Abstract
We describe a case of acute liver failure and myopericarditis due to herpes simplex virus–1 (HSV-1) in an immunocompetent adult. We estimate that, at the height of viremia, the patient contained a quantity of HSV-1 virions approaching that of human cells. The patient recovered with acyclovir that was dose-adjusted for neurotoxicity and developed a vigorous anti-HSV-1 T-cell response.
Keywords: hepatitis, herpes simplex virus, host response, HSV-1, HSV hepatitis, viral evolution
CLINICAL CASE
A 27-year-old man presented with a 1-week history of malaise, myalgia, and fever. He was previously healthy and took no medications including acetaminophen or dietary supplements. His physical exam was notable for scleral icterus and an absence of oral or genital lesions. He had acute kidney injury (creatinine 1.4 mg/dL), thrombocytopenia (platelets 86 000/µL), leukopenia (white blood cell count 1000/µL), and acute liver failure (aspartate aminotransferase (AST) 4969 U/L, alanine aminotransferase (ALT) 2111 U/L, international normalized ratio (INR) 1.5). Urine and serum toxicology screens were negative. He tested positive for hepatitis A IgG and hepatitis B surface antibody and negative for hepatitis B surface antigen/core antibody/DNA, hepatitis C antibody/RNA, and hepatitis E IgG and IgM. Plasma nucleic acid testing was negative for HIV, cytomegalovirus, Epstein-Barr virus, varicella-zoster virus, HHV-6, and adenovirus. Serum IgG antibodies were negative for herpes simplex virus (HSV)–1 and HSV-2. Antinuclear antibody, rheumatoid factor, and anti–smooth muscle antibody were negative. On hospital day 3, he developed worsening pancytopenia, progressively worsening liver function, and a ferritin of 69 000 ng/mL. He was given 20 mg of dexamethasone due to concern for hemophagocytic lymphohistiocytosis (HLH) and transferred to our center.
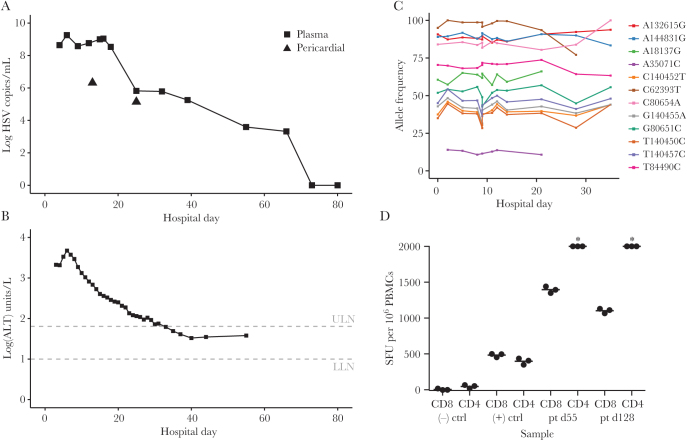
Dexamethasone was continued, and a dose of etoposide was given for possible HLH. A bone marrow biopsy demonstrated hypocellularity without evidence of hemophagocytosis. Polymerase chain reaction (PCR) from plasma on hospital day 4 returned with 440 000 000 copies/mL of HSV-1, which subsequently peaked at 1 800 000 000 copies/mL on hospital day 6 (Figure 1A). IgG antibodies for HSV-1 on hospital day 7 were weakly positive. Etoposide and dexamethasone were discontinued, and he was started on acyclovir 10 mg/kg intravenously every 8 hours. His liver function continue to worsen, with AST 15 045 U/L, ALT 4660 U/L, INR 2.5, total bilirubin 2.3 mg/dL, and ammonia 291 µg/dL on hospital day 6, with associated confusion attributed to hepatic encephalopathy. His acyclovir dose was increased to 15 mg/kg every 8 hours on hospital day 7, and he was evaluated for urgent liver transplantation. His liver function tests began to improve (Figure 1B), but his mental status deteriorated, with disorientation and echopraxia. Discordance between liver function and mental status as well as hyperammonemia suggested possible superimposed acyclovir neurotoxicity. His acyclovir dose was decreased to 10 mg/kg every 8 hours on hospital day 12, and his altered mental status improved over the next 3 days.
Figure 1.
A case of herpes simplex virus (HSV) hepatitis in an immunocompetent adult. Graph of plasma HSV-1 viral load in copies/mL (A) and alanine aminotransferase (B) by day of admission (or outpatient day). Half-life estimation of HSV DNA in plasma was measured for 2.7 days. Triangles indicate HSV-1 load in pericardial fluid. C, Examination of variants within unique long (UL)–unique short (excluding UL36) present at allele frequencies >10% over at least 3 time points shows extraordinarily limited evolution of HSV-1 genomes in vivo throughout the period of high viral load. D, Interferon-γ ELISPOT testing of anti-HSV-1 T-cell activity shows robust responses consistent with otherwise normal immune status. Asterisks indicate measurements above the dynamic range of the assay.
However, fevers with tachycardia persisted, and atrial fibrillation occurred intermittently. Troponin I on hospital day 9 was 3.74 ng/mL. Serial echocardiograms demonstrated a rapidly progressive pericardial effusion concerning for tamponade, requiring a pericardial drain on hospital day 12. The HSV DNA level in the pericardial fluid was 3 300 000 copies/mL. Colchicine and indomethacin were instituted.
He subsequently slowly improved and was discharged on hospital day 42, still receiving intravenous acyclovir (Figure 1B). His plasma HSV PCR was last detected on day 66 and undetectable by day 73. On day 73, he was transitioned to oral valacyclovir 1 g thrice daily. He remains on valacyclovir, has had no detectable HSV in plasma by PCR, and is clinically well 14 months after presentation.
LABORATORY INVESTIGATIONS AND DISCUSSION
HSV is a rare cause of fulminant hepatitis and pericarditis in immunocompetent hosts. This diagnosis is often delayed due to the lack of classic mucocutaneous findings [1]. Early initiation of antivirals is imperative, as the mortality rate for disseminated HSV leading to acute liver failure is high [2]. Liver transplantation has been used in some cases, though fatal relapse in the recipient has been reported. Most HSV hepatitis cases are thought to occur during primary HSV infection, suggesting defects in innate leukocyte or cell-intrinsic immunity as an underlying contributor. The paucity of hepatitis cases during HSV reactivation indicates that acquired T- or B-cell responses likely protect against severe visceral infection related to viral recurrence. Our patient had no known immune deficiency, and history, examination, and screening tests were unremarkable for immune deficiency. Whole-exome sequencing was performed on the patient, and variants identified were confirmed by targeted ImmunoPlex sequencing. These tests revealed 2 likely compound heterozygous variants of uncertain significance in CARD11, a scaffold protein associated with multiple primary immune disorders [3]. The first is a splicing variant in CARD11 intron 23, and the second is a missense variant in CARD11 exon 23, both of which are rare in public databases (Supplementary Data). Other rare heterozygous variants of uncertain significance were observed in UNC13D, PIK3CD, FOXK2, and IL7R, as well as a novel missense variant in TCF7. By analogy, with the requirement for cultured neurons to uncover molecular mechanisms for severe primary HSV encephalitis in persons with genetic variants in innate immunity genes, functional studies of cell-intrinsic antiviral immunity in genetically defined hepatocytes will be necessary to resolve liver-specific interactions between human genetic variants and HSV hepatitis [4, 5].
Given the extraordinarily high viral load in the blood, we performed HSV-1 whole-genome sequencing directly from patient plasma and pericardial fluid [6]. We detected no premature truncations of known open reading frames, and the overall genomic structure was similar to all other sequenced HSV-1 strains. Consensus genome alignments in the unique long (UL)–unique short (US) genome region containing the majority of protein-coding genes revealed no longitudinal changes. To screen for any minor longitudinal evolution in vivo, we examined all loci within the UL-US genome regions (excluding UL36) that had an allele frequency between 10% and 90% in any sample. These allele frequencies were stable, with no evidence of selective sweeps (Figure 1C). Examination of UL23 thymidine kinase and UL30 polymerase genes revealed no minor variants previously associated with acyclovir resistance [7]. A variant of unknown significance, R760Q, in UL30 relative to the HSV-1 strain 17 was found in all 12 samples tested.
We also performed metagenomic next-generation sequencing directly from plasma during hospital days 6–18. We found that up to 7.2% of the circulating DNA in plasma was of viral origin on day 16. Based on the relative genome size of HSV and human DNA combined with our qPCR viral load measurements, our results suggest that during the height of infection the patient had a total number of herpesvirus DNA-containing virions approaching that of human cells in the body.
Metagenomic data also demonstrated no minor variants in the UL23 and UL30 genes. Acyclovir resistance is rare (<0.5%) in the United States among immunocompetent hosts [8], but acyclovir resistance in HSV hepatitis is associated with poor outcomes [9], and the patient’s tenuous clinical course led to a desire to detect resistance as early as possible. The high viral load also provided the rare opportunity to study selective pressure directly in vivo in the setting of an enormous pool of virus. We performed Sanger sequencing of the UL23 thymidine kinase and UL30 polymerase on day 16 of hospitalization. Regardless of sequencing technology, no UL23 or UL30 variants previously associated with acyclovir resistance were detected in any sample. Intriguingly, despite the enormous population size of virus present and strong selective pressure afforded by acyclovir treatment, no alleles across the UL or US protein coding regions, including in genes UL23 and UL30, showed a >20% absolute change in allele frequency over the month in which sequencing was performed (Figure 1C). To ensure that we were not just detecting nonviable HSV-1 DNA, HSV-1 was also cultured on Vero cells from plasma on hospital day 4. Phenotypic acyclovir testing performed at a reference laboratory showed the viral isolate to be susceptible to acyclovir (ID50 0.29 µg/mL), consistent with the sequencing result and successful treatment.
HSV-specific immunity was assessed at the antibody and T-cell levels. Seroconversion for HSV-1-specific IgG occurred promptly, ruling out severe humoral immunodeficiency. For T cells, we have previously shown that T-cell responses among survivors of pediatric primary herpes encephalitis are similar to those of normal HSV-1 seropositive adults, consistent with the hypothesis that severe primary HSV infections are associated with defects in innate immunity, rather than in acquired T-cell immunity [10]. To our knowledge, T-cell immunity has not previously been investigated in HSV hepatitis. We tested the patient’s peripheral blood mononuclear cell (PBMC) CD4 and CD8 T-cell responses on days 55 and 128 after hospital admission via interferon-γ ELISPOT. We detected maximal HSV-specific CD4 T-cell responses, above the dynamic range of the assay used at both time points (Figure 1D). CD8 T-cell responses were also high (1397 and 1103 net HSV-1-specific CD8 T cells/million PBMCs at days 55 and 128, respectively). These values are at the top end of a series of healthy HSV-1 seropositive adults [10]. We conclude that the severe infection in this patient is unlikely to be related to T-cell immunodeficiency. Although the phenotype of the patient’s HSV-specific T cells could be atypical, their abundance at later time points in blood associated with memory T-cell function may inform the eventual reduction or cessation of chronic acyclovir therapy, by analogy with CMV immune reconstitution studies after hematopoietic stem cell transplant [11].
CONCLUSIONS
We describe a patient who developed severe hepatitis, pericarditis, and extraordinary levels of viremia due to primary HSV-1 infection. However, analysis of host genes known to predispose to severe HSV infections, T-cell responses to HSV, and the genotype of the infecting strain did not reveal a definitive explanation for the severe infection, although further characterization of the host variants identified is ongoing. No clinical, phenotypic, or genotypic evidence of acyclovir resistance developed, despite a viral population size measured in the trillions. This case illustrates the critical need to consider HSV as a potential cause of acute liver failure along with pericarditis, as well as the need for further studies to understand the virological and host underpinnings of this manifestation of HSV disease.
Supplementary Data
Supplementary materials are available at Open Forum Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Acknowledgments
We would like to thank the case patient, who provided written informed consent for clinical and research sequencing and this case report.
Potential conflicts of interest. All authors: no reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Kaufman B, Gandhi SA, Louie E, et al. . Herpes simplex virus hepatitis: case report and review. Clin Infect Dis 1997; 24:334–8. [DOI] [PubMed] [Google Scholar]
- 2. Moldovan B, Mentha G, Majno P, et al. . Demographics and outcomes of severe herpes simplex virus hepatitis: a registry-based study. J Hepatol 2011; 55:1222–6. [DOI] [PubMed] [Google Scholar]
- 3. Stepensky P, Keller B, Buchta M, et al. . Deficiency of caspase recruitment domain family, member 11 (CARD11), causes profound combined immunodeficiency in human subjects. J Allergy Clin Immunol 2013; 131:477–85.e1. [DOI] [PubMed] [Google Scholar]
- 4. Casanova JL. Severe infectious diseases of childhood as monogenic inborn errors of immunity. Proc Natl Acad Sci U S A 2015; 112:E7128–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dorjbal B, Stinson JR, Ma CA, et al. . Hypomorphic caspase activation and recruitment domain 11 (CARD11) mutations associated with diverse immunologic phenotypes with or without atopic disease. J Allergy Clin Immunol 2019; 143:1482–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Greninger AL, Roychoudhury P, Xie H, et al. . Ultrasensitive capture of human herpes simplex virus genomes directly from clinical samples reveals extraordinarily limited evolution in cell culture. mSphere. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sauerbrei A, Bohn-Wippert K, Kaspar M, et al. . Database on natural polymorphisms and resistance-related non-synonymous mutations in thymidine kinase and DNA polymerase genes of herpes simplex virus types 1 and 2. J Antimicrob Chemother 2016; 71:6–16. [DOI] [PubMed] [Google Scholar]
- 8. Danve-Szatanek C, Aymard M, Thouvenot D, et al. . Surveillance network for herpes simplex virus resistance to antiviral drugs: 3-year follow-up. J Clin Microbiol 2004; 42:242–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Czartoski T, Liu C, Koelle DM, et al. . Fulminant, acyclovir-resistant, herpes simplex virus type 2 hepatitis in an immunocompetent woman. J Clin Microbiol 2006; 44:1584–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jing L, Haas J, Chong TM, et al. . Cross-presentation and genome-wide screening reveal candidate T cells antigens for a herpes simplex virus type 1 vaccine. J Clin Invest 2012; 122:654–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. van der Heiden P, Marijt E, Falkenburg F, Jedema I. Control of cytomegalovirus viremia after allogeneic stem cell transplantation: a review on CMV-specific T cell reconstitution. Biol Blood Marrow Transplant 2018; 24:1776–82. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.