Abstract
Determining the genetic rearrangement and domestication footprints in Gossypium hirsutum cultivars and primitive race genotypes are essential for effective gene conservation efforts and the development of advanced breeding molecular markers for marker-assisted breeding. In this study, 94 accessions representing the 7 primitive races of G hirsutum, along with 9 G hirsutum and 12 Gossypium barbadense cultivated accessions were evaluated. The genotyping-by-sequencing (GBS) approach was employed and 146 558 single nucleotide polymorphisms (SNP) were generated. Distinct SNP signatures were identified through the combination of selection scans and association analyses. Phylogenetic analyses were also conducted, and we concluded that the Latifolium, Richmondi, and Marie-Galante race accessions were more genetically related to the G hirsutum cultivars and tend to cluster together. Fifty-four outlier SNP loci were identified by selection-scan analysis, and 3 SNPs were located in genes related to the processes of plant responding to stress conditions and confirmed through further genome-wide signals of marker-phenotype association analysis, which indicate a clear selection signature for such trait. These results identified useful candidate gene locus for cotton breeding programs.
Keywords: Gossypium hirsutum, genotyping-by-sequencing, single nucleotide polymorphisms, DNA markers
Introduction
Cotton is the most important fiber crop in the world and comprises 52 Gossypium species including 7 allotetraploid species (amphidiploids [AD] genome [2n = 52]) and 45 diploid species (2n = 26).1,2 The allopolyploid species, Gossypium hirsutum L (AD1 genome) and Gossypium barbadense (AD2 genome), have been domesticated and G hirsutum cultivars alone account for more than 90% of global cotton fiber production.3-5 However, the high genetic similarity among G hirsutum accessions has hindered opportunities for breeding new cotton cultivars with improved agricultural traits such as higher yield, ease of harvest, and stronger resistance to pest, diseases, and environmental stresses.6-10
Seven genetically related accessions of G hirsutum, including Latifolium, Palmeri, Marie-Galante, Richmondi, Yucatanense, Morrilli, and Punctatum, have been identified based on their locations of origin.11 These races have distinct characteristics that are common to wild cotton but not to cultivated G hirsutum, such as sensitivity to a short-term light cycle, greater disease resistance and drought tolerance, hard seed coats, and variable seed size, and are genetically compatible with domesticated cottons. All of these advantageous traits can be potentially applied to improve cotton yield and quality as well as tolerance to environmental stresses.8,12-14
In the past, the classification of G hirsutum has been primarily based on morphology, geographical distribution, and cytological markers.15,16 The classification of G hirsutum has now advanced significantly as a multitude of molecular markers have been identified,17,18 but simulation and empirical studies have shown that simple sequence repeat (SSR) markers are likely to result in a significant downward bias for FST estimation due to the mutational characteristics of highly polymorphic microsatellites.19-21 GBS (genotyping-by-sequencing) is a practical and low-cost single nucleotide polymorphism (SNP) marker identification platform which can be utilized for genetic variation screening, genome-wide association analyses, and genetic recombination studies. Application of a large number of genome-wide markers for genotyping across multiple populations enables the establishment of the adaptations that have taken place during evolution and the detection of novel trend during natural selection.22-25
The primary achievement of this study is the establishment of a fine scale genome-wide map of the distributions of SNPs and the determination of the phylogenetic relationships of 115 cotton genotypes including 94 G hirsutum primitive race accessions and 21 domesticated cotton cultivars. Selection-scan analyses and genome-wide association study (GWAS) signals were conducted, based on the phenotype association analysis, to correlate the linkage between the molecular markers for early seedling development with the evolutional and domestication of G hirsutum. The results obtained from this study will facilitate future investigations on the genetic structure of G hirsutum races and expand the marker resources available for breeding programs.
Materials and Methods
Plant materials and phenotypic evaluations
The study evaluated 115 cotton genotypes, including 94 accessions representing 7 G hirsutum primitive obtained from Wild Cotton Nursery located in Sanya City, Hainan Island, and supervised by the Institute of Cotton Research (28 Latifolium, 16 Marie-Galante, 14 Morrilli, 19 Punctatum, 8 Richmondi, 7 Palmeri, and 2 Yucatanense accessions; Supplemental Table S12), 9 G hirsutum cultivars were included to represent modern domesticated upland cotton genotypes, and 12 of the G barbadense cultivars as an outgroup.
The 115 genotypes were planted in the field at the Institute of Cotton Research of Chinese Academy of Agricultural Scientists in Sanya, in 5 m plots with 3 replications. All samples were planted on April 23, 2016, and the field was managed according to traditional production practices. During the harvest season, the number of fruiting branches and the number of bolls per plant were measured on 10 plants and then averaged. The single boll weight (average weight of 30 bolls), lint (lint weight obtained from the 30 bolls/weight of 30 bolls (g) × 100), and seed index (weight of 1000 seeds) were measured. Fiber quality (fiber length, uniformity index, strength, micronaire value, and elongation) were measured by HVI instrument. For the germination test, 30 de-linted seeds for each accession that had been stored for 3 months were placed in the germination box with 3 replicates, then cultured in a germination chamber at 28°C, and 10-hour daylight. The germination rate is the percentage of seeds that germinate at 8 days, the germination potential is the number of germinated seeds/30 seeds; the seedling weight, embryonic axis length, and root length were measured on 5 seedlings (Supplemental Table S1).
To determine phenotypic differences between G hirsutum races and G hirsutum cultivars and their association with genetic structure, phenotypic data comprising 15 morphological traits were subjected to principal components analysis (PCA) and agglomerative hierarchical cluster (AHC) analysis. In addition, differences between cultivars and G hirsutum races were determined by subjecting all morphological traits to analyses of random variance followed by the Tukey honest significant difference post hoc test at a significance level of P < .05. All calculations were performed with XLSTAT version 2013.
DNA extraction, GBS library preparation, sequencing, and data analysis
Young leaf tissues from a single plant for each genotype before flowering were collected, and DNA was extracted using a Qiagen DNeasy Plant Mini Kit following the manufacturer’s instructions. The concentration of DNA was determined by fluorimetry (Life Invitrogen Qubit 3.0, Qubit 3.0 Fluorometer; Thermo Fisher Scientific, Waltham, MA, USA) and confirmed by gel electrophoresis on a 1% (w/v) agarose gel. Genomic DNA at a concentration of at least 100 ng/µL was used to prepare the libraries for each genotype.
The library construction for GBS was conducted according to a previous report.26 In brief, genomic DNA was digested with the restriction enzyme ApeKI,27,28 followed by ligation with a barcode adaptor and a standard Illumina sequencing adaptor. DNA fragments were pooled for polymerase chain reaction (PCR) amplification. Finally, 100 bp fragments were single end-sequenced on an Illumina HiSeq 2000 platform.
The high-quality FASTQ read sequences generated for each accession were aligned to the reference TM-1 cotton genome.4 We applied Samtools29 to produce BAM files for removing unmapped reads based on the mapping outputs. Vcflib packages (https://github.com/vcflib/vcflib.git) were then used to filter SNPs with a mapping quality score <30.
Population structure analysis
Population structures were determined in 2 steps. First, we applied principal coordinate analysis (PcoA) to investigate genetic relationships using a dissimilarity matrix obtained by DARwin 6.0 (http://darwin.cirad.fr). The PcoA results were plotted using the ggplot2 package in R studio. We also applied the discriminant analysis of the principal components (DAPC) using adegenet package,30 which can determine relationships for redefined groups without requiring an a priori population genetics model.31 In brief, the data were first transformed using PCA, and then the number of genetic clusters was assessed using the find clusters function. The Bayesian information criterion (BIC) was calculated for K = 1 to 10. For K-means clustering, all of the principal components were retained, and the K value with the lowest BIC was selected as the optimal number of clusters. DAPC was implemented using the optimized number of principal components as determined by the optim.a.score function. We further used the fast STRUCTURE tool to determine the most probable number of genetic clusters, which was run at K = 1 and K = 10 with default parameters.32 Finally, we conducted the TreeMix (http://treemix.googlecode.com) to estimate population differentiation among all G hirsutum races and G hirsutum cultivar group by constructing the Maximum likelihood tree with m value (0-6) and block 1000, setting the G barbadense as the outgroup
Evidence of selection footprints in G hirsutum races
Population structure analysis prompted us to perform population genomic FST scans between G hirsutum cultivars and G hirsutum race groups to identify SNP-specific high FST outliers using both BAYESCAN version 2.133 and Arlequin v3.5.34 For BAYESCAN, the “snp” option was applied using SNP genotype matrix as input data. The analyses were run using default settings, including 20 pilot runs of 5000 steps each, followed by 50 000 burn-in and 5000 sampling steps with a thinning interval of 10. The prior odds parameters were set to the default of 10. The false discovery rate (FDR) was set to 0.1 with the PLOT_BAYESCAN R function for outlier detection. For Arlequin, 50 000 simulations were run on the same data set with default parameters, using both the “neutral mean FST” and “force mean FST” options. Loci outside the 95% confidence interval and those with FST = 1 were considered outliers. For Arlequin, 20 000 simulations were run with 10 simulated groups and 100 demes per group to identify candidate loci under selection. High FST outlier SNPs were considered candidates for evidence of positive selection under population divergence. We identified all genes containing outlier SNPs (outlier genes) based on the TM-1 reference genome annotation and analyzed their functions based on known functions of Arabidopsis thaliana orthologous genes.
The phenotype analysis (Table 1) indicated significant differences with respect to seed germination and lint traits between G hirsutum cultivars and most of the G hirsutum race accessions tested. Therefore, we further tested for genome-wide signals of marker-phenotype associations using these differing phenotypes (seed germination and lint) to determine the selective footprint during the evolution of G hirsutum races to hirsutum cultivars. A mixed linear model (MLM) was used to analyze marker-trait associations with TASSEL 5.0,35 which first discards heterozygous sites and then generates different sets of marker data. SNP data were further filtered using filter data with 0.05 site minor allele frequency (MAF) thresholds before association mapping. To correct the population structures, kinship analysis and PCA were carried out to obtain K and Q matrices, respectively, which were then used as a covariance matrix and integrated into the MLM. Correction for multiple tests was performed based on an FDR of 0.05 to identify significantly associated markers.36,37 The sequences of significant markers within genes were then used as queries for BLAST searches in the National Center of Biotechnology Information gene database based on the TM-1 genome sequence. Known genes linked to the significant loci were assigned as putative candidates based on the functions of A thaliana orthologous genes.
Table 1.
Phenotypic variations between Gossypium hirsutum races and cultivar populations.
| Trait | Sum of squares | Mean squares | P value | G barbadense | Yucatanense | G hirsutum | Morrilli | Latifolium | Richmondi | Punctatum | Marie-Galante | Palmeri |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Germination potential (%) | 49.427 | 5.492 | <.0001 | 0.892A | 0.875AB | 0.8AB | 0.64AB | 0.64AB | 0.63AB | 0.56AB | 0.49AB | 0.29BC |
| Germination rate (%) | 60.050 | 6.672 | <.0001 | 0.90A | 0.93A | 0.89A | 0.71B | 0.72B | 0.68B | 0.63B | 0.62B | 0.35C |
| Seedling weight (g) | 22.421 | 2.490 | <.0001 | 0.63A | 0.42AB | 0.54AB | 0.38BC | 0.44AB | 0.42AB | 0.36BC | 0.41B | 0.21C |
| Embryonic axis length (cm) | 3858.821 | 428.758 | <.0001 | 7.80A | 6.38A | 7.86A | 4.52A | 5.52A | 5.05A | 5.52A | 5.02A | 3.45AB |
| Root length | 3067.371 | 340.819 | <.0001 | 7.38A | 5.10A | 6.68B | 4.08BC | 4.67BC | 4.75BC | 5.09B | 4.29BC | 3.73C |
| Fruiting branches (no.) | 19 193.402 | 2132.600 | <.0001 | 10.64A | 14.50A | 10.17A | 13.72A | 11.29A | 13.95A | 13.47A | 12.89A | 15.00A |
| Bolls per plant (no.) | 30 891.448 | 3432.383 | <.0001 | 16.61BC | 25.50A | 9.81D | 17.19B | 9.88D | 24.15A | 17.92B | 13.39CD | 27.36A |
| Single boll weight (g) | 2060.994 | 228.999 | <.0001 | 3.31B | 2.67BC | 5.37A | 3.08BC | 5.58A | 3.39B | 3.03BC | 3.85B | 1.76C |
| Lint (%) | 94 654.862 | 10 517.207 | <.0001 | 33.56B | 20.54BC | 43.07A | 23.43C | 30.89B | 23.41C | 22.88C | 26.88BC | 22.62C |
| Seed index (100 seeds-g) | 11 827.508 | 1314.168 | <.0001 | 12.25A | 8.02AB | 10.24AB | 10.09AB | 11.33A | 10.46AB | 8.30AB | 10.79A | 7.68B |
| Fiber length | 75 159.012 | 8351.001 | <.0001 | 31.03A | 22.95A | 27.80A | 24.54A | 24.21A | 24.69A | 23.55A | 25.88A | 22.07A |
| Uniformity index | 769 007.099 | 85 445.233 | <.0001 | 84.20A | 79.55A | 84.28A | 80.85A | 81.52A | 80.94A | 80.79A | 81.86A | 78.81A |
| Strength | 75 782.633 | 8420.293 | <.0001 | 34.40A | 23.25B | 24.22B | 24.01B | 22.89B | 24.14B | 23.50B | 24.94B | 25.80B |
| Micronaire value | 2056.559 | 228.507 | .58 | 4.34A | 4.00A | 4.17A | 4.03A | 4.55A | 4.40A | 4.21A | 3.95A | 3.03A |
| Elongation (%) | 4964.156 | 551.573 | .34 | 7.03A | 6.45A | 6.64A | 6.42A | 6.49A | 6.44A | 6.46A | 6.48A | 6.63A |
Results
Phenotypic characterizations of G hirsutum races and G hirsutum cultivars
The biodiversity analysis demonstrated significant differences in several traits including seedling weight, embryonic axis length, bolls per plant, and single boll weight between the 7 races groups and cultivars (Table 1; Supplemental Table S2). Cultivars had a higher germination rate and lint percentage compared with races groups. However, no significant differences were detected for fiber quality traits (fiber length, uniformity index, and micronaire value) and fruiting branch traits. Only the Palmeri race genotypes were significantly different for the seed index from cultivars.
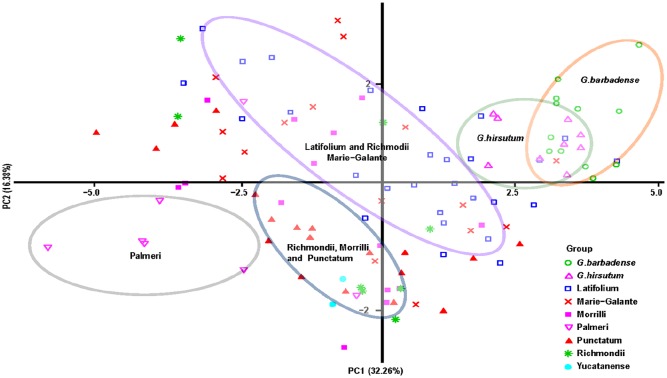
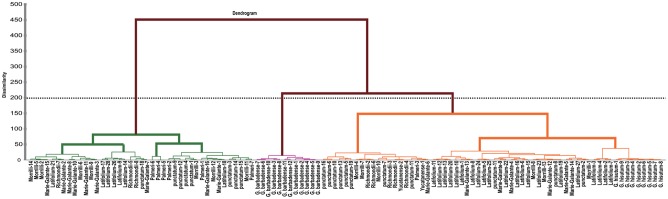
In the PCA, the first 2 components accounted for approximately 48.54% of the variation observed between G hirsutum races and G hirsutum cultivars (Figure 1; Supplemental Tables S3), and all accessions and cultivars could be simply clustered into 5 groups, which were the G barbadense cultivars group, G hirsutum cultivars group, Latifolium, Richmondi, and Marie-Galante group, Richmondi, Morrilliand, Punctatum, and Palmeri group along with the first component (32.26%). The second component (16.38%) separated Richmondi, Morrilliand, Punctatum, and Palmeri accessions group from the cultivar group. The PC analysis scatter plot suggests that most of these G hirsutum races occupy the most distant part of the ellipse including G hirsutum cultivars (Figure 1). The dendrogram of the AHC analysis further supported the PC analysis by classifying all samples into 3 major groups when the dissimilarity is at 200 (Figure 2), in which Punctatum, Latifolium, Morrilli, and Marie-Galante accounted for the more extended part of morphological variation along with G hirsutum. Furthermore, Latifolium, Marie-Galante, Morrilli, and Punctatum accessions were closer to G hirsutum cultivars group; meanwhile, these groups were clearly separated from the AD1 and AD2 samples that formed singular groups (Supplemental Table S4).
Figure 1.
Scatter plot distribution of the first and second principal components (PCs) based on 15 morphological characteristics of 94 Gossypium hirsutum races and 21 cotton cultivars. The different color circles represent different clusters. The orange circle mainly contained Gossypium barbadense cultivars, the green circle mainly contained G hirsutum cultivars, the purple circle mainly contained Latifolium and Marie-Galante, the blue circle mainly contained Richmondi, Morrilli, and punctatum, and the light brown circle mainly contained Palmeri.
Figure 2.
Agglomerative hierarchical clustering of 15 morphological characteristics of 94 Gossypium hirsutum races and 21 cultivated cotton accessions using the Ward’s agglomeration method and Euclidean distance dissimilarities; the horizontal dotted line represents the dissimilarity value is at 200; all cotton species can be divided into 3 groups.
Genome-wide detection of SNPs using GBS
A total of 877.41 million (98.4% of total reads) high-quality sequence reads (containing enzyme sites) were obtained. Sequence reads varied between 2.36 and 17.46 with a mean of 7.63 million reads per accession (Supplemental Figure S1, Supplemental Table S5). Approximately, 79.87% (75.7%-89.9%) of the sequenced nucleotides were evenly distributed across the 94 G hirsutum races and 21 cotton cultivars. The uniquely mapped sequence reads from the accessions or cultivars showed coverage from 8.27% (minimum) to 63.92% (maximum) of the G hirsutum acc.TM-1 reference genome (2189.14M) and the unique mapping ratios which ranged from 75.76% to 89.08% are presented in Supplemental Table S5.
A total of 146 558 SNPs was identified using Stacks tool (Supplemental Table S6). The MAF values varied from 0.005 to 0.5 with an average of 0.15 (Supplemental Table S6). According to the reference genome sequence, the detected SNPs were physically mapped to 26 chromosomes with an average density of 64 SNPs per Mb (Supplemental Table S6, Supplemental Figure S2). The leveraged SNP density is highest on chromosome 2 (117.25 Mb) with 8419 SNPs and lowest on chromosome 7 (23.27 Mb) with 1754 SNPs (Supplemental Figure S2). Tajima’s D, Theta, and Pi were calculated for the filtered SNPs with a mean of −0.22 (ranging from −0.97 to 0.28), 0.22 (ranging from −0.97 to 0.28), and 0.20 (ranging from 0.16 to 0.23), respectively (Supplemental Table S7). The transition/transversion ratio was calculated as 1.89. Most of the identified SNPs (62.9%) were transitions (A/G or T/C), whereas transversion events (A/C, A/T, C/G, or G/T) accounted for 37.1% of all SNPs. We also determined the physical locations of 146 558 SNPs based on the reference genome annotations, 35 499 SNPs were localized to 11 935 genes (Supplemental Table S8), and 111 316 (44.3%) SNPs were localized in the intergenic regions. Overall, 29 028 (11.6%) SNPs mapped to exons (coding sequences), 23 623 (9.4%) SNPs mapped to introns, and 42 261 (16.8%) mapped in the downstream regulatory regions (3′ untranslated region [UTR]). The SNPs detected in the upstream regulatory regions (promoter and 5′UTR) accounted for 14.4% (36 253) of all the SNPs (Supplemental Table S8).
Phylogenetic relationships of the cultivated and the wild relatives of G hirsutum
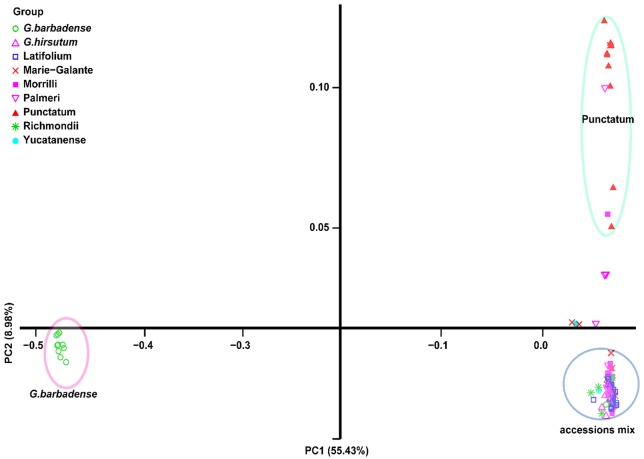
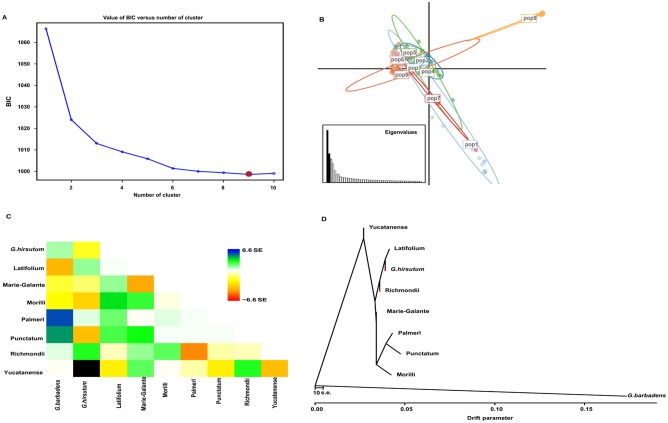
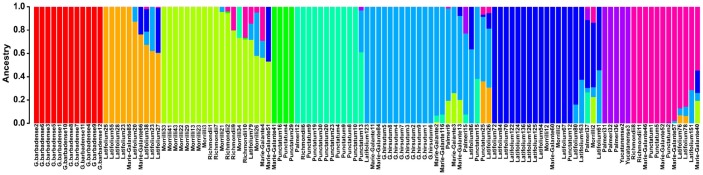
Initial assessment of the phylogenetic relationships was conducted using individual-based PC analysis with the identified 146 558 SNPs. PC1 analysis divided the selected cotton species into 2 groups associated with the AD1 and AD2 genomes (Figure 3). According to PC2 analysis, there were also 2 main groups: the first group mainly comprised Punctatum races and another group contains the cultivars and 6 G hirsutum races. However, the AD1 cultivars slightly overlapped with those 6 G hirsutum races except Punctatum (Figure 3; Supplemental Figure S3, Supplemental Table S9). Nine distinct clusters were identified as a result of the discriminant analysis of principal component (DAPC) analysis (Figure 4A). Visualization of DAPC results clearly clustered the first 50 principal components. AD2 samples were still in a separate cluster, whereas G hirsutum races were more diverse and could not be clearly separated, with some accessions (Latifolium, Richmondi and Marie-Galante) appearing to be more closely related to the AD1 cultivars (Figure 4B; Supplemental Table S10). The population splits and migration events from TreeMix analysis are shown in Figure 4C and D. In the model, the sampled populations in the selected cotton species were seemed to be related to their common ancestor through a graph of ancestral populations (Figure 4D). Using genome-wide allele frequency data and a Gaussian approximation to genetic drift, the historical distance among the population was as follows: Latifolium >Richmondi > Marie-Galante > Morrilli > Palmeri > Yucatanense > Punctatum (Figure 4D). The data also suggested that G hirsutum cultivars have the closest relationship with Latifolium, Richmondi, and Marie-Galante races and have the most distant relationship with Punctatum and Yucatanense races (Figure 4D; Supplemental Figure S3). A similar classification pattern was observed from the fast STRUCTURE analyses, 9 genetic clusters were visible, and AD1 cultivars showed a common genomic background with that of G hirsutum races (Figure 5; Supplemental Table S11).
Figure 3.
Principal components analysis of 115 cotton samples, including 94 Gossypium hirsutum race and 21 cultivar accessions based on 146 588 SNPs. The different color circles represent different clusters. The purple circle only contained Gossypium barbadense cultivars, and the green circle mainly contained Punctatum.
Figure 4.
Discriminant analyses of principal components (DAPC). (A) The optimal number of clusters (K) as determined by “K-means” clustering. The graph shows an apparent decrease of the Bayesian information criterion (BIC) until K = 9, red dot, which is the most likely value of K. After this value, BIC increases. (B) Scatter plot based on the DAPC output for 4 assigned genetic clusters indicated by different colors. Dots represent different individuals. pop1 represents group 1 containing almost all races accessions; pop2 represents group 2 containing mainly Latifolium accessions; pop3 represents group 3 containing mainly Punctatum accessions; pop4 represents group 4 containing mainly Punctatum and Marie-Galante accessions; pop5 represents group 5 containing mainly Latifolium, Richmondi, and all G hirsutum cultivars; pop 6 represents group 6 containing 2 Marie-Galante, 2 Palmeri accessions, Morrilli, and Marie-Galante accessions; pop 7 represents group 7 containing 2 Yucatanense accessions; pop 8 represents group 8 containing Richmondi, Morrilli, and Marie-Galante accessions; pop9 represents group 9 which only contained G barbadense cumainly cultivars (Table S10). (C, D) Plotted is the structure of the graph inferred by TreeMix for cotton populations, allowing 10 migration events. The scale bar shows 10 times the average standard error of the entries in the sample covariance matrix.
Figure 5.
Phylogenetic structures of 115 cotton species estimated using the fast structure admixture model at K = 9.
Footprints of positive selection during G hirsutum domestication
In FST outlier scans, Arlequin yielded a significant number of high outlier SNPs than did BAYESCAN (Table 2). Cultivars of G hirsutum and Latifolium race pair produced fewer outliers than other pairs, whereas G hirsutum cultivars and Punctatum race pair had the highest number of outliers. In general, 54 outlier SNPs were located in the coding sequence, consistent with the proportion (~88%) among all outlier SNPs tested (Table 2), with 2 of the same SNPs being located in each of the genes LOC107896563, LOC107963906, LOC107913656, and LOC107927053. Thus, a total of 50 genes with outlier SNPs (designated as outlier genes) were considered as possible footprints of positive selection during G hirsutum domestication. This conclusion was reached without the need to analyze the outliers from each group of G hirsutum races separately. Gene Ontology terms (biological process) and the established functions of A thaliana orthologous genes are presented in Supplemental Table S12 for the outlier SNPs. Several outlier genes are predicted to be associated with pollen germination and tube growth and 3 genes (LOC107896563, LOC107927053, and LOC107913656) with the regulation of flower development. Other processes shared by more than 1 pair of comparisons include hormone pathways, biotic and abiotic stress responses. Intriguingly, several candidate genes were closely clustered in a specific region on the chromosome. This suggests that some outlier SNPs reside in areas that may have undergone sweeps of selection, which would make it more difficult to identify the specific genes targeted by selection.
Table 2.
Summary of high FST SNP outliers from BAYESCAN and Arlequin analyses using 16 288 SNPs.
| Comparisons | Arlequin FST | BAYESCAN FST | No. of outliers detected by BAYESCAN | No. of outliers detected by Arlequin | Overlap outliers | Outlier SNPs contained in gene NO |
|---|---|---|---|---|---|---|
| G hirsutum cultivars vs Latifolium | 0.1054 | 0.1901 | 7 | 1571 | 3 | 3 |
| G hirsutum cultivars vs Marie-Galante | 0.0737 | 0.1327 | 17 | 1287 | 6 | 6 |
| G hirsutum cultivars vs Palmeri | 0.0798 | 0.1216 | 86 | 1626 | 37 | 31 |
| G hirsutum cultivars vs Punctatum | 0.1659 | 0.2183 | 23 | 1109 | 14 | 13 |
Abbreviations: SNP, single nucleotide polymorphism; G hirsutum, Gossypium hirsutum.
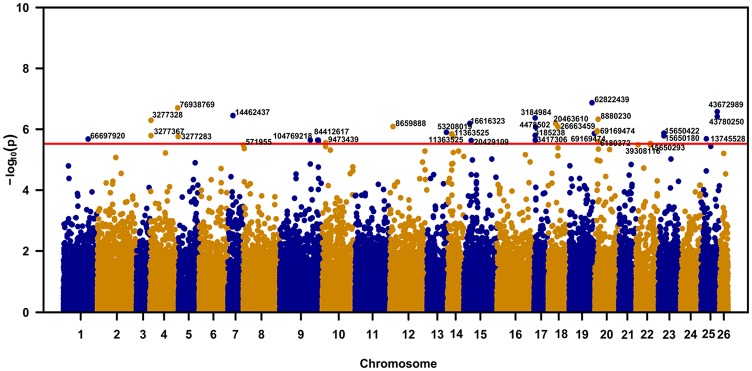
Footprints of positive selection were investigated based on the genome-wide-association phenotype of early seedling development (stress responses) using 92 G hirsutum races, which contained 98 436 SNPs. Although no significant signal could be detected throughout the whole genome or on a specific chromosome, this trait did correlate with an SNPs-per-chromosome with the largest −log10(3e−06) value (14 SNP-containing genes) being found on G hirsutum cultivars (Figure 6; Supplemental Table S13). Three of these genes (LOC107914109, LOC107922201, and LOC107921406 are predicted to be involved in the biological processes including cellular response to water deprivation, defense responses to bacterial attack, reductive pentose-phosphate cycle, response to cold, and response to nematode infection (Table 3).
Figure 6.
Genome-wide association study of early seedling development rate-related traits based on the genotyping-by-sequencing single nucleotide polymorphisms (SNPs), an SNPs-per-chromosome with the more than −log10(3e−06) value was selected to be involved in stress resistance–related traits. The red line means the cut off standard with −log10(3e−06); the number in X-axis represents chromosome number of Gossypium hirsutum.
Table 3.
Selective footprint of stress resistance–related traits between 94 Gossypium hirsutum races and 21 cultivated cotton populations.
| Marker | Chromosome | Position | Gene | Arabidopsis orthologous gene | Description in gene function |
|---|---|---|---|---|---|
| 155370 | NC_030086 | 53208019 | LOC107914109 | AT1G33240 | Cellular response to water deprivation, negative regulation of DNA endoreduplication, negative regulation of cell growth, negative regulation of transcription, DNA-templated, regulation of cell size, regulation of stomatal complex development, regulation of stomatal complex patterning, response to water deprivation, transcription, DNA-templated, trichome morphogenesis |
| 190679 | NC_030090 | 10079330 | LOC107922201 | AT1G32060 | Defense response to bacterium, phosphorylation, reductive pentose-phosphate cycle, response to cold |
| 218950 | NC_030093 | 9757870 | LOC107921406 | AT1G58360 | l-alanine transport, l-glutamate import across plasma membrane, amino acid import, amino acid transmembrane transport, neutral amino acid transport, response to nematode |
Discussion
The GBS assay is a robust, simple, and affordable tool for SNP discovery and genome mapping. By using appropriate restriction enzymes and PCR amplifications, it can sufficiently reduce genome complexity, avoid repetitive regions of genomes, and target lower copy regions. In addition, it has the advantages of minimizing alignments problems in genetically highly diverse species and dealing with a large number of genome samples at a lower cost than other methods. Theoretically, the method can be used to map any plant species, as it does not require a reference genome. It has been frequently used to analyze large segregating progenies and marker trait association studies based on linkage disequilibrium and even the evolutionally genomic selection.38 When the method was originally developed, it was used to analyze a high-resolution maize mapping population and doubled haploid barley lines for GBS accuracy and efficiency. The results demonstrated that 25 185 biallelic tags could be mapped to the maize genome and 24 186 sequence tags to the barley genome and the GBS reads were in 99% agreement with the reference markers. As a consequence, it has become a practical and convenient approach for genotyping in crop plants,39-43 including the analysis of cotton for genetic diversity, genetic mapping and GWAS analysis.44-47
In the current study, a total of 146 558 high-quality SNPs were identified in G hirsutum races and cultivated cotton accessions, which is consistent with the genome-wide SNP discovery capability previously reported in other crop plants.48-52 Moreover, 35 499 SNPs were located in 11 935 cotton genes correlating with some important agricultural traits such as fertility, germination, and fiber quality; thus, they can serve as useful marker-based resource or molecular design tools for genomic selection, genome-wide association analyses, and genetic diversity of G hirsutum. The obtained SNPs also have the potential for breeding programs aimed at improving cotton quality and yield.
Our results confirm those of others who have proposed that G hirsutum and other races were derived from a Yucatanense progenitor in the Old World.11 Similarly, the Punctatum, Palmeri, Marie-Galante, and, especially, Latifolium races are considered to be progenitors of the modern G hirsutum cultivars in the New World.53-56 Our genotyping results are also consistent with the classification based on phenotypic similarities, that Latifolium, Punctatum, and Marie-Galante should be included in the G hirsutum cultivars group.57 Overall, the results are consistent with the established history of cotton domestication and support the suggestion that G hirsutum cultivars originated from 2 distinct places in the Old World and New World from our TreeMix analysis.4,54,58
Genome scans and FST outlier SNP discovery approaches are effective strategies for identifying genes under evolutionary or domestication selection, and this approach is significantly different from phenotype-based GWAS analysis.59 Extreme differentiation in allele frequencies between genetic groups or populations in different geographic zones, as measured by the FST statistic, provides a signature of recent positive selection,60 and different populations of ancestral accessions/species can further serve as a useful biological model to study adaptive or directional selection in nature.61 Our study indicates that, during cotton evolution and domestication, a large portion of the outlier genes are involved in reproductive processes such as the regulation of pollen germination and tube growth, the regulation of flower development, hormone signaling processes, biotic and abiotic stress responses. Outlier genes were closely clustered in a specific region on the chromosome suggesting that the whole chromosome fragment, and not just specific genes, has undergone sweeps of selection. As a result, it is difficult to isolate or characterize the genes contributing to specific traits from this region. This finding revealed direct genetic evidence for a positive selection from G hirsutum races to G hirsutum cultivars. Indeed, these genes appear to be involved in the regulation of range of important agricultural traits; therefore, they may be able to serve as candidate molecular markers for G hirsutum cultivars breeding programs using G .hirsutum races.
The most strongly differentiated trait between G hirsutum races and G hirsutum cultivars was their resilience during the domestication. G hirsutum races underwent some phenotypic adaptations including exhibiting more sensitivity to photoperiod changes; loss of perennial growth habits; reduced seed dormancy; and greater resistance to various stress conditions.62,63 Three SNPs were found in genes corresponding to potential Arabidopsis orthologous involving in responses to stress. These results also suggest that genes with the functions contributing to abiotic and biotic stress responses are conserved in the G hirsutum cultivars as a consequence of artificial selection for improved environmental adaptations.
In our study, the outlier SNPs, as indicators of positive selection, did not associate closely with genome-wide marker-phenotype association signals. This may be due to the possibility that the GBS mapping approach may miss some sequences due to the requirement for genome cleavage by specific restriction enzymes. Alternatively, the outlier genes were detected using 146 588 SNPs, whereas the genome-wide marker association analysis was conducted using only 98 436 SNPs, and therefore, it might not reflect genomic coordination between the selection loci and SNP markers of the genome-wide association. Another explanation is that SNPs may not be able to adequately represent the major signature of selection on coding variants as comprehensively as gene expression analyses. Further studies using transcriptomics or higher SNP density or larger number of SNPs could help to improve the resolution of differentially selected loci and increase the concordance between the SNPs identified by the selection footprint analysis, transcription analysis, and genome-wide phenotype association studies.
Conclusions
The current study adopted a GBS approach to analyze 94 G hirsutum races accessions and 21 cotton domesticated cotton cultivars. We concluded that the Latifolium, Richmondi, and Marie-Galante accessions were more genetically related to the selectively domesticated G hirsutum cultivars, and 54 outlier SNPs were identified and 3 SNPs located in genes related to plant responding to stress were isolated based on their orthologous genes function. These findings provide a preliminary indication of adaptation and selection footprints during cotton domestication and offer candidate DNA markers that could be used for cotton breeding programs.
Supplemental Material
Supplemental material, Supplemental_File_Tables0908_xyz27463d4de7434 for Genotyping-by-Sequencing of Gossypium hirsutum Races and Cultivars Uncovers Novel Patterns of Genetic Relationships and Domestication Footprints by Shulin Zhang, Yaling Cai, Jinggong Guo, Kun Li, Renhai Peng, Fang Liu, Jeremy A Roberts, Yuchen Miao and Xuebin Zhang in Evolutionary Bioinformatics
Footnotes
Funding:The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by the National Natural Science Foundation of China (31671745), Innovation Scientists and Technicians Troop Construction Projects of Henan Province (184200510009), and The National Key Research and Development Program of China (2016YFD0100203-1-4, 2018YFD0100300).
Declaration of conflicting interests:The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Author Contributions: Y.M., X.Z., and R.P. contributed to the research design; S.Z. and J.G. conducted experiments and investigated the study; and S.Z. and X.Z. wrote the manuscript.
ORCID iD: Xuebin Zhang  https://orcid.org/0000-0002-6089-4339
https://orcid.org/0000-0002-6089-4339
Data Availability: All the genotyping-by-sequencing (GBS) data during the current study are available in the NCBI Sequence Read Archive (SRA) under project accession number PRJNA498359. The authors state that all data necessary for confirming the conclusions stated are represented fully within the article and in Supplemental Materials.
Supplemental material: Supplemental material for this article is available online.
References
- 1. Khadi BM, Santhy V, Yadav MS. Cotton: an introduction. In: Zehr UB, ed. Cotton: Biotechnological Advances. Berlin, Germany; Heidelberg, Germany: Springer; 2010:1-14. [Google Scholar]
- 2. Chen ZW, Cao JF, Zhang XF, et al. Cotton genome: challenge into the polyploidy. Sci Bull. 2017;62:1622-1623. [DOI] [PubMed] [Google Scholar]
- 3. Li F, Fan G, Wang K, et al. Genome sequence of the cultivated cotton Gossypium arboreum. Nat Genet. 2014;46:567-572. [DOI] [PubMed] [Google Scholar]
- 4. Li F, Fan G, Lu C, et al. Genome sequence of cultivated Upland cotton (Gossypium hirsutum TM-1) provides insights into genome evolution. Nat Biotechnol. 2015;33:524-530. [DOI] [PubMed] [Google Scholar]
- 5. Wendel JF, Grover CE. Taxonomy and evolution of the cotton genus. In: Fang DD, Percy RG, eds. Gossypium (Cotton Agronomy Monograph 57). 2nd ed Madison, WI: American Society of Agronomy, Crop Science Society of America and Soil Science Society of America; 2015;57:25-44. [Google Scholar]
- 6. Zhang JF, Lu Y, Cantrell RG, Hughs E. Molecular marker diversity and field performance in commercial cotton cultivars evaluated in the southwestern USA. Crop Sci. 2005;45:1483-1490. [Google Scholar]
- 7. Bertini C, Schuster I, Sediyama T, Barros EGD, Moreira MA. Characterization and genetic diversity analysis of cotton cultivars using microsatellites. Genet Mol Biol. 2006;29:321-329. [Google Scholar]
- 8. Abdurakhmonov IY, Kohel RJ, Yu JZ, et al. Molecular diversity and association mapping of fiber quality traits in exotic G. hirsutum L. germplasm. Genomics. 2008;92:478-487. [DOI] [PubMed] [Google Scholar]
- 9. Sethi K, Siwach P, Verma SK. Assessing genetic diversity among six populations of Gossypium arboreum L. using microsatellites markers. Physiol Mol Biol Plants. 2015;21:531-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Okubazghi KW, Li XN, Cai XY, et al. Genome-wide assessment of genetic diversity and fiber quality traits characterization in Gossypium hirsutum races. J Integr Agric. 2017;16:2402-2412. [Google Scholar]
- 11. Bibi AC, Oosterhuis DM, Gonias ED, Stewart JM. Comparison of responses of a ruderal Gossypium hirsutum L. with commercial cotton genotypes to high temperature stress. Am J Plant Sci Biotechnol. 2010;4:87-92. [Google Scholar]
- 12. Knutson A, Isaacs S, Campos C, Campos M, Smith CW. Resistance to cotton fleahopper feeding in primitive and converted race stocks of cotton, Gossypium hirsutum. J Cotton Sci. 2014;18:385-392. [Google Scholar]
- 13. Wu T, Weaver DB, Locy RD, McElroy S, van Santen E. Identification of vegetative heat-tolerant upland cotton (Gossypium hirsutum L.) germplasm utilizing chlorophyll fluorescence measurement during heat stress. Plant Breed. 2014;133:250-255. [Google Scholar]
- 14. Wendel JF, Brubaker CL, Percival AE. Genetic diversity in Gossypium hirsutum and the origin of upland cotton. Am J Bot. 1992;79:1291-1310. [Google Scholar]
- 15. Hanson RE, Zwick MS, Choi SD, et al. Fluorescent in-situ hybridization of a bacterial artificial chromosome. Genome. 1995;38:646-651. [DOI] [PubMed] [Google Scholar]
- 16. Wu YX, Daud MK, Chen L, Zhu SJ. Phylogenetic diversity and relationship among Gossypium germplasm using SSRs markers. Plant Syst Evol. 2007;268:199-208. [Google Scholar]
- 17. Jena SN, Srivastava A, Rai KM, et al. Development and characterization of genomic and expressed SSRs for levant cotton (Gossypium herbaceum L.). Theor Appl Genet. 2012;124:565-576. [DOI] [PubMed] [Google Scholar]
- 18. Wang S, Chen J, Zhang W, et al. Sequence-based ultra-dense genetic and physical maps reveal structural variations of allopolyploid cotton genomes. Genome Biol. 2015;16:108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kalinowski ST. Evolutionary and statistical properties of three genetic distances. Mol Ecol. 2002;11:1263-1273. [DOI] [PubMed] [Google Scholar]
- 20. O’Reilly PT, Canino MF, Bailey KM, Bentzen P. Inverse relationship between F-ST and microsatellite polymorphism in the marine fish, walleye pollock (Theragra chalcogramma): implications for resolving weak population structure. Mol Ecol. 2004;13:1799-1814. [DOI] [PubMed] [Google Scholar]
- 21. Zhang T, Hu Y, Jiang W, et al. Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nat Biotechnol. 2015;33:531-537. [DOI] [PubMed] [Google Scholar]
- 22. Yang H, Wei CL, Liu HW, et al. Genetic divergence between Camellia sinensis and its wild relatives revealed via genome-wide SNPs from RAD sequencing. PLoS ONE. 2016;11:e0151424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wei D, Cui Y, He Y, et al. A genome-wide survey with different rapeseed ecotypes uncovers footprints of domestication and breeding. J Exp Bot. 2017;68:4791-4801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Byrne RP, Martiniano R, Cassidy LM, et al. Insular Celtic population structure and genomic footprints of migration. PLoS Genet. 2018;14:e1007152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Liu H, Bayer M, Druka A, et al. An evaluation of genotyping by sequencing (GBS) to map the Breviaristatum-e (ari-e) locus in cultivated barley. BMC Genomics. 2014;15:104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Deschamps S, Llaca V, May GD. Genotyping-by-sequencing in plants. Biology. 2012;1:460-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Heim CB, Gillman JD. Genotyping-by-sequencing-based investigation of the genetic architecture responsible for a similar to sevenfold increase in soybean seed stearic acid. G3. 2017;7:299-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Li H, Handsaker B, Wysoker A, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078-2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jombart T. adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics. 2008;24:1403-1405. [DOI] [PubMed] [Google Scholar]
- 30. Jombart T, Devillard S, Balloux F. Discriminant analysis of principal components: a new method for the analysis of genetically structured populations. BMC Genet. 2010;11:94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Raj A, Stephens M, Pritchard JK. fastSTRUCTURE: variational inference of population structure in large SNP data sets. Genetics. 2014;197:573-589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Foll M, Gaggiotti O. A genome-scan method to identify selected loci appropriate for both dominant and codominant markers: a Bayesian perspective. Genetics. 2008;180:977-993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Excoffier L, Lischer HEL. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour. 2010;10:564-567. [DOI] [PubMed] [Google Scholar]
- 34. Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES. TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics. 2007;23:2633-2635. [DOI] [PubMed] [Google Scholar]
- 35. Benjamini Y, Hochberg Y. Controlling the false discovery rate a practical and powerful approach to multiple testing. J R Statist Soc B Methodol. 1995;57:289-300. [Google Scholar]
- 36. Yu LX, Zheng P, Bhamidimarri S, Liu XP, Main D. The impact of genotyping-by-sequencing pipelines on SNP discovery and identification of markers associated with verticillium wilt resistance in autotetraploid alfalfa (Medicago sativa L.). Front Plant Sci. 2017;8:89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Li H, Li K, Guo Y, et al. A transient transformation system for gene characterization in upland cotton (Gossypium hirsutum). Plant Methods. 2018;14:50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ahmed D, Comte A, Curk F, et al. Genotyping by sequencing can reveal the complex mosaic genomes in gene pools resulting from reticulate evolution: a case study in diploid and polyploid citrus. Ann Bot. 2019;123:1231-1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tardivel A, Sonah H, Belzile F, O’Donoughue LS. Rapid identification of alleles at the soybean maturity gene E3 using genotyping by sequencing and a haplotype-based approach. Plant Genome. 2014;7:1-9. [Google Scholar]
- 40. Sonah H, O’Donoughue L, Cober E, Rajcan I, Belzile F. Identification of loci governing eight agronomic traits using a GBS-GWAS approach and validation by QTL mapping in soya bean. Plant Biotechnol J. 2015;13:211-221. [DOI] [PubMed] [Google Scholar]
- 41. Kim C, Guo H, Kong W, Chandnani R, Shuang LS, Paterson AH. Application of genotyping by sequencing technology to a variety of crop breeding programs. Plant Sci. 2016;242:14-22. [DOI] [PubMed] [Google Scholar]
- 42. Diouf L, Pan Z, He SP, et al. High-density linkage map construction and mapping of salt-tolerant QTLs at seedling stage in upland cotton using genotyping by sequencing (GBS). Int J Mol Sci. 2017;18:E2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Agarwal G, Jhanwar S, Priya P, et al. Comparative analysis of kabuli chickpea transcriptome with desi and wild chickpea provides a rich resource for development of functional markers. PLoS ONE. 2012;7:e52443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Islam MS, Thyssen GN, Jenkins JN, Fang DD. Detection validation, and application of genotyping-by-sequencing based single nucleotide polymorphisms in upland cotton. Plant Genome. 2015;8:1-10. [DOI] [PubMed] [Google Scholar]
- 45. Li RJ, Erpelding JE, Stetina SR. Genome-wide association study of Gossypium arboreum resistance to reniform nematode. BMC Genet. 2018;19:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Liu GY, Pei WF, Li D, et al. A targeted QTL analysis for fiber length using a genetic population between two introgressed backcrossed inbred lines in upland cotton (Gossypium hirsutum). Crop J. 2019;7:273-282. [Google Scholar]
- 47. Qi HK, Wang N, Qiao WQ, et al. Construction of a high-density genetic map using genotyping by sequencing (GBS) for quantitative trait loci (QTL) analysis of three plant morphological traits in upland cotton (Gossypium hirsutum L.). Euphytica. 2017;213:17. [Google Scholar]
- 48. Parida SK, Mukerji M, Singh AK, Singh NK, Mohapatra T. SNPs in stress-responsive rice genes: validation, genotyping, functional relevance and population structure. BMC Genomics. 2012;13:426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Subbaiyan GK, Waters DL, Katiyar SK, Sadananda AR, Vaddadi S, Henry RJ. Genome-wide DNA polymorphisms in elite indica rice inbreds discovered by whole-genome sequencing. Plant Biotechnol J. 2012;10:623-634. [DOI] [PubMed] [Google Scholar]
- 50. Jain M, Misra G, Patel RK, et al. A draft genome sequence of the pulse crop chickpea (Cicer arietinum L.). Plant J. 2013;74:715-729. [DOI] [PubMed] [Google Scholar]
- 51. Varshney RK, Gaur PM, Chamarthi SK, et al. Fast-track introgression of “QTL-hotspot” for root traits and other drought tolerance traits in JG 11, an elite and leading variety of chickpea. Plant Genome. 2013;6:1-9. [Google Scholar]
- 52. May OL, Bowman DT, Calhoun DS. Genetic diversity of US upland cotton cultivars released between 1980 and 1990. Crop Sci. 1995;35:1570-1574. [Google Scholar]
- 53. Tyagi P, Gore MA, Bowman DT, Campbell BT, Udall JA, Kuraparthy V. Genetic diversity and population structure in the US Upland cotton (Gossypium hirsutum L.). Theor Appl Genet. 2014;127:283-295. [DOI] [PubMed] [Google Scholar]
- 54. Lubbers EL, Chee PW. The worldwide gene pool of G. hirsutum and its improvement. In: Paterson AH, ed. Genetics and Genomics of Cotton. New York, NY: Springer; 2009:23-52. [Google Scholar]
- 55. Fang XX, Wu DP, Chen JH, Zhu SJ. Diversity and genetic relationship among the semi-cultivars of G. hirsutum L. races using SSR markers. Cotton Sci. 2011;23:99-105. [Google Scholar]
- 56. d’Eeckenbrugge GC, Lacape JM. Distribution and differentiation of wild, feral, and cultivated populations of perennial upland cotton (Gossypium hirsutum L.) in Mesoamerica and the Caribbean. PLoS ONE. 2014;9:e107458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Akey JM, Zhang G, Zhang K, Jin L, Shriver MD, Shriver MD. Shriver interrogating a high-density SNP map for signatures of natural selection. Genome Res. 2002;12:1805-1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Gao W, Liu F, Li S, et al. Genetic diversity of allotetraploid cotton based on SSR markers. Acta Agronomica Sinica. 2010;36:1902-1909. [Google Scholar]
- 59. Song Z, Zhang M, Li F, et al. Genome scans for divergent selection in natural populations of the widespread hardwood species Eucalyptus grandis (Myrtaceae) using microsatellites. Sci Rep. 2016;6:34941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Gautier M, Naves M. Footprints of selection in the ancestral admixture of a New World Creole cattle breed. Mol Ecol. 2011;20:3128-3143. [DOI] [PubMed] [Google Scholar]
- 61. Evans LM, Slavov GT, Rodgers-Melnick E, et al. Population genomics of Populus trichocarpa identifies signatures of selection and adaptive trait associations. Nature Genet. 2014;46:1089-1096. [DOI] [PubMed] [Google Scholar]
- 62. Stephens SG. Some observations on photoperiodism and development of annual forms of domesticated cottons. Econ Bot. 1976;30:409-418. [Google Scholar]
- 63. Ertiro BT, Ogugo V, Worku M, et al. Comparison of Kompetitive Allele Specific PCR (KASP) and genotyping by sequencing (GBS) for quality control analysis in maize. BMC Genomics. 2015;16:908. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, Supplemental_File_Tables0908_xyz27463d4de7434 for Genotyping-by-Sequencing of Gossypium hirsutum Races and Cultivars Uncovers Novel Patterns of Genetic Relationships and Domestication Footprints by Shulin Zhang, Yaling Cai, Jinggong Guo, Kun Li, Renhai Peng, Fang Liu, Jeremy A Roberts, Yuchen Miao and Xuebin Zhang in Evolutionary Bioinformatics