Abstract
Background
Species diversity is likely to undergo a sharp decline in the next century. Perhaps as many as 33 % of all plant species may expire as a result of climate change. All parts of the globe will be impacted, and all groups of organisms will be affected. Hundreds of species throughout the world have already experienced local extinction
Perspectives
While thousands of species may become extinct in the next century and beyond, species formation will still occur. I consider which modes of plant species formation are likely to prevail in the next 500 years. I argue that speciation primarily will involve mechanisms that produce reproductively isolated lineages within less (often much less) than 100 generations. I will not especially consider the human element in promoting species formation, because it will continue and because the conclusions presented here are unaffected by it. The impact of climate change may be much more severe and widespread.
Conclusions
The most common modes of speciation likely to be operative in the next 500 years ostensibly will be auto- and allopolyploidy. Polyploid species or the antecedents thereof can arise within two generations. Moreover, polyploids often have broader ecological tolerances, and are likely to be more invasive than are their diploid relatives. Polyploid species may themselves spawn additional higher level polyploids either through crosses with diploid species or between pre-existing polyploids. The percentage of polyploid species is likely to exceed 50 % within the next 500 years vs. 35 % today. The stabilized hybrid derivatives (homoploid hybrid speciation) could emerge within a hundred generations after species contact, as could speciation involving chromosomal rearrangements (and perhaps number), but the number of such events is likely to be low. Speciation involving lineage splitting will be infrequent because the formation of substantive pre- and post-zygotic barriers typically takes many thousands of years.
Keywords: Allopolyploidy, autopolyploidy, chromosomal rearrangements, homoploid hybrid speciation, lineage splitting
INTRODUCTION
Climate change in the next century is likely to have a major effect on biodiversity (Bellard et al., 2012; Moritz and Agudo, 2013; Pacifici et al., 2015). As many as 33 % of all plant species may expire by the end of the century (Ceballos et al., 2015; Pimm and Joppa, 2015). This estimate exceeds the background rate of extinction by 1000 to 10 000 times. All parts of the globe will be impacted, and all groups of organisms will be affected (Urban, 2015). Over 900 species have become extinct globally since 1600 (IUCN, 2015); and hundreds of species throughout the world have already experienced local extinction (Wiens, 2016). Numerous species now persist at such low densities that they can be viewed as being practically extinct from an ecological point of view (Wiens, 2016).
Whereas much attention has been given to expected species extinctions, little is said about species formation in the coming few hundred years, which is likely to include major shifts in temperature and precipitation, sea levels and in land-cover due to human activity (Driesschaert et al., 2005). Here I ask which modes of plant species formation are likely to prevail in the next 500 years or so. I argue that rapid speciation primarily will involve mechanisms producing reproductively isolated lineages within <100 generations. I will not specifically consider the human element in promoting species formation, as have several publications focusing on rapid Anthropocene diversification related to habitat disturbance, transport and domestication (Thomas, 2015; Bull and Maron, 2016; Vellend et al., 2017; Otto 2018). Humans will continue to promote species formation as they have in the past 500 years.
CONDITIONS PROMOTING RAPID SPECIATION
Rapid speciation is promoted by environment-induced change in local plant communities (community disassembly), because species differ in their ability to tolerate or adapt to changing habitats and ecological associates (Stewart, 2009; Stewart et al., 2010). Correlatively, communities will be reconfigured because climate-related migration will be asynchronous (Urban et al., 2012; Blois et al., 2013). The nature of reconfiguration depends on its direct effects on the strength and climatic sensitivity of species interactions, direct effects on interacting species and the degree of species’ specialization (Gilman et al., 2010; Alexander et al., 2015). The altered interactions of ecological associates can affect the chances of species survival (CaraDonna et al., 2017; Saavedra et al., 2017; Cenci et al., 2018). Substantial opportunity for establishment may exist in transitional communities, because of the relative paucity of negative interspecific interactions (Wellborn and Langerhans, 2015).
Community disassembly is associated with the decline of some species’ populations and the growth of other species’ populations. The longer a climatic shift persists and the greater its magnitude, the greater will be the species turnover in a given community. Both contracting and expanding species would experience local bottlenecks. The demise of a species at one site may be accompanied by its colonization of a more suitable site. The longer the duration and magnitude of climate change, the greater the number of bottlenecks in a given species. Community disassembly provides opportunities for contact between previously isolated congeneric species (Anderson, 1948; Anderson and Stebbins, 1954).
MODES OF RAPID SPECIATION
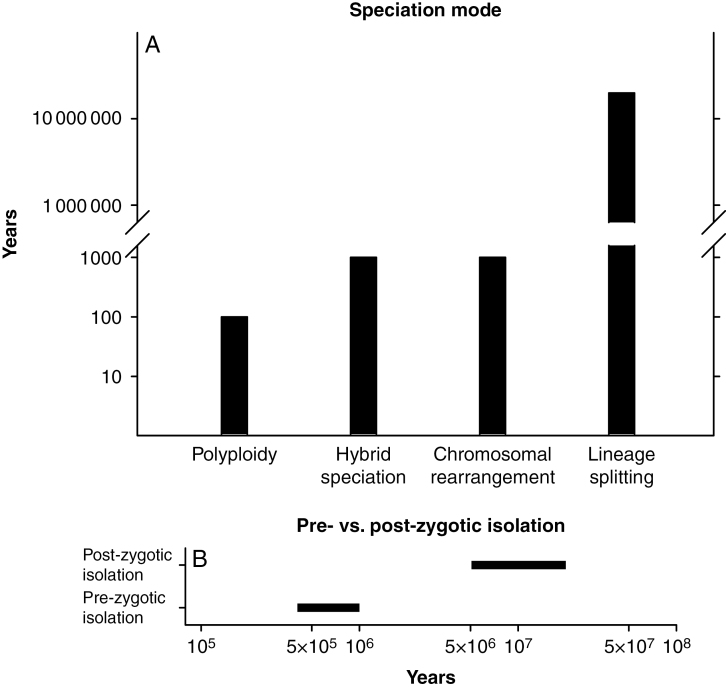
There are four modes of speciation which are expected to be operative in the next 500 years (Fig. 1). These are auto- and allopolyploidy, the stabilization of hybrid derivatives and speciation via chromosomal rearrangements (Abbott et al., 2013; Thomas, 2015; Bull and Maron, 2016; Vellend et al., 2017; Otto, 2018). All four modes yield entities that are reproductively isolated from their progenitors by virtue of post-zygotic barriers, especially hybrid sterility. Pre-zygotic barriers may or may not be present between any products of rapid speciation and their progenitors (Levin, 2000).
Fig. 1.
(A) Time intervals during which species arising by different modes are likely to become established; persistent entities. (B) Time intervals during which pre- and post-zygotic barriers are likely to become well established.
The most rapid modes of speciation involve polyploidy. Polyploidy can produce new species, or the antecedents thereof, in one generation (autopolyploidy) or two generations (allopolyploidy) through the production of unreduced gametes or doubled somatic cells (Ramsey and Schemske, 1998; Mason and Pires, 2015; Kreiner et al., 2017). Most polyploids, at least those that have been named, are allopolyploids. There may be many autopolyploids which have been unrecognized because of their undistinctive morphology or because of their small range (Soltis et al., 2007; Barker et al., 2016; Spoelhof et al., 2017). Since polyploidy is an ongoing process, it is not surprising that many instances of speciation via allopolyploidy have occurred during the past 200 years. For example, some species have arisen from diploids progenitors (e.g. Tragopogon miscellus and T. dubius; Ownbey, 1950) or from crosses of diploid and tetraploid species (e.g. Mimulus peregrinus; Vallejo-Marın, 2012), Autotetraploid populations of Mimulus guttatus also have arisen within the past 200 years (Simón-Porcar et al., 2017).
In contrast to hybridization followed by a ploidal increase, there are rather few examples of homoploid hybrid speciation and fewer yet within the past few hundred years (Abbott et al., 2013). However, the paucity may be due to our inability to recognize this process (Nieto-Feliner et al., 2017). In theory, a novel hybrid lineage could be established within tens of generations (Buerkle et al., 2000). One well-documented example of homoploid speciation within the past 300 years involves Senecio squalidus (Asteraceae) in the British Isles. This entity stabilized following long-distance dispersal of seeds from a hybrid zone between S. aethnensis and S. chrysanthemifolius (James and Abbott, 2005). Many generations of hybridization and selection for a narrow range of derivatives segregating for parental genomic segments are required for hybrid speciation (Buerkle and Rieseberg, 2007). Stabilization of the homoploid hybrid derivatives Helianthus anomalus, H. deserticola and H. paradoxus may have taken hundreds of generations (Buerkle and Rieseberg, 2007).
There are also relatively few examples of speciation via the fixation of novel chromosome rearrangements. The fixation of chromosomal novelty involves stochastic processes, wherein the greater the bottleneck or the fewer the number of founders of new populations, the greater is the fixation probability (Lande, 1985; Rieseberg, 2001; Jackson et al., 2016). Fixation could occur within 100 generations or considerably less, if a population bottleneck was severe. Chromosomal inversions are widespread in Mimulus guttatus, and contribute to local adaptation (Lowry and Willis, 2010; Twyford and Friedman, 2015).
Speciation via lineage splitting is not included here, because the time required for the origin of post-zygotic isolating barriers through the gradual accumulation of multiple genetic and/or chromosomal differences almost invariably is very much greater than 500 years (Levin, 2012, 2013). The waiting time for post-pollination barriers typically is at least hundreds of thousands of years; it may be millions of years. Ecological differences between lineages sufficient to warrant pre-zygotic isolation are also likely to take very much longer than 500 years to accumulate, as was the case (for example) during the Pleistocene climatic oscillations (Avise et al., 1998; Hewitt, 2001; Carstens and Knowles, 2007).
Although speciation via lineage splitting is quite unlikely during the next 500 years, lineage divergence may still proceed within species (Antonovics et al., 1971; Macnair et al., 2000; Hendry et al., 2007). For example, populations in some species can rapidly evolve tolerance to disparate edaphic conditions, especially when evolution is driven by major loci with large phenotypic effects (Macnair, 1983; Bratteler et al., 2006; Rajakaruna, 2018). Unco-ordinated plant and pollinator responses to climate change may yield divergence in floral attributes, leading to the formation of pollination ecotypes (e.g. Armbruster, 1985; Robertson and Wyatt, 1990; Johnson, 1997, 2010). The evolution of new adaptations is not equivalent to species formation, although it may be a step in that direction.
THE DYNAMICS OF RAPID SPECIATION
Autopolyploidy almost certainly will be prime mode of species formation during the next 500 years; and 2x to 4x the most frequent transition. I do not suggest that neoautopolyploids will persist as long as neoallopolyploids, which have a number of advantages over the former, as noted below. Autopolyploids can arise from single parents throughout a species’ range via the formation of unreduced gametes or through crosses between divergent conspecific autopolyploid lineages, thus bypassing the need for species contact and hybridization as required by allopolyploids. Unreduced gamete production would be increased by a variety of stressors including mineral and water limitation, temperature fluctuation, increased herbivory (Mason et al., 2011; Pécrix et al., 2011; Kreiner et al., 2017) and perhaps even hybridization. The higher the proportion of unreduced gametes produced within populations, the more often autopolyploids would be generated.
Autopolyploids may be better suited than their progenitors to changing environments (Levin, 2002; Parisod et al., 2010; Seagraves and Anneberg, 2016; Seagraves, 2017). Moreover, autopolyploids may have somewhat different ecological tolerances compared with their progenitors, which is reflected in differences in local or regional distributions (Ramsey, 2011; McIntyre, 2012; Laport and Ramsey, 2015; Visger et al., 2016; Gaynor et al., 2018b). However, some diploid–polyploid pairs have similar attributes (Godsoe et al., 2013; Glennon et al., 2014). A shift from diploid to tetraploid often is associated with numerous trait changes that include increased cell size, flower size and seed size (Ramsey and Schemske, 2002; Husband et al., 2013), increased drought tolerance (Maherali et al., 2009; del Pozo and Ramirez-Parra, 2015), increased salt tolerance, (Chao et al., 2013) delayed phenology (Levin, 1983) and slower growth rate (Parisod et al., 2010). Some polyploids derived from outcrossing progenitors may shift to partial self-fertility and apomixis (Alix et al., 2008; Husband et al., 2008; Robertson et al., 2010; Karunarathne et al., 2018). Many polyploids also reproduce vegetatively (Herben et al., 2017). Increased reproductive assurance and niche divergence allow autopolyploids to better persist in the presence of their progenitors and other ecological associates (Parisod et al., 2010; Seagraves and Anneberg, 2016).
Allopolyploidy ostensibly will be the second most common mode of species formation in the next 500 years. The reduction or dissolution of ecological and geographical barriers between previously isolated congeneric species may allow hybridization between numerous, previously isolated congeneric pairs (Mable, 2013; Chunco, 2014; Brennan et al., 2015; Taylor et al., 2015). The greater the disruption of the environment, by humans or otherwise, the greater the potential for species contact. A species may meet one congener in location X, another in location Y and another in location Z. The more congeneric species there are in a given area, the more different species combinations might hybridize. The greater the proximity of congeneric species, the greater the potential for interbreeding, because pollinators tend to move short distances between plants (Levin and Kerster, 1974).
Allopolyploids most probably would be generated following the formation of sterile diploid hybrids, which in turn produce unreduced gametes (Ramsey and Schemske, 1998). Allopolyploid species often are products of multiple independent origins in time and space (Soltis and Soltis, 1999; D. E. Soltis et al., 2014; Vallejo-Marin et al., 2016). Accordingly, if populations of their progenitors varied geographically and temporally, they may contain substantial genetic variation.
Although most changes in ploidal level involve the transition from diploids to tetraploids, higher level polyploids also may form from the production of unreduced gametes (Ramsey and Schemske, 1998). For example, hybridization between a tetraploid species and a diploid can yield sterile triploid hybrids producing triploid gametes, whose fusion yields fertile hexaploid offspring. Alternatively, hybridization between chromosomally differentiated tetraploid species can produce sterile tetraploids, whose unreduced tetraploid gametes may fuse to produce fertile octaploid individuals. If there are more diploid species within a genus than those of higher ploidal levels, ploidal change most probably will be from diploid to tetraploid.
Allopolyploids may be more ecologically divergent from their progenitors than are autopolyploids, thus facilitating their establishment (Levin, 2002; Parisod et al., 2010). Allopolyploids also may have broader ecological tolerances than diploid progenitors or autopolyploids (Glennon et al., 2014; P. S. Soltis et al., 2014; Soltis and Soltis, 2016; Visger et al., 2016). Allopolyploid species often are more common and have broader ranges than their diploid antecedents (te Beest et al., 2012). Allopolyploids also are likely to be more invasive than are their diploid relatives; and they tend to prefer drier and more open habitats than the latter (Pandit et al., 2011; te Beest et al., 2012). Conversely, some allopolyploids are intermediate in niche attributes to those of their progenitors (Marchant et al., 2016). The success of allopolyploids relative to their diploid progenitors and autopolyploids probably stems from their greater heterozygosity, more variable gene expression and greater plasticity (Paun et al., 2011; Hegarty et al., 2013; Doyle and Coate, 2019).
Whereas community disassembly provides opportunities for contact between previously isolated congeneric species, the frequency of homoploid hybrid speciation during the next 500 years is likely to be much lower than speciation via allopolyploidy. Species contact must be accompanied not only by hybridization but also by selection and stochastic processes that will stabilize recombined parts of their genomes, while erecting reproductive barriers between the new species and its progenitors (Buerkle and Rieseberg, 2007). Hybrids between diploid species may occupy more extreme edaphic niches than either parent (Abbott et al., 2010; Gramlich et al., 2016; Nieto Feliner et al., 2017).
Homoploid hybrid speciation differs from introgressive hybridization, which involves the selective incorporation of genetic material, usually from a closely related species. The enrichment of local gene pools may facilitate local adaptation or the colonization of novel habitats during periods of environmental change (Rieseberg et al., 2007; Abbott et al., 2013; Stelkens et al., 2014; Pfennig et al., 2016; Pierce et al., 2017). This process may also be accompanied by transgressive segregation, which produces novel phenotypes which exceed the phenotypic range of parental lineages (Rieseberg et al., 2003; Stelkens et al., 2009). Introgressive hybridization is not accompanied by reproductive isolation; and it is not a mode of speciation, although it certainly would promote divergence.
Speciation involving euploid chromosomal change in arrangement or number is also facilitated by community disassembly. The products of chromosomal change may emerge in marginal or contracting populations. Migration and founder episodes associated with community disassembly promote fixation. The closer the relatedness of founders, the greater the chances of a chromosomal novelty being fixed (Hedrick and Levin, 1984). The character of chromosomal change in one area is unlikely to be duplicated elsewhere, so species with a penchant for chromosomal rearrangement could generate a series of chromosomal novelties, each with a narrow geographical footprint. Inbreeding increases the incidence of chromosomal breakage in some species (Levin, 2002).
The fixation of novel chromosomal variants would be accompanied by a loss of genetic diversity, which would become more severe as the new entity spreads from one location to another (Excoffier et al., 2009; Yannic et al., 2014). Hybrids between the new entity and its progenitors will be sterile due to meiotic irregularities, as will hybrids between new species with different chromosome arrangements. Accordingly, these entities have no immediate source of genetic enrichment, and their adaptive potential is therefore minimal, at least in the short term. They are likely to be narrowly distributed, short-lived evolutionary entities.
The likelihood of an inversion or translocation being fixed in a population depends in part on how often a rearrangement occurs, and that usually is infrequent. A representative situation is that in Allium schoenoprasum, where of 23 of 1017 plants from 18 populations were heterozygous for translocations and 12 were heterozygous for inversions (Stevens and Bougourd, 1991). Fixation of novel rearrangements in this species is thus very unlikely. However, it is more likely, and indeed has occurred, in Clarkia unguiculata, where 35 % of the plants from 36 populations were translocation heterozygotes (Mooring, 1958). Four novel rearrangements were present as homozygotes in small populations near the ecological limit of the species. The fixation of rare translocations has also occurred in marginal populations of Clarkia exilis (Vasek, 1960).
Chromosomal change is not restricted to diploid species. There are many instances of euploid change in chromosome number that follow ploidal increase. Descending dysploidy (decline in the base number) is much more common than ascending dysploidy (Mandáková and Lysak, 2018). Translocations among non-homologous and homeologous chromosomes are frequent in polyploids, especially in younger species (Soltis et al., 2016; Mandáková and Lysak, 2018). For example, the tetraploid Tragopogon miscellus, which is only 40 generations old, displays extensive and repeated patterns of chromosomal variation including intergenomic translocations in all populations (Chester et al., 2012). Populations of the 80- to 90-year-old neotetraploid, Tragopogon mirus also contain chromosomal rearrangements, but not to the extent present in T. miscellus (Chester et al., 2015). Eventually, chromosomally variable lineages become diploidized, and meiotic pairing is normal (Mavrodiev et al., 2015; Mandakova and Lysek, 2018).
In general, polyploids have greater chromosome instability than diploids; and polyploids can better tolerate chromosome rearrangements than diploids (De Storme and Mason, 2014). Accordingly, in the next 500 years, chromosomally divergent lineages are more likely to emerge, and to contribute more to the speciation process within polyploid species than within diploid species. However, many of these species are apt to be short lived, as they are products of genetic bottlenecks, and may lack the genetic variation to respond to environmental change (Levin, 2019).
Conclusions
I propose that auto- and alloploidal change will be the primary modes of speciation in the next 500 years. Neopolyploid lineages are predisposed to conditions in new or transitional habitats (te Beest et al., 2012), where new contacts between previously isolated congeneric species may occur. Polyploid species evolving in the next 500 years may themselves spawn additional higher level polyploids either through crosses with diploid species (e.g. Mimulus peregrinus,Vallejo-Marın, 2012), or crosses between preexisting polyploids (e.g. Spartina anglica, Ainouche et al., 2004). Ploidal change is unidirectional. As a result, a genus will tend towards higher frequencies of polyploidy and higher ploidal levels as time passes (Meyers and Levin, 2006). Speciation involving lineage splitting via divergent evolution also will be less frequent than speciation via hybrid stabilization and chromosomal change because the former requires much more time.
Consider some possible consequences of polyploid persistence and genesis advantages. The genera with a higher percentage of polyploid species may survive disproportionally; and the genera with a penchant for the production of unreduced gametes may increase disproportionally in species number. Polyploid genera also may disproportionally increase in species numbers relative to diploids, because they have a greater penchant for chromosome rearrangement and aneuploidy than diploid genera. Since recent polyploids now account for about 35% of flowering plant species (Wood et al., 2009), a persistence and/or genesis advantage may mean that the percentage of polyploid species would substantially surpass 50% within the next few hundred years.
If tolerance to environmental change is independent of growth habit, the proportion of herbaceous species is likely to increase and that of woody species decline, because polyploid species are more prevalent in herbs than in woody species (Stebbins, 1971). The mean increase in chromosome number diversity per lineage per million years due to polyploidy was 0.05 in herbs compared to 0.01 in shrubs and 0.001 in trees (Levin and Wilson, 1976). Herbaceous species also have higher levels of aneuploidy than shrubs or trees (0.02 in herbs, 0.0005 in shrubs, 0.0003 in trees), which is another reason why herbaceous species may increase in proportional representation over the next few hundred years.
Whereas some diploid species may not persist during the abiotic and biotic stresses associated with community disassembly, it is noteworthy that neopolyploids may themselves contribute to the demise of their progenitors. If a polyploid species was better suited to novel conditions than its co-occurring diploid progenitor(s), the latter would be at a competitive disadvantage with regard to the polyploid. In the Brassicaceae and Rosaceae, polyploid species often are more distantly related to co-occurring diploids than diploids are to each other, possibly the result of polyploids outcompeting their progenitors (Gaynor et al., 2018a).
ACKNOWLEDGEMENTS
The author is grateful to Christian Parisod and two anonymous reviewers for their constructive comments. The authors also thanks Norma Fowler for preparing the figure.
LITERATURE CITED
- Abbott RJ, Hegarty MJ, Hiscock SJ, Brennan AC. 2010. Homoploid hybrid speciation in action. Taxon 59: 1375–1386. [Google Scholar]
- Abbott R, Albach D, Ansell S, et al. 2013. Hybridization and speciation. Journal of Evolutionary Biology 26: 229–246. [DOI] [PubMed] [Google Scholar]
- Alexander JM, Diez JM, Levine JM. 2015. Novel competitors shape species’ responses to climate change. Nature 525: 515–518. [DOI] [PubMed] [Google Scholar]
- Alix K, Joets J, Ryder CD, et al. 2008. The CACTA transposon Bot1 played a major role in Brassica genome divergence and gene proliferation. The Plant Journal 56: 1030–1044. [DOI] [PubMed] [Google Scholar]
- Anderson E. 1948. Hybridization of the habitat. Evolution 2: 1–9. [Google Scholar]
- Anderson E, Stebbins GL. 1954. Hybridization as an evolutionary stimulus. Evolution 8: 378–388. [Google Scholar]
- Antonovics J. 2006. Evolution in closely adjacent plant populations X: long-term persistence of pre-reproductive isolation at a mine boundary. Heredity 97: 33–37. [DOI] [PubMed] [Google Scholar]
- Antonovics J, Bradshaw AD, Turner RG. 1971. Heavy metal tolerance in plants. Advances in Ecological Research 7: 1–85. [Google Scholar]
- Armbruster WS. 1985. Patterns of character divergence and the evolution of reproductive ecotypes of Dalechampia scandens (Euphorbiaceae). Evolution 39: 733–752. [DOI] [PubMed] [Google Scholar]
- Avise JC, Walker D, Johns GC. 1998. Speciation durations and Pleistocene effects on vertebrate phylogeography. Proceedings of the Royal Society B: Biological Sciences 265: 1707–1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker MS, Arrigo N, Baniaga AE, Li Z, Levin DA. 2016. On the relative abundance of autopolyploids and allopolyploids. New Phytologist 210: 391–398. [DOI] [PubMed] [Google Scholar]
- te Beest M, Le Roux JJ, Richardson DM, et al. 2012. The more the better? The role of polyploidy in facilitating plant invasions. Annals of Botany 109: 19–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellard C, Bertelsmeier C, Leadley P, Thuiller W, Courchamp F. 2012. Impacts of climate change on the future of biodiversity. Ecology Letters 15: 365–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blois JL, Zarnetslke PL, Fitzpatrick MC, Finnegan S. 2013. Climate change and the past, present and future of biotic interactions. Science 341: 499–504. [DOI] [PubMed] [Google Scholar]
- Bratteler M, Lexer C, Widmer A. 2006. Genetic architecture of traits associated with serpentine adaptation of Silene vulgaris. Journal of Evolutionary Biology 19: 1149–1156. [DOI] [PubMed] [Google Scholar]
- Brennan AC, Woodward G, Seehausen O, et al. 2015. Hybridization due to changing species distributions: adding problems or solutions to conservation of biodiversity during global change? Evolutionary Ecology Research 16: 475–491. [Google Scholar]
- Buerkle CA, Rieseberg LH. 2007. The rate of genome stabilization in homoploid hybrid species. Evolution 62: 266–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buerkle CA, Morris RJ, Asmussen MA, Rieseberg LH. 2000. The likelihood of homoploid hybrid speciation. Heredity 84: 441–451. [DOI] [PubMed] [Google Scholar]
- Bull JW, Maron M. 2016. How humans drive speciation as well as extinction. Proceedings of the Royal Society B: Biological Sciences 283: 20160600. doi. 10.1098/rspb.2016.0600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- CaraDonna PJ, Petry WK, Brennan RM, et al. 2017. Interaction rewiring and the rapid turnover of plant–pollinator networks. Ecology Letters 20: 385–394. [DOI] [PubMed] [Google Scholar]
- Carstens BC, Knowles LL. 2007. Shifting distributions and speciation: species divergence during rapid climate change. Molecular Ecology 16: 619–627. [DOI] [PubMed] [Google Scholar]
- Ceballos G, Ehrlich PR, Barnosky AD, Garcia A, Pringle RM, Palmer T. 2015. Accelerated modern human induced species losses: entering the sixth mass extinction. Science Advances 1: e1400253. doi: 10.1126/sciadv.1400253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cenci S, Montero-Castano A, Saavedra S. 2018. Estimating the effect of the reorganization of interactions on the adaptability of species to changing environments. Journal of Theoretical Biology 437: 115–125. [DOI] [PubMed] [Google Scholar]
- Chao DY, Dilkes B, Luo H, et al. 2013. Polyploids exhibit higher potassium uptake and salinity tolerance in Arabidopsis. Science 341: 658–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chester M, Gallagher JP, Symonds VV, et al. 2012. Extensive chromosomal variation in a recently formed natural allopolyploid species, Tragopogon miscellus (Asteraceae). Proceedings of the National Academy of Sciences, USA 109: 1176–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chester M, Riley RK, Soltis PS, Soltis DE. 2015. Patterns of chromosomal variation in natural populations of the neoallotetraploid Tragopogon mirus (Asteraceae). Heredity 114: 309–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chunco AJ. 2014. Hybridization in a warmer world. Ecology and Evolution 4: 2019–2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Storme N, Mason A. 2014. Plant speciation through chromosome instability and ploidy change: cellular mechanisms, molecular factors and evolutionary relevance. Current Plant Biology 1: 10–33. [Google Scholar]
- Doyle JJ, Coate JE. 2019. Polyploidy, the nucleotype, and novelty: the impact of genome doubling on the biology of the cell. International Journal of Plant Sciences 180: 1–52. [Google Scholar]
- Driesschaert E, Brovkin V, Fichefet T, et al. 2005. Climate changes during the third millennium: a study with LOVECLIM, an Earth system model of intermediate complexity. In: The climate of the next millenia in the perspective of abrupt climate change during the late Pleistocene. Deklim/PAGES conference, Mainz, Germany, March 2005. [Google Scholar]
- Excoffier L, Foll M, Petit RJ. 2009. Genetic consequences of range expansions. Annual Review of Ecology, Evolution and Systematics 40: 481–501. [Google Scholar]
- Gaynor ML, Ng J, Laport RG. 2018a Phylogenetic structure of plant communities: are polyploids distantly related to co-occurring diploids? Frontiers of Ecology and Evolution 6: 1–14. [Google Scholar]
- Gaynor ML, Marchant DB, Soltis DE, Soltis PS. 2018b Climatic niche comparison among ploidal levels in the classic autopolyploid system, Galax urceolata. American Journal of Botany 105: 1631–1642. [DOI] [PubMed] [Google Scholar]
- Gilman SE, Urban MC, Tewksbury J, Gilchrist GW, Holt RD. 2010. A framework for community interactions under climate change. Trends in Ecology and Evolution 25: 325–331. [DOI] [PubMed] [Google Scholar]
- Glennon KL, Ritchie ME, Segraves KA. 2014. Evidence for shared broad-scale climatic niches of diploid and polyploid plants. Ecology Letters 17: 574–582. [DOI] [PubMed] [Google Scholar]
- Godsoe W, Larson MA, Glennon KL, Segraves KA. 2013. Polyploidization in Heuchera cylindrica (Saxifragaceae) did not result in a shift in climatic requirements. American Journal of Botany 100: 496–508. [DOI] [PubMed] [Google Scholar]
- Gramlich S, Sagmeister P, Dullinger S, Hadacek F, Hörandl E. 2016. Evolution in situ: hybrid origin and establishment of willows (Salix L.) on alpine glacier forefields. Heredity 116: 531–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedrick PW, Levin DA. 1984. Kin-founding and the fixation of chromosomal variants. The American Naturalist 124: 789–797. [Google Scholar]
- Hegarty M, Coate J, Sherman-Broyles S, Abbott R, Hiscock S, Doyle J. 2013. Lessons from natural and artificial polyploids in higher plants. Cytogenetic and Genome Research 140: 204–225. [DOI] [PubMed] [Google Scholar]
- Hendry AP, Nosil P, Rieseberg LH. 2007. The speed of ecological speciation. Functional Ecology 21: 455–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herben T, Suda J, Klimesova J. 2017. Polyploid species rely on vegetative reproduction more than diploids: a re-examination of the old hypothesis. Annals of Botany 120: 341–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewitt GM. 2001. Speciation, hybrid zones and phylogeography – or seeing genes in space and time. Molecular Ecology 10: 537–549. [DOI] [PubMed] [Google Scholar]
- Husband BC, Ozimec B, Martin SL, Pollock L. 2008. Mating consequences of polyploid evolution in flowering plants: current trends and insights from synthetic polyploids. International Journal of Plant Sciences 169: 195–206. [Google Scholar]
- Husband BC, Baldwin SJ, Suda J. 2013The incidence of polyploidy in natural plant populations: major patterns and evolutionary processes. In: Greilhuber J, Doležel J, Wendel JF, eds. Plant genome diversity, Vol. 2 Vienna: Springer, 255–276. [Google Scholar]
- IUCN 2015. http://www.iucnredlist.org The IUCN red list of threatened species.
- Jackson B, Butlin R, Navarro A, Fria R. 2016. Speciation, chromosomal rearrangements and. In: Kliman R, ed. Encyclopedia of evolutionary biology. Cambridge, MA: Elsevier, 149–158. [Google Scholar]
- James JK, Abbott RJ. 2005. Recent, allopatric, homoploid hybrid speciation: the origin of Senecio squalidus (Asteraceae) in the British Isles from a hybrid zone on Mount Etna, Sicily. Evolution 59: 2533–2547. [PubMed] [Google Scholar]
- Johnson SD. 1997. Pollination ecotypes of Satyrium hallackii (Orchidaceae) in South Africa. Botanical Journal of the Linnean Society 123: 225–235. [Google Scholar]
- Johnson SD. 2010. The pollination niche and its role in the diversification and maintenance of the southern African flora. Philosophical Transactions of the Royal Society B: Biological Sciences 365: 499–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karunarathne P, Schedler M, Martínez EJ, Honfi AI, Novichkova A, Hojsgaard D. 2018. Intraspecific ecological niche divergence and reproductive shifts foster cytotype displacement and provide ecological opportunity to polyploids. Annals of Botany 121: 1183–1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreiner JM, Kron P, Husband BC. 2017. Evolutionary dynamics of unreduced gametes. Trends in Genetics 33: 583–593. [DOI] [PubMed] [Google Scholar]
- Lande R. 1985. The fixation of chromosomal rearrangements in a subdivided population with local extinction and colonization. Heredity 54: 323–332. [DOI] [PubMed] [Google Scholar]
- Laport R, Ramsey J. 2015. Morphometric analysis of the North American creosote bush (Larrea tridentata, Zygophyllaceae) and the microspatial distribution of its chromosome races. Plant Systematics and Evolution 301: 1581–1599. [Google Scholar]
- Levin DA. 1983. Polyploidy and novelty in flowering plants. The American Naturalist 122: 1–25. [Google Scholar]
- Levin DA. 2000. The origin, expansion, and demise of plant species. New York: Oxford University Press. [Google Scholar]
- Levin DA. 2002. The role of chromosomal change in plant evolution. New York: Oxford University Press. [Google Scholar]
- Levin DA. 2012. The long wait for hybrid sterility in flowering plants. New Phytologist 196: 666–670. [DOI] [PubMed] [Google Scholar]
- Levin DA. 2013. The timetable for allopolyploidy in flowering plants. Annals of Botany 112: 1201–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin DA. 2019. Why polyploid exceptionalism is not accompanied by reduced extinction rates. Plant Systematics and Evolution 305: 1–11 [Google Scholar]
- Levin DA, Kerster HW. 1974. Gene flow in seed plants. In: Dobzhansky Th, Hecht MK, Steere WC, eds. Evolutionary biology, 7th edn.New York: Plenum Press, 139–220. [Google Scholar]
- Levin DA, Wilson AC. 1976. Rates of evolution in seed plants: net increase in diversity of chromosome numbers and species numbers through time. Proceedings of the National Academy of Sciences, USA 73: 2086–2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry DB, Willis JH. 2010. A widespread chromosomal inversion polymorphism contributes to a major life-history transition, local adaptation, and reproductive isolation. PLoS Biology 8: e1000500. doi: 10.1371/journal.pbio.1000500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mable BK. 2013. Polyploids and hybrids in changing environments: winners or losers in the struggle for adaptation? Heredity 110: 95–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macnair MR. 1983. The genetic control of copper tolerance in the yellow monkey flower, Mimulus guttatus. Heredity 50: 283–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macnair MR, Tilstone GH, Smith SE. 2000. The genetics of metal tolerance and accumulation in higher plants. In: Terry N, Bañuelos G, eds. Phytoremediation of contaminated soil and water. Boca Raton, FL: Lewis Publishers, 235–250 [Google Scholar]
- Maherali H, Walden AE, Husband BC. 2009. Genome duplication and the evolution of physiological responses to water stress. New Phytologist 184: 721–731. [DOI] [PubMed] [Google Scholar]
- Mandáková T, Lysak MA. 2018. Post-polyploid diploidization and diversification through dysploid changes. Current Opinion in Plant Biology 42: 55–65. [DOI] [PubMed] [Google Scholar]
- Marchant D, Soltis DE, Soltis PS. 2016. Patterns of abiotic niche shifts in allopolyploids relative to their progenitors. New Phytologist 212: 708–718. [DOI] [PubMed] [Google Scholar]
- Mason AS, Pires JC. 2015. Unreduced gametes: meiotic mishap or evolutionary mechanism? Trends in Genetics 31: 5–10. [DOI] [PubMed] [Google Scholar]
- Mason AS, Nelson MN, Yan G, Cowling WA. 2011. Production of viable male unreduced gametes in Brassica interspecific hybrids is genotype specific and stimulated by cold temperatures. BMC Plant Biology 11: 103. doi: 10.1186/1471-2229-11-103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavrodiev EV, Chester M, Suarez-Santiago VN, et al. 2015. Multiple origins and chromosomal novelty in the allotetraploid Tragopogon castellanus (Asteraceae). New Phytologist 206: 1172–1183. [DOI] [PubMed] [Google Scholar]
- McIntyre PJ. 2012. Polyploidy associated with altered and broader ecological niches in the Claytonia perfoliata (Portulacaceae) species complex. American Journal of Botany 99: 655–662. [DOI] [PubMed] [Google Scholar]
- Meyers L, Levin DA. 2006. On the abundance of polyploids in flowering plants. Evolution 60: 1198–1206. [PubMed] [Google Scholar]
- Mooring J. 1958. A cytogenetic study of Clarkia unguiculata. I. Translocations. American Journal of Botany 45: 233–242. [Google Scholar]
- Moritz C, Agudo R. 2013. The future of species under climate change: resilience or decline? Science 341: 504–508. [DOI] [PubMed] [Google Scholar]
- Nieto Feliner G, Álvarez I, Fuertes‐Aguilar J, et al. 2017. Is homoploid hybrid speciation that rare? An empiricist’s view. Heredity 118: 513–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto SP. 2018. Adaptation, speciation and extinction in the Anthropocene. Proceedings of the Royal Society B: Biological Sciences 285: 20182047. doi: 10.1098/rspb.2018.2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ownbey M. 1950. Natural hybridization and amphiploidy in the genus Tragopogon. American Journal of Botany 37: 487–499. [Google Scholar]
- Pacifici M, Foden WB, Visconti P, et al. 2015. Assessing species vulnerability to climate change. Nature Climate Change 5: 215–224. [Google Scholar]
- Pandit MK, Pocock MJO, Kunin WE. 2011. Ploidy influences rarity and invasiveness in plants. Journal of Ecology 99: 1108–1115. [Google Scholar]
- Parisod C, Holderegger R, Brochmann C. 2010. Evolutionary consequences of autopolyploidy. New Phytologist 186: 5–17. [DOI] [PubMed] [Google Scholar]
- Paun O, Forest F, Fay MF, Chase MW. 2011. Parental divergence and hybrid speciation in angiosperms revisited. Taxon 60: 1241–1244. [PMC free article] [PubMed] [Google Scholar]
- Pécrix Y, Rallo G, Folzer H, Cigna M, Gudin S, Le Bris M. 2011. Polyploidization mechanisms: temperature environment can induce diploid gamete formation in Rosa sp. Journal of Experimental Botany 62: 3587–3597. [DOI] [PubMed] [Google Scholar]
- Pfennig KS, Kelly AL, Pierce AA. 2016. Hybridization as a facilitator of species range expansion. Proceedings of the Royal Society B: Biological Sciences 283: 20161329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce AA, Gutierrez R, Rice AM, Pfennig KS. 2017. Genetic variation during range expansion: effects of habitat novelty and hybridization. Proceedings of the Royal Society B: Biological Sciences 284: 20170007. doi: 10.1098/rspb.2017.0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimm SL, Joppa LN. 2015. How many plant species are there, where are they, and at what rate are they going extinct? Annals of the Missouri Botanical Garden 100: 170–76. [Google Scholar]
- del Pozo JC, Ramirez-Parra E. 2015. Whole genome duplications in plants: an overview from Arabidopsis. Journal of Experimental Biology 66: 6991–7003. [DOI] [PubMed] [Google Scholar]
- Rajakaruna N. 2018. Lessons on evolution from the study of edaphic specialization. The Botanical Review 84: 39–78. [Google Scholar]
- Ramsey J. 2011. Polyploidy and ecological adaptation in wild yarrow. Proceedings of the National Academy of Sciences, USA 108: 7096–7101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey J, Schemske DW. 1998. Pathways, mechanisms, and rates of polyploid formation in flowering plants. Annual Review of Ecology and Systematics 29: 467–501. [Google Scholar]
- Ramsey J, Schemske DW. 2002. Neopolyploidy in flowering plants. Annual Review of Ecology and Systematics 33: 589–639. [Google Scholar]
- Rieseberg LH. 2001. Chromosomal rearrangements and speciation. Trends in Ecology and Evolution 16: 351–358. [DOI] [PubMed] [Google Scholar]
- Rieseberg LH, Kim SC, Randell RA, et al. 2007. Hybridization and the colonization of novel habitats by annual sunflowers. Genetica 129: 149– 165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieseberg LH, Widmer A, Arntz M, Burke B. 2003. The genetic architecture necessary for transgressive segregation is common in both natural and domesticated populations. Philosophical Transactions of the Royal Society B: Biological Sciences 358: 1141–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson A, Rich TCG, Allen AM, et al. 2010. Hybridization and polyploidy as drivers of continuing evolution and speciation in Sorbus. Molecular Ecology 19: 1675–1690. [DOI] [PubMed] [Google Scholar]
- Robertson JL, Wyatt R. 1990. Evidence for pollination ecotypes in the yellow-fringed orchid, Platanthera ciliaris. Evolution 44: 121–133. [DOI] [PubMed] [Google Scholar]
- Saavedra S, Cenci S, del Val E, Boege K, Rohr RP. 2017. Reorganization of interaction networks modulates the persistence of species in late successional stages. Journal of Animal Ecology 86: 1136–1146. [DOI] [PubMed] [Google Scholar]
- Seagraves KA. 2017. The effects of genome duplications in a community context. New Phytologist 215: 57–69. [DOI] [PubMed] [Google Scholar]
- Seagraves KA, Anneberg TJ. 2016. Species interactions and plant polyploidy. American Journal of Botany 103: 1326–1335. [DOI] [PubMed] [Google Scholar]
- Simón-Porcar VI, Silva JL, Higgins JD, Vallejo-Marin M. 2017. Recent autopolyploidisation in a wild population of Mimulus guttatus (Phrymaceae). Botanical Journal of the Linnean Society 185: 189–207. [Google Scholar]
- Soltis DE, Soltis PS. 1999. Polyploidy: recurrent formation and genome evolution. Trends in Ecology and Evolution 14: 348–352. [DOI] [PubMed] [Google Scholar]
- Soltis DE, Soltis PS, Schemske DW, et al. 2007. Autopolyploidy in angiosperms: have we grossly underestimated the number of species? Taxon 56: 13–30. [Google Scholar]
- Soltis DE, Visger CJ, Soltis PS. 2014. The polyploidy revolution then and now: Stebbins revisited. American Journal of Botany 101: 1057–1078. [DOI] [PubMed] [Google Scholar]
- Soltis DE, Visger CJ, Marchant DB, Soltis PS. 2016. Polyploidy: pitfalls and paths to a paradigm. American Journal of Botany 103: 1146–1166. [DOI] [PubMed] [Google Scholar]
- Soltis PS, Soltis DE. 2016. Ancient WGD events as drivers of key innovations in angiosperms. Current Opinion in Plant Biology 30: 159–165. [DOI] [PubMed] [Google Scholar]
- Soltis PS, Liu X, Marchant DB, C. Visger CJ, Soltis DE. 2014. Polyploidy and novelty: Gottlieb’s legacy. Philosophical Transactions of the Royal Society B: Biological Sciences 369: 20130351. doi: 10.1098/rstb.2013.0351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spoelhof JP, Soltis PS, Soltis DE. 2017. Pure polyploidy: closing the gaps in autopolyploid research. Journal of Systematics and Evolution 55: 340–352. [Google Scholar]
- Stebbins GL. 1971. Chromosomal evolution in higher plants. London: Addison-Wesley. [Google Scholar]
- Stelkens RB, Schmid C, Selz O, Seehausen O. 2009. Phenotypic novelty in experimental hybrids is predicted by the genetic distance between species of cichlid fish. BMC Evolutionary Biology 9: 283. doi: 10.1186/1471-2148-9-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelkens RB, Brockhurst MA, Hurst G, Greig D. 2014. Hybridization facilitates evolutionary rescue. Evolutionary Applications 7: 1209–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens JP, Bougourd SM. 1991. The frequency and meiotic behaviour of structural chromosome variants in natural populations of Allium schoeneprasum L. (wild chives) in Europe. Heredity 66: 391–401. [Google Scholar]
- Stewart JR. 2009. The evolutionary consequence of the individualistic response to climate change. Journal of Evolutionary Biology 22: 2363–2375. [DOI] [PubMed] [Google Scholar]
- Stewart JR, Lister AM, Barnes I, and Dalén L. 2010. Refugia revisited: individualistic responses of species in time and space. Proceedings of the Royal Society B: Biological Sciences 277: 661–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor SA, Larson EL, Harrison RG. 2015. Hybrid zones: windows on climate change. Trends in Ecology and Evolution 30: 198–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas CD. 2015. Rapid acceleration of plant speciation during the Anthropocene. Trends in Ecology and Evolution 30: 448–455. [DOI] [PubMed] [Google Scholar]
- Twyford AD, Friedman J. 2015. Adaptive divergence in the monkeyflower Mimulus guttatus is maintained by a chromosomal inversion. Evolution 69: 1476–1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban MC. 2015. Accelerating extinction risk from climate change. Science 348: 571–573. [DOI] [PubMed] [Google Scholar]
- Urban MC, Tewksbury JJ, Sheldon KS. 2012. On a collision course: competition and dispersal differences create no-analogue communities and cause extinctions during climate change. Proceedings of the Royal Society B: Biological Sciences 279: 2072–2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallejo-Marın M. 2012. Mimulus peregrinus (Phrymaceae): a new British allopolyploid species. PhytoKeys 14: 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallejo-Marín M, Hiscock SJ. 2016. Hybridization and hybrid speciation under global change. New Phytologist 211: 1170–1187. [DOI] [PubMed] [Google Scholar]
- Vasek FC. 1960. A cytogenetic study of Clarkia exilis. Evolution 14: 88–97. [Google Scholar]
- Vellend M, Baeten L, Becker-Scarpitta A, et al. 2017. Plant biodiversity change across scales during the Anthropocene. Annual Review of Plant Biology 68: 563–586. [DOI] [PubMed] [Google Scholar]
- Visger CJ, Germain-Aubrey CC, Patel M, Sessa EB, Soltis PS, Soltis DE. 2016. Niche divergence between diploid and autotetraploid Tolmiea. American Journal of Botany 103: 1396–1406. [DOI] [PubMed] [Google Scholar]
- Wellborn GA, Langerhans RB. 2015. Ecological opportunity and the adaptive diversification of lineages. Ecology and Evolution 5: 176–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiens JJ. 2016. Climate-related local extinctions are already wide-spread among plant and animal species. PLoS Biology 14: 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood TE, Takebayashi N, Barker MS, Mayrose I, Greenspoon PB, Rieseberg LH. 2009. The frequency of polyploid speciation in vascular plants. Proceedings of the National Academy of Sciences, USA 106: 13875–13879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yannic G, Pellissier L, Ortego J, et al. 2014. Genetic diversity in caribou linked to past and future climate change. Nature Climate Change 4: 132–137. [Google Scholar]