Abstract
Genetic variation in the serotonin transporter gene (SLC6A4) has been associated with psychopathology and aberrant brain functioning in a plethora of clinical and imaging studies. In contrast, the neurobiological correlates of epigenetic signatures in SLC6A4, such as DNA methylation profiles, have only recently been explored in human brain imaging research. The present study is the first to apply a resting state functional magnetic resonance imaging approach to identify changes in brain networks related to SLC6A4 promoter methylation (N = 74 healthy individuals). The amygdalae were defined as seed regions given that resting state functional connectivity in this brain area is under serotonergic control and relates to a broad range of psychiatric phenotypes. We further used bisulfite pyrosequencing to analyze quantitative methylation at 83 CpG sites within a promoter‐associated CpG island of SLC6A4 from blood‐derived DNA samples. The major finding of this study indicates a positive relation of SLC6A4 promoter methylation and amygdaloid resting state functional coupling with key nodes of the salience network (SN) including the anterior insulae and the dorsal anterior cingulate cortices. Increased intra‐network connectivity in the SN is thought to facilitate the detection and subsequent processing of potentially negative stimuli and reflects a core feature of psychopathology. As such, epigenetic changes within the SLC6A4 gene predict connectivity patterns in clinically and behaviorally relevant brain networks which may in turn convey increased disease susceptibility. Hum Brain Mapp 36:4361–4371, 2015. © 2015 Wiley Periodicals, Inc.
Keywords: serotonin transporter, methylation, resting state, connectivity, amygdala, salience network, epigenetics, imaging genetics, fMRI
INTRODUCTION
Genetic variation in the serotonin transporter gene (SLC6A4) is widely regarded as a potential risk marker for affective disorders [Caspi et al., 2010; Gatt et al., 2015]. By far most research has been dedicated to a 43bp insertion/deletion polymorphism in the promoter region of SLC6A4 (5‐HTTLPR). This genetic variant presumably influences the transcription rate of the gene with the short (S) allele being transcriptionally less efficient than the long (L) allele [Hu et al., 2006; Lesch et al., 1996]. Specifically, the S allele of 5‐HTTLPR has been linked to a broad range of psychiatric conditions such as post‐traumatic stress disorder (PTSD) and depression following exposure to environmental adversity [Caspi et al., 2003; Gressier et al., 2013; Karg et al., 2011; Sharpley et al., 2014], however overall results have been mixed [Munafo et al., 2009; Risch et al., 2009]. In parallel, molecular neuroimaging studies investigated putative mechanisms explaining how genetic variation in SLC6A4 may transfers into disease vulnerability. One of the best described neural correlate of 5‐HTTLPR is the finding of heightened amygdala activity at rest [Brockmann et al., 2011; Canli et al., 2006; El‐Hage et al., 2013; Rao et al., 2007] and in response to aversive stimuli [Hariri et al., 2002; Murphy et al., 2013] in S allele carriers. Although neural activity is thought to be more closely related to the genetic substrate compared to a psychiatric disease [Meyer‐Lindenberg, 2009], a recent meta‐analysis concluded that the effect of 5‐HTTLPR on amygdala reactivity is at best small [Bastiaansen et al., 2014].
Recent studies have promoted the idea that assessing epigenetic modification in addition to sequence variation is likely to constitute a more comprehensive approach in candidate gene studies [Meaney, 2010; Szyf, 2013]. With regard to SLC6A4 in specific, cytosine methylation within a promoter‐related region of the gene appears to be a biologically relevant marker associated with decreased SLC6A4 expression [Olsson et al., 2010; Philibert et al., 2008; Vijayendran et al., 2012; Wankerl et al., 2014; but also see Duman and Canli, 2015]. Moreover, individuals exposed to early life stress were found to have higher SLC6A4 promoter methylation levels [Beach et al., 2010, 2011; Duman and Canli, 2015; Kang et al., 2013; Ouellet‐Morin et al., 2013], which possibly reflect a molecular pathway from environmental adversity to disease susceptibility. In support of this notion, first studies have linked SLC6A4 promoter methylation to various psychiatric phenotypes such as lifetime diagnosis and severity of depression [Kang et al., 2013; Olsson et al., 2010; Philibert et al., 2008] depressive symptoms [Zhao et al., 2013] and unresolved trauma [van IJzendoorn et al., 2010], in some cases dependent on 5‐HTTLPR genotype [Olsson et al., 2010; van IJzendoorn et al., 2010]. Although there is some heterogeneity across studies concerning the specific SLC6A4 region investigated, they overall support the functional and clinical relevance of SLC6A4 methylation.
Surprisingly, very little is known about the neurobiological correlates of epigenetic signatures in SLC6A4 which possibly mediate differential vulnerability on a systemic level. A first imaging study using voxel‐based morphometry reported a significant link between SLC6A4 promoter methylation within an AluJb element and structural changes in mood‐related brain areas including the hippocampus, amygdala, and insula [Dannlowski et al., 2014]. With regard to functional measures, a recent seminal study provides strong evidence for a positive association of SLC6A4 promoter methylation and threat‐related amygdala reactivity [Nikolova et al., 2014]. In addition, a second task‐based fMRI study suggests that neural activity in the anterior insula elicited by negative emotional stimuli positively correlates with the degree of SLC6A4 methylation [Frodl et al., 2015]. These findings well‐align with neural alterations observed in 5‐HTTLPR S allele carriers [Hariri et al., 2002; Murphy et al., 2013].
Building on the seminal work conducted by Nikolova and coworkers, an important next step is to identify amygdala‐based functional connectivity networks associated with SLC6A4 methylation. These network‐based measures often better account for effects of genetic variation [Meyer‐Lindenberg, 2009] and behavioral variance [Vaidya and Gordon, 2013] than regional measures of activation. Aiming to address this question, we assessed resting state functional connectivity (rsFC) patterns with the amygdalae as seed regions in 74 healthy adults. This approach provides a reliable measure of functional coupling between brain regions unbiased by specific task demands [Fox and Raichle, 2007; Shehzad et al., 2009]. Aberrant amygdala rsFC has been linked to a broad range of clinically relevant phenotypes including fear learning [Feng et al., 2014; Schultz et al., 2012] as well as anxiety and mood disorders [Cullen et al., 2014; Patel et al., 2012; Peterson et al., 2014]. Moreover, amygdala rsFC appears to be responsive to pharmacological treatment directly targeting the serotonin transporter [Faria et al., 2014]. We further used bisulfite pyrosequencing to analyze quantitative methylation at 83 CpG sites within a 799‐bp promoter‐associated CpG island of SLC6A4 [Philibert et al., 2007] from blood‐derived DNA. Importantly, two independent post‐mortem studies indicate significant correlations between (site‐specific) SLC6A4 promoter methylation levels measured in amygdala tissue and peripheral blood cells [Nikolova et al., 2014; Riese et al., 2014]. These studies highlight the usefulness of peripheral epigenetic markers in SLC6A4 for the present study.
MATERIALS AND METHODS
Sample and Procedure
Seventy‐four healthy Caucasians [male: n = 45, age: 23.56 (± 2.36); female: n = 29, age: 23.17 (± 3.0)] participated in the present study. All participants were right handed and had normal or corrected to normal vision. Exclusion criteria were current or past mental and/or chronic physical diseases (for example, asthma, diabetes), medication intake (for example, psychotropic drugs), pregnancy, and failure to meet the MRI compatibility. After a structured telephone screening, participants were invited to a first appointment in order to complete the diagnostic interview for psychiatric disorders—short version (Mini‐DIPS) which asseses point and lifetime prevalence of axis I disorders on the basis of DSM IV criteria [Margraf, 1994]. Furthermore, blood samples were drawn into EDTA tubes (Sarstedt, Nümbrecht, Germany) for DNA extraction and stored at −20°C for no more than 6 months. For the scanning session, all participants were scheduled a second appointment between 11:00 am and 5:00 pm at the Neuroimaging Center and were instructed to arrive well‐rested. After entering the lab, participants were introduced to the study protocol and filled out the consent form prior to testing. The scanning session started with the acquisition of the structural image in order to acclimate participants to the scanning environment and to reduce stress reactions related to the scanner setting itself [Muehlhan et al., 2011, 2013]. During the RS acquisition, participants were instructed to close their eyes and let their mind wander but stay awake during the measurement. After completing the experiment, participants were explicitly asked whether they fell asleep during the scan which all participants had neglected. The study was conducted in accordance with the Declaration of Helsinki and approved by the Ethics Committee of the Technische Universität Dresden [EK: 152052012]. All participants received a monetary reward for participation.
Bisulfite Pyrosequencing and Genotyping
Quantitative methylation at 83 CpG sites within a 799‐bp promoter‐associated CpG island of SLC6A4 initially described by Philibert et al. [2007] was performed by Varionostic GmbH (Ulm, Germany, http://www.varionostic.de). Cytosine methylation within this region appears to be of functional relevance as it has been found to predict lower SLC6A4 mRNA expression in studies by our own [Wankerl et al., 2014] and other groups [Olsson et al., 2010; Philibert et al., 2008; Vijayendran et al., 2012]. For analysis, genomic DNA was extracted from EDTA whole blood samples and bisulfite‐treated using the EZ DNA Methylation Gold Kit (Zymo Research, Range, CA). Subsequent pyrosequencing was performed on the Q24/ID System and percent methylation at each CpG site was quantified using the PyroMark Q24 software (Qiagen). A detailed protocol containing amplicon and sequencing primers is provided elsewhere [Wankerl et al., 2014]. The percentages of methylation values passing quality control were > 95% for each individual CpG site. As previously reported, we observed significant interindividual variation regarding percentages of methylation at the CpG sites investigated (Supporting Information Fig. S3). Methylation levels were averaged across the 83 individual CpG sites to calculate a mean SLC6A4 promoter methylation score for each participant (referred to as SLC6A4 methylation). In accordance with previous studies, mean methylation scores were used for association analysis with fMRI data [Frodl et al., 2015; Nikolova et al., 2014]. As indicated by a Kaiser–Meyer–Olkin criterion larger than 0.5 (0.53), methylation across all CpG‐sites share a substantial portion of covariance (Cureton and D'Agostino, 1983), which is an important prerequisite for applying this procedure for our data. In addition, significant associations with functional connectivity values were reported for each of the 83 individual CpG sites separately to inform future studies regarding potential regions of interest.
In addition, participants were genotyped for 5‐HTTLPR according to a previously published protocol [Alexander et al., 2009] in order to control for genetic variation in SLC6A4. Genotyping included the analysis of the A/G single‐nucleotide polymorphism (rs25531) within the length polymorphism which results in the distinction of the genetic variants LA and LG, with the latter being functionally similar to the S allele [Hu et al., 2006]. This allows for conducting analyses based on the 5‐HTTLPR/rs25531 mini‐haplotype by comparing low [LG/S] and high [LA] expressing variants. Genotype frequencies (SS = 9, LGLG = 1, SLG = 4, SLA = 32, LGLA = 7, LALA = 21 were in Hardy–Weinberg equilibrium (bi‐allelic: = 0.18, P = 0.669, tri‐allelic: = 0.77, P = 0.855). As previously noted, mean SLC6A4 methylation levels did not differ as a function of 5‐HTTLPR genotype [Wankerl et al., 2014].
MRI Data Acquisition
MRI images were acquired using a 3‐Tesla Trio‐Tim MRI whole‐body scanner (Siemens, Erlangen, Germany). A standard 12 channel head coil and standard headphones were applied. Structural images were obtained by using a Magnetization Prepared Rapid Gradient Echo Imaging (MPRAGE) sequence (repetition time (TR) 1,900 ms, echo time (TE) 2.26 ms, flip angle α = 9°). Functional measurements were obtained using a T2* weighted gradient echo planar images (EPI) sequence (TR 2,200 ms, TE 25 ms, flip angle α = 80°). In each functional run, 200 whole brain volumes of 38 axial slices with a voxel size of 3.4 mm × 3.4 mm × 3 mm (25% gap) were acquired sequentially. Each slice had a matrix size of 64 × 64 voxels resulting in a field of view of 220 mm.
MRI Data Preprocessing and Connectivity Analysis
Prior to preprocessing the first four scans were discharged due to T1 equilibration effects. The remaining functional scans were spatially realigned and unwarped to correct for interscan movement. The structural image was segmented and normalized to the MNI (Montreal Neurological Institute, Quebec, Canada) reference brain. Functional images were normalized to the reference brain followed by outlier detection (ART‐based scrubbing). Finally, images were smoothed using an 8 mm Gaussian kernel. These steps were implemented in the conn toolbox pipeline [Whitfield‐Gabrieli and Nieto‐Castanon, 2012] and based on SPM12 (Wellcome Trust Center for Neuroimaging, UCL, London, UK). The conn toolbox (version 15a) was further used for functional connectivity analysis. Time series were extracted from the unsmoothed spatially normalized data to avoid confounding signals from surrounding regions. Prior to first‐level analyses, a denoising procedure including the component‐based correction method [CompCor; Behzadi et al., 2007] and temporal band‐pass filtering of 0.008 Hz–0.09 Hz was applied to remove motion artifacts, physiological and other artifactual effects from the fMRI‐signal. Moreover, the six movement parameters and a matrix containing the ART‐detected outliers were included as first level nuisance covariates. For time‐series extraction, binary masks from the left and right amygdala were created using the automated anatomical labeling atlas [Lancaster et al., 2000] from the WFU Pickatlas [Maldjian et al., 2003]. Then, seed‐to‐voxel analyses were performed for each participant and a general linear model was used to calculate positive and negative connectivity of both amygdalae on the second level. Additionally, SLC6A4 methylation was implemented as second level covariate to test for voxel wise correlations between the seed‐to‐voxel connectivity values and SLC6A4 methylation. For all connectivity analyses, the statistical threshold for each voxel was set at P < 0.001, cluster‐wise FWE‐corrected for multiple comparisons (P < 0.05). Finally, 6 mm spherical volumes were built around the peak voxel of the seed‐to‐voxel connectivity maps. Averaged time series from both amygdala masks and each spherical volume were further used to calculate Region of Interest ‐ to ‐ Region of Interest (ROI‐to‐ROI) connectivity values for a visual inspection of the cluster of points. Connectivity maps were automatically labeled by conn using the anatomy toolbox v2.0 [Eickhoff et al., 2007] and manually complemented by the WFU Pickatlas [Maldjian et al., 2003]. In addition, we calculated effect sizes for the correlation analyses between connectivity strength and SLC6A4 methylation. These β‐values represent changes in Fisher‐Z‐scores (z‐transformed correlation coefficients) for each unit change in SLC6A4 methylation. For additional analyses using distinct amygdala subregions as seeds, maximum probability maps for the basolateral (LB), the centromedial (CM), and the superficial (SF) divisions were created using the anatomy toolbox v2.0 [Eickhoff et al., 2007].
RESULTS
Resting State Seed‐to‐Voxel Connectivity
Seed‐to‐voxel analyses of amygdala rsFC revealed connectivity patterns which largely overlap with those identified by prior studies [Robinson et al., 2010; Roy et al., 2009]. Figure 1 depicts connectivity maps while Table 1 presents peak voxels and respective cluster sizes. Briefly, the amygdala seed regions showed positive rsFC to a network comprising large parts of the insular cortices, temporopolar areas, superior and middle temporal regions, the inferior frontal gyri as well as the dorsal/ventral anterior cingulate cortices (ACCs) and the dorsal posterior ACCs. Increased rsFC with the amygdala seeds was further observed for parahippocampal regions, the putamina and entorhinal cortices, the fusiform gyri, subgenual regions, premotor and primary motor cortices, somatosensory‐, primary auditory, and associative visual cortices as well as subcentral areas and piriform cortices. Negative correlations with the amygdaloid time series were observed for parts of the superior, middle and orbital prefrontal cortices, the inferior parietal lobules, parts of the precunei and the cerebellar crus 1 and 2.
Figure 1.
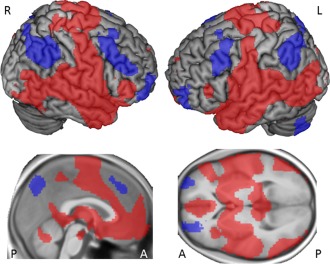
Seed‐to‐voxel connectivity maps. Red blobs depict regions positively coupled to the amygdalae. Blue blobs indicate regions negatively coupled to the amygdalae. Results are presented on a voxel‐level of P < 0.001, FWE cluster‐level corrected for multiple comparisons (P < 0.05). Connectivity maps are presented on a rendered brain surface (upper figure) from MRIcron (http://www.mccauslandcenter.sc.edu/mricro/mricron/) and on a medial and axial slice of an anatomically averaged image of all participants build with SPM 12 (lower figure). Anatomical labeling is reported in the text. [Color figure can be viewed in the online issue, which is available at http://wileyonlinelibrary.com.]
Table 1.
Peak voxels and cluster sizes from seed‐to‐voxel analyses
| Region | Hemisphere | Cluster size | MNI coordinates | z‐value | ||
|---|---|---|---|---|---|---|
| x | y | z | ||||
| Positive | ||||||
| Parahippocampal gyrus | L | 82110 | −24 | −02 | −20 | >8.00 |
| Parahippocampal gyrus | R | 25 | 00 | −18 | >8.00 | |
| Claustrum | R | 32 | −16 | 10 | >8.00 | |
| Cerebellum 8 L | L | 231 | −20 | −60 | 52 | 4.81 |
| Cerebellum 8 L | L | −25 | −52 | −58 | 4.43 | |
| Negative | ||||||
| Inferior parietal lobule | R | 3727 | 46 | −48 | 42 | 6.75 |
| Precuneus | L/R | 00 | −74 | 44 | 5.38 | |
| Precuneus | R | 08 | −70 | 44 | 5.32 | |
| Inferior parietal lobule | L | 2028 | −46 | −54 | 48 | 6.36 |
| Inferior parietal lobule | L | −28 | −70 | 46 | 4.48 | |
| Middle frontal gyrus | R | 1388 | 46 | 36 | 28 | 5.68 |
| Middle frontal gyrus | R | 48 | 24 | 42 | 4.98 | |
| Middle frontal gyrus | R | 42 | 20 | 50 | 4.89 | |
| SMA | L | 949 | −08 | 22 | 48 | 5.60 |
| Front. sup. medial gyrus | R | 08 | 32 | 42 | 5.32 | |
| Front. sup. medial gyrus | L | −04 | 36 | 38 | 4.62 | |
| Middle frontal gyrus | L | 745 | −42 | 22 | 44 | 5.49 |
| Middle frontal gyrus | L | −40 | 16 | 52 | 4.24 | |
| Middle frontal gyrus | L | −34 | 14 | 58 | 4.22 | |
| Sup. orbital cortex | L | 626 | −26 | 52 | −06 | 4.85 |
| Not labeled | L | −44 | 60 | −12 | 3.86 | |
| Not labeled | L | −4 | 58 | −18 | 3.72 | |
| Crus 1 L | L | 531 | −54 | −66 | −34 | 4.66 |
| Crus 2 L | L | −44 | −68 | −48 | 4.60 | |
| Crus 2 L | L | −34 | −76 | −52 | 4.29 | |
| Sup. orbital cortex | R | 664 | 22 | 50 | −12 | 4.39 |
| Front. sup. gyrus | R | 26 | 68 | 04 | 4.09 | |
Peak voxels are labeled according to the WFU Pickatlas. SMA: supplementary motor area. Results are reported using a statistical threshold of P < 0.001, FWE cluster‐level corrected (P < 0.05) for multiple comparisons.
Correlations of Amygdala Resting State Functional Connectivity and SLC6A4 Methylation
Connectivity values of three distinct clusters were found to vary with SLC6A4 methylation. As depicted in Figure 2A, correlational analyses revealed a positive relation between SLC6A4 methylation levels and amygdala resting state functional coupling with specific clusters which correspond to well‐known key nodes of the salience network [Menon and Uddin, 2010]. These associations were found for both hemispheres with slightly stronger effects being observed for the right amygdala network (Supporting Information Table S1 and Fig. S1). Specifically, a large medial cluster covered parts of the dorsal and ventral anterior cingulate cortices which extend to the premotor cortices. Furthermore, a lateral cluster comprised the left anterior insula, the inferior frontal gyrus, the operculum as well as the putamen. A second lateral cluster covered the right anterior insula, the operculum and the pars triangularis of the inferior frontal gyrus. These clusters largely overlap with those which were found to be positively coupled with the amygdala in the initial seed‐to‐voxel analyses of amygdala rsFC (Supporting Information Fig. S2). Table 2 presents effect sizes for respective correlational analyses with β‐values representing changes in Fisher‐Z‐scores (z‐transformed correlation coefficients) for each unit change in SLC6A4 methylation. To ensure the robustness of our findings with regard to potential confounds, respective analyses were repeated by entering sex, age, and 5‐HTTLPR/rs25531 genotype as covariates into the model. Controlling for these variables yielded largely comparable results with only small deviations in peak voxels and cluster sizes (see Table 2(B)).
Figure 2.
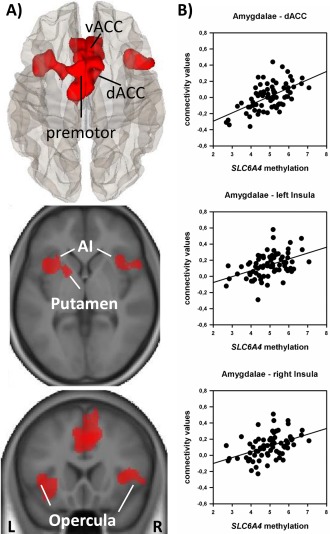
Positive correlations between amygdala resting state connectivity values and SLC6A4 methylation. A: Correlations between amygdala seed‐to‐voxel connectivity values and SLC6A4 methylation. Clusters are presented on a superior view of a glas brain build with conn toolbox (upper figure) and on axial and coronar slices of an anatomically averaged image of all participants build with SPM 12 (middle and lower figure). Results are presented on a voxel‐level of P < 0.001, FWE cluster corrected for multiple comparisons (P < 0.05). B: Scatter plots for visual inspection illustrate results from subsequent extracted mean connectivity values (ROI‐to‐ROI analyses): v/dACC = ventral/dorsal anterior cingulate cortices, AI = anterior insulae. [Color figure can be viewed in the online issue, which is available at http://wileyonlinelibrary.com.]
Table 2.
Peak voxels and cluster sizes derived from correlation analyses of amygdala seed‐to‐voxel connectivity and SLC6A4 methylation
| Region | Hemisphere | Cluster size | MNI coordinates | z‐value | β‐value | |||
|---|---|---|---|---|---|---|---|---|
| x | Y | z | ||||||
| A | dACC | R | 3266 | 06 | 22 | 40 | 5.45 | 0.08 |
| ACC | L | −10 | 28 | 26 | 4.98 | |||
| SMA | L | −08 | −12 | 62 | 4.64 | |||
| Insula | L | 832 | −40 | 08 | 06 | 4.09 | 0.07 | |
| Putamen | L | −20 | 06 | −06 | 4.04 | |||
| Insula | L | −40 | 16 | 00 | 3.98 | |||
| Insula | R | 468 | 42 | 16 | 08 | 4.18 | 0.08 | |
| Insula | R | 36 | 22 | 04 | 4.15 | |||
| Operculum | R | 54 | 14 | 02 | 3.72 | |||
| B | dACC | R | 3356 | 06 | 22 | 40 | 5.33 | 0.08 |
| SMA | L | −08 | −12 | 62 | 5.01 | |||
| SMA | L | 06 | 00 | 46 | 4.83 | |||
| Insula | L | 628 | −38 | 18 | −02 | 3.93 | 0.07 | |
| Operculum | L | −42 | 10 | 08 | 3.90 | |||
| Putamen | L | −20 | 06 | −06 | 3.89 | |||
| Insula | R | 405 | 36 | 22 | 04 | 4.17 | 0.08 | |
| Operculum | R | 42 | 16 | 08 | 4.04 | |||
| Operculum | R | 54 | 14 | 02 | 3.71 | |||
Peak voxels are labeled according to the WFU Pickatlas. Results are reported using a statistical threshold of P < 0.001, FWE cluster‐level corrected (P < 0.05) for multiple comparisons. dACC: dorsal anterior cingulate cortex; SMA: supplementary motor area. β = statistical strength, β‐values represent changes in Fisher‐Z‐scores (z‐transformed correlation coefficients) for each unit change in SLC6A4 methylation.
A: Main effects of SLC6A4 methylation, B: Main effect of SLC6A4 methylation corrected for 5‐HTTLPR genotype, sex and age.
For descriptive purposes, we further conducted ROI‐to‐ROI analyses by extracting mean connectivity values between the amygdala seeds and the spheres around the peak voxels of each cluster (Table 2, Fig. 2A). Visual inspection of the scatter plots confirmed that the strong positive correlations of SLC6A4 methylation and amygdala rsFC with key nodes of the salience network (dACC, left insula, right insula) are not driven by outliers. Additional analyses using distinct amygdala subregions as seeds indicated that both the BL and CM amygdala account for respective rsFC correlations with SLC6A4 methylation while no associations were observed for the SF division (Supporting Information Fig. S3). Supporting Information Table S2 further illustrates correlations of percentage methylation at specific CpG sites and amygdala rsFC with the dACC, the left and the right insula. Notably, these analyses revealed a cluster of individual CpG sites (CpG 76–83) with particularly strong associations. This cluster nicely corresponds to those sites with largest interindividual variation in overall methylation levels (Supporting Information Fig. S4).
For reasons of completeness, we further analyzed effects of 5‐HTTLPR genotype on amygdala rsFC. Both the seed‐to‐voxel and the ROI‐to‐ROI analyses revealed no significant differences in amygdala rsFC between carriers of at least one low‐expressing variant with carriers of two high‐expressing alleles.
DISCUSSION
The present study is the first to investigate the functional connectivity patterns associated with epigenetic variation in SLC6A4, a key candidate gene in psychiatric research. The major finding indicates a strong positive relation between SLC6A4 methylation and amygdaloid resting state functional coupling with key nodes of the salience network (SN) including the anterior insulae (AI) and the dorsal ACCs. These associations were primarily driven by rsFC patterns of the BL and CM amygdala and were particularly strong for a specific cluster of individual CpG sites in SLC6A4 (CpG 76–83). Combined with the positive correlation of SLC6A4 methylation and amygdala reactivity reported in a task‐based study [Nikolova et al., 2014], our findings suggest that neural substrates of epigenetic changes in SLC6A4 are not limited to regional amygdala activity but extend to connectivity patterns of larger brain networks. Moreover, our resting state data concurs with findings obtained in two previous task‐based studies, which suggest neural activity in both the amygdala and AI to be particularly sensitive to variability in SLC6A4 methylation [Frodl et al., 2015, Nikolova et al., 2014].
The pattern of amygdala rsFC observed with increased SLC6A4 methylation in the present study has been widely characterized as a clinically and behaviorally relevant neural marker. According to a rich literature, the SN serves to evaluate the relevance of internal and environmental stimuli, initiates their subsequent processing, and further guides behavioral responses by switching between other large‐scale neurocognitive networks [Menon and Uddin, 2010; Uddin, 2014] Increased intra‐network connectivity in the SN is thought to facilitate the detection and additional processing of potentially negative stimuli and may render the individual more prone to psychopathology, e.g., when salience is misattributed to trivial events [Menon and Uddin, 2010]. This notion is consistent with a growing number of task‐based and resting state studies suggesting that increased activation and stronger functional coupling of the SN is implicated in a broad range of anxiety disorders, most notably PTSD [Hayes et al., 2012; Patel et al., 2012; Peterson et al., 2014]. As confirmed by recent meta‐analyses, hyperactivation across core regions of the SN including the amygdala, the AI, and the dorsal ACC range among the most consistently identified neural features of PTSD [Hayes et al., 2012; Patel et al., 2012]. In addition, PTSD patients were repeatedly found to exhibit greater rsFC between the amygdala and the AI [Rabinak et al., 2011; Sripada et al., 2012a, 2012b]. These two regions are characterized by dense reciprocal connections [Reynolds and Zahm, 2005] broadly implicated in emotion generation, fear learning, and salience attribution [LeDoux, 1995; Menon and Uddin, 2010; Uddin, 2014]. As detailed in respective studies, behavioral manifestations of heightened amygdala–AI functional coupling may comprise symptoms of hypervigilance, a pervasive state of arousal irrespective of actual danger, as well as stronger anticipation of negative events [Rabinak et al., 2011; Sripada et al., 2012a, 2012b]. Regarding affective disorders, increased reactivity to negative stimuli of the SN, specifically of the amygdala, the AI, and the dorsal ACC, has proven a frequent and meta‐analytically confirmed correlate of Depression [Hamilton et al., 2012]. In contrast, analyses of rsFC in depressed patients applying both model‐free and seed‐based ROI approaches failed to identify a consistent pattern of altered functional coupling within the across individual studies [Avery et al., 2014; Connolly et al., 2013; Cullen et al., 2014; Pannekoek et al., 2014; Veer et al., 2010]. In conclusion, the pattern of enhanced SN intrinsic connectivity observed in individuals with higher SLC6A4 methylation appears to be of clinical significance and possibly reflects a premorbid risk factor for psychopathology in our sample of young adults.
In support of this idea, stronger functional coupling within core regions of the SN not only predicts psychiatric phenotypes, but has further been linked to increased self‐reported anxiety [Baur et al., 2013], enhanced fear learning [Feng et al., 2014; Schultz et al., 2012], and stress exposure [van Marle et al., 2010] in healthy populations. Referring to the assumption that enhanced sensitivity for salient events may underlie negative affect, a recent study identified amygdala‐AI rsFC as a promising biomarker for elevated state anxiety levels in healthy individuals [Baur et al., 2013]. Moreover, rsFC of the amygdala and dorsal ACC has been found to increase directly after stress exposure [van Marle et al., 2010] and Pavlovian fear conditioning [Feng et al., 2014; Schultz et al., 2012], with the magnitude of this change being predictive of a stronger conditioned fear response [Schultz et al., 2012]. The respective change in connectivity patterns has been suggested to reflect the process of fear memory storage, as the dorsal ACC presumably enhances fear expression and consolidation via excitation of the amygdala [Milad et al., 2007]. Consequently, the stronger amygdala–dorsal ACC functional coupling associated with increased SLC6A4 methylation may render the individual more sensitive to long‐term fear memories following adverse experiences.
In the light of these findings, one attractive hypothesis derived from animal models holds that epigenetic programming of neural networks reflects a systemic pathway explaining how environmental exposure may confer differential disease susceptibility [Szyf, 2013]. Besides genetic contributions, SLC6A4 methylation possibly captures the net effect of a broad range of those environmental influences relevant for long‐term changes in neural phenotypes under serotonergic control. Within the past years, growing evidence documents a link between increased SLC6A4 methylation and early adversity, such as sexual abuse [Beach et al., 2010, 2011; Vijayendran et al., 2012], different types of childhood trauma [Kang et al., 2013], and bullying victimization [Ouellet‐Morin et al., 2013], although individual findings vary regarding the specific CpG sites involved. In contrast, we failed to detect such associations for maternal prenatal stress and childhood maltreatment, indicating that these specific types of adversity unlikely account for variability in SLC6A4 methylation within the present sample [Wankerl et al., 2014]. Although the precise environmental correlates of SLC6A4 methylation are still largely unexplored, it is tempting to speculate that epigenetic modifications following adverse experiences shape neural circuitry in a way to facilitate salience detection and enhance fear learning. While this process is potentially adaptive under threatening environmental conditions, it may at the same time predispose the individual for the development of stress‐related psychiatric disorders. Indeed, several initial studies report associations of increased SLC6A4 promoter methylation with unresolved trauma [van IJzendoorn et al., 2010] and depressive symptoms [Olsson et al., 2010; Philibert et al., 2008; Zhao et al., 2013] in some cases dependent on genetic variation in SLC6A4 [Olsson et al., 2010; van IJzendoorn et al., 2010]. In sum, these findings concur with the idea that epigenetic programming of rsFC in the salience network might reflect a link between exposure to adversity and disease susceptibility.
Several limitations of the present study should be acknowledged. First, reported results rely on peripheral measures of SLC6A4 methylation which may not necessarily generalize to neural tissue. However, post‐mortem studies have provided a solid base for the use of peripheral markers by demonstrating substantial correlations across blood and neural cells for both genome‐wide [Byun et al., 2009] and SLC6A4 promoter [Nikolova et al., 2014; Riese et al., 2014] methylation in specific. Second, our study was underpowered to systematically evaluate joint contributions of genetic and epigenetic variation in the SLC6A4 gene. This potentially powerful approach should, however, be adopted in larger‐scale genetics imaging studies targeting the SLC6A4 gene as it may allow the detection of larger effects compared to those typically reported for 5‐HTTLPR. Indeed, SLC6A4 methylation has been recently found to moderate associations of 5‐HTTLPR on clinically relevant phenotypes, including depression [Olsson et al., 2010], unresolved trauma [van IJzendoorn et al., 2010], and cortisol stress reactivity [Alexander et al., 2014]. Third, our cross‐sectional findings provide no insights into causal relations or underlying mechanisms which have contributed to the observed association of SLC6A4 promoter methylation and rsFC. Fourth, our findings might be based on a generally more resilient sample as we had explicitly excluded individuals with current or past psychopathology. Lastly, a recent study raised the possibility that fMRI measurements of amygdala activation might be confounded by stimulus correlated signal fluctuation in nearby veins [Boubela et al., 2015].
In conclusion, our findings suggest that SLC6A4 methylation predicts aberrant rsFC of the amygdala with key regions of the SN, which has been described as a common feature of neuropsychiatric disorders. As methylation profiles appear to be responsive to behavioral [Roberts et al., 2014] and pharmacological [Korzus, 2010] interventions, uncovering their neural correlates may bear potential implications for the development of future treatment strategies targeting epigenetic mechanisms.
Supporting information
Supporting Information
ACKLOWLEDGMENT
Authors thank Matthis Wankerl and Janin Keitel for assisting in participant recruitment and Prof. Dr. Jürgen Hennig for 5‐HTTLPR genotyping.
Disclosure: MM, CK, and NA declare no conflict of interest. HUW has served on the advisory board and has received honoraria and travel reimbursement from industry during the past 5 years that might be perceived as constituting a conflict of interest: Sanofi, Aventis, Pfizer, Organon, Lilly, Lundbeck, Novartis and Essex Pharma.
REFERENCES
- Alexander N, Kuepper Y, Schmitz A, Osinsky R, Kozyra E, Hennig J (2009): Gene‐environment interactions predict cortisol responses after acute stress: Implications for the etiology of depression. Psychoneuroendocrinology 34:1294–1303. [DOI] [PubMed] [Google Scholar]
- Alexander N, Wankerl M, Hennig J, Miller R (2014): DNA methylation profiles within the serotonin transporter gene moderate the association of 5‐HTTLPR and cortisol stress reactivity. 4:e443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avery JA, Drevets WC, Moseman SE, Bodurka J, Barcalow JC, Simmons WK (2014): Major depressive disorder is associated with abnormal interoceptive activity and functional connectivity in the insula. Biol Psychiatry 76:258–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastiaansen JA, Servaas MN, Marsman JB, Ormel J, Nolte IM, Riese H, Aleman A (2014): Filling the gap: Relationship between the serotonin‐transporter‐linked polymorphic region and amygdala activation. Psychol Sci 25:2058–2066. [DOI] [PubMed] [Google Scholar]
- Baur V, Hanggi J, Langer N, Jancke L (2013): Resting‐state functional and structural connectivity within an insula‐amygdala route specifically index state and trait anxiety. Biol Psychiatry 73:85–92. [DOI] [PubMed] [Google Scholar]
- Beach SR, Brody GH, Todorov AA, Gunter TD, Philibert RA (2010): Methylation at SLC6A4 is linked to family history of child abuse: An examination of the Iowa Adoptee sample. Am J Med Genet B Neuropsychiatr Genet B 153:710–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beach SR, Brody GH, Todorov AA, Gunter TD, Philibert RA (2011): Methylation at 5HTT mediates the impact of child sex abuse on women's antisocial behavior: An examination of the Iowa adoptee sample. Psychosom Med 73:83–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behzadi Y, Restom K, Liau J, Liu TT (2007): A component based noise correction method (CompCor) for BOLD and perfusion based fMRI. Neuroimage 37:90–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boubela RN, Kalcher K, Huf W, Seidel EM, Derntl B, Pezawas L, Nasel C, Moser E (2015): fMRI measurements of amygdala activation are confounded by stimulus correlated signal fluctuation in nearby veins draining distant brain regions. Sci Rep 5:10499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockmann H, Zobel A, Schuhmacher A, Daamen M, Joe A, Biermann K, Schwab SG, Biersack HJ, Maier W, Boecker H (2011): Influence of 5‐HTTLPR polymorphism on resting state perfusion in patients with major depression. J Psychiatr Res 45:442–451. [DOI] [PubMed] [Google Scholar]
- Byun HM, Siegmund KD, Pan F, Weisenberger DJ, Kanel G, Laird PW, Yang AS (2009): Epigenetic profiling of somatic tissues from human autopsy specimens identifies tissue‐ and individual‐specific DNA methylation patterns. Hum Mol Genet 18:4808–4817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canli T, Qiu M, Omura K, Congdon E, Haas BW, Amin Z, Herrmann MJ, Constable RT, Lesch KP (2006): Neural correlates of epigenesis. Proc Natl Acad Sci USA 103:16033–16038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspi A, Sugden K, Moffitt TE, Taylor A, Craig IW, Harrington H, McClay J, Mill J, Martin J, Braithwaite A, Poulton R (2003): Influence of life stress on depression: Moderation by a polymorphism in the 5‐HTT gene. Science 301:386–389. [DOI] [PubMed] [Google Scholar]
- Caspi A, Hariri AR, Holmes A, Uher R, Moffitt TE (2010): Genetic sensitivity to the environment: The case of the serotonin transporter gene and its implications for studying complex diseases and traits. Am J Psychiatry 167:509–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly CG, Wu J, Ho TC, Hoeft F, Wolkowitz O, Eisendrath S, Frank G, Hendren R, Max JE, Paulus MP, et al. (2013): Resting‐state functional connectivity of subgenual anterior cingulate cortex in depressed adolescents. Biol Psychiatry 74:898–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen KR, Westlund MK, Klimes‐Dougan B, Mueller BA, Houri A, Eberly LE, Lim KO (2014): Abnormal amygdala resting‐state functional connectivity in adolescent depression. JAMA Psychiatry 71:1138–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EE Cureton, RB D'Agostino (1983): Factor Analysis: An Applied Approach. Hillsdale, NJ: Erlbaum. [Google Scholar]
- Dannlowski U, Kugel H, Redlich R, Halik A, Schneider I, Opel N, Grotegerd D, Schwarte K, Schettler C, Ambree O, et al. (2014): Serotonin transporter gene methylation is associated with hippocampal gray matter volume. Hum Brain Mapp 35:5356–5367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duman EA, Canli T (2015): Influence of life stress, 5‐HTTLPR genotype, and SLC6A4 methylation on gene expression and stress response in healthy Caucasian males. Biol Mood Anxiety Disord 5:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eickhoff SB, Paus T, Caspers S, Grosbras MH, Evans AC, Zilles K, Amunts K (2007): Assignment of functional activations to probabilistic cytoarchitectonic areas revisited. Neuroimage 36:511–521. [DOI] [PubMed] [Google Scholar]
- El‐Hage W, Zelaya F, Radua J, Gohier B, Alsop DC, Phillips ML, Surguladze SA (2013): Resting‐state cerebral blood flow in amygdala is modulated by sex and serotonin transporter genotype. Neuroimage 76:90–97. [DOI] [PubMed] [Google Scholar]
- Faria V, Ahs F, Appel L, Linnman C, Bani M, Bettica P, Pich EM, Wahlstedt K, Fredrikson M, Furmark T (2014): Amygdala‐frontal couplings characterizing SSRI and placebo response in social anxiety disorder. Int J Neuropsychopharmacol 17:1149–1157. [DOI] [PubMed] [Google Scholar]
- Feng P, Feng T, Chen Z, Lei X (2014): Memory consolidation of fear conditioning: Bi‐stable amygdala connectivity with dorsal anterior cingulate and medial prefrontal cortex. Soc Cogn Affect Neurosci 9:1730–1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox MD, Raichle ME (2007): Spontaneous fluctuations in brain activity observed with functional magnetic resonance imaging. Nat Rev Neurosci 8:700–711. [DOI] [PubMed] [Google Scholar]
- Frodl T, Szyf M, Carballedo A, Ly V, Dymov S, Vaisheva F, Morris D, Fahey C, Meaney J, Gill M, et al. (2015): DNA methylation of the serotonin transporter gene (SLC6A4) is associated with brain function involved in processing emotional stimuli. J Psychiatry Neurosci 40:140180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatt JM, Burton KL, Williams LM, Schofield PR (2015): Specific and common genes implicated across major mental disorders: A review of meta‐analysis studies. J Psychiatr Res C 60:1–13. [DOI] [PubMed] [Google Scholar]
- Gressier F, Calati R, Balestri M, Marsano A, Alberti S, Antypa N, Serretti A (2013): The 5‐HTTLPR polymorphism and posttraumatic stress disorder: A meta‐analysis. J Trauma Stress 26:645–653. [DOI] [PubMed] [Google Scholar]
- Hamilton JP, Etkin A, Furman DJ, Lemus MG, Johnson RF, Gotlib IH (2012): Functional neuroimaging of major depressive disorder: A meta‐analysis and new integration of base line activation and neural response data. Am J Psychiatry 169:693–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hariri AR, Mattay VS, Tessitore A, Kolachana B, Fera F, Goldman D, Egan MF, Weinberger DR (2002): Serotonin transporter genetic variation and the response of the human amygdala. Science 297:400–403. [DOI] [PubMed] [Google Scholar]
- Hayes JP, Hayes SM, Mikedis AM (2012): Quantitative meta‐analysis of neural activity in posttraumatic stress disorder. Biol Mood Anxiety Disord 2:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu XZ, Lipsky RH, Zhu G, Akhtar LA, Taubman J, Greenberg BD, Xu K, Arnold PD, Richter MA, Kennedy JL, et al. (2006): Serotonin transporter promoter gain‐of‐function genotypes are linked to obsessive‐compulsive disorder. Am J Hum Genet 78:815–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HJ, Kim JM, Stewart R, Kim SY, Bae KY, Kim SW, Shin IS, Shin MG, Yoon JS (2013): Association of SLC6A4 methylation with early adversity, characteristics and outcomes in depression. Prog Neuropsychopharmacol Biol Psychiatry 44:23–28. [DOI] [PubMed] [Google Scholar]
- Karg K, Burmeister M, Shedden K, Sen S (2011): The serotonin transporter promoter variant (5‐HTTLPR), stress, and depression meta‐analysis revisited: Evidence of genetic moderation. Arch Gen Psychiatry 68:444–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korzus E (2010): Manipulating the brain with epigenetics. Nat Neurosci 13:405–406. [DOI] [PubMed] [Google Scholar]
- Lancaster JL, Woldorff MG, Parsons LM, Liotti M, Freitas CS, Rainey L, Kochunov PV, Nickerson D, Mikiten SA, Fox PT (2000): Automated Talairach atlas labels for functional brain mapping. Hum Brain Mapp 10:120–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeDoux JE (1995): Emotion: Clues from the brain. Annu Rev Psychol 46:209–235. [DOI] [PubMed] [Google Scholar]
- Lesch KP, Bengel D, Heils A, Sabol SZ, Greenberg BD, Petri S, Benjamin J, Muller CR, Hamer DH, Murphy DL (1996): Association of anxiety‐related traits with a polymorphism in the serotonin transporter gene regulatory region. Science 274:1527–1531. [DOI] [PubMed] [Google Scholar]
- Maldjian JA, Laurienti PJ, Kraft RA, Burdette JH (2003): An automated method for neuroanatomic and cytoarchitectonic atlas‐based interrogation of fMRI data sets. Neuroimage 19:1233–1239. [DOI] [PubMed] [Google Scholar]
- Margraf J. (1994): Entstehung und Handhabung des Mini‐DIPS. Springer: Berlin, Heidelberg. [Google Scholar]
- Meaney MJ (2010): Epigenetics and the biological definition of gene × environment interactions. Child Dev 81:41–79. [DOI] [PubMed] [Google Scholar]
- Menon V, Uddin LQ (2010): Saliency, switching, attention and control: A network model of insula function. Brain Struct Funct 214:655–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer‐Lindenberg A (2009): Neural connectivity as an intermediate phenotype: Brain networks under genetic control. Hum Brain Mapp 30:1938–1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milad MR, Quirk GJ, Pitman RK, Orr SP, Fischl B, Rauch SL (2007): A role for the human dorsal anterior cingulate cortex in fear expression. Biol Psychiatry 62:1191–1194. [DOI] [PubMed] [Google Scholar]
- Muehlhan M, Lueken U, Wittchen HU, Kirschbaum C (2011): The scanner as a stressor: Evidence from subjective and neuroendocrine stress parameters in the time course of a functional magnetic resonance imaging session. Int J Psychophysiol 79:118–126. [DOI] [PubMed] [Google Scholar]
- Muehlhan M, Lueken U, Siegert J, Wittchen HU, Smolka MN, Kirschbaum C (2013): Enhanced sympathetic arousal in response to fMRI scanning correlates with task induced activations and deactivations. PLoS One 8:e72576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munafo MR, Freimer NB, Ng W, Ophoff R, Veijola J, Miettunen J, Jarvelin MR, Taanila A, Flint J (2009): 5‐HTTLPR genotype and anxiety‐related personality traits: A meta‐analysis and new data. Am J Med Genet B Neuropsychiatr Genet B 150:271–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy SE, Norbury R, Godlewska BR, Cowen PJ, Mannie ZM, Harmer CJ, Munafo MR (2013): The effect of the serotonin transporter polymorphism (5‐HTTLPR) on amygdala function: A meta‐analysis. Mol Psychiatry 18:512–520. [DOI] [PubMed] [Google Scholar]
- Nikolova YS, Koenen KC, Galea S, Wang CM, Seney ML, Sibille E, Williamson DE, Hariri AR (2014): Beyond genotype: Serotonin transporter epigenetic modification predicts human brain function. Nat Neurosci 17:1153–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsson CA, Foley DL, Parkinson‐Bates M, Byrnes G, McKenzie M, Patton GC, Morley R, Anney RJ, Craig JM, Saffery R (2010): Prospects for epigenetic research within cohort studies of psychological disorder: A pilot investigation of a peripheral cell marker of epigenetic risk for depression. Biol Psychol 83:159–165. [DOI] [PubMed] [Google Scholar]
- Ouellet‐Morin I, Wong CC, Danese A, Pariante CM, Papadopoulos AS, Mill J, Arseneault L (2013): Increased serotonin transporter gene (SERT) DNA methylation is associated with bullying victimization and blunted cortisol response to stress in childhood: A longitudinal study of discordant monozygotic twins. Psychol Med 43:1813–1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pannekoek JN, van der Werff SJ, Meens PH, van den Bulk BG, Jolles DD, Veer IM, van Lang ND, Rombouts SA, van der Wee NJ, Vermeiren RR (2014): Aberrant resting‐state functional connectivity in limbic and salience networks in treatment‐naive clinically depressed adolescents. J Child Psychol Psychiatry 55:1317–1327. [DOI] [PubMed] [Google Scholar]
- Patel R, Spreng RN, Shin LM, Girard TA (2012): Neurocircuitry models of posttraumatic stress disorder and beyond: A meta‐analysis of functional neuroimaging studies. Neurosci Biobehav Rev 36:2130–2142. [DOI] [PubMed] [Google Scholar]
- Peterson A, Thome J, Frewen P, Lanius RA (2014): Resting‐state neuroimaging studies: A new way of identifying differences and similarities among the anxiety disorders? Can J Psychiatry 59:294–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philibert R, Madan A, Andersen A, Cadoret R, Packer H, Sandhu H (2007): Serotonin transporter mRNA levels are associated with the methylation of an upstream CpG island. Am J Med Genet B Neuropsychiatr Genet B 144:101–105. [DOI] [PubMed] [Google Scholar]
- Philibert RA, Sandhu H, Hollenbeck N, Gunter T, Adams W, Madan A (2008): The relationship of 5HTT (SLC6A4) methylation and genotype on mRNA expression and liability to major depression and alcohol dependence in subjects from the Iowa Adoption Studies. Am J Med Genet B Neuropsychiatr Genet B 147:543–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinak CA, Angstadt M, Welsh RC, Kenndy AE, Lyubkin M, Martis B, Phan KL (2011): Altered amygdala resting‐state functional connectivity in post‐traumatic stress disorder. Front Psychiatry 2:62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao H, Gillihan SJ, Wang J, Korczykowski M, Sankoorikal GM, Kaercher KA, Brodkin ES, Detre JA, Farah MJ (2007): Genetic variation in serotonin transporter alters resting brain function in healthy individuals. Biol Psychiatry 62:600–606. [DOI] [PubMed] [Google Scholar]
- Reynolds SM, Zahm DS (2005): Specificity in the projections of prefrontal and insular cortex to ventral striatopallidum and the extended amygdala. J Neurosci 25:11757–11767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riese H, van den Heuvel ER, Snieder H, den Dunnen WF, Plosch T, Kema IP, Niezen‐Koning KE (2014): Association between methylation of the SLC6A4 promoter region in peripheral blood leukocytes and methylation in amygdala tissue. Psychosom Med 76:244–246. [DOI] [PubMed] [Google Scholar]
- Risch N, Herrell R, Lehner T, Liang KY, Eaves L, Hoh J, Griem A, Kovacs M, Ott J, Merikangas KR (2009): Interaction between the serotonin transporter gene (5‐HTTLPR), stressful life events, and risk of depression: A meta‐analysis. JAMA 301:2462–2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts S, Lester KJ, Hudson JL, Rapee RM, Creswell C, Cooper PJ, Thirlwall KJ, Coleman JR, Breen G, Wong CC, et al. (2014): Serotonin tranporter methylation and response to cognitive behaviour therapy in children with anxiety disorders. Transl Psychiatry 4:e444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JL, Laird AR, Glahn DC, Lovallo WR, Fox PT (2010): Metaanalytic connectivity modeling: Delineating the functional connectivity of the human amygdala. Hum Brain Mapp 31:173–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy AK, Shehzad Z, Margulies DS, Kelly AM, Uddin LQ, Gotimer K, Biswal BB, Castellanos FX, Milham MP (2009): Functional connectivity of the human amygdala using resting state fMRI. Neuroimage 45:614–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz DH, Balderston NL, Helmstetter FJ (2012): Resting‐state connectivity of the amygdala is altered following Pavlovian fear conditioning. Front Hum Neurosci 6:242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpley CF, Palanisamy SK, Glyde NS, Dillingham PW, Agnew LL (2014): An update on the interaction between the serotonin transporter promoter variant (5‐HTTLPR), stress and depression, plus an exploration of non‐confirming findings. Behav Brain Res 273:89–105. [DOI] [PubMed] [Google Scholar]
- Shehzad Z, Kelly AM, Reiss PT, Gee DG, Gotimer K, Uddin LQ, Lee SH, Margulies DS, Roy AK Biswal BB, et al. (2009): The resting brain: Unconstrained yet reliable. Cereb Cortex 19:2209–2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sripada RK, King AP, Garfinkel SN, Wang X, Sripada CS, Welsh RC, Liberzon I (2012a): Altered resting‐state amygdala functional connectivity in men with posttraumatic stress disorder. J Psychiatry Neurosci 37:241–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sripada RK, King AP, Welsh RC, Garfinkel SN, Wang X, Sripada CS, Liberzon I (2012b): Neural dysregulation in posttraumatic stress disorder: Evidence for disrupted equilibrium between salience and default mode brain networks. Psychosom Med 74:904–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szyf M (2013): DNA methylation, behavior and early life adversity. J Genet Genomics 40:331–338. [DOI] [PubMed] [Google Scholar]
- Uddin LQ (2014): Salience processing and insular cortical function and dysfunction. Nat Rev Neurosci 16:55–56. [DOI] [PubMed] [Google Scholar]
- Vaidya CJ, Gordon EM (2013): Phenotypic variability in resting‐state functional connectivity: Current status. Brain Connect 3:99–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van IJzendoorn MH, Caspers K, Bakermans‐Kranenburg MJ, Beach SR, Philibert R (2010): Methylation matters: Interaction between methylation density and serotonin transporter genotype predicts unresolved loss or trauma. Biol Psychiatry 68:405–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Marle HJ, Hermans EJ, Qin S, Fernandez G (2010): Enhanced resting‐state connectivity of amygdala in the immediate aftermath of acute psychological stress. Neuroimage 53:348–354. [DOI] [PubMed] [Google Scholar]
- Veer IM, Beckmann CF, van Tol M‐J, Ferrarini L, Milles J, Veltman DJ, Aleman A, van Buchem MA, van der Wee NJ, Rombouts SARB (2010): Whole brain resting‐state analysis reveals decreased functional connectivity in major depression. Front Syst Neurosci 4:41. doi: 10.3389/fnsys.2010.00041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijayendran M, Beach SR, Plume JM, Brody GH, Philibert RA (2012): Effects of genotype and child abuse on DNA methylation and gene expression at the serotonin transporter. Front Psychiatry 3:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wankerl M, Miller R, Kirschbaum C, Hennig J, Stalder T, Alexander N (2014): Effects of genetic and early environmental risk factors for depression on serotonin transporter expression and methylation profiles. 4:e402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield‐Gabrieli S, Nieto‐Castanon A (2012): Conn: A functional connectivity toolbox for correlated and anticorrelated brain networks. Brain Connect 2:125–141. [DOI] [PubMed] [Google Scholar]
- Zhao J, Goldberg J, Bremner JD, Vaccarino V (2013): Association between promoter methylation of serotonin transporter gene and depressive symptoms: A monozygotic twin study. Psychosom Med 75:523–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information


