Abstract
Background
Pneumonia, a leading cause of childhood mortality, is associated with household air pollution (HAP) exposure. Mechanisms between HAP and pneumonia are poorly understood, but studies suggest that HAP may increase the likelihood of bacterial, instead of viral, pneumonia. We assessed the relationship between HAP and infant microbial nasal carriage among 260 infants participating in the Ghana Randomized Air Pollution and Health Study (GRAPHS).
Methods
Data are from GRAPHS, a cluster-randomized controlled trial of cookstove interventions (improved biomass or LPG) versus the 3-stone (baseline) cookstove. Infants were surveyed for pneumonia during the first year of life and had routine personal exposure assessments. Nasopharyngeal swabs collected from pneumonia cases (n = 130) and healthy controls (n = 130) were analyzed for presence of 22 common respiratory microbes by MassTag polymerase chain reaction. Data analyses included intention-to-treat (ITT) comparisons of microbial species presence by study arm, and exposure-response relationships.
Results
In ITT analyses, 3-stone arm participants had a higher mean number of microbial species than the LPG (LPG: 2.71, 3-stone: 3.34, p < 0.0001, n = 260). This difference was driven by increased bacterial (p < 0.0001) rather than viral species presence (non-significant). Results were pronounced in pneumonia cases and attenuated in healthy controls. Higher prevalence bacterial species were Haemophilus influenzae, Streptococcus pneumoniae, and Moraxella catarrhalis. Exposure-response relationships did not yield significant associations between measured CO and nasal microbial carriage.
Conclusions
Our intention-to-treat findings are consistent with a link between HAP and bacterial nasal carriage. No relationships were found for viral carriage. Given the null results in exposure-response analysis, it is likely that a pollutant besides CO is driving these differences.
Keywords: Childhood pneumonia, Microbial carriage, Lower respiratory infections, Household air pollution, Biomass fuels
Highlights
-
•
Cookstove intervention showed lower bacterial species presence in upper airways.
-
•
No differences found in species presence for viruses.
-
•
Exposure-response analyses for carbon monoxide yield null results.
-
•
Differences likely due to other constituents of air pollution
1. Introduction
Approximately 3 billion people worldwide use biomass fuels for their cooking and heating needs, including wood, dung, charcoal, and crop residues (Bonjour et al., 2013). The combustion of these fuels produces a complex mixture of air pollutants collectively termed household air pollution (HAP). These air pollutants contributed to 59 million disability-adjusted life years and 1.6 million premature deaths worldwide in 2017 (IMHE, 2017). Of the diseases HAP contributes to, pneumonia has the largest impact on children (Gordon et al., 2014; Smith et al., 2014). Pneumonia is the second leading cause of mortality worldwide for children under five years of age (Stanaway et al., 2018), and almost half of that mortality is in Sub-Saharan Africa (Troeger et al., 2017). In Ghana, where our study is based, pneumonia is the second leading cause of death for children under five, and caused over 6000 childhood deaths in 2016 (Troeger et al., 2017; UNICEF, 2018).
There is strong evidence to support a link between air pollution and respiratory infections like pneumonia (Brauer et al., 2002; MacIntyre et al., 2014; Romieu et al., 2002; Troeger et al., 2017). A number of HAP-specific studies have found such an association (Dherani et al., 2008; Smith, 2000). However, evidence from randomized-controlled trials (RCTs) has been inconsistent with observational studies (Mortimer et al., 2017). Intention-to-treat results for RESPIRE in Guatemala yielded inconclusive results for pneumonia, but severe pneumonia showed significant results. Exposure-response analyses in RESPIRE, however, did find a significant relationship with pneumonia (Smith et al., 2011). A small subset of air pollution studies has specifically investigated the etiology of HAP-associated pneumonia. The RESPIRE study, for example, found a decrease in severe respiratory syncytial virus negative (RSV-) pneumonia with lower levels of HAP exposure, but no impact on severe RSV+ infections. Other studies suggest that air pollution exposure may increase susceptibility to bacterial pneumonias, but not viral pneumonias (Rylance et al., 2015; Smith et al., 2011; Zhou and Kobzik, 2007).
Broadly, pneumonia results from the inhalation of pathogenic microbes which colonize or infect the upper airways (Bosch et al., 2013). Pathogens migrate to the lower airways by aspiration or by contiguity (Barson et al., 2014). Infection of the lower airways ultimately leads to pneumonia. Mechanistic studies have shown that HAP is associated with higher bacterial abundance (Rylance et al., 2016), and impaired macrophage response (Rylance et al., 2015; Zhou and Kobzik, 2007) in the lower airways. Less is known about the relationship between HAP and microbes in the upper airways, which mechanistically precedes lower respiratory infections.
Pneumonia incidence has been on the decline worldwide, which has been attributed both to increased vaccine uptake and also to improved nutrition (Rudan, 2008; Walker et al., 2013). Development and uptake of the pneumococcal and Haemophilus influenzae type b vaccines have changed the distribution of attributable fractions of pneumonia by microbe, with higher viral proportions in vaccinated populations (Barson et al., 2014). Ghana has been particularly successful in its national childhood vaccination efforts, especially in rural areas (Asuman et al., 2018). In our Ghana cohort, we have found that 91.4% of our rural participants received the three scheduled pneumococcal vaccines by the child's first birthday. Therefore, Ghana serves as an opportunity to understand the role of HAP in pneumonia etiology in a high vaccination environment.
Leveraging the Ghana Randomized Air Pollution and Health Study (GRAPHS), we examined patterns of infant nasal microbial carriage in relation to HAP exposure (Jack et al., 2015). We focus on understanding the relationship between HAP and particular microbial agents known to cause pneumonia in clinician-diagnosed cases and healthy controls. We hypothesized that: 1) High and low HAP exposure groups will have similar viral carriage patterns, and 2) the high HAP exposure group will have higher bacterial carriage compared to the low exposure group. We then conducted exposure-response analyses to assess relationships between pre and postnatal HAP exposure on 1) species-specific bacterial carriage, and 2) the number of bacterial species present.
2. Methods
2.1. Study participants
Participants were drawn from GRAPHS, a cluster-randomized cookstove intervention trial in the rural area of Kintampo, Ghana (Jack et al., 2015). Starting in August 2013, 1414 non-smoking pregnant women were enrolled in GRAPHS. All women were enrolled by 24 weeks gestation, and were randomized to receive two improved biomass stoves (Biolite), or a dual-burner liquefied petroleum gas (LPG) stoves, or maintain use of their 3-stone fire, which is the traditional wood burning stove predominantly used in this part of Ghana. Mothers and infants were tracked until the infant's first birthday. LPG has been demonstrated to have the lowest air pollution emissions of the three types of stoves, whereas the 3-stone fire has the highest (Jetter et al., 2012).
2.2. Pneumonia surveillance, specimen collection and selection
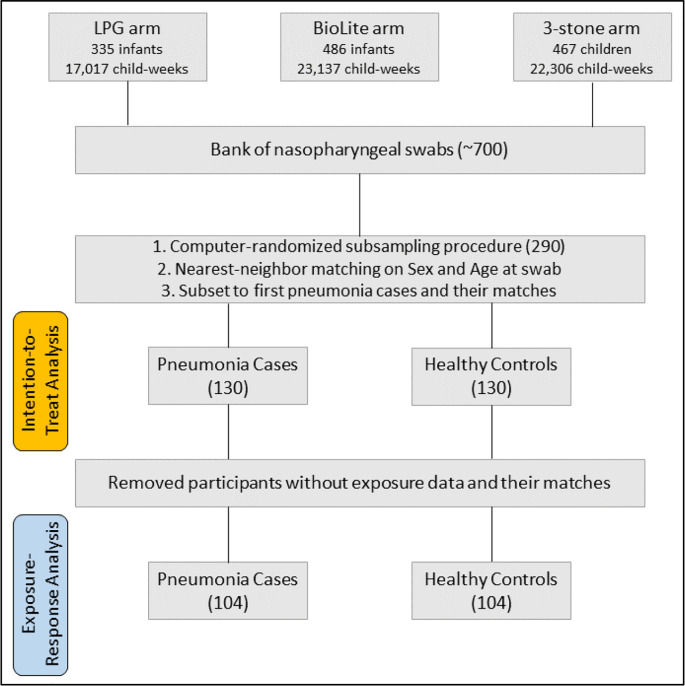
Trained fieldworkers conducted weekly pneumonia surveillance according to the World Health Organization's Integrated Management of Childhood Illness (IMCI) guidelines. Fieldworkers referred suspected cases to a study clinic for clinician diagnosis and treatment. Nasopharyngeal swabs were taken for all clinician-diagnosed cases. Fieldworkers then identified healthy infant controls, who were also sent for nasopharyngeal swabs. The fieldworkers chose healthy controls that were the same sex and close to the same age as the case, prioritizing those living in the same (or a neighboring) community. A total of 669 unique samples were catalogued and stored for future analysis. Samples were stored in a − 80 °C freezer until microbial profiling, except for transport to the Unites States when on dry ice. With resources to analyze 290 samples, a computer-generated random selection of swabs selected 145 pneumonia cases, and their matched controls, for PCR analysis (Fig. 1 ). Swabs were only selected from the LPG and 3-stone arms of the study to maximize the exposure contrast among samples. We determined that fieldworker selection of controls did not achieve sufficient balance on matching variables, so a “nearest neighbor” algorithm was used to perform a 1:1 match of each pneumonia case with its nearest control. The nearest neighbor algorithm finds the closest possible match between participants in the dataset based on chosen covariates, in our case, minimizing the difference between age and sex. We then removed all repeat pneumonia cases (swabs from reoccurring pneumonias), with their associated healthy controls, to preserve independence and given prior literature on altered microbiomal development with prior infection (Bosch et al., 2017). Our final sample consisted of 130 pneumonia cases and 130 healthy controls for our intention-to-treat analysis.
Fig. 1.
Sample selection and pneumonia case and healthy control matching.
2.3. Carbon monoxide exposure
Personal exposure monitoring was conducted for carbon monoxide (CO) using the Lascar EL-CO-USB Data Logger (Erie, PA). Mother-child pairs had a total of seven personal exposure monitoring sessions throughout the study period, four for the mother in the prenatal period, and three concurrent mother-child sessions in the postnatal period. The monitor measured CO every 10 s in parts per million. It was attached to the participants' clothing, close to the breathing zone, except during sleep or bathing, when participants were asked to keep the monitor off the ground and nearby. Fieldworkers visited daily to ensure wearing compliance and proper device functioning. Devices were checked against certified span gas every six weeks, and a data validation system addressed low-quality deployments. Final session values were based on a 48-h measurement period. See Quinn et al. (2016) for additional information. Two separate exposure variables were used to assess effects based on timing of exposure. First, a prenatal maternal exposure average was calculated using the data from the four prenatal sessions, herein referred to as the Mean Prenatal CO. Second, for the postnatal session, children's exposures were linearly interpolated between time points to provide regular estimates over time. The interpolation estimates the most recent CO value at the time of the nasopharyngeal swab, herein referred to as Recent Postnatal CO. We report R2 and intraclass correlation coefficients for CO measurements, for the entire GRAPHS cohort, by arm of the study and by individual repeated measures (Supplementary Table 2). These are provided to aid in the interpretation of results based on the exposure variables. Supplementary Tables 3 and 4 also report results of a sensitivity analysis based on a mean postnatal value, calculated from the 3 child-specific monitoring sessions.
2.4. Ethical approvals
Ethical approvals for GRAPHS were obtained from the Ghana Health Service Ethical Review Committee, the Kintampo Health Research Centre Institutional Ethics Committee, and the Institutional Review Board of Columbia University Medical Center.
2.5. Microbial identification
MassTag polymerase chain reaction (PCR) was used to determine binary presence or absence of 22 common causes of childhood respiratory illness in 290 randomly selected banked samples, see Table 1 . Total nucleic acid (TNA) from each sample was extracted with the EasyMag Extraction Platform (Biomerieux, Marcy l'Etoile, France). TNA (or cDNA, as appropriate) was used as template for MassTag PCR using two separate PCR multiplex assays. One assay targeted common respiratory RNA viruses, and the other targeted bacterial agents and adenovirus. Following the PCR, the products were purified and analyzed with a mass spectrometer for the presence of pathogen-specific tags (Briese et al., 2005; Lamson et al., 2006).
Table 1.
Microbes selected for MassTag PCR analysis.
| DNA agents | RNA agents |
|---|---|
| Adenonvirus | Influenza A |
| Chlamydia pneumoniae | Influenza B |
| Legionella pneumophilia | Respiratory Syncytial Virus A (RSVA) |
| Mycoplasma pneumoniae | Respiratory Syncytial Virus B (RSVB) |
| Neisseria meningitidis | Human Parainfluenza Virus 1 (HPIV1) |
| Haemophilus influenzae | Human Parainfluenza Virus 2 (HPIV2) |
| Streptococcus pneumoniae | Human Parainfluenza Virus 3 (HPIV3) |
| Mycobacteria tuberculosis | Human Parainfluenza Virus 4 (HPIV4) |
| Moraxella catarrahalis | Human metapneumovirus (MPV) |
| Bortadella pertussis | Coronavirus OC43 |
| Coronavirus 229E | |
| Enterovirus |
2.6. Stratification by disease status
Quantitative analyses were conducted stratified by disease status. Pneumonia cases were expected to have higher bacterial and/or viral presence than healthy controls given that upper respiratory colonization or infection characteristically precedes lower respiratory infection (Bogaert et al., 2004). Healthy controls, however, were still expected to have microbial carriage, but lower baseline levels (Dunne et al., 2018). We hypothesized that increased HAP exposure, or being in the 3-stone fire study arm, would be associated with a higher number of bacterial species present both in pneumonia cases and healthy controls. Cases and controls are stratified due to the potential for collider bias among cases (Fig. 2 ), whereby we hypothesize associations between HAP and carriage, but it is also likely that HAP operates on other elements of pneumonia etiology.
Fig. 2.
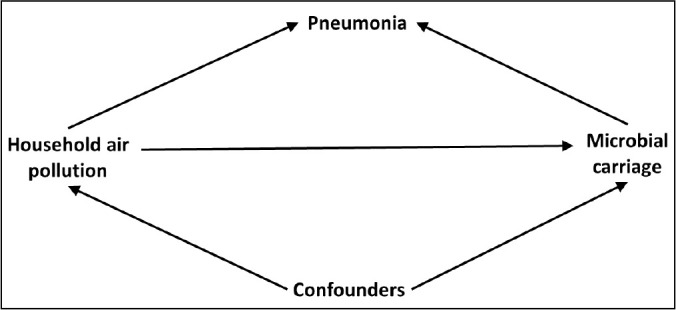
Directed acyclic graph representing the relationship between HAP and nasal microbial carriage for pneumonia cases.
2.7. Viral and bacterial carriage by study arm
We first analyzed results via pairwise comparisons of summed microbial species identifications by study arm, comparing the 3-stone fire arm to the intervention arm (LPG cookstove). Summed species totals could vary from zero to 22: 0 to 9 for bacteria, and 0 to 13 for viruses. Comparisons were then conducted specific to bacteria and viruses. Count data of microbes were not normally distributed; therefore, we used Wilcox rank sum tests to compare distributions.
Bacterial species presence by study arm was analyzed using contingency tables and Fisher's exact tests. Contingency tables were then used to calculate odds ratios, and confidence intervals were calculated.
2.8. Exposure-response relationships
Analyses using the study arm as a proxy for exposure have the potential for exposure misclassification, as within-arm exposures exhibit considerable heterogeneity. First, we present exposure distributions by arm of study. Then we conducted exposure-response analyses, leveraging the individual-level personal exposure data for mothers and infants in the GRAPHS study.
2.8.1. Microbial carriage
The outcome measures of interest were binary: testing positive or negative for each specific bacterium or virus. Bonferroni correction was used for multiple comparisons, one regression per bacterial species. The unadjusted alpha was 0.05, so a p-value of 0.017 was considered statistically significant.
2.8.2. Covariates and confounding
Several variables were assessed as potential confounders due to theorized relationships with household air pollution and bacterial nasal carriage. They included an Asset Index (a measure of socioeconomic status), population density, total household size, and the season an infant was swabbed. The Asset Index was constructed using a principal components analysis of variables including: type of housing materials, type of toilet facility, primary water source, type of home ownership, household ownership of livestock animals, and household ownership of consumer durables (Gunnsteinsson et al., 2010). The population density measure was constructed using mapped census information from the Kintampo Health and Demographic Surveillance System. A 100-meter radial buffer was created around each home to aggregate the number of individuals within 100 m of the reference household using a spatial join. The season of swab was based on the date of an infant's swab, comparing the Harmattan/dry season to the non-Harmattan/wet season. Ultimately, we found that the season of swab was the only potential confounder variable correlated with the exposure and the outcome, identified via univariable regressions with p-values < 0.05. Season of swab was included in the final regression analyses. Age at swab and sex were also included in the analysis given a priori knowledge of relationships with the exposure and outcome.
2.8.3. Regressions and data analyses
Two generalized linear models were used in these analyses. First, logistic regression was used to model the odds of species-specific microbial presence given exposure. Then we used multinomial logistic regression to model the number of bacterial positives, comparing to zero, represented as follows:
where KBS = the number of bacterial species (1, 2, or 3), 0BS = zero bacterial positives as the referent group, CO = measured carbon monoxide in parts per million, ChildAge = child's age in weeks, ChildSex = the child's biological sex at birth, SeasonSwabbed = whether the swab was during the wet or dry season. All exposure data had a substantial right skew and were transformed to the natural log scale for analysis.
The regression models the likelihood of testing positive for one, two, or three bacterial species as a function of exposure. Therefore, the model is, in effect, blind to the particular species.
Buffers and spatial joins were conducted in QGIS 2.1. All other quantitative analyses were conducted in R version 3.3.3 (Vienna, Austria). The nearest-neighbor matching was performed with the MatchIt package. Multinomial models were estimated with the nnet package. These packages are available on CRAN.
3. Results
3.1. Baseline demographics
Table 2 outlines the baseline demographics of the study participants, stratified by disease status. Variables included are those that might contribute to differences in microbial carriage, including breastfeeding, the child's age at the time of swab, household density, and season of birth. The only variable that which shows a marginal difference is the child's age at swab. This emphasizes the importance of including this variable in our adjusted exposure-response analyses. Cases and controls show balance on all relevant covariates for exposure-response analyses.
Table 2.
Baseline demographics, comparing pneumonia cases and healthy controls. p values derived from t-test if continuous or chi squared test if categorical.
| Intention-to-treat analyses |
Exposure-response analyses |
|||||
|---|---|---|---|---|---|---|
| Cases | Controls | p | Cases | Controls | p | |
| n | 130 | 130 | 104 | 104 | ||
| LPG/3-stone fire participants (n) | 63/67 | 75/55 | 41/63 | 60/44 | ||
| Postnatal CO Exposure in ppm (median (IQR)) | –a | –a | 0.64 (0.29–1.23) | 0.78 (0.28–1.42) | ||
| Mother's ethnicity (n (%)) | 0.73 | 0.56 | ||||
| Akan | 16 (12.3) | 24 (18.5) | 12 (11.5) | 21 (20.2) | ||
| Dagarti | 30 (23.1) | 26 (20.0) | 25 (24.0) | 22 (21.2) | ||
| Gonja | 15 (11.5) | 17 (13.1) | 12 (11.5) | 11 (10.6) | ||
| Konkonba | 18 (13.8) | 13 (10.0) | 15 (14.4) | 10 (9.6) | ||
| Mo | 17 (13.1) | 17 (13.1) | 16 (15.4) | 14 (13.5) | ||
| Other | 34 (26.2) | 33 (25.4) | 24 (23.1) | 26 (25.0) | ||
| Asset Index (mean (sd)) | 0.18 (2.14) | 0.43 (2.24) | 0.36 | 0.23 (2.14) | 0.59 (2.33) | 0.26 |
| Caesarean birth (n (%)) | 5 (3.9) | 10 (7.7) | 0.29 | 3 (2.9) | 7 (6.7) | 0.33 |
| Birth season = wet (n (%)) | 70 (53.8) | 69 (53.1) | 1.00 | 60 (57.7) | 56 (53.8) | 0.68 |
| Child's Sex = female (n (%)) | 62 (47.7) | 58 (44.6) | 0.71 | 46 (44.2) | 46 (44.2) | 1.00 |
| Breastfed within 4 days (n (%)) | 124 (96.1) | 123 (94.6) | 0.78 | 99 (95.2) | 99 (95.2) | 1.00 |
| Season swabbed = wet (n (%)) | 100 (76.9) | 105 (80.8) | 0.54 | 76 (73.1) | 86 (82.7) | 0.13 |
| Age at swab, in weeks (mean (sd)) | 21.16 (13.56) | 24.45 (12.72) | 0.06 | 22.66 (13.71) | 24.56 (13.02) | 0.31 |
| Children <5 in household (mean (sd))b | 1.12 (0.94) | 1.16 (0.97) | 0.74 | 1.09 (0.95) | 1.13 (0.86) | 0.75 |
| Persons in household (mean (sd)) | 6.82 (3.86) | 6.81 (3.20) | 0.98 | 6.53 (3.41) | 6.83 (3.22) | 0.51 |
| Population within 100 m (mean (sd)) | 180.3 (95.4) | 177.7 (96.3) | 0.83 | 182 (98.7) | 184 (101.2) | 0.89 |
Values were not calculated due to missing exposure data in some participants.
Not including participant child.
3.2. Microbial identification
MassTag PCR analysis yielded positives for 13 of the 22 microbes listed in Table 1. Three of the microbes were bacteria: M. catarrhalis, S. pneumoniae and H. influenzae. The positively identified viruses were: Rhinovirus, Influenza A, HPIV1, HPIV2, HPIV3, Metapneumovirus, Corona NL63, Corona OC43, RSVA, and Adenovirus (Supplementary Table 1).
3.3. Viral and bacterial carriage by study arm
Results from the Wilcox rank sum analyses show that infants in the 3-stone fire arm had a higher abundance of all microbe species compared to infants in the LPG arm (p < 0.0001) (Table 3 ). This relationship appears to be driven by more bacterial species (p < 0.0001) rather than viral species. Stratifying the analysis by pneumonia status shows that clinician-diagnosed cases maintain the same relationship, with higher overall microbial (p < 0.001) and higher bacterial (p < 0.0001) species abundance. Again, there were no differences in viral presence. Healthy controls show a similar pattern, with marginally significant differences for all microbes (p = 0.058), differences in bacterial species abundance (p = 0.011), and no differences in viral species abundance. These results warranted particular emphasis on bacterial species, rather than viruses, because there were no observed differences in viral abundance. There is also a temporal pattern of bacterial carriage in the whole cohort, whereby overall carriage increases with a child's age (Supplemental Fig. 1).
Table 3.
Mean (median) number of identified microbial species present in nasopharyngeal swabs from participants in the LPG arm vs. the 3 stone arm, stratified by disease status. p values calculated from Wilcox rank sum test.
| All participants (n = 260) |
Pneumonia cases (n = 130) |
Healthy controls (n = 130) |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| LPG (n = 138) | 3 Stone (n = 122) | p value | LPG (n = 63) | 3 Stone (n = 67) | p value | LPG (n = 75) | 3 Stone (n = 55) | p value | |
| All microbes | 2.71 (3) | 3.34 (4) | <0.0001 | 2.95 (3) | 3.70 (4) | <0.001 | 2.51 (2) | 2.91 (3) | 0.058 |
| Viruses | 0.97 (1) | 0.98 (1) | 0.964 | 1.22 (1) | 1.15 (1) | 0.457 | 0.76 (1) | 0.76 (1) | 0.83 |
| Bacteria | 1.74 (1) | 2.37 (1) | <0.0001 | 1.73 (2) | 2.55 (3) | <0.0001 | 1.75 (2) | 2.15 (2) | 0.011 |
Infants in the 3-stone arm were more likely to test positive for all of the bacterial species compared to those in the LPG arm of the study (Table 4 ). This association was consistent for cases as well. Among healthy controls there is a borderline significant difference for M.catarrhalis, and all associations are positive, in the same direction as cases. Given our a priori hypothesis of higher bacterial species presence, we limited species-specific pairwise statistical analysis to H.influenzae, S.pneumoniae, and M.catarrhalis, with Bonnferoni correction.
Table 4.
Odds ratios (98.34% confidence intervals) for the probability of testing positive for specific bacterial species among infants in the 3-stone arm compared to those in the LPG/intervention arm. Statistically significant values, calculated from Fischer's exact test, in bold.
| All infants (n = 260) | Cases (n = 130) | Controls (n = 130) | |
|---|---|---|---|
| S.pneumoniae | 2.42 (1.07–5.81) | 3.4 (1.01–13.46) | 1.73 (0.55–5.95) |
| M.catarrhalis | 3.06 (1.55–6.22) | 3.71 (1.27–11.96) | 2.46 (0.97–6.48) |
| H.influenzae | 3.1 (1.59–6.18) | 5.82 (2.15–17.09) | 1.69 (0.66–4.45) |
3.4. Exposure-response relationships
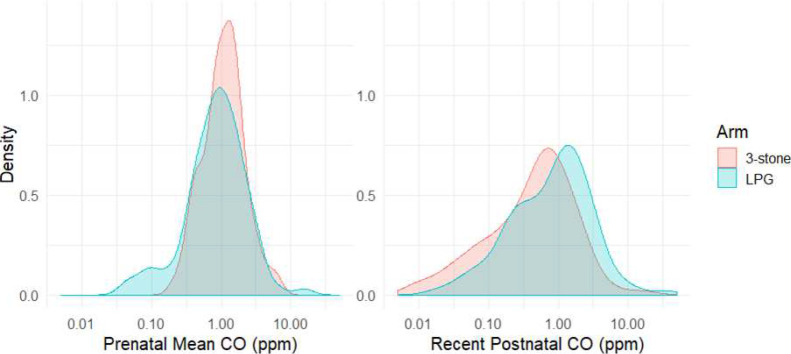
Exposure distributions vary by arm of study (Fig. 3 ). The median prenatal CO value for LPG arm was 0.87 ppm (IQR: 0.49–1.54), and 1.095 ppm (IQR: 0.71–1.65) for the 3-stone fire arm participants. Median CO values for the recent postnatal CO 1.03 ppm (IQR: 0.3–1.91) for LPG arm children and 0.53 (IQR: 0.15–1.00) for the 3-stone arm. We assessed the correlation between CO and PM2.5 and found that they have a low, but statistically significant, association (Spearman's ρ = 0.212, p value = 0.016).
Fig. 3.
Probability density distributions of exposure variables on the log10 scale. A) Prenatal Mean CO and B) Recent Postnatal CO (postnatal interpolated CO value).
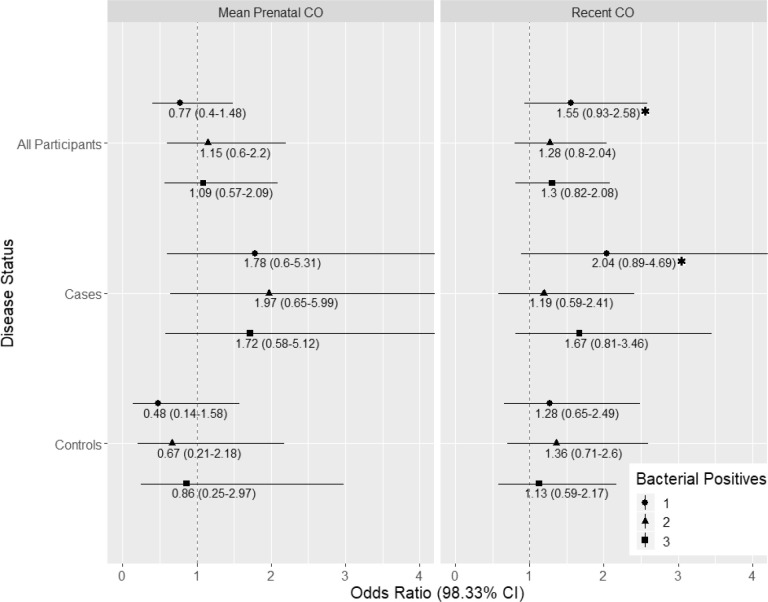
Multinomial logistic regressions were employed to model potential exposure-response relationships for increasing levels of bacterial carriage (Fig. 4 ). None of these models was statistically significant at the Bonferroni-adjusted confidence levels. However, the Recent postnatal CO regression has a suggestive finding whereby testing positive for one bacterial species increases with the child's most recent exposure (OR: 1.55, 98.33% CI: 0.93–2.58, p = 0.036). This trend appears to hold for cases (OR: 2.04, CI: 0.89–4.69, p = 0.038), but not controls. Bacterial species-specific logistic regression models produced null results for both exposure variables (Recent Postnatal CO and Mean Prenatal CO) of interest (Supplementary Tables 3 and 4).
Fig. 4.
Results from multinomial logistic regression examining the effect of a log-unit increase of CO on number of bacterial species present (referent = 0, max = 3). Point estimate odds ratios are indicated by periods, and 98.34% confidence intervals by horizontal lines. All models adjusted for age at swab, sex, and season swabbed. p values < 0.05 are starred, but the Bonferroni-adjusted p value is 0.017. N = 208.
4. Discussion
We conducted a study in a rural region of Ghana analyzing infant microbial carriage in relation to household air pollution exposures and cookstove interventions. Using MassTag PCR of nasopharyngeal swabs, we identified ten viruses and three bacteria present among our study participants. In an intention-to-treat analysis, we observed lower bacterial species presence among participants in the LPG study arm compared to the 3-stone fire study arm. This observation persisted among all three of the bacterial species analyzed. The difference was pronounced for pneumonia cases, and attenuated for healthy controls. We did not observe a difference in viral species presence between the LPG and 3-stone study arms. This finding is interesting given that colonization or infection of the upper airways is one of the first steps of pneumonia etiology (Bogaert et al., 2004). Other studies have shown that HAP is associated with increased bacterial abundance in the lower airways (Rylance et al., 2015) and impaired phagocytic response in the lower airways (Rylance et al., 2015; Zhou and Kobzik, 2007). Therefore, HAP may be operating on multiple parts of the pneumonia disease pathway.
Our models did not yield statistically significant findings in our exposure-response analyses of bacterial carriage for pre- or postnatal exposure to CO after adjusting for multiple comparisons. A suggestive observation in our multinomial model indicates that postnatal HAP-exposure may increase the likelihood of testing positive for one bacterial species compared to none – either H. influenzae, S. pneumoniae, or M. catarrhalis. More research is needed to understand the role of HAP in upper airway microbial carriage.
The timing of exposure is also emerging as an important variable in these investigations, as many air pollution studies indicate that risk changes over the life course (Goldizen et al., 2016; Lee et al., 2018). This can have important ramifications for policy. For example, if the antenatal period comprises a window of susceptibility, then policies that target exposure reductions to pregnant women may be particularly effective. We, however, find no evidence of an effect of prenatal air pollution exposure on nasal carriage.
This study has many strengths, including cookstove randomization, robust disease surveillance, and exposure monitoring. Past studies in this field either have lacked microbial specificity or have not employed a prospective study design, thus limiting the ability to account for confounding factors (Smith et al., 2011; Rylance et al., 2016). Our analysis is nested in a well-characterized longitudinal cohort tracked in the prenatal and postnatal periods. Therefore, we are able to assess the potential roles of antenatal exposures on microbial carriage, which has not been previously examined to our knowledge.
There are limitations to our analysis. PM from biomass combustion has been associated with components of pneumonia etiology, but no such evidence exists for CO (Rylance et al., 2015, Rylance et al., 2016; Zhou and Kobzik, 2007). With regard to exposure, CO was the only consistently monitored air pollutant. When designing the study, we intended to use CO as a proxy for fine particulate matter (PM2.5). Since then, studies have demonstrated that CO may not be an appropriate proxy (Carter et al., 2017; Klasen et al., 2015). PM2.5 was monitored in this trial, but due to the considerable cost of PM2.5 exposure monitoring relative to CO, it was only monitored on a subset of participants in fewer monitoring sessions. We investigated using PM exposures in the current analysis, but there was insufficient overlap between swabbed participants and those tracked for PM2.5 in order to conduct exposure-response analyses. Of the overlap we did have, there was a low, but significant, correlation between PM and CO. We are also limited by the fact that there was missing CO data among a subset of participants for our exposure-response analyses.
Another potential limitation to this study is the high prevalence of bacterial nasal carriage in the overall population, thus limiting our power to detect differences. Although we were unable to find literature about Ghana specifically, there is evidence of high bacterial carriage of pneumococcal bacteria and H.influenzae in West Africa, specifically in the Gambia and Nigeria (Adetifa et al., 2012; Goetghebuer et al., 2000; Hill et al., 2008). These high background levels mean that our study may not have been appropriately powered to detect differences in bacterial species abundance. Further, MassTag PCR, while a powerful tool, provides coarse measures of microbial diversity. Results from the MassTag PCR can only provide a binary outcome on a fixed library of microbes. Presence of a microbe at one time point may not provide adequate information to infer causal mechanisms; instead, it is preferred to confirm microbial presence over time as well as density of colonization. Finally, our analysis utilizes samples from the nasal epithelium whereas pneumonia occurs in the lower respiratory tract. However, growing evidence demonstrates the strong relationships between the composition of the upper respiratory tract and overall respiratory health (Biesbroek et al., 2014; Man et al., 2017; Teo et al., 2015).
5. Conclusion
To our knowledge, this is the first analysis assessing measured HAP-exposure and microbial carriage nested in a longitudinal cohort. Our findings support an association with HAP exposure and bacterial nasal carriage, which is on the pathway to pneumonia. This provides additional evidence on the ways in which HAP may be altering pneumonia susceptibility. This is an important area of investigation given the vast burden of disease caused by HAP, specifically on young children. However, given the null results in exposure-response analysis, it is likely that a pollutant besides CO is driving these differences. Identification of the etiologic relationships between HAP and pneumonia may spur advancements in vaccination, infection control, or allow for refined burden of disease estimates, which can support targeted public health initiatives.
Acknowledgments
Acknowledgements
The authors would like to thank community members in Kintampo, particularly those who participated in the study. We also wish to acknowledge Kenneth Wiru for producing and providing the geographic variables for the analysis. Finally, thanks to Carlos Gould for providing comments on an initial draft.
Funding sources
This work was supported by the National Institute of Environmental Health Sciences (NIH 1R01ES019547), the NIEHS Center for Environmental Health in Northern Manhattan (P30 ES009089), the Global Alliance for Clean Cookstoves, the Thrasher Research Fund, and the Ghana Ministry of Health. The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the U.S. NIH or Department of Health and Human Services.
Declaration of competing interest
The authors have no known conflicts of interest to report.
Handling Editor: Hanna Boogaard
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.envint.2019.105150.
Contributor Information
Daniel Carrión, Email: daniel.carrion@mssm.edu.
Darby W. Jack, Email: dj2183@columbia.edu.
Appendix A. Supplementary data
Supplementary material
References
- Adetifa I.M.O., Antonio M., Okoromah C.A.N., Ebruke C., Inem V., Nsekpong D.…Adegbola R.A. Pre-vaccination nasopharyngeal pneumococcal carriage in a Nigerian population: epidemiology and population biology. PLoS One. 2012;7(1) doi: 10.1371/journal.pone.0030548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asuman D., Ackah C.G., Enemark U. Inequalities in child immunization coverage in Ghana: evidence from a decomposition analysis. Heal. Econ. Rev. 2018;8 doi: 10.1186/s13561-018-0193-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barson, W. J., Kaplan, S., & Torchia, M. (2014). Pneumonia in Children: Epidemiology, Pathogenesis, and Etiology. UpToDate. Waltham, MA.(Accessed on January 28, 2018).
- Biesbroek G., Tsivtsivadze E., Sanders E.A.M., Montijn R., Veenhoven R.H., Keijser B.J.F., Bogaert D. Early respiratory microbiota composition determines bacterial succession patterns and respiratory health in children. Am. J. Respir. Crit. Care Med. 2014;190(11):1283–1292. doi: 10.1164/rccm.201407-1240OC. [DOI] [PubMed] [Google Scholar]
- Bogaert D., de Groot R., Hermans P. Streptococcus pneumoniae colonisation: the key to pneumococcal disease. Lancet Infect. Dis. 2004;4(3):144–154. doi: 10.1016/S1473-3099(04)00938-7. [DOI] [PubMed] [Google Scholar]
- Bonjour S., Adair-Rohani H., Wolf J., Bruce N.G., Mehta S., Prüss-Ustün A.…Smith K.R. Solid fuel use for household cooking: country and regional estimates for 1980–2010. Environ. Health Perspect. 2013;121(7):784–790. doi: 10.1289/ehp.1205987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch A.A.T.M., Biesbroek G., Trzcinski K., Sanders E.A.M., Bogaert D. Viral and bacterial interactions in the upper respiratory tract. PLoS Pathog. 2013;9(1) doi: 10.1371/journal.ppat.1003057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch A.A.T.M., Piters W.A.A. de S., van Houten M.A., Chu M.L.J.N., Biesbroek G., Kool J.…Bogaert D. Maturation of the infant respiratory microbiota, environmental drivers, and health consequences. A prospective cohort study. Am. J. Respir. Crit. Care Med. 2017;196(12):1582–1590. doi: 10.1164/rccm.201703-0554OC. [DOI] [PubMed] [Google Scholar]
- Brauer M., Hoek G., Van Vliet P., Meliefste K., Fischer P.H., Wijga A.…Brunekreef B. Air pollution from traffic and the development of respiratory infections and asthmatic and allergic symptoms in children. Am. J. Respir. Crit. Care Med. 2002;166(8):1092–1098. doi: 10.1164/rccm.200108-007OC. [DOI] [PubMed] [Google Scholar]
- Briese T., Palacios G., Kokoris M., Jabado O., Liu Z., Renwick N.…Lipkin W.I. Diagnostic system for rapid and sensitive differential detection of pathogens. Emerg. Infect. Dis. 2005;11(2):310–313. doi: 10.3201/eid1102.040492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter E., Norris C., Dionisio K.L., Balakrishnan K., Checkley W., Clark M.L.…Baumgartner J. Assessing exposure to household air pollution: a systematic review and pooled analysis of carbon monoxide as a surrogate measure of particulate matter. Environ. Health Perspect. 2017;125(7) doi: 10.1289/EHP767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dherani M., Pope D., Mascarenhas M., Smith K.R., Weber M., Bruce N. Indoor air pollution from unprocessed solid fuel use and pneumonia risk in children aged under five years: a systematic review and meta-analysis. Bull. World Health Organ. 2008;86:390–398. doi: 10.2471/BLT.07.044529. (C) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunne E.M., Murad C., Sudigdoadi S., Fadlyana E., Tarigan R., Indriyani S.A.K.…Hinds J. Carriage of Streptococcus pneumoniae, Haemophilus influenzae, Moraxella catarrhalis, and Staphylococcus aureus in Indonesian children: a cross-sectional study. PLoS One. 2018;13(4) doi: 10.1371/journal.pone.0195098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetghebuer T., West T.E., Wermenbol V., Cadbury A.L., Milligan P., Lloyd-Evans N.…Weber M.W. Outcome of meningitis caused by Streptococcus pneumoniae and Haemophilus influenzae type b in children in The Gambia. Tropical Med. Int. Health. 2000;5(3):207–213. doi: 10.1046/j.1365-3156.2000.00535.x. [DOI] [PubMed] [Google Scholar]
- Goldizen F.C., Sly P.D., Knibbs L.D. Respiratory effects of air pollution on children: respiratory effects of air pollution on children. Pediatr. Pulmonol. 2016;51(1):94–108. doi: 10.1002/ppul.23262. [DOI] [PubMed] [Google Scholar]
- Gordon S.B., Bruce N.G., Grigg J., Hibberd P.L., Kurmi O.P., Lam K.H.…Balmes J. Respiratory risks from household air pollution in low and middle income countries. Lancet Respir. Med. 2014;2(10):823–860. doi: 10.1016/S2213-2600(14)70168-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunnsteinsson S., Labrique A.B., West K.P., Christian P., Mehra S., Shamim A.A.…Klemm R.D. Constructing indices of rural living standards in northwestern Bangladesh. J. Health Popul. Nutr. 2010;28(5) doi: 10.3329/jhpn.v28i5.6160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill P.C., Cheung Y.B., Akisanya A., Sankareh K., Lahai G., Greenwood B.M., Adegbola R.A. Nasopharyngeal carriage of Streptococcus pneumoniae in Gambian infants: a longitudinal study. Clin. Infect. Dis. 2008;46(6):807–814. doi: 10.1086/528688. [DOI] [PubMed] [Google Scholar]
- Institute for Health Metrics and Evaluation Global Burden of Disease Study. 2017 http://ghdx.healthdata.org/gbd-2017 Retrieved November 13, 2018, from. [Google Scholar]
- Jack D.W., Asante K.P., Wylie B.J., Chillrud S.N., Whyatt R.M., Quinn A.K.…Mujtaba M. Ghana randomized air pollution and health study (GRAPHS): study protocol for a randomized controlled trial. Trials. 2015;16(1):420. doi: 10.1186/s13063-015-0930-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jetter J., Zhao Y., Smith K.R., Khan B., Yelverton T., DeCarlo P., Hays M.D. Pollutant emissions and energy efficiency under controlled conditions for household biomass cookstoves and implications for metrics useful in setting international test standards. Environmental Science & Technology. 2012;46(19):10827–10834. doi: 10.1021/es301693f. [DOI] [PubMed] [Google Scholar]
- Klasen E.M., Wills B., Naithani N., Gilman R.H., Tielsch J.M., Chiang M.…Apaka C. Low correlation between household carbon monoxide and particulate matter concentrations from biomass-related pollution in three resource-poor settings. Environ. Res. 2015;142:424–431. doi: 10.1016/j.envres.2015.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamson D., Renwick N., Kapoor V., Liu Z., Palacios G., Ju J.…Ian Lipkin W. MassTag polymerase-chain-reaction detection of respiratory pathogens, including a new rhinovirus genotype, that caused influenza-like illness in New York state during 2004–2005. J. Infect. Dis. 2006;194(10):1398–1402. doi: 10.1086/508551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee A.G., Kaali S., Quinn A., Delimini R., Burkart K., Opoku-Mensah J.…Asante K.P. Prenatal household air pollution is associated with impaired infant lung function with sex-specific effects: evidence from GRAPHS, a cluster randomized cookstove intervention trial. Am. J. Respir. Crit. Care Med. 2018 doi: 10.1164/rccm.201804-0694OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacIntyre E.A., Gehring U., Mölter A., Fuertes E., Klümper C., Krämer U.…Heinrich J. Air pollution and respiratory infections during early childhood: an analysis of 10 European birth cohorts within the ESCAPE project. Environ. Health Perspect. 2014;122(1):107–113. doi: 10.1289/ehp.1306755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Man W.H., de Steenhuijsen Piters W.A.A., Bogaert D. The microbiota of the respiratory tract: gatekeeper to respiratory health. Nat. Rev. Microbiol. 2017;15(5):259–270. doi: 10.1038/nrmicro.2017.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortimer K., Ndamala C.B., Naunje A.W., Malava J., Katundu C., Weston W.…Nyirenda M. A cleaner burning biomass-fuelled cookstove intervention to prevent pneumonia in children under 5 years old in rural Malawi (the Cooking and Pneumonia Study): a cluster randomised controlled trial. Lancet. 2017;389(10065):167–175. doi: 10.1016/S0140-6736(16)32507-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn A.K., Ae-Ngibise K.A., Jack D.W., Boamah E.A., Enuameh Y., Mujtaba M.N.…Asante K.P. Association of Carbon Monoxide exposure with blood pressure among pregnant women in rural Ghana: evidence from GRAPHS. Int. J. Hyg. Environ. Health. 2016;219(2):176–183. doi: 10.1016/j.ijheh.2015.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romieu I., Samet J.M., Smith K.R., Bruce N. Outdoor air pollution and acute respiratory infections among children in developing countries. J. Occup. Environ. Med. 2002;44(7) doi: 10.1097/00043764-200207000-00010. https://journals.lww.com/joem/Fulltext/2002/07000/Outdoor_Air_Pollution_and_Acute_Respiratory.10.aspx Retrieved from. [DOI] [PubMed] [Google Scholar]
- Rudan I. Epidemiology and etiology of childhood pneumonia. Bull. World Health Organ. 2008;86(5):408–416. doi: 10.2471/BLT.07.048769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rylance J., Chimpini C., Semple S., Russell D.G., Jackson M.J., Heyderman R.S., Gordon S.B. Chronic household air pollution exposure is associated with impaired alveolar macrophage function in Malawian non-smokers. PLoS One. 2015;10(9) doi: 10.1371/journal.pone.0138762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rylance J., Kankwatira A., Nelson D.E., Toh E., Day R.B., Lin H.…Gordon S.B. Household air pollution and the lung microbiome of healthy adults in Malawi: a cross-sectional study. BMC Microbiol. 2016;16(1) doi: 10.1186/s12866-016-0803-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith K.R. Indoor air pollution in developing countries and acute lower respiratory infections in children. Thorax. 2000;55(6):518–532. doi: 10.1136/thorax.55.6.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith Kirk R., McCracken J.P., Weber M.W., Hubbard A., Jenny A., Thompson L.M.…Bruce N. Effect of reduction in household air pollution on childhood pneumonia in Guatemala (RESPIRE): a randomised controlled trial. Lancet. 2011;378(9804):1717–1726. doi: 10.1016/S0140-6736(11)60921-5. [DOI] [PubMed] [Google Scholar]
- Smith Kirk R., Bruce N., Balakrishnan K., Adair-Rohani H., Balmes J., Chafe Z.…Rehfuess E. Millions dead: how do we know and what does it mean? Methods used in the comparative risk assessment of household air pollution. Annu. Rev. Public Health. 2014;35(1):185–206. doi: 10.1146/annurev-publhealth-032013-182356. [DOI] [PubMed] [Google Scholar]
- Stanaway J.D., Afshin A., Gakidou E., Lim S.S., Abate D., Abate K.H.…Murray C.J.L. Global, regional, and national comparative risk assessment of 84 behavioural, environmental and occupational, and metabolic risks or clusters of risks for 195 countries and territories, 1990–2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet. 2018;392(10159):1923–1994. doi: 10.1016/S0140-6736(18)32225-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teo S.M., Mok D., Pham K., Kusel M., Serralha M., Troy N.…Inouye M. The infant nasopharyngeal microbiome impacts severity of lower respiratory infection and risk of asthma development. Cell Host Microbe. 2015;17(5):704–715. doi: 10.1016/j.chom.2015.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troeger C., Forouzanfar M., Rao P.C., Khalil I., Brown A., Swartz S.…Mokdad A.H. Estimates of the global, regional, and national morbidity, mortality, and aetiologies of lower respiratory tract infections in 195 countries: a systematic analysis for the Global Burden of Disease Study 2015. Lancet Infect. Dis. 2017;17(11):1133–1161. doi: 10.1016/S1473-3099(17)30396-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UNICEF Estimates of child cause of death, acute respiratory infection 2018. 2018, February. https://data.unicef.org/topic/child-health/pneumonia/ Retrieved from.
- Walker C.L.F., Rudan I., Liu L., Nair H., Theodoratou E., Bhutta Z.A.…Black R.E. Global burden of childhood pneumonia and diarrhoea. Lancet. 2013;381(9875):1405–1416. doi: 10.1016/S0140-6736(13)60222-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H., Kobzik L. Effect of concentrated ambient particles on macrophage phagocytosis and killing of Streptococcus pneumoniae. Am. J. Respir. Cell Mol. Biol. 2007;36(4):460–465. doi: 10.1165/rcmb.2006-0293OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material