Abstract
Somatic DNMT3A mutations at R882 are frequently observed in AML patients including the very abundant R882H, but also R882C, R882P and R882S. Using deep enzymology, we show here that DNMT3A-R882H has more than 70-fold altered flanking sequence preferences when compared with wildtype DNMT3A. The R882H flanking sequence preferences mainly differ on the 3′ side of the CpG site, where they resemble DNMT3B, while 5′ flanking sequence preferences resemble wildtype DNMT3A, indicating that R882H behaves like a DNMT3A/DNMT3B chimera. Investigation of the activity and flanking sequence preferences of other mutations of R882 revealed that they cause similar effects. Bioinformatic analyses of genomic methylation patterns focusing on flanking sequence effects after expression of wildtype DNMT3A and R882H in human cells revealed that genomic methylation patterns reflect the details of the altered flanking sequence preferences of R882H. Concordantly, R882H specific hypermethylation in AML patients was strongly correlated with the R882H flanking sequence preferences. R882H specific DNA hypermethylation events in AML patients were accompanied by R882H specific mis-regulation of several genes with strong cancer connection, which are potential downstream targets of R882H. In conclusion, our data provide novel and detailed mechanistic understanding of the pathogenic mechanism of the DNMT3A R882H somatic cancer mutation.
INTRODUCTION
DNA cytosine-C5 methylation at CpG sites is a major chromatin regulator essential for development in mammals (1–3). DNA methylation is introduced by the family of DNA methyltransferases (DNMTs) in humans comprising DNMT1, DNMT3A and DNMT3B (4–6). It functions in concert with other epigenome modifications, most prominently histone tail modifications and represents one important part of the epigenome network (7,8). Aberrant DNA methylation has several connections to diseases including cancer (9–12). Strikingly, mutations in the DNMT3A enzyme occur in ∼25% of all Acute myeloid leukemia (AML) patients with a normal karyotype (13,14). The mutations show a strong enrichment of heterozygous missense mutations (73%) combined with an intact wildtype allele. Among them, R882 mutations are most abundant (about 60%), two third of them R882H, about one third R882C, and 3% R882S and R882P. The high prevalence of the R882H missense mutation, together with the low frequency of nonsense and frameshift mutations in DNMT3A suggests that this DNMT3A mutation has a specific molecular effect, which has been investigated by several groups.
The R882 residue is located at the central interface of the DNMT3A tetramer where it interacts with the DNA backbone (15). Different studies have determined the catalytic activity of the purified R882H mutant protein and mostly observed 50–70% residual activity (16–19). Kim et al. showed that the exogenously expressed murine R878H mutant (corresponding to human R882H) interacts with the wildtype enzyme, but it was less efficient in methylating major satellite repeats in mouse ES cells (20). Russler-Germain et al. described that enzyme preparations obtained after co-expression of wildtype and R882H in mammalian cells showed only 12% of residual activity leading to a model that R882H subunits inactivate the wildtype subunits in mixed complexes in a dominant negative effect (21). However, this model could not be validated biochemically (19).
It has been well documented that DNMT3A shows pronounced differences in the methylation activity of CpG sites depending on their flanking sequence (22–25) and CpG sites have been identified which could not be methylated by DNMT3A at all (25). We observed already in 2006 that the R882A mutation led to a change in the flanking sequence preferences of DNMT3A (26). Recently, we have extended this finding and showed that the flanking sequence preferences of DNMT3A are strongly altered by the R882H mutation leading to specific subsets of CpG sites which are methylated by R882H even better than by wildtype DNMT3A, while another subset of CpG sites shows a drop in activity of R882H that is more pronounced than on average sites (18). These changes in flanking sequence preferences of R882H may lead to hyper- and hypomethylation of genomic regions and by this play an important role in tumorigenesis by R882H (18). However, the previous analysis was based on activity data for only 56 CpG sites and, therefore, the derived profiles were not sufficiently detailed to correlate them with cellular methylation data.
It was the aim of this work to characterize the difference in flanking sequence preferences of DNMT3A and R882H in more detail using a newly developed deep enzymology approach. After confirming and extending the previous findings, we show that other mutations at R882 have similar effects. Moreover, after developing a novel method for analysis of DNA methylation patterns focusing on flanking sequence effects, we demonstrate that DNMT3A and R882H transfected in human cells methylate DNA with the same flanking sequence preferences as determined in vitro. We further demonstrate that R882H characteristic hypermethylation events occur in AML patients with R882H mutation and they are correlated with the mis-regulation of several genes with putative connection to AML, which could be part of the tumorigenic downstream signaling cascade emanating from R882H.
MATERIALS AND METHODS
Biochemical methods
For biochemical studies, the C-terminal domain of human DNMT3A (amino acids 612–912 of Q9Y6K1) and DNMT3B (amino acids 558–859 of O88509) were used. All enzymes were cloned into pET28+ (Novagen) containing an N-terminal His6-tag. Mutagenesis was performed using the megaprimer site-directed mutagenesis method (27) and confirmed by restriction marker analysis and DNA sequencing. Protein expression and purification was performed as described (28). Methylation activity was determined using radioactively labelled AdoMet (Perkin Elmer) and a biotinylated double stranded 30mer oligonucleotide (GAG AAG CTG GGA CTT CCG GGA GGA GAG TGC) as described (19). Specific methylation of one CpG site in the upper DNA strand of oligonucleotide substrates was determined using hemimethylated oligonucleotide substrates with methylation in the lower strand of the CpG site. In addition, the methylation of the same substrate in fully methylated state was determined and the rates subtracted from the methylation rates of the hemimethylated substrates as described (18,25). This approach allows to detect the specific methylation of the target site in one DNA strand ignoring additional methylation events in the remaining part of the substrate. This special DNMT assay is necessary for DNMT3 enzymes if methylation of individual sites should be measured, because of their high non-CpG methylation activity (29).
Deep enzymology experiments
Deep enzymology experiments were conducted as described (Gao et al., submitted for publication). In short, libraries of DNA substrates containing unmethylated and hemimethylated CpG sites embedded in a 10 nucleotide random context were prepared by primer extension using synthetic single stranded templates obtained from IDT. The pool of substrates was methylated at 37°C in the presence of 0.8 mM S-adenosyl-l-methionine (Sigma) in reaction buffer (20 mM HEPES pH 7.5, 1 mM EDTA, 50 mM KCl, 0.05 mg/ml bovine serum albumin) using enzyme concentrations and incubation times as indicated in the text. Stopping of the methylation was followed by hairpin ligation and bisulfite conversion using EZ DNA Methylation-Lightning kit (ZYMO RESEARCH). Libraries for Illumina Next Generation Sequencing (NGS) were produced in a two-step PCR approach, pooled in appropriate ratios and analyzed by Illumina NGS. Control experiments without enzyme revealed methylation levels of 0.3% confirming the efficiency of the bisulfite conversion (data not shown).
Bioinformatics
Bioinformatic analysis of NGS data was conducted with the tools available on the Usegalaxy.eu server (30) and with home written programs. First, fastq files were analyzed by FastQC, 3′ ends of the reads with a quality lower than 20 were trimmed and reads containing both full-length sense and antisense strands were selected (Suppl. Data). Next, using the information of both strands of the bisulfite-converted substrate, the original DNA sequence and methylation state of both CpG sites was reconstituted and average methylation levels of each NNCGNN and NNNCGNNN site were determined. Pearson correlation factors were calculated with Excel using the correl function. P-values were determined using the distribution of r-values from >200 correlation analyses with one data set shuffled. ERRBS DNA methylation data for 119 AML patients with annotated R882H mutational status were obtained from GSE86952 (31). Average NNCGNN and NNNCGNNN methylation levels in BED files obtained from 450k analyses and ERRBS data were determined using home written programs.
Generation of HCT116 cells expressing DNMT3AC and R882H
HTC116 DNMT1 hypomorphic cells (HCT116 DNMT1 hm) (kindly provided by Prof. Bert Vogelstein, HHMI, USA), were cultured in McCoy 5A Medium (Sigma) supplemented with 10% FCS, 2 mM l-glutamine, 100 U/ml penicillin and 100 μg/ml streptomycin at 37°C in 5% CO2. HTC116 DNMT1 hm cells were modified to express the ecotropic receptor and rtTA3 using retroviral transduction of pWPXLd-RIEP (pWPXLd-rtTA3-IRES-EcoR-PGK-Puro) followed by drug selection (0.8 ug/ml puromycin for 1 week) similarly as described (32). The resulting cell line was subsequently transduced with ecotropically packaged retroviruses containing the dnmt3a (or R882H mutant) gene fused to Venus under control of a TRE3G promoter. Retroviral gene transfer was performed as previously described (32). In brief, for each calcium phosphate transfection, 10–20 μg plasmid DNA and 5 μg helper plasmid (pCMV-Gag-Pol, Cell Biolabs) were used. Retroviral packaging was performed using PlatinumE cell line (Cell Biolabs). Transduction efficiencies of retroviral constructs (TRE3G-DNMT3A1-Venus-PGK-NEO, TRE3G-DNMT3A1(R882H)-Venus-PGK-NEO or TRE3G-Venus-PGK-NEO) (33) were measured 48 h post induction with 1 μg/ml Doxycycline (Dox) by flow cytometry. Transduced cell populations were selected 48 h post infection using 500 μg/ml G418 (Gibco Life technologies). After 7 days of induction about 1 million DNMT3A or R882H expressing cells (as judged by being Venus positive) were sorted for each replicate by FACS.
Infinium 450k DNA methylation analysis
To determine genome wide DNA methylation of HCT116 DNMT1 hm cells expressing DNMT3A wildtype or R882H, the Infinium 450k array was used according to the instructions of the DKFZ Genomics and Proteomics Core Facility. Two repeats of each data set were prepared and used for statistical analysis. From the raw intensity data (in total 485 577 sites), 35 654 CpG sites were removed during quality control, the remaining were background-subtracted with the methylumi (R package version 2.18.2) and normalized by the BMIQ method. Additional 1416 CpG sites were removed from analysis after normalization. For further analysis, the methylation levels of R882H and wildtype were compared and 92 823 sites with significant difference in methylation (P-value < 0.05) were extracted. In the next step, R882H hypermethylated sites were ranked by comparing the relative and absolute methylation difference, and the methylation data of the 1000 most significant sites were clustered (Euclidean distance, complete linkage) and visualized using pheatmap (Kolde R, R package version 1.0.8).
Amplicon-targeted bisulfite sequencing
To determine the methylation levels of selected CpG sites, the DNA samples used for 450k analysis were digested overnight at 37°C using BamHI and EcoRI and then bisulfite converted using EZ DNA Methylation-Lightning Kit (Zymo Research) following the manufacturer's instructions. The bisulfite treated DNA was then used for PCR amplification using the amplicon and primers with treatment specific barcodes and HotStarTaq DNA Polymerase (Qiagen). PCR products were resolved on an 1% agarose gel, followed by gel extraction and clean-up using the NucleoSpin Gel and PCR Clean-up (Macherey-Nagel). The products were mixed at an equimolar ratio and analyzed by paired-end NGS sequening. The high-throughput sequencing results were demultiplexed and analyzed using the CLC Genomic Workbench 10.0.1 (CLC Bios, MA, USA) following the manufacturer's standard data import protocol and the bisulfite sequencing plugin and the methylation levels normalized to the total read number where extracted.
RESULTS
Generation of flanking sequence preference profiles for DNMT3A, DNTM3B and R882H
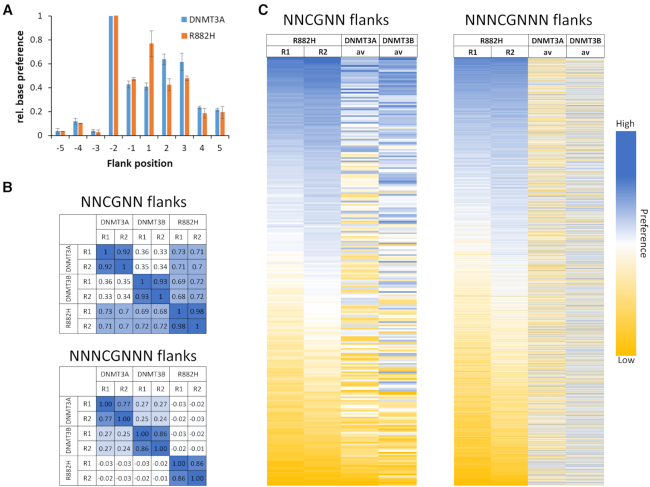
To determine the sequence preferences of DNA methyltransferases in great depth, we have developed a deep enzymology workflow (Supplementary Figure S1). In this approach, a pool of double-stranded DNA substrates is generated which contain one target CpG site flanked by 10 random nucleotides on each side. The substrate pool is enzymatically methylated and after hairpin ligation, bisulfite conversion is carried out which converts cytosine to uracil but leaves 5-methylcytosine intact (34). Afterwards, the bisulfite converted DNA is amplified by two PCRs stepwise adding barcodes and indices (35). Different substrates were then mixed and sequenced by NGS. In our previous work, duplicate reactions were carried out with the catalytic domains of DNMT3A and DNMT3B (Supplementary Table S1). Our results demonstrated that DNMT3A- and DNMT3B-mediated methylation reactions show clearly distinct flanking sequence preferences (Gao et al., submitted for publication). Here, the specificity of the R882H mutant of DNMT3A catalytic domain was studied with the same approach. Two independent methylation reactions were performed and sequenced at great depth (Supplementary Table S1). To determine the overall influence of each flanking position on the reaction rate, at each site the observed and expected frequencies of each nucleotide in the methylated products were determined. This analysis revealed that the reaction rates of R882H and DNMT3A are both significantly influenced by the CpG-flanking sequence from the –2 to the +3 site (Figure 1A). Based on this result, we focused on further analyzing the effect of the ±2 bp and the ±3 bp flanking positions on the activity of both enzymes and determined the average methylation for all 256 N2 (NNCGNN) and 4096 N3 (NNNCGNNN) flanks. Correlation analysis of these methylation profiles showed that the repetitions of experiments with the same enzyme always were highly correlated (Figure 1B). In contrast, the average methylation levels of CpG sites in different flanking context were only weakly correlated between DNMT3A, DNMT3B and R882H confirming the initial observation of altered flanking sequence preference of R882H (18). To compare the overall methylation activity of R882H with DNMT3A and DNMT3B across all different flanking sites, the DNMT3A results obtained at the lower enzyme concentrations were used and the DNMT3B data were combined. As shown in the heatmaps in Figure 1C, the R882H repeat 2 sample reached higher overall methylation than repeat 1, which is in agreement with the higher enzyme concentration used in this reaction. However, the relative preferences for different flanking sequences in both reactions were very similar but dissimilar from DNMT3A and DNMT3B (Figure 1B). Therefore, the R882H data were merged for further analyses and all data normalized to their average methylation.
Figure 1.
Compilation of the results of the random flank substrate methylation by DNMT3A and R882H. (A) Relative base preferences at the -5 to +5 flanking positions surrounding the CpG site of DNMT3A and R882H. The numbers refer to the standard deviations of the observed/expected base composition at each site among the methylated sequence reads, normalized to the highest number for each enzyme. (B) Methylation levels were collected and averaged for all two and three base pair flanking sites and the Pearson correlation coefficients (r-values) of the pairwise comparison of the data sets were determined. (C) Heatmaps of the methylation profiles of both R882H repeats and the DNMT3A and DNMT3B data. DNMT3A and DNMT3B data were taken from (Gao et al., submitted for publication).
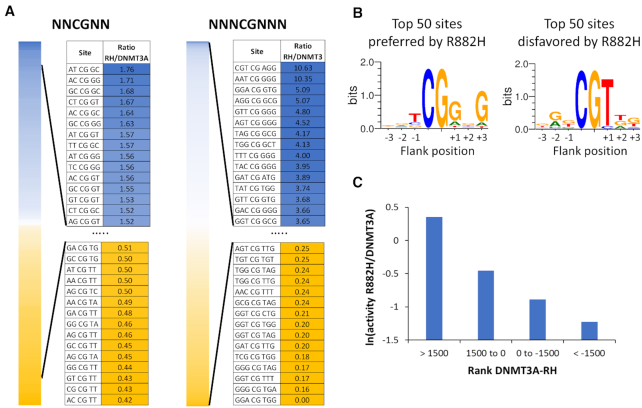
Using the averaged and normalized flanking sequence preferences of R882H and DNMT3A, the ratio of R882H/DNMT3A preferences was calculated to express the relative preference of R882H (Figure 2A). The data revealed a 4.2-fold difference in preferences for NNCGNN flanks. With NNNCGNNN flanks much larger differences in methylation rates were detected. For some of the NNNCGNNN flanks, no methylation was observed with R882H. At these sites the R882H/DNMT3A ratio was set to 0.15 reflecting the limits of detection. Based on this, the difference in preferences was >70-fold for the 3 bp flanks indicating drastic differences in methylation preferences between DNMT3A and R882H. For better visualization, Weblogos (http://weblogo.threeplusone.com/) were prepared of the 50 N3 flanks most preferred and most disfavored by R882H (Figure 2B). These images illustrate a very good agreement of the new flanking sequence preference profiles with the previously published data (18), although the older data set was based on methylation data of only 56 CpG sites. Specifically, in the previous data set a strong preference of R882H for a G at the +1 and +3 sites was observed together with weaker preference for T at –1 and disfavor for T at the +1 site (18). All these effects were recapitulated in the detailed analysis based on the deep enzymology data. In the previous publication, two substrates were designed on the basis of the preference profiles available at that time to be preferred and disfavored by R882H to validate the predicted flanking sequence effects (18). Using these and seven new substrates designed for different degrees of preference of R882H and DNMT3A (Supplementary Table S2), the rates of methylation of the central CpG site by DNMT3A and R882H were determined to validate the newly derived profiles. After arranging the substrates according to the differences in flanking sequence preferences of R882H and DNMT3A, the relative methylation rates of both enzymes were in a very good agreement with the flanking sequence data (Figure 2C), indicating that the deep enzymology results are in good agreement with methylation rates determined with these exemplary substrates.
Figure 2.
Ratio of methylation rates of R882H and DNMT3A in different flanking context. (A) NNCGNN and NNNCGNNN flanking sites ordered by decreasing R882H/DNMT3A preference ratio. (B) Sequence logos of the 50 flanking sites most favored or disfavored by R882H. (C) Validation of the deep enzymology data by in vitro methylation kinetics with nine defined substrates. Ratios of methylation activities were averaged for different substrates in defined intervals of the difference in the DNMT3A and R882H preference ranks (with a low rank indicating a high preference) (Supplementary Table S2).
R882H flanking effect on the 5′ and 3′ flank
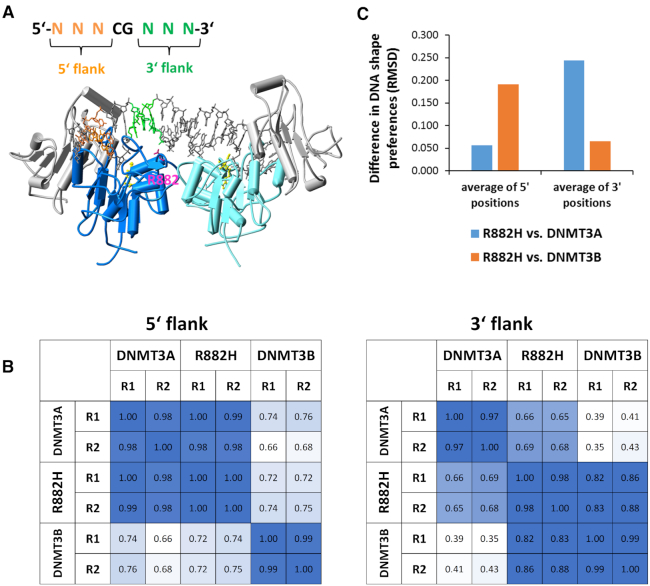
The structure of DNMT3A/3L in complex with DNA shows that R882 forms a backbone contact to the 3′ flank of the CpG site (15), but it does not contact the 5′ flank (Figure 3A). Therefore, we were interested to analyze the effect of R882H on the flanking sequence preference separately for the 5′ and 3′ flanks (Figure 3B). In agreement with the structural data, the 5′ flanking sequence preferences of R882H are highly correlated with those of DNMT3A, while the 3′ preferences differ.
Figure 3.
R882H has an effect on the DNA interaction at the 3′ flank. (A) Structure of the DNMT3A/3L-DNA complex (15). DNMT3A is shown in dark blue and cyan, DNMT3L in gray. The DNA is shown in grey, with the 5′ flank colored orange and the 3′ flank colored green. R882 is shown for the dark blue DNMT3A subunit in pink. AdoMet is shown in yellow. (B) Correlation analysis of flanking sequence preferences of DNMT3A, DNMT3B and R882H for the 5′ and 3′ flanks. (C) DNA shape readout by DNMT3A, DNMT3B and R882H. Structural parameters of the DNA were determined for all NNNCGNNN sites with the DNA shape prediction server (http://rohslab.cmb.usc.edu/DNAshape/) (38) and correlated with the activities levels of DNMT3A, R882H and DNMT3B determined in the deep enzymology experiments (Supplementary Figure S2). For all properties and positions, the RMSD differences of R882H correlation factors with DNMTA and DNMT3B correlation factors were determined and the values averaged for the 5′ and 3′ flanking positions.
We have shown in our previous paper that the loop containing R882 is a hotspot of amino acid sequence deviation between DNMT3A and DNMT3B, because the DNMT3A residues S881, L883 and R887 are replaced by G822, G824 and K826 in DNMT3B. Moreover, DNMT3B residue R823 (corresponding to R882 in DNMT3A) adopts a different conformation and it does not engage in a backbone DNA contact suggesting that this loop region is a major structural cause for the differences in flanking sequence preferences between DNMT3A and DNMT3B (Gao et al., submitted for publication). Therefore, it was interesting to compare the DNMT3A and R882H profiles also with DNMT3B (Figure 3B). The 5′ flank profiles showed high similarity of DNMT3A and R882H, which both differed from DNMT3B. In contrast, the 3′ flank preferences of R882H cluster with DNMT3B much better than with DNMT3A. We conclude that the sites from +1 to +3 are responsible for the differences in the flanking sequence preferences of DNMT3A and R882H. The –1 and –2 sites do influence the reaction rates of both enzyme (Figure 1A), but they act in the same way in DNMT3A and R882H. Strikingly, these data demonstrate that R882H behaves like a chimera of DNMT3A and DNMT3B with respect to flanking sequence preference, it shows DNMT3A preferences at the 5′ flank, but DNMT3B preferences at the 3′ flank. Structurally this result suggests that the loss of the R882 backbone contact in R882H is the main reason for the flanking sequence change.
Readout of DNA shape
Contacts of DNA binding proteins to the phosphodiester backbone of the DNA are known to influence protein–DNA recognition by indirect readout, because they can change the protein–DNA interaction depending on the static or dynamic conformational preferences of the DNA (36,37). This leads to a complex readout of the DNA sequence dependent structural parameters of the DNA. To investigate the potential influence of the DNA conformation on DNA methylation by DNMT3A, DNMT3B and R882H, the DNA shape prediction server has been used (http://rohslab.cmb.usc.edu/DNAshape/) (38) to determine the structural parameters for all NNNCGNNN sequences. The predicted structural features include minor groove width, base pair roll, propeller twist, and helix twist. For all NNNCGNNN sites, these parameters were then correlated with the corresponding catalytic activities of DNMT3A, DNMT3B and R882H from the deep enzymology analysis revealing strong and enzyme dependent correlations for some positions and properties (Supplementary Figure S2). These overall correlations suggest that shape related properties of the DNA are an important basis for DNA recognition by the DNMT3 enzymes. To determine whether the preferences of R882H resemble DNMT3A or DNMT3B, the root mean square deviation of the correlation factors observed for the corresponding pairs of enzymes were determined. Strikingly, averaging these numbers for the 5′ flank and the 3′ flank clearly shows that the structure dependent preferences of R882H resemble DNMT3A for the 5′ flanks, but they resemble DNMT3B for the 3′ flanks, again confirming that R882H behaves like a chimera of both enzymes (Figure 3C).
Investigation of other R882 mutants
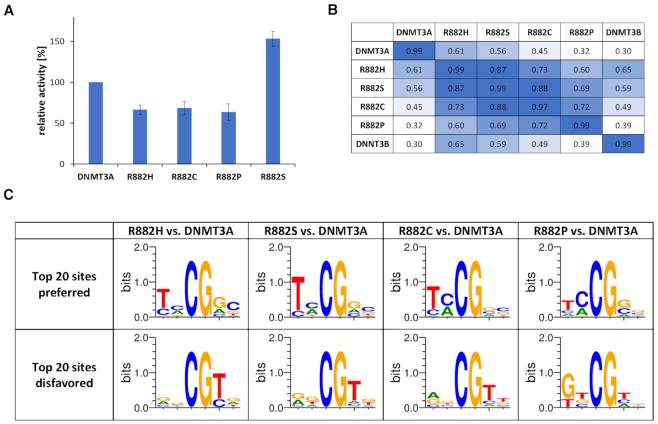
As mentioned above, beside R882H also other mutations are occurring at R882 in AML, namely R882C, R882S and R882P. We were interested to find out, if these mutations also lead to changes in the flanking sequence preference of DNMT3A. We therefore cloned and purified the mutated proteins in the context of the catalytic domain of DNMT3A. Initially, enzyme activity was analyzed using a 30mer oligonucleotide substrate. Our data showed that R882C and R882P caused 30–40% reduction in activity (Figure 4A), similarly as previously found for R882H with the same substrate (35% reduction) (19). Interestingly, R882S showed an overall increased activity.
Figure 4.
Investigation of other R882 mutants. (A) Catalytic activities of R882 mutations determined with a 30mer substrate. The figure shows averages of at least 3 independent experiments. Error bars display the standard deviation. R882H data were taken from Emperle et al. (19). (B) Correlation of NNCGNN methylation profiles determined by deep enzymology using the data obtained with unmethylated substrates. For comparison, all data sets were reduced to equal read counts of 38 000. (C) Weblogos of the 20 most preferred and disfavored flanks by the R882 mutants based on the NNCGNN profiles.
We next tested if flanking sequence preferences of DNMT3A, DNMT3B or R882H differ depending on the methylation state of the substrate library pool (unmethylated or hemimethylated). To this end additional deep enzymology reactions were performed with DNMT3A, DNMT3B and R882H in several experimental repeats (Supplementary Table S3). In both settings, methylation of the upper DNA strand was analyzed. We obtained a medium coverage allowing to analyze N2 flanking sequence preferences, which revealed that the preferences obtained with unmethylated or hemimethylated substrates were highly similar (Supplementary Figure S3). Therefore, we were using unmethylated substrates for the further analysis, which are more convenient.
We conducted deep enzymology analyses with the R882C, S and P mutants using unmethylated substrates and obtained a coverage sufficient for the analysis of N2 flanking sequence preferences (Supplementary Table S3). Afterwards, the flanking sequence preferences of all mutants were compared by correlation analysis (Figure 4B). In addition, Weblogos of the 20 most preferred and most disfavored flanks when compared with wildtype DNMT3A were prepared (Figure 4C). Strikingly, these analyses showed that the flanking sequence preferences of R882C, R882S and R882P are very similar to R882H. This result suggests that the removal of the arginine side chain at position 882 resulting in the loss of its backbone contact to the DNA is the main cause for the changes in the flanking sequence preferences, rather than specific effects associated with the amino acid introduced instead of arginine at position 882. The R882P profile showed the largest deviations suggesting some proline specific effects. This result can be rationalized by the strongly reduced regions of allowed ψ and ϕ angles in the Ramachandran plot of proline. Therefore, the introduction of proline in place of an arginine has a high probability to cause larger changes in the loop geometry than all other mutations, which would explain unique changes in flanking sequence preferences.
Analysis of DNA methylation patterns generated by DNMT3A R882H in human HCT116 cells
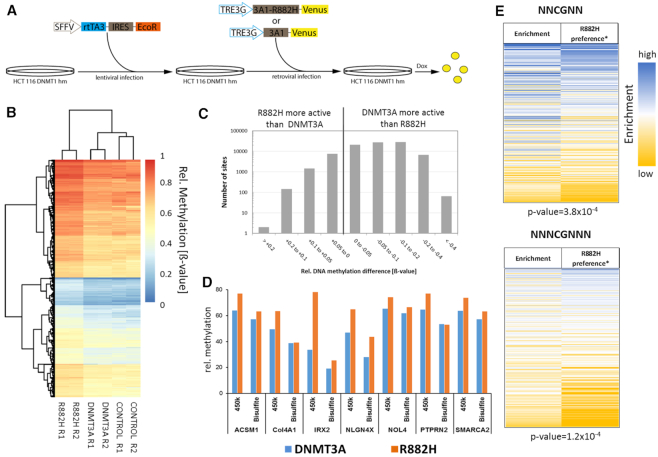
To study the effect of the DNMT3A R882H mutation on DNA methylation in human cells, we used the HCT116 DNMT1 hypomorphic cell line (HCT116 DNMT1 hm), which contains a truncated DNMT1 with reduced activity, but active copies of DNMT3A and DNMT3B (39,40). Because of the impaired maintenance DNA methylation activity, these cells have an about 20% reduced level of overall DNA methylation. To facilitate inducible transgene expression, we initially generated a ‘tet-competent’ HCT116 DNMT1 hm cell line via stable integration of the reverse tet transactivator (rtTA3) together with the ecotropic receptor. Subsequently, a retroviral vector was used to generate two cell lines with integrated genes encoding for the full length human DNMT3A-Venus fusion protein or its R882H mutant under the control of a TRE3G promoter (Figure 5A). A third cell line only expressing the fluorophore served as control. Transgene expression was induced by addition of Dox and after 3 days comparable expression levels of DNMT3A and R882H were observed (Supplementary Figure S4). After 7 days, Venus positive cells were isolated by flow cytometry from all three cell lines. Genomic DNA was isolated and DNA methylation patterns analyzed with Illumina 450k arrays. Heatmap analysis confirmed the similarity of the repeated data sets (Figure 5B). Filtering of sites showing a significant difference in methylation between wildtype and R882H (P-value < 0.05) revealed that ∼80 000 sites displayed reduced methylation levels after expression of R882H when compared with cells expressing wildtype DNMT3A (Figure 5C). Most of them showed up to 20% reduced methylation in agreement with the reduced overall activity of R882H described above. However, ∼100 sites showed a stronger (>40%) reduction of methylation activity. In addition, >9000 sites were identified at which R882H was more active than DNMT3A. Hypermethylation of CpG sites in the R882H sample was confirmed at selected sites by amplicon based bisulfite analysis (Figure 5D). We conclude that altered flanking sequence preferences of R882H are influencing DNA methylation patterns in human cells.
Figure 5.
DNA methylation analysis in DNMT1 hypomorphic HCT116 cells. (A) Generation of DNMT1 hypomorphic HCT116 cells which express DNMT3A or R882H under Dox regulation. In the first step, the genomic integration and stable expression of the ecotrophic receptor and the reverse tet-transactivator (rtTA3) was archived using lentiviral infection. In the second step, genomic integration of DNMT3A-Venus or R882H-Venus under the control of a Dox inducible TRE3G promotor was achieved using retroviral infection. Six days after addition of Dox, the cells expressing DNMT3A or R882H were isolated by FACS (Supplementary Figure S4). (B) Clustering of 1000 CpG sites with the strongest increase in DNA methylation with the R882H mutant. R1 and R2 denotes the two repeats of the experiment. CONTROL denotes for the untreated cells. (C) Overview of all CpG sites showing a significant difference (P< 0.05) in the methylation levels of wildtype DNMT3A and R882H. (D) Methylation of exemplary CpG sites showing hypermethylation with the R882H mutant on the 450k array was analyzed by bisulfite conversion followed by PCR and amplicon sequencing (Supplementary Table S4). The figure shows the relative methylation (%) of target CpG sites in 450k data (based on ß-values) and the bisulfite data. (E) Correlation of DNA methylation profiles in HCT116 cells expressing R882H or wildtype DNMT3A with the R882H* flanking sequence preferences. The enrichment of flanks in 8507 CpG sites that were hypermethylated after expression of R882H when compared to cells after expression of wildtype DNMT3A were compared with the R882H/DNMT3A* flanking sequence preference profile. For both flank sizes highly significant correlations were observed. P-values determined by randomization of the data sets are indicated.
When comparing the in vitro preference data with cellular data, it needs to be considered that DNMT1 is present in cells and the DNMT1 enzyme has a high activity on hemimethylated CpG sites. Therefore, in cells a DNMT3 enzyme needs to methylate just one DNA strand in order to trigger the final appearance of a fully methylated CpG site. To account for this, we have defined symmetrical R882H/DNMT3A preference profiles by averaging the preferences of pairs of corresponding complementary flanks (called R882H/DNMT3A* preference) (Supplementary Figure S5). To correlate the R882H specific hypermethylation profiles in HCT116 DNMT1 hm cells with the R882H/DNMT3A* preference, all hypermethylated sites with P<0.05 and at least 0.5-fold higher methylation in the R882H data set were selected (8507 CpGs) and these sites were analyzed focusing on potential influences of their flanking sequence on DNA methylation. To this end, the enrichment of two and three bp flanks within the data set was determined using the distribution of flanking sequences among the entire 450k data set as reference. Then, the overrepresentation of flanks in the hypermethylated data set was compared with the R882H/DNMT3A* preference (Figure 5E) revealing clear correlations between the enrichment of flanks in the hypermethylated data set with the R882H/DNMT3A* flanking sequence preferences for both flank lengths. To determine the statistical significance of these correlations, the correlation analyses were repeated with randomly shuffled enrichment data. Based on the distribution of the r-values in these random correlations, P-values were determined, indicating that all correlations are highly significant with P-values of 3.8 × 10−4 for N2 flanks and 1.2 × 10−4 for N3 flanks, respectively. These results clearly indicate that DNA methylation patterns generated in human HCT116 DNMT1 hm cells by wildtype DNMT3A and the R882H mutant reflect the flanking sequence preferences of both enzymes, indicating that the flanking sequence preferences influence the DNA methylation activity of DNMT3A in human cells.
Correlation of major satellite methylation with the R882H/DNMT3A* flanking sequence preferences
As described above, Kim et al. demonstrated a dominant negative effect of R882H expression on DNA methylation of major satellite repeat sequences in murine ES cells (20). They conducted DNA methylation analysis by Southern blot hybridization after digestion of the DNA with the methylation specific restriction enzyme MaeII (ACGT) probing 3 CpG sites and by bisulfite analysis of 6 CpG sites. We investigated the preference of R882H for these sites and strikingly observed that the majority of the sites on the major satellite were strongly disfavored by R882H (Supplementary Figure S6). Hence our data are in good agreement with the experimental observations reported in this paper.
Correlation of AML patient DNA methylation profiles with the R882H* flanking sequence preferences
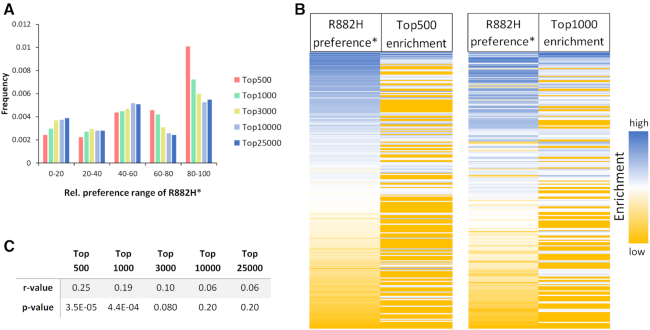
Next we aimed to compare global DNA methylation patterns from AML patients carrying the R882H mutation with AML patients not containing it. Glass et al. reported enhanced reduced representation bisulfite sequencing (ERRBS) DNA methylation profiles of 119 AML samples, 27 of them with R882H mutation (31). The data include CpGs covered between 10× and 400×, methylation changes were called requiring at least three samples to support the difference. In total, 184589 differentially methylated CpGs with a q-value ≤0.01 and ≥25% methylation change were identified for AML with R882H mutations as compared to AML with wildtype DNMT3A, 56870 of them were hypermethylated in R882H AML and 127719 were hypomethylated (31). To analyze the distribution of flanking sequences in sites specifically hypermethylated in R882H containing AMLs, the methylation levels of these sites were binned into different categories depending on the degree of hypermethylation (Supplementary Figure S7) and for each group the distribution of NNCGNN flanks was determined. Next, the frequency of flanks within certain preference ranges of R882H were determined in all groups (Figure 6A). Strikingly, the Top-500 group containing the 500 sites most strongly hypermethylated in R882H containing AML across the entire data set of 119 AML cases showed a very strong enrichment of flanks, which are highly preferred by R882H, while flanks disfavored by R882H were depleted. A similar but less pronounced effect was observed in the Top-1000 group. The strong enrichment of R882H preferred flanks in the Top-500 and Top-1000 groups is also clearly visible in the corresponding heatmaps (Figure 6B). To determine the statistical significance of these correlations, the correlation analyses were repeated with randomly shuffled R882H* data. Based on the distribution of the r-values in these random correlations, P-values were determined, indicating that the correlations observed with the Top-500 (P-value = 3.5 × 10−5) and Top-1000 (P-value = 4.4 × 10−4) groups are highly significant (Figure 6C). This finding clearly indicates that the altered flanking sequence preferences of the R882H mutant shape the global DNA methylation profile in AML patients. It is very striking that these effects were detectable in a global data set obtained by averaging 119 AML patient data (27 of them with R882H mutation and 92 without) despite the high background of DNA methylation changes related to AML progression (41). These data suggest that corresponding alterations in the DNA methylation might cause disease relevant expression changes of genes with oncogenic or tumor suppressive roles.
Figure 6.
Correlation of R882H* flanking sequence preference with the flanking sequence context of CpG sites specifically hypermethylated in R882H containing AML (31). (A) Enrichment or depletion of R882H*preferred flanks in the different hypermethylated groups. Flanks strongly preferred by R882H have a relative preference of 100 disfavored flanks have a preference of 0. (B) Correlation of the enrichment of flanks in the Top-500 and Top-1000 group with the R882H* preference. (C) Compilation of r-values and corresponding P-values of the correlation of the R882H* flanking sequence preferences with the enrichment of flanks in the different groups of hypermethylated CpG sites.
Identification of potential AML target genes regulated by R882H
Next we aimed to identify if DNA hypermethylation in the R882H patient AML data set was correlated with R882H specific expression changes reported by Glass et al. (2017) (31). For this we focused on the hypermethylated CpG sites which were assigned to gene promoters, gene bodies or close to genes with a distance of 2–50 kb. The rationale behind this approach was that hypermethylation of gene promoters often leads to repression of genes, but some transcription factors have also been identified to preferentially bind methylated CpG sites (42,43). Hypermethylation of gene bodies often increases gene expression (44,45). In addition, methylation of enhancers in gene bodies and close to genes can influence expression in both directions, depending on the effect of the regulated transcription factors (stimulatory or repressive) and their methylation specificity. In total, 136 genes were identified in which R882H specific DNA hypermethylation was co-occurring with an R882H specific expression change. These initial targets were further analyzed for a potential role in cancer and AML in particular, finally leading to a list of 17 top-candidates, several of them associated with more than one hypermethylated CpG site (Table 1). Twelve of the genes contain at least one hypermethylated CpG site in a flanking context that is among the 10% most preferred flanks of the R882H* profile, the remaining five genes contained a CpG in the 40% preferred context. Many of the downregulated genes were known to be tumor suppressor genes and many of the upregulated ones oncogenes. The strong correlation of PRDM16 over-expression with DNMT3A mutations has already been reported (46) and PRDM16 is a known oncogene and its expression is correlated with poor clinical outcome in AML (47). Overexpression of WT1 is generally observed in AML and it has been associated with poor clinical outcome in many studies (48). Additional literature evidence suggests roles of TP53INP1 and PER3 in AML, because down-regulation of any of them is a negative prognostic marker in AML (49,50). We conclude that these genes represent strong candidates for mis-regulated targets downstream of R882H, which could have a direct influence on AML onset and progression.
Table 1.
List of potential R882H AML target genes. Gene part refers to the region of hypermethylation (P – promoter, I – Intron, EX – Exon, D – Distance) as defined in (31). CpGs: number of hypermethylated CpGs. Expression: ln2 ratio of expression level in R882H and non-R882H AML. Comment: Oncogene and TSG (Tumor suppressor gene) as listed in COSMIC Cancer gene census (52) or the indicated references. AML connection is based on Harmonizome AML gene set (53)
| Category | Gene | Gene symbol | Gene part | CpGs | Expression | Comment |
|---|---|---|---|---|---|---|
| Hypermet. promoter | NM_014629 | ARHGEF10 | P | 1 | –0.85 | TSG |
| NM_001135733 | TP53INP1 | P | 1 | –0.91 | Negative clinical marker (50) | |
| NM_016831 | PER3 | P | 1 | –0.55 | (49) | |
| Hypermet. gene body | NM_022114 | PRDM16 | I | 4 | 1.04 | Ongogene, AML connection |
| NM_000378 | WT1 | I | 4 | 0.94 | Oncogene, TSG, AML connection | |
| NM_024083 | ASPSCR1 | I | 3 | 0.57 | ||
| NM_000368 | TSC1 | I | 1 | 0.64 | TSG | |
| NM_032173 | ZNRF3 | I | 1 | –0.58 | TSG | |
| NM_000179 | MSH6 | I | 1 | –1.16 | TSG | |
| NM_181523 | PIK3R1 | I | 1 | –1.49 | TSG | |
| NM_001136239 | PRDM6 | I | 3 | –0.52 | TSG (54) | |
| NM_001535 | PRMT2 | EX | 1 | 0.53 | Oncogene (55) | |
| Hypermet. close to gene | NM_007084 | SOX21 | D | 1 | –0.54 | TSG |
| NM_004496 | FOXA1 | D | 3 | –0.52 | Oncogene, TSG | |
| NM_001013736 | FAM47C | D | 1 | –0.35 | ||
| NM_015274 | MAN2B2 | D | 1 | 0.58 | Listed in AML gene set | |
| NM_002609 | PDGFRB | D | 1 | 0.66 | Oncogene |
DISCUSSION
Cancer is caused by genetic mutations and epigenetic changes (epimutations) which in combination lead to the characteristic physiological and morphological features of tumor cells, most importantly unregulated cell division. These effects are connected in several instances, for example mutations or epigenetic silencing of DNA repair genes leads to a mutator phenotype stimulating the generation of genetic changes in tumor cells and, conversely, the mutation of epigenetic factors can perturb the epigenetic signaling system leading to the increased occurrence of epimutations (11,51). One example of the latter case are mutations in the DNA methyltransferase DNMT3A, which are observed in ∼25% of all AML patients with normal karyotype. About 60% of all missense mutations in DNMT3A occur at R882 and in about two-thirds of them are R882H exchanges. The high frequency of this specific mutation has stimulated several studies to investigate the pathogenic mechanism of R882 mutations, in particular R882H, but its role has not been finally settled. Previous studies have provided evidence for a dominant negative effect of R882H (21), but this result has remained controversial (19). We have recently demonstrated that the R882H mutation causes a strong shift in the flanking sequence preference of DNMT3A affecting three base pairs of DNA sequence surrounding the CpG target site (18). This finding was in agreement with structural data showing that R882 is located at the DNA binding site of the enzyme where it forms a backbone phosphate contact to the flanking base pair at the +3 position (15). However, the flanking sequence preference data in the previous paper were based on a limited set of kinetic data referring to only 56 different CpG sites. Therefore, these data were insufficient for the correlation of R882H specific changes of flanking sequence preferences with cellular methylation pattern, which would have to consider target sites embedded into 4096 different three base flanking contexts (NNNCGNNN).
In this work, we have exploited a recently developed deep enzymology approach to analyze flanking sequence preferences of R882H and other R882 mutations in great detail. In this technique, a pool of DNA substrates is generated in which the CpG target site is embedded in a 10 bp random sequence context on either side. The mixture of substrates is methylated and methylation levels are measured by bisulfite conversion coupled to NGS. Depending on the read depth, average methylation levels of all N2 or N3 flanking sequences can be determined, allowing to derive detailed flanking sequence preference profiles. We previously conducted deep enzymology flanking sequence analysis for DNMT3A and DNMT3B (Gao et al., submitted for publication) and now added detailed data on R882H. The new results clearly confirm the previous observation of altered flanking sequence preferences of R882H (18) now showing up to 70-fold differences in the methylation rates of CpG sites in different N3 flanking environments. Strikingly, a comparison of the flanking sequence preferences of R882H with DNMT3A and DNMT3B revealed that R882H behaves like a chimera of DNMT3A and DNMT3B. With respect to the 5′ flank, the flanking sequence preferences of R882H and DNMT3A were very similar and distinct from DNTM3B. At the 3′ flank, however, the profiles of R882H and DNMT3A differed and R882H was more similar to DNMT3B. This result is in agreement with our previous finding, that the loop containing R882 is one key determinant for the divergence of the flanking sequence preferences of DNMT3A and DNMT3B (Gao et al., manuscript submitted for publication). We also prepared the R882C, S and P mutations and investigated their catalytic activity showing that the activities of R882C and R882P are similar to R882H, but R882S is even more active than wildtype DNMT3A, arguing against a loss-of-function mechanism for this mutation. We determined the flanking sequence preferences of all three mutants showing that the profiles of R882C, R882S and R882H are very similar to R882H, suggesting a common pathomechanism. This result also indicates, that the shift in the flanking sequence preferences most likely is caused by the loss of the arginine at positon 882 and its contact to the DNA, and it is not due to the presence of the new amino acid at this site. Future studies may investigate the effect of an R882K mutant, the pH dependence of the flanking sequence effects of R882H and DNA binding preferences in order to elucidate the molecular mechanism of how R882 mutations alter the interaction of DNMT3A with the 5′ flanking sequences.
Next, we aimed to compare the in vitro flanking sequence preferences of R882H with cellular methylation data. To this end, a new bioinformatic method was developed allowing to analyze DNA methylation patterns in the context of the flanking sequences of CpG sites instead of considering genome locations or genomic features. As a first step, symmetrical R882H/Dnmt3A* preference profiles were calculated from the biochemical profiles to take into account of the effect of DNMT1 in human cells. Then, these profiles were compared with methylation data obtained after expression of DNMT3A and R882H in HCT116 cells containing a hypomorphic DNMT1. A clear and highly significant correlation of flanking sequence preferences with genomic methylation levels was observed demonstrating that flanking sequence effects guide the activity of DNMT3A and R882H in human cells. It has been shown previously that the expression of R882H in mouse ES cells reduced the methylation of major satellite repeats (20). Strikingly, the flanking sequence preferences of R882H readily explain this finding, because the CpG sites present in the mouse major satellite repeats are strongly disfavored by R882H. We next aimed to analyze patient DNA methylation data to determine if an effect of the altered flanking sequence preferences of R882H is detectable in them as well. To this end, we used a comprehensive analysis of DNA methylation profiles of 119 AML patients, 27 of them with R882H (31). Basing our newly developed flanking sequence analysis on the CpG sites consistently hypermethylated in R882H containing tumors, we find that the sites with strongest hypermethylation show a striking and statistically highly significant correlation with R882H* flanking sequence preferences, indicating that even in this highly aggregated patient data analysis, the specific flanking sequence preferences of the R882H mutation are still detectable. This result strongly suggests that the flanking sequence preferences of DNMT3A and R882H are an important determinant of the global genome wide DNA methylation patterns in AML cells. To investigate potential downstream effect of the R882H induced hypermethylation, we combined the DNA methylation data with gene expression data (31) and functional gene information and compiled a list of 17 genes with strong tumor connection that are connected to R882H specific hypermethylated CpG sites and that show R882H specific changes in gene expression. These genes are strong downstream target candidates potentially mis-regulated by R882H and then promoting tumor formation.
CONCLUSIONS
In this work, we describe details of a gain-of-function effect caused by the R882H mutation in DNMT3A. By changing the flanking sequence preferences of DNMT3A, the mutation leads to altered DNA methylation patterns in human cells and patient samples, including the hypermethylation of about one third of the differentially methylated CpG sites. By this our data provide mechanistic understanding of the pathogenic mechanism of one abundant mutation in AML patients. Our data are in agreement with the genetic finding of the massive enrichment of heterozygous R882H point mutations in patients and the co-occurrence of them with IDH1/2 and TET2 mutations, both leading to reduced DNA demethylation. By analysis of published patient DNA methylation and gene expression data, we identify a group of genes with strong cancer connection, which are specifically hypermethylated and mis-regulated in R882H AML. These genes are strong downstream target candidates of R882H and they represent potential targets for further treatment of R882H containing AML.
Supplementary Material
ACKNOWLEDGEMENTS
We gratefully acknowledge sharing of primary expression data published in Glass et al. (2017) by Drs Maria Figueroa and Jacob Glass.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Deutsche Forschungsgemeinschaft [JE252/20, JE252/36]; Sander Foundation [2019.058.1]. The open access publication charge for this paper has been waived by Oxford University Press – NAR Editorial Board members are entitled to one free paper per year in recognition of their work on behalf of the journal.
Conflict of interest statement. None declared.
REFERENCES
- 1. Jeltsch A., Broche J., Bashtrykov P.. Molecular processes connecting DNA methylation patterns with DNA methyltransferases and histone modifications in mammalian genomes. Genes. 2018; 9:E566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ambrosi C., Manzo M., Baubec T.. Dynamics and context-dependent roles of DNA methylation. J. Mol. Biol. 2017; 429:1459–1475. [DOI] [PubMed] [Google Scholar]
- 3. Schubeler D. Function and information content of DNA methylation. Nature. 2015; 517:321–326. [DOI] [PubMed] [Google Scholar]
- 4. Gowher H., Jeltsch A.. Mammalian DNA methyltransferases: new discoveries and open questions. Biochem. Soc. Trans. 2018; 46:1191–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jeltsch A., Jurkowska R.Z.. Allosteric control of mammalian DNA methyltransferases - a new regulatory paradigm. Nucleic Acids Res. 2016; 44:8556–8575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jurkowska R.Z., Jurkowski T.P., Jeltsch A.. Structure and function of mammalian DNA methyltransferases. Chembiochem. 2011; 12:206–222. [DOI] [PubMed] [Google Scholar]
- 7. Allis C.D., Jenuwein T.. The molecular hallmarks of epigenetic control. Nat. Rev. Genet. 2016; 17:487–500. [DOI] [PubMed] [Google Scholar]
- 8. Soshnev A.A., Josefowicz S.Z., Allis C.D.. Greater than the sum of parts: complexity of the dynamic epigenome. Mol. Cell. 2018; 69:533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Baylin S.B., Jones P.A.. A decade of exploring the cancer epigenome - biological and translational implications. Nat. Rev. Cancer. 2011; 11:726–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bergman Y., Cedar H.. DNA methylation dynamics in health and disease. Nat. Struct. Mol. Biol. 2013; 20:274–281. [DOI] [PubMed] [Google Scholar]
- 11. Feinberg A.P., Koldobskiy M.A., Gondor A.. Epigenetic modulators, modifiers and mediators in cancer aetiology and progression. Nat. Rev. Genet. 2016; 17:284–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Norvil A.B., Saha D., Saleem Dar M., Gowher H.. Effect of disease-associated germline mutations on structure function relationship of DNA methyltransferases. Genes. 2019; 10:E369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yang L., Rau R., Goodell M.A.. DNMT3A in haematological malignancies. Nat. Rev. Cancer. 2015; 15:152–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hamidi T., Singh A.K., Chen T.. Genetic alterations of DNA methylation machinery in human diseases. Epigenomics. 2015; 7:247–265. [DOI] [PubMed] [Google Scholar]
- 15. Zhang Z.M., Lu R., Wang P., Yu Y., Chen D., Gao L., Liu S., Ji D., Rothbart S.B., Wang Y. et al.. Structural basis for DNMT3A-mediated de novo DNA methylation. Nature. 2018; 554:387–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yan X.J., Xu J., Gu Z.H., Pan C.M., Lu G., Shen Y., Shi J.Y., Zhu Y.M., Tang L., Zhang X.W. et al.. Exome sequencing identifies somatic mutations of DNA methyltransferase gene DNMT3A in acute monocytic leukemia. Nat. Genet. 2011; 43:309–315. [DOI] [PubMed] [Google Scholar]
- 17. Holz-Schietinger C., Matje D.M., Reich N.O.. Mutations in DNA methyltransferase (DNMT3A) observed in acute myeloid leukemia patients disrupt processive methylation. J. Biol. Chem. 2012; 287:30941–30951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Emperle M., Rajavelu A., Kunert S., Arimondo P.B., Reinhardt R., Jurkowska R.Z., Jeltsch A.. The DNMT3A R882H mutant displays altered flanking sequence preferences. Nucleic Acids Res. 2018; 46:3130–3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Emperle M., Dukatz M., Kunert S., Holzer K., Rajavelu A., Jurkowska R.Z., Jeltsch A.. The DNMT3A R882H mutation does not cause dominant negative effects in purified mixed DNMT3A/R882H complexes. Sci. Rep. 2018; 8:13242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kim S.J., Zhao H., Hardikar S., Singh A.K., Goodell M.A., Chen T.. A DNMT3A mutation common in AML exhibits dominant-negative effects in murine ES cells. Blood. 2013; 122:4086–4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Russler-Germain D.A., Spencer D.H., Young M.A., Lamprecht T.L., Miller C.A., Fulton R., Meyer M.R., Erdmann-Gilmore P., Townsend R.R., Wilson R.K. et al.. The R882H DNMT3A mutation associated with AML dominantly inhibits wild-type DNMT3A by blocking its ability to form active tetramers. Cancer Cell. 2014; 25:442–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lin I.G., Han L., Taghva A., O’Brien L.E., Hsieh C.L.. Murine de novo methyltransferase Dnmt3a demonstrates strand asymmetry and site preference in the methylation of DNA in vitro. Mol. Cell Biol. 2002; 22:704–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Handa V., Jeltsch A.. Profound flanking sequence preference of Dnmt3a and Dnmt3b mammalian DNA methyltransferases shape the human epigenome. J. Mol. Biol. 2005; 348:1103–1112. [DOI] [PubMed] [Google Scholar]
- 24. Wienholz B.L., Kareta M.S., Moarefi A.H., Gordon C.A., Ginno P.A., Chedin F.. DNMT3L modulates significant and distinct flanking sequence preference for DNA methylation by DNMT3A and DNMT3B in vivo. PLos Genet. 2010; 6:e1001106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jurkowska R.Z., Siddique A.N., Jurkowski T.P., Jeltsch A.. Approaches to enzyme and substrate design of the murine Dnmt3a DNA methyltransferase. Chembiochem. 2011; 12:1589–1594. [DOI] [PubMed] [Google Scholar]
- 26. Gowher H., Loutchanwoot P., Vorobjeva O., Handa V., Jurkowska R.Z., Jurkowski T.P., Jeltsch A.. Mutational analysis of the catalytic domain of the murine Dnmt3a DNA-(cytosine C5)-methyltransferase. J. Mol. Biol. 2006; 357:928–941. [DOI] [PubMed] [Google Scholar]
- 27. Jeltsch A., Lanio T.. Site-directed mutagenesis by polymerase chain reaction. Methods Mol. Biol. 2002; 182:85–94. [DOI] [PubMed] [Google Scholar]
- 28. Emperle M., Rajavelu A., Reinhardt R., Jurkowska R.Z., Jeltsch A.. Cooperative DNA binding and protein/DNA fiber formation increases the activity of the Dnmt3a DNA methyltransferase. J. Biol. Chem. 2014; 289:29602–29613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gowher H., Jeltsch A.. Enzymatic properties of recombinant Dnmt3a DNA methyltransferase from mouse: the enzyme modifies DNA in a non-processive manner and also methylates non-CpG [correction of non-CpA] sites. J. Mol. Biol. 2001; 309:1201–1208. [DOI] [PubMed] [Google Scholar]
- 30. Afgan E., Baker D., Batut B., van den Beek M., Bouvier D., Cech M., Chilton J., Clements D., Coraor N., Gruning B.A. et al.. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res. 2018; 46:W537–W544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Glass J.L., Hassane D., Wouters B.J., Kunimoto H., Avellino R., Garrett-Bakelman F.E., Guryanova O.A., Bowman R., Redlich S., Intlekofer A.M. et al.. Epigenetic Identity in AML Depends on Disruption of Nonpromoter Regulatory Elements and Is Affected by Antagonistic Effects of Mutations in Epigenetic Modifiers. Cancer Discov. 2017; 7:868–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rathert P., Roth M., Neumann T., Muerdter F., Roe J.S., Muhar M., Deswal S., Cerny-Reiterer S., Peter B., Jude J. et al.. Transcriptional plasticity promotes primary and acquired resistance to BET inhibition. Nature. 2015; 525:543–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Liu G.J., Cimmino L., Jude J.G., Hu Y., Witkowski M.T., McKenzie M.D., Kartal-Kaess M., Best S.A., Tuohey L., Liao Y. et al.. Pax5 loss imposes a reversible differentiation block in B-progenitor acute lymphoblastic leukemia. Genes Dev. 2014; 28:1337–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhang Y., Rohde C., Tierling S., Stamerjohanns H., Reinhardt R., Walter J., Jeltsch A.. DNA methylation analysis by bisulfite conversion, cloning, and sequencing of individual clones. DNA Methylation: Methods Protoc. 2009; 507:177–187. [DOI] [PubMed] [Google Scholar]
- 35. Bashtrykov P., Jeltsch A.. Jeltsch A, Rots MG. Epigenome Editing. 2018; Springer-Nature. [Google Scholar]
- 36. Garvie C.W., Wolberger C.. Recognition of specific DNA sequences. Mol. Cell. 2001; 8:937–946. [DOI] [PubMed] [Google Scholar]
- 37. Rohs R., Jin X., West S.M., Joshi R., Honig B., Mann R.S.. Origins of specificity in protein–DNA recognition. Annu. Rev. Biochem. 2010; 79:233–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhou T., Yang L., Lu Y., Dror I., Dantas Machado A.C., Ghane T., Di Felice R., Rohs R.. DNAshape: a method for the high-throughput prediction of DNA structural features on a genomic scale. Nucleic Acids Res. 2013; 41:W56–W62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Egger G., Jeong S., Escobar S.G., Cortez C.C., Li T.W., Saito Y., Yoo C.B., Jones P.A., Liang G.. Identification of DNMT1 (DNA methyltransferase 1) hypomorphs in somatic knockouts suggests an essential role for DNMT1 in cell survival. PNAS. 2006; 103:14080–14085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rhee I., Bachman K.E., Park B.H., Jair K.W., Yen R.W., Schuebel K.E., Cui H., Feinberg A.P., Lengauer C., Kinzler K.W. et al.. DNMT1 and DNMT3b cooperate to silence genes in human cancer cells. Nature. 2002; 416:552–556. [DOI] [PubMed] [Google Scholar]
- 41. Spencer D.H., Russler-Germain D.A., Ketkar S., Helton N.M., Lamprecht T.L., Fulton R.S., Fronick C.C., O’Laughlin M., Heath S.E., Shinawi M. et al.. CpG island hypermethylation mediated by DNMT3A is a consequence of AML progression. Cell. 2017; 168:801–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yin Y., Morgunova E., Jolma A., Kaasinen E., Sahu B., Khund-Sayeed S., Das P.K., Kivioja T., Dave K., Zhong F. et al.. Impact of cytosine methylation on DNA binding specificities of human transcription factors. Science. 2017; 356:eaaj2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kribelbauer J.F., Laptenko O., Chen S., Martini G.D., Freed-Pastor W.A., Prives C., Mann R.S., Bussemaker H.J.. Quantitative analysis of the DNA methylation sensitivity of transcription factor complexes. Cell Rep. 2017; 19:2383–2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yang X., Han H., De Carvalho D.D., Lay F.D., Jones P.A., Liang G.. Gene body methylation can alter gene expression and is a therapeutic target in cancer. Cancer Cell. 2014; 26:577–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Su J., Huang Y.H., Cui X., Wang X., Zhang X., Lei Y., Xu J., Lin X., Chen K., Lv J. et al.. Homeobox oncogene activation by pan-cancer DNA hypermethylation. Genome Biol. 2018; 19:108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yamato G., Yamaguchi H., Handa H., Shiba N., Kawamura M., Wakita S., Inokuchi K., Hara Y., Ohki K., Okubo J. et al.. Clinical features and prognostic impact of PRDM16 expression in adult acute myeloid leukemia. Genes Chromosomes Cancer. 2017; 56:800–809. [DOI] [PubMed] [Google Scholar]
- 47. Shiba N., Ohki K., Kobayashi T., Hara Y., Yamato G., Tanoshima R., Ichikawa H., Tomizawa D., Park M.J., Shimada A. et al.. High PRDM16 expression identifies a prognostic subgroup of pediatric acute myeloid leukaemia correlated to FLT3-ITD, KMT2A-PTD, and NUP98-NSD1: the results of the Japanese Paediatric Leukaemia/Lymphoma Study Group AML-05 trial. Br. J. Haematol. 2016; 172:581–591. [DOI] [PubMed] [Google Scholar]
- 48. Rampal R., Figueroa M.E.. Wilms tumor 1 mutations in the pathogenesis of acute myeloid leukemia. Haematologica. 2016; 101:672–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Yang M.Y., Lin P.M., Hsiao H.H., Hsu J.F., Lin H.Y., Hsu C.M., Chen I.Y., Su S.W., Liu Y.C., Lin S.F.. Up-regulation of PER3 expression Is correlated with better clinical outcome in acute leukemia. Anticancer Res. 2015; 35:6615–6622. [PubMed] [Google Scholar]
- 50. Torrebadell M., Diaz-Beya M., Kalko S.G., Pratcorona M., Nomdedeu J., Navarro A., Gel B., Brunet S., Sierra J., Camos M. et al.. A 4-gene expression prognostic signature might guide post-remission therapy in patients with intermediate-risk cytogenetic acute myeloid leukemia. Leuk. Lymphoma. 2018; 59:2394–2404. [DOI] [PubMed] [Google Scholar]
- 51. Plass C., Pfister S.M., Lindroth A.M., Bogatyrova O., Claus R., Lichter P.. Mutations in regulators of the epigenome and their connections to global chromatin patterns in cancer. Nat. Rev. Genet. 2013; 14:765–780. [DOI] [PubMed] [Google Scholar]
- 52. Sondka Z., Bamford S., Cole C.G., Ward S.A., Dunham I., Forbes S.A.. The COSMIC Cancer Gene Census: describing genetic dysfunction across all human cancers. Nat. Rev. Cancer. 2018; 18:696–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Rouillard A.D., Gundersen G.W., Fernandez N.F., Wang Z., Monteiro C.D., McDermott M.G., Ma’ayan A.. The harmonizome: a collection of processed datasets gathered to serve and mine knowledge about genes and proteins. Database. 2016; 2016:baw100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wu Y., Ferguson J.E. 3rd, Wang H., Kelley R., Ren R., McDonough H., Meeker J., Charles P.C., Patterson C.. PRDM6 is enriched in vascular precursors during development and inhibits endothelial cell proliferation, survival, and differentiation. J. Mol. Cell Cardiol. 2008; 44:47–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Dong F., Li Q., Yang C., Huo D., Wang X., Ai C., Kong Y., Sun X., Wang W., Zhou Y. et al.. PRMT2 links histone H3R8 asymmetric dimethylation to oncogenic activation and tumorigenesis of glioblastoma. Nat. Commun. 2018; 9:4552. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.