Abstract
A serological study was carried out in two Senegalese villages located in the Sine‐Saloum region in order to estimate the presence of anti‐leptospiral antibodies in humans and animals, and to identify the predominant serogroups. Seven hundred and forty‐nine serum samples were collected from humans (n = 545), dogs (n = 33), donkeys (n = 20), goats (n = 52), sheep (n = 43) and N’Dama cattle (n = 56), all originated from Dielmo and Ndiop villages. All samples were tested for different serovars of pathogenic Leptospira species by the microscopic agglutination test. Considering titres ≥ 1:100, 7.7% [CI 95:5.5 to 9.9] on the 545 human blood samples tested and 42.2% [CI95:35.4 to 48.9] on the 204 animal blood samples tested were found to be positive to one or more serovars. The results obtained indicate that the Australis serogroup is the most prevalent serogroup in human (67.3%) and cattle (27.3%). Serogroup Icterohaemorhagiae is the most frequent serogroup in goat (55.6%) and donkey (37.5%). Canicola (23.4%), Icterohaemorhagiae (21.1%) and Australis (12.5%) serogroups are the most prevalent serogroups in dogs. This study shows that diverse Leptospira serovars occur in a wide range of wild and domestic mammal species, as well as in humans in Senegal. However, further studies are needed to better understand the complexity of Leptospira epidemiology in Africa, identify the reservoirs of different serogroups and estimate its impact on livestock. Understanding the multi‐host epidemiology of leptospirosis is essential to control and prevent the disease.
Keywords: animal, epidemiology, human, Leptospira spp., leptospirosis, Senegal
A serological study was carried out in two Senegalese villages located in the Sine‐Saloum region and 8% on the 545 human blood samples tested and 42% on the 204 animal blood samples tested were found to be positive.The Australis serogroup is the most prevalent serogroup in human (67%) and cattle (27%). Serogroup Icterohaemorhagiae is the most frequent serogroup in goat (56%) and donkey (37%); Canicola (23%), Icterohaemorhagiae (21%) and Australis (12%) serogroups are the most prevalent serogroups in dogs.

1. INTRODUCTION
Leptospirosis is a worldwide zoonotic disease caused by spirochaetes of the genus Leptospira. More than 300 distinct leptospiral serovars of pathogenic Leptospira species are now recognized (Adler & de la Pena 2010). These serovars are grouped into 25 serogroups, according to the presence of agglutinating antibodies in analysed sera (Picardeau 2013). Recent species determination by DNA homology has identified 13 pathogenic Leptospira spp., and seven of them (L. interrogans, L. borgpetersenii, L. santarosai, L. noguchii, L. weilli, L. kirschneri and L. alexanderi) are considered to be the principal agents of human and animal disease (Cerqueira & Picardeau 2009; Evangelista & Coburn 2010). All these pathogenic species were previously considered as different serovars of one same species, Leptospira interrogans. Leptospirosis is considered an emerging or re‐emerging disease in several countries. Therefore, although present on all continents except Antarctica, leptospirosis is more prevalent in tropical or subtropical areas of the world. This disease is often linked to climate changes, poor living conditions in urban slums, but also to hobbies, pet ownership or recreational activities in the wild. Human are infected by contact with infected animal tissues or urine, contaminated water or soil. Many reservoirs belonging to wild and domestic animals are described but the most notorious are rodents. Leptospira serovars often demonstrate a degree of animal host preference, and some common relationships between serovars and their hosts are reported (Bharti et al. 2003; Adler & de la Pena 2010).
Leptospirosis is an important but neglected disease. Because of the lack of data, the global impact of this disease is unknown. Estimated leptospirosis incidence ranges from 0.1 to 1/100 000/year in temperate countries to over than 100/100 000/year during epidemics outbreaks in tropical areas. Between 300,000 and 500,000 severe cases are estimated to occur each year. More than 50% of severe cases may cause death (Bharti et al. 2003; Lau et al. 2010). More recently, a study based on a systematic review of published morbidity and mortality studies and databases estimated, using modelling, that there were annually 1.03 million cases (95% CI 434,000–1,750,000) and 58,900 deaths (95% CI 23,800‐95,900) due to leptospirosis worldwide (Costa et al. 2015).
The highest median annual incidence of leptospirosis is in Africa, standing at 95.5 per 100,000 people. Africa is followed by Western Pacific (66.4), the Americas (12.5), South‐East Asia (4.8) and Europe (0.5) (WHO 2011). Leptospirosis is likely endemic in Sub‐Saharan Africa. In West‐Africa, according to recent reviews, serological data are only available for five countries: Nigeria, Ghana, Senegal, Mali and Burkina‐Faso (Zida et al. 2018). In Senegal, only three surveys, conducted during the 1970s, were conducted and concerned only Dakar and its district. Two of them have included studies in animals (de Vries et al. 2014). Few data are available to compare Leptospira infection in related human and animal populations. A good knowledge of Leptospira serovars circulating in local animal populations is important to determine the sources and transmission routes of human infection (Allan et al., 2015), and for establishing disease control procedures. The objectives of this study were to estimate the prevalence of anti‐leptospiral antibodies among humans and animals living in a rural Senegal, and to identify the predominant serogroups. This knowledge will help to rationally design control and prevention measures.
2. MATERIAL AND METHODS
2.1. Study area
The study was conducted in two Senegalese villages located in the Sine‐Saloum region: Dielmo (13°43′26″N;16°24′38″W), 280 km south of Dakar and 10 km from the Gambia, located near the Nema river, and Ndiop (13°41′07″N;16°23′01″W), 5 km from Dielmo (Figure 1). The two villages are situated in an area of Sudan type savanna area with rainfall over a period of 4 months, from mid‐June to mid‐October. Annual rainfall is about 600–700 mm (Trape et al. 1994). Dielmo and Ndiop are two typical Senegalese villages inhabited by Serers (Dielmo) and Wolof (Ndiop) people, who are settled agricultural workers. Pearl millet (Pennisetum glaucum) and peanuts are cultivated during the rainy season. Most of the household possess different domestic animals: dogs, cats, donkeys, cattle, sheep and goats. There are also many poultry (mainly chickens and ducks).
Figure 1.
Location of the study area in Senegal (copyright of map: Wikimedia Commons Atlas)
2.2. Human sample collection
For serological studies, we used the serum samples collected in 2014 from the serological bank created for the longitudinal study mentioned above (Trape et al. 1994). In total, 266 serum samples collected in Dielmo (129 men and 137 women, mean age 21.8 ranged from 0.6 to 92.3) and 279 samples from Ndiop (115 men and 164 women, mean age 19.5 ranged from 0.3 to 88.6) were tested.
2.3. Domestic animals sampling
Two hundred and four blood samples were collected in 2012 from 33 dogs (Canis lupus familiaris Linnaeus, 1758), 20 donkeys (Equus asinus Linnaeus, 1758), 52 goats (Capra aegagrus hircus Linnaeus, 1758), 43 sheep (Ovis aries Linnaeus, 1758) and 56 N'Dama cattle (Bos taurus Linnaeus, 1758), all originated from Dielmo and Ndiop villages. The sampled animals represented 76% of the dogs, 34% of the cattle, 43% of the sheep, 30% of the goats and 29% of the donkeys present in these two villages according to the information collected from each family. The animals collected were chosen according to the consent of the owners and especially the limited times spent in the field and were not vaccinated against leptospirosis. Dogs were collected in the radial vein and other species in the jugular vein.
2.4. Sampling of wild small mammals
Small mammals were captured alive in 2013 using locally made wire‐mesh traps baited with peanut butter and/or onions. The traps were installed inside and outside the houses (two traps per room) in Dielmo and Ndiop villages. The animals trapped were sacrificed by means of cervical dislocation and necropsied in the field. For most animals, a thick blood film was immediately prepared in the field. Spleens of rodents and insectivores were collected (spleens were initially collected for a study of Bartonella spp.) and were stored at −80°C. Species of rodents and insectivores caught were identified using morphological method (Granjon & Duplantier 2009). In total, 36 rodents were captured and sampled.
2.5. Laboratory diagnostics
2.5.1. Serological studies
After centrifugation, human and animal sera were stored at −20°C before being sent to France for microscopic agglutination tests (MAT) in the veterinary school of Lyon. MAT is the reference serological test, particularly appropriate for epidemiological studies, since it can be applied to sera of any animal species, and because the range of antigens used can be expanded or decreased as required. The choice of the serovars used for each serogroup was based upon the experience of the laboratory. These serogroups were more extended than those previously described in epidemiological studies carried in continental Africa (Allan et al. 2015).
The MAT was performed using respectively 14 and 24 serovars of pathogenic Leptospira species, for human and for animals’ samples: L. interrogans (L. i.). Icterohaemorrhagiae (IH), L. i. Copenhageni (COP), L. i. Australis (AUS), L. i. Muenchen (MUN), L. i. Bratislava (BRAT), L. i. Autumnalis (AUT) L. i. Bim (BIM), L. i. Ballum BAL, L. i. Bataviae (BAT), L. i. Canicola (CAN), L. kirshneri Grippotyphosa (GRIP), L. i. Grippotyphosa Vanderhoedoni (VAN), L. i. Hebdomadis (HEB), L. i. Panama (PAN), L. i. Mangus (MAN), L. i. Pomona (POM), L. i. Mozdok (MOZ), L. i. Pyrogenes (PYR), L. i. Sejroe (SJ), L. i. Saxkoebing (SAX), L. borgpetersenii Hardjo (HJ), L. i. Wolfii (WOLF), L. i. Tarassovi (TAR) and L. i. Cynopteri (CYN). The end point is the highest dilution of serum in which 50% agglutination occurs. According to observations recorded in the French Leptospira laboratory in Lyon, titres higher than 1:100 have been considered positive for humans and all animal species.
2.5.2. Molecular biology
PCR assays were used to demonstrate Leptospira spp. infection from DNA extracts of rodent spleens (Mérien et al. 1992; Zilber et al. 2016). Thirty‐six DNA samples were delivered to the French Leptospira laboratory in Lyon. The analysis was carried out in three steps: Identification of positive DNAs by PCR on 16S DNA/ Typing of Leptospira species on positive DNAs/ Trying to identify the serovar involved by Variable Number Tandem (VNTR).The samples were identified by number and tested in pure concentration PCR according to the standardized protocol of the laboratory. The presence of an amplifiat of between 300 and 400 bp on 1.5% agarose gel after migration is considered a positive signal. This method has been validly used in previous studies to explore the renal carrier state in a very large variety of wildlife mammals (Ayral et al. 2016).
2.6. Statistical analyses
The prevalence was calculated as the number of animals with positive serum devised by the total number of animals studied in each group. Prevalence rates were compared between species, location and sex using Chi2 when the number of observation was sufficient, or the Fisher exact test when it was not. Analysis of variance (ANOVA) was used to evaluate associations between positivity to leptospiral antibodies and age. Odds ratios (OR), 95% confidence interval (CI) and p values were calculated separately for each variable. A p value ≤ .05 was considered significant. All statistical tests were carried out using the Epi Info Software (7.1.3.0 version, CDC Atlanta, USA).
3. RESULTS
Of the 545 human blood samples tested, 42 (7.7% [CI95:5.5 to 9.9]) were found to be positive to one or more serovars when a cut off of 1:100 was applied (Data S1 and S2). Leptospiral prevalence according to respective location was shown in Figure 2. The prevalence rate was not significantly different between the two villages: 6.4% Dielmo (17/266) and 9% Ndiop (25/279). Among the 42 human positive samples there is no significant difference (p > .5; OR: 0.88 [0.44–1.75]) between female (22/301) and male villagers (20/244).
Figure 2.
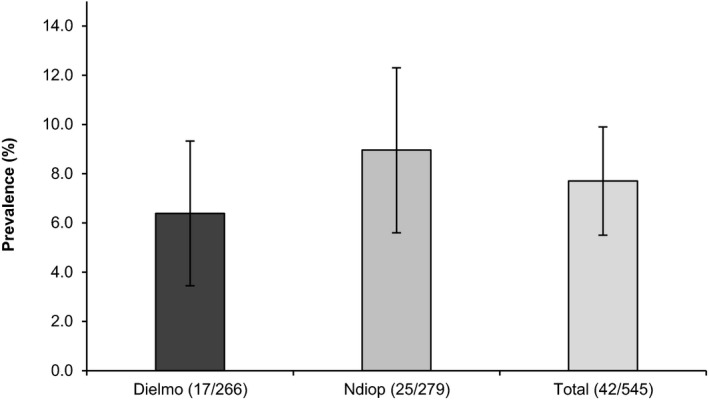
Percentage of Human positive samples for leptospirosis (MAT), according to location (error bars indicate 95% confidence intervals)
Correlation was found between seroprevalence and age (p < .01). Positive individuals were older (27.9 ± 20) than seronegative (mean age 20 ± 19). According to location, no correlation was found between seroprevalence and age in Ndiop (p > .1), but there is a statistically significant difference due to age in Dielmo (p < .01). There is no statistically significant difference (p > .5) between the age of seropositive women (mean age 29.1 ± 25) and seropositive men (mean age 26.5 ± 15.4).
Positivity can be observed for one or more leptospiral antigens, so we have recorded 60 positive serological reactions with different serovars for the 42 positive samples. The results of positive MAT according to leptospiral serovars in human (and different animal species) are shown in Table 1.
Table 1.
Results of positive MAT per pathogenic leptospiral serovars in human and different animal species
| Serogroup | Human (n = 545) | Donkey (n = 20) | Goat (n = 52) | Dog (n = 33) | Sheep (n = 43) | N’Dama (n = 56) | |
|---|---|---|---|---|---|---|---|
| Icterohemorrhagiae | IH | 2 | 12 | 2 | 27 | 1 | 0 |
| COP | 0 | 11 | 13 | 27 | 2 | 0 | |
| Australis | MUN | 0 | 5 | 1 | 16 | 0 | 3 |
| AUS | 11 | 4 | 0 | 9 | 0 | 0 | |
| BRAT | 33 | 7 | 0 | 16 | 0 | 3 | |
| Autumnalis | AUT | X | 1 | 0 | 9 | 0 | 0 |
| BIM | 0 | 2 | 9 | 7 | 1 | 0 | |
| Ballum | BAL | 7 | 0 | 0 | 0 | 0 | 4 |
| Bataviae | BAT | X | 0 | 0 | 0 | 0 | 0 |
| Canicola | CAN | 0 | 3 | 1 | 30 | 1 | 0 |
| Grippothyphosa | GRIP | 4 | 2 | 0 | 8 | 0 | 4 |
| VAN | X | 0 | 0 | 6 | 0 | 0 | |
| Hebdomadis | HEB | 0 | 0 | 0 | 2 | 0 | 3 |
| Panama | PAN | X | 0 | 0 | 2 | 0 | 0 |
| MAN | X | 0 | 0 | 5 | 0 | 0 | |
| Pomona | POM | X | 0 | 0 | 3 | 0 | 0 |
| MOZ | X | 0 | 0 | 4 | 0 | 1 | |
| Pyrogenes | PYR | 0 | 4 | 1 | 3 | 1 | 1 |
| Sejroe | SJ | X | 1 | 0 | 6 | 0 | 0 |
| SAX | 3 | 0 | 0 | 2 | 0 | 2 | |
| HJ | 0 | 0 | 0 | 2 | 0 | 1 | |
| WOLF | X | 0 | 0 | 2 | 0 | 0 | |
| Tarassovi | TAR | X | 0 | 0 | 6 | 0 | 0 |
| Cynopteri | CYN | 0 | 1 | 0 | 12 | 0 | 0 |
| 60 | 53 | 27 | 204 | 6 | 22 |
X: unintended serovar
Of a total number of 204 animal blood samples, 86 (42.2% [CI95:35.4 to 48.9]) tested positive with one or several serovars of pathogenic leptospiras when a cut off of 1:100 was applied (Data S3). According to the species, the positive sera were obtained from 16 of 20 donkeys (80% [CI95:56 to 94]), 18 of 52 goats (34.6% [CI95:22 to 49]), 3 of 43 sheep's (7% [CI95:1 to 19]), 17 of 56 cattle (30.4% [CI95:19 to 44]) and 32 of 33 dogs (97% [CI95:91 to 100]) tested. The prevalence rate is significantly lower (p < .001) in sheep than in any other animal species tested. There is no statistically significant difference between dogs and donkeys (p > .05), as well as between goats and cattle (p > .05). The prevalence rate is significantly higher (p < .001) in dogs and donkeys.
Positivity can be observed for one or more leptospiral antigens, so we have recorded 312 positive serological reactions with different serovars for the 86 positive samples. This phenomenon is due in general to the presence of co‐agglutinins in sera, but does not exclude the possibility that the animal is infected simultaneously with several strains or keeps a serological trace of prior(s) infection(s).
The results of positive MAT according to leptospiral serovars in different animal species (and human) are shown in Table 1. The prevalence rates (%) of the different serovars according to species are presented in Figure 3. The distribution of different serogroups among the seropositive animals, according to species is presented in Figure 4. Figure 5 illustrates the distribution of animal hosts by serogroups. Trapped rodents have all been identified. They belonged to four species and were distributed as follows: Gerbilliscus gambianus (Thomas, 1910) (n = 17), Mastomys erythroleucus (Temminck, 1853) (n = 15), Mus musculus (Linnaeus, 1758) (n = 3) and Arvicanthis niloticus (Desmarest, 1822) (n = 1). Two of 36 rodents were positive for 16S PCR for leptospirosis (Data S4). Sequences of amplified DNA fragment by the 16S rDNA PCR allowed species identification after alignment with all 16S rDNA sequences of all known Leptospira species and result permit the identification of L. interrogans for G. gambianus and L. kirschneri for M. erythroleucus. The identification of the corresponding serovars could not be completed due to a too weak and simultaneously non‐specific signal (VNTR).
Figure 3.
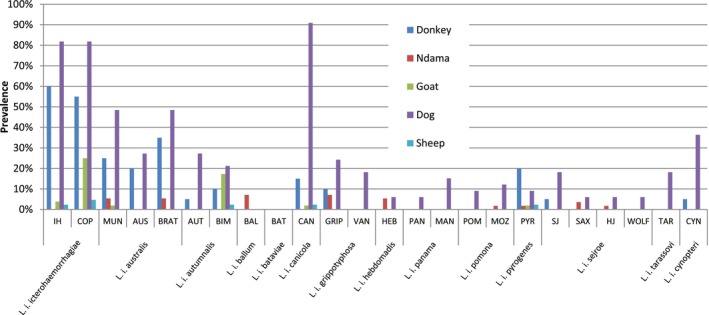
Prevalence rates (%) of the different serovars according to species
Figure 4.
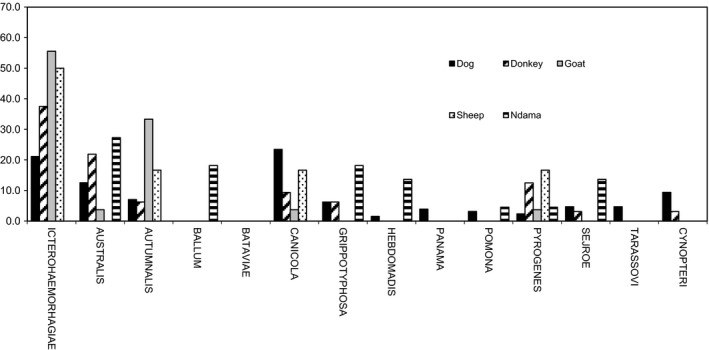
Distribution of different serogroups of pathogenic Leptospira species in seropostives animals
Figure 5.
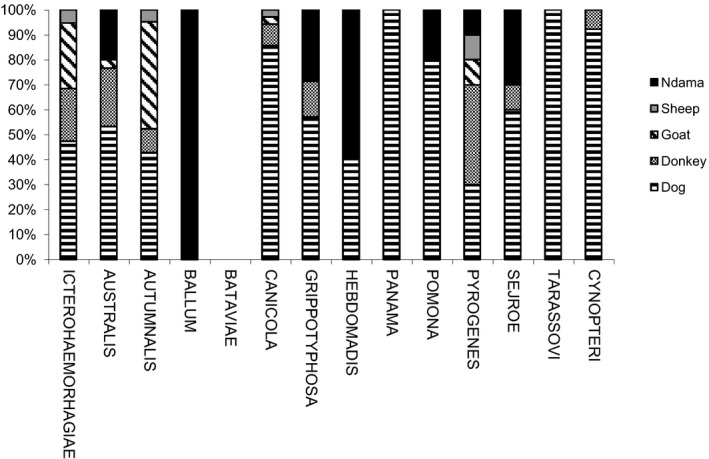
Distribution of animal hosts by serogroup. Each histogram stands for the percent contribution of each host on the total positive hosts for the whole given serogroup. All serogroups were considered on a basis of a similar scale.
4. DISCUSSION
4.1. Prevalence
The results obtained show that 7.7% of apparently healthy villagers and 42.2% of domestic animals from the same households are serologically positive with one or several serovars of pathogenic leptospira, without evidence of clinical leptospirosis.
Leptospirosis is probably endemic in West Africa, where recent epidemiological data are lacking. The last studies in Senegal date back to the 1970s (Sarrat et al. 1973; Sankale et al. 1976).
4.2. Serogroups
The MAT is the reference test for the diagnosis and screening of leptospiral infection. However, the existence of cross‐reactions between several serovars of Leptospira limits the accuracy of this test for serotyping. Therefore, the results were analysed at the serogroup level. Samples that were positive for several serovars of the same serogroup were considered positive for this serogroup, and in case several serogroups were positive, the infecting serogroup was affected to the one with the highest titre. The results obtained indicate serogroup Australis as the most frequent serogroup in human (67.3%) and cattle (27.3%). Serogroup Icterohaemorhagiae is the most frequent serogroup in goat (55.6%) and donkey (37.5%). Rats are usually considered the reservoir host of this serogroup (Bharti et al. 2003). Ninety‐seven percent of 33 dogs tested are positive. Serogroups Canicola (23.4%), Icterohaemorhagiae (21.1%) and Australis (12.5%) are the most prevalent serogroups in dogs. The dogs collected in the two villages were not vaccinated against leptospirosis, so the results indicate the real prevalence and the circulation of strains of serogroups Icterohaemorrhagiae and Canicola among dogs. Dogs are not usually considered as a reservoir for Leptospira, except for Canicola (André‐Fontaine 2006), thus the high prevalence found in this study results from a heavy exposure of dogs as incidental hosts, thus revealing the pressure of infection from the environment. The dog seems to be a good indicator species for exposure, due to its extreme sensitivity to leptospirosis and the fact that it shares the close human environment. This particularly high prevalence in dogs living in a rural environment has already been described in Gabon where the seroprevalence rate was higher in rural areas compared to an urban environment (Roqueplo et al. 2015). This difference could be a consequence of their way of life, because dogs in villages are more often used for hunting in the forests and therefore have more contact with wildlife (Roqueplo et al. 2015). The prevalence rate observed in our study is higher to the one observed in dogs in different studies conducted in Africa (Nigeria, Zimabwe, Ugandan and South Africa). The prevalence observed in these studies varies between 4.7% and 26.7%, with a cut‐off ≥ 1:100 (Agunloye et al. 2002; Gatley 2009; Dhliwayo et al. 2012; Millán et al. 2013).
Although the small number of individuals per group did not allow statistical comparative tests to be applied, it is possible to indicate that, except in sheep, the Australis serogroup circulates in all hosts studied.
Sheep are considered naturally insensitive and relatively resistant to leptospiras (Ellis 1994), which may explain the low level of seropositivity found in our study.
Results indicate a high biological diversity as, apart serogroup Bataviae, all serogroups tested were identified. This high biodiversity has already been reported in tropical or equatorial areas and is supposed to be related to a wide range of mammalian reservoirs (Bharti et al. 2003). The infection is endemic in domestic animals and concerns all the species. The leptospirosis in ruminants, although clinically not important, causes huge economic losses and is considered a major plague of livestock farming. Indeed, the involvement of the leptospirosis in the abortion of ruminants is largely recognized (Smith & Easmon 1990), venereal transmission of the bacterium has been demonstrated and the occurrence of mastitis, associated with leptospirosis, has been described (Simegnew 2016; Garoussi et al. 2017).
Our results confirm that the majority of domestic animals which were positive have survived to leptospiral infections or are asymptomatic. The presence of antibodies indicates that animals were exposed to the pathogen. However, the cut‐off points selected (low in epidemiological investigation) and the absence of kinetic serology do not, in most cases, lead to the conclusion of a current active infection. Clinical signs are quite variable and most cases are probably unapparent and associated with host‐adapted serovars, such as Canicola in dogs and Hardjo in cattle (André‐Fontaine 2006; Grooms 2006).
4.3. Rodents
Pathogenic leptospiral DNAs were identified in 2/36 samples belonging to G. gambianus and M. erythroleucus. G. gambianus is known from the Sahel and Sudan savannas of Senegal, Mali and Niger. This is a common species that is locally abundant and is occasionally considered to be an agricultural pest. This species is found in woodlands on sandy and clay soils, and also in fallow lands and cultivated fields. It has also been recorded on sandy and mangrove islands of the Sine‐Saloum Delta (Musser & Carleton 2005; Granjon 2017).
Mastomys erythroleucus occurs mostly in sub‐Saharan northern Africa. The main range is Gambia, Senegal, Guinea, Ghana, Sierra Leone, Côte d'Ivoire, Burkina Faso, Mali, Niger, Benin, Nigeria, Cameroon, Central African Republic, Sudan, Ethiopia, eastern Democratic Republic of the Congo, Burundi, western Uganda and northern Kenya. It is mainly a lowland species. This species is found in a wide range of habitats, mainly moist and dry savannas and dry forest in the west, as well as moist and dry scrublands. It is a commensal species found in close association with human habitation but also in gardens and cultivated lands (Granjon 2016).
The relatively low prevalence of leptospirosis infection in rodents is certainly due to the fact that the research was conducted in the spleen and not in the kidneys (natural persistence site for Leptospira species) of rodents which, unfortunately, were not available for this study. However, the presence of leptospiras in rodents’ spleens may indicate that these two rodents were very heavily loaded with leptospiras or had only a current or recent acute infection.
5. CONCLUSION
This study shows that diverse Leptospira serovars occur in a wide range of wild and domestic mammal species, and humans in Senegal. This study also highlights the high prevalence of different leptospiral serogroups, particularly among donkeys and dogs. The main serogroup identified in human (Australis) is also significantly found in animals. While virulent sources are essentially represented by the inexhaustible reservoir of small wild mammals, it should not be forgotten that domestic animals can in turn become a source of contamination.
As the key‐point in the control of leptospirosis is based in the control of rodents, in these villages, a fight against rodents with burrows in the houses was carried out. Burrow entrances have been cemented. This measure prevents occurrence of leptospirosis and borreliosis in both human and domestic animal (dog and production animal's herds). In addition, screening for human leptospirosis has become more regular during fever.
Dogs can be considered as sentinels for human exposure to this zoonotic organism. Their use as a sentinel animal for leptospirosis in a national (or local) disease surveillance system in Africa could be explored. The impact of this zoonosis on the health and livelihoods of rural Senegalese living in these villages would merit further study.
Neglected tropical diseases are now a priority for global public health. Leptospirosis may be a real public health problem in Africa, for both human and animals. Virtual lack of data about leptospirosis in West Africa should not hinder the possible impact of this disease on public health and economy. Using an integrated “One Health” approach to explore the relationship between human and animal Leptospira infection in areas where human disease is identified, would also provide invaluable evidence to quantify the direct and indirect impacts of leptospirosis on human and animal populations in Africa (Allan et al. 2015).
CONFLICT OF INTEREST
All of the authors declare no conflict of interest related to this article.
SOURCE OF FUNDING
This study was supported by the Institut Hospitalo‐Universitaire (IHU) Méditerranée Infection, the National Research Agency under the program « Investissementsd’avenir », reference ANR‐10‐IAHU‐03, the Région Provence‐Alpes‐Côte d’Azur and European funding FEDER PRIMI.
AUTHOR CONTRIBUTIONS
BD, OM, CS and DR designed the study; BD, PS, JPD, GD, HB and OM collected the samples; AK conducted the laboratory analysis; AK, JPD and CR analysed the data; and CR, OM, AK and BD prepared the manuscript.
ETHICAL STATEMENT
Collection and handling of wild and domestic animals has followed the guidelines of the American Society of Mammalogists (Gannon 2007). The Dielmo project was approved by the National Ethics Committee of Senegal (Trape et al. 1994), and Local Ethics Committee (Marseille, France). Individual written and informed consent were obtained from each participant, including the parents or legal guardians of all children at the start of the Dielmo project. All participants were subjected to a questionnaire and were examined before sampling.
Supporting information
ACKNOWLEDGEMENTS
We thank Charles Bouganali and Tristan Ulivieri for their kind cooperation.
Roqueplo C, Kodjo A, Demoncheaux J‐P, et al. Leptospirosis, one neglected disease in rural Senegal. Vet Med Sci. 2019;5:536–544. 10.1002/vms3.186
Funding information
This study was supported by the Institut Hospitalo‐Universitaire (IHU) Méditerranée Infection, the National Research Agency under the program « Investissements d'avenir», reference ANR‐10‐IAHU‐03, the Région Provence Alpes Côte d’Azur and European funding FEDER PRIMI.
REFERENCES
- Adler B. & de la Pena M. A. (2010) Leptospira and leptospirosis. Veterinary Microbiology 140, 287–296. 10.1016/j.vetmic.2009.03.012 [DOI] [PubMed] [Google Scholar]
- Agunloye C. A., Ajuwape A. T. P. & Nottidge H. O. (2002) Comparative study of the prevalence of leptospirosis in vaccinated and unvaccinated dogs in Ibadan, Nigeria. Tropical Veterinary 20(1), 22–26. 10.4314/tv.v20i1.4505 [DOI] [Google Scholar]
- Allan K. J., Biggs H. M., Halliday J. E. B., Kazwala R. R., Maro V. P., Cleaveland S. & Crump J. A. (2015) Epidemiology of leptospirosis in Africa: A systematic review of a neglected zoonosis and a paradigm for ‘One Health’ in Africa. PLoS Neglected Tropical Diseases 9(9), e0003899 10.1371/journal.pntd.0003899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- André‐Fontaine G. (2006) Canine leptospirosis: Do we have a problem? Veterinary Microbiology 117, 19–24. 10.1016/j.vetmic.2006.04.005 [DOI] [PubMed] [Google Scholar]
- Ayral F., Djelouadji Z., Raton V., Zilber A. L., Gasqui P., Faure E., et al. (2016) Hedgehogs and mustelid species: Major carriers of pathogenic Leptospira, a survey in 28 animal species in France (2012, 2015). PLoS ONE 11(9), e0162549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharti A. R., Nally J. E., Ricaldi J. N., Matthias M. A., Diaz M. M., Lovett M. A., et al (2003) Leptospirosis: A zoonotic disease of global importance. Lancet Infectious Diseases 3, 757–771. 10.1016/S1473-3099(03)00830-2 [DOI] [PubMed] [Google Scholar]
- Cerqueira G. M. & Picardeau M. (2009) A century of Leptospira strain typing. Infection, Genetics and Evolution 9(5), 760–768. 10.1016/j.meegid.2009.06.009 [DOI] [PubMed] [Google Scholar]
- Costa F., Hagan J. E., Calcagno J., Kane M., Torgerson P., Martinez‐Silveira M. S., et al (2015) Global morbidity and mortality of leptospirosis: A systematic review. PLoS Neglected Tropical Diseases 9(9), e0003898 10.1371/journal.pntd.0003898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhliwayo S., Matope G., Marabini L., Dutlow K. & Pfukenyi D. M. (2012) Sero‐prevalence of leptospirosis in dogs in urban Harare and selected rural communities in Zimbabwe Onderstepoort. Journal of Veterinary Research 79(1), 1–6. [DOI] [PubMed] [Google Scholar]
- Ellis W. A. (1994) Leptospirosis as a cause of reproductive failure. Veterinary Clinics of North America: Food Animal Practice 10(3), 463–478. [DOI] [PubMed] [Google Scholar]
- Evangelista K. V. & Coburn J. (2010) Leptospira as an emerging pathogen: A review of its biology, pathogenesis and host immune responses. Future Microbiology 5(9), 1413–1425. 10.2217/fmb.10.102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gannon W. L. & Sikes R. S. (2007) Guidelines of the American Society of Mammalogists for the use of wild mammals in research. Journal of Mammalogy 88(3), 809–823. 10.1644/06-MAMM-F-185R1.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garoussi M. T., Atarie S., Vodjgani M., Gharagozloo F. & Vand e Ussefi, J. (2017) The prevalence and control of bovine mastitis in Leptospira outbreak. Comparative Clinical Pathology 26, 189–192. 10.1007/s00580-016-2367-1 [DOI] [Google Scholar]
- Gatley J. M. (2009) The prevalence of Leptospira serovars causing infection in dogs in South Africa Thesis for the degree of magister scientiae, University of Pretoria, p 60. [Google Scholar]
- Granjon L. (2016) Mastomys erythroleucus. The IUCN Red List of Threatened Species 2017, 10.2305/IUCN.UK.2016‐3.RLTS.T12866A22424945.en. Downloaded on 15 November.
- Granjon L. (2017) Gerbilliscus gambiana. The IUCN Red List of Threatened Species 2017, 10.2305/IUCN.UK.2017‐2.RLTS.T45076A22427087.en. Downloaded on 15 November
- Granjon L. & Duplantier J. M. (2009) Les Rongeurs de l’Afrique sahélo‐soudanienne. IRD/MNHN Editions, Collection Faune et Flore tropicale, no 43, Marseille, p 215.
- Grooms D. L. (2006) Reproductive losses caused by bovine viral diarrhea virus and leptospirosis. Theriogenology 66, 624–628. 10.1016/j.theriogenology.2006.04.016 [DOI] [PubMed] [Google Scholar]
- Lau C., Smythe L. & Weinstein P. (2010) Leptospirosis: An emerging disease in travellers. Travel Medicine and Infectious Diseases 8, 33–39. 10.1016/j.tmaid.2009.12.002 [DOI] [PubMed] [Google Scholar]
- Mérien F., Amouriaux P., Perolat P., Baranton G. & Saint G. I. (1992) Polymerase chain reaction for detection of Leptospira spp. in clinical samples. Journal of Clinical Microbiology 30, 2219–2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millán J., Chirife A. D., Kalema‐Zikusoka G., Cabezón O., Muro J., Marco I., et al (2013) Serosurvey of dogs for human, livestock, and wildlife pathogens, Uganda. Emerging Infectious Diseases 19(4), 680–682. 10.3201/eid1904.121143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musser G. G. & Carleton M. D. (2005) Superfamily muroidea In: Wilson D. E. & Reeder D. A. (Eds), Mammal species of the world: A geographic and taxonomic reference (pp. 894–1531). Baltimore, USA: The John Hopkins University Press. [Google Scholar]
- Ndiaye P., Trape J.‐F., Druilhe P., Diagne N., Faye O., Legros F., et al (1994) The Dielmo project: A longitudinal study of natural malaria infection and the mechanisms of protective immunity in a community living in a holoendemic area of Senegal. The American Journal of Tropical Medicine and Hygiene 51, 123–137. 10.4269/ajtmh.1994.51.123 [DOI] [PubMed] [Google Scholar]
- Picardeau M. (2013) Diagnosis and epidemiology of leptospirosis. Médecine Et Maladies Infectieuses 43(1), 1–9. 10.1016/j.medmal.2012.11.005 [DOI] [PubMed] [Google Scholar]
- Roqueplo C., Marié J. L., André‐Fontaine G., Kodjo A. & Davoust B. (2015) Serological survey of canine leptospirosis in three countries of tropical Africa: Sudan, Gabon and Ivory Coast. Comparative Immunology Microbiology and Infectious Diseases 38, 57–61. 10.1016/j.cimid.2014.10.006 [DOI] [PubMed] [Google Scholar]
- Sankale M., Sow A. M., Ruscher H. & Sarrat H. (1976) Leptospirosis in a Dakar hospital: Results of a new survey. African Journal of Medicine and Medical Sciences 5(3), 213–220. [PubMed] [Google Scholar]
- Sarrat H., Doutre M. P. & Ruscher H. (1973) On the epidemiology of leptospirosis in the region of Cap‐Vert (Senegal). Bulletin De La Société De Médecine D’afrique Noire 18(2), 236–239. [PubMed] [Google Scholar]
- Simegnew A. (2016) A review of bovine leptospirosis. European Journal of Applied Sciences 8(6), 347–355. [Google Scholar]
- Smith G. R. & Easmon C. S. F. (1990) Leptospirosis In Parker M. T. & Collier L. H. (Eds.), Topley and Wilson's principles of bacteriology, virology and immunity, (8th edn, Vol. 3), Bacterial diseases (pp. 619–640). London, UK: Edward Arnold Division of Hodder & Stoughton. [Google Scholar]
- de Vries S. G., Visser B. J., Nagel I. M., Goris M. G. A., Hartskeerl R. A. & Grobusch M. P. (2014) Leptospirosis in Sub‐Saharan Africa: A systematic review. International Journal of Infectious Diseases 28, 47–64. 10.1016/j.ijid.2014.06.013 [DOI] [PubMed] [Google Scholar]
- World Health Organization . (2011) Report of the second meeting of leptospirosis burden epidemiology reference group Geneva, Switzerland. [Google Scholar]
- Zida S., Kania D., Sotto A., Brun M., Picardeau M., Castéra J., et al (2018) Leptospirosis as cause of febrile icteric illness, Burkina Faso. Emerging Infectious Diseases 24(8), 1569–1572. 10.3201/eid2408.170436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilber A. L., Belli P., Grezel D., Artois M., Kodjo A. & Djelouadji Z. (2016) Comparison of mucosal, subcutaneous and intraperitoneal routes of rat Leptospira infection. PLoS Neglected Tropical Diseases 10(3), e0004569 10.1371/journal.pntd.0004569 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


