Abstract
Ring-shaped replicative helicases are hexameric and play a key role in cellular DNA replication. Despite their importance, our understanding of the unwinding mechanism of replicative helicases is far from perfect. Bovine papillomavirus E1 is one of the best-known model systems for replicative helicases. E1 is a multifunctional initiator that senses and melts the viral origin and unwinds DNA. Here, we study the unwinding mechanism of E1 at the single-molecule level using magnetic tweezers. The result reveals that E1 as a single hexamer is a poorly processive helicase with a low unwinding rate. Tension on the DNA strands impedes unwinding, indicating that the helicase interacts strongly with both DNA strands at the junction. While investigating the interaction at a high force (26–30 pN), we discovered that E1 encircles dsDNA. By comparing with the E1 construct without a DNA binding domain, we propose two possible encircling modes of E1 during active unwinding.
INTRODUCTION
DNA replication is the most essential biological process for proliferation of life. Replicative helicases play a key role in the unwinding of DNA to initiate replication and lead a parade of proteins required for downstream DNA synthesis. Because of the importance of replicative helicases, their structure and unwinding mechanism have been subject to intensive investigation. Bovine papillomavirus E1 is one of the best-known model systems for studying replicative helicases in eukaryotic cells with SV40 Large-T-antigen (LTag) (1,2). E1 is a multifunctional initiator protein that is required for the replication of viral episomes (3). During replication, E1 senses and melts the viral origin (ori) and unwinds DNA in the 3′-5′ direction (2–4). In contrast to prokaryotic replicative helicases, E1 and LTag have the same AAA+ adenosine triphosphatase (ATPase) domain as archaeal and eukaryotic replicative helicases (1). E1 is a homohexamer that consists solely of identical monomers, whereas eukaryotic replicative helicases are complexes composed of different types of subunits (5). Although E2, a well-known interaction partner of E1, is required for recruiting E1 to ori invivo, it is dispensable in invitro unwinding assays (6,7). The simpler constitution of this unwinding complex makes E1 useful as a model system for eukaryotic replicative helicases.
The domain of E1 is divided into an N-terminal regulatory region, a DNA binding domain (DBD) and a helicase domain (HD) (1,2). The N-terminal regulatory domain provides regulation invivo. It contains several sequence motifs necessary for nucleo-cytoplasmic shuttling of E1. The DBD recognizes and binds to the ori sequence. The HD is further divided into an oligomerization domain (OD) and an ATPase domain containing a flexible C-terminal tail. HD induces assembly into an oligomeric form and unwinds DNA by hydrolyzing ATP. E1 assembles either into a double hexamer (DH) in the presence of ori sequences and ATP or into a single hexamer (SH) in the presence of ssDNA (8–10). Both DH and SH have unwinding activity. Unlike DH, SH did not have sequence specificity. Unwinding efficiency of dsDNA without ori is, in general, very low because an active DH could not be formed (11). To unwind dsDNA without ori, a 3′ ssDNA tail onto which the SH can load is required (9,10). Active unwinding and translocation occur only when coupled with ATP hydrolysis (12).
All replicative helicases for cellular DNA replication adopt a hexameric ring shape (5,13–14). Although the constituent structural motifs differ across the hexameric helicases, the homology in three-dimensional (3D) structural architecture suggests a similar unwinding process (1,15–16). Roughly speaking, the unwinding process is initiated by the assembly of a hexameric helicase to the ori. After undergoing a conformational transition to an active form, it starts to unwind the DNA. Biochemical and structural studies of hexameric helicases have provided a wealth of information on the detailed mechanism; however, some key aspects are still elusive. Most archaeal and eukaryotic hexameric helicases are assembled as DHs at the ori (5,11). E1 and LTag can be assembled as either the SH or DH depending on the DNA substrate (5,11). Although the DH seems to be a major form upon assembly, it was not clear whether DH was maintained or separated into SHs during the active unwinding phase invivo. In fact, the DH and SH forms coexisted in the EM structure of the unwinding-active LTag complex (17). However, the flow-stretching experiment by Yardimci et al. clearly demonstrated that LTag hexamers separate during DNA replication by visualizing replication forks being apart (18). The evidence regarding E1 is still inconclusive because experimental techniques that can monitor oligomeric states in real time have not yet been applied and, in contrast to LTag, only the DH form was observed in similar EM studies (19). Regardless of whether hexameric units separate, there has been a unified consensus for unwinding mechanisms in a single hexameric unit. Most hexameric helicases have been believed to unwind DNA through a steric exclusion (SE) model (5,15,20). In the SE model, the helicase encircles one strand and separates it from the other strand by SE. An important breakthrough in understanding the unwinding mechanism of E1 was provided by the X-ray structure of E1 cocrystallized with ssDNA reported in 2006 (21). The structure revealed that ssDNA is indeed encircled by the helicase and passes through the central channel, which strongly supports the SE model. Single-molecule FRET assay results also suggested that E1 followed the SE model (22). The SE model treats the strand along which the helicase does not translocate [the non-translocating (NTL) strand] as a passive component resulting from unwinding. However, the possibility that the NTL strand interacts with helicases and thus affects unwinding has been raised over the past decade (23–26). A representative example is Saccharolobussolfataricus MCM (SsoMCM), whose structure revealed that the NTL strand was wrapped around the exterior of the helicase (27). This mode of unwinding was termed the steric exclusion and wrapping (SEW) model (20,28). On the other hand, central channels of some hexameric helicases are known to be too wide to accommodate only ssDNA, which implies that dsDNA can pass through the central channel (29,30). Recently, O’Donnell group proposed a modified SE (MSE) model after finding that some dsDNA in a replication fork penetrated into the central channel of the eukaryotic CMG complex (20,31). This finding indicates that the NTL strand not only interacts with the helicase but also is encircled by the helicase along with the translocating (TL) strand. This model differs from the SE model in that the duplex separates inside the helicase. The crystal structure of E1 with ssDNA (21) was obtained using the E1 construct without the N-terminal half, whereas a similar structure encircling dsDNA was found when full-length E1 was used with replication fork substrates (32). In addition, the NTL strand that entered the helicase appeared to be extruded from a side channel on the helicase wall. In fact, hexameric helicases have been known to possess positively charged side channels that can be used as an extrusion outlet for ssDNA (16,29,32–34). Langston et al. referred to the unwinding mode in which ssDNA is extruded from the side channel as a side channel extrusion (SCE) model (35). Both the MSE and SCE models posit dsDNA encircling, however, the NTL strand in MSE model is extruded from the central channel, not from the side channel. Although the possibility of an unwinding mechanism other than the SE model is being recognized, it has not been experimentally confirmed for E1, especially during the active unwinding of DNA.
Here, we investigate the unwinding mechanism of E1 at the single-molecule level using a magnetic tweezers assay. The results obtained with replication-fork-mimicking substrates reveal that the SH form of E1 has poor unwinding activity. Force negatively affects the unwinding activity, indicating that there is an interaction between the helicase and DNA strands. While investigating the interaction, we found that the dsDNA is encircled by E1. By comparing the results of E1HD lacking the DBD, we propose two possible structures of E1 that differ mainly in their encircling behavior.
MATERIALS AND METHODS
Purification of papillomavirus E1
E1 (DBD+HD, 128–605) and E1HD (HD only, 308–605) from bovine papillomavirus type 1 (Figure 1A) were expressed through Escherichiacoli strain BL21(DE3) as N-terminal GST fusions and purified by glutathione agarose affinity chromatography. The GST portion was cleaved with thrombin and purified by ion exchange chromatography. The detailed purification is described (10,36).
Figure 1.
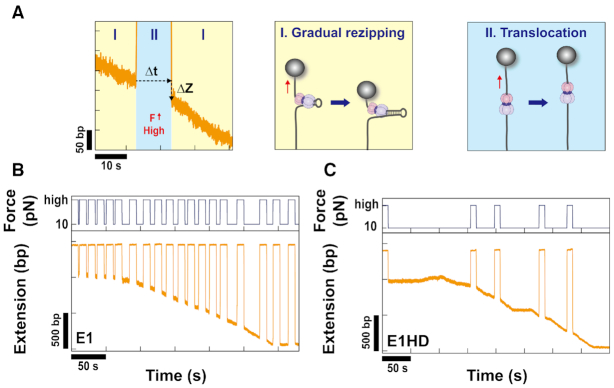
The characteristic unwinding dynamics of E1. (A) Two kinds of E1 constructs were used. One is E1 (residue 128–605), which consists of a DBD and an HD. The other is E1HD (residue 308–605), which contains only an HD. (B) Schematics of the magnetic tweezers assay to monitor the unwinding of E1. While HP is pulled by a pair of magnets at a force lower than Funzip, E1 monomers and ATP-Mg2+ are flushed into the chamber. E1 assembles into an active hexameric form and then begins to unwind HP. The resultant extension increase (Δz) is monitored and converted into the number of unwound base pairs. (C) A representative unwinding trace of E1 on HP50bp at 5 pN. E1 could fully unwind HP50bp. After reaching the maximum, HP was gradually rewound because of E1 translocating past the hairpin apex. (D) E1 showed diverse dynamics from monotonic single unwinding (upper two panel) to repetitive unwinding (lower two panel). Between two unwinding motions, repetitive motion is more dominant, comprising 65%. In repetitive motion, E1 moved back through either abrupt slippage or gradual rewinding. (E) To assess the unwinding processivity of E1, we performed the experiment with a longer HP (HP1kbp). The number of base pairs unwound by E1 within 500 s was broadly distributed from several tens (upper two panels) to more than 100 bp (lower panel) at 5 pN. However, the maximum number of unwound base pairs was, in general, below 180 bp.
Preparation of DNA hairpin constructs
The DNA hairpin construct (HP) was prepared as described previously (37). Two HPs with 50 and 1038 bp stems (HP50bp and HP1kbp, respectively) were used in the experiments. Each HP consists of a digoxigenin handle, an insert, and a core hairpin (Supplementary Figure S1). The digoxigenin handle is for immobilization to the anti-digoxigenin-coated glass surface. The insert between the digoxigenin handle and the core hairpin prevents magnetic beads from approaching the glass surface too closely. The core hairpin contains a single-stranded region (dT30) for helicase loading and a duplex stem that the helicase unwinds. At the 5′ end of the core hairpin, three biotin molecules are labeled for binding to streptavidin-coated magnetic beads, and at the other end, there is a 10 base overhang for ligation with the insert. The digoxigenin handle and the insert were prepared by digesting polymerase chain reaction (PCR) fragments with restriction endonucleases. In the case of the digoxigenin handle, PCR was carried out in the presence of digoxigenin-dUTP (Roche Diagnostics). The core hairpin of the HP50bp was made by annealing two oligonucleotides (Integrated DNA Technologies, Coralville, IA, USA). The GC content of the HP50bp stem is 32%. The core hairpin of HP1kbp was prepared by the ligation of a pre-annealed oligonucleotide, an ∼1 kbp PCR fragment and a self-annealed loop. The GC content of the HP1kbp stem is 47%. The whole construct was assembled by ligating the three parts with an appropriate complementary overhang and purified. All the sequences of the oligonucleotides are given in Supplementary Table S1.
Magnetic tweezers assay
Flow cells were assembled with two cover glasses and a sheet of parafilm and sealed by melting the parafilm. The upper glass had two holes for buffer exchange, and the bottom glass was functionalized with nitrocellulose (0.1% wt/vol in amyl acetate). Polystyrene beads 3.2 μm in diameter (Invitrogen), which served as a reference for drift correction, were injected into the flow cell and immobilized to the surface by heating at 100°C for 1 min. Then, the flow cells were incubated with 100 μg/ml anti-digoxigenin (Roche Diagnostics) at 4°C overnight. To reduce non-specific binding, the mixture was further incubated with 10 mg/ml bovine serum albumin (BSA) (New England Biolabs) for at least 1 h at room temperature. The flow cells were washed with 1× phosphate-buffered saline after each incubation step. Streptavidin-coated magnetic beads 2.8 μm in diameter (M280, Invitrogen) were mixed with the HP and incubated for 10 min. Unbound DNA was removed through pull-down steps. The bead–DNA hairpin complex was flushed into the flow cell. After incubation for 5 min, the unbound beads were thoroughly washed out with E1 binding buffer (E1BS buffer; 50 mM HEPES (pH 7.6), 100 mM NaCl, 5 mM MgCl2, 1 mM DTT, 0.005% Triton X-100, 0.1 mg/ml BSA).
A magnetic tweezers assay was performed at a home-built setup, which was constructed similarly to the one described in the previous paper (38). A pair of magnets made of NdFeB (W-05-N50-G, Supermagnete) were mounted on a linear motorized stage (M-126, Physik Instrumente) and placed above the flow cell. The force was controlled by changing the distance from the flow cell. To obtain the diffraction pattern, beads were illuminated with a visible LED light source (CR5111A-WY, Roithner Lasertechnik). The light collected through an oil-immersion objective lens (Olympus, N.A. 1.25) was imaged with a CCD camera (EoSens CL MC1362, Mikrotron) at 120 Hz. The x and y positions of a bead were obtained from the center of the diffraction ring. The Z position was determined by correlating a measured diffraction pattern with a series of reference patterns generated while changing the objective lens. The position of the objective lens was controlled by a piezoelectric stage (P-721, Physik Instrumente). Drift was corrected using the positions of reference beads. The force was calibrated as a function of distance between the magnets and flow cell for 11 kbp dsDNA molecules.
Helicase unwinding experiments
Before the unwinding experiments, all DNA hairpin molecules were tested to determine whether a sharp unzipping transition occurred near the unzipping force, Funzip (Supplementary Figure S2). Only molecules showing proper unzipping behavior were measured (Supplementary Figure S3). Unwinding was initiated by flushing E1BS buffer containing E1 helicase (140 nM) and ATP (2 mM) into HP molecules (Figure 1B). We occasionally injected E1 without ATP to examine only the assembly process. The concentration of E1 was kept as low as possible to avoid multiple-protein binding and non-specific association with DNA. The extension of a DNA bead was recorded at a force of 5–10 pN. The extension increase upon unwinding was converted to the number of unwound base pairs by using the elastic property of ssDNA calculated with the extensible FJC model (See Supplementary Figure S4). To calculate unwinding (UW) rates, raw data were filtered through a moving average with 120 window points. Only periods in which the unwinding was continuous were selected for calculation. We determined the processivity from the maximum number of base pairs that E1 unwound within ∼500 s. In the experiment to measure translocation activity, E1 or E1HD was injected while the HP was mechanically unzipped. After lowering the force, traces were recorded. When the helicase assembled to ssDNA past the HP loop, the HP was gradually rezipped at the wake of the translocating helicase. We periodically unzipped HP to compare gradual rezipping and translocation (TR) rates. The TR rate was calculated from the downward slopes. To investigate the interaction between the DNA strands and E1, we repetitively increased the force to a very high value (26 or 30 pN) while recording unwinding traces at 10 pN. The high force lasted ∼10 s.
RESULTS
Heterogeneous unwinding dynamics of E1 helicase
To initiate unwinding, E1 was injected into HP molecules with ATP at a force lower than Funzip (Figure 1B). If ori is present, E1 first binds to ori as a double trimer (DT). After locally melting ori, the DT is converted to an active DH and begins unwinding (39). In order to avoid interference by ori-melting and to focus the activity of SH, we used HP that does not contain BPV ori. Without ori, E1 will preferentially assembled around the single-stranded region of HP into a SH form. E1 SH has been known to occupy ∼30 bases (12). Therefore, each HP was able to accommodate only one hexamer. When E1 started to unwind HP, the extension gradually increased, as shown in Figure 1C. Without ATP, such increases were not observed. We first performed experiments with E1 and HP50bp to determine the overall characteristics of unwinding dynamics. Representative traces obtained at 5 pN are plotted in Figure 1D. The unwinding dynamics were diverse, ranging from a single monotonic unwinding (upper two panels in Figure 1D) to repetitive unwinding (lower two panels in Figure 1D). Because the repetitive unwinding continued after the removal of residual helicase, it was not due to dissociation and reassociation of E1. In the second panel of Figure 1D, the duplex was fully unwound in the first unwinding phase. E1 continuously translocated toward the 5′ end and the duplex was gradually rezipped behind E1. In this case, the unwinding did not resume and ended in a single monotonic event. Occasionally, E1 moved back to the original position before reaching full unwinding and resumed unwinding. Then repetitive unwinding as shown in lower two panels of Figure 1D appeared. Interestingly, the repetitive unwinding ceased once E1 fully unwound the duplex and reached a position at 5′ side through continuous translocation. It may be because E1 at 5′ side could not jump back to the original position at once. This diverse and heterogeneous dynamics is consistent with a previous single-molecule FRET assay result (22). To analyze the dynamics quantitatively, we calculated UW rates in the slopes during which unwinding was continuous. The UW rate at 5 pN was 0.90 ± 0.57 bp/s. While the UW rate was markedly lower than those of prokaryotic hexameric helicases such as T7 gp4, T4 gp41 and DnaB (30–50 bp/s) (40–42), it was comparable to that of LTag (1–2 bp/s) (43,44). In repetitive unwinding, E1 moved back either gradually or abruptly. Such backward movement is often observed when helicases fully unwind DNA hairpins because ds/ssDNA junctions to which helicases are stably anchored disappears (45). However, the backward movement of E1 occurred regardless of reaching full unwinding (see the first and the third panels in Figure 1D). This behavior is similar to that of RecQ helicases that continue unwinding, regressing back and resuming (37,45). The processivity was measured with the longer HP (HP1kbp). Although it was somewhat broadly distributed as presented in the traces in Figure 1E, the processivity of E1 did not exceed 180 bp, which was much lower than that of LTag with the similar UW rate. Under similar experimental configuration, LTag could unwind a few kilo-base pairs without dissociation (43,44). Archaeal MCMs are also known to be able to unwind at least 500 bp (46,47). This result reveals that E1, as an SH, is a poor helicase.
Force impedes the unwinding of E1
To understand the unwinding mechanism, we carried out experiments at various forces. The typical traces obtained with HP50bp are presented in Figure 2A. The UW rates and the processivity obtained at the various forces are summarized in Figure 2B and C. Helicases have been classified into active or passive helicases depending on the mechanism by which they separate a duplex. Passive helicases unwind DNA simply trapping transiently melted junctions by thermal fluctuation, whereas active helicases unwind DNA by lowering the energy barrier. Most hexameric helicases are passive or partially passive helicases (48,49). In passive helicases, the UW rate greatly increases as force increases because force facilitates the melting of the junction. To determine which type E1 is, we examined the unwinding under various forces. Unexpectedly, we found the opposite trend: the UW rate decreased as the force increased (Figure 2B). UW rate at 10 pN was 0.23 ± 0.16 bp/s, which is a fourth of that at 5 pN (0.90 ± 0.57 bp/s). Large error bars in the data were due to the heterogeneity of the dynamics. Considering the dependence of the UW rate on force, E1 could not be classified as a passive helicase. Next, we examined whether E1 is an active helicase. According to the criterion that Manosas et al. proposed (48), helicases whose UW rate is higher than a quarter of the TR rate are considered active helicases. The TR rate determined from the downward slope in Figure 3A and B was 4.19 ± 1.88 bp/s at 10 pN. Because the UW rate was lower than a quarter of the TR rate at all forces, E1 did not fit the active model either. In general, the UW rate increases as force increases, although the sensitivity is different depending on whether they are active helicases or passive helicases. For example, Mode I of E1HD, which will be discussed later, shows the behavior of active helicases. The UW rate and processivity of E1HD Mode I increased moderately as the force increased, as shown in Figure 2B and C. Although the data on E1 alone seemed somewhat scattered due to large error, the negative effect of force was apparent when the data were compared to those of E1HD Mode I. A similar trend was also observed in the RW rate (Supplementary Figure S5A) and overall time for full unwinding (Supplementary Figure S5B). The fact that force applied to two DNA strands in the opposite direction impeded unwinding indicated that the helicase interacted with the NTL strand which was not encircled by the helicase. A trial to classify E1 as an active or passive helicase failed, probably because the model does not take into account such interaction. The negative effect of force was also reported for the T4 gp41 and DnaB hexameric helicases, which reached the same conclusion (26,42).
Figure 2.
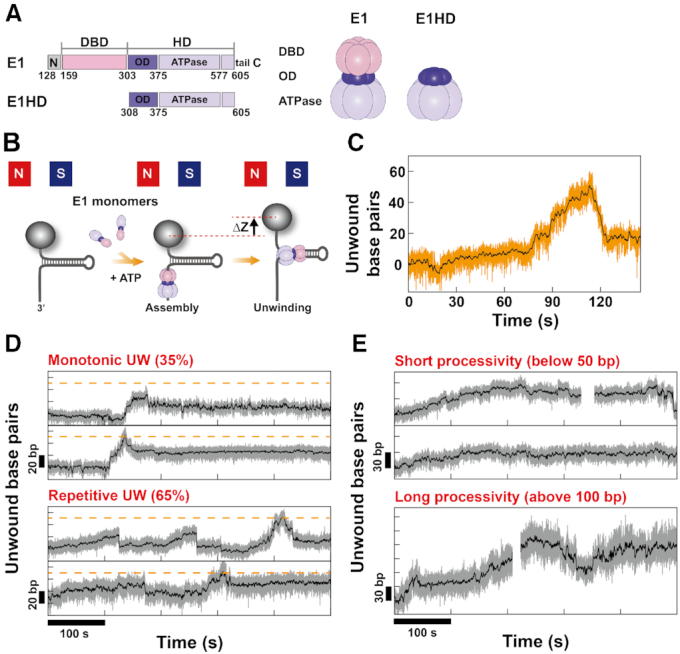
Effect of force on the unwinding activity of E1. (A) Representative unwinding traces of E1 obtained with HP50bp at various forces. (B) The unwinding (UW) rates of E1 for HP50bp (solid gray squares) and HP1kbp (solid blue triangles) were measured as a function of force. For comparison, the UW rates of E1HD for HP1kbp (solid orange circles for Mode I and a solid pink circle for Mode II) were plotted. For calculation of the UW rate, a total of 205 slopes from 96 HP molecules were analyzed, and more than eight events were used per data point. The UW rate of E1 decreased as the force increased regardless of the length of HP, which indicates the negative effect of force. Specifically, the UW rate for HP50bp was 0.90 ± 0.57 bp/s (S.D. for N = 20 events) at 5 pN and 0.23 ± 0.16 bp/s (S.D. for N = 11 events) at 10 pN. The UW rates of E1HD Mode I (solid orange circles) were larger than those of E1 at all force ranges. In contrast to E1, the UW rate of E1HD Mode I increased as force increased, although the sensitivity was not very high due to the active character of the helicase. The UW rate of E1HD Mode II (a solid pink circle) measured at 10 pN was similar to that of E1. (C) The maximum unwound base pairs within a given time (processivity) of E1 and E1HD on HP1kbp were collected at various forces for 29 and 14 HP molecules, respectively. The processivity of E1 was poor at all forces, whereas that of E1HD was very high and further enhanced by increasing force, reaching 1 kbp at 10 pN.
Figure 3.
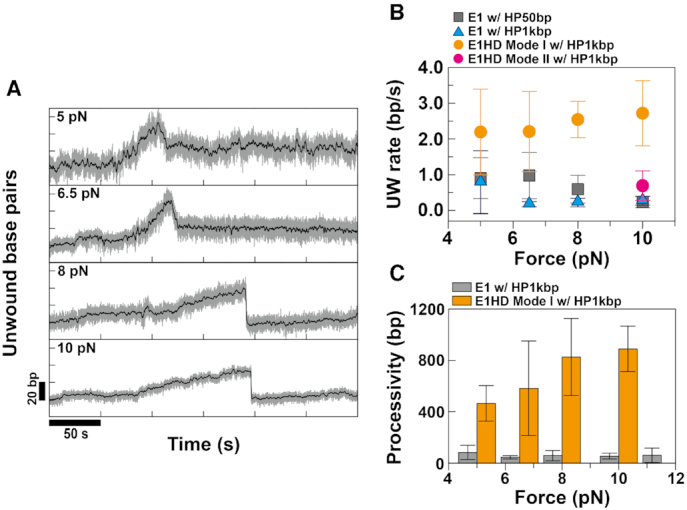
The translocation of E1 and E1HD on single-stranded DNA. (A) To monitor translocation activity, E1 was injected while HP1kbp was mechanically unzipped. After waiting for E1 to assemble onto single-stranded DNA, we lowered the force below Funzip and recorded traces. When E1 was bound to the DNA past the hairpin loop, a gradual decrease in extension was observed because of rezipping following the TL helicase as in phase (I). To compare gradual rezipping (I) with translocation along ssDNA (II), we periodically unzipped HP for ∼10 s. Displacement in z (Δz) during the unzipping period is an indication of the translocation of E1 along ssDNA. TR rates were calculated either from the downward slopes in phase (I) or from |Δz|/Δt in phase (II). TR rates calculated from phases (I) and (II) were almost identical. Translocation traces of (B) E1 and (C) E1HD. The TR rates of E1 and E1HD calculated from the traces were 4.19 ± 1.88 and 4.47 ± 1.29 bp/s, respectively, which are consistent within the error.
Double-stranded DNA is encircled by E1
From the negative effect of force on unwinding, we found that there was an interaction between the NTL strand and the helicase. To elucidate the nature of the interaction, we applied a high force to HP while recording traces at 10 pN. In this experiment, E1 alone was injected, and later ATP was added to separately monitor assembly and unwinding processes. Without E1, bare HP (HP1kbp) was rapidly unzipped at a higher force than Funzip and re-zipped at a lower force (Supplementary Figure S3). However, once E1 was injected, the HP was never unzipped, even at 26 pN, which is much higher than Funzip. HP remained closed while active unwinding occurred by the addition of ATP (see Figure 4A). The fact that E1 completely blocked the separation of the junction indicates that E1 bound strongly to the NTL strand in addition to the encircled TL strand. A slight increase in extension at high force was due to the elastic elongation of dsDNA part inserted between core hairpin and digoxigenin handle (see Supplementary Figure S1). As mentioned previously, the SEW model based on SsoMCM results proposed that the unwound NTL strand wraps around the helicase. In SsoMCM, the wrapping was found to be held via electrostatic interaction, which was presumed to be weak to enable unwound strand to slide over the exterior of the helicase (27). If the NTL strand is bound by wrapping as in the SEW model, it would not withstand such a high force. Another basis for this reasoning is that the strong gripping behavior of E1 was observed without ATP and therefore without substantial unwinding. Then, the only available ssDNA was 5 bases at the 5′ end of the HP core, which was too short to wrap around the helicase. Therefore, it is unlikely that the NTL strand was grabbed by the helicase through exterior wrapping. In the MSE model mentioned in the ‘Introduction' section, one strand of the entered dsDNA (the NTL strand) is re-extruded from the entrance (31). There may be an interaction between the NTL strand and the inner surface of the helicase. Similar to the SEW model, the interaction should not be strong for the NTL strand to continuously escape from the helicase. In cryo-EM of CMG helicase that follows the MSE model, the extruded strand could not be visualized due to highly mobility (31). The highly mobile NTL strand is not compatible with the strong interaction observed in this work. Therefore, the remaining possibility is that the NTL strand is encircled by E1 and extruded through a side channel on the helicase wall, as suggested by Chaban et al. (32). Once the NTL strand is encircled in this way, mechanical force cannot easily pull it out of the helicase without rupturing the helicase ring.
Figure 4.
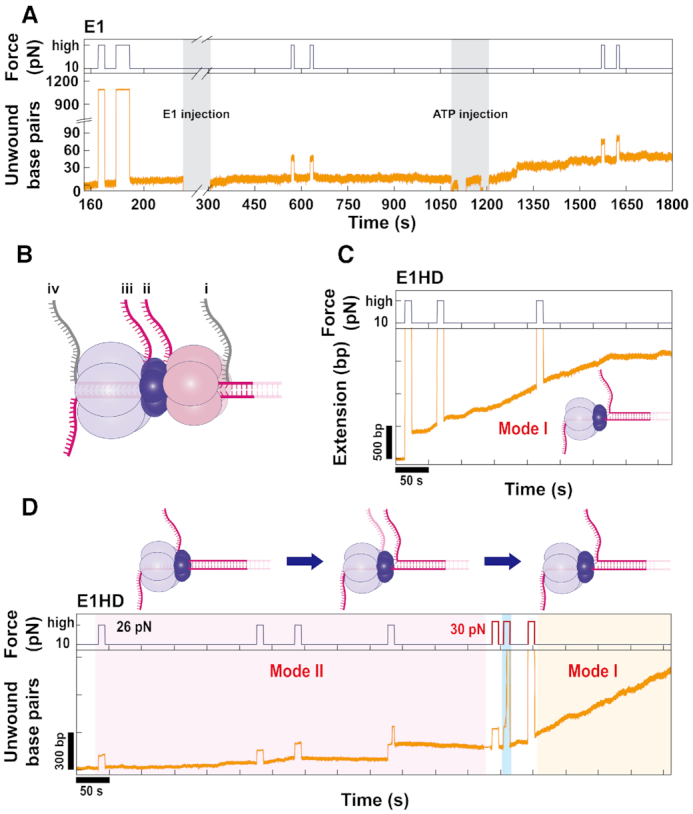
Encircling of dsDNA by E1 observed during the periodic application of a high force (>Funzip). (A) A high force (26 pN) was frequently applied for 10 s while recording unwinding traces at 10 pN for HP1kbp. Before the injection of E1, HP was fully unzipped at high force in a reversible way. However, after the injection of E1, HP could not be unzipped at the high force without ATP and remained closed after the addition of ATP. This strong grip on both strands of HP indicates dsDNA encircling by E1. (B) All possible extrusion pathways of the NTL strand from E1 during unwinding. Pathway (i) is before entering the helicase, while pathways (ii) and (iii) are from a side channel between the DBD and OD tiers and before the ATPase tier, respectively. Pathway (iv) shows that the NTL strand passes all through the central channel together with the TL strand. Pathway (i) was excluded on the basis of observation of strong grip behavior, and pathway (iv) was excluded because of the lack of dsDNA translocation activity of E1. Therefore, the most likely pathway would be either (ii) or (iii), which are drawn in pink. (C) A representative trace of the first mode (Mode I) between two different unwinding modes observed in experiments of E1HD. In contrast to E1, E1HD did not prevent full unzipping of HP at the high force. The UW rate was 2.72 ± 0.91 bp/s, which is higher than that of E1. E1HD in Mode I could processively unwind HP1kbp without pauses. (D) A trace of E1HD in the second mode (Mode II) was observed at first and converted into Mode I later. Mode II of E1HD showed very similar characteristics to those of E1, such as a low UW rate and strong grip at high force. Occasionally, sudden jumps in extension as shown in the middle of the trace were observed during the application of the high force. We suspect that DNA was dragged out of the helicase because of the high force. Interestingly, E1HD in the Mode II state changed to that in the Mode I state, showing fast unwinding and full unzipping when slightly higher force of 30 pN was repeatedly applied.
Two unwinding modes by E1HD reveal the unwinding mechanism of E1
The strong grip behavior indicated that dsDNA was encircled by E1 and that the NTL strand was extruded through a side channel. According to a single-molecule FRET study, the ds/ssDNA junction is located at the N-terminal side of helicase (22). Since our E1 construct did not contain the N-terminal regulatory domain, the dsDNA should enter the DBD tier. While the entry position seemed clear, the position from which the NTL strand is extruded was still elusive. All possible extrusion pathways are illustrated in Figure 4B. Re-extrusion from the entrance as in the MSE model could be excluded as mentioned above. A strong grip can be observed when the NTL strand passes through the central channel to exit at the ATPase side (Figure 4B, pathway (iv)). Although some hexameric helicases possess dsDNA translocation activity (50,51), it has not been reported for E1. In addition, if both strands exited from the ATPase side, unwinding could not be observed in our experiment because the DNA would immediately rehybridize in the wake of the helicase. Therefore, it is highly unlikely that the NTL strand exits from the ATPase side. Considering the overall 3D structure, the position from which the NTL strand can be extruded is a side channel between the DBD and ATPase domains (Figure 4B).
To further investigate dsDNA encircling, we performed measurements for E1HD lacking the DBD. When E1HD alone was injected into HP1kbp without ATP, strong grip behavior was observed at a high E1HD concentration of 1.3 μM (Supplementary Figure S6). Unfortunately, we could not observe active unwinding after adding ATP. Therefore, it was not certain whether the E1HD that strongly grabbed HP was an unwinding-competent form. However, when E1HD was injected simultaneously with ATP, active unwinding occurred, as shown in Figure 4C. The unwinding dynamics and grip behavior of E1HD were more complex than those of E1. After careful analysis, we found that two different unwinding modes coexisted in E1HD with similar probability. We divided them into ‘Mode I’ and ‘Mode II’ according to grip behavior. In Mode I, HP underwent reversible unzipping at 26 pN (Figure 4C). This behavior indicated that there was no strong interaction between the NTL strand and E1HD. To further characterize Mode I, we analyzed the UW and TR rates, and the processivity. The UW rate of Mode I was 2.72 ± 0.91 bp/s at 10 pN, which was 10 times higher than that of E1 (Figure 2B). In addition, Mode I was much more processive, being able to fully unwind HP1kbp (Figure 2C). The TR rates calculated from the slopes shown in Figure 3C were 4.47 ± 1.29 bp/s at 10 pN. Comparing UW and TR rates, Mode I can be classified as an active helicase. The disappearance of strong grip behavior upon removal of the DBD means that the NTL strand was encircled by the DBD in E1. Confusingly, E1HD often showed another dynamic, Mode II. In Mode II, HP was not unzipped at 26 pN (Figure 4D), indicating dsDNA encirclement. Although the UW rate (0.69 ± 0.41 bp/s) was slightly higher (Figure 2B), the overall characteristics of Mode II were very similar to those of E1. Comparing the results of E1HD Mode II and E1, we reached the completely different conclusion that the effect of the DBD on unwinding was minimal, and the NTL strand was still encircled by E1HD. The observation that the removal of DBD resulted in both Mode I and Mode II implies that there are two different conformations in E1, which differ mainly in the encircling structure. Interestingly, we found that HP showing Mode II dynamics could convert to Mode I, losing its strong grip on the NTL strand, when 30 pN was repetitively applied (Figure 4D). This result demonstrates that the two modes originated from a species with the same chemical composition. Based on our results, high force seemed to release the NTL strand from the helicase by partially rupturing the helicase ring.
DISCUSSION
Using a magnetic tweezers assay, we investigated the unwinding dynamics of E1 for replication fork DNA substrates. It was found that E1, as an SH, is a poor helicase with a low unwinding rate and low processivity. In particular, the processivity of E1 was at most ∼180 bp, which is in stark contrast to the highly processive LTag that can continuously unwind ∼2 kbp (43,44). To efficiently replicate a viral episome of 6–7 kbp, other factors to boost unwinding activity are probably needed. E1 is assembled to viral ori in a DH form. Dimerization into DH might be such a boosting factor. This is in line with recent observations that a change of oligomerization is an important regulatory mechanism to modulate the activity and functionality of helicases (49). Because SH does not have sequence specificity, it can randomly unwind DNA wherever there is a forked junction. The poor unwinding activity of E1 SH seems to suppress unwinding occurring at sites other than ori without proper coordination of two E1 hexamers and related proteins. This would be especially important because in its latent stage, the number of viral episomes is at most ∼100 copies (52), which is extremely small compared to cellular DNA. Dimerization of LTag has been known to be essential for DNA replication (53,54). However, the strong processivity of LTag as a SH form implies that dimerization is not a prerequisite for robust DNA replication once ori is melted and two forks are established (18). Aside from dimerization, interaction with other proteins can be a stimulatory factor, as shown in elF4A DEAD box helicase and WRN (37,55). E2 is a representative protein that interacts with E1. Although E2 is required for recruiting E1 to ori invivo, it does not itself increase unwinding activity (7,56). Replication protein A (RPA), a eukaryotic ssDNA binding protein, interacts with many helicases, including E1 (57). One striking example is the WRN helicase, whose processivity is dramatically increased by RPA (37). However, E1 has been known not to be stimulated by RPA (58). Instead, E1 and RPA compete for ssDNA binding. For efficient DNA replication, the activity of replicative helicase is, in general, linked to that of DNA polymerase (59). Association with polymerase can increase the processivity of the replicative helicase. In fact, the processivity of polymerase ϵ has been known to be stimulated by E1 (60). However, the contrary, namely, the stimulation of E1 by polymerases, has not yet been reported.
From the negative force effect, we speculated that there was an interaction between the NTL strand and the helicase. While examining the interaction, we discovered that both DNA strands were encircled by the helicase. Although E1 DH is known to encircle dsDNA during ori-melting (11,16), it has been unclear whether the dsDNA remains encircled after active unwinding is initiated. The EM structure of full-length E1 by Chaban et al. (32) revealed that a short stretch of dsDNA in a replication fork was encircled by the helicase. The authors also suggested that the encircled NTL strand is extruded through a side channel between the DBD and OD tiers. On the basis of the structure, they predicted that E1 would follow the SE model if the N-terminal half was removed (Figure 5A, left). Our experiment using E1HD yielded mixed results, half of which agreed with the prediction, and the other half did not. There were two unwinding modes in E1HD: Mode I exhibited reversible HP unzipping and fast unwinding, and Mode II showed similar characteristics to E1. The absence of strong grip interaction in Mode I seemed to agree with the prediction by Chaban et al. Therefore, it is reasonable to posit that E1 adapts the structure the authors suggested (Figure 5B, left). In contrast, Mode II, which resembles E1, indicated that the DBD only slightly affected unwinding dynamics and that dsDNA was encircled by other domain tiers in E1HD. The first N-tier that dsDNA should enter in E1HD is OD tier (22). However, according to the crystal structure of apo-E1, the channel diameter at the OD tier is ∼13 Å (61), which is small to accommodate dsDNA of ∼20 Å. The recent high-resolution structures of LTag and MCM with dsDNA have revealed a few interesting features (62,63). In particular, it should be noted that LTag was able to encircle dsDNA throughout the central channel despite a small channel diameter (14–19 Å) at the ATPase tier. Surprisingly, the narrow channel appeared to squeeze and deform the dsDNA, breaking the Watson–Crick base pairing. We speculate that the OD tier of E1HD can encircle dsDNA via a similar mechanism; OD domains assembled around dsDNA partially melt dsDNA by forming a rigid collar. Because the NTL strand will not completely pass through the central channel, as explained in the ‘Results’ section, it is reasonable to assume that the melted NTL strand is extruded through a side channel before the ATPase tiers (Figure 5A, right). In summary, the second possible structure of E1 based on E1HD Mode II will be as follows. First, dsDNA is encircled by DBD and OD tiers, partially melts at the OD tier and then one of the melted strands is extruded through a side channel before ATPase tier (Figure 5B, right). The coexistence of Mode I and Mode II suggests that E1 can adapt either of the two structures described above. However, we think that the second (Figure 5B, right) is more probable considering the close similarity in unwinding dynamics between E1 and Mode II of E1HD. Another basis for our presumption is that if dsDNA was encircled only by the DBD, it could not withstand such high forces because the intersubunit interaction between six DBDs is known to be quite weak (64). However, the DNA binding ability of the DBD could help relatively small OD domains encircle dsDNA by bringing two DNA strands closer. Therefore, removal of the DBD would increase the population in Mode I.
Figure 5.
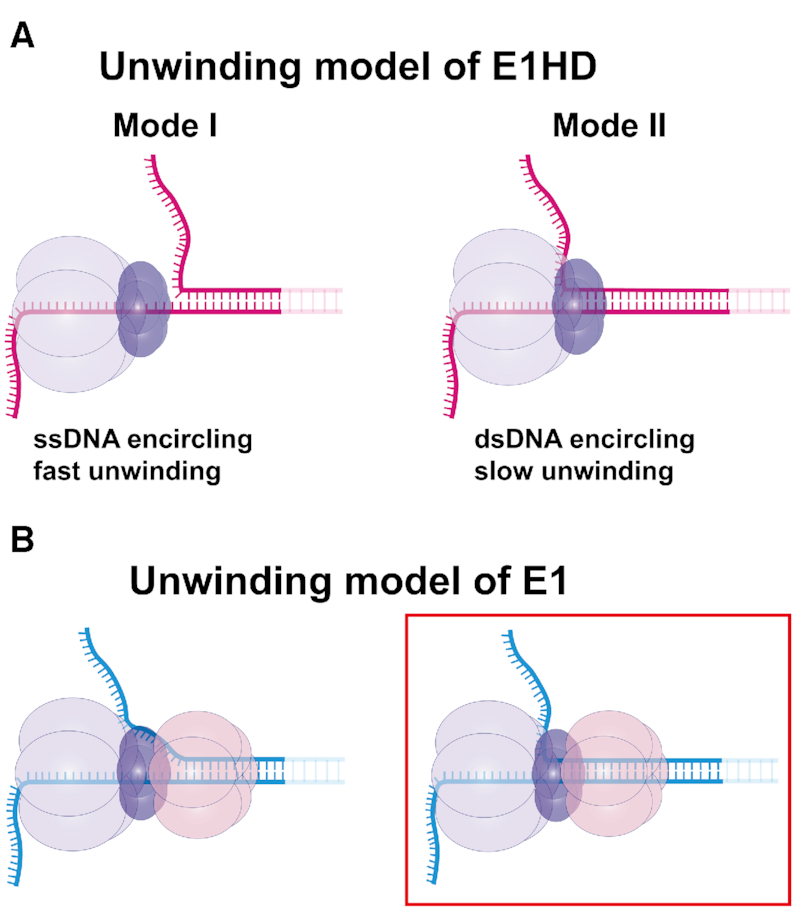
Expected unwinding models of E1 and E1HD. (A) Two unwinding models of E1HD based on characteristics of two different unwinding modes observed in this study. In the structure based on Mode I (left), the NTL strand is excluded before the helicase. Because the OD collar is rigid and has a small hollow diameter, unwinding is likely to follow the SE model in which the OD tier excludes the NTL strand. In contrast, in the structure based on Mode II (right), dsDNA is encircled by the OD tier and the NTL strand is extruded from a side channel before the ATPase tier. At the assembly of a hexamer, dsDNA seems to be partially melted by a strong oligomerization force between ODs. This model is different from the SE model in the point at which duplex separation occurs inside the helicase. (B) Proposed unwinding models of E1. The strong grip behavior observed in E1 indicates that at least one domain tier should encircle dsDNA. Considering the encircling of dsDNA and the structures of E1HD, we propose two different unwinding structures of E1. The main difference between the structures is where the NTL strand is extruded. In the left structure, the NTL strand is extruded before entering the OD tier. In the right structure, the NTL strand is extruded after the OD tier. We think that the right model is more probable considering the similarity between the dynamics of E1 and of E1HD Mode II.
It should be noted that the unwinding was fast and processive when ssDNA is encircled (E1HD Mode I), whereas it was slow and poorly processive when dsDNA was encircled (E1 and E1HD Mode II). However, the differences in UW rate and processivity might not be as large in invivo where force is absent or low, given the tendency shown in Figure 2B and C. Biochemical assays reported that processivity of E1 and E1HD was significantly different; E1HD unwound longer than 1 kbp, whereas E1 unwound at most 0.2 kbp (12,22). Mode I of E1HD is probably responsible for the strong processivity observed with E1HD. As revealed in biochemical assays performed without force, E1 was poorer in activity than E1HD Mode I even at zero-force. The poor unwinding activity of E1 seems to be due to the difficulty with which dsDNA passes through the narrow central channel, especially at the OD tier. Encircling dsDNA can act as a regulatory mechanism to decelerate unwinding activity. We imagine that the partial opening of the N-tier will be a switch to convert from a poor unwinding mode to a robust unwinding mode as the force of 30 pN does by rupturing the helicase ring and pulling a strand out of it. Mcm10 has been reported to greatly stimulate the unwinding activity of the CMG complex, and at the same time, it enabled the CMG complex that stalled at a block on the NTL strand to bypass the block (65). Based on our results, we presume that Mcm10 promoted the unwinding activity of the CMG complex by converting it from the slow dsDNA encircling mode to the fast SE mode. Like RecQ helicases, E1 often underwent repetitive unwinding, although the degree of repetition was much lower than that of RecQ. RecQ catalyzes various secondary structures of DNA in a multimeric form. The repetitive unwinding of RecQ helicases was attributed to synchronization with other subunits, waiting for others to bind to DNA (37). Analogously, that of E1 could be an action to harmonize with another hexamer or a replication complex. Regardless of which one E1 adopts between the two structures we proposed, dsDNA is separated inside the helicase, which does not agree with the generally accepted SE model. We believe that dsDNA encircling is not limited to viral helicases because high-resolution structural data and refined biochemical assays on eukaryotic hexameric helicases have also been prompting modifications to existing SE models.
Supplementary Material
ACKNOWLEDGEMENTS
Prof. Bong Hyun Chung devoted his whole life to being a passionate principal investigator. The authors would like to dedicate this paper to Prof. Bong Hyun Chung who passed away on 24 February 2016 in the middle of this project. Additionally, we appreciate Prof. Arne Stenlund (Cold Spring Harbor Laboratory) for providing E1 and E1HD proteins and Dr Seung-Jae Lee for valuable advices about preliminary experiments.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Development of Platform Technology for Innovative Medical Measurements Program [KRISS-2019-GP2019-0013]; Research Initiative Program [KRIBB]; R&D Convergence Program [CAP-14-3-KRISS, NST]. Funding for open access charge: R&D Convergence Program [CAP-14-3-KRISS, NST].
Conflict of interest statement. None declared.
REFERENCES
- 1. Hickman A.B., Dyda F.. Binding and unwinding: SF3 viral helicases. Curr. Opin. Struct. Biol. 2005; 15:77–85. [DOI] [PubMed] [Google Scholar]
- 2. Bergvall M., Melendy T., Archambault J.. The E1 proteins. Virology. 2013; 445:35–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Seo Y.S., Müller F., Lusky M., Hurwitz J.. Bovine papilloma virus (BPV)-encoded E1 protein contains multiple activities required for BPV DNA replication. Proc. Natl. Acad. Sci. U.S.A. 1993; 90:702–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yang L., Mohr I., Fouts E., Lim D.A., Nohaile M., Botchan M.. The E1 protein of bovine papilloma virus 1 is an ATP-dependent DNA helicase. Proc. Natl. Acad. Sci. U.S.A. 1993; 90:5086–5090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. O’Donnell M.E., Li H.. The ring-shaped hexameric helicases that function at DNA replication forks. Nat. Struct. Mol. Biol. 2018; 25:122–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yang L., Li R., Mohr I.J., Clark R., Botchan M.R.. Activation of BPV-1 replication in vitro by the transcription factor E2. Nature. 1991; 353:628–632. [DOI] [PubMed] [Google Scholar]
- 7. Gopalakrishnan V., Khan S.A.. E1 protein of human papillomavirus type 1a is sufficient for initiation of viral DNA replication. Proc. Natl. Acad. Sci. U.S.A. 1994; 91:9597–9601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fouts E.T., Yu X., Egelman E.H., Botchan M.R.. Biochemical and electron microscopic image analysis of the hexameric E1 helicase. J. Biol. Chem. 1999; 274:4447–4458. [DOI] [PubMed] [Google Scholar]
- 9. Titolo S., Pelletier A., Pulichino A.-M., Brault K., Wardrop E., White P.W., Cordingley M.G., Archambault J.. Identification of domains of the human papillomavirus type 11 E1 helicase involved in oligomerization and binding to the viral origin. J. Virol. 2000; 74:7349–7361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sedman J., Stenlund A.. The papillomavirus E1 protein forms a DNA-dependent hexameric complex with ATPase and DNA helicase activities. J. Virol. 1998; 72:6893–6897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Schuck S., Stenlund A.. Assembly of a double hexameric helicase. Mol. Cell. 2005; 20:377–389. [DOI] [PubMed] [Google Scholar]
- 12. Castella S., Burgin D., Sanders C.M.. Role of ATP hydrolysis in the DNA translocase activity of the bovine papillomavirus (BPV-1) E1 helicase. Nucleic Acids Res. 2006; 34:3731–3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Patel S.S., Picha K.M.. Structure and function of hexameric helicases. Annu. Rev. Biochem. 2000; 69:651–697. [DOI] [PubMed] [Google Scholar]
- 14. Trakselis M.A. Structural mechanisms of hexameric helicase loading, assembly, and unwinding [version 1; peer review: 3 approved]. F1000Res. 2016; 5:111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Brewster A.S., Chen X.S.. Insights into the MCM functional mechanism: lessons learned from the archaeal MCM complex. Crit. Rev. Biochem. Mol. Biol. 2010; 45:243–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Stenlund A. Initiation of DNA replication: lessons from viral initiator proteins. Nat. Rev. Mol. Cell Biol. 2003; 4:777–785. [DOI] [PubMed] [Google Scholar]
- 17. Wessel R., Schweizer J., Stahl H.. Simian virus 40 T-antigen DNA helicase is a hexamer which forms a binary complex during bidirectional unwinding from the viral origin of DNA replication. J. Virol. 1992; 66:804–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yardimci H., Wang X., Loveland A.B., Tappin I., Rudner D.Z., Hurwitz J., Oijen A.M.V., Walter J.C.. Bypass of a protein barrier by a replicative DNA helicase. Nature. 2012; 492:205–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lin B.Y., Makhov A.M., Griffith J.D., Broker T.R., Chow L.T.. Chaperone proteins abrogate inhibition of the human papillomavirus (HPV) E1 replicative helicase by the HPV E2 protein. Mol. Cell Biol. 2002; 22:6592–6604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Trakselis M.A., Seidman M.M., Brosh R.M.. Mechanistic insights into how CMG helicase facilitates replication past DNA roadblocks. DNA Repair. 2017; 55:76–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Enemark E.J., Joshua-Tor L.. Mechanism of DNA translocation in a replicative hexameric helicase. Nature. 2006; 442:270–275. [DOI] [PubMed] [Google Scholar]
- 22. Lee S.-J., Syed S., Enemark E.J., Schuck S., Stenlund A., Ha T., Joshua-Tor L.. Dynamic look at DNA unwinding by a replicative helicase. Proc. Natl Acad. Sci. U.S.A. 2014; 111:E827–E835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fu Y.V., Yardimci H., Long D.T., Ho T.V., Guainazzi A., Bermudez V.P., Hurwitz J., Oijen A.v., Schärer O.D., Walter J.C.. Selective bypass of a lagging strand roadblock by the eukaryotic replicative DNA helicase. Cell. 2011; 146:931–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rothenberg E., Trakselis M.A., Bell S.D., Ha T.. MCM forked substrate specificity involves dynamic interaction with the 5′-Tail. J. Biol. Chem. 2007; 282:34229–34234. [DOI] [PubMed] [Google Scholar]
- 25. Rabhi M., Tuma R., Boudvillain M.. RNA remodeling by hexameric RNA helicases. RNA Biol. 2010; 7:655–666. [DOI] [PubMed] [Google Scholar]
- 26. Ribeck N., Saleh O.A.. DNA unwinding by Ring-Shaped T4 helicase gp41 is hindered by tension on the occluded strand. PLoS One. 2013; 8:e79237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Graham B.W., Schauer G.D., Leuba S.H., Trakselis M.A.. Steric exclusion and wrapping of the excluded DNA strand occurs along discrete external binding paths during MCM helicase unwinding. Nucleic Acids Res. 2011; 39:6585–6595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Carney S.M., Trakselis M.A.. The excluded DNA strand is SEW important for hexameric helicase unwinding. Methods. 2016; 108:79–91. [DOI] [PubMed] [Google Scholar]
- 29. Li D., Zhao R., Lilyestrom W., Gai D., Zhang R., DeCaprio J.A., Fanning E., Jochimiak A., Szakonyi G., Chen X.S.. Structure of the replicative helicase of the oncoprotein SV40 large tumour antigen. Nature. 2003; 423:512–518. [DOI] [PubMed] [Google Scholar]
- 30. Cuesta I., Núñez-Ramírez R., Scheres S.H.W., Gai D., Chen X.S., Fanning E., Carazo J.M.. Conformational rearrangements of SV40 large T antigen during early replication events. J. Mol. Biol. 2010; 397:1276–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Georgescu R., Yuan Z., Bai L., Santos R.d.L.A., Sun J., Zhang D., Yurieva O., Li H., O’Donnell M.E.. Structure of eukaryotic CMG helicase at a replication fork and implications to replisome architecture and origin initiation. Proc. Natl. Acad. Sci. U.S.A. 2017; 114:E697–E706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chaban Y., Stead J.A., Ryzhenkova K., Whelan F., Lamber E.P., Antson A., Sanders C.M., Orlova E.V.. Structural basis for DNA strand separation by a hexameric replicative helicase. Nucleic Acids Res. 2015; 43:8551–8563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Brewster A.S., Wang G., Yu X., Greenleaf W.B., Carazo J.M., Tjajadi M., Klein M.G., Chen X.S.. Crystal structure of a near-full-length archaeal MCM: Functional insights for an AAA+ hexameric helicase. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:20191–20196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Li N., Zhai Y., Zhang Y., Li W., Yang M., Lei J., Tye B.-K., Gao N.. Structure of the eukaryotic MCM complex at 3.8 Å. Nature. 2015; 524:186–191. [DOI] [PubMed] [Google Scholar]
- 35. Langston L., O’Donnell M.. Action of CMG with strand-specific DNA blocks supports an internal unwinding mode for the eukaryotic replicative helicase. Elife. 2017; 6:e23449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sedman T., Sedman J., Stenlund A.. Binding of the E1 and E2 proteins to the origin of replication of bovine papillomavirus. J. Virol. 1997; 71:2887–2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lee M., Shin S., Uhm H., Hong H., Kirk J., Hyun K., Kulikowicz T., Kim J., Ahn B., Bohr V.A. et al.. Multiple RPAs make WRN syndrome protein a superhelicase. Nucleic Acids Res. 2018; 46:4689–4698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Brutzer H., Luzzietti N., Klaue D., Seidel R.. Energetics at the DNA supercoiling transition. Biophys. J. 2010; 98:1267–1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Schuck S., Stenlund A.. Mechanistic analysis of local ori melting and helicase assembly by the papillomavirus E1 Protein. Mol. Cell. 2011; 43:776–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lionnet T., Spiering M.M., Benkovic S.J., Bensimon D., Croquette V.. Real-time observation of bacteriophage T4 gp41 helicase reveals an unwinding mechanism. Proc. Natl. Acad. Sci. U.S.A. 2007; 104:19790–19795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Johnson D.S., Bai L., Smith B.Y., Patel S.S., Wang M.D.. Single-Molecule studies reveal dynamics of DNA unwinding by the Ring-Shaped T7 helicase. Cell. 2007; 129:1299–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ribeck N., Kaplan D.L., Bruck I., Saleh O.A.. DnaB helicase activity is modulated by DNA geometry and force. Biophys. J. 2010; 99:2170–2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Klaue D. DNA Unwinding by Helicases Investigated on the Single Molecule Level. 2012; PhD ThesisTU Dresden: Department of Physics. [Google Scholar]
- 44. Burnham D.R., Kose H.B., Hoyle R.B., Yardimci H.. The mechanism of DNA unwinding by the eukaryotic replicative helicase. Nat. Commun. 2019; 10:2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Klaue D., Kobbe D., Kemmerich F., Kozikowska A., Puchta H., Seidel R.. Fork sensing and strand switching control antagonistic activities of RecQ helicases. Nat. Commun. 2013; 4:2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Chong J.P.J., Hayashi M.K., Simon M.N., Xu R.-M., Stillman B.. A double-hexamer archaeal minichromosome maintenance protein is an ATP-dependent DNA helicase. Proc. Natl. Acad. Sci. U.S.A. 2000; 97:1530–1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Shechter D.F., Ying C.Y., Gautier J.. The Intrinsic DNA Helicase activity of methanobacterium thermoautotrophicum ΔH minichromosome maintenance protein. J. Biol. Chem. 2000; 275:15049–15059. [DOI] [PubMed] [Google Scholar]
- 48. Manosas M., Xi X.G., Bensimon D., Croquette V.. Active and passive mechanisms of helicases. Nucleic Acids Res. 2010; 38:5518–5526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Patel S.S., Donmez I.. Mechanisms of helicases. J. Biol. Chem. 2006; 281:18265–18268. [DOI] [PubMed] [Google Scholar]
- 50. Kaplan D.L., O’Donnell M.. DnaB Drives DNA Branch Migration and dislodges proteins while encircling two DNA strands. Mol. Cell. 2002; 10:647–657. [DOI] [PubMed] [Google Scholar]
- 51. Shin J.-H., Jiang Y., Grabowski B., Hurwitz J., Kelman Z.. Substrate requirements for duplex DNA translocation by the eukaryal and archaeal minichromosome maintenance helicases. J. Biol. Chem. 2003; 278:49053–49062. [DOI] [PubMed] [Google Scholar]
- 52. Mecsas J., Sugden B.. Replication of plasmids derived from bovine papilloma virus type 1 and Epstein-Barr virus in cells in culture. Annu. Rev. Cell Biol. 1987; 3:87–108. [DOI] [PubMed] [Google Scholar]
- 53. Weisshart K., Taneja P., Jenne A., Herbig U., Simmons D.T., Fanning E.. Two regions of simian virus 40 T antigen determine cooperativity of Double-Hexamer assembly on the viral origin of DNA replication and promote hexamer interactions during bidirectional origin DNA unwinding. J. Virol. 1999; 73:2201–2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Sreekumar K.R., Prack A.E., Winters D.R., Barbaro B.A., Bullock P.A.. The simian virus 40 core origin contains two separate sequence modules that support T-Antigen Double-Hexamer assembly. J. Virol. 2000; 74:8589–8600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Garcia-Garcia C., Frieda K.L., Feoktistova K., Fraser C.S., Block S.M.. Factor-dependent processivity in human eIF4A DEAD-box helicase. Science. 2015; 348:1486–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Bonne-Andrea C., Santucci S., Clertant P.. Bovine papillomavirus E1 protein can, by itself, efficiently drive multiple rounds of DNA synthesis in vitro. J. Virol. 1995; 69:3201–3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Fanning E., Klimovich V., Nager A.R.. A dynamic model for replication protein A (RPA) function in DNA processing pathways. Nucleic Acids Res. 2006; 34:4126–4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Loo Y.M., Melendy T.. Recruitment of replication protein A by the papillomavirus E1 protein and modulation by single-stranded DNA. J. Virol. 2004; 78:1605–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Patel S.S., Pandey M., Nandakumar D.. Dynamic coupling between the motors of DNA replication: hexameric helicase, DNA polymerase, and primase. Curr. Opin. Chem. Biol. 2011; 15:595–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Chojnacki M., Melendy T.. The human papillomavirus DNA helicase E1 binds, stimulates, and confers processivity to cellular DNA polymerase epsilon. Nucleic Acids Res. 2018; 46:229–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Sanders C.M., Kovalevskiy O.V., Sizov D., Lebedev A.A., Isupov M.N., Antson A.A.. Papillomavirus E1 helicase assembly maintains an asymmetric state in the absence of DNA and nucleotide cofactors. Nucleic Acids Res. 2007; 35:6451–6457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Gai D., Wang D., Li S.-X., Chen X.S.. The structure of SV40 large T hexameric helicase in complex with AT-rich origin DNA. Elife. 2016; 5:e18129. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 63. Ali F.A., Douglas M.E., Locke J., Pye V.E., Nans A., Diffley J.F.X., Costa A.. Cryo-EM structure of a licensed DNA replication origin. Nat. Commun. 2017; 8:2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Enemark E. Crystal structure of the DNA binding domain of the replication initiation protein E1 from papillomavirus. Mol. Cell. 2000; 6:149–158. [PubMed] [Google Scholar]
- 65. Langston L.D., Mayle R., Schauer G.D., Yurieva O., Zhang D., Yao N.Y., Georgescu R.E., O’Donnell M.E.. Mcm10 promotes rapid isomerization of CMG-DNA for replisome bypass of lagging strand DNA blocks. Elife. 2017; 6:e29118. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.