Abstract
Brain‐derived neurotrophic factor (BDNF) has been implicated in multiple aspects of brain function including regulation of serotonin signaling. The BDNF val66met polymorphism (rs6265) has been linked to aspects of serotonin signaling in humans but its effects are not well understood. To address this, we evaluated whether BDNF val66met was predictive of a putative marker of brain serotonin levels, serotonin 4 receptor (5‐HT4) binding assessed with [11C]SB207145 positron emission tomography, which has also been associated with the serotonin‐transporter‐linked polymorphic region (5‐HTTLPR) polymorphism. We applied a linear latent variable model (LVM) using regional 5‐HT4 binding values (neocortex, amygdala, caudate, hippocampus, and putamen) from 68 healthy humans, allowing us to explicitly model brain‐wide and region‐specific genotype effects on 5‐HT4 binding. Our data supported an LVM wherein BDNF val66met significantly predicted a LV reflecting [11C]SB207145 binding across regions (P = 0.005). BDNF val66met met‐carriers showed 2–9% higher binding relative to val/val homozygotes. In contrast, 5‐HTTLPR did not predict the LV but S‐carriers showed 7% lower neocortical binding relative to LL homozygotes (P = 7.3 × 10−6). We observed no evidence for genetic interaction. Our findings indicate that BDNF val66met significantly predicts a common regulator of brain [11C]SB207145 binding, which we hypothesize reflects brain serotonin levels. In contrast, our data indicate that 5‐HTTLPR specifically affects 5‐HT4 binding in the neocortex. These findings implicate serotonin signaling as an important molecular mediator underlying the effects of BDNF val66met and 5‐HTTLPR on behavior and related risk for neuropsychiatric illness in humans. Hum Brain Mapp, 36:313–323, 2015. © 2014 Wiley Periodicals, Inc.
Keywords: serotonin, 5‐HT4 receptor, positron emission tomography, BDNF, 5‐HTTLPR
INTRODUCTION
Serotonin (5‐HT) critically affects many aspects of brain function including emotional behavior, memory, and cognition. Dysfunction of serotonin‐dependent signaling is thought to contribute to the pathophysiology of neuropsychiatric disorders including depression [Albert et al., 2012]. Therefore, it is beneficial to identify factors that contribute to variability in serotonin signaling because knowledge of how such variability emerges may inform risk for related illnesses. Brain‐derived neurotrophic factor (BDNF) is a neurotrophin that affects development and plasticity in the central nervous system [Waterhouse and Xu, 2009]. Genetic and pharmacological manipulation studies in rodents indicate that BDNF affects brain serotonin receptor levels and can positively affect brain serotonin levels [Lyons et al., 1999; Klein et al., 2010a; Mattson et al., 2004; Guiard et al., 2008; Deltheil et al., 2008; Siuciak et al., 1998], but the effects of BDNF‐related genetic variation on serotonin signaling in humans are less clear.
A genetic polymorphism (rs6265, BDNF val66met) in the BDNF gene substitutes a valine (val) for methionine (met) within the “proBDNF” sequence. The met‐allele is associated with decreased proBDNF intracellular trafficking and activity‐dependent BDNF release [Egan et al., 2003]. Furthermore, a recent meta‐analysis reported that the met‐allele confers vulnerability to depression in the presence of stressful life events [Hosang et al., 2014]. Functional magnetic resonance imaging studies reported reduced memory‐related hippocampal activity, disrupted fear extinction‐related amygdala and prefrontal reactivity and related behavioral phenotypes in met‐carriers (i.e., possessing one or two met‐alleles) [Hariri et al., 2003; Egan et al., 2003; Soliman et al., 2010]. Convergent evidence that serotonin signaling also modulates these pathways suggests serotonin may represent a molecular mechanism mediating BDNF val66met effects [Holmes, 2008; Bigos et al., 2008; Haahr et al., 2013]. BDNF val66met genotype has also been associated with brain volume and cortical thickness including evidence for an interaction with the serotonin‐transporter‐linked polymorphic region (5‐HTTLPR) polymorphism within the gene encoding the serotonin‐transporter (5‐HTT), suggesting these polymorphisms may interact in shaping neural pathways [Pezawas et al., 2008; Molendijk et al., 2012; Voineskos et al., 2011].
A limited number of in vivo molecular neuroimaging genetics studies have evaluated BDNF val66met effects on aspects of serotonin signaling in humans. A positron emission tomography (PET) study from our lab reported no BDNF val66met effect on neocortical serotonin 2A receptor binding with [18F]altanserin PET [Klein et al., 2010b]. One study reported no effect on raphe serotonin 1A receptor (5‐HT1A) binding whereas a recent study reported decreased 5‐HT1A binding throughout the brain of met‐carriers [Henningsson et al., 2009; Lan et al., 2014]. Henningsson et al. [2009] also reported lower 5‐HTT binding in male, but not female, met‐carriers using [11C]MADAM PET whereas we found no genotype effect using [11C]DASB PET [Klein et al., 2010b]. Thus, although there is some evidence linking BDNF val66met and serotonin signaling in vivo, evaluating BDNF‐related genetic effects on additional aspects of brain serotonin signaling (e.g., serotonin levels) would benefit our understanding of this association in humans.
Recently, we reported that serotonin 4 receptor (5‐HT4) binding with [11C]SB207145 PET significantly decreased following a three‐week fluoxetine intervention [Haahr et al., 2014], consistent with studies in animal models suggesting that 5‐HT4 levels are negatively regulated with respect to prolonged alterations in serotonin signaling and, therefore, represent a useful proxy for brain serotonin levels [Licht et al., 2009; Jennings et al., 2011]. Consistent with this model, we recently reported that 5‐HTTLPR S‐carriers showed 9% lower [11C]SB207145 binding in neocortex relative to LL homozygotes [Fisher et al., 2012]. Here, we evaluated whether this proxy for brain serotonin levels was predicted by BDNF val66met, accounting for effects of 5‐HTTLPR genotype and evaluated possible genetic interactions. We estimated genetic effects using a linear latent variable model (LVM), a flexible structural equation model that allows for explicitly testing global and region‐specific genotype effects on binding within a single model [Holst and Budtz‐Jørgensen, 2013]. We hypothesized that the low‐expressing met‐allele would be associated with increased 5‐HT4 binding across brain regions, reflecting decreased brain serotonin levels.
MATERIALS AND METHODS
Participants
Data were included from a cross‐sectional group of healthy participants (Table 1) recruited by advertisement for different research protocols approved by the Ethics Committee of Copenhagen and Frederiksberg, Denmark [KF‐01‐274821, KF‐11‐282837, HKF‐274821, H‐1‐2010‐085]. After a complete description of the study, written informed consent was obtained from all participants for the respective studies. Although inclusion criteria varied slightly across studies (e.g., some studies recruited only males), exclusion criteria for all participants included (1) primary psychiatric disease, (2) substance or drug abuse, and (3) severe systemic or neurological disease based on history and physical/neurological examination. Data from one participant was excluded because of a relatively high body‐mass index (BMI > 40 kg/m2) and two were excluded due to high [11C]SB207145 injected mass (>6 µg). Our final dataset included 73 scans acquired from 68 individuals (five participants scanned twice). Some PET datasets included here have been used in previous studies including all datasets from our previous molecular neuroimaging genetics study relating 5‐HTTLPR and [11C]SB207145 binding [Madsen et al., 2011a; Madsen et al., 2011b; Marner et al., 2009; Marner et al., 2010; Fisher et al., 2012; Haahr et al., 2013; Haahr et al., 2014].
Table 1.
Demographic, genotype, and [11C]SB207145 PET information
| BDNF val66met | 5‐HTTLPR | ||||
|---|---|---|---|---|---|
| Total | Val/val | Met‐carrier | LL | S‐carrier | |
| N | 73(68) | 31(30) | 42(38) | 30(27) | 43(41) |
| Age (years) | 34.2 ± 16.1 | 29.4 ± 13.1 | 37.6 ± 17.3 | 39.3 ± 18.3 | 30.5 ± 13.4 |
| Sex (male/female) | 53/15 | 26/4 | 27/11 | 22/5 | 31/10 |
| Scanner (A/H) | 25/48 | 8/23 | 17/25 | 13/17 | 12/31 |
| BMI (kg/m2) | 23.6 ± 2.5 | 23.6 ± 2.9 | 23.6 ± 2.2 | 24.2 ± 2.4 | 23.2 ± 2.5 |
| Injected mass (μg) | 2.14 ± 1.58 | 1.92 ± 1.52 | 2.30 ± 1.62 | 2.45 ± 1.55 | 1.93 ± 1.58 |
| w.a. injected mass (μg/kg) | 0.029 ± 0.023 | 0.026 ± 0.020 | 0.032 ± 0.024 | 0.032 ± 0.022 | 0.027 ± 0.024 |
| Injected dose (MBq) | 529 ± 120 | 522 ± 126 | 533 ± 117 | 523 ± 119 | 533 ± 133 |
Seventy‐three scans were acquired from 68 participants. Values are reported as mean ± standard deviation. Scanner types are Advance and HRRT PET scanners. BDNF val66met: val/val = 30, val/met = 34, met/met = 4. 5‐HTTLPR: LL = 27, LS = 32, SS = 9. A/H, Advance/HRRT PET scanner; BMI, body‐mass index; kg, kilograms; m, meters; µg, micrograms; w.a., weight‐adjusted; MBq, megabequerels.
DNA Extraction and Genotyping
Genomic DNA was extracted from blood using QIAamp DNA Blood Mini Kit (Qiagen, Valencia, CA). DNA quality and concentration were measured using a UV‐Vis spectrophotometer (NanoDrop2000, Thermo Scientific), which enabled the calculation of a target DNA concentration of 10–50 nanograms per microliter.
Genotype for the 5‐HTTLPR was determined as described previously [Kalbitzer et al., 2010]. We report only the “biallelic” classification (i.e., L vs. S alleles) to be consistent with our previous study [Fisher et al., 2012] and because considering the “triallelic” classification including rs25531 (i.e., LA, LG, SA/G) did not substantively affect the results.
BDNF val66met (rs6265) genotyping was performed using TaqMan SNP Genotyping Assay (Applied Biosystems, Foster City, CA). PCR amplification for allelic discrimination was performed using the LightCycler 480 Real‐Time PCR System (Roche Applied Science) using 40× Probe (C_11592758_10), TaqMan 2× PCR Master Mix (TaqMan Universal PCR Master Mix, Applied Biosystems), and standard thermal cycling. BDNF val66met genotype was verified with results visualized using Chromas Lite (http://www.technelysium.com.au) software, version 2.01, and compared to the reference sequence of the human BDNF gene (Ensembl: Transcript ID: ENST00000525528) using NCBI Align Sequences Nucleotide BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi).
MRI Data Acquisition
High‐resolution structural MR images were acquired using a Siemens Magnetom Trio 3T MR scanner. A 3D T1‐weighted MPRAGE and a T2‐weighted turbo spin echo (TSE) structural image were acquired and used for segmentation (MPRAGE) and brain masking (TSE) as previously described [Fisher et al., 2012].
[11C]SB207145 PET Data Acquisition
[11C]SB207145 was synthesized using a fully automated radiosynthesis system, as described previously [Marner et al., 2009]. An intravenous bolus injection of [11C]SB207145 was given over 20 s. Immediately following injection, a 120‐min dynamic emission scan including 38 time frames (length: 6 × 5, 10 × 15, 4 × 30, 5 × 120, 5 × 300, and 8 × 600 s) was acquired using one of two scanners: (1) an eighteen‐ring GE‐Advance scanner (GE, Milwaukee, WI) operating in 3D‐acquisition mode with an approximate in‐plane resolution of 6 mm or (2) a Siemens ECAT HRRT scanner operating in 3D‐acquisition mode with an approximate in‐plane resolution of 2 mm. All participants included in analyses received an injected mass within previously determined tracer dose limits [Madsen et al., 2011b]. Dynamic PET images acquired on the GE‐Advance scanner were reconstructed using filtered back projection and corrected for attenuation, dead‐time, and scatter. Dynamic PET images acquired on the HRRT scanner were reconstructed using an iterative OP‐OSEM3D method with resolution modeling (10 iterations and 16 subsets) [Sureau et al., 2008; Comtat et al., 2008; Hong et al., 2007].
Single‐subject PET data were aligned using AIR 5.2.5 to estimate in‐scan motion [Woods et al., 1992]. PET scans acquired on the GE‐Advance and HRRT scanners were smoothed using a within‐frame Gaussian filter before alignment (12 and 10 mm filter, respectively). We estimated rigid translation/rotation parameters aligning each PET frame to a single PET frame with sufficient structural information using the scaled least squares cost‐function in AIR (frame 26: 15–20 min post‐injection). Nonfiltered PET images were resliced using these parameters. Coregistration of the high‐resolution MR and PET images was performed using SPM5 including the mean of frames 10–26 of the PET scan (1–20 min post‐injection), corresponding to a flow‐weighted/nonspecific binding image with good delineation of cortical and subcortical structures. Accurate coregistration was confirmed by visual inspection across all planes. Regions were automatically delineated on the participant's structural MRI scan using Pvelab and time‐activity curves (TACs) reflecting the mean of gray‐matter voxels within each region were determined [Svarer et al., 2005]. Kinetic modeling of regional TACs was performed using the simplified reference tissue model with cerebellum as the reference region [Marner et al., 2009]. From this method, we obtain regional binding potential values, BPND = f ND*B avail*(1/K D), where f ND is the free fraction of ligand in the nondisplaceable tissue compartment, B avail is the concentration of receptors available for binding, and K D is the dissociation constant [Innis et al., 2007]. Kinetic modeling was performed using PMOD (PMOD, Zurich, Switzerland). Bilateral regional binding values were calculated by computing a volume‐weighted mean of the BPND values from the left and the right hemisphere.
Data Analysis
Statistical analyses were carried out in R v3.0.2 [R Core Team, 2013]. The primary outcome measures were log‐transformed regional BPND values from neocortex (i.e., occipital‐, orbital frontal‐, parietal‐cortex, pre/post central‐, medial/inferior frontal‐, medial/inferior temporal‐, superior frontal‐, superior temporal‐gyrus), amygdala, caudate, hippocampus, and putamen [Fisher et al., 2012]. Multiple observations from the five participants scanned twice were accounted for in all analyses using cluster robust standard errors (i.e., a Generalized Estimating Equation approach) [Liang and Zeger, 1986]. Genotype groupings for all analyses were BDNF val66met: val/val versus met‐carriers (val/met and met/met individuals) and 5‐HTTLPR: LL versus S‐carriers (LS and SS individuals). Sex, scanner type (i.e., Advance or HRRT), age, BMI, and weight‐adjusted injected mass were included as covariates in all models. Genotype effects were evaluated using a structural equation model including a single latent variable (LV). We also report effects from a univariate linear regression analysis of individual regions to highlight the strengths of this multivariate approach. Genotype differences in binding are reported as relative differences of met‐carriers or S‐carriers to val/val or LL individuals, respectively, which is directly related to the parameter estimate, β, when using log‐transformed regional binding values (exp(β)).
Latent variable model (structural equation model)
Model development and parameter estimates were determined using the Lava package [Holst and Budtz‐Jørgensen, 2013] in R. The addition of model paths was considered based on Wald tests with a false‐discovery rate (FDR) of q < 0.05 (Benjamini‐Hochberg FDR‐corrected). The Wald test is similar to a likelihood ratio test in that it tests whether the addition of a specific path significantly benefits overall model fit. This allowed us to effectively identify model paths supported by the data while controlling the rate of false‐positive model paths. An identifiable model was chosen such that covariate effects we report can be interpreted in terms of effects on neocortical binding (reference scale). All model path estimates/significance values were estimated simultaneously, and P‐values < 0.05 were considered statistically significant.
Linear regression analysis
Separately for each region, BDNF val66met and 5‐HTTLPR effects were determined within the same model to identify independent genotype effects. Genetic interaction effects were assessed in a model including the gene–gene interaction term. Two‐tailed P‐values (uncorrected and Bonferroni–Holm family‐wise error corrected, P FWE) are reported for all t‐tests.
RESULTS
Demographic Information
5‐HTTLPR and BDNF val66met status did not deviate from Hardy–Weinberg equilibrium (Table 1, P = 0.92 and 0.16, respectively), and there was no strong association between genotypes (Χ 2 = 0.21, P = 0.65).
Genetic Effects on [11C]SB207145 Binding: LVM
Model development
We developed a LVM structure starting with the a priori assumption that an underlying LV would predict binding across regions, consistent with a common regulatory mechanism. This basic model predicted a high correlation between regions captured by the LV (all factor loadings: P < 3.7 × 10−11). To this model, we added BDNF val66met, 5‐HTTLPR, sex, age, BMI, and weight‐adjusted injected mass as predictors of the LV. Additionally, scanner type was modeled to predict each region independently because of evidence for region‐specific effects. This model indicated a significant BDNF val66met effect on the LV (parameter estimate and 95% confidence interval: 0.070 [0.018; 0.122], P = 0.008) but not 5‐HTTLPR (−0.028 [−0.097; 0.040], P = 0.41). Wald tests supported a direct 5‐HTTLPR effect on neocortical binding without an effect on the LV and additional partial correlation between (1) amygdala and hippocampus and (2) caudate and putamen (all q < 0.02). Wald tests following these modifications supported an additional direct age effect on caudate (q = 0.02). Wald tests following this addition to the model indicated no support for additional model paths (q > 0.15), consistent with sufficient model fitting. This final model is depicted in Figure 1. Notably, the effect size of BDNF val66met on the LV was consistent throughout these model modifications (range of BDNF val66met on LV parameter estimate: 0.069–0.074).
Figure 1.
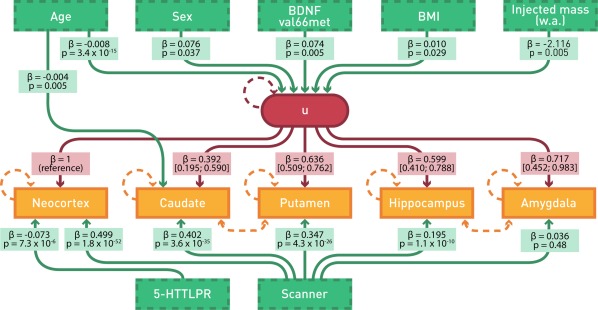
Latent variable model of genetic effects on [11C]SB207145 binding. Our final LVM is depicted with model paths. Green boxes indicate observed predictors. The red circle reflects the latent variable (u). Orange boxes represent measured log‐transformed regional [11C]SB207145 binding potential values. Age, sex, BMI, weight‐adjusted (w.a.) injected mass, and BDNF val66met genotype all map onto the latent variable whereas 5‐HTTLPR, scanner type, and an age effect on caudate directly affect regional binding potential values. BDNF val66met val/val and 5‐HTTLPR LL groups are reference for respective genotype parameter estimates. Orange hatched lines between (1) caudate and putamen and (2) amygdala and hippocampus indicate additional shared correlation; circular red and orange hatched lines reflect error estimates included in the model. The parameter estimate, β, for each model path is noted in respective boxes (95% confidence intervals indicated for estimates between latent variable and regional binding estimates). Significance of parameters estimates, P, is also noted. All regions loaded significantly onto the latent variable (all factor loadings: P < 9.8 × 10−5).
Summary of final model
Within our final model BDNF val66met was significantly predictive of the LV (0.074 [0.022, 0.13], P = 0.005) that significantly predicted [11C]SB207145 binding in all regions (P < 9.8 × 10−5; Fig. 1). Additionally, our model supported a specific effect of 7.0% lower neocortical binding in 5‐HTTLPR S‐carriers relative to LL individuals (−0.073 [−0.10, −0.04], P = 7.3 × 10−6). Additional observations from our final model included a statistically significant sex effect (male > female: 0.076 [0.0047, 0.15], P = 0.04), negative age effect (−0.0082 [−0.010, −0.0061], P = 3.5 × 10−15) and weight‐adjusted injected mass effect (−2.12 [−3.60, −0.63], P = 0.005), and positive BMI effect (0.010 [0.0010, 0.018], P = 0.03) on the LV. We observed an additional direct negative age effect on caudate binding (−0.0041 [−0.0069, −0.0012], P = 0.005) and the higher resolution HRRT PET scanner predicted significantly higher binding in all regions (P < 1.1 × 10−10) except the amygdala (P = 0.48).
Genetic Effects on [11C]SB207145 Binding: Linear Regression Models
Summary statistics of region‐specific linear regression models for genetic effects on [11C]SB207145 binding can be found in Table 2 and data are plotted in Figures 2 and 3. Met‐carriers showed generally higher [11C]SB207145 binding relative to val/val individuals but only the neocortex effect was statistically significant (percent difference of met‐carriers relative to val/val: amygdala: 8.7%, P FWE = 0.16; hippocampus: 4.1%, P FWE = 0.30; caudate: 2.5%, P FWE = 0.35; putamen: 5.5%, P FWE = 0.09; neocortex: 7.6%, P FWE = 0.03). 5‐HTTLPR S‐carriers showed 6.8% lower neocortical [11C]SB207145 binding relative to LL individuals (P FWE = 0.03) but nonsignificant differences in all other regions (percent difference of S‐carriers relative to LL: amygdala: +5.4%, P FWE = 0.88; hippocampus: +2.5%, P FWE = 1; caudate: −1.7%, P FWE = 1; putamen: +0.3%, P FWE = 1). We did not observe any statistically significant interaction effects between genotypes (P FWE > 0.99, Table 2).
Table 2.
Genetic effects on regional [11C]SB207145 binding values
| BDNF val66met | 5‐HTTLPR | Gene–gene interaction | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Region | % Diff | 95% CI | P | P FWE | % Diff | 95% CI | P | P FWE | Estimate | 95% CI | P | P FWE |
| Amygdala | 8.71 | [−0.1, 18.3] | 0.053 | 0.16 | 5.37 | [−3.1, 14.5] | 0.219 | 0.88 | −0.035 | [−0.21, 0.14] | 0.688 | 1 |
| Hippocampus | 4.13 | [−1.5, 10.0] | 0.151 | 0.30 | 2.53 | [−4.4, 10.0] | 0.485 | 1 | −0.058 | [−0.18, 0.07] | 0.371 | 1 |
| Caudate | 2.45 | [−2.6, 7.8] | 0.353 | 0.35 | −1.73 | [−7.4, 4.4] | 0.570 | 1 | −0.050 | [−0.15, 0.05] | 0.328 | 1 |
| Putamen | 5.45 | [0.7, 10.4] | 0.023 | 0.09 | 0.33 | [−4.4, 5.3] | 0.894 | 1 | −0.011 | [−0.10, 0.08] | 0.807 | 1 |
| Neocortex | 7.64 | [2.3, 13.3] | 0.005 | 0.03 | −6.77 | [−11.2, −2.1] | 0.005 | 0.03 | 0.003 | [−0.10, 0.11] | 0.956 | 1 |
Genotype main effects were determined within the same model, excluding the interaction term. Main effects are expressed as percent difference (% Diff) of minor allele (i.e., met‐carrier or S‐carrier) relative to major allele (i.e., val/val or LL). Gene–gene interaction effects are expressed as model parameter estimates. CI, confidence interval; P, P‐value (uncorrected); pFWE, Bonferroni–Holm family‐wise error rate corrected P‐value.
Figure 2.
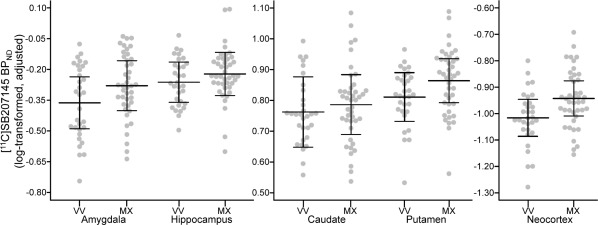
BDNF val66met effects on [11C]SB207145 binding. BDNF met‐carriers showed higher [11C]SB207145 binding potential (BPND) in all regions evaluated. However, only effects in neocortex were statistically significant (see Table 2 for model estimates). Gray dots represent individual residuals of log‐transformed BPND values, adjusted for age, sex, weight‐adjusted injected mass, scanner, BMI, and 5‐HTTLPR genotype. Black lines represent group mean and corresponding 95% confidence intervals. VV = BDNF val66met val/val genotype, MX = BDNF val66met met‐carrier.
Figure 3.
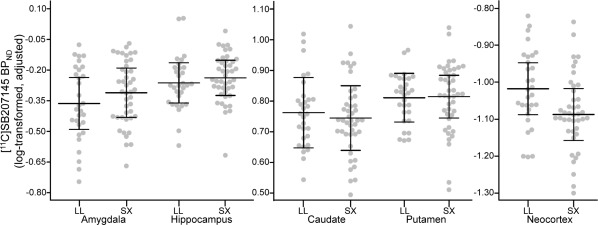
5‐HTTLPR effects on [11C]SB207145 binding. 5‐HTTLPR S‐carriers showed significantly lower [11C]SB207145 binding potential (BPND) within neocortex compared to LL homozygotes. However, effects in all other regions were not statistically significant (see Table 2 for model estimates). Gray dots represent individual residuals of log‐transformed BPND values, adjusted for age, sex, weight‐adjusted injected mass, scanner, BMI, and BDNF val66met genotype. Black lines represent group mean and corresponding 95% confidence intervals. LL = 5‐HTTLPR LL genotype, SX = 5‐HTTLPR S‐carriers.
DISCUSSION
Here, we determined the association between 5‐HT4 binding, assessed with [11C]SB207145 PET, and two commonly studied genetic polymorphisms: BDNF val66met and 5‐HTTLPR. The LVM supported a LV that was predictive of binding in all regions, suggesting a “global regulator” of [11C]SB207145 binding and that BDNF val66met significantly predicted variability in this regulator such that met‐carriers showed 2–9% higher binding across brain regions relative to val/val individuals. 5‐HTTLPR S‐carriers showed 7% lower binding in neocortex but nonsignificant effects in other regions. Based on a recent report that [11C]SB207145 binding reflects endogenous serotonin levels [Haahr et al., 2014], our data suggest that met‐carriers show lower serotonin levels (i.e., higher [11C]SB207145 binding) across brain regions, compared to val/val individuals, and S‐carriers show higher serotonin levels (i.e., lower binding) within neocortex, compared to LL individuals. Notably, our findings support significant and independent effects of these polymorphisms on serotonin levels. This represents novel in vivo evidence that BDNF val66met affects serotonin signaling in the healthy adult human brain, providing a molecular mechanism through which it may affect behavioral phenotypes and influence the risk architecture for neuropsychiatric illness.
Rodent and human studies indicate that 5‐HT4 receptor binding shows a monotonic inverse relation to serotonin levels [Jennings et al., 2011; Haahr et al., 2014; Licht et al., 2009; Vidal et al., 2009]. Considering this and evidence that alterations in BDNF signaling modulate serotonin signaling, we hypothesize that our LV reflects differences in central serotonin levels. Although studies genetically manipulating BDNF signaling in mice have reported effects on serotonin signaling, these precise mechanisms are unclear as the region and directionality of effects vary [Daftary et al., 2012; Deltheil et al., 2008; Guiard et al., 2008; Yu et al., 2012]. This variability may be due to methodological differences but nevertheless supports a link between BDNF and serotonin signaling. This link is further supported in humans by a recent study reporting that met‐carriers show significantly decreased 5‐HT1A binding across brain regions [Lan et al., 2014]. Notably, other studies have reported mixed effects of BDNF val66met on 5‐HTT, 5‐HT1A, and serotonin 2A receptor binding in humans using PET [Klein et al., 2010b; Henningsson et al., 2009]. Taken together, these findings indicate that BDNF val66met may affect certain features of serotonin signaling more strongly than others, which is relevant for more accurately utilizing this polymorphism as a model for differences in molecular signaling pathways.
An alternative interpretation of our findings is that BDNF signaling may act as an upstream transcriptional regulator of 5‐HT4 receptor gene expression. This is plausible considering general evidence in mice that genetic manipulation of BDNF modulates gene transcription [Mattson et al., 2004; Wang et al., 2014]. We are unaware of evidence directly linking BDNF signaling and regulation of 5‐HT4 gene expression in humans. However, we cannot rule out this possibility based on our current data. Future expression studies evaluating these possibilities would shed light on the molecular pathways underlying our observed effects. Regardless, our data support a significant BDNF val66met effect on 5‐HT4 binding.
The neocortical‐specific 5‐HTTLPR effect on 5‐HT4 binding is intriguing considering evidence that 5‐HT4 signaling in prefrontal cortex regulates serotonergic neurons in the dorsal raphe [Lucas et al., 2005]. The finding that S‐carriers showed decreased [11C]SB207145 binding, putatively reflecting increased serotonin levels, is consistent with a model based on in vitro evidence that the S‐allele shows lower 5‐HTT expression resulting in relatively increased serotonin signaling (Lesch et al., 1996; Hu et al., 2006]. However, it is not clear why this effect was restricted to only the neocortex. Previous studies have reported a 5‐HTTLPR effect on 5‐HTT binding within subcortical structures [Praschak‐Rieder et al., 2007; Reimold et al., 2007; Kalbitzer et al., 2010] but effects of 5‐HTTLPR on brain function and volume have been reported in neocortical regions and may be mediated by similar mechanisms affecting [11C]SB207145 binding [Surguladze et al., 2008; Heinz et al., 2005]. Our finding indicates that 5‐HTTLPR effects on serotonin signaling are present in adulthood, which may stem from genetic effects on neuro‐developmental processes [Sibille and Lewis, 2006].
We did not find evidence for a gene–gene interaction between these two polymorphisms. Previous studies have reported such effects on brain volume [Pezawas et al., 2008] and neural responses to emotionally salient stimuli [Wang et al., 2012; Outhred et al., 2012]. Behaviorally, an interaction between these variants has been linked to neuroticism, anxiety, rumination, and vulnerability markers of depression [Arias et al., 2012; Clasen et al., 2011; Terracciano et al., 2010; Kourmouli et al., 2013]. It is possible that these effects stem from a gene–gene interaction phenomenon modulating molecular mechanisms other than what is captured by our PET measure. However, Terracciano et al. [2010] reported a small interaction effect on neuroticism scores (∼1–3 points) from a very large population (N > 2000). Thus, although our molecular neuroimaging measure is likely more proximal to direct biological effects of these polymorphisms and, therefore, more sensitive to its effects, our sample size may be such that we are underpowered to detect gene–gene interaction effects within our current dataset.
LVMs provide a flexible framework for modeling multivariate effects [Sanchez et al., 2005; Holst and Budtz‐Jørgensen, 2013], which benefits statistical power by utilizing shared correlation in binding across regions. LVMs explicitly test for effects shared across regions, meaningfully affecting how results can be interpreted relative to a set of univariate models. Our observation of a BDNF val66met effect on the LV is distinct from a statistically significant effect in only neocortex, which the univariate approach supported. We regard this as important and highlighting the value of LVMs in their ability to more effectively model global versus region‐specific effects, allowing for a more nuanced interpretation of the data. The univariate equivalent would be each test reaching statistical significance, after multiple comparisons correction, but we argue this can be an unnecessarily onerous threshold given multivariate alternatives. Additionally, LVMs agree with the biological framework of a molecular neuroimaging genetics study, where genetic effects are likely to affect receptor binding across multiple brain regions but can also effectively account for polymorphisms that have region‐specific effects. For example, a polymorphism (rs6265, HTR1A C(‐1019)G) in the 5‐HT1A gene has been shown to affect 5‐HT1A expression in a cell‐specific manner [Czesak et al., 2006]. Our results highlight these strengths within a molecular neuroimaging genetics framework.
There is evidence for an interactive effect between BDNF val66met and age on neurobiological features including cortical thickness and brain volume [Voineskos et al., 2011]. However, whether this involves effects on the serotonin system is not clear. As a post hoc analysis, we evaluated whether BDNF val66met moderated the age effect on our LV. Interestingly, we observed evidence for an interaction such that met‐carriers decreased 5‐HT4 binding less quickly than val/val individuals (Fig. 4). This suggests serotonin may be an important molecular mechanism mediating the effects of BDNF val66met on age‐related processes [Mattson et al., 2004]. However, this was a post hoc evaluation of our model, which we interpret with caution and suggest future studies evaluate more closely.
Figure 4.
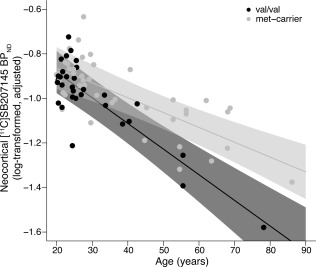
BDNF val66met by age interaction effect on [11C]SB20715 binding. Within a post hoc analysis, we observed evidence that BDNF val66met moderated the age effect on [11C]SB207145 binding. This effect is visualized here for neocortex (interaction effect: 0.0049 [0.0024, 0.0073], P = 1.1 × 10−4). Black and gray dots represent val/val individuals and met‐carriers, respectively. Lines and shading represent genotype slope estimates and 95% confidence intervals, respectively. Data are plotted as log‐transformed neocortex [11C]SB207145 binding values, adjusted for sex, weight‐adjusted injected mass, scanner, BMI and 5‐HTTLPR genotype. Notably, our data suggest that met‐carriers show higher binding through adulthood compared to val/val individuals.
Our study is not without its limitations. The interpretation of our PET measure is based on a recent study indicating that [11C]SB207145 binding is a useful proxy for brain serotonin levels [Haahr et al., 2014]. Although future studies investigating this association would be beneficial, our current findings provide compelling support for a genetic effect on this measure. Our data are heterogeneous, reflecting a cross‐sectional design, pooling data from multiple studies. Scanner effects were large but regionally variable. However, we observed a similar LVM fit and genotype effect sizes in a post hoc model considering only scans from the high‐resolution HRRT scanner (N = 48, BDNF val66met on LV: 0.065 [7.1 × 10−4, 0.13], P = 0.048, 5‐HTTLPR on neocortical binding: −0.078 [−0.12, −0.037], P = 2.1 × 10−4). Although injected masses differed between genotypes, it is unlikely this confounded our findings because the direction of these differences would undermine our observed genetic effects. Injected mass correlated negatively with binding, despite excluding observations with injected masses outside estimated tracer doses [Madsen et al., 2011b]. Genotype effects remained when using a more conservative threshold (<4.5 µg) indicating that this did not confound our findings (N = 64, BDNF val66met on LV: 0.068 [0.012, 0.12], P = 0.017, 5‐HTTLPR on neocortical binding: −0.081 [−0.12, −0.047], P = 2.5 × 10−6). However, future studies should correct for injected mass to account for any effects. We used a composite neocortex because of no clear a priori hypothesis about neocortex subregional effects. Post hoc univariate analysis of these regions showed some regional variability in percent difference between BDNF val66met (range: [4.2%, 14.9%], met‐carriers relative to val/val) and 5‐HTTLPR (range: [−13.3%, 0.4%], S‐carriers relative to LL). However, future studies would be needed to more appropriately evaluate possible subregion‐specific effects. We could not evaluate a “dose‐dependent” met‐allele effect due to its low frequency in our cohort (N = 4), consistent with its low frequency in those of European ancestry. However, a post hoc LVM separating LS and SS individuals showed that neocortical binding was significantly lower in both groups compared to LL individuals (Wald test for significance of LS and SS subgroups: X 2 = 21.1, P = 2.5 × 10−5) and did not significantly differ from each other (Wald test comparing LS and SS groups: X 2 = 0.68, P = 0.41). These observations suggest that observed genotype effects are not a result of heterogeneity of our data.
In summary, we find evidence using a LVM that BDNF val66met met‐carriers show higher 5‐HT4 binding across brain regions, which we hypothesize reflects central serotonin levels. Thus, our data indicate met‐carriers show decreased serotonin levels across cortical and subcortical brain regions. Additionally, 5‐HTTLPR S‐carriers showed significantly lower neocortical binding. Taken together, our findings indicate that these polymorphisms independently contribute to the regulation of serotonin signaling in healthy humans, which may underlie effects on behavior and contribute to risk for neuropsychiatric illness.
ACKNOWLEDGMENTS
We would like to thank B. Dall, G. Thomsen, S. Larsen, A. Dyssegaard, and L. Freyr for their assistance in scheduling and data collection at both the MR and PET centers. We thank J. Frederking for her contribution to the figures. We would like to gratefully acknowledge The John and Birthe Meyer Foundation for the donation of the Cyclotron and PET scanner.
Conflicts of Interest: The authors declare no competing financial interests.
REFERENCES
- Albert PR, Benkelfat C, Descarries L (2012): The neurobiology of depression—revisiting the serotonin hypothesis. I. Cellular and molecular mechanisms. Philos Trans R Soc B Biol Sci 367:2378–2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arias B, Aguilera M, Moya J, Saiz PA, Villa H, Ibanez MI, Garcia‐Portillo MP, Bobes J, Ortet G, Fananas L (2012): The role of genetic variability in the SLC6A4, BDNF and GABRA6 genes in anxiety‐related traits. Acta Psychiatr Scand 125:194–202. [DOI] [PubMed] [Google Scholar]
- Bigos KL, Pollock BG, Aizenstein HJ, Fisher PM, Bies RR, Hariri AR (2008): Acute 5‐HT Reuptake Blockade Potentiates Human Amygdala Reactivity. Neuropsychopharmacology 33:3221–3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clasen PC, Wells TT, Knopik VS, McGeary JE, Beevers CG (2011): 5‐HTTLPR and BDNF Val66Met polymorphisms moderate effects of stress on rumination. Genes Brain Behav 10:740–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comtat C, Sureau FC, Sibomana M, Hong IK, Sjoholm N, Trebossen R (2008) Image based resolution modeling for the HRRT OSEM reconstructions software. In: IEEE Nuclear Science Symposium Conference Record, NSS′08. Dresden, Germany. pp 4120–4123.
- Czesak M, Lemonde S, Peterson EA, Rogaeva A, Albert PR (2006): Cell‐specific repressor or enhancer activities of Deaf‐1 at a serotonin 1A receptor gene polymorphism. J Neurosci 26:1864–1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daftary SS, Calderon G, Rios M (2012): Essential role of brain‐derived neurotrophic factor in the regulation of serotonin transmission in the basolateral amygdala. Neuroscience 224:125–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deltheil T, Guiard BP, Cerdan J, David DJ, Tanaka KF, Reperant C, Guilloux JP, Coudore F, Hen R, Gardier AM (2008): Behavioral and serotonergic consequences of decreasing or increasing hippocampus brain‐derived neurotrophic factor protein levels in mice. Neuropharmacology 55:1006–1014. [DOI] [PubMed] [Google Scholar]
- Egan MF, Kojima M, Callicott JH, Goldberg TE, Kolachana BS, Bertolino A, Zaitsev E, Gold B, Goldman D, Dean M, Lu B, Weinberger DR (2003): The BDNF val66met polymorphism affects activity‐dependent secretion of BDNF and human memory and hippocampal function. Cell 112:257–269. [DOI] [PubMed] [Google Scholar]
- Fisher PM, Holst KK, McMahon B., Haahr ME, Madsen K, Gillings N, Baare WF, Jensen PS, Knudsen GM (2012): 5‐HTTLPR status predictive of neocortical 5‐HT4 binding assessed with [(11)C]SB207145 PET in humans. Neuroimage 62:130–136. [DOI] [PubMed] [Google Scholar]
- Guiard BP, David DJ, Deltheil T, Chenu F, Le ME, Renoir T, Leroux‐Nicollet I, Sokoloff P, Lanfumey L, Hamon M, Andrews AM, Hen R, Gardier AM (2008): Brain‐derived neurotrophic factor‐deficient mice exhibit a hippocampal hyperserotonergic phenotype. Int J Neuropsychopharmacol 11:79–92. [DOI] [PubMed] [Google Scholar]
- Haahr ME, Fisher P, Holst K, Madsen K, Jensen CG, Marner L, Lehel S, Baare W, Knudsen G, Hasselbalch S (2013): The 5‐HT4 receptor levels in hippocampus correlates inversely with memory test performance in humans. Hum Brain Mapp 34:3066–3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haahr ME, Fisher PM, Jensen CG, Frokjaer VG, Mahon BM, Madsen K, Baare WF, Lehel S, Norremolle A, Rabiner EA, Knudsen GM (2014): Central 5‐HT4 receptor binding as biomarker of serotonergic tonus in humans: A [(11)C]SB207145 PET study. Mol Psychiatry 19:427–432. [DOI] [PubMed] [Google Scholar]
- Hariri AR, Goldberg TE, Mattay VS, Kolachana BS, Callicott JH, Egan MF, Weinberger DR (2003): Brain‐derived neurotrophic factor val66met polymorphism affects human memory‐related hippocampal activity and predicts memory performance. J Neuroscience 23:6690–6694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz A, Braus DF, Smolka MN, Wrase J, Puls I, Hermann D, Klein S, Grusser SM, Flor H, Schumann G, Mann K, Buchel C (2005): Amygdala‐prefrontal coupling depends on a genetic variation of the serotonin transporter. Nat Neurosci 8:20–21. [DOI] [PubMed] [Google Scholar]
- Henningsson S, Borg J, Lundberg J, Bah J, Lindström M, Ryding E, Jovanovic H, Saijo T, Inoue M, Rosén I, Träskman‐Bendz L, Farde L, Eriksson E (2009): Genetic variation in brain‐derived neurotrophic factor is associated with serotonin transporter but not serotonin‐1A receptor availability in men. Biol Psychiatry 66:477–485. [DOI] [PubMed] [Google Scholar]
- Holmes A (2008): Genetic variation in cortico‐amygdala serotonin function and risk for stress‐related disease. Neurosci Biobehav Rev 32:1293–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holst KK, Budtz‐Jørgensen E (2013): Linear latent variable models: The lava‐package. Comput Stat 28:1385–1452. [Google Scholar]
- Hong IK, Chung ST, Kim HK, Kim YB, Son YD, Cho ZH (2007): Ultra fast symmetry and SIMD‐based projection‐backprojection (SSP) algorithm for 3‐D PET image reconstruction. IEEE Trans Med Imaging 26:789–803. [DOI] [PubMed] [Google Scholar]
- Hosang GM, Shiles C, Tansey KE, McGuffin P, Uher R (2014): Interaction between stress and the BDNF Val66Met polymorphism in depression: A systematic review and meta‐analysis. BMC Med 12:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu XZ, Lipsky RH, Zhu G, Akhtar LA, Taubman J, Greenberg BD, Xu K, Arnold PD, Richter MA, Kennedy JL, Murphy DL, Goldman D (2006): Serotonin transporter promoter gain‐of‐function genotypes are linked to obsessive‐compulsive disorder. Am J Hum Genet 78:815–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innis RB, Cunningham VJ, Delforge J, Fujita M, Gjedde A, Gunn RN, Holden J, Houle S, Huang SC, Ichise M, Iida H, Ito H, Kimura Y, Koeppe RA, Knudsen GM, Knuuti J, Lammertsma AA, Laruelle M, Logan J, Maguire RP, Mintun MA, Morris ED, Parsey R, Price JC, Slifstein M, Sossi V, Suhara T, Votaw JR, Wong DF, Carson RE (2007): Consensus nomenclature for in vivo imaging of reversibly binding radioligands. J Cereb Blood Flow Metab 27:1533–1539. [DOI] [PubMed] [Google Scholar]
- Jennings KA, Licht CL, Bruce A, Lesch KP, Knudsen GM, Sharp T (2011): Genetic variation in 5‐hydroxytryptamine transporter expression causes adaptive changes in 5‐HT4 receptor levels. Int J Neuropsychopharmacol 2011/08/19:1–9. [DOI] [PubMed] [Google Scholar]
- Kalbitzer J, Erritzoe D, Holst KK, Nielsen FA, Marner L, Lehel S, Arentzen T, Jernigan TL, Knudsen GM (2010): Seasonal changes in brain serotonin transporter binding in short serotonin transporter linked polymorphic region‐allele carriers but not in long‐allele homozygotes. Biol Psychiatry 67:1033–1039. [DOI] [PubMed] [Google Scholar]
- Klein AB, Santini MA, Aznar S, Knudsen GM, Rios M (2010a): Changes in 5‐HT2A‐mediated behavior and 5‐HT2A‐ and 5‐HT1A receptor binding and expression in conditional brain‐derived neurotrophic factor knock‐out mice. Neuroscience 169:1007–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein AB, Trajkovska V, Erritzoe D, Haugbol S, Madsen J, Baare W, Aznar S, Knudsen GM (2010b): Cerebral 5‐HT2A receptor and serotonin transporter binding in humans are not affected by the val66met BDNF polymorphism status or blood BDNF levels. J Cereb Blood Flow Metab 30:e1–e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kourmouli N, Samakouri M, Mamatsiou A, Trypsianis G, Livaditis M, Veletza S (2013): Effect of BDNF Val66Met and serotonin transporter 5‐HTTLPR polymorphisms on psychopathological characteristics in a sample of university students. Psychiatr Genet 23:188–197. [DOI] [PubMed] [Google Scholar]
- Lan MJ, Ogden RT, Huang YY, Oquendo MA, Sullivan GM, Miller J, Milak M, Mann JJ, Parsey RV (2014): Genetic variation in brain‐derived neurotrophic factor val66met allele is associated with altered serotonin‐1A receptor binding in human brain. Neuroimage 94:33–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesch KP, Bengel D, Heils A, Sabol SZ, Greenberg BD, Petri S, Benjamin J, Muller CR, Hamer DH, Murphy DL (1996): Association of anxiety‐related traits with a polymorphism in the serotonin transporter gene regulatory region. Science 274:1527–1531. [DOI] [PubMed] [Google Scholar]
- Liang KY, Zeger SL (1986): Longitudinal data analysis using generalized linear models. Biometrika 73:13–22. [Google Scholar]
- Licht CL, Marcussen AB, Wegener G, Overstreet DH, Aznar S, Knudsen GM (2009): The brain 5‐HT4 receptor binding is down‐regulated in the Flinders Sensitive Line depression model and in response to paroxetine administration. J Neurochem 109:1363–1374. [DOI] [PubMed] [Google Scholar]
- Lucas G, Compan V, Charnay Y, Neve RL, Nestler EJ, Bockaert J, Barrot M, Debonnel G (2005): Frontocortical 5‐HT4 receptors exert positive feedback on serotonergic activity: Viral transfections, subacute and chronic treatments with 5‐HT4 agonists. Biol Psychiatry 57:918–925. [DOI] [PubMed] [Google Scholar]
- Lyons WE, Mamounas LA, Ricaurte GA, Coppola V, Reid SW, Bora SH, Wihler C, Koliatsos VE, Tessarollo L (1999): Brain‐derived neurotrophic factor‐deficient mice develop aggressiveness and hyperphagia in conjunction with brain serotonergic abnormalities. Proc Natl Acad Sci USA 96:15239–15244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen K, Haahr MT, Marner L, Keller SH, Baare WF, Svarer C, Hasselbalch SG, Knudsen GM (2011a): Age and sex effects on 5‐HT(4) receptors in the human brain: A [(11)C]SB207145 PET study. J Cereb Blood Flow Metab 2011/03/03:1475–1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen K, Marner L, Haahr M, Gillings N, Knudsen GM (2011b): Mass dose effects and in vivo affinity in brain PET receptor studies–a study of cerebral 5‐HT4 receptor binding with [11C]SB207145. Nucl Med Biol 38:1085–1091. [DOI] [PubMed] [Google Scholar]
- Marner L, Gillings N, Comley RA, Baare WF, Rabiner EA, Wilson AA, Houle S, Hasselbalch SG, Svarer C, Gunn RN, Laruelle M, Knudsen GM (2009): Kinetic modeling of 11C‐SB207145 binding to 5‐HT4 receptors in the human brain in vivo. J Nucl Med 50:900–908. [DOI] [PubMed] [Google Scholar]
- Marner L, Gillings N, Madsen K, Erritzoe D, Baaré WFC, Svarer C, Hasselbalch SG, Knudsen GM (2010): Brain imaging of serotonin 4 receptors in humans with [11C]SB207145‐PET. Neuroimage 50:855–861. [DOI] [PubMed] [Google Scholar]
- Mattson MP, Maudsley S, Martin B (2004): BDNF and 5‐HT: A dynamic duo in age‐related neuronal plasticity and neurodegenerative disorders. Trends Neurosci 27:589–594. [DOI] [PubMed] [Google Scholar]
- Molendijk ML, Bus BAA, Spinhoven P, Kaimatzoglou A, Voshaar RCO, Penninx BWJH, van IJzendoorn MH, Elzinga BM (2012): A systematic review and meta‐analysis on the association between BDNF val66met and hippocampal volume: genuine effect or a winners curse? Am J Med Genet 159B:731–740. [DOI] [PubMed] [Google Scholar]
- Outhred T, Das P, Dobson‐Stone C, Griffiths K, Felmingham KL, Bryant RA, Malhi G, Kemp AH (2012): The functional epistasis of 5‐HTTLPR and BDNF Val66Met on emotion processing: A preliminary study. Brain Behav 2:778–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pezawas L, Meyer‐Lindenberg A, Goldman AL, Verchinski BA, Chen G, Kolachana BS, Egan MF, Mattay VS, Hariri AR, Weinberger DR (2008): Evidence of biologic epistasis between BDNF and SLC6A4 and implications for depression. Mol Psychiatry 13:709–716. [DOI] [PubMed] [Google Scholar]
- Praschak‐Rieder N, Kennedy J, Wilson AA, Hussey D, Boovariwala A, Willeit M, Ginovart N, Tharmalingam S, Masellis M, Houle S, Meyer JH (2007): Novel 5‐HTTLPR allele associates with higher serotonin transporter binding in putamen: A [(11)C] DASB positron emission tomography study. Biol Psychiatry 62:327–331. [DOI] [PubMed] [Google Scholar]
- R Core Team (2013): A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria.
- Reimold M, Smolka MN, Schumann G, Zimmer A, Wrase J, Mann K, Hu XZ, Goldman D, Reischl G, Solbach C, Machulla HJ, Bares R, Heinz A (2007): Midbrain serotonin transporter binding potential measured with [11C]DASB is affected by serotonin transporter genotype. J Neural Transm 114:635–639. [DOI] [PubMed] [Google Scholar]
- Sanchez BN, Budtz‐Jørgensen E, Ryan LM, Hu H (2005): Structural equation models. J Am Stat Assoc 100:1443–1455. [Google Scholar]
- Sibille E, Lewis DA (2006): SERT‐ainly involved in depression, but when? Am J Psychiatry 163:8–11. [DOI] [PubMed] [Google Scholar]
- Siuciak JA, Clark MS, Rind HB, Whittemore SR, Russo AF (1998): BDNF induction of tryptophan hydroxylase mRNA levels in the rat brain. J Neurosci Res 52:149–158. [DOI] [PubMed] [Google Scholar]
- Soliman F, Glatt CE, Bath KG, Levita L, Jones RM, Pattwell SS, Jing D, Tottenham N, Amso D, Somerville LH, Voss HU, Glover G, Ballon DJ, Liston C, Teslovich T, Van Kempen T, Lee FS, Casey BJ (2010): A genetic variant BDNF polymorphism alters extinction learning in both mouse and human. Science 327:863–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sureau FC, Reader AJ, Comtat C, Leroy C, Ribeiro MJ, Buvat I, Trebossen R (2008): Impact of image‐space resolution modeling for studies with the high‐resolution research tomograph. J Nucl Med 49:1000–1008. [DOI] [PubMed] [Google Scholar]
- Surguladze SA, Elkin A, Ecker C, Kalidindi S, Corsico A, Giampietro V, Lawrence N, Deeley Q, Murphy DG, Kucharska‐Pietura K, Russell TA, McGuffin P, Murray R, Phillips ML (2008): Genetic variation in the serotonin transporter modulates neural system‐wide response to fearful faces. Genes Brain Behav 7:543–551. [DOI] [PubMed] [Google Scholar]
- Svarer C, Madsen K, Hasselbalch SG, Pinborg LH, Haugbol S, Frokjaer VG, Holm S, Paulson OB, Knudsen GM (2005): MR‐based automatic delineation of volumes of interest in human brain PET images using probability maps. Neuroimage 24:969–979. [DOI] [PubMed] [Google Scholar]
- Terracciano A, Tanaka T, Sutin AR, Deiana B, Balaci L, Sanna S, Olla N, Maschio A, Uda M, Ferrucci L, Schlessinger D, Costa PT Jr (2010): BDNF Val66Met is Associated with Introversion and Interacts with 5‐HTTLPR to Influence Neuroticism. Neuropsychopharmacology 35:1083–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal R, Valdizan EM, Mostany R, Pazos A, Castro E (2009): Long‐term treatment with fluoxetine induces desensitization of 5‐HT4 receptor‐dependent signalling and functionality in rat brain. J Neurochem 2009/06/16:1120–1127. [DOI] [PubMed] [Google Scholar]
- Voineskos AN, Lerch JP, Felsky D, Shaikh S, Rajji TK, Miranda D, Lobaugh NJ, Mulsant BH, Pollock BG, Kennedy JL (2011): The brain‐derived neurotrophic factor Val66Met polymorphism and prediction of neural risk for Alzheimer disease. Arch Gen Psychiatry 68:198–206. [DOI] [PubMed] [Google Scholar]
- Wang DD, Tian T, Dong Q, Xu XF, Yu H, Wang Y, Chen ZY (2014): Transcriptome profiling analysis of the mechanisms underlying the BDNF Val66Met polymorphism induced dysfunctions of the central nervous system. Hippocampus 24:65–78. [DOI] [PubMed] [Google Scholar]
- Wang L, Ashley‐Koch A, Steffens DC, Krishnan KR, Taylor WD (2012): Impact of BDNF Val66Met and 5‐HTTLPR polymorphism variants on neural substrates related to sadness and executive function. Genes Brain Behav 11:352–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse EG, Xu B (2009): New insights into the role of brain‐derived neurotrophic factor in synaptic plasticity. Mol Cell Neurosci 42:81–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods RP, Cherry SR, Mazziotta JC (1992): Rapid automated algorithm for aligning and reslicing PET images. J Comput Assist Tomogr 16:620–633. [DOI] [PubMed] [Google Scholar]
- Yu H, Wang DD, Wang Y, Liu T, Lee FS, Chen ZY (2012): Variant brain‐derived neurotrophic factor Val66Met polymorphism alters vulnerability to stress and response to antidepressants. J Neurosci 32:4092–4101. [DOI] [PMC free article] [PubMed] [Google Scholar]


