Abstract
Inactivation of the β-cell transcription factor NEUROD1 causes diabetes in mice and humans. In this study, we uncovered novel functions of NEUROD1 during murine islet cell development and during the differentiation of human embryonic stem cells (HESCs) into insulin-producing cells. In mice, we determined that Neurod1 is required for perinatal proliferation of α- and β-cells. Surprisingly, apoptosis only makes a minor contribution to β-cell loss when Neurod1 is deleted. Inactivation of NEUROD1 in HESCs severely impaired their differentiation from pancreatic progenitors into insulin-expressing (HESC-β) cells; however, survival or proliferation was not affected at the time points analyzed. NEUROD1 was also required in HESC-β cells for the full activation of an essential β-cell transcription factor network. These data reveal conserved and distinct functions of NEUROD1 during mouse and human β-cell development and maturation, with important implications about the function of NEUROD1 in diabetes.
Introduction
Studies in rodents have identified networks of transcription factors that are essential for the differentiation and maintenance of functionally mature pancreatic islet cells (reviewed in Romer and Sussel [1]). To date, most monogenic forms of diabetes have been linked to disruptive mutations in transcription factors essential for the development of pancreatic islet cells (2). Accordingly, homozygous and heterozygous inactivating mutations in the basic helix-loop-helix (bHLH) transcription factor NEUROD1 can cause permanent neonatal diabetes mellitus and maturity-onset diabetes of the young 6, respectively (3,4). NEUROD1 was initially identified as a transactivator of the insulin gene (5), and subsequent studies demonstrated that NEUROD1 has a broader role in regulating β-cell function through the direct activation of genes important for β-cell maturation and function (6–10).
Loss-of-function studies in mice have revealed distinct cell type– and age-dependent requirements for NEUROD1 in the development of functional α- and β-cells. During murine pancreas development, NEUROD1 is expressed by differentiating endocrine cells and is subsequently restricted to insulin-producing β-cells and a subset of glucagon-expressing α-cells (11–13). Neurod1 knockout (KO) mice develop lethal neonatal diabetes that is at least partly due to a severe reduction in the numbers of α- and β-cells (12). Despite the transactivating functions of NEUROD1 for β-cell–specific gene expression, loss of Neurod1 in mice did not appear to affect the formation of the normal complement of insulin-expressing cells (12). Rather, the lack of α- and β-cells in Neurod1 KO mice was reported to result from perinatal loss due to apoptosis (12). Unexpectedly, while the Neurod1 KO phenotype suggested that NEUROD1 was a β-cell survival factor, β-cell–specific deletion of Neurod1 did not affect β-cell numbers (11,14). However, β-cell deletion of Neurod1 did impair the functional maturation of β-cells and blunted glucose-stimulated insulin secretion (14). Recent clinical studies as well as functional studies using the differentiation of human embryonic stem cells (HESCs) to model human pancreas development have demonstrated both conserved and divergent functions for several islet transcription factors between mice and humans (15–18). For example, NEUROG3, another bHLH transcription factor closely related to NEUROD1, is absolutely essential for the differentiation of endocrine cells in the mouse pancreas while only being partially required for the differentiation of human β-cells (16,17).
In this study, we revisited the embryonic phenotype of the Neurod1 KO mice to determine whether the discrepant phenotypes associated with global versus β-cell–specific deletion of Neurod1 were due to 1) timing of Neurod1 inactivation, 2) previously unappreciated functions of NEUROD1, and/or 3) cell nonautonomous effects. In addition, we performed detailed phenotypic analysis of NEUROD1 deletion in mouse and human model systems to discover novel requirements for NEUROD1 during the development of murine islet cells and during the differentiation of HESC-derived insulin-producing (HESC-β) cells. We discovered that Neurod1 KO mice develop fewer α- and β-cells due to major defects in embryonic α- and β-cell proliferation prior to any appreciable cell loss from apoptosis. Alternatively, disruption of NEUROD1 in HESCs severely impaired the differentiation of pancreatic progenitors into HESC-β cells, without impacting proliferation or survival, revealing the importance of NEUROD1 for the differentiation of human insulin-producing cells. On the other hand, NEUROD1 appears to be essential for maintaining the maturation state of both mouse and human β-cells.
Research Design and Methods
Mice
Details about the Neurod1LacZ (mouse genomics informatics [MGI]: 2385826) (19), Neurod1Floxed (MGI: 2385826) (20), Neurog3Cre (MGI: 3052639) (21), InsCre (MGI: 2387567) (22), and R26RTomato (MGI: 3809523) (23) mice and genotyping protocols used in these experiments have been previously published (14,19,21,24). All mice were maintained on a C57BL/J background. All experimental procedures and husbandry of mice were performed according to Columbia University–approved Institutional Animal Care and Use Committee protocols. Embryonic stages were counted as days after verification of insemination. Blood glucose of perinatal mice was measured using a glucometer (Freestyle Lite). Perinatal mouse studies did not consider sex as a factor in the statistical analysis of the data.
Culturing of HESCs
The HESCs used in these experiments are a National Institutes of Health–approved male line (MEL-1) that was previously modified to have a GFP expression cassette knocked into the insulin locus INSGFP/wt (research resource identifier [RRID] MEL-1 INSGFP/wt) (25). Cells of passage number 13–20 were maintained as previously described (26).
CRISPR Mutagenesis of HESCs
To target frameshift indels to the bHLH coding sequence of NEUROD1, a single guide (sg)RNA 5′-ACGCATGAAGGCTAACGCCC-3′ was designed with minimal off targets using the online CRISPR design tool (crispr.mit.edu). An sgRNA expression fragment was designed and synthesized using a previously published protocol (27). HESCs were nucleofected with this sgRNA fragment along with a Cas9-GFP plasmid (catalog no. 44719; Addgene) using a Human Stem Cell H9 Nucleofector kit (catalog no. VPH-5012; Lonza). GFP+ cells were sorted 48 h postnucleofection using a BD FACSAria II cell sorter and plated at 5,000 cells per 10-cm dish. Individual colonies were isolated by microdissection. QuickExtract DNA Extraction Solution (catalog no. QE09050; Epicentre) was used to isolate genomic DNA. Clonal lines were genotyped by Sanger sequencing of PCR products amplified using the following primers: forward 5′-CCAAGCTGGACAGACGAGTG-3′ and reverse 5′-AGTGTCGCTGCAGGATAGTG-3′. The genotypes of clonal lines with indels were then verified by cloning of PCR products using a TOPO TA cloning kit (catalog no. K4575J10; Invitrogen) followed by Sanger sequencing of at least four individual bacterial clones. Karyotyping was performed by Cell Line Genetics.
Differentiation of HESC-β Cells
After CRISPR mutagenesis, four clonal HESC lines homozygous with NEUROD1 bHLH–disrupting indels and four NEUROD1 wild-type control lines were differentiated into HESC-β cells using a published protocol (26).
Measurements of Insulin Secretion and Content
Insulin secretion and content were measured using 20 HESC-β cell clusters. The clusters were first rinsed in 0.5 mL Krebs-Ringer buffer (KRB) and then preincubated for 90 min in 0.2 mL 2.8 mmol/L glucose KRB inside a 1.5-mL tube with the cap open in a 37°C cell culture incubator. The clusters were then sequentially incubated in 200 μL of 2.8 mmol/L glucose KRB and then with 20 mmol/L glucose KRB for 30 min each. The insulin secreted by the clusters was measured using samples of media after each incubation. Insulin content was extracted by sonicating cells in 50 μL Tris EDTA buffer with 0.1% SDS followed by mixing the lysate in a 1:3 ratio with 0.18 mol/L HCl 96% ethanol. Insulin secretion and content were measured using a human insulin ELISA kit (catalog no. 10-1113-01; Mercodia).
Immunofluorescence Analysis
HESC-derived cells and frozen sections of mouse pancreas were prepared and stained for immunofluorescence (IF) analysis using previously described protocols (26,28). Primary and secondary antibodies are listed in Supplementary Table 1. Apoptotic cells were labeled using a TUNEL Assay Apoptosis Detection Kit (catalog no. 30064; Biotium). Entire sections of mouse pancreas or day 27 HESC clusters were imaged using a Zeiss LSM 710 confocal scope. HESC day 12 pancreatic progenitors were imaged using an Olympus IX53 fluorescent microscope. Fluorescently labeled cells and pancreas area were manually quantified using ImageJ software. Cells were quantified on at least three sections of mouse pancreas spaced 200 μm apart and on four sections of different day 27 HESC–derived clusters. Pancreatic progenitors were quantified on four random images in the well.
Flow Cytometry Analysis
Cells were prepared and stained for flow cytometry analysis using previously described protocols (26). Primary and secondary antibodies used for flow cytometry analysis are listed in Supplementary Table 1. To measure 5-ethynyl-2′-deoxyuridine (EDU) incorporation, cells were incubated in media with 10 μmol/L EDU 4 h prior to their dissociation. A Click-iT EDU Cell Proliferation Kit (catalog no. C10338; Thermo Fisher Scientific) was used to stain cells that had incorporated EDU. Automated cell counting was performed using a BD Biosciences LSRFortessa cytometer, and data were analyzed using FCS Express 6 software.
RNA Sequencing
Single-cell suspensions of dissected embryonic day 17 (E17) mouse pancreas and HESC-β cell clusters were prepared by a 30-min 37°C digestion with Accutase (catalog no. A6964; Sigma-Aldrich) supplemented with 25 μg/mL DNASE1 (catalog no. 10104159001; Sigma-Aldrich) and TrypLE Express (catalog no. 12605036; Gibco) for 10 min at room temperature, respectively, followed by dissociation with pipetting. The mouse pancreas and HESC-β cell digestions were quenched by adding 2- and 10-fold the volume of RPMI 1640 media (catalog no. 11875119; Gibco) supplemented with 10% FBS (catalog no. 900-108; Gemini Bioproducts) and 10 μmol/L Y27632 rock inhibitor (catalog no. S1049; Selleckchem), respectively, filtered through a 40-μm mesh, pelleted by centrifugation, and suspended in quench media. Human INS-GFP+ and E17 mouse Neurog3:Cre; R26R:Tomato+ lineage–labeled cells were sorted using a BD FACSAria II cell sorter. Pancreatic islets were isolated from Neurod1fl/fl;InsCre+ and control (Neurod1fl/fl or InsCre+) 12-week-old female mice by the perfusion and digestion of pancreata using collagenase P (catalog no. 11 213 857 001; Roche), followed by the separation of islets using a Histopaque (catalog no. 10831; Sigma-Aldrich) gradient as previously described (14). RNA was purified using RNAeasy Micro Kits (catalog no. 74004; QIAGEN). RNA quality and concentration were determined using an Agilent Bioanlyzer. Due to low yield of E17 mouse Neurog3:Cre; R26R:Tomato+ lineage–labeled cells, 2.315 ng RNA (RNA integrity number >8) was converted to cDNA and amplified using an Ovation RNA Amplification System V2 (catalog no. 3100-12; NuGen) according to the manufacture’s protocol. Other cDNA libraries were prepared from ∼70–700 ng total RNA (RNA integrity number >8) using a TruSeq RNA prep kit (catalog no. RS-122-2001; Illumina). RNA Sequencing (RNA-Seq) and data processing were performed by the Columbia Genome Center. Multiplexed libraries of cDNA were sequenced using an Illumina HiSeq2000 to yield ∼30 million single-end 100-bp reads. Base calling was performed using RTA (Illumina) software, and bcl2fastq (version 1.8.4) coupled with adaptor trimming was used to convert binary base call format to fastq format. Tophat (version 2.1.0) was used to map reads to the reference genome (human: NCBI/build37.2; mouse: UCSC/mm9), allowing for four mismatches and 10 maximum multiple hits (29). Cufflinks (version 2.0.2) with default settings was used to estimate the relative expression level of genes and splice isoforms (30). Statistics for differential gene expression were calculated using the R package DEseq (31).
Real-Time Quantitative PCR
Total RNA was extracted from cells at days 13, 15, 17, 20, and 27 of differentiation using an RNeasy Mini Kit (catalog no. 74106; QIAGEN). Total RNA was reverse transcribed into cDNA using iScript Reverse Transcription Supermix (catalog no. 170-8841; Bio-Rad). Quantitative PCR reactions were set up by mixing SsoFast EvaGreen Supermix (catalog no. 172-5202; Bio-Rad) with primers and cDNA templates. Quantitative PCR was performed using a CFX96 Touch Real-Time PCR Detection System. Primer sequences were as follows: INSULIN (INS) (TTCTACACACCCAAGACCCG and CAATGCCACGCTTCTGC), GLUCAGON (GCG) (AAGTTCCCAAAGAGGGCTTG and AGCTGCCTTGTACCAGCATT), and TBP (TGTGCACAGGAGCCAAGAGT and ATTTTCTTGCTGCCAGTCTGG). NEUROD1 primer sequences were published by Zengrong et al. (17).
Determination of Genes With Nearby Neurod1 Binding Sites
The identification of genes with nearby NEUROD1 binding sites in adult mouse islets was performed by the Herbert Irving Comprehensive Cancer Center Bioinformatics Shared Resource. ChIP-Seq data from Tennant et al. (6) for NEUROD1 binding in adult mouse islets was obtained as a BAM file from GSE30298 (32). Peaks were identified with MACS 14.0 (33) using default parameters and associated with genes 3 kb in either direction with ChipPeakAnno (34).
Statistics
The P value of significance for data sets of indicated genotypes was calculated using two-tailed, unequal variance Student t tests. Adjusted P values were calculated as false discovery rate using the Benjamini-Hochberg procedure.
Data and Resource Availability
RNA-Seq data sets are available at the National Center for Biotechnology Information’s Gene Expression Omnibus database under the accession number GSE30298. All other data sets generated and/or analyzed for this study are available from the corresponding author upon reasonable request. NEUROD1mut/mut HESC lines [RRID: MEL1INSGFP/wt NEUROD1(−/−)] are available from the corresponding author upon reasonable request.
Results
Murine NEUROD1 Is Required in the NEUROG3 Lineage for the Proliferative Expansion of Embryonic α- and β-Cells
To elucidate the Neurod1-dependent mechanisms that are important for the development of fully functional complements of α- and β-cells, we first needed to precisely determine the developmental stage and cell type in which NEUROD1 is required for α- and β-cell development. Previous studies reported that NEUROD1 was dispensable for the differentiation of endocrine islet cells but was subsequently required for their perinatal survival and maturation (12,14). Specific deletion of Neurod1 in β-cells, however, had no effect on β-cell numbers, suggesting that NEUROD1 has stage-specific functions and/or is required prior to endocrine cell differentiation to establish and/or maintain normal numbers of islet cells (11,14).
Consistent with previous reports, we found that Neurod1 KO pancreata develop a nearly 3.0- to 4.5-fold reduction in the numbers of insulin-expressing β-cells and glucagon-expressing α-cells between E17 and birth (P0) (Supplementary Fig. 1A and B). There were no detectable defects in the formation of Neurog3+ endocrine progenitor cells (Supplementary Fig. 1C), and the reduction in α- and β-cell numbers occurred after the majority of endocrine islet cell differentiation was completed, suggesting that loss of Neurod1 did not disrupt endocrine progenitor differentiation. This finding was supported by deletion of Neurod1 in the NEUROG3 endocrine lineage using Neurod1fl/fl; Neurog3:Cre mice (Neurod1Δendo); these mice phenocopied Neurod1 KO mice, resulting in a similar reduction in the numbers of Ins+ and Gcg+ cells in the pancreas by birth, as well as lethal perinatal hyperglycemia (11) (Supplementary Fig. 2A–C).
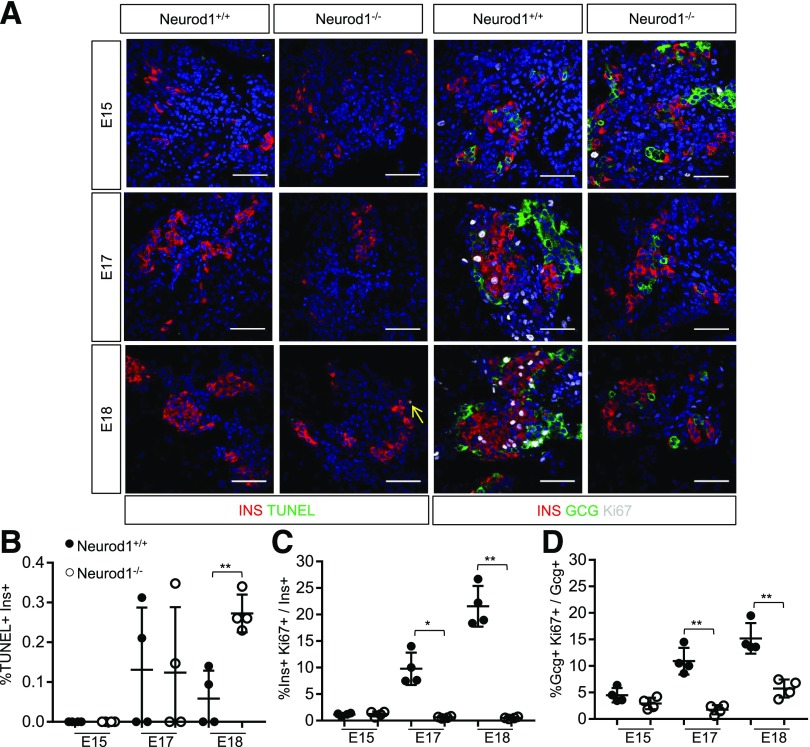
We also wanted to confirm that the observed reduction in β-cell numbers was due primarily to apoptosis, as previously described (12). Surprisingly, although Neurod1 KO pancreata had significantly elevated numbers of apoptotic TUNEL+ insulin-expressing cells at E18, these apoptotic cells were extremely rare (0.25% of insulin+ cells) and were unlikely to explain the 3.0- to 4.5-fold reduction in the numbers of α- and β-cells that occurred between E17 and birth (Fig. 1A and B). We also did not detect measurable apoptosis at earlier developmental time points, suggesting that we had not missed an earlier wave of cell death that would account for the loss of α- and β-cells in Neurod1 KO pancreata. Since the reduction in α- and β-cells could not be explained by defects in differentiation or elevated cell death, we examined the proliferation of α- and β-cells in Neurod1 KO mice at E15, E17, and E18, prior to and during the stages of robust endocrine cell proliferation. Beginning at E17, Neurod1 KO pancreata had a severe reduction of insulin+ and glucagon+ cells expressing the proliferation marker Ki67 (Fig. 1A, C, and D). This analysis suggests that the perinatal reduction of α- and β-cells in the Neurod1 KO pancreas is primarily due to defects in their proliferative expansion, with only minor contribution from apoptosis.
Figure 1.
Murine NEUROD1 is required for the late fetal expansion of pancreatic INSULIN+ and GLUCAGON+ cells. A: IF expression analysis of indicated markers and TUNEL staining performed on sections of E15, E17, and E18 pancreata. Scale bar = 50 μm. B: Quantification of the percent of insulin+ cells that are TUNEL positive. C: Quantification of INSULIN+ cells expressing Ki67. The total number of Ins+ cells counted per Neurod1+/+ sample was E18 = 554 ± 162, E17 = 347 ± 184, and E15 = 308 ± 161 and per Neurod1−/− was E18 = 548 ± 102, E17 = 344 ± 100, and E15 = 293 ± 36. D: Quantification of GLUCAGON+ cells expressing Ki67. The total number of GCG+ cells counted per Neurod1+/+ sample was E18 = 207 ± 89, E17 = 94 ± 49, and E15 = 220 ± 121 and per Neurod1−/− was E18 = 112 ± 45, E17 = 108 ± 42, and E15 = 111 ± 57. All error bars indicate SD. Student t test P value compares values of controls to those of indicated genotype. *P < 0.01; **P < 0.005.
Murine NEUROD1 Regulates a β-Cell Differentiation and Proliferation Program
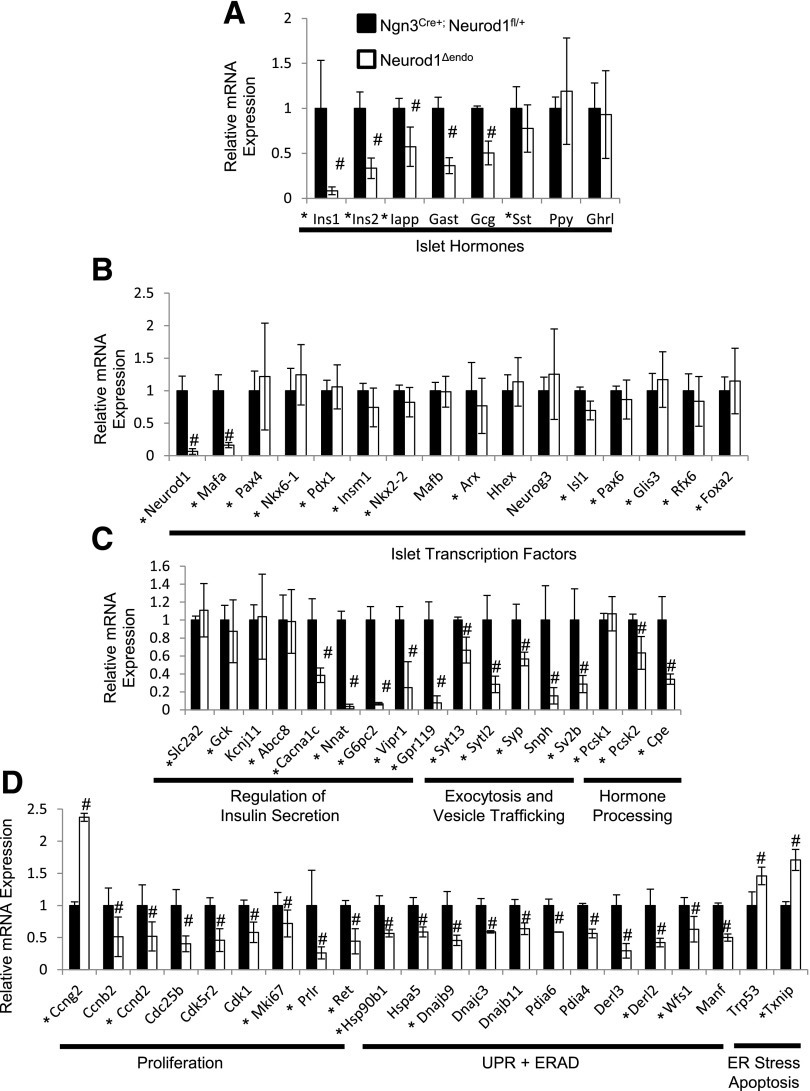
The surprising discovery that α- and β-cell loss in the Neurod1 KO mice was largely due to proliferation defects prompted us to revisit the molecular pathways regulated by NEUROD1 in the endocrine lineage. We performed RNA-Seq transcriptome analysis on sorted pancreatic Neurod1fl/fl; Neurog3:Cre; R26R:Tomato+ lineage–labeled endocrine cells and compared them to littermate controls at E17, prior to the reported major α- and β-cell deficiency caused by Neurod1 ablation (Supplementary Table 2). As expected from previous studies, gene ontology analysis indicated that a significant number of genes downregulated by Neurod1 depletion are involved in the regulation of insulin secretion, hormone secretion, and hormone levels (Supplementary Table 3). Importantly, the β-cells maintained expression of definitive β-cell transcription factors, including Nkx6-1, Pdx1, and Nkx2-2, suggesting that β identity was not compromised (Fig. 2B and Supplementary Fig. 3). However, Neurod1-deficient endocrine cells had reduced expression of genes that encoded proteins specifically important for the maturation and function of β-cells, including Ins1 and Ins2, the β-cell maturation transcription factor gene Mafa, as well as several regulators of insulin secretion and insulin processing (Fig. 2A–C). In addition, the expression of Gcg and Gast was reduced (Fig. 2), agreeing with previous reports showing that pancreatic mouse Gcg and Gast expression is dependent on NEUROD1 regulation (24,35).
Figure 2.
Murine NEUROD1 binds near and is required to regulate the transcription of genes important for the differentiation of functional β-cells, proliferation, and survival. A–D: RNA-Seq expression data for Neurod1Δendo and Ngn3Cre+; Neurod1fl/+ E17-sorted pancreatic endocrine cells. An asterisk indicates Neurod1-bound genes in 12-week-old mouse islets determined using a published ChIP-Seq data set (Tennant et al. [6]). #P value <0.01. n = 4 for each genotype. Error bars indicate SD.
The expression profile of cell cycle regulators in Neurod1-deficient cells was consistent with the severe proliferation defects we observed in the endocrine cells. Notably, Neurod1-deficient cells had elevated expression of Ccng2, a negative regulator of cell cycle progression, and reduced expression of Ccnb2, Ccnd2, Cdc25b, and Cdk1, which promote cell cycle progression (Fig. 2D). In addition, Neurod1-deficient cells had reduced expression of Prlr and Ret, which are receptors for the potent secreted β-cell mitogens PROLACTIN and GDNF, respectively (36–39) (Fig. 2D). Both Prlr lactogens and GDNF have been shown to be elevated during embryogenesis at the stage where we observe β-cell proliferation defects in the Neurod1 KO and Neurod1Δendo mice (40,41). Furthermore, Prlr is required for the proliferation of β-cells specifically during the perinatal stages (42). These data support a role for Neurod1 in inducing a program essential for the proliferative expansion of embryonic α- and β-cells.
Interestingly, transcriptome analysis also indicated that Neurod1-deficient E17 endocrine cells had a significant number of downregulated genes that are involved in the unfolded protein response (UPR) and endoplasmic reticulum (ER)–associated degradation (Fig. 2D and Supplementary Table 3). UPR and ER-associated degradation genes remediate ER stress by facilitating the folding and degradation of misfolded proteins, respectively. Prolonged ER stress can blunt insulin secretion and induce β-cell apoptosis via induction of TXNIP, which had elevated expression in Neurod1 KO cells (43,44) (Fig. 2D). These data suggest that in addition to its proliferation and maturation functions, NEUROD1 regulates genes important for the remediation of ER stress.
Distinct Neurod1-Dependent Gene Regulation Programs in Embryonic and Adult Mouse Islet Cells
Since our studies identified novel perinatal defects associated with Neurod1 KO and Neurod1Δendo mice, we wished to readdress the similarities and differences of NEUROD1 function in the embryo versus adult animals that have an impaired maturation phenotype (12,14). We generated RNA-Seq transcriptome data for adult (12 weeks old) islets from β-cell–specific Neurod1 KO (Neurod1fl/fl; Ins:Cre; hereafter referred to as Neurod1Δβ) and control (Ins:Cre or Neurod1fl/fl) female mice (Supplementary Table 4) and compared NEUROD1-dependent gene regulation in embryonic pancreatic endocrine lineages versus adult islet β-cells (Fig. 3A). As expected, NEUROD1 is required in both embryonic endocrine lineages and adult β-cells for the expression of numerous genes important for β-cell function (Fig. 3B). Importantly, genes specifically dysregulated by loss of Neurod1 in embryonic NEUROG3 lineages included many genes important for proliferation, which is consistent with the perinatal proliferation defects observed in these mice (Fig. 3B). Alternatively, β-cell–specific deletion of Neurod1 preferentially altered the expression of genes involved in the functional maturation of β-cells, as previously described (14) (Fig. 3B). Interestingly, loss of Neurod1 in embryonic endocrine cells compared with its loss in adult islet β-cells had the opposite effect on the regulation of several genes, including the β-cell immaturity marker Npy, proliferation gene Prlr, and several UPR genes. These data indicate that the gene programs and transcriptional regulatory activity of NEUROD1 are stage and context dependent.
Figure 3.
Stage-specific NEUROD1-dependent gene regulation in murine E17 endocrine cells and adult β-cells. A: Venn diagrams depicting overlap of genes bound by NEUROD1 in adult islets and either down- or upregulated (P < 0.01, fold change > 1.5×) by NEUROD1 depletion in E17-sorted pancreatic endocrine cells vs. adult islet β-cells (beta). NEUROD1-bound genes in adult mouse islets determined using a published ChIP-seq data set (Tennant et al. [6]) B: Tables of Neurod1-dependent genes in E17 endocrine cells or 12-week-old adult β-cells or both. Genes down- or upregulated in NEUROD1-deficient cells are in red and blue, respectively. An asterisk indicates genes bound by NEUROD1 in adult mouse islets.
NEUROD1 Binds Near Proliferation and β-Cell Functional Genes in Adult Murine Islets
Since there appears to be stage-specific requirements for NEUROD1 regulation of the proliferation and maturation of islet cells, we wanted to determine which genes were directly regulated by NEUROD1. To identify direct regulated targets of Neurod1, we used a previously published ChIP-Seq data set in adult murine islets that identified NEUROD1 binding at islet-specific enhancers (6). We found that Neurod1 binds near many of the shared genes known to require NEUROD1 for their regulation during embryonic endocrine cell differentiation and in adult islet β-cells (Figs. 2 and 3). NEUROD1 also bound several proliferation genes that had altered expression in Neurod1-deficient cells specifically during embryonic endocrine differentiation but were unchanged in adult β-cells (Figs. 2D and 3B). Conversely, NEUROD1 bound and regulated a distinct set of β-cell maturation and function genes in the adult that were either unchanged or differentially regulated in the embryo, again suggesting that there are important context-dependent factors influencing NEUROD1 function. Similar to what has been reported for several other transcription factors, many genes bound by NEUROD1 in adult islets do not appear to be dependent on NEUROD1 for their regulation in adult islet β-cells. For example, NEUROD1 only appears to regulate a subset of the β-cell transcription factors that it binds to in adult islets (Figs. 2B and 3A). Hence, NEUROD1 is dispensable for the transcriptional regulation of many of its targets, indicating either compensatory mechanisms for NEUROD1-regulated gene expression or functional redundancy between Neurod1 and other islet transcription factors. The latter explanation is supported by the co-occupancy of NEUROD1 with other β-cell transcription factors at islet cell–specific enhancers (6,7).
Mutation of NEUROD1 in HESCs Specifically Impairs β-Cell Differentiation
Aspects of murine and human pancreas development have been shown to differ in their requirements for several pancreatic transcription factors (15,16). Therefore, we wanted to determine whether NEUROD1 has conserved essential functions in human β-cell development, which can now be modeled by the differentiation of HESCs into HESC-β cells (Supplementary Fig. 4A). To disrupt NEUROD1 in HESCs, we used CRISPR/Cas9-mediated mutagenesis to target frameshift indels into the essential bHLH domain of NEUROD1 (Supplementary Fig. 4B). We used a previously described HESC line (MEL1 INSGFP/wt) that has a GFP expression cassette knocked into a single copy of the insulin gene (25). We differentiated four mutated (NEUROD1mut/mut) HESC lines (three to four experimental replicates per line) into insulin-producing cells using a protocol that recapitulates the major stages of in vivo β-cell development, including the differentiation of definitive endoderm and pancreatic progenitors (26). All four lines produced similar results, and we confirmed that targeted line NEUROD1mut/mut clone 12 had reduced NEUROD1 protein expression at the β-cell stage of differentiation (day 27) and a normal karyotype (Supplementary Fig. 4C and D). This line was used for all subsequent analyses.
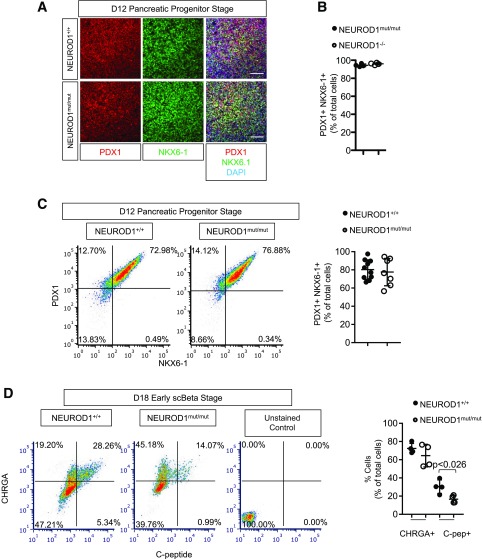
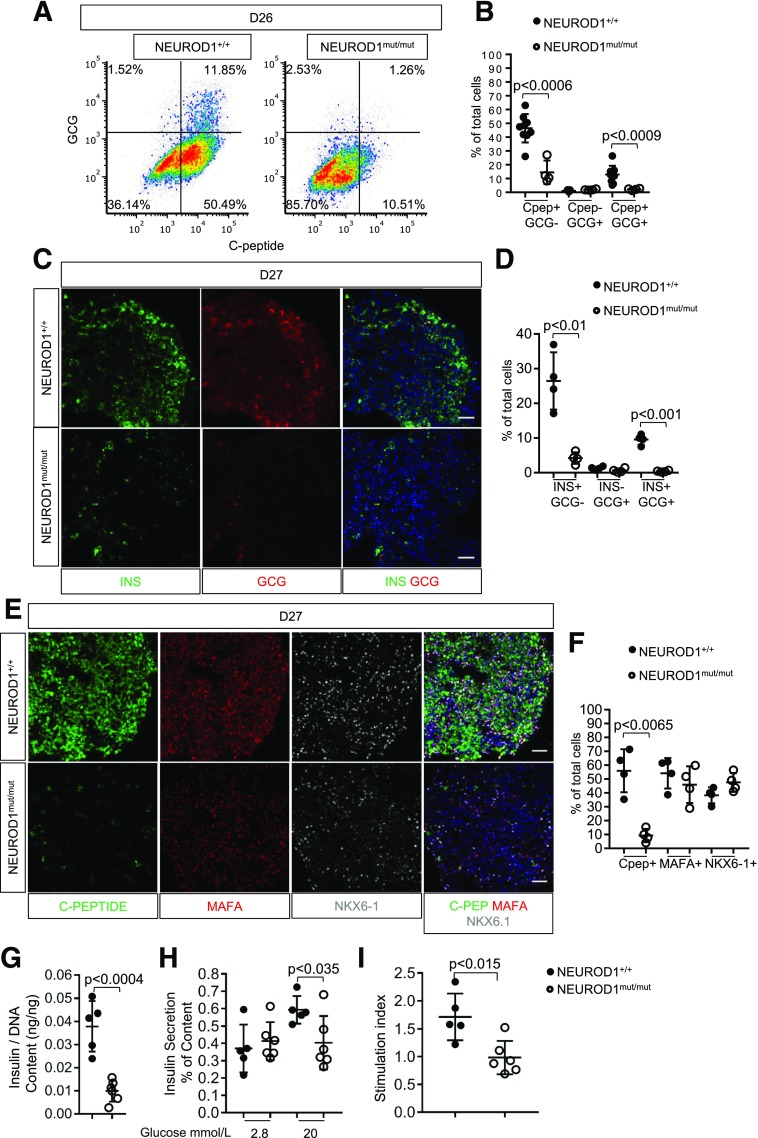
During HESC-β cell differentiation, NEUROD1 mRNA expression was induced at the pancreatic endocrine precursor stages (day 15) just prior to hormone induction (Supplementary Fig. 5A). At day 12 of the differentiation process, almost all NEUROD1mut/mut and control cells differentiated into cells expressing the endocrine-biased pancreatic progenitor markers PDX1+ NKX6-1+ (Fig. 4A–C). At the HESC-β cell stages (days 18–27) of differentiation, NEUROD1mut/mut cell populations had normal numbers of CHRGA+ endocrine cells; however, there was a striking reduction in the number of cells expressing INSULIN and/or the INSULIN-GFP reporter (Figs. 4D and 5A–F, Supplementary Fig. 5B and C, and Supplementary Fig. 6A and B). NEUROD1mut/mut cells did have similar numbers of cells expressing the β-cell differentiation and maturation transcription factors NKX6-1 and MAFA compared with NEUROD1 wild-type controls; however, there was a noticeable reduction in their expression levels per cell, based on IF intensity (Fig. 5E and F). Loss of NEUROD1 during HESC-β cell differentiation did not appear to direct cells to an α-cell or polyhormonal cell fate: NEUROD1mut/mut cell populations had similar low numbers of GLUCAGON+ C-PEPTIDE− cells compared with controls, as well as reduced numbers of GLUCAGON and C-PEPTIDE coexpressing cells (Fig. 5A–D and Supplementary Fig. 5B and C). Consistent with the reduction in C-peptide+ cells, NEUROD1mut/mut cells displayed a reduction in insulin content compared with control cells, in addition to defective glucose-stimulated insulin secretion (Fig. 5G–I). Collectively, these data indicate that NEUROD1 is required downstream of NKX-6-1+ PDX1+ HESC-derived endocrine progenitors to produce normal numbers of HESC-β cells.
Figure 4.
Impaired differentiation of NEUROD1mut/mut HESC-derived pancreatic progenitors into insulin-expressing cells. A: IF of pancreatic progenitor markers PDX1 and NKX6.1 at day 12 of differentiation. Scale bar = 100 μm. B: Quantification of PDX1+, NKX6-1+ cells at day 12 of differentiation. C: Flow cytometry expression analysis and quantification of lineage markers at day 12 of HESC-β differentiation. D. Flow cytometry expression analysis of lineage markers at day 18 of HESC-β differentiation. Scale bar = 50 μm. n = 4 for each genotype. Error bars indicate SD. Student t test P value compares values of controls to NEUROD1mut/mut. scBeta, stem cell–derived β-cells. *P < 0.01.
Figure 5.
NEUROD1 is required for the differentiation HESC-β cells. A: Flow cytometry expression analysis of lineage markers for day 26 (D26) cells of HESC-β differentiation. B: Quantification of hormone-expressing cells determined by flow cytometry. C: IF expression of INSULIN and GLUCAGON by day 27 HESC–derived endocrine clusters, nuclei stained with DAPI. Scale bar = 50 μm. D: Quantification of hormone-expressing cells determined by IF analysis. E: IF expression analysis revealed reduced expression of C-peptide, MAFA, and NKX6-1 by NEUROD1mut/mut day 27 endocrine clusters compared with controls. Nuclei are stained with DAPI. Scale bar = 50 μm. F: Quantification of C-peptide, MAFA, and NKX-6-1–expressing cells as a percentage of total nuclei. G: Insulin content normalized to DNA content H: Secreted insulin content by day 27 endocrine clusters differentiated from HESCs in response to 2 and 20 mmol/L glucose. I: Fold change in insulin secretion of HESC-β cells in response to 20 mmol/L glucose relative to insulin secretion at 2 mmol/L glucose. Error bars indicate SD. Student t test P value compares values of controls to NEUROD1mut/mut.
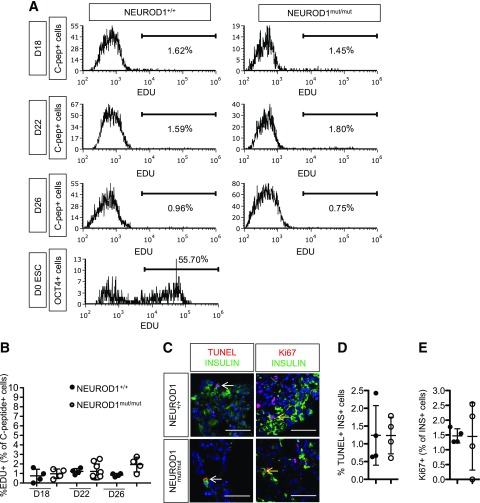
Since loss of NEUROD1 impairs the proliferation and survival of embryonic β-cells in mice, we looked for similar phenotypes in NEUROD1-deficient HESC-β cells. We measured EDU incorporation at several time points during the differentiation protocol but could not identify proliferation defects (Fig. 6A and B). Furthermore, IF staining for Ki67 and TUNEL analysis at the end of the differentiation protocol detected similarly low percentages of either proliferating or apoptotic HESC-β cells between control and NEUROD1mut/mut cells (Fig. 6C and D). Therefore, under these culture conditions, we found no requirement of NEUROD1 for the proliferation or survival of HESC-β cells.
Figure 6.
NEUROD1mut/mut and control HESC-β cells have similar low rates of proliferation and apoptosis. A: Flow cytometry histograms of EDU incorporation for C-peptide+ cells at different stages of HESC-β cell differentiation as well as for OCT4+ cells in ESCs. B: Quantification of proliferating EDU+ C-peptide+ cells determined by flow cytometry analysis. C: TUNEL and IF staining for Ki67 and insulin on sections of day 27 (D27) HESC–derived endocrine clusters. Yellow arrow points to Ki67+ insulin+ cells. White arrow points to TUNEL+ INSULIN+ cell D: Quantification of apoptotic TUNEL+ INSULIN+ cells. E: Quantification of INSULIN+ cells expressing proliferation marker Ki67.
Conserved and Distinct Gene Regulation by NEUROD1 in Mouse and HESC-β Cells
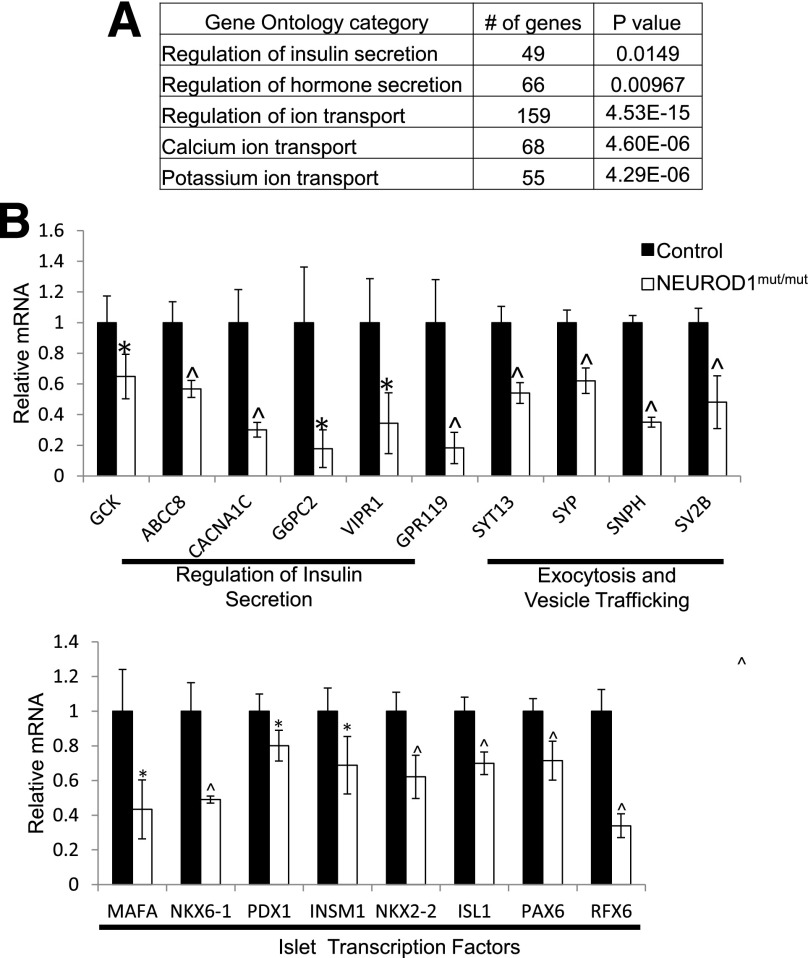
To identify the molecular defects associated with the dysfunctional NEUROD1mut/mut HESC-β cells, we performed RNA-Seq expression analysis on NEUROD1mut/mut and control FACS-sorted day 27 INSULIN-GFP+ cells at the end of the differentiation protocol (Supplementary Fig. 6A). Consistent with the observed defect in β-cell function (Fig. 5H and I), gene ontology analysis indicated that many of the genes downregulated by NEUROD1 ablation are involved in the regulation of insulin secretion, including GLUCOKINASE (GCK), ATP-gated K+ efflux channel ABCC8 (SUR1), and the calcium channel CACNA1a (Fig. 7A and B). To identify conserved functions for NEUROD1 between mice and humans, we compared the dysregulated genes due to loss of NEUROD1 in HESC-β cells and murine E17 endocrine cells (Neurod1Δendo) as well as adult murine islet β-cells (Neurod1Δβ) (Supplementary Fig. 7). Although each population displayed distinct gene expression profiles, likely due to differences in developmental time points (embryonic vs. adult) and artifacts associated with the in vitro differentiation program, NEUROD1mut/mut cells shared a large proportion of dysregulated genes with Neurod1-deficient adult mouse islets, suggesting that NEUROD1 has a number of conserved β-cell functions. Interestingly, however, unlike in mice, mutation of NEUROD1 in HESC-β cells caused reduced expression of many of the essential β-cell transcription factors, including MAFA, NKX6-1, PDX1, INSM1, NKX2-2, ISL1, PAX6, and RFX6 (Fig. 7B and Supplementary Fig. 7), suggesting that NEUROD1 may have an expanded and/or less redundant role in regulating the essential β-cell transcription factor network in humans.
Figure 7.
Reduced expression of β-cell genes in NEUROD1mut/mut INSULIN-GFP+ cells differentiated from HESCs. A: PANTHER gene ontology categories with significant enrichment of genes downregulated (adjusted P value = 0.05) in NEUROD1mut/mut compared with control day 27-sorted INSULIN-GFP+ cells. B: RNA-Seq data of β-cell genes downregulated in NEUROD1mut/mut relative to control D27-sorted INSULIN-GFP+ cells. *P value <0.05; ^P value <0.01. n = 4 for each genotype. Error bars indicate SD.
Discussion
Previous studies using mouse models suggested that NEUROD1 was a survival factor for neonatal β-cells but played a predominant role in regulating β-cell maturation in adult β-cells (12,14). In this study, we used detailed phenotypic and transcriptome analysis to identify a prenatal requirement of Neurod1 in the regulation of β-cell proliferation. We also identified conserved and distinct functions of NEUROD1 in HESC-β cells. Collectively, these findings provide valuable mechanistic insight to how inactivating mutations in NEUROD1 can cause defects in the development and function of β-cells.
In mice, NEUROD1 directly regulates a broad scope of genes important for the formation and maturation of β-cells. Consistent with previous reports, we found that loss of NEUROD1 in the NEUROG3 lineage prior to endocrine cell differentiation drastically reduced the number of α- and β-cells at birth (11). However, our study determined that deletion of Neurod1 severely impairs the perinatal proliferative expansion of α- and β-cell populations while causing a relatively minor loss of β-cells by apoptosis. Therefore, we propose that the major cause of α- and β-cell deficiency in Neurod1 KO mice is primarily due to α- and β-cell proliferation defects and not from apoptosis. This novel requirement of NEUROD1 for β-cell proliferation during embryonic development may have been previously overlooked because of the limited markers and assays and/or strain background differences that are known to influence Neurod1 mutant phenotypes (12,45).
Our data using an HESC differentiation protocol also provide support for NEUROD1 inactivation causing permanent neonatal diabetes mellitus and defective β-cell function. Patients with NEUROD1-inactivating mutations have been reported to have normal pancreas size and exocrine function but reduced circulating insulin (3,4). These clinical features of NEUROD1-linked diabetes are consistent with our finding that NEUROD1 has conserved functions in mice and humans for inducing and/or maintaining insulin expression and the expression of other genes that are important for β-cell function. Consistent with Neurod1 loss-of-function phenotypes in mice, NEUROD1 is dispensable in HESCs for pancreatic progenitor cell differentiation, but it is critical for the subsequent differentiation of HESC-β cells. However, unlike Neurod1 KO mice, the low yield of NEUROD1 mutant HESC-β cells in culture appeared to result not from defects in proliferation or apoptotic loss but rather from a failure of pancreatic progenitors to differentiate into insulin-expressing HESC-β cells. Furthermore, the few remaining NEUROD1-deficient HESC-β cells that did form displayed reduced expression of a β-cell transcription factor network necessary for the formation of fully functional β-cells. These data suggest there is a more robust requirement of NEUROD1 for the differentiation of fully functional insulin-expressing β-cells.
Although we were surprised by the lack of proliferation and survival defects in the HESC-derived endocrine cells, there are several possible explanations. Differences in the functional properties of human and murine NEUROD1 protein are unlikely since their amino acid sequences are 98% identical; however, differences in genetic modifiers between mice and humans could possibly influence the effects of NEUROD1 deficiency. Genetic background, for instance, has already been shown to profoundly influence the penetrance and expressivity of diabetes caused by Neurod1 deletion in mice (45). Similarly, varying penetrance of maturity-onset diabetes of the young 6 has been observed within pedigrees (4). Some of the observed differences in phenotypes and gene regulation could also reflect inherent differences in the HESC-β cell culture conditions, which may not accurately mimic the burst of proliferation observed in mice. We were also surprised that the NEUROD1-deficient HESC-β cells displayed much more severe impairment of many essential β-cell transcription factors than observed in mice. In this case, the difference in phenotypes may indicate a more critical role for NEUROD1 in regulating the β-cell regulatory network, or may suggest that the transcriptional program of HESC-β cells is more susceptible to minor transcriptional perturbations.
In summary, these studies clarify the roles for NEUROD1 in the regulation of β-cell formation and function. Whereas NEUROD1 appears to be essential for perinatal expansion of the β-cell population in mice, NEUROD1 may play a more important role in the differentiation of human β-cells. Alternatively, NEUROD1 has conserved functions in its regulation of key β-cell genes that are essential for maintaining β-cell maturation and function.
Supplementary Material
Article Information
Funding. The research of this study was funded by National Institute of Diabetes and Digestive and Kidney Disorders National Service Research Awards F32-DK-103506 (A.I.R.), R01-DK-082590, and U01-DK-072504 (L.S.) and the American Diabetes Association (1-16-ICTS-029 to D.E.). These studies used the resources of the Herbert Irving Comprehensive Cancer Center Flow Cytometry Shared Resources funded in part through Center grant P30-CA-013696 and the Diabetes and Endocrinology Research Center Flow Core Facility funded in part through Center grant 5P30-DK-063608.
Duality of Interest. No potential conflicts of interest relevant to this article were reported.
Author Contributions. A.I.R. wrote the manuscript. A.I.R. designed and conducted the experiments with advice from D.E. and L.S. Experiments were conceived by A.I.R., D.E., and L.S. R.A.S. provided computational analysis of the RNA-Seq data with help from A.I.R., D.E., and L.S. in the interpretation of results. R.A.S. and L.S. reviewed and edited the manuscript. A.I.R. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Footnotes
This article contains Supplementary Data online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db19-0117/-/DC1.
References
- 1.Romer AI, Sussel L. Pancreatic islet cell development and regeneration. Curr Opin Endocrinol Diabetes Obes 2015;22:255–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McCarthy MI, Hattersley AT. Learning from molecular genetics: novel insights arising from the definition of genes for monogenic and type 2 diabetes. Diabetes 2008;57:2889–2898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rubio-Cabezas O, Minton JA, Kantor I, Williams D, Ellard S, Hattersley AT. Homozygous mutations in NEUROD1 are responsible for a novel syndrome of permanent neonatal diabetes and neurological abnormalities. Diabetes 2010;59:2326–2331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Malecki MT, Jhala US, Antonellis A, et al. . Mutations in NEUROD1 are associated with the development of type 2 diabetes mellitus. Nat Genet 1999;23:323–328 [DOI] [PubMed] [Google Scholar]
- 5.Naya FJ, Stellrecht CM, Tsai MJ. Tissue-specific regulation of the insulin gene by a novel basic helix-loop-helix transcription factor. Genes Dev 1995;9:1009–1019 [DOI] [PubMed] [Google Scholar]
- 6.Tennant BR, Robertson AG, Kramer M, et al. . Identification and analysis of murine pancreatic islet enhancers. Diabetologia 2013;56:542–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jia S, Ivanov A, Blasevic D, et al. . Insm1 cooperates with Neurod1 and Foxa2 to maintain mature pancreatic β-cell function. EMBO J 2015;34:1417–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moates JM, Nanda S, Cissell MA, Tsai MJ, Stein R. BETA2 activates transcription from the upstream glucokinase gene promoter in islet beta-cells and gut endocrine cells. Diabetes 2003;52:403–408 [DOI] [PubMed] [Google Scholar]
- 9.Kim JW, Seghers V, Cho JH, et al. . Transactivation of the mouse sulfonylurea receptor I gene by BETA2/NeuroD. Mol Endocrinol 2002;16:1097–1107 [DOI] [PubMed] [Google Scholar]
- 10.Marsich E, Vetere A, Di Piazza M, Tell G, Paoletti S. The PAX6 gene is activated by the basic helix-loop-helix transcription factor NeuroD/BETA2. Biochem J 2003;376:707–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mastracci TL, Anderson KR, Papizan JB, Sussel L. Regulation of Neurod1 contributes to the lineage potential of Neurogenin3+ endocrine precursor cells in the pancreas. PLoS Genet 2013;9:e1003278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Naya FJ, Huang HP, Qiu Y, et al. . Diabetes, defective pancreatic morphogenesis, and abnormal enteroendocrine differentiation in BETA2/neuroD-deficient mice. Genes Dev 1997;11:2323–2334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li HJ, Kapoor A, Giel-Moloney M, Rindi G, Leiter AB. Notch signaling differentially regulates the cell fate of early endocrine precursor cells and their maturing descendants in the mouse pancreas and intestine. Dev Biol 2012;371:156–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gu C, Stein GH, Pan N, et al. . Pancreatic beta cells require NeuroD to achieve and maintain functional maturity. Cell Metab 2010;11:298–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xuan S, Borok MJ, Decker KJ, et al. . Pancreas-specific deletion of mouse Gata4 and Gata6 causes pancreatic agenesis. J Clin Invest 2012;122:3516–3528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rubio-Cabezas O, Codner E, Flanagan SE, Gómez JL, Ellard S, Hattersley AT. Neurogenin 3 is important but not essential for pancreatic islet development in humans. Diabetologia 2014;57:2421–2424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu Z, Li QV, Lee K, et al. . Genome editing of lineage determinants in human pluripotent stem cells reveals mechanisms of pancreatic development and diabetes. Cell Stem Cell 2016;18:755–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shi ZD, Lee K, Yang D, et al. . Genome editing in hPSCs reveals GATA6 haploinsufficiency and a genetic interaction with GATA4 in human pancreatic development. Cell Stem Cell 2017;20:675–688.e6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miyata T, Maeda T, Lee JE. NeuroD is required for differentiation of the granule cells in the cerebellum and hippocampus. Genes Dev 1999;13:1647–1652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goebbels S, Bode U, Pieper A, Funfschilling U, Schwab MH, Nave KA. Cre/loxP-mediated inactivation of the bHLH transcription factor gene NeuroD/BETA2. Genesis 2005;42:247–252 [DOI] [PubMed] [Google Scholar]
- 21.Schonhoff SE, Giel-Moloney M, Leiter AB. Neurogenin 3-expressing progenitor cells in the gastrointestinal tract differentiate into both endocrine and non-endocrine cell types. Dev Biol 2004;270:443–454 [DOI] [PubMed] [Google Scholar]
- 22.Herrera PL. Adult insulin- and glucagon-producing cells differentiate from two independent cell lineages. Development 2000;127:2317–2322 [DOI] [PubMed] [Google Scholar]
- 23.Madisen L, Zwingman TA, Sunkin SM, et al. . A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci 2010;13:133–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chao CS, Loomis ZL, Lee JE, Sussel L. Genetic identification of a novel NeuroD1 function in the early differentiation of islet alpha, PP and epsilon cells. Dev Biol 2007;312:523–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Micallef SJ, Li X, Schiesser JV, et al. . INS(GFP/w) human embryonic stem cells facilitate isolation of in vitro derived insulin-producing cells. Diabetologia 2012;55:694–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sui L, Danzl N, Campbell SR, et al. . β-cell replacement in mice using human type 1 diabetes nuclear transfer embryonic stem cells. Diabetes 2018;67:26–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mali P, Yang L, Esvelt KM, et al. . RNA-guided human genome engineering via Cas9. Science 2013;339:823–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Churchill AJ, Gutiérrez GD, Singer RA, Lorberbaum DS, Fischer KA, Sussel L. Genetic evidence that Nkx2.2 acts primarily downstream of Neurog3 in pancreatic endocrine lineage development. eLife 2017;6:pii:e20010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 2009;25:1105–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trapnell C, Williams BA, Pertea G, et al. . Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 2010;28:511–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol 2010;11:R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barrett T, Wilhite SE, Ledoux P, et al. . NCBI GEO: archive for functional genomics data sets--update. Nucleic Acids Res 2013;41:D991–D995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Y, Liu T, Meyer CA, et al. . Model-based analysis of ChIP-Seq (MACS). Genome Biol 2008;9:R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhu LJ, Gazin C, Lawson ND, et al. . ChIPpeakAnno: a Bioconductor package to annotate ChIP-seq and ChIP-chip data. BMC Bioinformatics 2010;11:237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Suissa Y, Magenheim J, Stolovich-Rain M, et al. . Gastrin: a distinct fate of neurogenin3 positive progenitor cells in the embryonic pancreas. PLoS One 2013;8:e70397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brelje TC, Scharp DW, Lacy PE, et al. . Effect of homologous placental lactogens, prolactins, and growth hormones on islet B-cell division and insulin secretion in rat, mouse, and human islets: implication for placental lactogen regulation of islet function during pregnancy. Endocrinology 1993;132:879–887 [DOI] [PubMed] [Google Scholar]
- 37.Vasavada RC, Garcia-Ocaña A, Zawalich WS, et al. . Targeted expression of placental lactogen in the beta cells of transgenic mice results in beta cell proliferation, islet mass augmentation, and hypoglycemia. J Biol Chem 2000;275:15399–15406 [DOI] [PubMed] [Google Scholar]
- 38.Mwangi S, Anitha M, Mallikarjun C, et al. . Glial cell line-derived neurotrophic factor increases beta-cell mass and improves glucose tolerance. Gastroenterology 2008;134:727–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mwangi SM, Usta Y, Raja SM, et al. . Glial cell line-derived neurotrophic factor enhances neurogenin3 gene expression and beta-cell proliferation in the developing mouse pancreas. Am J Physiol Gastrointest Liver Physiol 2010;299:G283–G292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Muñoz-Bravo JL, Hidalgo-Figueroa M, Pascual A, López-Barneo J, Leal-Cerro A, Cano DA. GDNF is required for neural colonization of the pancreas. Development 2013;140:3669–3679 [DOI] [PubMed] [Google Scholar]
- 41.Parsons JA, Brelje TC, Sorenson RL. Adaptation of islets of Langerhans to pregnancy: increased islet cell proliferation and insulin secretion correlates with the onset of placental lactogen secretion. Endocrinology 1992;130:1459–1466 [DOI] [PubMed] [Google Scholar]
- 42.Auffret J, Freemark M, Carré N, et al. . Defective prolactin signaling impairs pancreatic β-cell development during the perinatal period. Am J Physiol Endocrinol Metab 2013;305:E1309–E1318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lerner AG, Upton JP, Praveen PV, et al. . IRE1α induces thioredoxin-interacting protein to activate the NLRP3 inflammasome and promote programmed cell death under irremediable ER stress. Cell Metab 2012;16:250–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xu G, Chen J, Jing G, Shalev A. Thioredoxin-interacting protein regulates insulin transcription through microRNA-204. Nat Med 2013;19:1141–1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang HP, Chu K, Nemoz-Gaillard E, Elberg D, Tsai MJ. Neogenesis of beta-cells in adult BETA2/NeuroD-deficient mice. Mol Endocrinol 2002;16:541–551 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.