Abstract
Because hypoperfusion of brain tissue precedes atrophy in dementia, the detection of dementia may be advanced by the use of perfusion information. Such information can be obtained noninvasively with arterial spin labeling (ASL), a relatively new MR technique quantifying cerebral blood flow (CBF). Using ASL and structural MRI, we evaluated diagnostic classification in 32 prospectively included presenile early stage dementia patients and 32 healthy controls. Patients were suspected of Alzheimer's disease (AD) or frontotemporal dementia. Classification was based on CBF as perfusion marker, gray matter (GM) volume as atrophy marker, and their combination. These markers were each examined using six feature extraction methods: a voxel‐wise method and a region of interest (ROI)‐wise approach using five ROI‐sets in the GM. These ROI‐sets ranged in number from 72 brain regions to a single ROI for the entire supratentorial brain. Classification was performed with a linear support vector machine classifier. For validation of the classification method on the basis of GM features, a reference dataset from the AD Neuroimaging Initiative database was used consisting of AD patients and healthy controls. In our early stage dementia population, the voxelwise feature‐extraction approach achieved more accurate results (area under the curve (AUC) range = 86 − 91%) than all other approaches (AUC = 57 − 84%). Used in isolation, CBF quantified with ASL was a good diagnostic marker for dementia. However, our findings indicated only little added diagnostic value when combining ASL with the structural MRI data (AUC = 91%), which did not significantly improve over accuracy of structural MRI atrophy marker by itself. Hum Brain Mapp 35:4916–4931, 2014. © 2014 Wiley Periodicals, Inc.
Keywords: Alzheimer's disease, arterial spin labeling, classification, diagnostic imaging, frontotemporal dementia, magnetic resonance imaging, presenile dementia, support vector machines
Introduction
The growing prevalence of dementia is an increasing health problem [Alzheimer's Association, 2011]. Early and accurate diagnosis is beneficial for patient care, aiding the planning of care and living arrangements, and preserving function and independence for as long as possible [Paquerault, 2012; Prince et al., 2011]. In addition, an early and accurate diagnosis increases research opportunities into understanding the disease process and into the development of treatments. However, early stage diagnosis can be very difficult, as clinical symptoms and the loss of brain tissue, atrophy, may not yet be marked. To aid the diagnosis of dementia, machine‐learning techniques applied to imaging and associated data are of interest. These techniques may improve diagnosis of individual patients, since they are trained on group differences, which may not be noted from qualitative visual inspection of brain imaging data. The machine‐learning techniques use labeled data to train a classifier to categorize two groups (e.g., patients and controls) based on features derived from brain imaging or other data. Several studies demonstrated the successful classification of dementia based on atrophy derived from structural MRI using such machine‐learning methods, [e.g., Cuingnet et al., 2011; Davatzikos et al., 2008; Duchesne et al., 2008; Fan et al., 2008a, b; Klöppel et al., 2008; Koikkalainen et al., 2012; Magnin et al., 2009; Vemuri et al., 2008; Wolz et al., 2011].
Because hypoperfusion of brain tissue precedes atrophy in dementia [Jack et al., 2010; Sperling et al., 2011], early diagnosis may be advanced by the use of perfusion information. Such information can be obtained with arterial spin labeling (ASL), an MRI technique, which measures brain perfusion noninvasively, without the need for injecting contrast media [Detre et al., 1992; Williams et al., 1992]. ASL uses inversion labeling of arterial blood to quantify the cerebral blood flow (CBF).
Although previous studies have indicated that perfusion information may be valuable for diagnosing early stage dementia [Binnewijzend et al., 2013; Wang et al., 2013; Wolk and Detre, 2012], to the best of our knowledge only three studies have applied machine‐learning techniques to ASL data showing the diagnostic value of ASL for Alzheimer's disease (AD) using linear discriminant analysis [Dashjamts et al., 2011], for frontotemporal dementia (FTD) using logistic regression methods [Du et al., 2006], and for mild cognitive impairment (MCI) using regression preceded by local linear embedding [Schuff et al., [Link]].
In this work, we studied the value of CBF as quantified with ASL for differentiation of dementia patients from healthy controls using machine‐learning techniques. This was studied on a patient group consisting of presenile (disease onset <65 years), early stage dementia patients suspected of AD or FTD and a matched control group (Group I). For comparison of the structural‐MRI‐based classifications with previous work [e.g., Cuingnet et al., 2011; Davatzikos et al., 2008; Duchesne et al., 2008; Fan et al., 2008a, b; Klöppel et al., 2008; Koikkalainen et al., 2012; Magnin et al., 2009; Vemuri et al., 2008; Wolz et al., 2011], we also included a reference dataset from the AD neuroimaging initiative (ADNI) database (Group II). We evaluated several linear support vector machine (SVM) classification methods. Two aspects of the classification model were examined: (1) the type of data and (2) the feature‐extraction approach. For the first aspect, we included three groups of data in the analysis: CBF as perfusion marker on its own, gray matter (GM) volume as an atrophy marker, obtained from high‐resolution structural T1‐weighted (T1w) MRI, and their combination. CBF and GM features were combined using four methods: feature concatenation, feature multiplication, and classifier combination using both the product rule and the mean rule [Tax et al., 2000]. For the second aspect regarding feature extraction, we examined the two main approaches that were used in previously published dementia‐classification papers: voxel‐wise [e.g., Klöppel et al., 2008] and region of interest (ROI)‐wise feature extraction [e.g., Magnin et al., 2009].
Materials and Methods
Participants
Group I consisted of participants from the Iris study, which was approved by the review board at our institution. Informed consent was obtained from all participants. For this group, 32 presenile patients with early stage dementia (17 male, age = 62.8 ± 4.1 years) were recruited from the outpatient clinic. As presenile dementia is defined by the age at disease onset (<65 years), this does not exclude a 69‐year‐old patient to suffer from a presenile form of dementia. Therefore, we considered patients in the age range of 45–70 years and with a Mini Mental State Examination (MMSE) score ≥ 20 for inclusion. Exclusion criteria were normal pressure hydrocephalus, Huntington's disease, cerebral vascular disease, psychiatric disease, alcohol abuse, brain tumor, epilepsy, or encephalitis. All patients underwent neurological and neuropsychological examination as part of their routine diagnostic work up, and diagnosis of dementia was established in a multidisciplinary clinical meeting on the basis of neurological, neuropsychological, and conventional‐imaging criteria. Patients who were subsequently suspected of having either AD [Dubois et al., 2007, 2010; McKhann et al., 2011] or FTD [Rascovsky et al., 2011] were asked to participate in this study. The participating patients had a MMSE score of 26.6 ± 2.9 (mean ± standard deviation) out of 30. This indicated that cognitive function was not yet much impaired, and confirmed that dementia was still at an early stage. Based on patient history and neuropsychological testing, every patient was assigned a provisional diagnostic label in the multidisciplinary meeting. These labels were probable AD (n = 8), possible AD (n = 3), AD/FTD (n = 9), possible FTD (n = 8), and probable FTD (n = 3). We additionally included 32 age‐matched healthy controls (18 male, age = 62.0 ± 4.4 years). Control subjects had no history of neurological or psychiatric disease and did not have contraindications for MRI. An MMSE score was obtained from 23 of the controls, which was 29.0 ± 1.0 on average.
Group II consisted of participants from the ADNI and was used as reference dataset for validation of the pipeline for classification based on GM features. This group was included to enable comparison with results from previous articles. The ADNI was launched in 2003 by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, the Food and Drug Administration, private pharmaceutical companies and non‐profit organizations, as a $60 million, 5‐year public‐private partnership. The primary goal of ADNI has been to test whether serial MRI, PET, other biological markers, and clinical and neuropsychological assessment can be combined to measure the progression of MCI and early AD. The ADNI cohort used in this article is adopted from the study of Cuingnet et al. [2011], from which we selected the AD patient group and the elderly control group. The inclusion criteria for participants were defined in the ADNI GO protocol (http://www.adni-info.org/Scientists/Pdfs/ADNI_Go_Protocol.pdf). The patient group consisted of 137 patients (67 male, age = 76.0 ± 7.3 years, MMSE = 23.2 ± 2.0), and the control group of 162 participants (76 male, age = 76.3 ± 5.4 years, MMSE = 29.2 ± 1.0).
MR Imaging
For Group I, images were acquired on a 3T MR scanner (Discovery MR750, GE Healthcare, Milwaukee, WI) using a dedicated 8‐channel brain coil. For each participant, a T1w image and a pseudo‐continuous ASL image [Dai et al., 2008; Wu et al., 2007] were acquired. T1w images were acquired with a 3D inversion recovery fast spoiled gradient‐recalled echo sequence with the following parameters: inversion time (TI) = 450 ms, repetition time (TR) = 7.9 ms, and echo time (TE) = 3.1 ms. These T1w images had a resolution of 0.94 × 0.94 mm in the sagittal plane and a slice thickness of 1.0 mm. For 10 of the controls, T1w images were acquired axially with a resolution of 0.94 × 0.94 × 0.8 mm and acquisition parameters of TI = 450 ms, TR = 6.1 ms, and TE = 2.1 ms. Acquisition time was around 4 min. The ASL data were acquired with a postlabeling delay time of 1.53 s using background suppression. 3D acquisition was performed with an interleaved stack of spiral readouts using 512 sampling points on 8 spirals, resulting in an isotropic 3.3 mm resolution in a 24 cm field of view. Other imaging parameters were TR = 4.6 s, TE = 10.5 ms, number of excitations = 3, labeling pulse duration = 1.45 s. The reconstructed voxel size was 1.9 × 1.9 × 4 mm. For the ASL data, the acquisition time was 4:30 min.
For Group II, T1w imaging data were acquired at 1.5T. Acquisition had been performed according to the ADNI acquisition protocol [Jack et al., 2008].
Image Processing
Probabilistic tissue segmentations were obtained for white matter (WM), GM, and cerebrospinal fluid on the T1w image using the unified tissue segmentation method [Ashburner and Friston, 2005] of SPM8 (Statistical Parametric Mapping, London, UK). To minimize errors in the image processing, visual inspections of the tissue maps were performed after specific image processing steps. The tissue segmentation procedures did not compensate for WM lesions and infarcts, but this was not necessary as patients with a history of cerebrovascular accidents (CVA) or CVA reported in their MRI examination were excluded from our study. Accordingly, since the study population was quite young and vascular dementia patients were not included, only few WM lesions were present.
For Group I, ASL imaging data consisted of a difference image (ΔA) and a control image (A 0) [Buxton et al., 1998]. To obtain an indication of the image quality, we estimated the signal‐to‐noise ratio (SNR) of the ΔA images of five randomly chosen patients and five controls. SNR was defined as,
| (1) |
in which is the mean ΔA in a small ROI in the brain, and is the standard deviation of the signal in a small ROI in the background. For the patients the SNR was 20.3 ± 7.7 (mean ± std), and for the controls 27.0 ± 5.4. Figure 1A shows an example ΔA scan for a patient with SNR = 24.4.
Figure 1.
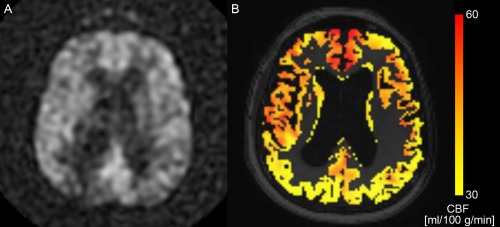
(A) ASL difference scan (ΔA) of a dementia patient (SNR = 24.4), and (B) the corresponding CBF map in the GM after partial volume correction in color overlay. The background image in (B) is the T1w image. [Color figure can be viewed in the online issue, which is available at http://wileyonlinelibrary.com.]
For each subject, T1w images were rigidly registered to the A 0 images using Elastix registration software [Klein et al., 2010] by maximizing mutual information [Thévenaz and Unser, 2000] within a mask. For the T1w images, a dilated brain mask obtained with the brain extraction tool [Smith, 2002] was used, and for the A 0 image, voxels with zero intensity, outside the brain, were masked out. All registrations were visually checked. Tissue maps and brain masks were transformed to ASL space accordingly. In the ASL space, ΔA and A 0 were corrected for partial volume effects using local linear regression based on the tissue probability maps using a 3D kernel of 3 × 3 × 3 voxels [Asllani et al., 2008; Oliver et al., 2012]. CBF maps were quantified using the single‐compartment model by Buxton et al. [1998] as implemented by the scanner manufacturer. Figure 1B shows the partial volume corrected CBF map, which corresponds to the ΔA image in Figure 1A. After quantification, CBF maps were transformed to T1w space. In the analysis only the CBF in the GM was used, as cortical CBF is of primary interest in the disease processes studied here. In addition, quantification of CBF with ASL in WM is less reliable than in GM [Van Gelderen et al., 2008].
For partial volume correction of the ASL images and for estimation of intracranial volume, a brain mask was required for each subject. This brain mask was constructed using a multiatlas segmentation approach. We performed brain extraction [Smith, 2002] on the T1w images associated with a set of 30 atlases [Gousias et al., 2008; Hammers et al., 2003], checked the brain extractions visually, and adjusted extraction parameters if needed. The extracted brains were transformed to each subject's T1w image and the labels were fused, resulting in a brain mask for each subject. The multiatlas segmentation approach is explained in more detail the next section.
Common Template Space and Individual Regions‐of‐Interest (ROIs)
For each subject, we defined two image spaces, which refer to the coordinate systems of the subject's ASL and T1w scan respectively: an ASL‐space (Ω ASL) and a T1w‐space (Ω T1w). Additionally, a common template space (Ω Template) was defined on the basis of the T1w images of all subjects. For registration of images, the following notation is used: a transformation T is applied to an image (moving image, M) to optimally fit another image (fixed image, F). The deformed moving image can be written as M(T). Figure 2 illustrates the image spaces and the transformations between them.
Figure 2.
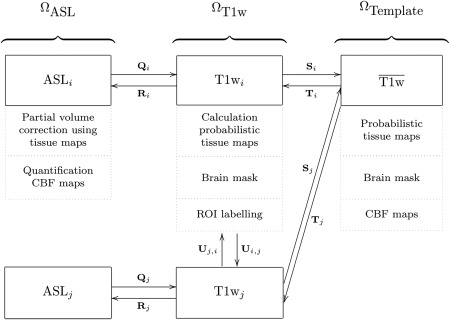
Image spaces including processed images in these spaces: ASL space (Ω ASL), T1w space (Ω T1w), and the template space (Ω Template). Transformations between the image spaces are indicated by Q, R, S, T, and U. The arrows are pointing from the fixed to the moving domain. Different subjects are represented by i and j. From all T1wi, a template space image (T1wj) is calculated. In each image space, the dotted boxes represent the processed images.
The template space (Ω Template) was constructed based on the T1w images of all subjects using a procedure that avoids bias towards any of the individual T1w images [Seghers et al., 2004]. In this approach, the coordinate transformations from the template space to the subject's T1w space (T i: Ω Template → Ω T1w i) were derived from pairwise image registrations. For computation of T i, the T1w image of an individual subject (T1wi) was registered to all other subjects' images (T1wj) using T1wi as the fixed image. This resulted in a set of transformations U i,j: Ω T1w i → Ω T1w j . By averaging the transformations U i,j, the transformation S i: Ω T1w i → Ω Template was calculated:
| (2) |
The transformation T i was calculated as an inversion of S i: T i = S i −1. Note that the identity transformation U i,i is also included in [2]. The pairwise registrations were performed using a similarity, affine, and nonrigid B‐spline transformation model consecutively. A similarity transformation is a rigid transformation including isotropic scaling. The nonrigid B‐spline registration used a three‐level multiresolution framework with isotropic control‐point spacing of 24, 12, and 6 mm in the three resolutions, respectively. A T1w template image was created by averaging the deformed individual T1w images. This template was thresholded and dilated to create a dilated brain mask for this population. To prevent background information in the T1w images from influencing the process, the complete pairwise registration procedure was repeated masking the T1wi images with these dilated brain masks in Ω T1w i. To check if subjects were properly registered to the template space, the final T1w template image was visually inspected. Processed images (Pi) were transformed to template space using Pi(T i) for the brain masks and tissue maps, and using Pi(R i(T i)) for the CBF maps with R i: Ω T1w i → Ω ASL i . We defined a common GM mask in template space by combining the GM segmentations of all subjects using majority vote. The voxel‐wise CBF features included only voxels within this common GM mask.
Five sets of ROIs in the GM were constructed for every subject individually in T1w space (Ω T1w) differing in the number and size of ROIs (Fig. 3): (a) regional labeling of the supratentorial brain (region; 72 features), (b) selection of brain regions affected by AD or FTD based on the literature (selection; 28 features) [Foster et al., 2008; Fukuyama et al., 1994; Herholz et al., 2007; Ishii et al., 1996, 1998, 1997a, b, 2000; Johannsen et al., 2000; Minoshima et al., 1997; Santens et al., 2001; Scarmeas et al., 2004; Womack et al., 2011], (c) brain lobes (lobe; occipital, temporal, parietal, frontal lobes and central structures in both hemispheres; 10 features), (d) hemispheres (hemisphere; 2 features), and (e) the total GM in the entire supratentorial brain (brain; 1 feature). The ROI‐sets were constructed using a multiatlas segmentation procedure. Thirty labeled T1w images containing 83 ROIs each [Gousias et al., 2008; Hammers et al., 2003] were used as atlas images. The atlas images were registered to the subject's T1w image using a rigid, affine, and nonrigid B‐spline transformation model consecutively. A rigid transformation model was used instead of the similarity transformation model that was used in the template space registrations. The rigid model was used because the similarity transformation failed here, probably due to the cropping around the brain, which had been performed in the atlas images to remove most nonbrain tissue. Registration was performed by maximization of mutual information [Thévenaz and Unser, 2000] within dilated brain masks [Smith, 2002]. For initialization, the dilated brain masks were rigidly registered. For nonrigid registration, the same multiresolution settings were used as in the template‐space construction. The subjects' T1w images were corrected for inhomogeneities to improve registrations [Tustison et al., 2010]. Labels were fused using a majority voting algorithm [Heckemann et al., 2006]. All final region segmentations were visually inspected. The brain stem, corpus callosum, third ventricle, lateral ventricles, cerebellum, and substantia nigra were excluded. For construction of the lobe, hemisphere, and brain GM ROIs, the regions were fused in the original atlas images before transformation to Ω T1w.
Figure 3.
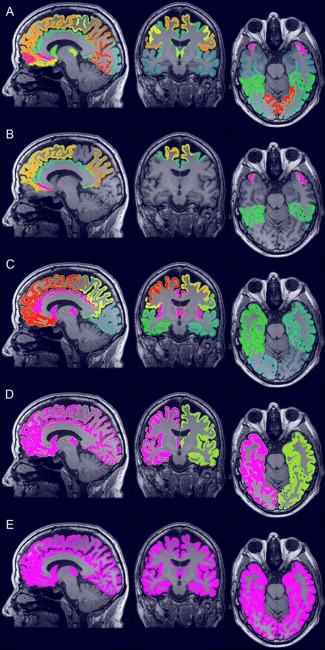
The five ROI‐sets for ROI‐wise feature extraction of the GM: (A) region (72 features), (B) selection (28 features), (C) lobe (10 features), (D) hemisphere (2 features), and (E) brain (1 feature). [Color figure can be viewed in the online issue, which is available at http://wileyonlinelibrary.com.]
Classification Methods
We evaluated two aspects of dementia classification, which are discussed in this section: (1) the type of data and (2) the method used to extract features. For the first aspect, classifications were performed using three types of data: CBF values quantified with ASL, GM volumes derived from the T1w images, and their combination. Four combination strategies were explored. In the first strategy, the feature vectors for CBF and GM were concatenated into one large feature vector, which was used to train the classifier ([CBF GM], feature concatenation). In the second strategy, we multiplied the CBF and GM features element‐wise (CBF × GM, feature multiplication). In the third and fourth strategy, two separate SVM models for CBF and GM were combined by respectively the product rule , and the mean rule [Tax et al., 2000]. In these approaches, the combined classifier was obtained by multiplication or averaging of the posterior class probabilities (ω) of the single modality classifiers and by renormalizing the posterior probabilities. As an SVM does not naturally output posterior probabilities, these were obtained from the distance between the sample and the classifier by applying a logistic function [Duin and Tax, 1998]. For the second aspect, six methods were used to extract features from the data: a voxel‐wise method (voxel) and a ROI‐wise approach using the five previously defined ROI‐sets (region, selection, lobe, hemisphere, and brain). These methods were applied in turn to the T1w data, ASL data and combined data. Voxel‐wise features were defined as CBF intensities and GM probabilistic segmentations in the template space (Ω Template) [Cuingnet et al., 2011; Klöppel et al., 2008]. For the CBF features, only voxels within the common GM mask were included. For the GM segmentations, we performed a modulation step, that is, multiplication by the Jacobian determinant of the deformation field (Fig. 1, transformation T i), to take account of compression and expansion [Ashburner and Friston, 2000]. This modulation step ensures that the overall GM volume was not changed by the transformation to template space. The ROI‐wise features were calculated in subject T1w space (Ω T1w) for the five ROI‐sets. The CBF features were defined as the mean CBF intensity in the GM, and the GM features as the GM volume obtained from the probabilistic GM maps [Cuingnet et al., 2011; Magnin et al., 2009]. To correct for head size, the GM features were divided by intracranial volume. All features were normalized to have zero mean and unit variance.
Analysis and Statistics
For classification, linear SVM classifiers [Vapnik, 1995] were applied using the LibSVM software package [Chang and Lin, 2011]. Classification performance was quantified by the area under the curve (AUC). The SVM C‐parameter was optimized using grid search on the training set with LOO cross‐validation.
On Group I, the SVM classifiers were trained and tested using both LOO cross‐validation and iterated four‐fold cross‐validation. LOO cross‐validation was used for calculation of classification performance because it uses the maximum number of available data for training of the classifier, resulting in the best possible classifier using those data and features. In four‐fold cross‐validation, however, only a part of the available training data is used, which allows for calculation of the standard deviations on the AUC. These standard deviations provide an indication of the robustness of the classifier, that is, the dependence of the performance on the sampling of training and test sets. For the iterated four‐fold cross‐validation, classification was performed iteratively on four groups, each consisting of eight patients and eight control subjects, using repeatedly three groups for training and one group for testing. The total number of iterations was 50. To assess whether ASL improved the performance of the classifications relative to those based on structural GM features only, we performed McNemar's binomial exact test.
For detection of features associated with group differences using the SVM classifier, we calculated statistical significance maps (P‐maps). Using permutation testing, a null distribution for the features was obtained using 5000 permutations [Mourão‐Miranda et al., 2005; Wang et al., 2007]. The P‐maps were calculated for every feature extraction method on both the CBF and GM data. We used a P‐value threshold of α = 0.05 and we did not correct for multiple comparisons, as permutation testing has a low false positive detection rate [Gaonkar and Davatzikos, 2013]. Voxel‐wise P‐maps were visually inspected to identify clusters of significant voxels.
On Group II, we evaluated the classifications based on GM features. Instead of cross‐validation, separate training and test sets were used for classification. The participants were randomly split into two groups of the same size, a training set and a test set, while preserving the age and sex distribution [Cuingnet et al., 2011]. All postprocessing and classification methods were identical to those of Group I, except for the construction of the template space, which is for Group II only based on the training set. In Cuingnet et al. [2011], classification results are presented as the highest sum of sensitivity and specificity. For comparison, we also included this measure for Group II.
Results
Group I
Figure 4 shows the classification results for (a) the LOO cross‐validation and (b) the iterated four‐fold cross‐validation. The voxelwise feature‐extraction approach (AUC range = 86–91%) resulted in higher performance than all other approaches (AUC range = 57–84%). CBF and GM single modality classifications performed similarly in the voxel‐wise approach, but in the ROI‐wise approaches the AUC for the CBF classification declined with decreasing feature numbers.
Figure 4.
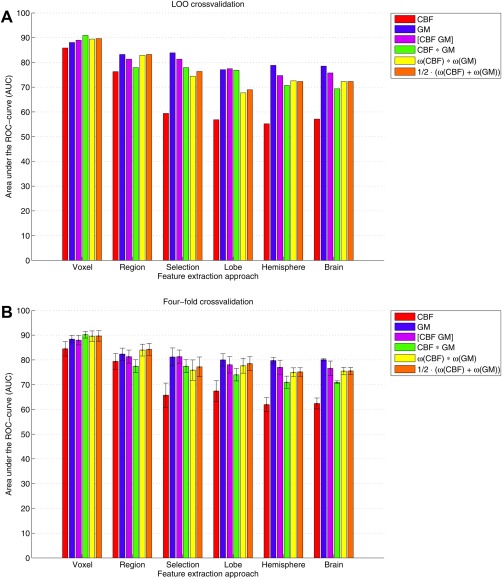
Classification performances quantified by the area under the ROC‐curve (AUC) determined using (A) leave‐one‐out and (B) four‐fold cross‐validation. For the four‐fold cross‐validation, the bars represent mean AUC and the standard deviations are shown as error bars. Features were extracted using two approaches: voxel‐wise and ROI‐wise using 5 GM ROI‐sets (region, selection, lobe, hemisphere, and brain). We included CBF data, GM data, and their combination using (1) feature concatenation ([CBF GM]), (2) feature multiplication (CBF × GM), (3) the product rule , and (4) the mean rule . [Color figure can be viewed in the online issue, which is available at http://wileyonlinelibrary.com.]
For the voxel‐wise method, the combination of CBF and GM data (AUC range = 89–91%) performed somewhat better than classification based on a single modality (AUC = 86–88%) as can be appreciated from the ROC‐curves shown in Figure 5. For the other approaches, the GM classification performed best (AUC range = 77–84%) and this was not improved by adding the CBF data (AUC range = 73–83%). In the voxel‐wise approach, the feature multiplication method had a slightly higher performance than the other approaches, but overall the performances of the four combination methods were similar. For the region‐wise method, combination of CBF and GM by the product and mean combination methods (AUC = 83%) performed better than feature concatenation or multiplication (AUC range = 78–81%), while in the other ROI‐wise approaches with fewer ROIs, feature concatenation was the best performing combination method.
Figure 5.
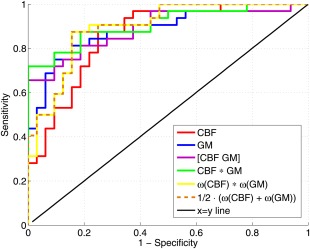
Receiver operator characteristic (ROC) curves for the voxel‐wise classifications using LOO cross‐validation: based on CBF features, GM features, and the combination of both using feature concatenation ([CBF GM]), feature multiplication (CBF × GM), the product rule , and the mean rule . [Color figure can be viewed in the online issue, which is available at http://wileyonlinelibrary.com.]
The McNemar tests showed no significant differences between the performance of the voxel‐wise classification based on GM features and the other voxel‐wise classifications: CBF (P = 0.38), the mean rule (P = 0.38), and the other combination methods (all P = 1.0).
Generally, the mean classification performances for the iterated four‐fold cross‐validation were similar to those obtained with LOO cross‐validation (Fig. 4B). The standard deviations, indicated by the error bars in Figure 4B, showed that the classifications had a relatively small variance and were rather robust.
Posterior probabilities for the voxel‐wise classifications are shown in Figure 6 and do not indicate that the type of dementia influences the success for patients of being correctly classified, as AD and FTD patients cannot be clearly separated in the plot. It should be noted that the classifiers were not trained for this specific differentiation.
Figure 6.
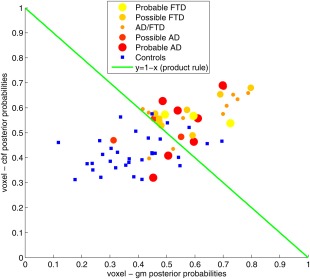
Scatter plot of the posterior probabilities for the voxel‐wise classifications based on GM features (x‐axis) and CBF features (y‐axis). Patients are represented by dots colored and sized according to the assigned provisional diagnostic label. Controls are represented by blue squares. The green line (y = 1 − x) shows the decision boundary for the product rule and mean rule combination methods (for a threshold of 0.5 on the combined posterior probability). [Color figure can be viewed in the online issue, which is available at http://wileyonlinelibrary.com.]
P‐maps for the voxel‐wise classifications are shown in Figure 7. For CBF (Fig. 7A), several clusters of significantly different voxels were observed, located mainly in the thalamus, amygdala, and anterior and posterior cingulate gyrus. For GM (Fig. 7B), clusters of significantly different voxels were seen in the hippocampus, insula, posterior cingulate gyrus and thalamus. We also observed significantly different voxels in regions with a low GM probability, around the ventricles and corpus callosum. Table 1 lists all regions with visually observed clusters of significantly different voxels in the P‐map. Within these regions, as defined by Hammers et al. [2003] and Gousias et al. [2008], only a small percentage of voxels was significantly different. For CBF, the highest percentage of significantly different voxels was observed in the amygdala (20%), and for GM in the hippocampus (18%), see Figure 8.
Figure 7.
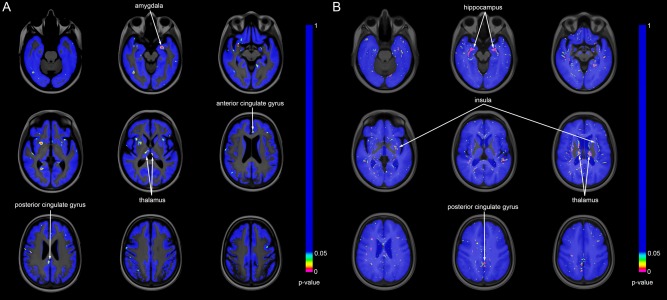
Statistical significance maps (P‐maps) for the voxel‐wise classifications: (A) CBF, (B) GM. Nonblue voxels are significantly different (P < 0.05) between patient and control groups based on SVM classification. [Color figure can be viewed in the online issue, which is available at http://wileyonlinelibrary.com.]
Table 1.
Regions with clusters of significant voxels in the P‐maps
| CBF | GM |
|---|---|
| Amygdala (left > right) | Hippocampus (bilateral) |
| Cingulate gyrus, anterior part (left) | Insula (bilateral) |
| Cingulate gyrus, posterior part (right) | Cingulate gyrus, posterior part (bilateral) |
| Thalamus (bilateral) | Thalamus (bilateral) |
| Postcentral gyrus (right > left) | Medial temporal gyrus (bilateral) |
| Inferior frontal gyrus (bilateral) | Inferior temporal gyrus (bilateral) |
| Putamen (right > left) | Lingual gyrus (bilateral) |
| Insula (left) | Superior frontal gyrus (bilateral) |
| Medial frontal gyrus (bilateral) | |
| Superior frontal gyrus (left) | |
| Caudate nucleus (left) | |
| Occipital gyrus (left) | |
| Gyrus parahippocampalis (bilateral) | |
| Medial temporal gyrus (bilateral) |
Figure 8.
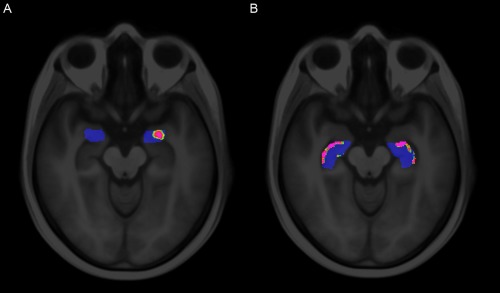
Voxel‐wise P‐maps (A) within the amygdala for CBF and (B) within the hippocampus for GM. These two regions showed the highest percentage of significant voxels. The regions were based on the region labeling in template space. Nonblue voxels are significantly different (P < 0.05). [Color figure can be viewed in the online issue, which is available at http://wileyonlinelibrary.com.]
In the Supporting Information, the P‐values for the region classification are listed. For CBF, two significantly different regions were found, and for GM one region. For CBF, one of the significantly different regions was also clearly found in the voxel‐wise P‐maps. However, for GM this correspondence was less clear since the only significantly different ROI (right Subgenual anterior cingulate gyrus) was not shown in the voxel‐wise P‐map. The regions with the most clear clusters of significantly different voxels in the voxel‐wise P‐map (hippocampus, insula, and thalamus) were not found to be significantly different in the region‐wise approach. In the selection and lobe ROI‐wise approaches, two significantly different ROIs were found for both CBF (selection: superior parietal gyrus left and presubgenual anterior cingulate gyrus right; lobe: occipital lobe left and frontal lobe right) and GM (selection: subgenual anterior cingulate gyrus right and presubgenual anterior cingulate gyrus left; lobe: temporal lobe left and right). For hemisphere and brain, no significant ROIs were found.
Group II
Classification performances based on GM features for the ADNI reference data are shown in Figure 9. For both voxel‐ and ROI‐wise approaches, we obtained an AUC of about 90%. For the voxel‐wise method, the performance reported by Cuingnet et al. was somewhat higher than what we found (Table 2). For the region‐wise method, performances were similar: we obtained a slightly higher sum of sensitivity and specificity, and Cuingnet et al. obtained a slightly higher AUC.
Figure 9.
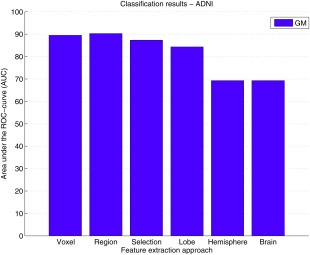
Classification performances for the ADNI data quantified by the area under the ROC‐curve (AUC). GM features were extracted using two approaches: voxel‐wise, and ROI‐wise using 5 GM ROI sets (region, selection, lobe, hemisphere, and brain). [Color figure can be viewed in the online issue, which is available at http://wileyonlinelibrary.com.]
Table 2.
Classification performance on the ADNI reference data for the voxel‐ and region‐wise approaches compared with the performances on the same data reported by Cuingnet et al. [2011]
| Study | Method | AUC (%) | Sens. (%) | Spec. (%) | Sum |
|---|---|---|---|---|---|
| This study | Voxel | 89 | 85 | 79 | 165 |
| Cuingnet et al. | Voxel‐Direct‐D‐gm | 95 | 81 | 95 | 176 |
| This study | Region | 90 | 83 | 90 | 172 |
| Cuingnet et al. | Voxel‐Atlas‐D‐gm | 92 | 78 | 91 | 169 |
The method Voxel‐Direct‐D‐gm is similar to our voxel‐wise method, using modulated GM maps, and the method Voxel‐Atlas‐D‐gm is similar to our method region, using features for a set of ROIs. Performance measures were area under the ROC curve (AUC), sensitivity (Sens.), specificity (Spec.), and the sum of sensitivity and specificity (Sum).
Discussion
We evaluated different approaches for classification of early stage presenile dementia patients and controls. These approaches included different types of MRI data, and both voxel‐wise and ROI‐wise methods for feature extraction. In this section we first discuss the classification performances on Group I. Second, the added value of ASL for diagnosis of dementia is discussed. Finally, we discuss the validation of methods using the reference dataset of Group II.
Classification Performance
The voxel‐wise classification methods showed a high diagnostic performance with an AUC of up to 91% for early stage presenile dementia (Group I). We can consider this a high accuracy for this patient population, because the patients were still at an early stage of the disease, when both clinical symptomatology and GM atrophy are known to be less pronounced than at more advanced stages of the disease. Additionally, our patient population was relatively young, since we only included presenile dementia patients. The group was also rather heterogeneous, as patients were included when they were suspected of suffering from either AD or FTD, in which different regions of the brain are affected. In AD, hypoperfusion and atrophy are expected mainly in the medial temporal and parietal lobes, while in FTD this is mainly seen in the frontal and temporal lobes [Hu et al., 2010]. Such heterogeneity of affected brain regions makes the classification of dementia more difficult. Due to these issues, diagnostic performance in this group may be expected to be lower than that in homogeneous patient populations at more advanced stages of disease. However, one can also argue that a young patient group and therefore also a young control group, may have positively influenced the diagnostic performance as the younger control group is not so much affected by age‐related atrophy and might therefore be better distinguishable. For Group I, cross‐validation was used for estimating classifier performance. This technique is frequently used and mainly applied when a relatively small amount of data is available (for classification of dementia. The voxel‐wise methods overall provided higher performance than the ROI‐based techniques, which indicates that important diagnostic information was lost by averaging over the ROIs. This is illustrated by the P‐maps obtained with permutation testing (Figs. 7 and 8), which showed that the voxel‐wise classifiers mainly rely on small clusters of voxels within the anatomically defined regions used here. These clusters only maximally covered 20% of the voxels within such a region. Therefore, we can assume that the used anatomical region labeling was not optimal for the ROI‐wise classifications, as the regions may have been too large to be sensitive to information from a small proportion of significantly different voxels.
For the voxel‐wise and region methods, the feature concatenation method was outperformed by the other combination methods, possibly due to the large number of features relative to the small amount of data. However, for the other ROI‐wise approaches, feature concatenation was the best performing combination methods. The relatively small standard deviations obtained with the four‐fold cross‐validation indicated that the classifications were rather robust.
When using one feature only, that is, whole brain measures, ROI‐wise methods for GM still gave a relatively good performance (AUC = 73%). However for CBF, the classification performance declined with decreasing number of features. Especially remarkable was the reduction in AUC for CBF after selection of 28 dementia‐related brain regions. For the GM classifications, we did not find this dramatic decrease in performance. This might be due to the fact that the regions were selected on the basis of the literature reporting either focal atrophy or hypoperfusion/hypometabolism. Such regions may not coincide, particularly not in the early stage of dementia. For instance, in fluoro‐deoxyglucose positron emission tomography (FDG‐PET) studies no significant hypoperfusion is found in specific brain regions which are known to have volume loss in AD, for example the hippocampus [La Joie et al., 2012; Maldjian et al., 2012], or vice versa. For assessing the diagnostic performance of CBF classification methods, the selection classification may have reduced performance because certain regions may have been included that only exhibited atrophy but not perfusion changes.
Using the P‐maps, we visualized which features were significant for classification. For CBF, we mainly found clusters of significantly different voxels in the amygdala, thalamus and cingulate gyrus, corresponding to findings from the literature on AD reporting hypoperfusion in the cingulate gyrus and prefrontal cortex. Hypoperfusion in the parietal lobe is also reported, but was not found here [Wolk and Detre, 2012]. For GM, significantly different voxels were found in the hippocampus, insula, thalamus and cingulate gyrus, corresponding to the literature [Chételat and Baron, 2003; Karas et al., 2003, 2004]. GM P‐maps were mostly symmetrical, showing similar clusters of significantly different voxels bilaterally, whereas CBF P‐maps where more asymmetrical. Some asymmetry is expected particularly in FTD patients [McKhann et al., 2001]. Cuingnet et al. [2011] did not calculate P‐maps, but evaluated the optimal margin hyperplane (w‐map), which provides qualitative information on the classifiers showing regions in which atrophy increased the likelihood of being classified as AD. These regions were the medial temporal lobe (including hippocampus), thalamus, posterior cingulate gyrus, inferior and middle temporal gyri, posterior middle frontal gyrus, and fusiform gyrus. This corresponds well to our P‐maps as we found the same regions except the last two. In addition, we detected clusters of significantly different voxels in the insula.
Because in AD and FTD different brain regions are affected, atrophy and hypoperfusion information could be used to make a differential diagnosis. A future aim of this work is to perform a multiclass classification to distinguish the two groups of patients. One year after inclusion, follow‐up information will be used to establish a definitive diagnosis, which is needed for the multiclass classification.
A minor limitation of this work is that a different T1w protocol was used for 10 of the control subjects. We believe that the impact of this is minor, because the used sequences are very similar, both are near isotropic with a resolution ≤ 1 mm, and both sequences allow for good differentiation between white and GM.
The Added Value of ASL
CBF‐based classification yielded high diagnostic performance for the voxelwise and region‐wise approaches with AUCs of 87 and 76%, respectively. For the voxel‐wise classification, this was similar to the diagnostic performance based on GM features (P = 0.38, McNemar's test). This indicates that CBF quantified with ASL is a good diagnostic marker for early stage dementia, in concordance with previous studies [Binnewijzend et al., 2013; Wang et al., 2013; Wolk and Detre, 2012].
Although CBF may be a good diagnostic marker by itself, our results showed no added value over atrophy markers based on structural MRI. The four different combination methods — feature concatenation, feature multiplication, the product rule, and the mean rule — showed a slight improvement in AUC for the voxel‐wise approaches, but the McNemar tests showed no significant increase in diagnostic performance by using any methods (P ≥ 0.38). For ASL to add value, other combination methods than these four may need to be explored to more efficiently combine the CBF and GM features. In addition, one should note that the limited added value of ASL over structural MRI found in this work may be partly attributed to the specific methodology used, both in ASL acquisition and analysis. A potential confounder in this study is the arterial transit time (ATT), which could conceivably be different between patient and control group. However, we expect these differences to be small, since on the one hand patients with cerebral vascular disease were excluded and on the other hand the patients and control groups were age‐matched. We compared our results to those of three previously published articles or abstracts studying the added value of ASL for the diagnosis of dementia. Du et al. [2006] based classification of FTD patients and controls on logistic regression. The mean CBF and GM volume in certain regions in the frontal and parietal lobes were used as features. Performance was evaluated on the training data. Classification based on GM volume only showed no significant separation between the groups, but including CBF yielded an AUC of 80% (P < 0.01). The second study by Dashjamts et al. [2011] performed linear discriminant analysis to discriminate between AD patients and controls using LOO cross‐validation. Features were defined for the whole brain as the normalized CBF intensities and the GM segmentation in DARTEL template space [Ashburner, 2007]. For the GM features no modulation step was performed. The number of features was reduced using a VBM approach, which performs voxel‐wise t‐tests at different significance levels. The classification AUCs were 78% for GM, 89% for CBF, and 92% for the combination of both using concatenation. These findings are similar to our results, except for the AUC for GM, which in their study was lower than our results and lower than the values reported by Cuingnet et al. [2011] and Klöppel et al. [2008]. The classifiers may have been over‐trained since the feature reduction was performed on the complete set and since optimal significance levels for the classification on both CBF and GM were selected using the labels of the test data. The third study, an abstract by Schuff et al. [2012], studied the classification of early MCI using local linear embedding and logistic regressions. Features were defined as the mean CBF or tissue volume for a set of ROIs. The accuracies of the classification were 67% based on the volume features, 58% based on CBF, and 71% for the combination of both.
These studies on classification using ASL [Dashjamts et al., 2011; Du et al., 2006; Schuff et al., 2012] conclude that ASL improves the classification of dementia over structural MRI. Although in our dataset we also observed a small increase in performance by combining CBF and GM, we could not conclude that this significantly improves classification, as classifications on the basis of GM features alone already had a high performance. For early stage dementia lower performances were expected, as for instance Klöppel et al. [2008] reported a GM‐based classification accuracy of 81.1% in a mild AD group (age ≤ 80 years, MMSE range = 20–30). The relatively high performances for the GM‐based classifications we found here may be attributed to the presenile patient and control population, as addressed in the previous section. We therefore assume that the added value of ASL in this study was limited by the relatively high performance of the classifications based on structural MRI.
In addition, the small samples sizes of each of these studies may hinder a reliable comparison. Similar to the studies mentioned above, we used a relatively small dataset (32 patients/32 controls; Du et al.: 21 FTD/24 AD/25 controls; Dashjamts et al.: 23 AD/23 controls; Schuff et al.: 7 AD/44 early MCI/17 MCI/29 controls). To our knowledge, the added value of ASL for classification of dementia has not been assessed with larger sample size studies, but for further verification of our conclusion larger sample size studies would be preferred.
Comparison with Related Work
The GM image‐processing and classification methods were evaluated on an AD patient group and a healthy control group from the ADNI database (Group II) to enable comparison with related work. The classification performances we obtained were generally comparable (Table 2) to those of Cuingnet et al. [2011], from which the subject groups were adopted. However, some performance differences could be observed, which we think may be largely attributed to three differences in the methodology. The first difference is in the region approach, in which we used 72 regions constructed with multi‐atlas registration, whereas the Voxel‐Atlas‐D‐gm of Cuingnet et al. uses 119 regions from a single atlas [Tzourio‐Mazoyer et al., 2002]. Although our atlas contains fewer ROIs, which could impact the performance either positively, as fewer features reduce the risk of overtraining, or negatively, as fewer features contain less information, we chose this atlas because multi‐atlas‐based segmentation is more accurate and robust than single‐atlas‐based segmentation [Heckemann et al., 2006]. Second, the data used for template‐space construction differs. We based the template space for Group II on the training data only, whereas Cuingnet's Voxel‐Direct‐D‐gm method uses the complete set. Our approach requires less computation time, which is practical for clinical use, but may perform slightly worse as the testing subjects are not included in the template space. Third, we used a different method for template‐space construction. Cuingnet et al. uses the DARTEL algorithm [Ashburner, 2007], which differs from our method in three main ways: (1) DARTEL iteratively maps the scans to their average, instead of using the pairwise registrations of our approach; (2) DARTEL uses tissue segmentations instead of directly registering T1w images; and (3) DARTEL uses a large‐deformation diffeomorphic algorithm, while our approach uses a small‐deformation parametric (B‐spline) transformation model assuming small deformations. Although the methods use different approaches, both aim to find the group mean image.
Although some steps in our method differed from the method of Cuingnet et al., classification performances on the same dataset were very similar, indicating that our methodology is valid and providing context for our findings in the presenile early stage dementia patients (Group I).
Conclusion
Of the different classification methods, voxel‐wise classifications provided the best classification performance for early stage presenile dementia and controls with an AUC of about 91%. This can be considered a high diagnostic accuracy in this presenile patient population in the very early stage of either of two different types of dementia.
Although CBF quantified with ASL was found to be a good diagnostic marker of dementia, with similar diagnostic accuracy as GM in the voxel‐based classifications, its added value over structural MRI was not significant.
Supporting information
Supplementary Information Table 3.
ACKNOWLEDGMENTS
The authors would like to thank Carolina Mendez and Jaap van Dijke (Department of Radiology, Erasmus MC, Netherlands) for providing data and data acquisition of healthy control participants, and Rémi Cuingnet (Philips Research MediSys, France) for providing the classification results of his article for extraction of AUC values.
Conflicts of interest: W.J. Niessen is cofounder, part‐time Chief Scientific Officer, and stock holder of Quantib BV. Other authors had no conflicts of interest to declare.
Data used in preparation of this article were obtained from the Alzheimer's Disease Neuroimaging Initiative (ADNI) database (http://adni.loni.ucla.edu). As such, the investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data but did not participate in analysis or writing of this report. A complete listing of ADNI investigators can be found at: http://adni.loni.ucla.edu/wpcontent/uploads/how_to_apply/ADNI_Acknowledgement_List.pdf
REFERENCES
- Alzheimer's Association (2011): 2011 Alzheimer's disease facts and figures. Alzheimers Dement 7:208–244. [DOI] [PubMed] [Google Scholar]
- Ashburner J (2007): A fast diffeomorphic image registration algorithm. Neuroimage 38:95–113. [DOI] [PubMed] [Google Scholar]
- Ashburner J, Friston KJ (2000): Voxel‐based morphometry ‐ the methods. Neuroimage 11:805–821. [DOI] [PubMed] [Google Scholar]
- Ashburner J, Friston KJ (2005): Unified segmentation. Neuroimage 26:839–851. [DOI] [PubMed] [Google Scholar]
- Asllani I, Borogovac A, Brown TR (2008): Regression algorithm correcting for partial volume effects in arterial spin labeling MRI. Magn Reson Med 60:1362–1371. [DOI] [PubMed] [Google Scholar]
- Binnewijzend MAA, Kuijer JPA, Benedictus MR, van der Flier WM, Wink AM, Wattjes MP, van Berckel BNM, Scheltens P, Barkhof F (2013): Cerebral blood flow measured with 3D pseudocontinuous arterial spin labeling MR imaging in Alzheimer disease and mild cognitive impairment: A marker for disease severity. Radiology 267:221–230. [DOI] [PubMed] [Google Scholar]
- Buxton RB, Frank LR, Wong EC, Siewert B, Warach S, Edelman RR (1998): A general kinetic model for quantitative perfusion imaging with arterial spin labeling. Magn. Reson Med 40:383–396. [DOI] [PubMed] [Google Scholar]
- Chang C‐C, Lin C‐J (2011): LIBSVM: A library for support vector machines. ACM TIST 2:27–27. [Google Scholar]
- Chételat G, Baron J‐C (2003): Early diagnosis of Alzheimer's disease: Contribution of structural neuroimaging. Neuroimage 18:525–541. [DOI] [PubMed] [Google Scholar]
- Cuingnet R, Gerardin E, Tessieras J, Auzias G, Lehéricy S, Habert MO, Chupin M, Benali H, Colliot O (2011): Automatic classification of patients with Alzheimer's disease from structural MRI: A comparison of ten methods using the ADNI database. Neuroimage 56:766–781. [DOI] [PubMed] [Google Scholar]
- Dai W, Garcia D, de Bazelaire C, Alsop DC (2008): Continuous flow‐driven inversion for arterial spin labeling using pulsed radio frequency and gradient fields. Magn Reson Med 60:1488–1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dashjamts T, Yoshiura T, Hiwatashi A, Yamashita K, Monji A, Ohyagi Y, Kamano H, Kawashima T, Kira J, Honda H (2011): Simultaneous arterial spin labeling cerebral blood flow and morphological assessments for detection of Alzheimer's disease. Acad Radiol 18:1492–1499. [DOI] [PubMed] [Google Scholar]
- Davatzikos C, Fan Y, Wu X, Shen D, Resnick SM (2008): Detection of prodromal Alzheimer's disease via pattern classification of magnetic resonance imaging. Neurobiol Aging 29:514–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Detre JA, Leigh JS, Williams DS, Koretsky AP (1992): Perfusion imaging. Magn Reson Med 23:37–45. [DOI] [PubMed] [Google Scholar]
- Du AT, Jahng GH, Hayasaka S, Kramer JH, Rosen HJ, Gorno‐Tempini ML, Rankin KP, Miller BL, Weiner MW, Schuff N (2006): Hypoperfusion in frontotemporal dementia and Alzheimer disease by arterial spin labeling MRI. Neurology 67:1215–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois B, Feldman HH, Jacova C, DeKosky ST, Barberger‐Gateau P, Cummings J, Delacourte A, Galasko D, Gauthier S, Jicha G, Meguro K, O'Brien J, Pasquier F, Robert P, Rossor M, Salloway S, Stern Y, Visser PJ, Scheltens P (2007): Research criteria for the diagnosis of Alzheimer's disease: Revising the NINCDS‐ADRDA criteria. Lancet Neurol 6:734–746. [DOI] [PubMed] [Google Scholar]
- Dubois B, Feldman HH, Jacova C, Cummings JL, DeKosky ST, Barberger‐Gateau P, Delacourte A, Frisoni G, Fox NC, Galasko D, Gauthier S, Hampel H, Jicha GA, Meguro K, O'Brien J, Pasquier F, Robert P, Rossor M, Salloway S, Sarazin M, de Souza LC, Stern Y, Visser PJ, Scheltens P (2010): Revising the definition of Alzheimer's disease: A new lexicon. Lancet Neurol 9:1118–1127. [DOI] [PubMed] [Google Scholar]
- Duchesne S, Caroli A, Geroldi C, Barillot C, Frisoni GB, Collins DL (2008): MRI‐based automated computer classification of probable AD versus normal controls. IEEE Trans Med Imaging 27:509–520. [DOI] [PubMed] [Google Scholar]
- Duin RPW, Tax DMJ (1998): Classifier conditional posterior probabilities, Advances in Pattern Recognition. Springer, Berlin‐Heidelberg, Germany: p 611–619. [Google Scholar]
- Fan Y, Batmanghelich N, Clark CM, Davatzikos C, the ADNI (2008a): Spatial patterns of brain atrophy in MCI patients, identified via high‐dimensional pattern classification, predict subsequent cognitive decline. Neuroimage 39:1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y, Resnick SM, Wu X, Davatzikos C (2008b): Structural and functional biomarkers of prodromal Alzheimer's disease: A high‐dimensional pattern classification study. Neuroimage 41:277–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster NL, Wang AY, Tasdizen T, Fletcher PT, Hoffman JM, Koeppe RA (2008): Realizing the potential of positron emission tomography with 18F‐fluorodeoxyglucose to improve the treatment of Alzheimer's disease. Alzheimers Dement 4:S29–36. [DOI] [PubMed] [Google Scholar]
- Fukuyama H, Ogawa M, Yamauchi H, Yamaguchi S, Kimura J, Yonekura Y, Konishi J (1994): Altered cerebral energy metabolism in Alzheimer's disease: A PET study. J Nucl Med 35:1–6. [PubMed] [Google Scholar]
- Gaonkar B, Davatzikos C (2013): Analytic estimation of statistical significance maps for support vector machine based multi‐variate image analysis and classification. Neuroimage 78:270–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gousias IS, Rueckert D, Heckemann RA, Dyet LE, Boardman JP, Edwards AD, Hammers A (2008): Automatic segmentation of brain MRIs of 2‐year‐olds into 83 regions of interest. Neuroimage 40:672–684. [DOI] [PubMed] [Google Scholar]
- Hammers A, Allom R, Koepp MJ, Free SL, Myers R, Lemieux L, Mitchell TN, Brooks DJ, Duncan JS (2003): Three‐dimensional maximum probability atlas of the human brain, with particular reference to the temporal lobe. Hum Brain Mapp 19:224–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckemann RA, Hajnal J V, Aljabar P, Rueckert D, Hammers A (2006): Automatic anatomical brain MRI segmentation combining label propagation and decision fusion. Neuroimage 33:115–126. [DOI] [PubMed] [Google Scholar]
- Herholz K, Carter SF, Jones M (2007): Positron emission tomography imaging in dementia. Br J Radiol 80:S160–S167. [DOI] [PubMed] [Google Scholar]
- Hu WT, Wang Z, Lee V‐Y, Trojanowski JQ, Detre JA, Grossman M (2010): Distinct cerebral perfusion patterns in FTLD and AD. Neurology 75:881–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii K, Kitagaki H, Kono M, Mori E (1996): Decreased medial temporal oxygen metabolism in Alzheimer's disease shown by PET. J Nucl Med 37:1159–1165. [PubMed] [Google Scholar]
- Ishii K, Sasaki M, Kitagaki H, Yamaji S, Sakamoto S, Matsuda K, Mori E (1997a): Reduction of cerebellar glucose metabolism in advanced Alzheimer's disease. J Nucl Med 38:925–928. [PubMed] [Google Scholar]
- Ishii K, Sasaki M, Yamaji S, Sakamoto S, Kitagaki H, Mori E (1997b): Demonstration of decreased posterior cingulate perfusion in mild Alzheimer's disease by means of H215O positron emission tomography. Eur J Nucl Med 24:670–673. [DOI] [PubMed] [Google Scholar]
- Ishii K, Sakamoto S, Sasaki M, Kitagaki H, Yamaji S, Hashimoto M, Imamura T, Shimomura T, Hirono N, Mori E (1998): Cerebral glucose metabolism in patients with frontotemporal dementia. J Nucl Med 39:1875–1878. [PubMed] [Google Scholar]
- Ishii K, Sasaki M, Matsui M, Sakamoto S, Yamaji S, Hayashi N, Mori T, Kitagaki H, Hirono N, Mori E (2000): A diagnostic method for suspected Alzheimer's disease using H(2)15O positron emission tomography perfusion Z score. Neuroradiology 42:787–794. [DOI] [PubMed] [Google Scholar]
- Jack CR, Bernstein M, Fox NC, Thompson P, Alexander G, Harvey D, Borowski B, Britson PJL, Whitwell J, Ward C, Dale AM, Felmlee JP, Gunter JL, Hill DLG, Killiany R, Schuff N, Fox‐Bosetti S, Lin C, Studholme C, DeCarli CS, Krueger G, Ward H, Metzger GJ, Scott KT, Mallozzi R, Blezek D, Levy J, Debbins JP, Fleisher AS, Albert M, Green R, Bartzokis G, Glover G, Mugler J, Weiner MW (2008): The Alzheimer's Disease Neuroimaging Initiative (ADNI): MRI methods. J Magn Reson Imaging 27:685–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CR Jr, Knopman DS, Jagust WJ, Shaw LM, Aisen PS, Weiner MW, Petersen RC, Trojanowski JQ (2010): Hypothetical model of dynamic biomarkers of the Alzheimer's pathological cascade. Lancet Neurol 9:119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannsen P, Jakobsen J, Gjedde A (2000): Statistical maps of cerebral blood flow deficits in Alzheimer's disease. Eur J Neurol 7:385–392. [DOI] [PubMed] [Google Scholar]
- Karas GB, Burton EJ, Rombouts S, Van Schijndel RA, O'Brien JT, Scheltens PH, McKeith IG, Williams D, Ballard C, Barkhof F (2003): A comprehensive study of gray matter loss in patients with Alzheimer's disease using optimized voxel‐based morphometry. Neuroimage 18:895–907. [DOI] [PubMed] [Google Scholar]
- Karas GB, Scheltens P, Rombouts S, Visser PJ, Van Schijndel RA, Fox NC, Barkhof F (2004): Global and local gray matter loss in mild cognitive impairment and Alzheimer's disease. Neuroimage 23:708–716. [DOI] [PubMed] [Google Scholar]
- Klein S, Staring M, Murphy K, Viergever MA, Pluim JPW (2010): Elastix: a toolbox for intensity‐based medical image registration. IEEE Trans Med Imaging 29:196–205. [DOI] [PubMed] [Google Scholar]
- Klöppel S, Stonnington CM, Chu C, Draganski B, Scahill RI, Rohrer JD, Fox NC, Jack CR Jr, Ashburner J, Frackowiak RSJ (2008): Automatic classification of MR scans in Alzheimer's disease. Brain 131:681–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koikkalainen J, Pölönen H, Mattila J, van Gils M, Soininen H, Lötjönen J, the ADNI (2012): Improved classification of Alzheimer's disease data via removal of nuisance variability. PLoS One 7:e31112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Joie R, Perrotin A, Barré L, Hommet C, Mézenge F, Ibazizene M, Camus V, Abbas A, Landeau B, Guilloteau D, de La Sayette V, Eustache F, Desgranges B, Chételat G (2012): Region‐specific hierarchy between atrophy, hypometabolism, and β‐amyloid (Aβ) load in Alzheimer's disease dementia. J Neurosci 32:16265–16273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnin B, Mesrob L, Kinkingnéhun S, Pélégrini‐Issac M, Colliot O, Sarazin M, Dubois B, Lehéricy S, Benali H (2009): Support vector machine‐based classification of Alzheimer's disease from whole‐brain anatomical MRI. Neuroradiology 51:73–83. [DOI] [PubMed] [Google Scholar]
- Maldjian JA, Whitlow CT, the ADNI. (2012): Whither the hippocampus? FDG‐PET hippocampal hypometabolism in Alzheimer disease revisited. Am J Neuroradiol 33:1975–1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKhann GM, Albert MS, Grossman M, Miller B, Dickson D, Trojanowski JQ (2001): Clinical and pathological diagnosis of frontotemporal dementia: Report of the Work Group on Frontotemporal Dementia and Pick's Disease. Arch Neurol 58:1803. [DOI] [PubMed] [Google Scholar]
- McKhann GM, Knopman DS, Chertkow H, Hyman BT, Jack CR Jr., Kawas CH, Klunk WE, Koroshetz WJ, Manly JJ, Mayeux R, Mohs RC, Morris JC, Rossor MN, Scheltens P, Carillo MC, Thies B, Weintraub S, Phelps CH (2011): The diagnosis of dementia due to Alzheimer's disease: Recommendations from the National Institute on Aging‐Alzheimer's Association workgroups on diagnostic guidelines for Alzheimer's disease. Alzheimers Dement 7:263–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minoshima S, Giordani B, Berent S, Frey KA, Foster NL, Kuhl DE (1997): Metabolic reduction in the posterior cingulate cortex in very early Alzheimer's disease. Ann Neurol 42:85–94. [DOI] [PubMed] [Google Scholar]
- Mourão‐Miranda J, Bokde ALW, Born C, Hampel H, Stetter M (2005): Classifying brain states and determining the discriminating activation patterns: Support Vector Machine on functional MRI data. Neuroimage 28:980. [DOI] [PubMed] [Google Scholar]
- Oliver RA, Thomas DL, Golay X (2012): Improved partial volume correction of ASL images using 3D kernels. ISMRM British Chapter, Cambridge, UK. [Google Scholar]
- Paquerault S (2012): Battle against Alzheimer's disease: The scope and potential value of magnetic resonance imaging biomarkers. Acad Radiol 19:509–511. [DOI] [PubMed] [Google Scholar]
- Prince M, Bryce R, Ferri C (2011): World Alzheimer Report 2011. The benefits of early diagnosis and intervention. Alzheimer's Disease International, London, UK. [Google Scholar]
- Rascovsky K, Hodges JR, Knopman D, Mendez MF, Kramer JH, Neuhaus J, van Swieten JC, Seelaar H, Dopper EGP, Onyike CU, Hillis AE, Josephs KA, Boeve BF, Kertesz A, Seeley WW, Rankin KP, Johnson JK, Gorno‐Tempini ML, Rosen HJ, Prioleau‐Latham CE, Lee A, Kipps CM, Lillo P, Piguet O, Rohrer JD, Rossor MN, Warren JD, Fox NC, Galasko D, Salmon DP, Black SE, Mesulam M, Weintraub S, Dickerson BC, Diehl‐Schmid J, Pasquier F, Deramecourt V, Lebert F, Pijnenburg Y, Chow TW, Manes F, Grafman J, Cappa SF, Freedman M, Grossman M, Miller BL (2011): Sensitivity of revised diagnostic criteria for the behavioural variant of frontotemporal dementia. Brain 134:2456–2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santens P, De Bleecker J, Goethals P, Strijckmans K, Lemahieu I, Slegers G, Dierckx R, De Reuck J (2001): Differential regional cerebral uptake of 18F‐fluoro‐2‐deoxy‐D‐glucose in Alzheimer's disease and frontotemporal dementia at initial diagnosis. Eur Neurol 45:19–27. [DOI] [PubMed] [Google Scholar]
- Scarmeas N, Habeck CG, Zarahn E, Anderson KE, Park A, Hilton J, Pelton GH, Tabert MH, Honig LS, Moeller JR, Devanand DP, Stern Y (2004): Covariance PET patterns in early Alzheimer's disease and subjects with cognitive impairment but no dementia: Utility in group discrimination and correlations with functional performance. Neuroimage 23:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuff N, Liu X, Weiner MW, the ADNI (2012): Regional abnormalities of cerebral blood flow in early mild cognitive impairment: Insights from the ASL‐MRI study of ADNI. ISMRM Scient Worksh Perf MRI. p 5500. [Google Scholar]
- Seghers D, D'Agostino E, Maes F, Vandermeulen D, Suetens P (2004): Construction of a brain template from MR images using state‐of‐the‐art registration and segmentation techniques. Proc Intl Conf Med Image Comput Comp Ass Intervent. Springer, Berlin‐Heidelberg, Germany: p 696–703. [Google Scholar]
- Smith SM (2002): Fast robust automated brain extraction. Hum Brain Map 17:143–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperling RA, Aisen PS, Beckett LA, Bennett DA, Craft S, Fagan AM, Iwatsubo T, Jack CR, Kaye J, Montine TJ, et al. (2011): Toward defining the preclinical stages of Alzheimer's disease: Recommendations from the National Institute on Aging‐Alzheimer's Association workgroups on diagnostic guidelines for Alzheimer's disease. Alzheimers Dement 7:280–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tax DMJ, Van Breukelen M, Duin RPW, Kittler J (2000): Combining multiple classifiers by averaging or by multiplying? Pattern Recogn 33:1475–1485. [Google Scholar]
- Thévenaz P, Unser M (2000): Optimization of mutual information for multiresolution image registration. IEEE Trans Image Proc 9:2083–2099. [DOI] [PubMed] [Google Scholar]
- Tustison NJ, Avants BB, Cook PA, Zheng Y, Egan A, Yushkevich PA, Gee JC (2010): N4ITK: Improved N3 bias correction. IEEE Trans Med Imaging 29:1310–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzourio‐Mazoyer N, Landeau B, Papathanassiou D, Crivello F, Etard O, Delcroix N, Mazoyer B, Joliot M (2002): Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single‐subject brain. Neuroimage 15:273–289. [DOI] [PubMed] [Google Scholar]
- Van Gelderen P, de Zwart JA, Duyn JH (2008): Pittfalls of MRI measurement of white matter perfusion based on arterial spin labeling. Magn. Reson Med 59:788–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vapnik VN (1995): The nature of statistical learning theory. New York: Springer‐Verlag. [Google Scholar]
- Vemuri P, Gunter JL, Senjem ML, Whitwell JL, Kantarci K, Knopman DS, Boeve BF, Petersen RC, Jack CR Jr. (2008): Alzheimer's disease diagnosis in individual subjects using structural MR images: Validation studies. Neuroimage 39:1186–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Childress AR, Wang J, Detre JA (2007): Support vector machine learning‐based fMRI data group analysis. Neuroimage 36:1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Das SR, Xie SX, Arnold SE, Detre JA, Wolk DA (2013): Arterial Spin Labeled MRI in Prodromal Alzheimer's Disease: A Multi‐Site Study. Neuroimage Clin 2:630–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams DS, Detre JA, Leigh JS, Koretsky AP (1992): Magnetic resonance imaging of perfusion using spin inversion of arterial water. Proc Nat Acad Sci USA 89:212–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolk DA, Detre JA (2012): Arterial spin labeling MRI: An emerging biomarker for Alzheimer's disease and other neurodegenerative conditions. Curr Opin Neurol 25:421–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolz R, Julkunen V, Koikkalainen J, Niskanen E, Zhang DP, Rueckert D, Soininen H, Lötjönen J (2011): Multi‐method analysis of MRI images in early diagnostics of Alzheimer's disease. PLoS One 6:e25446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Womack KB, Diaz‐Arrastia R, Aizenstein HJ, Arnold SE, Barbas NR, Boeve BF, Clark CM, DeCarli CS, Jagust WJ, Leverenz JB, et al. (2011): Temporoparietal hypometabolism in frontotemporal lobar degeneration and associated imaging diagnostic errors. Arch Neurol 68:329–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W‐C, Fernández‐Seara M, Detre JA, Wehrli FW, Wang J (2007): A theoretical and experimental investigation of the tagging efficiency of pseudocontinuous ASL. Magn Reson Med 58:1020–1027. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information Table 3.


