Abstract
Interstitial deletions involving 2q24 have been associated with a wide range of phenotypes including intellectual disability and short stature. To date, the smallest common region among reported cases of deletions in this region is approximately 2.65 Mb and contains 15 genes. In the present case report, we describe an 18-year-old male with mild intellectual disability, short stature, and mosaicism for a 0.422 Mb deletion on 2q24.2 that was diagnosed by comparative genomic hybridization and confirmed with fluorescent in situ hybridization (FISH). This deletion, which is present in approximately 61% of cells, includes three genes: TBR1, TANK, and PSMD14. The findings suggest that the critical region for intellectual disability and short stature in 2q24.2 can be narrowed to a 0.422 Mb segment. TBR1, a transcription factor involved in early cortical development, is a strong candidate for the intellectual disability phenotype seen in our patient and in patients with larger deletions in this region of the genome.
Keywords: 2q24, deletion, intellectual disability, mosaicism, comparative genomic hybridization, chromosome 2
INTRODUCTION
Interstitial deletions of 2q have been associated with a variety of phenotypes including intellectual disability and developmental delay [Pescucci et al., 2007; Magri et al., 2011]. The most commonly described deletions of 2q involve 2q24–31. However, smaller deletions within this region have also been described. A 5.3 Mb deletion at 2q24.2–24.3 involving 20 genes was described in a 3-year-old boy with intellectual disability, dysmorphic features, joint laxity, and hypotonia [Magri et al., 2011]. Another recent report described a patient with intellectual disability and generalized hypotonia with a 7.5 Mb deletion involving 2q24.1q24.2 [Palumbo et al., 2012]. Using other cases from the literature, the authors of the latter report suggested that the candidate region could be narrowed to a 2.65 Mb interval containing 15 genes. In the present report, we describe an 18-year-old male with intellectual disability and short stature who is mosaic for a 0.422 Mb deletion in 2q24.2 that encompasses only three genes: TANK, PSMD14, and TBR1.
CLINICAL REPORT
The patient was born full term via vaginal delivery requiring forceps. The pregnancy was uncomplicated, and he was discharged from the newborn nursery with his mother soon after delivery. The mother first noted developmental delay at 8–9 months of age, when he had persistent head lag. He sat with support at approximately 9 months and walked at 12 months. His first words were at 3 years of age, and he continued to have speech difficulties requiring therapy during his school-age years. Learning problems were noted at age 4 years, and he was eventually referred to developmental pediatrics for evaluation. He was followed for several years by developmental pediatrics with a diagnosis of mild intellectual disability. His most recent school evaluations were performed at the age of 14 years. This testing included the Woodcock–Johnson tests of achievement, which showed the following standard scores: 48 for oral language, 53 for oral expression, 55 for listening comprehension, 69 for story recall, 65 for understanding directions, 51 for picture vocabulary, and 56 for oral comprehension. In addition, Bateria III Woodcock–Munoz showed the following standard scores: 56 for oral language, 53 for oral expression, 69 for auditory comprehension, 40 for story recall, 71 for understanding directions, 59 for picture vocabulary, and 73 for oral comprehension. Informal testing (Gesell Milestones, Durrell Oral Reading Test, Ayres Handwriting Test) in the office at the age of 17 years revealed overall language development for concrete and inference at a 9-year-old level, overall visual perceptual/fine motor ability at a 10-year-old level, reading of 4th grade material in English at high 3rd grade speed level but with ability to correctly answer from memory only one of seven of concrete questions asked regarding the text and ability to perform math at the early 3rd grade level.
Because of his history of short stature and mild intellectual disability, array comparative genomic hybridization was performed and a sub-microscopic deletion was discovered. He was subsequently referred to the adult genetics clinic for further work-up and counseling.
At his initial genetics visit (age 18 years, 7 months), physical examination was notable for short stature. His height was 158.8 cm (0.74 centile, z-score = −2.44) and his weight was 52.2 kg (2.56 centile, z-score = −1.95). His adult height is significantly below the expected height, based on a mid-parental height of 167.2 cm. His head circumference was 53 cm (<3rd centile) [Bushby et al., 1992]. Aside from a high palate and 5th finger clinodactyly, no dysmorphic features were noted, and his physical examination was otherwise unremarkable. An MRI of the brain and skeletal survey were also normal.
MATERIALS AND METHODS
Array Comparative Genomic Hybridization
BCM V8 OLIGO is a custom-designed array with approximately 180,000 interrogating oligonucleotides, manufactured by Agilent Technologies, Inc. (Santa Clara, CA). Array coverage details are available at https://www.bcm.edu/geneticlabs/.
FISH Analysis
BAC clones for fluorescent in situ hybridization (FISH) analysis were selected using the UCSC genome browser (https://genome.ucsc.edu). TB media with 20 mcg/ml chloramphenicol was used to grow the BAC clone of interest. DNA was extracted from BAC clones (Eppendorf Plasmid Mini Prep kit, Hamburg, Germany) and directly labeled with SpectrumOrangeTM dUTP by nick-translation (Vysis, Downer Grove, IL). After hybridization, a Power Macintosh G3 System using MacProbe software version 4.4 (Applied Imaging, San Jose, CA) was used to capture the FISH images.
RESULTS
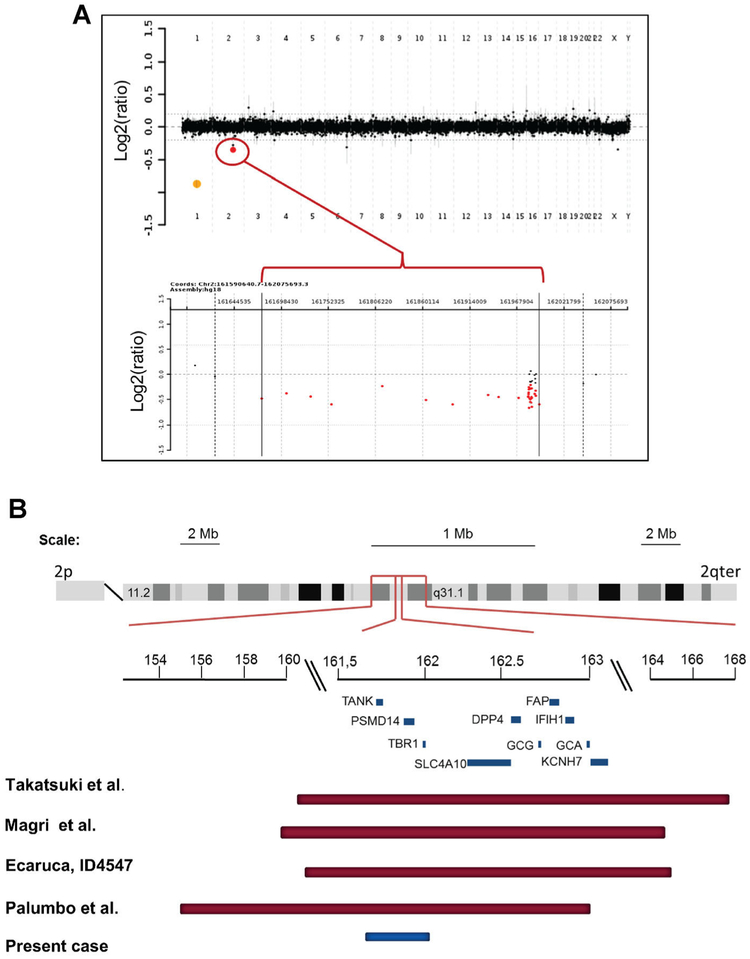
A deletion at 2q24.2 was identified by comparative genomic hybridization and confirmed by FISH. The maximum interval for this deletion was 0.422 Mb, extending from 161,622,412 to 162,043,922 bp (March 2006 assembly, NCBI36, hg18, minimum interval: 161,675,879–161,993,592 bp). This region contains three genes: TANK [OMIM 603893], PSMD14 [OMIM 607173], and TBR1 [OMIM 604616]. The log ratio of the loss was suggestive of mosaicism (Fig. 1). FISH analysis in a peripheral blood sample confirmed mosaicism: the 2q24.2 deletion was identified in 61% of cells analyzed (Supplementary eFig. 1-see Supporting Information online). The deletion was not detected in parental samples. A review of over 25,000 studies performed to date at our institution using custom-designed whole-genome oligonucleotide arrays (V7 and V8) identified four patients with deletions in this region with minimum sizes ranging between 4.7 and 19.6 Mb. The Database of Genomic Variants does not presently report deletions or duplications encompassing the entire region, but small intragenic deletions in PSMD14 (DGV Variation_9968 and Variation_1358) and in TANK (DGV Variation_1357) have been reported [Conrad et al., 2006; Wang et al., 2007]. A review of the 1000 Genomes Project Integrated Phase 1 variant set revealed no frameshift, essential splice-site (within 2 bp of an intron–exon boundary), or stop codon (gain or loss) mutations in TBR1 or PSMD14, while a coding frameshift insertion has been reported in TANK.
FIG. 1.
The deleted region at 2q24 encompasses three genes. A: Array comparative genomic hybridization (aCGH) plots. Each dot represents one oligonucleotide reporter on the array. In the upper panel (whole-genome plot), the area in red represents the deletion and the area in yellow represents an internal control. In the lower panel (close-up plot), reporters with a loss of copy number are represented as red dots. In both panels, the y-axis represents log2-converted ratios (sample: control). The maximum interval for this deletion was 0.422 Mb, extending from 161,622,412 to 162,043,922 bp (minimum interval: 161,675,879–161,993,592 bp). B: Comparison of the deletion documented in our patient with those of previously published reports [Magri et al., 2011; Palumbo et al., 2012; Takatsuki et al., 2010]. Coordinates are based on NCBI36/hg18.
DISCUSSION
Loss of 2q24.2 has been associated with intellectual disability, hypotonia, short stature, and dysmorphic features [Takatsuki et al., 2010; Magri et al., 2011; Palumbo et al., 2012]. The smallest common segment among previously published patients is approximately 2.65 Mb (160,408,000–163,058,894 bp, based on NCBI36/hg18) and includes 15 genes described in OMIM [Palumbo et al., 2012]. The deletion we report encompasses a 0.422 Mb interval containing the genes TANK, PSMD14, and TBR1 (Fig. 1). The intellectual disability documented in our patient is milder than what has been reported in patients with larger deletions. In addition, he does not have the dysmorphic features or hypotonia described in some patients with larger deletions. His milder presentation could be due to mosaicism. Alternatively, the more severe presentation in patients with larger deletions could result from the concomitant deletion of adjacent regions. The report of a patient with a more distal 2.8 Mb deletion (2q24.2–24.3) and an autistic phenotype provides support for the latter explanation [Chen et al., 2010].
Of the three genes in the region we report, TBR1 is in our view the most likely candidate for the intellectual disability phenotype. TBR1 is a transcription factor expressed early in cortical development and is known to promote frontal cortex identity [Bedogni et al., 2010a]. TBR1 knockout mice, which die by postnatal day 0–3, have a cortical migration disorder [Bulfone et al., 1998; Hevner et al., 2001]. Furthermore, Auts2, a gene associated with intellectual disability and autism, has been identified as a target of TBR1 [Bedogni et al., 2010b].
Of the two other genes in the deleted region, PSMD14 is the next best candidate for the intellectual disability phenotype. PSMD14 is also expressed in many tissues and functions as a component of the 26S proteasome, which plays a role in protein degradation via deubiquitination [Verma et al., 2002; Yao and Cohen, 2002]. Knock-down of PSMD14 expression in human cell lines revealed decreased cell viability [Gallery et al., 2007] and in carcinoma cells it leads to cell cycle arrest [Byrne et al., 2010]. Furthermore, knock-down of this gene in postmitotic neurons resulted in apoptosis [Staropoli and Abeliovich, 2005]. Lastly, TANK (tumor necrosis factor receptor-associated factor) is a ubiquitously expressed gene whose protein product functions to activate NF-κβ with TRAF2 (tumor necrosis factor receptor-associated factor) [Cheng and Baltimore, 1996; Kaye et al., 1996].
Short stature is a common feature noted in our patient and in several previously described patients with larger deletions [Takatsuki et al., 2010; Magri et al., 2011]. It is unclear which of the three genes in the deleted region contributes to the short stature phenotype. However, the association of PSMD14 with decreased cell viability and cell cycle arrest makes it an interesting candidate gene for this component of the phenotype.
In summary, we describe the case of an 18-year-old male with intellectual disability, short stature, and mosaicism for a 0.422 Mb deletion at 2q24.2. Deletions of such small size in this region appear to be very uncommon, even among patients with clinical indications for comparative genomic hybridization. This finding suggests that the critical region for intellectual disability and short stature in 2q24.2 can be narrowed to an interval that encompasses only three genes: TBR1, PSMD14, and TANK.
Supplementary Material
Footnotes
Additional supporting information may be found in the online version of this article.
REFERENCES
- Bedogni F, Hodge RD, Elsen GE, Nelson BR, Daza RA, Beyer RP, Bammler TK, Rubenstein JL, Hevner RF. 2010. Tbr1 regulates regional and laminar identity of postmitotic neurons in developing neocortex. Proc Natl Acad Sci USA 107:13129–13134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedogni F, Hodge RD, Nelson BR, Frederick EA, Shiba N, Daza RA, Hevner RF. 2010. Autism susceptibility candidate 2 (Auts2) encodes a nuclear protein expressed in developing brain regions implicated in autism neuropathology. Gene Expr Patterns 10:9–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulfone A, Wang F, Hevner R, Anderson S, Cutforth T, Chen S, Meneses J, Pedersen R, Axel R, Rubenstein JL. 1998. An olfactory sensory map develops in the absence of normal projection neurons or GABAergic interneurons. Neuron 21:1273–1282. [DOI] [PubMed] [Google Scholar]
- Bushby KM, Cole T, Matthews JN, Goodship JA. 1992. Centiles for adult head circumference. Arch Dis Child 67:1286–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne A, McLaren RP, Mason P, Chai L, Dufault MR, Huang Y, Liang B, Gans JD, Zhang M, Carter K, Gladysheva TB, Teicher BA, Biemann HP, Booker M, Goldberg MA, Klinger KW, Lillie J, Madden SL, Jiang Y. 2010. Knockdown of human deubiquitinase PSMD14 induces cell cycle arrest and senescence. Exp Cell Res 316:258–271. [DOI] [PubMed] [Google Scholar]
- Chen CP, Lin SP, Chern SR, Chen YJ, Tsai FJ, Wu PC, Wang W. 2010. Array-CGH detection of a de novo 2.8 Mb deletion in 2q24.2 → q24.3 in a girl with autistic features and developmental delay. Eur J Med Genet 53:217–220. [DOI] [PubMed] [Google Scholar]
- Cheng G, Baltimore D. 1996. TANK, a co-inducer with TRAF2 of TNF- and CD 40L-mediated NF-kappaB activation. Genes Dev 10:963–973. [DOI] [PubMed] [Google Scholar]
- Conrad DF, Andrews TD, Carter NP, Hurles ME, Pritchard JK. 2006. A high-resolution survey of deletion polymorphism in the human genome. Nat Genet 38:75–81. [DOI] [PubMed] [Google Scholar]
- Gallery M, Blank JL, Lin Y, Gutierrez JA, Pulido JC, Rappoli D, Badola S, Rolfe M, Macbeth KJ. 2007. The JAMM motif of human deubiquitinase Poh1 is essential for cell viability. Mol Cancer Ther 6:262–268. [DOI] [PubMed] [Google Scholar]
- Hevner RF, Shi L, Justice N, Hsueh Y, Sheng M, Smiga S, Bulfone A, Goffinet AM, Campagnoni AT, Rubenstein JL. 2001. Tbr1 regulates differentiation of the preplate and layer 6. Neuron 29:353–366. [DOI] [PubMed] [Google Scholar]
- Kaye KM, Devergne O, Harada JN, Izumi KM, Yalamanchili R, Kieff E, Mosialos G. 1996. Tumor necrosis factor receptor associated factor 2 is a mediator of NF-kappa B activation by latent infection membrane protein 1, the Epstein–Barr virus transforming protein. Proc Natl Acad Sci USA 93:11085–11090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magri C, Piovani G, Pilotta A, Michele T, Buzi F, Barlati S. 2011. De novo deletion of chromosome 2q24.2 region in a mentally retarded boy with muscular hypotonia. Eur J Med Genet 54:361–364. [DOI] [PubMed] [Google Scholar]
- Palumbo O, Palumbo P, Palladino T, Stallone R, Zelante L, Carella M 2012. A novel deletion in 2q24.1q24.2 in a girl with mental retardation and generalized hypotonia: A case report. Mol Cytogenet 5:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pescucci C, Caselli R, Grosso S, Mencarelli MA, Mari F, Farnetani MA, Piccini B, Artuso R, Bruttini M, Priolo M, Zuffardi O, Gimelli S, Balestri P, Renieri A. 2007. 2q24-q31 deletion: Report of a case and review of the literature. Eur J Med Genet 50:21–32. [DOI] [PubMed] [Google Scholar]
- Staropoli JF, Abeliovich A. 2005. The ubiquitin-proteasome pathway is necessary for maintenance of the postmitotic status of neurons. J Mol Neurosci 27:175–183. [DOI] [PubMed] [Google Scholar]
- Takatsuki S, Nakamura R, Haga Y, Mitsui K, Hashimoto T, Shimojima K, Saji T, Yamamoto T. 2010. Severe pulmonary emphysema in a girl with interstitial deletion of 2q24.2q24.3 including ITGB6. Am J Med Genet Part A 152A:1020–1025. [DOI] [PubMed] [Google Scholar]
- Verma R, Aravind L, Oania R, McDonald WH, Yates JR III, Koonin EV, Deshaies RJ. 2002. Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science 298:611–615. [DOI] [PubMed] [Google Scholar]
- Wang K, Li M, Hadley D, Liu R, Glessner J, Grant SF, Hakonarson H, Bucan M. 2007. PennCNV: An integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res 17:1665–1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao T, Cohen RE. 2002. A cryptic protease couples deubiquitination and degradation by the proteasome. Nature 419:403–407. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.