 interaction helps maintain the H3K27me3-mediated silent chromatin during DNA replication (26). Recently, it has been shown that depletion of CAF-1 components in murine ESCs promotes the emergence of totipotent-like cells that resemble two-cell stage (2C) blastomeres (27). Moreover, reduction of CAF-1 levels in somatic cells accelerates the reprogramming process (28), indicating the importance of CAF-1 in maintaining the identity of somatic cells. These studies highlight the potential role of CAF-1 and other factors involved in DNA replication-coupled nucleosome assembly in the maintenance of chromatin states and cell identity during mitotic cell divisions.
In addition to maintaining parental chromatin states, chromatin replication during the S phase in principle also offers a window-of-opportunity to facilitate changes in chromatin states. However, to what extent CAF-1 and PCNA contribute to such changes remains underexplored. Here, we investigate the function of CAF-1 and PCNA during in vitro differentiation of mouse ESCs. We show that the large subunit of CAF-1, p150, plays an important role in mouse ESC differentiation in vitro. We observed that deletion of p150 or disruption of the p150–PCNA interaction impairs mouse ESC differentiation and the silencing of many genes including pluripotency genes. Moreover, deletion of p150, while having little effect on the global levels of H3K27me3, compromises the gain of H3K27me3 at the promoters of this group of genes during differentiation. These results support a role for CAF-1-mediated DNA replication-coupled nucleosome assembly in the establishment of the facultative heterochromatin during ESC differentiation.
MATERIALS AND METHODS
Embryonic stem cell culture
Male TL1 ESCs were generously donated by Dr. Jan van Deursen at Mayo Clinic. ESCs were cultured under 5% CO2 at 37°C in regular Dulbecco's modified Eagle's medium supplemented with 15% fetal bovine serum (FBS), 1% glutamine (Cellgro), 1% Pen/Strep (Invitrogen), 1% sodium pyruvate(Cellgro), 1% of nonessential amino acids (Invitrogen), 0.1 mM 2-mercaptoethanol (Sigma) and 10 ng/ml leukemia inhibitory factor (LIF). Mouse embryonic fibroblasts inactivated by irradiation were used as feeder cells.
Generation of p150 deletion and site-specific PCNA mutations in murine ESCs using CRISPR/Cas9
To knock out the p150 gene, sgRNAs targeting the 5′ end of the p150 gene were designed according to the website http://www.genome-engineering.org/crispr/. Oligos were synthesized by Integrated DNA technologies, Inc. (IDT, Coralville, IA, USA) and inserted into pX459 vector according to published protocol (29). Plasmids were transfected into ESCs by transfection using Lipo-3000 (Invitrogen). After Puromycin (2 μg/ml) selection for 2 days, single-cell clones were picked and expanded for further characterization. Genetic mutations were confirmed by Surveyor nuclease assay and Sanger sequencing. Knockout of p150 was confirmed by western blot (WB) using two antibodies raised against different antigens.
To generate murine ESCs with PCNA point mutation, sgRNAs targeting PCNA sites were cloned into pX459 vector. Repair template single-stranded DNA oligonucleotides (ssODN) were synthesized by IDT. Targeting plasmid and ssODN were introduced into ESCs via nucleofection according to the manufacturer's protocol (Lonza, Basel, Switzerland). After Puromycin selection, single-cell clones were picked and screened for PCNA point mutation by restriction enzyme digestion and validated by Sanger sequencing.
Generation of PCNA mutation mouse
All animal experimental protocols were approved by the Columbia University's Institutional Animal Care and Use Committee (IACUC) and are in compliance with national and institutional guidelines. To generate PCNA mutation allele in ESCs, we introduced PCNA point mutation though homologous recombination via CRISPR/Cas9 system as described above. Correctly targeted ES clones were confirmed with Sanger sequencing. Correctly targeted clones were injected to blastocyst to generate chimeras and subsequently establish founder knock-in mice.
Embryoid body assay
To perform embryoid body (EB) in vitro differentiation assay, ESCs were disaggregated with TrypLE (Invitrogen) and EBs were formed using the hanging drop method with 300 cells per each 30 μl drop in ESC culture medium without LIF (30). After 3 days in hanging drops, EBs were collected from the lids and cultured in 10-cm low-attachment Petri dish in ESC culture medium without LIF. Samples were collected at the indicated time points for analysis.
H&E staining of EB
EBs were fixed with 4% of PFA at 4°C for 3 h and washed with 1× phosphate buffered saline (PBS) twice. EBs then is mixed with Matrigel (Corning). After solidification of the Matrigel/EB mixture, mixture is dehydrated with 95% ethanol (30 min), 100% ethanol (30 min, three times), 100% ethanol (overnight), histo-clear (30 min twice, National diagnostics) and embed in paraffin. 5 μm sections were prepared for H&E staining. Sample slides was incubated with histo-clear (15 min), second histo-clear (15 min), 100% ethanol (2 min), second 100% ethanol (2 min), 95% ethanol (2 min), 70% ethanol (2 min), distilled water (2 min), Mayer's hematoxylin (1 min), ammonium hydroxide (30 s; 1:200 in distilled water, Fisher scientific), distilled water (1 min; four times), 95% ethanol (2 min; twice), eosin (2 min), 95% ethanol (1 min), second 95% ethanol (1 min), 100% ethanol (2 min), second 100% ethanol (2 min), histo-clear (5 min), second histo-clear (5 min). The slides were covered by Fisherbrand Supreslip cover glass with Permount and dried overnight.
Teratoma assay
8 × 105 ESCs were resuspended in 200 μl of chilled 1:1 mixture of ESC culture medium without Lif and Matrigel Growth Factor Reduced Basement Membrane Matrix (BD Biosciences). Cells were then injected subcutaneously into both dorsal flank of NOD.CB17-Prkdcscid/Jimmunodeficient mice. Ten mice were used for each cell line. After one month, teratomas were excised and weighed. A piece of specimen (<50 mg) was cut from each teratoma and grinded in chilled TRI Reagent (Sigma) for RNA purification. The residual part of each teratoma was fixed in 4% paraformaldehyde overnight at 4°C. Fixed teratomas were then paraffin-embedded and sections were used to do Hematoxylin & Eosin (H&E) staining according to standard procedures.
Acid extraction of histone
5 × 106 cells are collected and washed with 1× PBS once. After washing with 1 ml hypotonic lysis buffer (10 mM Tris–HCl, pH8.0, 1 mM KCl, 1.5 mM MgCl2, 1 mM dithiothreitol (DTT) and protease inhibitor), pellet is incubated with 1ml hypotonic buffer rocking at 4°C for 30 min. After washing three times with hypotonic buffer, nuclei pellet is resuspended in 400 μl 0.2N HCl and rock at 4°C for 30 min. After spin down, 100 μl TCA is added to the supernatant and incubate on ice for 30 min. Histone pellet is washed by cold acetone twice. After air dry, histone pellet is dissolved by boiling in 1× sodium dodecyl sulphate (SDS) sampling buffer.
Chromatin fraction of ESCs
1 × 106 ESCs were collected and washed with PBS. ESCs were resuspended with 200 μl CSK buffer (10 mM PIPES-KOH pH7.0, 100 mM NaCl, 300 mM sucrose, 3 mM MgCl2, 0.5% Triton X-100, 0.5 mM PMSF and proteinase inhibitor) and incubated on ice for 5 min. A total of 100 μl of the cell suspension was removed as whole cell lysates (WCL). The rest samples were centrifuged (1300 g, 5 min, 4°C) and the supernatant was collected as the cytoplasmic fraction. The pellet was washed with CSK buffer twice and used as the chromatin fraction.
iPOND-SILAC-MS
iPOND was performed as described previously (31). Briefly, ESCs were cultured in light isotope growth media (p150 WT) and heavy isotope growth media (p150 KO) for 10 generations. Incorporation percentage was confirmed to be > 97% for each of these two isotopically labeled amino acid with LC/MS analysis. 40 × 107 ES cells for each cell line were pulsed with 10 μM EdU for 10 min at 37°C incubator. After pulse, cells were immediately fixed by 1% formaldehyde in PBS for 10 min at RT and quenched with 125 mM glycine for 5 min. Cells were collected by scrap from the plates and washed three times with 1× PBS. Cells were resuspended with permeabilization buffer (0.25% of Triton X-100 in PBS; 1ml/1 × 107 cells) and incubated at RT for 30 min. After permeabilization, cells were washed with cold 0.5% (wt/vol) bovine serum albumin (BSA) in PBS and once with PBS. The light and heavy labeled cells were mixed 1:1 before Click reaction. The mixed cells were suspended with Click reaction cocktails (biotin-azide-PEG4 10 μM, sodium ascorbate 10 mM and CuSO4 2 mM in PBS; 5 ml/1 × 108 cells) and rotate at RT for 2 h. After Click reaction, cells were washed with cold 0.5% (wt/vol) BSA in PBS and once with PBS. Cells were suspended with lysis buffer (1% SDS in 50 mM Tris pH 8.0 with 1 μg/ml of aprotinin and leupeptin; 100 μl/1.5 × 107cells) and sonicated for 15 cycles (30 s on and 30 s off) in Bioruptor (Diagenode, Inc., Danville, NJ, USA). After sonication, samples were collected by centrifuge and supernatant was filtered through 40 μm cell strainer. Lysate was mixed 1:1(vol/vol) with cold PBS containing 1 μg/ml of aprotinin and leupeptin. Lysate was incubated with streptavidin Myone C1 beads (Thermofisher; 100μl/10 × 107 cells) and rotate at RT for 1 h. Beads were washed with lysis buffer (1% SDS in 50 mM Tris pH 8.0), low salt buffer (1% Triton-X100, 20 mM Tris pH8.0, 2 mM ethylenediaminetetraacetic acid (EDTA), 150 mM NaCl), high salt buffer (1% Triton-X100, 20 mM Tris pH8.0, 2 mM EDTA, 500 mM NaCl), lithium chloride washing buffer (100 mM Tris pH8.0, 500 mM LiCl, 1% Igepal-CA630) and twice with lysis buffer. The beads were resuspended with 25 μl of 2× sampling buffer and incubate at 95°C for 30 min to revers cross-link. The supernatant was collected by spin down and the eluted protein were analyzed by immunoblotting and Mass spectrometry (Harvard Medical School Taplin Mass Spectrometry Facility).
Micrococcal nuclease (MNase) digestion assay
1 × 106 ES cells were collected and cross-linked with 1% formaldehyde for 10 min at RT and quenched with 125 mM glycine for 5 min. Cells were collected and washed once with cold Tris-buffered saline (TBS). Fixed cells were re-suspended in lysis buffer (10 mM Tris–HCl, pH7.5, 10 mM NaCl, 0.5% IGEPAL-CA630) and incubated on ice for 10 min. After centrifugation, the cell pellet was washed once with MNase digestion buffer (20 mM Tris–HCl, pH7.5, 15 mM NaCl, 60 mM KCl, 1 mM CaCl2). After re-suspended in 250 μl MNase digestion buffer with proteinase inhibitors, 500, 250, 125, 62.5, 31, 16, 0 units per 1 × 106 cells MNase (M0247S; New England Biolabs, Ipswitch, MA, USA) was added to same aliquoted sample. After incubation at 37°C for 20 min, 250 μl of STOP buffer (100 mM Tris–HCl, pH8.1, 20 mM EDTA, 200 mM NaCl, 2% Triton X-100, 0.2% sodium deoxycholate) was added. Lysates were sonicated for 15 cycles (30 s on and 30 s off) in Bioruptor (Diagenode, Inc., Danville, NJ, USA) and supernatant was collected after centrifugation. The supernatant was reverse-crosslinked at 65°C overnight and DNA was purified by ethanol precipitation and analyzed by agarose gel electrophoresis.
RT-PCR analysis
Total RNA was isolated from 1 × 106 cells using RNeasy Plus kit (Qiagen). A total of 1 μg of RNA was used for reverse transcription with random hexamers (Invitrogen). Quantitative polymerase chain reaction (PCR) was performed in triplicates for each sample with SYBR Green PCR Master Mix on CFX96 platform (Bio-Rad Laboratories). Primers used are listed in Supplementary Table S1.
Immunofluorescence
ESCs were fixed in 4% of formaldehyde for 10 min at room temperature (RT), then washed three times with PBS and blocked for 1 h with 5% normal goat serum (NGS) in PBS containing 0.1% Triton X-100 (PBST) at RT. Primary antibody was diluted in PBST containing 1% NGS and incubated with cells overnight at 4°C. After incubation, cells were washed once with PBST and incubated with fluorophore-labeled secondary antibodies for 1 h at RT. DNA/nuclei were counterstained with DAPI. Immunofluorescence images were recorded by Olympus FV1000 confocal system. The primary antibodies used for immunofluorescence were anti-Oct4 (1:100, sc-5279; Santa Cruz Biotechnology, Inc., Dallas, TX, USA) and anti-MERVL-Gag (1:100, A-2801, Epigentek, Farmingdale, NY, USA).
Cell-cycle analysis
ESCs cultured in 6-well plate were pulsed with 1 μM EdU for 15 min at 37°C. Flow cytometry assay was carried out using Click-iT EdU Flow Cytometry Assay kits (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions. Briefly, after EdU labeling, ESCs were collected and fixed with Click-iT fixative for 15min at room temperature. After fixation, ESCs were washed with 1% BSA in PBS and Click-iT wash buffer. Click-iT reaction was performed at room temperature for 30 min. Cells were then stained with propidium iodide (PI) and analyzed by an LSR II flow cytometer (Becton Dickinson, Franklin Lakes, NJ, USA).
Cell proliferation assay
7 × 104 ESCs were seeded on immortalized feeder cells. ESCs were collected and passaged every 3 days for 12 days. Cell numbers were counted by Bio-Rad TC10 automated cell counter. The number of feeder cells was subtracted before calculation.
Expression of p150 and EGFP in ESC
For exogenous expression in p150 KO cells, full-length p150 cDNA and domain-truncated mutations were cloned into the lentivirus-based vector pWPXL (Addgene) with one EGFP tag at the C-terminus. To generate pluripotency EGFP reporter ESC line, the lentivirus-based EGFP reporter vector PL-SIN-EOS-C(3+)-EGFP plasmid (Addgene) was used. Lentivirus expressing p150 or EGFP infected ESC. Expected cell line is selected by single cell clone and characterized by exogenous protein expression.
Immunoprecipitation
To immunoprecipitate GFP-p150, 1 × 107 cells were collected and suspended in 1 ml of lysis buffer (50 mM HEPES-KOH, pH7.4, 150 mM NaCl, 1% NP40, 10% glycerol, 1 mM EDTA and proteinase inhibitor) and dounce-homogenized for 40 times (D8938-1SET; MilliporeSigma). DNase I (2 μg/ml) and EtBr (20 μg/ml) was added to lysates and rotated at 4°C for 30 min. After two consecutive centrifugations at 13 000 rpm for 15 min each time, soluble lysate was incubated with GBP beads at 4°C for 1 h. The beads were washed with washing buffer (50 mM HEPES-KOH, pH7.4, 100 mM NaCl, 0.01% NP40, 10% glycerol, 1 mM EDTA and 0.02 μm PMSF), and proteins were eluted using 1× SDS sampling buffer and analyzed by WB.
ChIP-qPCR analysis
1 × 107 ES cells were collected and cross-linked with 1% formaldehyde for 10 min at RT and quenched with 125 mM glycine for 5 min. Cells were collected and washed once with cold TBS. EBs were digested by 0.25% trypsin (Gibco, Gaithersburg, MD, USA) at 37°C before fixation using 1% formaldehyde for 10 min at RT. Fixed cells were re-suspended in lysis buffer (10 mM Tris–HCl, pH7.5, 10 mM NaCl, 0.5% IGEPAL-CA630) and incubated on ice for 10 min. After centrifugation, the cell pellet was washed once with MNase digestion buffer (20 mM Tris–HCl, pH7.5, 15 mM NaCl, 60 mM KCl, 1 mM CaCl2). After re-suspended in 250 μl MNase digestion buffer with proteinase inhibitors, 2000 units per 4 × 106 cells MNase (M0247S; New England Biolabs, Ipswitch, MA, USA) was added. After incubation at 37°C for 20 min, 250 μl of STOP buffer (100 mM Tris–HCl, pH8.1, 20 mM EDTA, 200 mM NaCl, 2% Triton X-100, 0.2% sodium deoxycholate) was added. Lysates were sonicated for 15 cycles (30 s on and 30 s off) in Bioruptor (Diagenode, Inc., Danville, NJ, USA) and supernatant was collected after centrifugation. The supernatant was incubated with 2 μg antibody at 4°C overnight on a rotator. A total of 30 μl Protein-G Mag Sepharose beads (GE Healthcare, Little Chalfont, UK) was added and incubated for additional 3 h at 4°C. Beads were washed with ChIP buffer (50 mM Tris–HCl, pH8.0, 10 mM EDTA, 100 mM NaCl, 1% Triton X-100, 0.1% sodium deoxycholate), high salt buffer (ChIP buffer with 500 mM NaCl), LiCl buffer (10 mM Tris–HCl, pH8.0, 0.25 M LiCl2, 0.5% NP-40, 0.5% sodium deoxycholate, 1 mM EDTA) and TE buffer. Chromatin on beads was eluted and reverse-crosslinked at 65°C overnight and was purified by PCR purification kit (Qiagen). DNA was analyzed by real-time PCR. Primers used are listed in Supplementary Table S1.
ChIP-seq
ChIP DNA was prepared the same way as for ChIP-qPCR. A total of 10 ng of ChIP and input DNA were processed for library preparations by following Ovation Ultralow DR Multiplex kit (NuGEN Technologies, San Carlos, CA, USA). The ChIP-seq library DNA was sequenced using an Illumina NextSeq 500 instrument.
ChIP-seq data analysis
Sequencing reads from H3K4me3 and H3K27me3 ChIP-seq were aligned to the mouse genome (mm9) using the Bowtie2 software (32). After removal of PCR duplication reads by SAMtools (33), genome-wide read coverage was calculated by BEDTools (34). In order to calculate the ChIP-seq read density at promoters, normalized read densities (RPKM, Reads Per Kilobase per Million mapped reads) were calculated across gene promoter regions. The promoters used in this study were defined as 2 kb upstream and 500 bp downstream of the transcription start site determined based on the UCSC gene annotation.
RNA-seq
Total RNA was extracted the same way as for RT-PCR. RNA-seq libraries were prepared and deep sequencing was performed by Novogene Corp., Inc. (San Diego, CA, USA). Two replicates for each sample were sequenced. Raw data were aligned to the mouse genome version mm9 and to gene annotations from RefSeq using TopHat v2.05. Cufflinks v2.0.2 was used to quantify FPKM values (35,36). Differential expression was determined by Cuffdiff at FDR < 0.01. Gene ontology analysis was conducted using GO TermFinder.
Statistical analysis
Unless otherwise specified, values are depicted as mean ± SEM. Parameters including statistical significance, P-value and statistical analysis methods are reported in the figure legends and Supplementary Figure legends. Statistical analysis was performed using the two-tailed Student's t-test, with the exception of Figure 6B-I, which were analyzed using the Wilcoxon test. In all cases, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 were considered statistically significant.
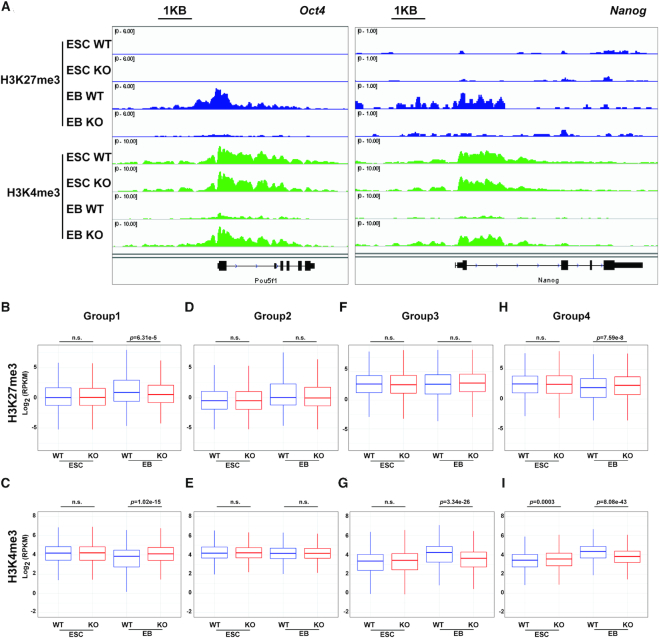
Figure 6.
Deletion of p150 impairs chromatin dynamics during ESC differentiation. (A) Representative browser tracks showing H3K27me3 and H3K4me3 at the Oct4 and Nanog loci before (ESC) and after (EB, day 7 EB) differentiation. (B–G) Relative levels of H3K27me3 and H3K4me3 between p150 WT and KO cells at the promoters of gene groups identified in Figure 2C. H3K27me3 for Group 1 (B), Group 2 (D), Group 3 (F) and Group 4 (H); H3K4me3 for Group 1 (C), Group 2 (E), Group 3 (G) and Group 4 (I). The y-axis represents the log2 ratio of ChIP-seq reads. The P-values were calculated using Wilcoxon test.
RESULTS
Mouse CAF-1 p150 is dispensable for ESC self-renewal and viability
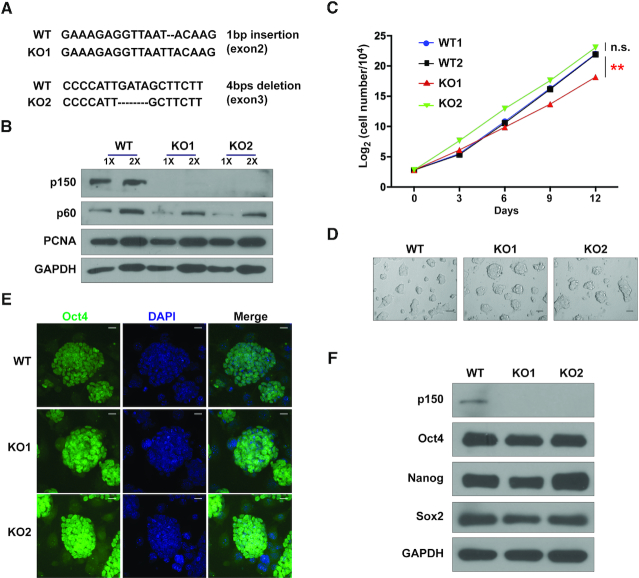
We deleted p150 (p150 KO), the large subunit of CAF-1 complex, in mouse ESCs using CRISPR (clustered regularly interspaced short palindromic repeats)/Cas9 (CRISPR-associated 9) (Supplementary Figure S1A and B). Clones with indels introduced by CRISPR/Cas9 were confirmed by Sanger sequencing (Figure 1A). Moreover, p150 proteins were not detectable by WB using two different primary antibodies raised from antigens at either the C-terminus or the N-terminus of p150 (Figure 1B and Supplementary Figure S1C). The p150 KO cells exhibited a slightly reduced proliferation rate (Figure 1C) and a similar cell-cycle profiles compared to p150 wild-type (WT) cells (Supplementary Figure S1D). Consistent with these results, p150 KO ES cells and day 7 embryonic bodies (EB) did not show much increase in DNA damage as detected by H2AX phosphorylation (Supplementary Figure S1E and F), which in contrast to human cancer cells with p150 depletion where a dramatic change in S phase progression and an increase in DNA damage were observed (37). Moreover, p150 KO cells had normal karyotypes (Supplementary Figure S1G and H). The p150 KO cells could form dome-shaped cell clones (Figure 1D) and were positive for alkaline phosphatase (AP) staining (Supplementary Figure S1I). At last, compared to WT cells, the p150 KO cells expressed similar levels of three pluripotency genes (Nanog, Oct4 and Sox2) (Figure 1E and F). Thus, the p150 KO ESCs are, surprisingly, largely normal compared to WT ESCs. However, consistent with previous reports (27,38), cells lacking p150 had fewer chromocenters and had more 2C-like cells (Supplementary Figure S2A–D). 2C-like cells expressed MERVL retroelement-derived Gag protein (MERVL-Gag), but did not express Oct4. However, the percentage of 2C-like cells in p150 KO cells was <5% (Supplementary Figure S2E). These results indicate that most of the p150 KO ES cells remain in the ESC state, and that p150 is likely not essential for the viability and stemness of mouse ESCs.
Figure 1.
Mouse CAF-1 p150 is dispensable for ESC self-renewal. (A) Sanger sequencing analysis of two different p150 KO clones generated by two different sgRNAs. Sequence alignments show frame-shift mutations in KO clones. (B) WB analysis of p150 proteins in WT and two different p150 KO lines. GAPDH (bottom) was used as loading control. (C) Growth curves of two p150 WT and two p150 KO lines. The results are from three independent experiments and bars represent means ± SEM (**P < 0.01, two-tailed Student's t-test). (D) Bright field images show the morphology of spheres derived from p150 WT and KO clones. Scale bar: 50 μM. (E) Representative confocal images of p150 WT and KO lines seeded on feeder cells and stained for Oct4. Scale bar: 20 μM. (F) WB analysis of Oct4, Sox2 and Nanog expression in p150 WT and KO lines. GAPDH was used as loading control.
Deletion of p150 impairs mouse ESC differentiation
Mice with p150 KO die in early embryogenesis (38). Therefore, we analyzed whether p150 KO had any effect on the differentiation of mouse ESCs using the in vitro EB differentiation assay (Supplementary Figure S2F), which mimics the formation of three germ layers during early embryonic development (30,39). The p150 KO EB failed to form cavities observed in WT cells (Supplementary Figure S2G), a feature of EB maturation, and exhibited a more compact structure (Supplementary Figure S2H), indicating a defect in differentiation. Consistent with this observation, gene expression analysis using RT-qPCR showed that compared to WT cells, Oct4, Nanog and Sox2 were not silenced in both p150 KO clones (Figure 2A). The high expression level of Oct4 and Nanog in p150 KO cells compared to WT cells during differentiation was confirmed by WB analysis (Figure 2B). In addition, the induction of several genes that represent the three embryonic lineages during differentiation was also compromised in p150 KO cells compared to WT cells (Supplementary Figure S3A). Together, these results indicate that deletion of p150 compromises the differentiation of mouse ES cells in the in vitro EB assay.
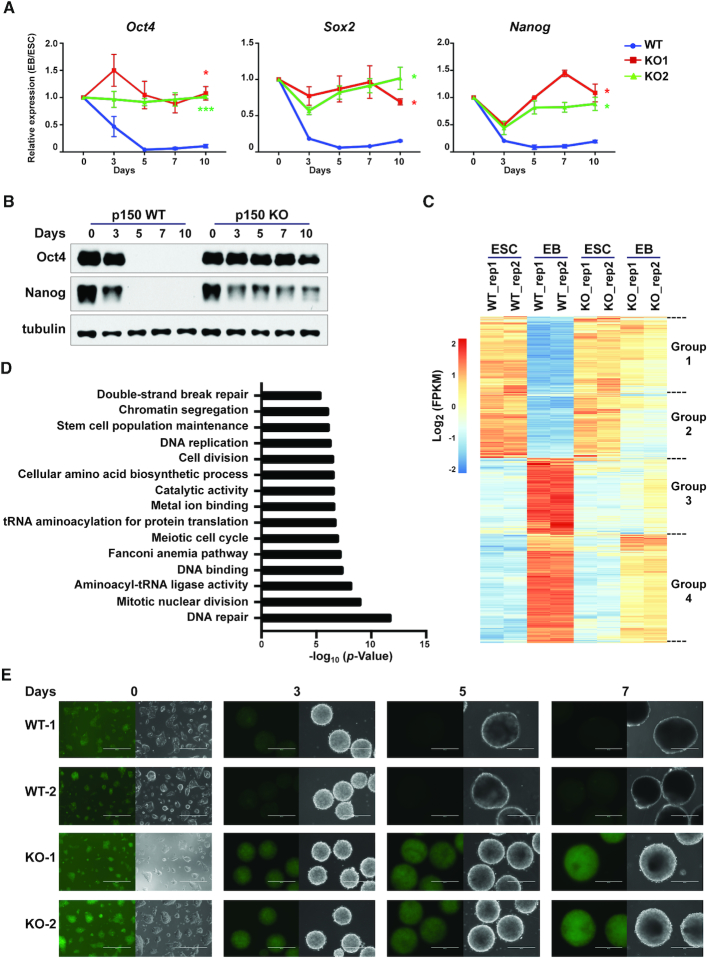
Figure 2.
Mouse CAF-1 p150 is required for silencing of pluripotency genes during ESC differentiation. (A) RT-qPCR analysis of expression of three pluripotency genes during EB formation. Results are from three independent experiments. Error bar represents means ± SEM. *P < 0.05, ***P < 0.001 (two-tailed Student's t-tests between p150 WT and each KO line). (B) WB analysis of Oct4 and Nanog during EB formation. Tubulin (bottom) was used as loading control. (C) Hierarchical cluster analysis of differentially expressed genes before (ESC, day 0) and after (EB, day 7) differentiation of p150 WT and KO cells identified by RNA-seq. Results from two independent repeats (rep1 and rep2) are shown. (D) GO analysis of the Group 1 genes identified in C. (E) Representative EGFP fluorescence images of two independent reporter lines of p150 WT and KO ESCs during EB formation. The expression of EGFP is driven by the Oct4 distal enhancer. Scale bar: 400 μM.
To determine the impact of p150 depletion on gene expression in mouse ESCs and during differentiation in an unbiased manner, we analyzed the transcriptome of p150 WT and KO ESCs and EBs collected after 7 days differentiation using RNA-seq. Compared to WT ESCs, deletion of p150 affected the expression of 587 genes in ESCs. Pathway analysis of the 587 genes indicates that these genes are enriched at plasma membrane and cell surface (Supplementary Figure S3B). However, none of these proteins are receptors known to be involved in the regulation of stem cell fate, consistent with our previous observation that the p150 KO has a minor impact on ESC proliferation and stemness.
Compared to WT ES cells, the expression of 2253 genes decreased, whereas that of 2997 genes increased at day 7 WT EBs. Unsupervised cluster analysis indicated that the 2253 genes that were silenced during the differentiation of WT ESCs could be clearly separated into two groups based on the impact of p150 KO on gene expression (Group1 and Group2, Figure 2C). Group 1 genes were not silenced in KO cells during differentiation (Figure 2C), whereas silencing of group 2 genes was affected less dramatically. Gene Ontology (GO) analysis revealed that Group 1 genes were enriched in genes involved in stem cell population maintenance, DNA replication and repair (Figure 2D), whereas group 2 genes included those involved in ribosome biogenesis and mRNA processing (Supplementary Figure S3C). It is known that genes involved in DNA repair including homologous recombination are expressed highly in ES cells compared to differentiated cells. It is proposed that high expression of these genes in ES cells helps maintain genome integrity of these cells (40,41).
The 2997 upregulated genes during differentiation of WT ES cells could also be separated into two groups (Group3 and Group4, Figure 2C), with Group 3 genes enriched in germ layer differentiation (Supplementary Figure S3D) and Group 4 genes enriched in cell migration (Supplementary Figure S3E). We noticed, however, that the effect of p150 KO on the Group 3 genes were not so consistent among two repeats, likely due to the variations in differentiations. At last, the expression of both Group 3 and Group 4 genes in p150 KO EBs was also compromised compared to WT EB (Supplementary Figure S3F and G). Together, these results indicate that p150 plays an important role in regulating gene expression critical for ESC differentiation.
Impact of p150 deletion on the silencing of Oct4 during differentiation
Silencing of pluripotency regulatory network precedes lineage commitment (42–44). Moreover, CAF-1 in other organisms such as yeast is known to be involved in gene silencing (23). Therefore, we focused on our analysis on the role of CAF-1 in gene silencing. First, we monitored the dynamics of silencing of Oct4 during EB differentiation using the EGFP reporter driven by the Oct4 distal enhancer in WT and p150 KO cell lines (Supplementary Figure S3H-I) (45). Following EB differentiation, EGFP signals in WT cells diminished, whereas EGFP signals in p150 KO cells were preserved (Figure 2E). Flow cytometry analysis of the disaggregated cells from Day 7 EBs showed that EGFP expression was lost in almost all WT cells. In contrast, the majority of p150 KO cells had similar levels of EGFP as ESCs based on analysis of GFP fluorescence (Supplementary Figure S3J) and expression by RT-PCR (Supplementary Figure S3K). These results indicate that p150 deletion compromises the silencing of Oct4 and potentially other pluripotency genes in almost all cells. We also found that the expression of EGFP-tagged full-length p150 in KO ESCs two independent clones fully rescued the defects in silencing of three pluripotent genes (Oct4, Nanog and Sox2) tested (Figure 3A), indicating that defects in silencing of these genes in p150 KO cells during EB differentiation are due to the absence of p150 proteins.
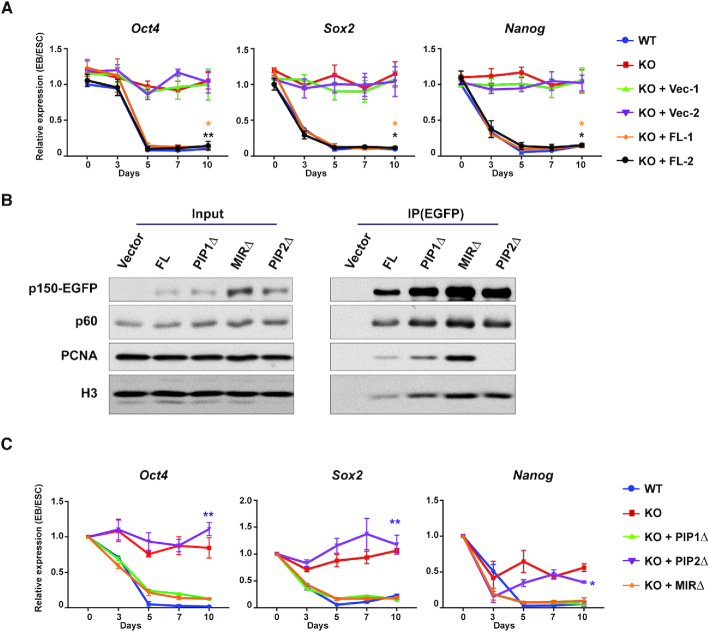
Figure 3.
The PIP2 region of p150 is important for silencing of pluripotency genes. (A) RT-qPCR analysis of expression of three pluripotency genes (Oct4, Sox2 and Nanog) in p150 KO cells expressing either full-length p150 or empty vector during EB differentiation. Two independent lines of full-length (FL) or empty vector (Vec) were used for the analysis. Results were from three independent experiments (means ± SEM, *P < 0.05, **P < 0.01, two-tailed Student's t-test between p150 KO and KO + FL lines). (B) Analysis of interaction of p150 and p150 mutants with p60, PCNA and histone H3. EGFP-tagged full-length p150 and p150 mutants with deletions of the indicated regions were expressed in p150 KO cells and immunoprecipitated with antibodies against GFP. Co-purified proteins were analyzed by WB. PIP1Δ, PIP1 domain deletion; PIP2Δ, PIP2 domain deletion; MIRΔ, HP1-interacting domain (MIR) deletion. (C) RT-qPCR analysis of expression of three pluripotency genes (Oct4, Sox2 and Nanog) from p150 KO lines expressing WT p150 and p150 mutants during EB differentiation. Results represent means ± SEM of three independent experiments (*P < 0.05, **P < 0.01, two-tailed Student's t-test between WT and KO + PIP2Δ lines). Results from additional lines are shown in Supplementary Figure S4.
The PIP2 domain of p150 is required for gene silencing
CAF-1 p150 contains two regions that interacts with PCNA (PIP1 and PIP2) and one region (the MIR domain) for HP1 (Supplementary Figure S4A) (20,25,46). To understand how p150 is involved in gene silencing during ESC differentiation, we analyzed the impact of deleting each of these regions of p150 (PIP1Δ, PIP2Δ, MIRΔ) on cell differentiation. Each p150 deletion mutant was expressed in p150 KO cells to a level similar to the endogenous p150 (Supplementary Figure S4B). Moreover, the p150 PIP2Δ mutant exhibited a reduced interaction with PCNA, whereas the p150 PIP1Δ mutant had no apparent effects (Figure 3B). At last, all three p150 deletion mutants bound the p60 subunit of CAF-1 and histone H3 to the same extent as WT p150 (Figure 3B). These results indicate that these p150 mutants did not affect CAF-1 complex formation and/or their ability to bind histones.
Upon differentiation, expression of p150 PIP1Δ and MIRΔ mutants in p150 KO cells rescued the defect in the silencing of three pluripotency genes tested (Figure 3C), whereas expression of the p150 PIP2Δ mutant failed do so (Figure 3C and Supplementary Figure S4C). The same conclusion could be obtained when examining the morphology of EB expressing WT and various p150 mutants (Supplementary Figure S4D). These results indicate that the PIP2 domain of p150 is essential for differentiation of ESC including silencing of pluripotency genes, whereas the PIP1 and the HP1-interacting regions are dispensable.
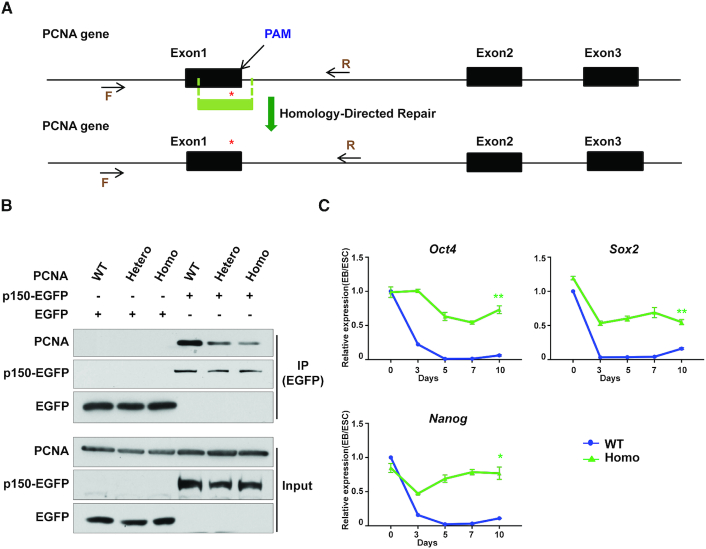
Cells containing the PCNA mutation defective in p150 interaction phenocopy p150 deletion mutant cells
The results presented above strongly support the idea that targeting CAF-1 to the DNA replication fork through its interaction with PCNA is important for the gene silencing during differentiation. This observation is reminiscent of our early study in yeast cells expressing the PCNA point mutations Arg61Ala and Asp63Ala (R61A, D63A) (21). This mutation, while having no apparent effect on DNA synthesis, is defective in the PCNA–p150 interaction and transcriptional silencing at telomeres and silent mating-type loci. Therefore, we introduced the same PCNA mutation into a mouse ESC line using the CRISPR/Cas9-mediated gene editing technology (Figure 4A and Supplementary Figure S5A) (29). As in yeast cells (21), the PCNA mutation exhibited reduced association with CAF-1 p150 compared to WT PCNA (Figure 4B). Moreover, the PCNA mutant cells maintained ESC identity and self-renewal based on the expression of Oct4 (Supplementary Figure S5B) and AP staining (Supplementary Figure S5C). Therefore, similarly to the CAF-1 p150 KO, the PCNA (R61A, D63A) mutant has little impact on mouse ESC.
Figure 4.
The PCNA mutant defective in CAF-1 interaction is also defective in silencing of pluripotency genes. (A) A schematic representation of generating PCNA R61A D63A mutations using CRISPR/Cas9-mediated homologous recombination. Red asterisk indicates the PCNA mutation sites. The target region was amplified by PCR and sequenced by Sanger sequencing. F, forward primer. R, reverse primer. PAM, protospacer adjacent motif. (B) Analysis of p150–PCNA interaction in mouse ESCs expressing WT and mutant PCNA. EGFP-tagged p150 was expressed in PCNA WT and mutant cells and immunoprecipitated using antibodies against GFP. Co-immunoprecipitated proteins were analyzed by WB. Hetero, one allele with the R61A D63A mutation and one with WT PCNA; Homo, both alleles mutated to R61A, D63A. (C) RT-qPCR analysis of expression of Oct4, Sox2 and Nanog in PCNA WT and mutant lines during EB differentiation. Homo, PCNA homozygous mutation. Results represent means ± SEM from three independent experiments (*P < 0.05, **P < 0.01, two-tailed Student's t-test between PCNA WT and Homo mutant lines).
Upon differentiation, we observed that, similarly to p150 KO cells, EBs from the PCNA mutant cells failed to form cavities after culturing 10 days (Supplementary Figure S5D and E). Moreover, Oct4, Nanog and Sox2 were not properly silenced in PCNA mutant cells compared to WT ESCs during differentiation (Figure 4C). Moreover, the induction of genes involved in germ layer specification was compromised (Supplementary Figure S5F). Interestingly, we noticed that the effect of p150 KO and PCNA mutants had a differential impact on the expression of Brachyury (T) involved in formation of the posterior mesoderm, possibly due to the fact that PCNA (R61A, D63A) mutation only reduces, but not abolish PCNA–CAF-1 interaction (Figure 4B). Together, these results strongly support the idea that disrupting the PCNA–CAF-1 interaction, through either the deletion of the p150 PIP2 domain or PCNA point mutations, impairs silencing of genes including pluripotent genes during differentiation, highlighting the importance of CAF-1–PCNA-mediated nucleosome assembly pathway in this process.
PCNA homozygous mutation impairs mouse embryonic development
CAF-1 p150 is essential for early embryonic development (38). To test whether the PCNA (R61A, D63A) mutations also affect mouse embryonic development, we introduced heterozygous PCNA (R61A, D63A) mutation in ES cells by CRISPR/Cas9 and generated heterozygous PCNA mutation mouse (PCNA +/Mut). PCNA+/Mut mice were born at a normal Mendelian Frequency. Moreover, PCNA+/Mut mice grew to normal size, were fertile and displayed no obvious abnormalities. However, crossing heterozygous PCNA (+/Mut) mice failed to generate viable homozygous PCNA (Mut/Mut) mice (Supplementary Figure S5G). Further analysis showed that embryo development is terminated before E6.5. Thus, the CAF-1 binding deficient PCNA mutant, also exhibits a similar defect in early embryonic development as p150 KO, indicating that the CAF-1–PCNA interaction is important for early embryonic development.
CAF-1 p150 is important for formation of silent chromatin at the promoters of pluripotency genes during differentiation
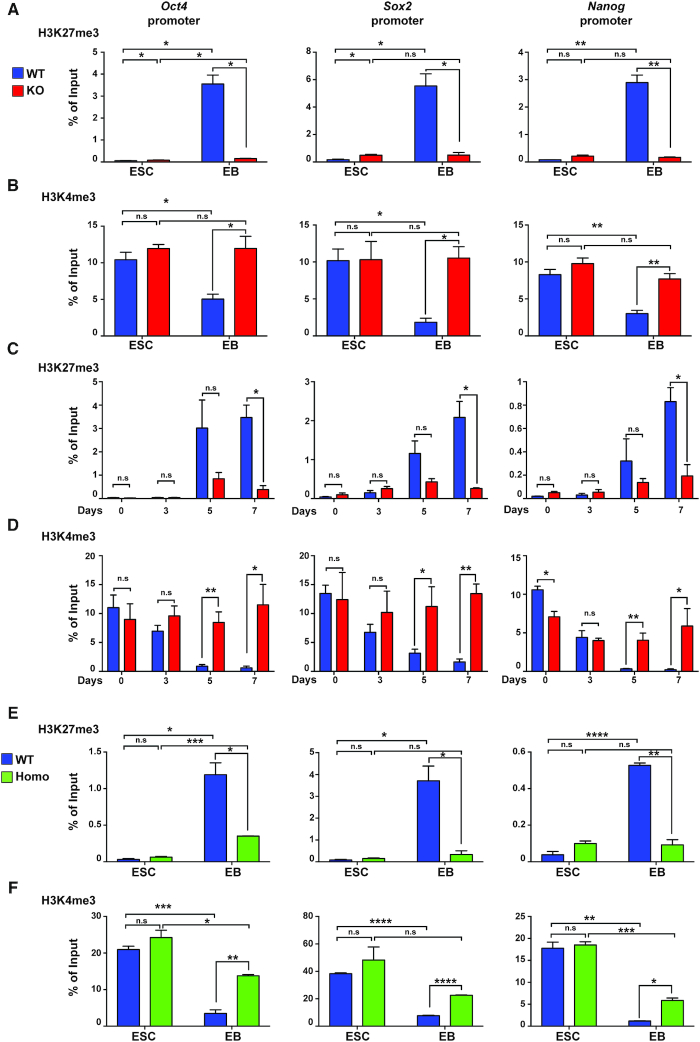
The histone modifications H3K4me3 and H3K27me3 are associated with opposite gene activity. While H3K4me3 at promoters is associated to active gene transcription, H3K27me3 at promoters is linked to gene repression (47,48). To understand how p150 impacts gene silencing during differentiation, we first employed chromatin immunoprecipitation (ChIP)-qPCR and analyzed changes of H3K4me3 and H3K27me3 levels at promoter regions of Oct4, Sox2 and Nanog before and after differentiation. After 10 days of differentiation of WT ESCs into EBs, H3K27me3 levels increased substantially at promoter regions of these three genes (Figure 5A). In contrast, when p150 KO cells were subjected to the same differentiation protocol, H3K27me3 occupancy of the Oct4, Nanog and Sox2 promoters showed little, if any, increase compared to undifferentiated mouse ESCs. Moreover, we found that the levels of Ezh2 at the promoter of Oct4 and Sox2 increased in WT EB compared to ES cells, and this increase was compromised in p150 KO ES cells (Supplementary Figure S6A). In contrast, p150 KO had no apparent effects on the Ezh2 levels at genes (Il13 and Crhr2) that were silenced in both ES and EB by the presence of H3K27me3 (Supplementary Figure S6B and C). At last, the H3K27me3 levels at two lineage specific genes (Gata4 and Gata6) were not reduced in p150 KO EB compared to ES cells (Supplementary Figure S6D). These results are consistent with gene expression analysis that silencing of these three pluripotent genes is compromised in p150 KO EBs.
Figure 5.
CAF-1–PCNA-mediated nucleosome assembly is important for histone modifications at the promoters of pluripotency genes during ESC differentiation. (A andB) ChIP-qPCR analysis of H3K27me3 (A) and H3K4me3 (B) occupancy of promoter regions of Oct4, Sox2 and Nanog in p150 WT and p150 KO lines during EB differentiation. Both ESC and day 10 EB were used for analysis. (C andD) ChIP-qPCR analysis of H3K27me3 (C) and H3K4me3 (D) levels at promoter regions of Oct4, Sox2 and Nanog at different time points during differentiation in p150 WT and p150 KO lines. Results are means ± SEM (n = 3, *P < 0.05, **P < 0.01, two-tailed Student's t-test). (E andF) ChIP-qPCR analysis of H3K27me3 (E) and H3K4me3 (F) occupancy at promoter regions of Oct4, Sox2 and Nanog in PCNA WT and Homo mutant ESC and day10 EB. Results are from three independent experiments (means ± SEM, n = 3, *P < 0.05, **P < 0.01, ****P < 0.0001, two-tailed Student's t-test).
We also analyzed H3K4me3 levels at these genes and found that H3K4me3 occupancy in WT EBs was reduced compared to ES cells, and the reduction of H3K4me3 at these genes was not detectable in p150 KO EB (Figure 5B). Furthermore, we monitored the changes of H3K27me3 and H3K4me3 during earlier time points of EB formation (Figure 5C and D). The increase of H3K27me3 at the promoters of Oct4, Nanog and Sox2 in WT cells could first be detected on day 5 of EB differentiation. The effect of p150 KO on H3K27me3 was evident at the same time point but became much more pronounced by day 7 of EB differentiation (Figure 5C). The H3K4me3 levels at promoters of Oct4, Nanog and Sox2 in p150 WT cells were reduced slightly 3 days after differentiation, were significantly reduced at 5 days of differentiation. The impact of p150 deletion on H3K4me3 levels at these gene promoter regions was observed at a kinetic similar to or slightly prior to the impact of p150 depletion on H3K27me3 (Figure 5D). However, CAF-1 p150 deletion had no apparent effect on the total levels of H3K27me3 and H3K4me3 in both ES cells and in EB (Supplementary Figure S6E). This result suggests that p150 affects pluripotency gene silencing through regulating histone modifications at these genes during differentiation.
Next, we analyzed the effect of the PCNA mutant with CAF-1 binding defects on changes of histone modifications at the promoter regions of three pluripotency genes during differentiation. In contrast to the dramatic increase of H3K27me3 in WT EBs (Figure 5E), H3K27me3 levels in the PCNA mutant EBs only showed a slight increase. Moreover, H3K4me3 levels at the promoters of these three genes in the PCNA mutant EBs were not reduced to a similar degree as in WT EBs during differentiation (Figure 5F). Thus, the PCNA mutant defective in its interaction with CAF-1 also shows a defect similar to the p150 KO mutant in establishing the H3K27me3 mark at chromatin loci destined for silencing during differentiation. Together, these data strongly support the idea that the CAF-1–PCNA-mediated nucleosome assembly pathway is likely involved in this process.
CAF-1 p150 is important for changes in chromatin landscape at a subset of genes during differentiation
To analyze the impact of p150 KO on H3K27me3 and H3K4me3 at gene promoters in an unbiased manner, we performed H3K27me3 and H3K4me3 ChIP-seq using WT and p150 KO ESCs and day 7 EBs. Consistent with the ChIP-qPCR results, upon differentiation of WT ES cells, H3K27me3 levels at the promoters of Oct4 and Nanog increased in day 7 EBs compared to ESCs, whereas this increase was barely detectable in p150 KO EBs. Similarly, H3K4me3 ChIP-seq peaks at promoters of these two pluripotency genes were reduced in WT but not in p150 KO EBs (Figure 6A). Next, we compared the levels of H3K27me3 and H3K4me3 at the promoters of the groups of genes identified in Figure 2C based on RNA-seq. In ESCs, p150 KO had no apparent effect on H3K27me3 and H3K4me3 at either of these four groups of genes except slight changes of H3K4me3 at the Group 4 genes (Figure 6B–I). Upon differentiation, H3K27me3 levels at the promoters of Group 1 genes, which are silenced in WT, but not in p150KO EBs, increased, and this increase was significantly compromised in p150 KO EBs (Figure 6B). Similarly, the differentiation-associated reduction of H3K4me3 at the promoters of Group 1 genes was also compromised in p150 KO EBs (Figure 6C). In contrast, the effect of p150 KO on H3K27me3 and H3K4me3 levels at the promoters of Group 2 genes, which are silenced in both WT and p150 KO EBs, was not significant (Figure 6D and E). After 7 days of differentiation, H3K27me3 levels at the promoters of Group 3 were not altered dramatically upon differentiation of either WT or p150 KO ES into EB. In contrast, H3K27me3 levels at the promoters of Group 4 genes were reduced upon differentiation of WT ES cells, and this reduction was compromised in p150 KO EB (Figure 6F and H). The H3K4me3 levels at the promoters of both Group 3 and 4 genes increased after differentiation of WT cells, which was compromised in p150KO EBs (Figure 6G and I). These results are consistent with the gene expression changes and indicate that p150 depletion impairs the remodeling of epigenetic landscape including formation of facultative heterochromatin marked by H3K27me3 at a subgroup of genes downregulated during ESC differentiation.
The nucleosome occupancy is compromised in p150 knockout ES cells
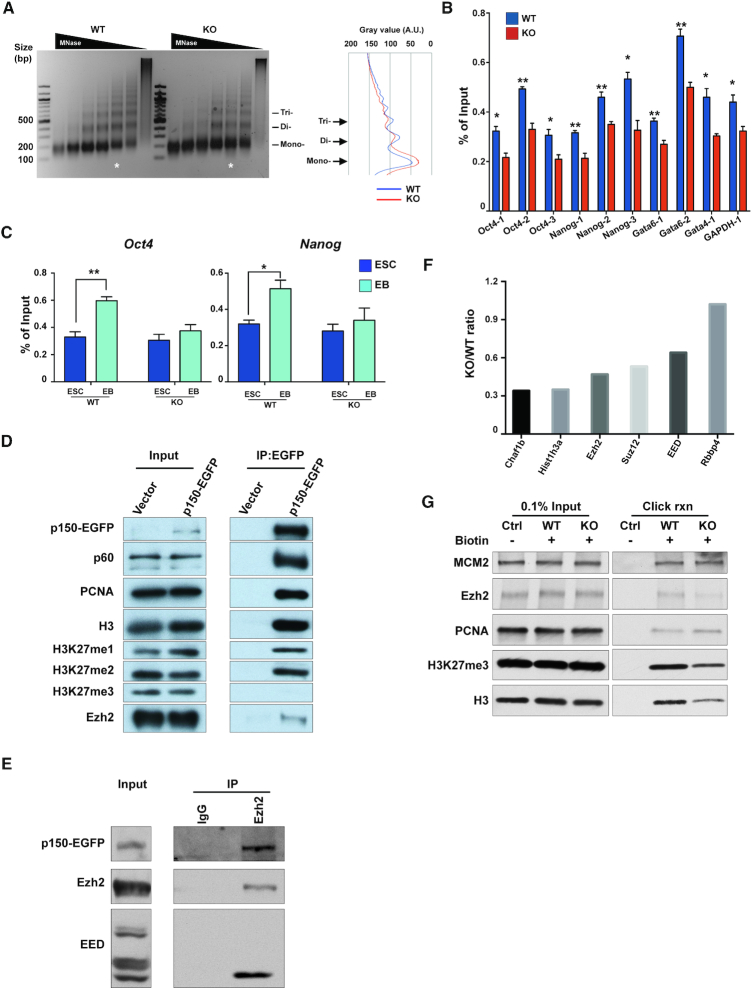
Chromatin compaction facilitates the activation of the PRC2 complex to establish H3K27me3-mediated gene silencing (49). Given that CAF-1 is a histone chaperone and deletion of yeast CAF-1 impacts nucleosome occupancy (50). We hypothesized that CAF-1 affects establishment of silent chromatin through formation of a compact chromatin. To test this idea, we first compared the chromatin structure in p150 WT and KO ES cells using digestion by micrococcal nuclease (MNase), which cleaves the linker DNA between nucleosomes. We observed that chromatin in p150 KO ES cells are more susceptible to MNase digestion than WT cells, suggesting that chromatin in p150 KO cells is less compact than that in WT cells (Figure 7A). Consistent with the idea, we found that nucleosome occupancy, as measured by histone H3 ChIP-qPCR, at the promoters of several group of genes was reduced in p150 KO ES cells compared to WT ES cells (Figure 7B). However, the total levels of H3 and H4 in ES cells were not affected (Supplementary Figure S7A). Upon differentiation, nucleosome occupancy at the promoter of two pluripotent genes (Oct4 and Nanog) tested increased in p150 WT EBs compared to ESCs. However, the increase of nucleosome occupancy at the promoters of these two genes was compromised in p150 KO EB (Figure 7C). Similarly, the nucleosome occupancy at the promoters of Gata4 and Gata6 genes was reduced in WT EB, and this reduction was compromised in p150 KO EBs (Supplementary Figure S7B). Together, these results support the idea that defects in DNA replication-coupled nucleosome assembly in p150 KO cells lead to a reduced nucleosome occupancy, which contributes to the defects in establishment of silent chromatin during the differentiation of p150 KO ES cells.
Figure 7.
Deficiency of p150 disrupts the formation of facultative silent chromatin. (A) DNA fragment distribution after MNase digestion on p150 WT and p150 KO ESC. Chromatin from fixed p150 WT and KO ESC was digested by titrated MNase. After purification, DNA was analyzed by agarose gel electrophoresis and DNA size corresponding to mono-, di- and tri-nucleosome is indicated. Image is representative from three independent experiments. Quantification of the signal intensity over indicated lane is shown on the right by ImageJ/plot profiler. A.U., arbitrary units. (B) Nucleosome occupancy at two pluripotent genes (Oct4 and Nanog), two lineage-specific genes (Gata4 and Gata6) and housekeeping gene (GAPDH). H3 ChIP was performed using chromatin from p150 WT and KO cells. Quantitative PCR was performed targeting the nucleosomes upstream of transcription start sited (TSS) of the indicated genes. Results are from three independent experiments. (means ± SEM, n = 3, *P < 0.05, **P < 0.01, two-tailed Student's t-test) (C) Nucleosome occupancy at two pluripotent genes (Oct4 and Nanog). H3 ChIP was performed using chromatin from ESC and day7 EB of p150 WT and KO cell line. Quantitative PCR was performed targeting the −1 nucleosomes upstream of TSS of the indicated genes. Results are from three independent experiments (means ± SEM, n = 3, *P < 0.05, **P < 0.01, two-tailed Student's t-test). (D and E) CAF-1 interacts with Ezh2. (D) EGFP-tagged full-length p150 expressed in p150 KO cells was immunoprecipitated using antibodies against GFP. Co-purified proteins were analyzed by WB using the indicated antibodies. (E) Ezh2 was immunoprecipitated using antibodies against Ezh2 and extracts prepared from p150 KO cells expressing EGFP-tagged p150. Co-purified proteins were analyzed by WB using the indicated antibodies. Immunoprecipitation using IgG was used as a control. (F and G) Impacts of p150 KO on histone and Ezh2 binding at replicating chromatin. (F) Peptides ratio of indicated proteins between p150 WT and KO detected from SILAC-iPOND-MS. ES cells cultured with light isotope growth media (p150 WT) and heavy isotope growth media (p150 KO) were pulsed with EdU to label nascent chromatin and fixed with formaldehyde. Heavy and light labeled cells were mixed 1:1 before click chemistry reaction. After Click reaction to conjugate EdU with biotin, DNA–protein complex on nascent chromatin was purified and eluting proteins were analyzed by mass spectrometry. (G) Eluted proteins from iPOND procedures were analyzed by SDS-polyacrylamide gel electrophoresis followed by immunoblotting for indicated antibodies. Representative blots from three independent iPOND experiments were shown.
CAF-1 is a histone chaperone for histone H3.1, it is possible that the relative amount of H3.1 and H3.3 on chromatin may change. To test this idea, we tagged H3.1 (Hist1h3g) and H3.3 (H3f3b) with the Flag epitope and analyzed levels of H3.1 and H3.3 in WT and p150 KO ES cells using chromatin fractionation assays. We observed a reduction of H3.1 and an increase of H3.3, slightly and consistently, on chromatin in p150 KO ES cells compared to WT ES cells (Supplementary Figure S7C and D). This result raises the possibility that the change in H3.1/H3.3 ratio on chromatin in p150 KO ES cells may affect the establishment of H3K27 methylation during differentiation.
CAF-1 interacts with Ezh2 as well as H3-H4 with methylation at H3K27 in ESCs
CAF-1 is known to deposit newly synthesized H3-H4 onto replicating DNA for nucleosome formation. H4 co-purified with CAF-1 is acetylated at lysine residues, 5 and 12, a mark of newly synthesized H4 (51). Moreover, in plants, it has been shown that CAF-1 interacts with Ezh2 (26). Therefore, we identified modifications on histones co-purified with CAF-1 in ES cells and tested whether CAF-1 interacts with Ezh2. As expected, the p60 subunit of the CAF-1 complex, PCNA and histone H3 co-purified with CAF-1 p150 in ESCs. Interestingly, we observed that histone H3 co-purified with CAF-1 was mono- and di-methylated at H3K27 (H3K27me1 and H3K27me2) (Figure 7D). Mass spectrometry analysis indicate that in addition to H3K27me1 and H3K27me2, H3K27me3 was also detected, but at a lower abundance compared to H3K27me2 (Supplementary Figure S8B and C). Interestingly, we also observed that H3 co-purified with CAF-1 is methylated at lysine 36. Histone H4 co-purified with CAF-1 is acetylated at lysine 5, 8, 12 and 16. These results indicate that a fraction of H3-H4 co-purified with CAF-1 is from newly synthesized H3-H4 in ES cells. It has been reported that newly synthesized histone H3 is mono-methylated at lysine 9 (52,53). However, H3 peptides from residues 1 to 9 were barely detectable in mass spectrometry analysis, in part likely due to the digestion of peptides by Trypsin.
At last, we found that Ezh2, the catalytic subunit of H3K27 methyltransferase PRC2, also interacted with p150 (Figure 7D and E), and this interaction was also detected in PCNA mutant cells with CAF-1 binding defect (Supplementary Figure S8D). These results indicate that CAF-1–PRC2 interaction is conserved from plant to mammalian cells.
To gain insight into the functional significance of CAF-1–Ezh2 interaction in ES cells, we analyzed how p150 KO affected the binding of Ezh2 with replicating chromatin using iPOND coupled with mass spectrometry analysis. As reported, the PCR2 complex bound to replicating chromatin (31,54). Interestingly, the association of three core subunits of PRC2 complex (Ezh2, Suz12 and EED) with replicating chromatin is reduced in p150KO ES cells, whereas the binding of RbAp48, a protein that is also subunit of other chromatin complexes including NURD (55), was not (Figure 7F). The reduction of Ezh2 with replicating DNA was confirmed by WB analysis (Figure 7G). Moreover, we observed that the association of H3.1 and H3K27me3 with replicating chromatin were also reduced (Figure 7F and G). Together, these results also raise the possibility that CAF-1 may also contribute to the establishment of H3K27me3-mediated silencing through recruiting the PRC2 complex as well as modified histones to replication forks.
DISCUSSION
Here we show that in mouse ESCs, deletion of the p150 subunit of CAF-1, and mutations at CAF-1 p150 and PCNA that compromise the CAF-1–PCNA interaction, all dramatically impair stem cell differentiation in the in vitro EB formation assay, while having minimal effect on ESC proliferation and maintenance. Specifically, we show that in these mutant cells, silencing of a group of genes including those involved in ESC maintenance such as Oct4, Nanog and Sox2 is compromised upon differentiation. Moreover, H3K27me3, a mark associated with gene repression, is not established at the promoter of these genes. In ESCs, CAF-1 interacts with Ezh2 as well as with H3 bearing H3K27 methylations. Depletion of p150 leads to reduced nucleosome occupancy and impaired association of the PRC2 complex with replication forks. These results suggest that CAF-1 p150 is dispensable for the maintenance of heterochromatin state marked by H3K27me3 that is already established. Instead, our results indicate that CAF-1 facilitates the formation of facultative heterochromatin for gene silencing during early stages of ESC differentiation, likely through multiple mechanisms.
CAF-1 likely functions with other histone chaperones to promote DNA replication-coupled nucleosome assembly in mouse ESCs
CAF-1 is classic histone chaperone that regulates the deposition of newly synthesized histone H3-H4 on replicated DNA (16). Depletion of CAF-1 in human cell lines results in defects in DNA synthesis and increased DNA replication stress (37). Mice lacking p150 die in early development (38). Moreover, CAF-1 plays an important role in maintenance of chromatin states of somatic cells (28). Therefore, it was surprising that mouse ESCs lacking p150 showed little defects in cell-cycle progression, DNA synthesis and genome integrity. In addition to p150, CAF-1 has two other subunits, p60 and p48. Interestingly, we could not obtain ESC clones with the deletion of p60, suggesting that unlike p150, CAF-1 p60 is essential for cell viability. It is known that CAF-1 p48 also exists in other protein complexes including histone deacetylase complex and NuRD (55–57). Therefore, it is possible that CAF-1 p60 is also a subunit of other protein complexes besides CAF-1. In budding yeast, deletion of CAF-1 also has a mild impact on cell proliferation and cell viability (22,23). These results indicate that the impact of p150 deletion in mouse ES cells is similar to yeast cells. Chromatin in mouse ES cells is less compact than that of somatic cells, which could provide an explanation for the non-essentiality of p150 in both in yeast and mouse ES cells.
In budding yeast, it is known that CAF-1, Rtt106 and FACT coordinate to promote nucleosome assembly during DNA replication (58–60). FACT is a histone chaperone conserved from yeast to human cells, and is important for both gene transcription and DNA replication (61,62). Rtt106 shares limited sequence homology with DAXX, a histone chaperone for histone H3 variant H3.3 (63,64). DAXX also has a role during S phase (65). In the future, it would be interesting to determine to what extent these histone chaperones cooperate with CAF-1 to promote nucleosome assembly during the S phase in ESCs.
CAF-1 plays an important role for the establishment of gene silencing during mouse ESC differentiation
Using two independent approaches (deleting the PCNA interacting motif in CAF-1 p150 subunit and mutating residues in PCNA that mediate PCNA–CAF-1 interaction), we show that H3K27me3-mediated silencing of a subset of genes including pluripotency genes is compromised when CAF-1’s function in nucleosome assembly are impaired. In budding yeast, mutations at the same PCNA residues have little apparent effect on DNA synthesis but result in defects in transcriptional silencing in the same genetic pathway as deletion of yeast CAF-1 (21). PCNA is involved in recruiting p150 to DNA replication forks in both yeast and mammalian cells (20,66). These results indicate that the CAF-1–PCNA-mediated nucleosome assembly pathway plays an important role for silencing of a subset of genes including pluripotency genes in mouse ESC differentiation.
The CAF-1-mediated nucleosome assembly pathway is best known for its role in maintenance of chromatin states from yeast to human cells. For instance, CAF-1 has a role in maintaining transcriptional silencing at silent chromatin loci in yeast cells (22,23). More recently, it has been found that CAF-1-mediated nucleosome assembly is critical for safeguarding somatic cell identity as depletion of CAF-1 in somatic cells leads to a dramatic increase in reprograming efficiency (28). Moreover, depletion of CAF-1 in ESCs leads to an increase in 2C-like cells (27). We confirmed the latter observation as we also observed that in p150 KO mouse ESCs, there is an increase in cells with 2C-like properties. In these rare 2C-like cells, Oct4 is silenced. However, the majority of p150 KO ESCs retain stem cell properties as they express Oct4. This situation is reminiscent of yeast cells lacking SIR1 (67,68). In sir1Δ cells, while a fraction of cells maintains the silencing at the silent mating type locus, other cells lose silencing of this locus. In a manner analogous to Sir1’s role in the establishment of silent chromatin in budding yeast, we propose that CAF-1 in ESCs also has a role in establishing facultative heterochromatin at the Oct4 locus as well as at a subset of genes that must be silenced during differentiation. Supporting this idea, p150 knockout has little effects on H3K27 methylation on two genes that are silenced in ES cells and remained silenced in EB. This explanation is also consistent with our observations that p150 is largely dispensable for the maintenance of chromatin state in mouse ES cells.
How does p150 function in establishment of facultative heterochromatin during differentiation? It is known that CAF-1 p150 interacts with LSD1/CoREST/HDAC1 complex (69). LSD1 is a demethylase for histone H3K4 methylation. We observed that removal of H3K4me3 mark during differentiation is compromised in p150 KO cells. Therefore, it is possible that defects in establishment of facultative heterochromatin is due to a failure in the recruitment of LSD1 to these loci during differentiation. Arguing against this possibility, it has been shown that silencing of Oct4 and Nanog is not compromised during differentiation of LSD1 knockout mouse ESC or LSD1 depleted human ESCs (70,71). Instead, we present several lines of evidence indicating that CAF-1 may play a more direct role in establishing the H3K27me3-mediated silencing. First, we show that chromatin in p150 KO ESC cells is more susceptible for MNase digestion, indicating a more open chromatin state in p150 KO ES cells compared to WT ES cells. Moreover, nucleosome occupancy at regions close to the promoters of Nanog and Oct4 was less compact than that of WT cells during differentiation. Previously, it has been shown that a compact nucleosome density is important to activate the PRC2 complex for the establishment of H3K37me3 (49). Therefore, it is likely that reduced nucleosome occupancy in p150 KO cells contributes to the defects in the establishment of H3K27me3. In addition to nucleosome occupancy, we show that CAF-1 interacts with the PRC2 complex. Moreover, the association of PRC2 complex with DNA replication forks is compromised in p150 KO ES cells. Therefore, it is also possible that CAF-1 contributes to establishment of H3K27me3 through recruitment of the PRC2 complex during S phase. Consistent with this idea, in Arabidopsis, CAF-1 interacts with PRC2 complex (26). In addition to maintaining silencing, they also reported that H3.1, which is deposited by CAF-1, is also required for the establishment of silencing at the FLC locus during vernalization in part through spreading of H3K27me3 (26). Therefore, it is likely that CAF-1 also facilitates the establishment of H3K27me3-mediated silencing through its recruitment of the PRC2 complex to replication forks. At last, we observed that histone H3 co-purified with CAF-1 was methylated at H3K27. In addition to H3K27 methylation, methylation of H3K36 and acetylation of histone H4 lysine 5 and 12 were also detected on histones co-purified with CAF-1. Acetylation of H4 lysine 5 and 12 residues is a mark for newly synthesized H4. Currently, we do not have evidence that histone H3 methylated at 36 and 27 co-purified with the CAF-1 complex from mouse ESC cells is newly synthesized histone. Nonetheless, this raises the possibility that CAF-1 can deposit H3-H4 with containing H3K27 methylation to chromatin. Therefore, CAF-1 facilitates the establishment of H3K27me3-mediated silencing through at least three non-exclusive direct mechanisms: nucleosome density for activation of the PRC2 complex, recruitment of the PRC2 complex to replication forks and deposition of histones with H3K27 methylation. In addition to these H3K27me3-linked possibilities, a recent study shows that p60, another subunit of CAF-1, can bind to specific chromatin loci. Moreover, CAF-1 displaces specific TFs to inhibit differentiation of leukemia cells and it does so through its function in DNA replication-coupled nucleosome assembly (72). Therefore, it is also possible that CAF-1-mediated nucleosome assembly helps displace TF network that is needed for the maintenance of ESC and thereby facilitates the establishment of facultative heterochromatin. Future studies are needed to test these and other models on the involvement of CAF-1 in the establishing facultative silent chromatin.
DATA AVAILABILITY
Raw data have been deposited in the GEO database with accession number GSE130706.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr. Jan van Deursen for the ESC lines, Dr. Bruce Stillman for p150 antibodies, Dr. Thomas Fazzio for LIF-expression construct and protocols, Gabriella B. Gajdos and Jagneet Kaur at Mayo Clinic, Rochester, Minnesota for their help with the flow cytometry experiments and Dr. Simon J. Gibbons for his help with confocal microscopy. We thank Dr. David Cortez and his lab member for help on iPOND and all the lab members of Dr. Zhang's group for critical discussions.
Authors’ contribution: L.C., X.Z. and Z.Z. designed research; L.C., X.Z.,Y.W., X.L., X.X. and J.Q. performed experiments. L.C., H.G., X.H. and Z.Z. analyzed the data; Z.Z. and T.O. supervised this project. L.C. and Z.Z. wrote the paper. All authors read and edit the paper.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institute of Health (NIH) [R35 GM119015 to Z.Z.]; NIH/NCI Cancer Center Support Grant [p30 CA013696, R01 DK058185 to T.O., P01 DK068055, P30 DK084567]; Mayo Clinic Graduate School of Biomedical Sciences (to L.C.). Funding for open access charge: NIH.
Conflict of interest statement. None declared.
REFERENCES
- 1. Evans M.J., Kaufman M.H.. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981; 292:154–156. [DOI] [PubMed] [Google Scholar]
- 2. Martin G.R. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc. Natl. Acad. Sci. U.S.A. 1981; 78:7634–7638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. De Los Angeles A., Ferrari F., Xi R., Fujiwara Y., Benvenisty N., Deng H., Hochedlinger K., Jaenisch R., Lee S., Leitch H.G. et al.. Hallmarks of pluripotency. Nature. 2015; 525:469–478. [DOI] [PubMed] [Google Scholar]
- 4. Serra-Cardona A., Zhang Z.. Replication-coupled nucleosome assembly in the passage of epigenetic information and cell identity. Trends Biochem. Sci. 2018; 43:136–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Young R.A. Control of the embryonic stem cell state. Cell. 2011; 144:940–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jaenisch R., Young R.. Stem cells, the molecular circuitry of pluripotency and nuclear reprogramming. Cell. 2008; 132:567–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chen X., Xu H., Yuan P., Fang F., Huss M., Vega V.B., Wong E., Orlov Y.L., Zhang W., Jiang J. et al.. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008; 133:1106–1117. [DOI] [PubMed] [Google Scholar]
- 8. Chew J.L., Loh Y.H., Zhang W., Chen X., Tam W.L., Yeap L.S., Li P., Ang Y.S., Lim B., Robson P. et al.. Reciprocal transcriptional regulation of Pou5f1 and Sox2 via the Oct4/Sox2 complex in embryonic stem cells. Mol. Cell Biol. 2005; 25:6031–6046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Loh Y.H., Wu Q., Chew J.L., Vega V.B., Zhang W., Chen X., Bourque G., George J., Leong B., Liu J. et al.. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat. Genet. 2006; 38:431–440. [DOI] [PubMed] [Google Scholar]
- 10. Atlasi Y., Stunnenberg H.G.. The interplay of epigenetic marks during stem cell differentiation and development. Nat. Rev. Genet. 2017; 18:643–658. [DOI] [PubMed] [Google Scholar]
- 11. Luger K., Mader A.W., Richmond R.K., Sargent D.F., Richmond T.J.. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997; 389:251–260. [DOI] [PubMed] [Google Scholar]
- 12. Santos-Rosa H., Schneider R., Bannister A.J., Sherriff J., Bernstein B.E., Emre N.C., Schreiber S.L., Mellor J., Kouzarides T.. Active genes are tri-methylated at K4 of histone H3. Nature. 2002; 419:407–411. [DOI] [PubMed] [Google Scholar]
- 13. Schuettengruber B., Bourbon H.M., Di Croce L., Cavalli G.. Genome Regulation by Polycomb and Trithorax: 70 Years and Counting. Cell. 2017; 171:34–57. [DOI] [PubMed] [Google Scholar]
- 14. Mikkelsen T.S., Ku M., Jaffe D.B., Issac B., Lieberman E., Giannoukos G., Alvarez P., Brockman W., Kim T.K., Koche R.P. et al.. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007; 448:553–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Creyghton M.P., Cheng A.W., Welstead G.G., Kooistra T., Carey B.W., Steine E.J., Hanna J., Lodato M.A., Frampton G.M., Sharp P.A. et al.. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:21931–21936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Burgess R.J., Zhang Z.. Histone chaperones in nucleosome assembly and human disease. Nat. Struct. Mol. Biol. 2013; 20:14–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Stillman B. Chromatin assembly during SV40 DNA replication in vitro. Cell. 1986; 45:555–565. [DOI] [PubMed] [Google Scholar]
- 18. Kaufman P.D., Kobayashi R., Kessler N., Stillman B.. The p150 and p60 subunits of chromatin assembly factor I: a molecular link between newly synthesized histones and DNA replication. Cell. 1995; 81:1105–1114. [DOI] [PubMed] [Google Scholar]
- 19. Shibahara K., Stillman B.. Replication-dependent marking of DNA by PCNA facilitates CAF-1-coupled inheritance of chromatin. Cell. 1999; 96:575–585. [DOI] [PubMed] [Google Scholar]
- 20. Rolef Ben-Shahar T., Castillo A.G., Osborne M.J., Borden K.L., Kornblatt J., Verreault A.. Two fundamentally distinct PCNA interaction peptides contribute to chromatin assembly factor 1 function. Mol. Cell Biol. 2009; 29:6353–6365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhang Z., Shibahara K., Stillman B.. PCNA connects DNA replication to epigenetic inheritance in yeast. Nature. 2000; 408:221–225. [DOI] [PubMed] [Google Scholar]
- 22. Enomoto S., Berman J.. Chromatin assembly factor I contributes to the maintenance, but not the re-establishment, of silencing at the yeast silent mating loci. Genes Dev. 1998; 12:219–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kaufman P.D., Kobayashi R., Stillman B.. Ultraviolet radiation sensitivity and reduction of telomeric silencing in Saccharomyces cerevisiae cells lacking chromatin assembly factor-I. Genes Dev. 1997; 11:345–357. [DOI] [PubMed] [Google Scholar]
- 24. Quivy J.P., Gerard A., Cook A.J., Roche D., Almouzni G.. The HP1-p150/CAF-1 interaction is required for pericentric heterochromatin replication and S-phase progression in mouse cells. Nat. Struct. Mol. Biol. 2008; 15:972–979. [DOI] [PubMed] [Google Scholar]
- 25. Murzina N., Verreault A., Laue E., Stillman B.. Heterochromatin dynamics in mouse cells: interaction between chromatin assembly factor 1 and HP1 proteins. Mol. Cell. 1999; 4:529–540. [DOI] [PubMed] [Google Scholar]
- 26. Jiang D., Berger F.. DNA replication-coupled histone modification maintains Polycomb gene silencing in plants. Science. 2017; 357:1146–1149. [DOI] [PubMed] [Google Scholar]
- 27. Ishiuchi T., Enriquez-Gasca R., Mizutani E., Boskovic A., Ziegler-Birling C., Rodriguez-Terrones D., Wakayama T., Vaquerizas J.M., Torres-Padilla M.E.. Early embryonic-like cells are induced by downregulating replication-dependent chromatin assembly. Nat. Struct. Mol. Biol. 2015; 22:662–671. [DOI] [PubMed] [Google Scholar]
- 28. Cheloufi S., Elling U., Hopfgartner B., Jung Y.L., Murn J., Ninova M., Hubmann M., Badeaux A.I., Euong Ang C., Tenen D. et al.. The histone chaperone CAF-1 safeguards somatic cell identity. Nature. 2015; 528:218–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., Zhang F.. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013; 8:2281–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Behringer R., Gertsenstein M., Nagy K.V., Nagy A.. Differentiating mouse embryonic stem cells into embryoid bodies by Hanging-Drop cultures. Cold Spring Harb. Protoc. 2016; 2016:1073–1076. [DOI] [PubMed] [Google Scholar]
- 31. Dungrawala H., Rose K.L., Bhat K.P., Mohni K.N., Glick G.G., Couch F.B., Cortez D.. The replication checkpoint prevents two types of fork collapse without regulating replisome stability. Mol. Cell. 2015; 59:998–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Langmead B., Salzberg S.L.. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012; 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R. Genome Project Data Processing, S. . The sequence alignment/map format and SAMtools. Bioinformatics. 2009; 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Quinlan A.R., Hall I.M.. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010; 26:841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D.R., Pimentel H., Salzberg S.L., Rinn J.L., Pachter L.. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012; 7:562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S.L.. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013; 14:R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hoek M., Stillman B.. Chromatin assembly factor 1 is essential and couples chromatin assembly to DNA replication in vivo. Proc. Natl. Acad. Sci. U.S.A. 2003; 100:12183–12188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Houlard M., Berlivet S., Probst A.V., Quivy J.P., Hery P., Almouzni G., Gerard M.. CAF-1 is essential for heterochromatin organization in pluripotent embryonic cells. PLoS Genet. 2006; 2:e181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Martin G.R., Evans M.J.. Differentiation of clonal lines of teratocarcinoma cells: formation of embryoid bodies in vitro. Proc. Natl. Acad. Sci. U.S.A. 1975; 72:1441–1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Momcilovic O., Knobloch L., Fornsaglio J., Varum S., Easley C., Schatten G.. DNA damage responses in human induced pluripotent stem cells and embryonic stem cells. PLoS One. 2010; 5:e13410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tichy E.D., Pillai R., Deng L., Liang L., Tischfield J., Schwemberger S.J., Babcock G.F., Stambrook P.J.. Mouse embryonic stem cells, but not somatic cells, predominantly use homologous recombination to repair double-strand DNA breaks. Stem Cells Dev. 2010; 19:1699–1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Semrau S., Goldmann J.E., Soumillon M., Mikkelsen T.S., Jaenisch R., van Oudenaarden A.. Dynamics of lineage commitment revealed by single-cell transcriptomics of differentiating embryonic stem cells. Nat. Commun. 2017; 8:1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kalkan T., Olova N., Roode M., Mulas C., Lee H.J., Nett I., Marks H., Walker R., Stunnenberg H.G., Lilley K.S. et al.. Tracking the embryonic stem cell transition from ground state pluripotency. Development. 2017; 144:1221–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kalkan T., Smith A.. Mapping the route from naive pluripotency to lineage specification. Philos.Trans. R. Soc. Lond. B Biol. Sci. 2014; 369:20130540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hotta A., Cheung A.Y., Farra N., Vijayaragavan K., Seguin C.A., Draper J.S., Pasceri P., Maksakova I.A., Mager D.L., Rossant J. et al.. Isolation of human iPS cells using EOS lentiviral vectors to select for pluripotency. Nat. Methods. 2009; 6:370–376. [DOI] [PubMed] [Google Scholar]
- 46. Ye X., Franco A.A., Santos H., Nelson D.M., Kaufman P.D., Adams P.D.. Defective S phase chromatin assembly causes DNA damage, activation of the S phase checkpoint, and S phase arrest. Mol. Cell. 2003; 11:341–351. [DOI] [PubMed] [Google Scholar]
- 47. Herz H.M., Garruss A., Shilatifard A.. SET for life: biochemical activities and biological functions of SET domain-containing proteins. Trends Biochem. Sci. 2013; 38:621–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Cao R., Wang L., Wang H., Xia L., Erdjument-Bromage H., Tempst P., Jones R.S., Zhang Y.. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002; 298:1039–1043. [DOI] [PubMed] [Google Scholar]
- 49. Yuan W., Wu T., Fu H., Dai C., Wu H., Liu N., Li X., Xu M., Zhang Z., Niu T. et al.. Dense chromatin activates Polycomb repressive complex 2 to regulate H3 lysine 27 methylation. Science. 2012; 337:971–975. [DOI] [PubMed] [Google Scholar]
- 50. Tamburini B.A., Carson J.J., Linger J.G., Tyler J.K.. Dominant mutants of the Saccharomyces cerevisiae ASF1 histone chaperone bypass the need for CAF-1 in transcriptional silencing by altering histone and Sir protein recruitment. Genetics. 2006; 173:599–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Verreault A., Kaufman P.D., Kobayashi R., Stillman B.. Nucleosome assembly by a complex of CAF-1 and acetylated histones H3/H4. Cell. 1996; 87:95–104. [DOI] [PubMed] [Google Scholar]
- 52. Loyola A., Tagami H., Bonaldi T., Roche D., Quivy J.P., Imhof A., Nakatani Y., Dent S.Y., Almouzni G.. The HP1alpha-CAF1-SetDB1-containing complex provides H3K9me1 for Suv39-mediated K9me3 in pericentric heterochromatin. EMBO Rep. 2009; 10:769–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Alabert C., Barth T.K., Reveron-Gomez N., Sidoli S., Schmidt A., Jensen O.N., Imhof A., Groth A.. Two distinct modes for propagation of histone PTMs across the cell cycle. Genes Dev. 2015; 29:585–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hansen K.H., Bracken A.P., Pasini D., Dietrich N., Gehani S.S., Monrad A., Rappsilber J., Lerdrup M., Helin K.. A model for transmission of the H3K27me3 epigenetic mark. Nat. Cell Biol. 2008; 10:1291–1300. [DOI] [PubMed] [Google Scholar]
- 55. Zhang Y., Ng H.H., Erdjument-Bromage H., Tempst P., Bird A., Reinberg D.. Analysis of the NuRD subunits reveals a histone deacetylase core complex and a connection with DNA methylation. Genes Dev. 1999; 13:1924–1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zhang Y., LeRoy G., Seelig H.P., Lane W.S., Reinberg D.. The dermatomyositis-specific autoantigen Mi2 is a component of a complex containing histone deacetylase and nucleosome remodeling activities. Cell. 1998; 95:279–289. [DOI] [PubMed] [Google Scholar]
- 57. Zhang Y., Sun Z.W., Iratni R., Erdjument-Bromage H., Tempst P., Hampsey M., Reinberg D.. SAP30, a novel protein conserved between human and yeast, is a component of a histone deacetylase complex. Mol. Cell. 1998; 1:1021–1031. [DOI] [PubMed] [Google Scholar]
- 58. Li Q., Zhou H., Wurtele H., Davies B., Horazdovsky B., Verreault A., Zhang Z.. Acetylation of histone H3 lysine 56 regulates replication-coupled nucleosome assembly. Cell. 2008; 134:244–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Hondele M., Stuwe T., Hassler M., Halbach F., Bowman A., Zhang E.T., Nijmeijer B., Kotthoff C., Rybin V., Amlacher S. et al.. Structural basis of histone H2A-H2B recognition by the essential chaperone FACT. Nature. 2013; 499:111–114. [DOI] [PubMed] [Google Scholar]
- 60. Yang J., Zhang X., Feng J., Leng H., Li S., Xiao J., Liu S., Xu Z., Xu J., Li D. et al.. The histone chaperone FACT contributes to DNA replication-coupled nucleosome assembly. Cell Rep. 2016; 14:1128–1141. [DOI] [PubMed] [Google Scholar]
- 61. Belotserkovskaya R., Oh S., Bondarenko V.A., Orphanides G., Studitsky V.M., Reinberg D.. FACT facilitates transcription-dependent nucleosome alteration. Science. 2003; 301:1090–1093. [DOI] [PubMed] [Google Scholar]
- 62. Winkler D.D., Muthurajan U.M., Hieb A.R., Luger K.. Histone chaperone FACT coordinates nucleosome interaction through multiple synergistic binding events. J. Biol. Chem. 2011; 286:41883–41892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Drane P., Ouararhni K., Depaux A., Shuaib M., Hamiche A.. The death-associated protein DAXX is a novel histone chaperone involved in the replication-independent deposition of H3.3. Genes Dev. 2010; 24:1253–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Goldberg A.D., Banaszynski L.A., Noh K.M., Lewis P.W., Elsaesser S.J., Stadler S., Dewell S., Law M., Guo X., Li X. et al.. Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell. 2010; 140:678–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Ishov A.M., Vladimirova O.V., Maul G.G.. Heterochromatin and ND10 are cell-cycle regulated and phosphorylation-dependent alternate nuclear sites of the transcription repressor Daxx and SWI/SNF protein ATRX. J. Cell Sci. 2004; 117:3807–3820. [DOI] [PubMed] [Google Scholar]
- 66. Moggs J.G., Grandi P., Quivy J.P., Jonsson Z.O., Hubscher U., Becker P.B., Almouzni G.. A CAF-1-PCNA-mediated chromatin assembly pathway triggered by sensing DNA damage. Mol. Cell Biol. 2000; 20:1206–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Pillus L., Rine J.. Epigenetic inheritance of transcriptional states in S. cerevisiae. Cell. 1989; 59:637–647. [DOI] [PubMed] [Google Scholar]
- 68. Zhang Z., Hayashi M.K., Merkel O., Stillman B., Xu R.M.. Structure and function of the BAH-containing domain of Orc1p in epigenetic silencing. EMBO J. 2002; 21:4600–4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Yang B.X., El Farran C.A., Guo H.C., Yu T., Fang H.T., Wang H.F., Schlesinger S., Seah Y.F., Goh G.Y., Neo S.P. et al.. Systematic identification of factors for provirus silencing in embryonic stem cells. Cell. 2015; 163:230–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Adamo A., Sese B., Boue S., Castano J., Paramonov I., Barrero M.J., Izpisua Belmonte J.C.. LSD1 regulates the balance between self-renewal and differentiation in human embryonic stem cells. Nat. Cell Biol. 2011; 13:652–659. [DOI] [PubMed] [Google Scholar]
- 71. Foster C.T., Dovey O.M., Lezina L., Luo J.L., Gant T.W., Barlev N., Bradley A., Cowley S.M.. Lysine-specific demethylase 1 regulates the embryonic transcriptome and CoREST stability. Mol. Cell Biol. 2010; 30:4851–4863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Volk A., Liang K., Suraneni P., Li X., Zhao J., Bulic M., Marshall S., Pulakanti K., Malinge S., Taub J. et al.. A CHAF1B-dependent molecular switch in hematopoiesis and leukemia pathogenesis. Cancer Cell. 2018; 34:707–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw data have been deposited in the GEO database with accession number GSE130706.