Abstract
Extracellular vesicles (EVs), a heterogeneous group of vesicles differing in size and shape, cargo content and function, are membrane‐bound and nano‐sized vesicles that could be released by nearly all variations of cells. EVs have gained considerable attention in the past decades for their functions in modulating intercellular signalling and roles as potential pools for the novel diagnostic and prognostic biomarkers, as well as therapeutic targets in several cancers including urological neoplasms. In general, human and animal cells both can release distinct types of EVs, including exosomes, microvesicles, oncosomes and large oncosomes, and apoptotic bodies, while the content of EVs can be divided into proteins, lipids and nucleic acids. However, the lack of standard methods for isolation and detection platforms rein the widespread usage in clinical applications warranted furthermore investigations in the development of reliable, specific and sensitive isolation techniques. Whether and how the EVs work has become pertinent issues. With the aid of high‐throughput proteomics or genomics methods, a fully understanding of contents contained in EVs from urogenital tumours, beyond all doubt, will improve our ability to identify the complex genomic alterations in the process of cancer and, in turn, contribute to detect potential therapeutic target and then provide personalization strategy for patient.
Keywords: bladder cancer, exosomes, extracellular vesicles, kidney cancer, microvesicles, prostate cancer
1. INTRODUCTION
Urogenital carcinomas are mainly leading to morbidity and mortality worldwide.1 Although therapeutic strategies (surgery, biotherapy and chemotherapy) for patients suffering from urinary cancers have improved, the curative and monitoring efficacy for these cancers still remains poor, as existing tests (general examinations and biopsies) are not a sufficiently sensitive or specific and heterogeneous peculiarity of these malignancies.2 Moreover, the deep location of urogenital cancers makes them hard to access to be diagnosed at early stages. Crosstalk between cells and their microenvironment is a fundamental principle under the normal and pathological condition.3 EVs, small membrane‐bound vesicles, serve as important players of bidirectional communication,4, 5 which released from almost eukaryotic and prokaryotic cells with transmitting complicate messages from donor cells towards anchored cells, and have been discovered in various types of body fluids including urine, blood and bile.6 According to the ways of production and secretion, EVs are produced by inward budding of intercellular endosomes, which are defined as exosomes (40‐1200 nm), EVs straightly shed by budding from the cell membrane, which are almost recognized as large oncosomes or microvesicles, with 1‐10 μm or 50‐1500 nm, and apoptotic vesicles are released during cell undergoing apoptosis ranged from 50 to 2000 nm, respectively.7, 8, 9
Extracellular vesicles serve as an appealing source for the development of biomarkers as their membrane‐bound structure to protect against exogenous proteases and RNases.10, 11 The biological function of EVs is performed by cytosolic lipids, proteins, DNA, mRNA, miRNA, lincRNA and other non‐coding RNAs, as well as cell membrane.12 In addition, cancer cell releases more EVs than that normal one does.13 Herein, we introduce EVs briefly and provide a comprehensive overview of their biophysical properties, roles and applications in the most common urologic neoplasms, including kidney, prostate and bladder, and discuss potential clinical applications in the future.
2. EV CLASSES, BIOGENESIS AND CONTENTS
Our current understanding of EVs indicates that at least four heterogeneous types of EVs have been identified based on their mechanism of formation and distinguished size: microvesicles, exosomes, oncosomes or large oncosomes, and apoptotic bodies (Table 1). In general, the formation of exosomes and microvesicles is two completely different approaches, but they function similarly. Oncosomes or large oncosomes resemble the way of microvesicles via membrane budding. Apoptotic bodies specifically arise resulting in indiscriminate membrane bubbling during apoptosis (Figure 1).
Table 1.
Details of different extracellular vesicles
| Exosomes | Microvesicles | Oncosomes | Large oncosomes | Apoptotic bodies | |
|---|---|---|---|---|---|
| Size | 40‐120 nm | 50‐1500 nm | 100‐500 nm | 1‐10 μm | 50‐2000 nm |
| Intracellular origin | Endosomes | Plasma membrane | Plasma membrane | Plasma membrane | Plasma membrane |
| Electron microscopy | Round shape | Irregular shape | Irregular shape | Amoeboid phenotype | Heterogenous |
| Release | Endolysosomal pathway, internal budding, exocytosis | Membrane budding | Membrane budding | Membrane budding | Generated as a result of apoptotic disintegration, resulting vesicles become part of the extracellular milieu |
| Marker proteins |
Membrane‐associated proteins: tetraspanin (CD9, CD63, CD81, CD82). Endosomal sorting complex required for Transport‐associate protein: Tsg101, ALIX. Cytoplasmic proteins: Hsp70, Hsp90. Membrane transport and fusion proteins: Rab GTPases, annexins |
Integrins, selectins, CD40 ligand | Integrins, selectins, Membrane‐associated proteins |
Integrins, selectins, VDAC ½ SLC25A3/5/6 ITGA5/6 |
Histones, C3b, Annexin V, Caspase 3 |
| Contents | Proteins, lipids, mRNA, miRNA and cytosol | Proteins, lipids, mRNA, miRNA and cytosol | Proteins, lipids, mRNA, miRNA and cytosol | Proteins, lipids, mRNA, miRNA and cytosol | Proteins, lipids, DNA, rRNA, organelles and cytosol |
Abbreviations: C3b, complement component 3; HSP, heat shock proteins; ITGA5/6, human integrin alpha 5/6; miRNA, microRNA; SLC25A3/5/6, solute carrier family 25 member3/5/6; VDACs, Voltage‐dependent anion channels.
Figure 1.
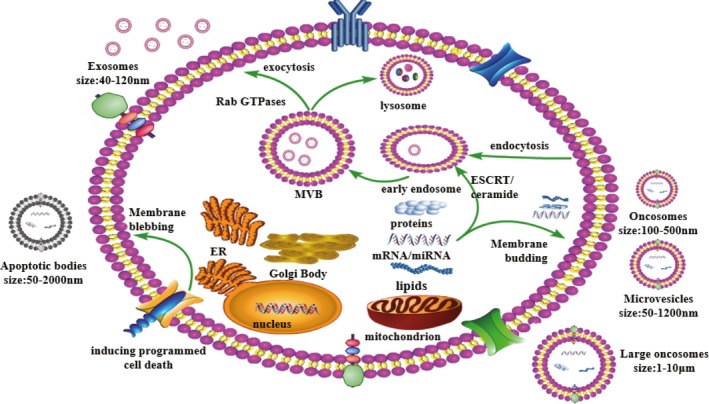
Release and uptake mechanisms of extracellular vesicles. Extracellular vesicles can be classed as exosomes, microvesicles and apoptotic bodies, based on the mechanism by which they are released from cells and differentiated based on their size and content. MVs, and oncosomes or large oncosomes are directly shed or bud from the plasma membrane. Apoptotic bodies are released from the cell undergoing programmed cell death. Exosomes are formed by inward budding of multivesicular bodies
Exosomes generally form an early endosome by the endocytosis and internalization of cell‐surface receptors into membrane‐bound vacuoles in the first step,14 which then matures to generate a late endosome within undergoing several changes, such as the limiting membrane of the late endosome then buds inward and pinches off as the result of the formation of intraluminal vesicles (ILVs), also known as multivesicular bodies, and ILVs traffick to and fuse with the plasma membrane leading in releasing exosomes eventually.15 ESCRT‐0‐III plays significant roles in driving exosome formation16, 17; in addition, multivesicular bodies could intermediate in the lysosomal degradation pathway.18 However, the mechanisms related to the fusion of multivesicular bodies with the cellular membrane are uncovered, which may be regulated by several factors including lipid ceramide and Rab GTPase (including Rab5 and Rab7) proteins and ESCRT.19, 20, 21 Numerous literatures have indicated that several biomarkers expressed in the exosome differentially compared with another types of EVs, including heat shock proteins (eg HSP60, 70 and 90), tetraspanins (eg CD9, CD63 and CD81), membrane transporters, fusion proteins, ALG‐2‐interacting protein X (Alix) and tumour susceptibility gene 101 protein (TSG101).20, 22
In contrast, microvesicles with nano‐sized with 100‐1500 nm are straightforwardly shed from the cellular membrane responding to stimuli or physiological conditions.23 It is believed that ADP ribosylation factor 6 (ARF6) can meditate the freeing of protease‐loaded vesicles from the cellular membrane due to the crosstalk with Rho signalling pathways.24, 25, 26 Moreover, microvesicles are specifically produced in the cellular membrane regions that are linked to be enriched in cholesterol, ceramide and lipid rafts.27 TSG101 is also known to interact with accessory proteins Alix and arresting domain‐containing protein‐1 (ARRDC1) during releasing microvesicles, illustrating that microvesicles sharing some same characteristics with exosomal biogenesis.28 As described for microvesicles, oncosomes and large oncosomes are generated by plasma membrane budding, with amoeboid‐like phenotype. Notably, oncosomes and large oncosomes specially derived from cancerous cells are indicated to play vital roles in malignancies invasion. The term “oncosome” is firstly referred that the EVs with size range from 100 to 500 nm. Subsequently, large non‐apoptotic EVs were detected in prostate tumours with their unusual size in 1‐10 mm, called large oncosomes. Currently, some studies demonstrate that oncosomes and large oncosomes are definitely different variety of EVs respecting to their size, cargo contents and target effects, and additional studies are therefore of the essence to clarify differences between oncosomes and large oncosomes.
Apoptotic bodies, imposing an effect on the cellular response by transmitting their substance towards receptor cells, are generated during undergoing programmed cell death with 500‐4000 nm, and their content contains fragmented cytoplasmic organelles as well as destructive nuclei.29, 30
3. ISOLATION TECHNIQUES OF EVS
No remarkable consensus is in existence of the best approach for isolation, qualitative and quantitative analysis of EVs. There are listing several methods for the isolation of EVs (Figure 2) and demonstrate the available disadvantages and advantages as well (Table 2).31
Figure 2.
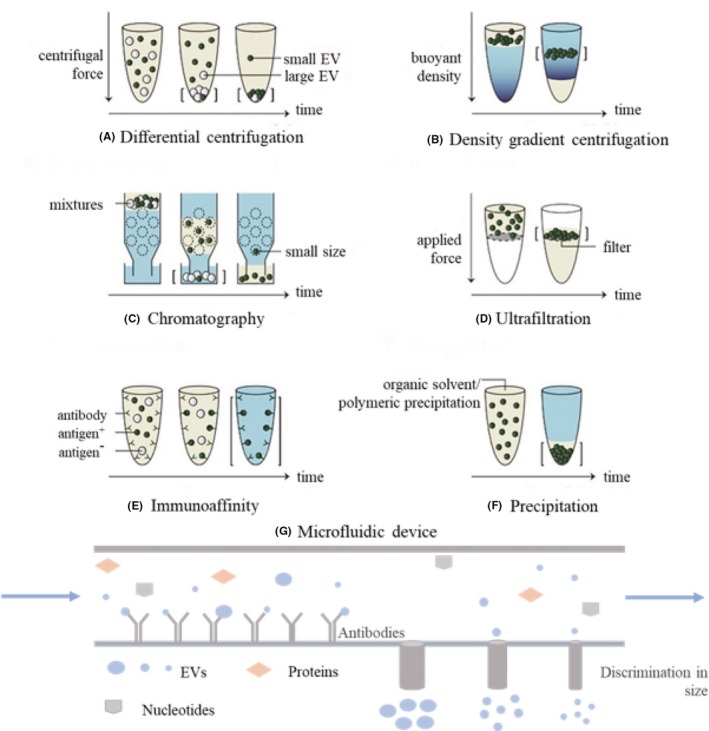
The common methods to isolate EVs. Straight brackets: isolated EVs; yellow: soluble components; and blue: buffer. A, In differential centrifugation, separation is based on sedimentation velocity, largely depended by size; B, in density gradient centrifugation, separation is relied on buoyant and density; C, size exclusion chromatography uses a porous matrix (dotted circles) that separates on size; D, in ultrafiltration, separation is based on size; E, in immunocapture assays, EVs are captured based on the presence of specific EVs surface molecules. F, in precipitation, EVs isolation via adding some water‐excluding polymers to sample to force the precipitation of small EVs out. G, In microfluidic device, EVs isolation via combining several methods such as immunoaffinity and filtration systems. Copyright 2017, University of Helsinki, Frank AW Coumans31
Table 2.
Summary of EVs isolation techniques
| Methods | Isolation method | Isolation Principle | Advantages | Limitations |
|---|---|---|---|---|
| Centrifugation | Differential centrifugation | Sedimentation velocity |
|
|
| Density gradient centrifugation | Buoyant density |
|
|
|
| Filtration | Ultrafiltration | Size |
|
|
| Chromatography | Size/charge |
|
|
|
| Immunoaffinity | Immunological separation | Presence of specific EVs surface molecules |
|
|
| Precipitation based | Polymeric precipitation | PEG precipitation |
|
|
| Protein organic solvent precipitation | The ion‐pairing effect |
|
|
|
| Microfluidics based | Microfluidics | Presence of specific molecules, Physical properties such as size, Microfluidic filtration |
|
|
Abbreviations: PEG, polyethylene glycol.
4. CENTRIFUGATION
In recent, differential centrifugation is the most common technique in responding to isolating EVs, and this approach is consisting of three main centrifugation processes: low speed to eliminate a main portion of the cells, then intermediate speed to subside cell debris and aggregate biopolymers and the other structures with density higher than that of EVs and finally high speed to pellet extracellular vesicles. The advent of density gradient ultracentrifugation increases the efficiency of particle separation according to their buoyant density. However, an important disadvantage of differential centrifugation cannot thoroughly separate protein and other non‐exosomal particles from EVs, limiting its efficacy and usage in clinical studies for diagnosis, to large extent, the advent of density gradient ultracentrifugation that reverses the poor separation efficiency due to their buoyant density, and it is frequently used for EV isolation though with a considerable loss of EVs.31, 32
5. FILTRATION
Due to the micropores or nanopores, EVs can also be isolated by numerous protocols (eg ultrafiltration and hydrostatic dialysis). Ultrafiltration is currently available method used for EV isolation, which represents an efficient alternative to ultracentrifugation. It involves the use of membrane filters with a narrow range of pore size distribution to deplete the proteins with molecular weight over 100 kDa, cell debris and floating cells. However, the size of filters is especially suitable for cell culture media and urine samples, which limit their applications to clinical routine tests. Hydrostatic filtration dialysis (HFD), another approach developed for isolating EVs from urine samples, shows its advantages on the removal of ultracentrifugation via multiple steps and the possibility of isolating EVs from highly diluted solutions. Gel filtration, also called size exclusion chromatography, is one of the methods to collect EVs based on the basis of the size differences, and the disadvantages of this method are its low yield and rather expensive chromatographic sorbent.33, 34
6. PRECIPITATION METHODS
The current research provides numerous protocols for EV isolation via adding some water‐excluding polymers to sample, and subsequently force the precipitation of small EVs out, its time‐saving and easy usage make it suitable for clinical use, though show unavoidable contamination of the isolated EVs with proteins, protein complexes, lipoproteins and nucleoproteins, as well as viral and other particles.35, 36 There are several polymer kits available that have already applied for purifying the EVs, such as hydrophilic polymers, protamine, sodium acetate and proteins with organic solvent (PROSPR) with along their disadvantages and advantages demonstrating in Table 2.
7. IMMUNOAFFINITY ISOLATION METHODS
Immunoaffinity isolation is another approach to isolate the EVs with increasing purity, owing to selectively exploiting the presence of specific molecules in the small EV surface37; for example, the lipids, proteins and polysaccharides are common substances that exposed on the surface of EVs, as a result, showing potency in being ligands for manifold molecules. Generally, there are five main methods for the isolation of EVs based on immunoaffinity, including antibodies to EV receptors,38 phosphatidylserine‐binding proteins,39 heparin‐modified sorbents40 and binding of heat shock proteins,34 as well as lectins.41 Although along with evident advantages of the EV purified isolation, the expensive costs, and the insufficient efficiency of isolation, and difficulties encountering in the process of isolation the large volumes of EVs, which substantially limits the applicability of immunoaffinity isolation methods.
8. MICROFLUIDIC DEVICES
Microfluidic devices are composed of a network of microchannels with different sizes, which have been implicated for EV isolation from cell culture and various tissue fluids on the basis of the immunoaffinity principle, as well as systems. However, some issues are yet to be removed; for instance, the inputted sample shows great possibility to block channels and the efficacy of isolation of EVs is extremely slow, consequently, decreasing their diagnostic potential,38 and overcome some of the challenges involving in EVs detection, such as the problem of the small size and lacking in distinct biomarkers, which contributing to get a comprehensive understanding function of their contents (eg protein, RNA and lipid).
Several qualitative and quantitative analysis techniques are currently available (Table 3). For instance, transmission electron microscopy (TEM) could be combined with immunogold staining to represent structural details and delineate the subpopulations of EVs.42 A study indicates that cryo‐electron microscopy might be more suitable for depicting the morphology of EVs as its no fixation or staining.43 The size, morphology and intactness of EVs also could be determined by scanning electron microscopy (SEM) and atomic force microscopy (AFM).44 Measuring the size and number distribution of single EVs can be made by dynamic light scattering (DLS) and nanoparticle tracking analysis (NTA).45 Both the conventional flow cytometry and novel fluorescence‐based flow cytometry could be promising tools to qualitatively and quantitatively analyse the EVs.46 Western blot, enzyme‐linked immunosorbent assay (ELISA) and EVs arrays are used to present purity and enrichment. Micronuclear magnetic resonance [μNMR] system and a photosensitizer‐bead detection system (ExoScreen) are other sensitive qualitative and quantitative approaches.47, 48, 49
Table 3.
Techniques for extracellular vesicle detection and characterization
| Methods | Size detection range/detection limit | Size distribution | Concentration | Marker detection |
|---|---|---|---|---|
| Quantitative methods | ||||
| DLS | 1 nm‐6 μm | + | − | − |
| qNano | 70 nm‐10 μm | + | + | − |
| Qualitative methods | ||||
| Western blot and ELISA | NA | − | − | + |
| Extracellular vesicle array | NA | − | − | + |
| TEM | <1 nm | + | − | + |
| SEM | ~1 nm | + | − | + |
| Cryo‐EM | <1 nm | + | − | + |
| AFM | <1 nm | + | − | − |
| Quantitative and qualitative methods | ||||
| NTA | 50 nm‐1 μm | + | + | + |
| Conventional flow cytometry | ≥300 nm | − | + | + |
| <300 nm | − | − | + | |
| TRPS | 70 nm‐10 μm | + | + | − |
| Fluorescence high‑resolution flow cytometry | ~100 nm | − | + | + |
| μNMR system | 50‐150 nm | − | + | + |
| nPLEX assay | NA | − | + | + |
| ExoScreen | NA | − | + | + |
“+” indicates variable can be measured, “−” indicates it cannot.
Abbreviations: AFM, atomic force microscopy; Cryo‐EM, Cryo‐electron microscopy; DLS, dynamic light scattering; nPLEX, nanoplasmonic exosome; NTA, nanoparticle tracking analysis; SEM, scanning electron microscopy; TEM, transmission electron microscopy; TRPS, Tunable resistive pulse sensing; μNMR, micronuclear magnetic resonance.
As mentioned above (Table 2), each type of isolation approach has intrinsic advantages and restrictions with respect to cost‐efficacy, complexity, purity, yield and functionality of the EVs. In urological neoplasms similarly, one of the current challenges is how to develop the sensitive detection platforms and robust and remarkable isolation techniques, with promising potency in the identification of EVs and their subpopulations to trigger reliable prognosis and precise prediction of treatment response, or to provide novel neoplasm grading and staging via analysing of easier accessible and minimal‐invasive body fluids. Therefore, many standards should be established to develop such interesting EVs isolation and capture tools. Initially, it is highlighted that the optimal specimen pools should be determined, such as give priority to the sample safety and accessibility, target EVs quantity, and simple and convenient manoeuvrability, especially urine for urogenital tumours; second, the preservation conditions such as temperature, time and additives should be standardized because the quantity and variation of EVs could diminish differential preservation conditions, consequentially giving rise to the deflection of results but also difficulty in continued supervision of cancer development; and third, promising isolation and detection techniques that satisfy the clinical application in a hospital setting should be established.
9. GENERAL FUNCTIONS OF EVS IN MALIGNANCIES
Bioactive molecules of EVs secreted by both cancer cells and tumour‐associated cells provide the essential signals for favouring tumour growth via remodelling the architectures in tumour microenvironments and forming pre‐metastatic niches. Different mechanisms of EVs‐mediated tumour proliferation and progression will be discussed in the following sections (Figure 3).
Figure 3.
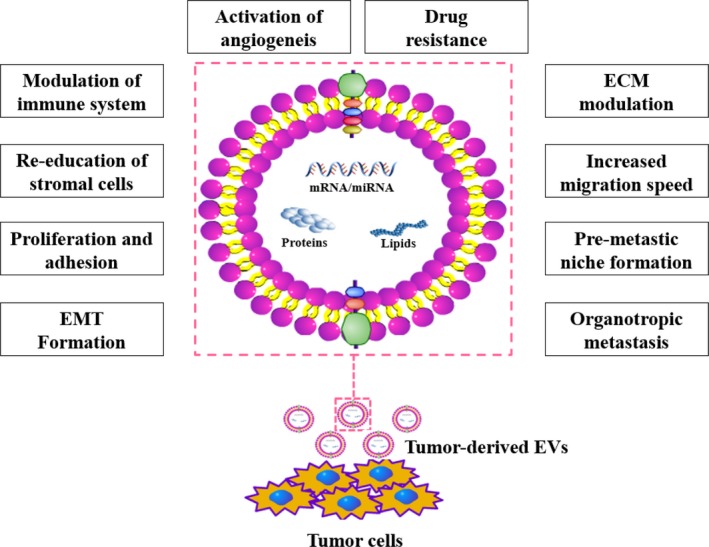
Physiological processes influenced by EVs. Extracellular vesicles are involved in most physiological processes that are associated with intercellular communication, and the content of extracellular vesicles, including mRNAs, microRNAs (miRNAs), lipids and proteins, is depicted
10. PROMOTION OF ANGIOGENESIS
Tumour progression is a dynamic and multistep process requiring continuous nutrient and oxygen supplied by sufficient blood conducts, while also serving to remove waste materials. The advent of cancer stem cells (CSCs) has provided a novel mechanism for the development and progression of the tumour via differentiating into endothelial cells to contribute to the angiogenesis.50, 51 In addition, a research indicates, for example, that miRNAs, secreted from exosomes, regulate transcription, proliferation, metabolic processing and mRNAs encode vascular endothelial growth factor (VEGF), fibroblast growth factor (FGF), angiopoietin1, Ephrin A3, matrix metalloproteinase‐2 (MMP‐2), and MMP‐9 and growth factors in contrast to CD105‐negative CSCs.19
11. EPITHELIAL‐TO‐MESENCHYMAL TRANSITION
Epithelial‐to‐mesenchymal transition (EMT) is a developmentally vital reversible process of which fully differentiated cells lose their epithelial features (eg E‐cadherin, β‐catenin and plakoglobin), acquiring a migratory mesenchymal phenotype (eg N‐cadherin and vimentin). EMT also contributes to the metastatic potential of tumours.52 Exosome mediates that growth in migration and invasion by the way of EMT has been observed in many other studies.53, 54, 55 Urothelial cells exhibit the EMT after exposing to tumour‐derived exosomes.56
12. FORMATION OF PRE‐METASTATIC NICHES
Primary tumours can release some biological factors that migrate to preferred metastatic regions and dynamically remodel these sites before spreading to a distant organ, which means that form predetermined metastatic microenvironments, also referred to as pre‐metastatic niches.57 In general, exosomes display the characteristics of organ tropism, and the process of the construction of pre‐metastatic niche involves with initial tumour‐derived exosomes releasing into the circulation system and then escaping from the vascular beds to migrate to distant secondary organ.58, 59 During the process, the crucial initial step is how vascular leaking exosomes can target organ tissues; nowadays, it is induced by complicated processes involved combination of stromal cells and released cancer cell‐derived exosomes resulting in reprogramming of these cells60, 61, 62, 63 and activation of several vital signalling pathways,64, 65 which alter the local chemokine repertoire of the tumour microenvironment (TME) and remodel the components of the extracellular matrix (ECM) in turns.61, 62, 66 Moreover, the vast researches indicate that there are exerting cooperations between bone marrow‐derived cells and exosomes in the primary phase of pre‐metastatic niche that can stimulate the mobilization of these cells into the circulatory system circulation and disseminate to distant sites, sequentially generate a local pro‐inflammatory focus with pro‐tumorigenic immunosuppression.60, 61, 62
13. MODULATION OF THE TUMOUR MICROENVIRONMENT
Accumulating evidence support tumour progression is the consequence of communication between tumour cells and cells within the tumour microenvironment via paracrine or autocrine, such as adipocytes, fibroblasts, immune cells and cells of the vascular. It has been shown that tumour cells have a higher propensity to secrete larger quantity of exosome67, 68; for instance, cancer‐associated fibroblasts (CAFs) support tumour cells in proliferation, delaying senescence and resisting against drugs, which induced by exosome‐secreted miR‐9 and the human telomerase reverse transcriptase (hTERT).69, 70 Activation of signalling pathways by exosomes is one of the way to modulate the tumour microenvironment, such as irritation of the TGF‐β/Smad pathway by transferring TGF‐β to human umbilical cord‐derived mesenchymal cells (hucMSCs), subsequently differentiating into CAFs.71 Moreover, recent research shows that transferring TGF‐β also contributes fibroblasts converse to myofibroblasts, which could secrete insulin‐like growth factor 1 (IGF‐1), activin A and VEGF to induce tumour progression.72 The irritation of another signalling pathway by exosomes in the bidirectional crosstalk between cancer cells and normal stromal cells, such as nuclear factor kappa B (NF‐κB) and epidermal growth factor receptor (EGFR) signalling, also plays vital roles in the proliferation and migration of tumour. Endothelial cells show enhanced cell motility and tube formation ability after re‐educated by tumour‐derived exosomes73; moreover, RNA secreted from EVs develops hepatocyte growth factor synthesis through the activation of ERK1/2 and AKT signalling pathways.74 It is widely believed that tumour‐derived EVs impose significant effects in mediating communication between immune and cancer cells of renal cell cancer (RCC),75 such as immune evasion of tumours.76, 77 MiR‐222‐3p induces polarization of tumour‐associated macrophages by the activation of SOCS3/STAT3 pathway to facilitate tumourigenesis and cancer progression.78 Additionally, Rab27a supports exosome could modify the tumour microenvironment via advancing recruitment and differentiation of bone marrow‐derived neutrophils to cancer cells.79 Furthermore, a few studies suggest that tumour‐derived EVs facilitate cancer progression by attenuating immune and more specific EVs could diminish the cytotoxicity of natural killer cells and T cell in immunoreaction.80, 81, 82 Tumour‐derived EVs also could influence the cancer cells themselves via autocrine to irritate the invasion and migration, and reduce adhesion abilities as well via enhancing MMP‐9 or chemokine receptor type 4 (CXCR4).74
14. MANAGEMENT OF UROLOGIC MALIGNANCIES
In the past several decades, although with the increasing development of renewable treatments for urological tumours, for instance chemotherapies and molecular targeted therapies and renowned immunotherapies, the prognosis remains poor. Recently, sufficient researches on EVs in urinary tumours provide a deeper understanding of biogenesis and pathogenesis and might be offered underlying therapeutic targets in urologic cancers. Herein, we review the current published research on EV for commonly urogenital carcinomas including bladder cancer (BCa), kidney cancer (RCC) and prostate cancer (PCa).
15. KIDNEY CANCER (RENAL CELL CANCER)
Renal cell cancers (RCCs) represent 2%‐3% of all cancers.83, 84 Many kidney cancers remain asymptomatic until the late disease stages with 50% of patients are detected incidentally by non‐invasive imaging investigating, and approximately 30% of patients with metastasis in the primary time of diagnosis.85, 86 The utility of urinary EVs could be recognized as potential diagnostic and prognostic markers in RCCs (Table 4).87 Proteomic analysis of urinary EVs differs from the patients and healthy groups with showing an RCC‐specific signature of the effectiveness of proteins.58, 88 Another proteins, for instance, podocalyxin (PODXL), WNT signalling pathway inhibitor 4 (DKK4), ceruloplasmin, carbonic anhydrase IX (CAIX) and MMP‐9, were validated by using immunoblotting method.89 In addition, among RCC patients, the mRNA levels of glutathione S‐transferase alpha 1 (GSTA1), CCAAT enhancer binding protein alpha (CEBPA) and PCBD1 in EVs decrease compared with healthy groups.90 Moreover, a report demonstrates that the lipids within urinary EV show difference between RCC patients and healthy groups.91 Based on microRNA expression screening, the cluster of miRNAs including miR‐449a, miR‐126‐3p, miR‐486‐5p and miR‐34b‐5p could differentiate clear cell RCCs from benign subjects92 and could be recognized as potential diagnostic and prognostic markers.58, 93, 94 Serum‐derived EVs have also been recognized as prominently diagnostic and prognostic tools for clear cell RCCs.93, 94 Recently, azurocidin as a permeabilizer for vascular endothelial cells has been isolated from serum‐free medium within incubating clear cell RCC tissues samples.95
Table 4.
Candidate biomarkers for kidney cancer derived from EVs
| Source | Methodologies | End point | Type of marker | Markers | Reference |
|---|---|---|---|---|---|
| Urine | Ultracentrifugation | Diagnosis | mRNA | GSTA1, CEBPA and PCBD1 | De Palma et al89 |
| Urine | Density gradient ultracentrifugation | Diagnosis | Proteins | MMP‐9, PODXL, DKK4, CAIX and ceruloplasmin | Raimondo et al88 |
| Cancer stem cells | Ultracentrifugation, Flow cytometry immunohistochemistry | Diagnosis | Proteins | VEGF, FGF, angiopoietin 1, Ephrin‑A3, MMP‑2, MMP‑9 | Grange et al57 |
| RCC cells |
Centrifugation Filtration Flow cytometry Western blot ELISA |
Diagnosis | Proteins | Fas ligand, Bcl2‑L‑4 | Yang et al87 |
| Viable human tissue |
Ultracentrifugation Mass spectrometry Western blot |
Diagnosis | Proteins | Azurocidin 1 | Jingushi et al93 |
| Serum | Immunoaffinity magnetic beads | Diagnosis | miRNA | miR‐210 and miR‐1233 | Zhang et al92 |
| Serum | Total Exosome isolation kit | Prognosis | miRNA | miR‐224 | Fujii et al93 |
| Cancer stem cells |
Ultracentrifugation Microarray analysis qRT‑PCR |
Diagnosis | miRNA | miR‑200c, miR‑92, miR‑141, miR‑19b, miR‑29a, miR‑29c, miR‑650, miR‑151 | Grange et al94 |
| Urine |
Centrifugation Urine exosome RNA isolation kit |
Diagnosis | miRNA | miR‐126‐3p, miR‐449a, miR‐34b‐5p, miR‐486‐5p | Butz et al91 |
| Urine |
Ultracentrifugation Mass spectrometry |
Diagnosis | Lipids | Lysophosphatidylethanolamine metabolite | Del Boccio et al90 |
Renal cell cancers generate EVs that could educate endothelial95 and immune cells,96 with promoting angiogenesis and immunosuppressive activity, respectively. As for immune systems, increasing evidence shows that tumour‐derived EVs result in immune evasion of tumours partly via the activation of caspase pathway to trigger apoptosis in T lymphocytes.97 In addition, both EVs‐derived antigens and Hsp70 can inhabit the immunoreaction though induction tumour growth factors of and pro‐inflammatory cytokines similarly.98, 99 Moreover, EVs also can be applied for cancer immunotherapy, mainly due to promoting cytotoxic effects and proliferation of T cells via releasing interferons.100 Furthermore, RCCs also could secrete EVs to interact with endothelial cells to promote lymphopoiesis and angiogenesis and thereby metastasis. A study shows that EVs derived from RCC cell line (eg 786‐O) increase the expression of vascular endothelial growth factor in human umbilical vein endothelial cells (HUVECs) with resulting in tubular formation of HUVECs,97 involving the downregulation of hepatocyte cell adhesion molecule by upregulating phosphorylated AKT in RCCs.101 What is more, CD105+ stem cells of RCC release EVs that promote and trigger the formation of a pre‐metastatic niche by upregulating MMP2 and VEGF58, 102; although sunitinib is a first‐line targeted regimen for metastatic RCCs,103 its efficacy of biotherapy for the long time is controversial in respect to drug resistance, which has been confirmed that regulated by lncARSR by promoting MET and AXL expression in RCC cells.104, 105 Therefore, all those support that EV‐based targets display a promising potency for the development of novel cancer therapies.
EVs also can be served as a vaccine for RCCs. A report reveals that RCCs derived from EVs could increase immunogenicity by proliferating T cells and releasing interferons subsequently100; the effectiveness of vaccines when dendritic cells (DCs) load with EVs is higher rather than whole tumour lysate.98
16. THE BLADDER CANCER
Bladder cancer (BCa) is the seventh most commonly diagnosed cancer in the male population worldwide, and the diagnosis for BCa is usually on the basis of cytology, urinalysis and cystoscopy. Cytology is a highly specific test, but low in the sensitivity for the diagnosis of BCa.106 Cystoscopy is the gold standard to diagnose the BCa, while this method is expensive and invasive, even for flexible cystoscopy, and the risk of developing urinary infections is up to 10%107; non‐invasive and reliable biomarkers are therefore required in the future. Given that, urine is an excellently suitable fluid for biomarkers discovery in BCa. The biomarkers (mainly including proteins, mRNA, lncRNA and miRNA) within EVs isolated from BCa were investigated by different research groups, which could be promising molecules to identify the BCa and predict the progression of the BCa (Table 5). Based on proteomic analysis of urinary EVs, several studies have been identified cargoes of possible biomarkers for BC patient, but not in healthy volunteers.108, 109 Among them, research shows that the levels of tumour‐associated calcium‐signal transducer 2 (TACSTD2) are correlative with BCa, compared with high‐grade BCa. The author identified seven proteins differentially expressed in the low‐risk group (Table 5).109 Proteome profiling of urinary exosomes indicates H2B1K and alpha 1‐antitrypsin as prognostic and diagnostic biomarkers for urothelial bladder cancer, which could be verified in immunohistochemistry (IHC).110 Additionally, HEXB, S100A4 and SND1 significantly identified in EV derived from the MIBC cell line also are upregulating in urinary EV from MIBC patients when vs to normal groups.111 There are other proteins could be recognized as potential diagnostic and prognostic markers for BCa.112, 113, 114, 115 Using a whole transcriptome array, a study reports the potential application of mRNAs in urinary EVs for diagnoses, such as LASS2 and GALT1 involving progression with only in BCa patients, and ARHGEF39 and FOXO3 while only expressing in healthy controls to suppress the tumour.116 In addition, based on the microarray analysis of miRNA, great studies pay their attention on the roles of diagnostic and prognostic.117, 118, 119, 120, 121 Interestingly, research shows that several microRNAs from urinary EVs significantly upregulate in BCa, but not in plasma from same patients, which suggests that different biofluids may harbour different molecules.122
Table 5.
Candidate biomarkers for bladder cancer derived from EVs
| Source | Methodologies | End point | Type of marker | Markers | Reference |
|---|---|---|---|---|---|
| BCC/urine |
Ultracentrifugation Flow cytometry In‑gel digestion Mass spectrometry |
Diagnosis | Proteins | β1 and α6 integrins, CD36, CD44, CD73, CD10, MUC1, basigin, 5T4 | Welton et al107 |
| Urine | Ultracentrifugation | Diagnosis | Proteins | APOA1, CD5L, FGA, FGB, FGG, HPR, HP | Chen et al108 |
| Urine | Differential ultracentrifugation | Diagnosis Prognosis | Proteins | Alpha‐1 antitrypsin, histone H2B1K | Lin et al109 |
| Urine | Differential ultracentrifugation | Diagnosis | Proteins | HEXB, S100A4, SND1 | Silvers et al110 |
| BCC/urine |
Sucrose/D2O cushion Ultracentrifugation |
Diagnosis | Proteins | EDIL‐3 | Beckham et al111 |
| Urine |
Ultracentrifugation In‑gel digestion Mass spectrometry |
Diagnosis | Proteins | Resistin, GTPase NRas, MUC4, EPS8L1, EPS8L2, EHD4, G3BP, RAI3, GSA | Smalley et al112 |
| Urine |
Centrifugation Filtration Integrated double‐filtration Microfluidic device |
Prognosis | Proteins | CD63 + EV signal intensity | Liang et al113 |
| Urine | Ultracentrifugation | Prognosis | Proteins | Periostin | Silvers et al114 |
| Urine |
Ultracentrifugation NanoSight microarray PCR |
Diagnosis | mRNA | LASS2, GALNT1 | Perez et al115 |
| Urine | Differential ultracentrifugation | Diagnosis | miRNA | miR‐21‐5p | Matsuzaki et al116 |
| Urine |
Differential ultracentrifugation Filtration |
Diagnosis Prognosis |
miRNA proteins |
miR‐375, miR‐146a, apoB | Andreu et al117 |
| Urine |
Differential centrifugation Total exosome isolation kit |
Prognosis | miRNA | miR‐141‐3p, miR‐200a‐3p, miR‐205‐5p | Baumgart et al118 |
| Urine | Differential ultracentrifugation | Prognosis | miRNA | miR‐940 | Long et al119 |
| Urine |
Nanostring miRNA assays Droplet digital PCR |
Diagnosis | miRNA |
miR‐205, miR‐200c‐3p, miR‐29b‐3p; miR‐921, miR‐23b |
Ostenfeld et al120 Berrondo et al122 |
| Urine |
Centrifugation Exosome RNA isolation kit |
Diagnosis | miRNA | miR‐4454, miR‐21, miR‐720 | Armstrong et al121 |
| Urine | Differential ultracentrifugation | Diagnosis Prognosis |
mRNA; lncRNA |
HOTAIR, HOX‐AS‐2, MALAT1, SOX2, OCT4, HYMA1, LINC00477, LOC 100506688, OTX2‐AS1 | Berrondo et al122 |
Furthermore, to unravel the roles of lncRNA in BCa, recent researches show that lncRNAs, such as HOX‐AS‐2, HOTAIR, ANRIL and lnc‐RoR, are enriched in BCs cancer cell line EVs as well as urinary EVs from high‐grade BCa patients, demonstrating that lncRNAs have potential as biomarkers for BCa.123 Additionally, recent reports demonstrate that the CD63+ urinary EVs could be a biomarker for the detection of BCa.114, 124 EVs derived from BCa cell line also influence local regions microenvironments or distant cells by transferring their content, as result of facilitating proliferation, angiogenesis, invasion, and migration and the inhibition of apoptosis56, 112, 125; for example, EGF‐like repeat and discoidin I‐like domain‐containing protein 3 (EDIL‑3) from invasive BCa cell lines could stimulate the migration and angiogenesis of urothelial and endothelial cells.112 Periostin, another factor from invasive BCa cell lines, could contribute low‐grade BCa cancer cells to gain the aggressiveness within via activating ERK oncogenic pathway.115 Urothelial cells exposed to EVs from cancer cell lines or patient specimen show the phenomena of EMT.56 Similarly, lncRNA‐UCA1, which may be play an important role in causing intratumoural hypoxia, could irritate tumour progression via the EMT as well.126 However, exosomes also can discard tumour‐suppressive miRNAs contributed to BCa progression, such as miR‐23b and miR‐921,121 all those could provide underlying targets for the future therapies for the BCa. Recently, a report demonstrates the effectiveness of EVs as a vector to carry siRNAs in BCa; therefore, in the future, EVs have the potential functions to stably deliver substantial therapeutic cargoes including miRNAs and siRNAs, to anchor organs with the development of tissue engineering technology.127
17. PROSTATE CANCER
Prostate cancer (PCa) is the second most commonly diagnosed cancer in men, accounting for 15% of all cancers diagnosed.128 Although PSA testing contributes to identify and manage PCa in the early phase, it still has some limitations, for example, the specificity of discrimination of benign prostate diseases, such as acute prostatitis and benign hyperplasia.129 Thus, the more specific and ideal substrate (eg urine, prostatic plasmas and blood samples) for PCa are urgently developed rather than invasive prostate biopsies.130 Some studies have presented the usefulness of urinary EVs as diagnostic factors (Table 6).131, 132, 133, 134 EV‐involved transmembrane proteins CD63 and CD9 are sufficient in urine from PCa.135 Integrin α3, δ‐Catenin, integrin β1 and FABP5 proteins are identified in urinary EVs of PC patients with the significantly increased levels of PCa patients during the process of the investigation of the proteomic cargo of urinary PC‐derived EV.136, 137, 138, 139 Moreover, on the basis of the mass spectrometry proteome analysis, thousands of proteins encapsulated on and within vesicles are identified as biomarker candidates from urinary EVs or cell lines of PCa,139, 140, 141, 142, 143, 144, 145, 146 whereas the value as biomarkers is still controversial, and several types of research regenerate the previous biomarkers by the using targeted proteomics and immuno‐assays.134 Based on a proximity ligation assay, PC‐derived EVs in blood has also shown to contain proteins specific to PCa, such as phosphatase and tensin homolog gene (PTEN), survivin and other factors with decreased level compared to benign prostatic hyperplasia or health subjects.133, 147, 148, 149 PC‐based EVs show great value in comprehensively mapping nucleic acid changes in PCa via urine‐ or blood‐based liquid biopsies, and it has been known that the EVs is important pool resource for circulating‐free DNA (cfDNA)150; as such, some factors demonstrated good clinical usefulness and diagnostic value in predicting for high‐grade PC, including TP53 mutations, ERG and PCA3, can be detected in the EVs.150 ExoDx Prostate IntelliScore urine exosome assay has been developed as a non‐invasive detective tool to distinguish the high‐grade PCa from low‐grade groups and benign diseases at initial biopsy.151 Several mRNAs could be recognized as promising diagnostic and prognostic tools for the PCa152, 153, 154, 155, 156; for instance, the transcripts of CDH3 from EVs were significantly decreased compared with benign hyperplasia156; contrarily, the mRNA level of PTEN gene can only be detected in the patient of PC,147 and nevertheless, both of them are expected to be powerful for the diagnosis and monitoring of PCa.152 RT‐PCR, microarray and RNA sequencing technologies have focused on the non‐coding RNA content within EVs so far; for example, in the urinary sample, the levels of lncRNA‐p21, a suppressor of p53 signalling, contribute to detecting PC from benign disease.157 Next‐generation sequencing reveals the potential values for miRNA served as diagnostic and prognostic biomarkers for PCa within serum or plasma EVs,41, 131, 132, 158, 159, 160, 161, 162 such as miR‐141 and miR‐375 in serum, have been correlated with metastatic PCa.159, 163 Another study indicates that exosomal miR‐1290 and miR‐375 could be as prognostic markers in castration‐resistant prostate cancer (CRPC).164 In recent, several research works demonstrate that the lipids including diacylglycerol and triacylglycerol are differentially enriched in PCa rather than healthy groups.165, 166 Glycomic and metabolomic profiling of urinary EV reveal several small molecule metabolites could be novel biomarkers to predict the development of PCa, for example levels of N‐linked glycans, glucuronate, adenosine, d‐ribose‐5‐phosphate and isobutyryl‐l‐carnitine.167, 168
Table 6.
Candidate biomarkers for prostate cancer derived from EVs
| Source | Methodologies | End point | Type of marker | Markers | Reference |
|---|---|---|---|---|---|
| Plasma |
Ultracentrifugation Western blot ELISA |
Diagnosis | Proteins | Survivin | Khan et al132 |
| Urine | Differential ultracentrifugation Filtration | Diagnosis Prognosis | Proteins |
TGM4, ADSV, PPAP, PSA, CD63, SPHM, GLPK5 TMEM256, flotillin 2, Rab3B, PARK7, LAMTOR1 TM256, LAMTOR1, ADIRF TMEM256, flotillin 2, Rab3B, PARK7, LAMTOR1 |
Wang et al133 Sequeiros et al145 |
| Urine | Differential ultracentrifugation |
Diagnosis Prognosis |
Proteins |
δ‑catenin Integrin α3, Integrin β1 FABP5 |
Liu et al135 Lu et al136 Fujita et al138 |
|
Tissue Urine |
Differential ultracentrifugation Filtration |
Diagnosis Prognosis |
Proteins | CD63, ANXA1‐3, FASN, FOLH1, GDF15, MDR1, XPO1, TGM4, TIMP1, SFN, TMEM256, LAMTOR1, ADIRF, ITGA3, and ITGB1 | Bijnsdorp et al137 |
| Tissue |
Ultracentrifugation Gel filtration Chromatography, 2D‐PAGE Mass spectrometry |
Diagnosis Prognosis |
Proteins | ANXA1, ANXA3, ANXA5, DDAH1 | Ronquist et al140 |
| Cell lines |
Ultracentrifugation Mass spectrometry Bead immuno‑isolation Western blot |
Diagnosis Prognosis |
Proteins | CDCP1, CD151, CD147 | Sandvig et al141 |
|
Cell lines Urine |
Ultracentrifugation immunoprecipitation Western blot Electron microscopy |
Diagnosis Prognosis |
Proteins |
ANXA2, CLSTN1, FASN, FLNC, FOLH1, GDF15ACPP, LTF, DDP4, TGM4, MME, PSA, SEMG1, AZGP1, ANPEP, G3BP, PSMA, TMPRSS2, FASN, LGALS3, PSCA, KLK2, KLK11, TIMP1 PDCD6IP, XPO‑1, ENO1 |
Duijvesz et al142 Utleg et al143 Principe et al144 |
| Plasma |
Ultracentrifugation Western blot immunofluorescence |
Diagnosis | Proteins | PTEN | Gabriel et al134 |
| Serum | Differential centrifugation |
Predictive Monitoring |
Proteins | ABCB1, ABCB4, PABPC4 | Kato et al147 |
|
Urine Plasma |
Differential ultracentrifugation Filtration Chromatography |
Diagnosis | Proteins | Afamin, cardiotrophin‐1, CDON, endoplasmic reticulum aminopeptidase 1, FGF19, IL17RC, NAMPT, IL1RAPL2, CD226, IGFBP2, CCL16, TNFSF18, IGFBP5; Aromatic‐l‐amino‐acid decarboxylase | Welton et al148 |
| Urine |
Centrifugation Filtration Ultrafiltration |
Diagnosis Prognosis |
mRNA |
PCA3, TMPRSS2‐ERG AGR2, SV‐G, AGR2 SV‐H CDH3 |
Neeb et al151 Donovan et al152 Hendriks et al153 Motamedinia et al154 Royo et al155 |
| Plasma/Serum/Urine |
ExoMiR extraction Filtration qRT‑PCR |
Diagnosis | miRNA |
miR‑107, miR‑130b, miR‑181a‑2, miR141, miR‑301a, miR‑326, miR‑331‑3p, miR‑375, miR‑432, miR‑574‑3p, miR‑22110, miR‑625 miR‐1290, miR‐375, miR‐574‐3p, miR‐141‐5p, and miR‐21‐5p miRNA‐21, let‐7c, miR‐196a‐5p, miR‐501‐3p, miR‐19b, miR‐145 |
Samsonov et al41 Foj et al130 Bryant et al158 Bryzgunova et al159 Rodríguez et al131 Wani et al160 Xu et al161 |
| Urine |
Differential centrifugation Urine exosome RNA isolation kit |
Diagnosis | lincRNA | lincRNA‐p21 | Işin et al156 |
| Urine |
Differential ultracentrifugation Filtration |
Diagnosis | Lipids | Lactosylceramide, phosphatidylserine, phosphatidylglycerol, diacylglycerol, triacylglycerol |
Skotland et al164 Yang et al165 |
| Urine |
Differential ultracentrifugation Filtration |
Diagnosis | Metabolites | Adenosine, glucuronate, isobutyryl‐l‐carnitine, d‐ribose 5‐phosphate | Puhka et al166 |
| Urine | Differential ultracentrifugation | Diagnosis Prognosis | Glycomic | N‐linked glycans | Nyalwidhe et al167 |
The intercellular crosstalk through EVs could stimulate tumour progression. Several proteins presenting on and in EVs from PCa cell lines are recognized as significant meditators for the biological communication between cancer cells and tumour microenvironment or surrounding cell, including cytokine CX3CL1, MMPs and transforming growth factor B, play significant roles in the proliferation and differentiation of fibroblasts.169, 170 In addition, integrins ITGA3 and ITGB1 can affect invasion and migration of normal prostate epithelial cells.138 Several studies suggest that complicate intercellular interactions between cancer cells, osteoclasts and osteoblasts contribute to bone metastasis.171, 172 It is the first protein that has been reported in the EVs originated from human hormone‐refractory PCa cells to facilitate mouse pre‐osteoblast differentiation.171 Another molecular miR‐141‐3p is involved with the osteoblastic metastasis of prostate cancer via reducing the expression of Deleted in Liver Cancer 1 (DLC1) and activating the p38MAPK and OPG/RANKL pathway in osteoblasts173; however, another research fixes their attention on the role of EVs derived from PCa cell line in the osteoclast for the bone metastasis promoting osteoblast proliferation and identifies that tumour cell‐derived EVs play important roles in impairing the osteoclast formation and differentiation through the underlying mechanism is unknown yet.172 Moreover, the effect of biological crosstalk between PCa cell and immune cells though EVs causes the induction immune suppression via down‐regulating the NKG2D cytotoxicity receptor and diminishing the IL‐2 response.174, 175
PCa‐derived EVs also involve drug resistance. ABCB1, ABCB4, PABPC4 and SH3GL1 are much more sufficient in EVs from docetaxel‐resistant prostate cancer cell lines and potentially higher in serum EVs in men with docetaxel‐resistant PCa.148 AR‐V7 is correlated with resistance to enzalutamide and abiraterone in metastatic CRPC patients, which could be a biomarker to predict the CRPC.176, 177 Finally, EVs‐derived PCs can be used as vaccine vesicles that present prostate‐associated antigens such as PSA and PAP on their membrane to exert an anti‐tumour immune response.98, 178, 179
18. DISCUSSION
With the advent of novel concepts involving EVs in many physiologic as well as pathologic conditions, the field of EV research develops much excitement in the urologic malignancies. Unlike conventional biopsies of that only consist of a small amount of tumour solid masses with ignoring heterogeneity, EVs, as liquid biopsies, could capture and obtain overall tumour heterogeneity owing to directly releasing from all cells in the cancer tissue and its microenvironment. Beyond a doubt, specific bioactive contents contained in circulating EVs have great promise as reliable surrogates of urological cancers; therefore, their molecular cargoes including nucleic acids, protein and lipid composition, as well as their numbers, are representing as diagnostic, prognostic biomarkers for urinary tract diseases, and immense promising for therapeutic advancements. To conclude, we have a faith for the implement of EVs in urological cancers diagnosis and therapeutics owing to their enormous potencies in several aspects, as described below (Figure 4).19
Figure 4.
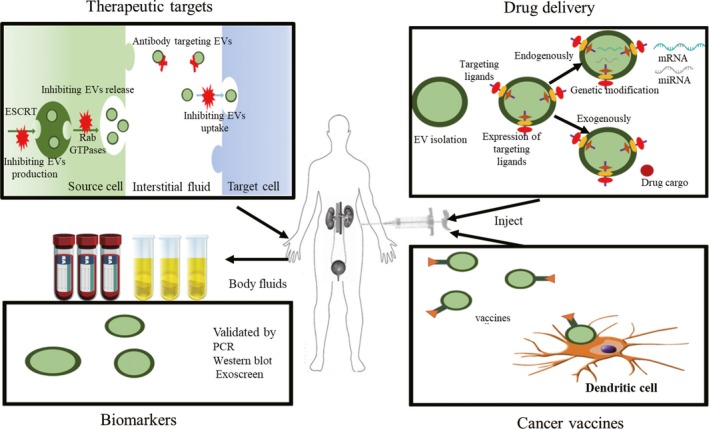
Future implements for EVs in urological cancer. EVs impact the multistep process of cancer; therefore, EVs should be a novel treatment strategy by inhibiting intercellular crosstalk. EVs could serve as promising diagnostic and prognostic biomarkers to dynamically trace the changes in cancer due to their high specificity and sensitivity. In addition, EVs have the potential functions to stably deliver substantial therapeutic cargoes liking miRNAs and siRNAs with stability, few side effects and organ specificity. Furthermore, several studies have reported the potential of EVs derived from dendritic cells used as vaccine vesicles. Copyright 2018, The Jikei University School of Medicine, Fumihiko Urabe19
19. ROLES IN DIAGNOSTIC AND PREDICTIVE BIOMARKERS
As more is understood about the fundamentals of EVs biology and roles involved in tumorigenesis and therapy resistance, EVs‐based analytical methods are increasingly interesting targets for clinical application. EVs are directly released from heterogeneous tumorous and reflect a snapshot of the current state of the neoplasm; therefore, EVs have great potential as remarkable, specific and sensitive biomarkers of oncogenesis, treatment response and therapy resistance. In urogenital cancers, it is thought that increased exosomes are produced by more advanced cancers, and it thus has been suggested that total circulating exosome burden may serve as indicators for disease surveillance. Exosomal contents can also identify disease or predict treatment response, such as several proteins (eg PD‐1, PD‐L1) or some nucleic acids (eg miRNA) with the roles as diagnostic biomarkers for cancer, indicators for therapeutics, worse still, research to date strongly indicate EVs involve treatment irresponsiveness. Such phenomenon was observed in various malignancies also including urogenital cancers; for example, docetaxel‐sensitive cell lines of prostate carcinoma undesirably acquire drug resistance again when co‐culture with EVs derived from the drug‐resistant cells.180 To date, increased pumping agents out of tumorous cells or omics alternations induced by the cargo of EVs are two most common opinions, but the underlying mechanisms are still entirely unknown. Further investigations that tailored clinical studies are now warranted to determine how best to prevent this occurring, in the interest of patients and also for economic benefit. Additionally, endothelial cell‐derived EVs can reflect transient cellular stress conditions and could be useful as predictors of anti‐angiogenic therapy effectiveness and cancer cell status. However, issues with interpretation of studies and reproducibility have arisen due to the deficit of standard isolation and characterization, and nomenclature employed, and as the result of the publication of the Minimal Information for Studies of Extracellular Vesicles 2014 (MISEV 2014) with emphasizing that build the set of biophysical, biochemical and functional standard that help in detecting particular biological cargo or functions in extracellular vesicles.181 Until 2018, the updated MISEV guidelines were published, and it continued to standardize the experimental parameters for EV isolation and characterization to provide more reproducible and robust outcomes.182 Continued efforts to systematically catalogue the protein, nucleic acid and lipid constituents of EVs isolated from richly annotated specimens could ultimately attribute to rapidly evolve and expand the development of selective and sensitive capture platforms directed towards specific EVs.
20. ROLES IN THERAPEUTICS
Directly or indirectly, EVs derived from tumour or TME can influence urogenital neoplasm via intercellular crosstalk or modification of TME, respectively. As discussed above, cargoes of tumour‐derived EVs attribute to cancer development. Thus, the blockage of exosome production, secretion and ablation of specific active exosomal contents, as well as exosome‐mediated cell‐cell communication between cancer and TME, have been proposed as alternative therapeutic strategies. Importantly, it is essential to note that EVs show promising potency for immunotherapy. As we all known, immunotherapy have Immunotherapy has revolutionized cancer therapy, especially the advent of immune checkpoint blockades (ie PD‐1/PD‐L1). Recently, it has been demonstrated that cancer‐derived exosomes transfer functional PD‐L1 and inhibit immune responses,183 while suppression of exosomal PD‐L1 induces systemic anti‐tumour immunity and memory in urological carcinomas,184 and clinical trials have already been initiated to explore their safety and efficacy in humans. Interestingly, EVs‐based vaccines can serve as new candidates that have shown their potential as novel cancer intervention in some clinical trials, indicating that rely on their role as tumour antigens and facilitate an anti‐tumour immunity in turns. The popular EVs‐based vaccines mainly derive from dendritic cell (Dex immunotherapy), but the clinical efficacy is not ideal to date; further research will be required to reassess clinical applications with taking the defects in current prospective designs into considerations such as lack of preselection criteria and small sample size. EVs, on the other hand, have garnered much attention as several characteristics of an optimal delivery system. First of all, the nanometric‐sized EVs confer the effective assimilation and intracellular trafficking for recipient cells. Second, the EV bioactive molecules are protected from degradation in the extracellular milieu and circulation due to lipid bilayer‐membrane structure of EVs.185 Third, autologous EVs show lower immunogenicity and toxicity than other conventional drug‐delivery platforms.186 Furthermore, EVs possessing specific surface proteins (eg integrins) could bear intrinsic targeting properties that are able to interact with target cells or organs.187 Although EVs have many advantages as described above, this enormous promise therapeutic delivery tool requires further study for clinical applications. These include the identification of the optimal EV donor cell type, large‐scale EVs isolation, preservation of EV structural integrity during drug loading, scalable manufacture and storage. A further challenge remains improving methods to shift in vivo biodistribution of EVs from non‐specific sites towards accumulation in desired tissues. Although considerable efforts by engineering EVs to present cell type‐specific ligands have been made in guaranteeing rich accumulation in target tissues, one of the major obstacles remains low delivery efficacy. The elucidation to these questions will enhance rationality and reliability to irritate the utility of EV‐involving molecular cargoes as cancer diagnostics in the clinical practices.
21. CONCLUSION
In recent, EVs have gained rocketing interest in the field of urological tumour research owing to their multifaceted role in the development and treatment of cancer, and their perspective as a weapon to the armoury for cancer treatment. Since EVs play pivotal effects on intercellular interactions in variable biological fluids, their numbers, protein, nucleic acids, lipid and signalling/epigenetic regulators components could be transfer to recipient cells and subsequently affect the pathologic process of the receptor cells, eventually result in abnormal proliferation, EMT, angiogenesis and metastasis by regulation of the TME to cause drug resistance, and preparation of pre‐metastatic niches, to enhance dissemination of cancer cells and cause relapse after a prolonged period of dormancy; thereby, targeting this communication will offer a novel therapeutic strategy for urologic cancer eradication. Apart from that, EVs could serve as a non‐invasive liquid biopsy and have been emerged as new potential diagnostic/prognostic biomarkers, as well as playing provoking roles in predicting anti‐cancer drug responses. In additions, we could use EVs as cancer vaccines or as drug delivery modules with promising therapeutic applications, unless more researches are required for clinical applications. However, the research in EVs is encountered with urgent challenges, including the standardization of approaches for the isolation, quantification and analysis of EVs from complicated tissues sample (mainly from cancer line cell medium or low numbers of patient urine samples). Great efforts have been made to precisely determine EV particles and, nevertheless, still have a great way to standardize EVs enumeration in some particular specimens, such as blood. Moreover, a further challenge is what EVs, their contents or their ratio should best be quantified as robust biomarkers in the surveillance of urological diseases staging is yet unknown; thus, in the future studies, we need pay more attention to develop stereospecific antibodies to map the topography of EVs. Furthermore, another big problem in the field is their half‐life in human samples are yet unexplored. With the increasing knowledge of their roles and development of the next‐generation sequencing, mass spectrometry‐based metabolomics and proteomics, we are enthusiastically sure that EVs will contribute to play clinical applications for urological cancer treatment and management in the near future.
CONFLICT OF INTEREST
The authors declare that they have no competing interests.
AUTHOR CONTRIBUTIONS
Z.W., S.W. and Z.Z. designed the review and made a retrieval strategy; Z.W. and Y.L. drafted the review text; W.X. and J.C. drafted the tables and figures; and both authors contributed to revision and finalization of the manuscript.
CONSENT FOR PUBLICATION
The patient has given his consent for his case report to be published.
ACKNOWLEDGEMENTS
We gratefully acknowledged the help from staffs at the Department of Urology of the Third Affiliated Hospital of Shenzhen University for the data assistance.
Wu Z, Zhang Z, Xia W, Cai J, Li Y, Wu S. Extracellular vesicles in urologic malignancies—Implementations for future cancer care. Cell Prolif. 2019;52:e12659 10.1111/cpr.12659
Funding information
This work was financially supported by a scholarship from The National Key Research and Development Program of China (No. 2017YTA0105900).
DATA ACCESSIBILITY
Research data are not shared.
REFERENCES
- 1. Siegel L, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7‐30. [DOI] [PubMed] [Google Scholar]
- 2. Dy GW, Gore JL, Forouzanfar MH, Naghavi M, Fitzmaurice C. Global burden of urologic cancers, 1990–2013. Eur Urol. 2017;71(3):437‐446. [DOI] [PubMed] [Google Scholar]
- 3. Pitt JM, Kroemer G, Zitvogel L. Extracellular vesicles: masters of intercellular communication and potential clinical interventions. J Clin Invest. 2016;126(4):1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Colombo M, Raposo G, Théry C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu Rev Cell Dev Biol. 2014;30(1):255‐289. [DOI] [PubMed] [Google Scholar]
- 5. Xu R, Greening DW, Zhu H‐J, Takahashi N, Simpson RJ. Extracellular vesicle isolation and characterization: toward clinical application. J Clin Invest. 2016;126(4):1152‐1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. de Pablos Torró LM, Retana Moreira L, Osuna A. Extracellular vesicles in Chagas disease: a new passenger for an old disease. Front Microbiol. 2018;9:1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Li W, Wang H, Zhao Z, et al. Emerging nanotechnologies for liquid biopsy: the detection of circulating tumor cells and extracellular vesicles. Adv Mater. 2018;e1805344. [DOI] [PubMed] [Google Scholar]
- 8. Shao H, Im H, Castro CM, Breakefield X, Weissleder R, Lee H. New technologies for analysis of extracellular vesicles. Chem Rev. 2018;118(4):1917‐1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Nguyen H, Simpson RJ, Salamonsen LA, Greening DW. Extracellular vesicles in the intrauterine environment: challenges and potential functions. Biol Reprod. 2016;95(5):109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Vickers KC, Palmisano BT, Shoucri BM, Shamburek RD, Remaley AT. Corrigendum: microRNAs are transported in plasma and delivered to recipient cells by high‐density lipoproteins. Nat Cell Biol. 2014;17(1):423‐433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nawaz M, Camussi G, Valadi H, et al. The emerging role of extracellular vesicles as biomarkers for urogenital cancers. Nature Reviews Urology. 2014;11(12):688‐701. [DOI] [PubMed] [Google Scholar]
- 12. Zaborowski MP, Balaj L, Breakefield XO, Lai CP. Extracellular vesicles: composition, biological relevance, and methods of study. Bioscience. 2015;65(8):783‐797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Latifkar A, Cerione RA, Antonyak MA. Probing the mechanisms of extracellular vesicle biogenesis and function in cancer. Biochem Soc Trans. 2018;46(5):1137‐1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cocucci E, Meldolesi J. Ectosomes and exosomes: shedding the confusion between extracellular vesicles. Trends Cell Biol. 2015;25(6):364‐372. [DOI] [PubMed] [Google Scholar]
- 15. Yanez‐Mo M, Siljander PR, Andreu Z, et al. Biological properties of extracellular vesicles and their physiological functions. J Extracell Vesicles. 2015;4:27066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hurley JH. ESCRTs are everywhere. EMBO J. 2015;34(19):2398‐2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tai Y‐L, Chen K‐C, Hsieh J‐T, Shen T‐L. Exosomes in cancer development and clinical applications. Cancer Sci. 2018;109(8):2364‐2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhao G‐N, Zhang P, Gong J, et al. Tmbim1 is a multivesicular body regulator that protects against non‐alcoholic fatty liver disease in mice and monkeys by targeting the lysosomal degradation of Tlr4. Nat Med. 2017;23(6):742‐752. [DOI] [PubMed] [Google Scholar]
- 19. Urabe F, Kosaka N, Kimura T, Egawa S, Ochiya T. Extracellular vesicles: toward a clinical application in urological cancer treatment. Int J Urol. 2018;25(6):533‐543. [DOI] [PubMed] [Google Scholar]
- 20. Xu R, Rai A, Chen M, Suwakulsiri W, Greening DW, Simpson RJ. Extracellular vesicles in cancer – implications for future improvements in cancer care. Nat Rev Clin Oncol. 2018;15(10):617‐638. [DOI] [PubMed] [Google Scholar]
- 21. Ostrowski M, Carmo NB, Krumeich S, et al. Rab27a and Rab27b control different steps of the exosome secretion pathway. Nat Cell Biol. 2010;12(1):19‐30. [DOI] [PubMed] [Google Scholar]
- 22. Junker K, Heinzelmann J, Beckham C, Ochiya T, Jenster G. Extracellular vesicles and their role in urologic malignancies. Eur Urol. 2016;70(2):323‐331. [DOI] [PubMed] [Google Scholar]
- 23. Wilhelm EN, Mourot L, Rakobowchuk M. Exercise‐derived microvesicles: a review of the literature. Sports Med. 2018;48(9):2025‐2039. [DOI] [PubMed] [Google Scholar]
- 24. Li R, Peng C, Zhang X, Wu Y, Pan S, Xiao Y. Roles of Arf6 in cancer cell invasion, metastasis and proliferation. Life Sci. 2017;182:80‐84. [DOI] [PubMed] [Google Scholar]
- 25. Yoo JH, Shi DS, Grossmann AH, et al. ARF6 is an actionable node that orchestrates oncogenic GNAQ signaling in uveal melanoma. Cancer Cell. 2016;29(6):889‐904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Li B, Antonyak MA, Zhang J, Cerione RA. RhoA triggers a specific signaling pathway that generates transforming microvesicles in cancer cells. Oncogene. 2012;3(4):219‐224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Becker A, Thakur BK, Weiss JM, Kim HS, Peinado H, Lyden D. Extracellular vesicles in cancer: cell‐to‐cell mediators of metastasis. Cancer Cell. 2016;30(6):836‐848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nabhan JF, Hu R, Oh RS, Cohen SN, Lu Q. Formation and release of arrestin domain‐containing protein 1‐mediated microvesicles (ARMMs) at plasma membrane by recruitment of TSG101 protein. Proc Natl Acad Sci USA. 2012;109(11):4146‐4151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bergsmedh A, Szeles A, Henriksson M, et al. Horizontal transfer of oncogenes by uptake of apoptotic bodies. Proc Natl Acad Sci USA. 2001;98(11):6407‐6411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhu Z, Zhang D, Lee H, et al. Macrophage‐derived apoptotic bodies promote the proliferation of the recipient cells via shuttling microRNA‐221/222. J Leukoc Biol. 2017;101(6):1349‐1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Coumans F, Brisson AR, Buzas EI, et al. Methodological guidelines to study extracellular vesicles. Circ Res. 2017;120(10):1632‐1648. [DOI] [PubMed] [Google Scholar]
- 32. Merchant ML, Rood IM, Deegens J, Klein JB. Isolation and characterization of urinary extracellular vesicles: implications for biomarker discovery. Nat Rev Nephrol. 2017;13(12):731‐749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Woo H‐K, Sunkara V, Park J, et al. Exodisc for rapid, size‐selective, and efficient isolation and analysis of nanoscale extracellular vesicles from biological samples. ACS Nano. 2017;11(2):1360‐1370. [DOI] [PubMed] [Google Scholar]
- 34. Konoshenko MY, Lekchnov EA, Vlassov AV, Laktionov PP. Isolation of extracellular vesicles: general methodologies and latest trends. Biomed Res Int. 2018;2018:8545347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Van Deun J, Mestdagh P, Sormunen R, et al. The impact of disparate isolation methods for extracellular vesicles on downstream RNA profiling. J Extracell Vesicles. 2014;3(1):24858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lobb RJ, Becker M, Wen Wen S, et al. Optimized exosome isolation protocol for cell culture supernatant and human plasma. J Extracell Vesicles. 2015;4:27031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Salih M, Fenton RA, Knipscheer J, et al. An immunoassay for urinary extracellular vesicles. Am J Physiol Renal Physiol. 2016;310(8):F796‐F801. [DOI] [PubMed] [Google Scholar]
- 38. Ingato D, Lee JU, Sim SJ, Kwon YJ. Good things come in small packages: overcoming challenges to harness extracellular vesicles for therapeutic delivery. J Control Release. 2016;241:174‐185. [DOI] [PubMed] [Google Scholar]
- 39. Nakai W, Yoshida T, Diez D, et al. A novel affinity‐based method for the isolation of highly purified extracellular vesicles. Sci Rep. 2016;6:33935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Balaj L, Atai NA, Chen W, et al. Heparin affinity purification of extracellular vesicles. Sci Rep. 2015;5:10266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Samsonov R, Shtam T, Burdakov V, et al. Lectin‐induced agglutination method of urinary exosomes isolation followed by mi‐RNA analysis: application for prostate cancer diagnostic. Prostate. 2016;76(1):68‐79. [DOI] [PubMed] [Google Scholar]
- 42. Jung MK, Mun JY. Sample preparation and imaging of exosomes by transmission electron microscopy. J Vis Exp. 2018;131:e56482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Brisson AR, Tan S, Linares R, Gounou C, Arraud N. Extracellular vesicles from activated platelets: a semiquantitative cryo‐electron microscopy and immuno‐gold labeling study. Platelets. 2017;28(3):263‐271. [DOI] [PubMed] [Google Scholar]
- 44. Chuo S‐Y, Chien J‐Y, Lai C‐K. Imaging extracellular vesicles: current and emerging methods. J Biomed Sci. 2018;25(1):91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Szatanek R, Baj‐Krzyworzeka M, Zimoch J, Lekka M, Siedlar M, Baran J. The methods of choice for extracellular vesicles (EVs) characterization. Int J Mol Sci. 2017;18(6):1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Libregts S, Arkesteijn G, Németh A, Nolte‐’t Hoen E, Wauben M. Flow cytometric analysis of extracellular vesicle subsets in plasma: impact of swarm by particles of non‐interest. J Thromb Haemost. 2018;16(7):1423‐1436. [DOI] [PubMed] [Google Scholar]
- 47. Im H, Shao H, Park YI, et al. Label‐free detection and molecular profiling of exosomes with a nano‐plasmonic sensor. Nat Biotechnol. 2014;32(5):490‐495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Shao H, Chung J, Balaj L, et al. 443 Protein typing of circulating microvesicles allows real‐time monitoring of glioblastoma therapy. Nat Med. 2012;48(12):137‐138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Yoshioka Y, Kosaka N, Konishi Y, et al. Ultra‐sensitive liquid biopsy of circulating extracellular vesicles using ExoScreen. Nat Commun. 2014;5:3591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ricci‐Vitiani L, Pallini R, Biffoni M, et al. Tumour vascularization via endothelial differentiation of glioblastoma stem‐like cells. Nature. 2010;468(7325):824‐828. [DOI] [PubMed] [Google Scholar]
- 51. Audia A, Conroy S, Glass R, Bhat K. The impact of the tumor microenvironment on the properties of glioma stem‐like cells. Front Oncol. 2017;7:143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Singh M, Yelle N, Venugopal C, Singh SK. EMT: Mechanisms and therapeutic implications. Pharmacol Ther. 2018;182:80‐94. [DOI] [PubMed] [Google Scholar]
- 53. Aga M, Bentz GL, Raffa S, et al. Exosomal HIF1α supports invasive potential of nasopharyngeal carcinoma‐associated LMP1‐positive exosomes. Oncogene. 2014;33(37):4613‐4622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Gopal SK, Greening DW, Rai A, et al. Extracellular vesicles: their role in cancer biology and epithelial‐mesenchymal transition. Biochem J. 2017;474(1):21‐45. [DOI] [PubMed] [Google Scholar]
- 55. Greening DW, Gopal SK, Mathias RA, et al. Emerging roles of exosomes during epithelial‐mesenchymal transition and cancer progression. Semin Cell Dev Biol. 2015;40:60‐71. [DOI] [PubMed] [Google Scholar]
- 56. Franzen CA, Blackwell RH, Todorovic V, et al. Urothelial cells undergo epithelial‐to‐mesenchymal transition after exposure to muscle invasive bladder cancer exosomes. Oncogenesis. 2015;4:e163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Peinado H, Zhang H, Matei IR, et al. Pre‐metastatic niches: organ‐specific homes for metastases. Nat Rev Cancer. 2017;17(5):302. [DOI] [PubMed] [Google Scholar]
- 58. Grange C, Tapparo M, Collino F, et al. Microvesicles released from human renal cancer stem cells stimulate angiogenesis and formation of lung premetastatic niche. Cancer Res. 2011;71(15):5346‐5356. [DOI] [PubMed] [Google Scholar]
- 59. Smyth T, Kullberg M, Malik N, Smith‐Jones P, Graner MW, Anchordoquy TJ. Biodistribution and delivery efficiency of unmodified tumor‐derived exosomes. J Control Release. 2015;199:145‐155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Peinado H, Alečković M, Lavotshkin S, et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro‐metastatic phenotype through MET. Nat Med. 2012;18(6):883‐891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Liu Y, Gu Y, Han Y, et al. Tumor exosomal RNAs promote lung pre‐metastatic niche formation by activating alveolar epithelial TLR3 to recruit neutrophils. Cancer Cell. 2016;30(2):243‐256. [DOI] [PubMed] [Google Scholar]
- 62. Costa‐Silva B, Aiello NM, Ocean AJ, et al. Pancreatic cancer exosomes initiate pre‐metastatic niche formation in the liver. Nat Cell Biol. 2015;17(6):816‐826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Fong MY, Zhou W, Liu L, et al. Breast‐cancer‐secreted miR‐122 reprograms glucose metabolism in premetastatic niche to promote metastasis. Nat Cell Biol. 2015;17(2):183‐194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Choi D, Lee TH, Spinelli C, Chennakrishnaiah S, D’Asti E, Rak J. Extracellular vesicle communication pathways as regulatory targets of oncogenic transformation. Semin Cell Dev Biol. 2017;67:11‐22. [DOI] [PubMed] [Google Scholar]
- 65. Nabet BY, Qiu YU, Shabason JE, et al. Exosome RNA unshielding couples stromal activation to pattern recognition receptor signaling in cancer. Cell. 2017;170(2):352‐366.e313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Hood JL, San RS, Wickline SA. Exosomes released by melanoma cells prepare sentinel lymph nodes for tumor metastasis. Cancer Res. 2011;71(11):3792. [DOI] [PubMed] [Google Scholar]
- 67. Ruivo CF, Adem B, Silva M, Melo SA. The biology of cancer exosomes: insights and new perspectives. Cancer Res. 2017;77(23):6480‐6488. [DOI] [PubMed] [Google Scholar]
- 68. Sung BH, Weaver AM. Exosome secretion promotes chemotaxis of cancer cells. Cell Adh Migr. 2017;11(2):187‐195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Baroni S, Romero‐Cordoba S, Plantamura I, et al. Exosome‐mediated delivery of miR‐9 induces cancer‐associated fibroblast‐like properties in human breast fibroblasts. Cell Death Dis. 2016;7(7):e2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Mishra VK, Subramaniam M, Kari V, et al. Kruppel‐like transcription factor KLF10 suppresses TGFbeta‐induced epithelial‐to‐mesenchymal transition via a negative feedback mechanism. Cancer Res. 2017;77(9):2387‐2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Gu J, Qian H, Shen LI, et al. Gastric cancer exosomes trigger differentiation of umbilical cord derived mesenchymal stem cells to carcinoma‐associated fibroblasts through TGF‐β/Smad pathway. PLoS ONE. 2012;7(12):e52465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Corrado C, Saieva L, Raimondo S, Santoro A, De Leo G, Alessandro R. Chronic myelogenous leukaemia exosomes modulate bone marrow microenvironment through activation of epidermal growth factor receptor. J Cell Mol Med. 2016;20(10):1829‐1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Todorova D, Simoncini S, Lacroix R, Sabatier F, Dignat‐George F. Extracellular vesicles in angiogenesis. Circ Res. 2017;120(10):1658‐1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Du T, Ju G, Wu S, et al. Microvesicles derived from human Wharton's jelly mesenchymal stem cells promote human renal cancer cell growth and aggressiveness through induction of hepatocyte growth factor. PLoS ONE. 2014;9(5):e96836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Berlato C, Khan MN, Schioppa T, et al. A CCR76 antagonist reverses the tumor‐promoting microenvironment of renal cancer. J Clin Invest. 2017;127(3):801‐813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Katlinski KV, Gui J, Katlinskaya YV, et al. Inactivation of interferon receptor promotes the establishment of immune privileged tumor microenvironment. Cancer Cell. 2017;31(2):194‐207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Saha D, Martuza RL, Rabkin SD. Macrophage polarization contributes to glioblastoma eradication by combination immunovirotherapy and immune checkpoint blockade. Cancer Cell. 2017;32(2):253‐267.e255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Li X, Wang X. The emerging roles and therapeutic potential of exosomes in epithelial ovarian cancer. Mol Cancer. 2017;16(1):92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Guo D, Lui G, Lai SL, et al. RAB27A promotes melanoma cell invasion and metastasis via regulation of pro‐invasive exosomes. Int J Cancer. 2019;144:3070‐3085. [DOI] [PubMed] [Google Scholar]
- 80. Seo N, Shirakura Y, Tahara Y, et al. Activated CD8(+) T cell extracellular vesicles prevent tumour progression by targeting of lesional mesenchymal cells. Nat Commun. 2018;9(1):435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Sun Z, Yang S, Zhou Q, et al. Emerging role of exosome‐derived long non‐coding RNAs in tumor microenvironment. Mol Cancer. 2018;17(1):82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Zhou R, Chen KK, Zhang J, et al. The decade of exosomal long RNA species: an emerging cancer antagonist. Mol Cancer. 2018;17(1):75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet‐Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):69‐90. [DOI] [PubMed] [Google Scholar]
- 84. Barata PC, Rini BI. Treatment of renal cell carcinoma: current status and future directions. CA Cancer J Clin. 2017;67(6):507‐524. [DOI] [PubMed] [Google Scholar]
- 85. Novara G, Ficarra V, Antonelli A, et al. Validation of the 2009 TNM version in a large multi‐institutional cohort of patients treated for renal cell carcinoma: are further improvements needed? Eur Urol. 2010;58(4):588‐595. [DOI] [PubMed] [Google Scholar]
- 86. Fisher R, Gore M, Larkin J. Current and future systemic treatments for renal cell carcinoma. Semin Cancer Biol. 2013;23(1):38‐45. [DOI] [PubMed] [Google Scholar]
- 87. Schubert M, Junker K, Heinzelmann J. Prognostic and predictive miRNA biomarkers in bladder, kidney and prostate cancer: Where do we stand in biomarker development? J Cancer Res Clin Oncol. 2016;142(8):1673‐1695. [DOI] [PubMed] [Google Scholar]
- 88. Yang L, Wu X, Wang D, Luo C, Chen L. Renal carcinoma cell‐derived exosomes induce human immortalized line of Jurkat T lymphocyte apoptosis in vitro. Urol Int. 2013;91(3):363‐369. [DOI] [PubMed] [Google Scholar]
- 89. Raimondo F, Morosi L, Corbetta S, et al. Differential protein profiling of renal cell carcinoma urinary exosomes. Mol Biosyst. 2013;9(6):1220‐1233. [DOI] [PubMed] [Google Scholar]
- 90. De Palma G, Sallustio F, Curci C, et al. The three‐gene signature in urinary extracellular vesicles from patients with clear cell renal cell carcinoma. J Cancer. 2016;7(14):1960‐1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Del BP, Raimondo F, Pieragostino D, et al. A hyphenated microLC‐Q‐TOF‐MS platform for exosomal lipidomics investigations: application to RCC urinary exosomes. Electrophoresis. 2012;33(4):689‐696. [DOI] [PubMed] [Google Scholar]
- 92. Butz H, Nofech‐Mozes R, Ding Q, et al. Exosomal microRNAs are diagnostic biomarkers and can mediate cell‐cell communication in renal cell carcinoma. Eur Urol Focus. 2016;2(2):210‐218. [DOI] [PubMed] [Google Scholar]
- 93. Zhang W, Ni M, Su Y, et al. MicroRNAs in serum exosomes as potential biomarkers in clear‐cell renal cell carcinoma. Eur Urol Focus. 2018;4:412‐419. [DOI] [PubMed] [Google Scholar]
- 94. Fujii N, Hirata H, Ueno K, et al. Extracellular miR‐224 as a prognostic marker for clear cell renal cell carcinoma. Oncotarget. 2017;8(66):109877‐109888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Jingushi K, Uemura M, Ohnishi N, et al. Extracellular vesicles isolated from human renal cell carcinoma tissues disrupt vascular endothelial cell morphology via azurocidin. Int J Cancer. 2018;142(3):607‐617. [DOI] [PubMed] [Google Scholar]
- 96. Milella M, Felici A. Biology of metastatic renal cell carcinoma. J Cancer. 2011;2:369‐373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Zhang L, Wu X, Luo C, et al. The 786–0 renal cancer cell‐derived exosomes promote angiogenesis by downregulating the expression of hepatocyte cell adhesion molecule. Mol Med Report. 2013;8(1):272‐276. [DOI] [PubMed] [Google Scholar]
- 98. Gu X, Erb U, Büchler MW, Zöller M. Improved vaccine efficacy of tumor exosome compared to tumor lysate loaded dendritic cells in mice. Int J Cancer. 2015;136(4):E74‐E84. [DOI] [PubMed] [Google Scholar]
- 99. Diao J, Yang X, Song X, et al. Exosomal Hsp70 mediates immunosuppressive activity of the myeloid‐derived suppressor cells via phosphorylation of Stat3. Med Oncol. 2015;32(2):453. [DOI] [PubMed] [Google Scholar]
- 100. Zhang Y, Luo CL, He BC, Zhang JM, Cheng G, Wu XH. Exosomes derived from IL‐12‐anchored renal cancer cells increase induction of specific antitumor response in vitro: a novel vaccine for renal cell carcinoma. Int J Oncol. 2010;36(1):133‐140. [PubMed] [Google Scholar]
- 101. Jiang XL, Zhang Y, Tan B, Luo CL, Wu XH. Renal tumor‐derived exosomes inhibit hepaCAM expression of renal carcinoma cells in a p‐AKT‐dependent manner. Neoplasma. 2014;61(4):416. [DOI] [PubMed] [Google Scholar]
- 102. Bussolati B, Bruno S, Grange C, Ferrando U, Camussi G. Identification of a tumor‐initiating stem cell population in human renal carcinomas. FASEB J. 2008;22(10):3696‐3705. [DOI] [PubMed] [Google Scholar]
- 103. Posadas EM, Limvorasak S, Figlin RA. Targeted therapies for renal cell carcinoma. Nat Rev Nephrol. 2017;13(8):496‐511. [DOI] [PubMed] [Google Scholar]
- 104. Molina AM, Lin X, Korytowsky B, et al. Sunitinib objective response in metastatic renal cell carcinoma: analysis of 1059 patients treated on clinical trials. Eur J Cancer. 2014;50(2):351‐358. [DOI] [PubMed] [Google Scholar]
- 105. Qu LE, Ding J, Chen C, et al. Exosome‐transmitted lncARSR promotes sunitinib resistance in renal cancer by acting as a competing endogenous RNA. Cancer Cell. 2016;29(5):653‐668. [DOI] [PubMed] [Google Scholar]
- 106. Babjuk M, Böhle A, Burger M, et al. EAU Guidelines on non‐muscle‐invasive urothelial carcinoma of the bladder: update 2016. Eur Urol. 2017;71(3):447‐461. [DOI] [PubMed] [Google Scholar]
- 107. Almallah YZ, Rennie CD, Stone J, Lancashire M. Urinary tract infection and patient satisfaction after flexible cystoscopy and urodynamic evaluation. Urology. 2000;56(1):37‐39. [DOI] [PubMed] [Google Scholar]
- 108. Welton JL, Khanna S, Giles PJ, et al. Proteomics analysis of bladder cancer exosomes. Mol Cell Proteomics. 2010;9(6):1324‐1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Chen C‐L, Lai Y‐F, Tang P, et al. Comparative and targeted proteomic analyses of urinary microparticles from bladder cancer and hernia patients. J Proteome Res. 2012;11(12):5611. [DOI] [PubMed] [Google Scholar]
- 110. Lin S‐Y, Chang C‐H, Wu H‐C, et al. Proteome profiling of urinary exosomes identifies alpha 1‐antitrypsin and H2B1K as diagnostic and prognostic biomarkers for urothelial carcinoma. Sci Rep. 2016;6:34446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Silvers CR, Miyamoto H, Messing EM, Netto GJ, Lee Y‐F. Characterization of urinary extracellular vesicle proteins in muscle‐invasive bladder cancer. Oncotarget. 2017;8(53):91199‐91208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Beckham CJ, Olsen J, Yin P‐N, et al. Bladder cancer exosomes contain EDIL‐3/Del1 and facilitate cancer progression. J Urol. 2014;192(2):583‐592. [DOI] [PubMed] [Google Scholar]
- 113. Smalley DM, Sheman NE, Nelson K, Theodorescu D. Isolation and identification of potential urinary microparticle biomarkers of bladder cancer. J Proteome Res. 2008;7(5):2088‐2096. [DOI] [PubMed] [Google Scholar]
- 114. Liang K, Liu F, Fan J, et al. Nanoplasmonic quantification of tumour‐derived extracellular vesicles in plasma microsamples for diagnosis and treatment monitoring. Nat Biomed Eng. 2017;1(4):21‐21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Silvers CR, Liu Y‐R, Wu C‐H, Miyamoto H, Messing EM, Lee Y‐F. Identification of extracellular vesicle‐borne periostin as a feature of muscle‐invasive bladder cancer. Oncotarget. 2016;7(17):23335‐23345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Perez A, Loizaga A, Arceo R, et al. A pilot study on the potential of RNA‐associated to urinary vesicles as a suitable non‐invasive source for diagnostic purposes in bladder cancer. Cancers (Basel). 2014;6(1):179‐192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Matsuzaki K, Fujita K, Jingushi K, et al. MiR‐21‐5p in urinary extracellular vesicles is a novel biomarker of urothelial carcinoma. Oncotarget. 2017;8(15):24668‐24678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Andreu Z, Otta Oshiro R, Redruello A, et al. Extracellular vesicles as a source for non‐invasive biomarkers in bladder cancer progression. Eur J Pharm Sci. 2017;98:70‐79. [DOI] [PubMed] [Google Scholar]
- 119. Baumgart S, Hölters S, Ohlmann C‐H, et al. Exosomes of invasive urothelial carcinoma cells are characterized by a specific miRNA expression signature. Oncotarget. 2017;8(35):58278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Long JD,Sullivan TB, Humphrey J, et al. A non‐invasive miRNA based assay to detect bladder cancer in cell‐free urine. Am J Transl Res. 2015;7(11):2500‐2509. [PMC free article] [PubMed] [Google Scholar]
- 121. Ostenfeld MS, Jeppesen DK, Laurberg JR, et al. Cellular disposal of miR23b by RAB27‐dependent exosome release is linked to acquisition of metastatic properties. Cancer Res. 2014;74(20):5758‐5771. [DOI] [PubMed] [Google Scholar]
- 122. Armstrong DA, Green BB, Seigne JD, Schned AR, Marsit CJ. MicroRNA molecular profiling from matched tumor and bio‐fluids in bladder cancer. Mol Cancer. 2015;14(1):194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Berrondo C, Flax J, Kucherov V, et al. Expression of the long non‐coding RNA HOTAIR correlates with disease progression in bladder cancer and is contained in bladder cancer patient urinary exosomes. PLoS ONE. 2016;11(1):e0147236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Liang L‐G, Kong M‐Q, Zhou S, et al. An integrated double‐filtration microfluidic device for isolation, enrichment and quantification of urinary extracellular vesicles for detection of bladder cancer. Sci Rep. 2017;7:46224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Yang L, Wu X‐H, Wang D, Luo C‐L, Chen L‐X. Bladder cancer cell‐derived exosomes inhibit tumor cell apoptosis and induce cell proliferation in vitro. Mol Med Report. 2013;8(4):1272‐1278. [DOI] [PubMed] [Google Scholar]
- 126. Xue M, Chen W, Xiang AN, et al. Hypoxic exosomes facilitate bladder tumor growth and development through transferring long non‐coding RNA‐UCA1. Mol Cancer. 2017;16(1):143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Greco KA, Franzen CA, Foreman KE, Flanigan RC, Kuo PC, Gupta GN. PLK‐1 silencing in bladder cancer by siRNA delivered with exosomes. Urology. 2016;91(241):e241‐247. [DOI] [PubMed] [Google Scholar]
- 128. Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136(5):E359‐386. [DOI] [PubMed] [Google Scholar]
- 129. Rao AR, Motiwala HG, Karim OM, et al. The discovery of prostate‐specific antigen. BJU Int. 2008;101(1):5‐10. [DOI] [PubMed] [Google Scholar]
- 130. Crawford ED, Abrahamsson P‐A. PSA‐based screening for prostate cancer: how does it compare with other cancer screening tests? Eur Urol. 2008;54(2):262‐273. [DOI] [PubMed] [Google Scholar]
- 131. Foj L, Ferrer F, Serra M, et al. Exosomal and non‐exosomal urinary miRNAs in prostate cancer detection and prognosis. Prostate. 2017;77(6):573‐583. [DOI] [PubMed] [Google Scholar]
- 132. Rodríguez M, Bajo‐Santos C, Hessvik NP, et al. Identification of non‐invasive miRNAs biomarkers for prostate cancer by deep sequencing analysis of urinary exosomes. Mol Cancer. 2017;16(1):156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Khan S, Jutzy J, Valenzuela M, et al. Plasma‐derived exosomal survivin, a plausible biomarker for early detection of prostate cancer. PLoS ONE. 2012;7(10):e46737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Wang L, Skotland T, Berge V, Sandvig K, Llorente A. Exosomal proteins as prostate cancer biomarkers in urine: from mass spectrometry discovery to immunoassay‐based validation. Eur J Pharm Sci. 2016;98:80. [DOI] [PubMed] [Google Scholar]
- 135. Duijvesz D, Versluis C, van der Fels C, et al. Immuno‐based detection of extracellular vesicles in urine as diagnostic marker for prostate cancer. Int J Cancer. 2015;137(12):2869‐2878. [DOI] [PubMed] [Google Scholar]
- 136. Liu C, Guo J, Tian F, et al. Field‐free isolation of exosomes from extracellular vesicles by microfluidic viscoelastic flows. ACS Nano. 2017;11(7):6968‐6976. [DOI] [PubMed] [Google Scholar]
- 137. Lu Q, Zhang J, Allison R, et al. Identification of extracellular delta‐catenin accumulation for prostate cancer detection. Prostate. 2009;69(4):411‐418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Bijnsdorp IV, Geldof AA, Lavaei M, Piersma SR, van Moorselaar R, Jimenez CR. Exosomal ITGA3 interferes with non‐cancerous prostate cell functions and is increased in urine exosomes of metastatic prostate cancer patients. J Extracell Vesicles. 2013;2:22097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Fujita K, Kume H, Matsuzaki K, et al. Proteomic analysis of urinary extracellular vesicles from high Gleason score prostate cancer. Sci Rep. 2017;7:42961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Øverbye A, Skotland T, Koehler CJ, et al. Identification of prostate cancer biomarkers in urinary exosomes. Oncotarget. 2015;6(30):30357‐30376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Ronquist KG, Ronquist G, Larsson A, Carlsson L. Proteomic analysis of prostate cancer metastasis‐derived prostasomes. Anticancer Res. 2010;30(2):285‐290. [PubMed] [Google Scholar]
- 142. Sandvig K, Llorente A. Proteomic analysis of microvesicles released by the human prostate cancer cell line PC‐3. Mol Cell Proteomics. 2012;11(7):M111.012914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Duijvesz D, Burnum‐Johnson KE, Gritsenko MA, et al. Proteomic profiling of exosomes leads to the identification of novel biomarkers for prostate cancer. PLoS ONE. 2013;8(12):e82589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Utleg AG, Yi EC, Xie T, et al. Proteomic analysis of human prostasomes. Prostate. 2010;56(2):150‐161. [DOI] [PubMed] [Google Scholar]
- 145. Simona Principe E, Kim Y, Sinha A, et al. In‐depth proteomic analyses of exosomes isolated from expressed prostatic secretions in urine. Proteomics. 2013;13(10‐11):1667‐1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Sequeiros T, Rigau M, Chiva C, et al. Targeted proteomics in urinary extracellular vesicles identifies biomarkers for diagnosis and prognosis of prostate cancer. Oncotarget. 2016;8(3):4960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Gabriel K, Ingram A, Austin R, et al. Regulation of the tumor suppressor PTEN through exosomes: a diagnostic potential for prostate cancer. PLoS ONE. 2013;8(7):e70047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Kato T, Mizutani K, Kameyama K, et al. Serum exosomal P‐glycoprotein is a potential marker to diagnose docetaxel resistance and select a taxoid for patients with prostate cancer. Urol Oncol. 2015;33(9):385.e315‐320. [DOI] [PubMed] [Google Scholar]
- 149. Welton JL, Brennan P, Gurney M, et al. Proteomics analysis of vesicles isolated from plasma and urine of prostate cancer patients using a multiplex, aptamer‐based protein array. J Extracell Vesicles. 2016;5(1):31209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Lázaro‐Ibáñez E, Sanz‐Garcia A, Visakorpi T, et al. Different gDNA content in the subpopulations of prostate cancer extracellular vesicles: apoptotic bodies, microvesicles, and exosomes. Prostate. 2014;74(14):1379‐1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. McKiernan J, Donovan MJ, O’Neill V, et al. A novel urine exosome gene expression assay to predict high‐grade prostate cancer at initial biopsy. JAMA Oncol. 2016;2(7):882‐889. [DOI] [PubMed] [Google Scholar]
- 152. Neeb A, Hefele S, Bormann S, et al. Splice variant transcripts of the anterior gradient 2 gene as a marker of prostate cancer. Oncotarget. 2014;5(18):8681‐8689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Donovan MJ, Noerholm M, Bentink S, et al. A molecular signature of PCA3 and ERG exosomal RNA from non‐DRE urine is predictive of initial prostate biopsy result. Prostate Cancer Prostatic Dis. 2015;18(4):370‐375. [DOI] [PubMed] [Google Scholar]
- 154. Hendriks RJ, Dijkstra S, Jannink SA, et al. Comparative analysis of prostate cancer specific biomarkers PCA3 and ERG in whole urine, urinary sediments and exosomes. Clin Chem Lab Med. 2016;54(3):483‐492. [DOI] [PubMed] [Google Scholar]
- 155. Motamedinia P, Scott AN, Bate KL, et al. Urine exosomes for non‐invasive assessment of gene expression and mutations of prostate cancer. PLoS ONE. 2016;11(5):e0154507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Royo F, Zuñiga‐Garcia P, Torrano V, et al. Transcriptomic profiling of urine extracellular vesicles reveals alterations of CDH3 in prostate cancer. Oncotarget. 2016;7(6):6835‐6846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Işın M, Uysaler E, Özgür E, et al. Exosomal lncRNA‐p21 levels may help to distinguish prostate cancer from benign disease. Front Genet. 2015;6:168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Koppers‐Lalic D, Hackenberg M, deMenezes R, et al. Noninvasive prostate cancer detection by measuring miRNA variants (isomiRs) in urine extracellular vesicles. Oncotarget. 2016;7(16):22566‐22578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Bryant RJ, Pawlowski T, Catto J, et al. Changes in circulating microRNA levels associated with prostate cancer. Br J Cancer. 2012;106(4):768‐774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Bryzgunova OE, Zaripov MM, Skvortsova TE, et al. Comparative study of extracellular vesicles from the urine of healthy individuals and prostate cancer patients. PLoS ONE. 2016;11(6):e0157566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Wani S, Kaul D, Mavuduru RS, Kakkar N, Bhatia A. Urinary‐exosomal miR‐2909: a novel pathognomonic trait of prostate cancer severity. J Biotechnol. 2017;259:135‐139. [DOI] [PubMed] [Google Scholar]
- 162. Xu Y, Qin S, An T, Tang Y, Huang Y, Zheng L. MiR‐145 detection in urinary extracellular vesicles increase diagnostic efficiency of prostate cancer based on hydrostatic filtration dialysis method. Prostate. 2017;77(10):1167‐1175. [DOI] [PubMed] [Google Scholar]
- 163. Hao X‐K, Li Z, Ma Y‐Y, et al. Exosomal microRNA‐141 is upregulated in the serum of prostate cancer patients. Onco Targets Ther. 2016;9(1):139‐148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Huang X, Yuan T, Liang M, et al. Exosomal miR‐1290 and miR‐375 as prognostic markers in castration‐resistant prostate cancer. Eur Urol. 2015;67(1):33‐41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Skotland T, Ekroos K, Kauhanen D, et al. Molecular lipid species in urinary exosomes as potential prostate cancer biomarkers. Eur J Cancer. 2017;70:122‐132. [DOI] [PubMed] [Google Scholar]
- 166. Yang JS, Lee JC, Byeon SK, Rha KH, Moon MH. Size dependent lipidomic analysis of urinary exosomes from patients with prostate cancer by flow field‐flow fractionation and nanoflow liquid chromatography‐tandem mass spectrometry. Anal Chem. 2017;89(4):2488‐2496. [DOI] [PubMed] [Google Scholar]
- 167. Puhka M, Takatalo M, Nordberg M‐E, et al. Metabolomic profiling of extracellular vesicles and alternative normalization methods reveal enriched metabolites and strategies to study prostate cancer‐related changes. Theranostics. 2017;7(16):3824‐3841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Nyalwidhe JO, Betesh LR, Powers TW, et al. Increased bisecting N‐acetylglucosamine and decreased branched chain glycans of N‐linked glycoproteins in expressed prostatic secretions associated with prostate cancer progression. Proteomics Clin Appl. 2013;7(9‐10):677‐689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Webber JP, Spary LK, Sanders AJ, et al. Differentiation of tumour‐promoting stromal myofibroblasts by cancer exosomes. Oncogene. 2015;34(3):290‐302. [DOI] [PubMed] [Google Scholar]
- 170. Castellana D, Zobairi F, Martinez MC, et al. Membrane microvesicles as actors in the establishment of a favorable prostatic tumoral niche: a role for activated fibroblasts and CX3CL1‐CX3CR176 axis. Cancer Res. 2009;69(3):785‐793. [DOI] [PubMed] [Google Scholar]
- 171. Itoh T, Ito Y, Ohtsuki Y, et al. Microvesicles released from hormone‐refractory prostate cancer cells facilitate mouse pre‐osteoblast differentiation. J Mol Histol. 2012;43(5):509‐515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Karlsson T, Lundholm M, Widmark A, Persson E. Tumor cell‐derived exosomes from the prostate cancer cell line TRAMP‐C1 impair osteoclast formation and differentiation. PLoS ONE. 2016;11(11):e0166284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Ye Y, Li S‐L, Ma Y‐Y, et al. Exosomal miR‐141‐3p regulates osteoblast activity to promote the osteoblastic metastasis of prostate cancer. Oncotarget. 2017;8(55):94834‐94849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Lundholm M, Schröder M, Nagaeva O, et al. Prostate tumor‐derived exosomes down‐regulate NKG2D expression on natural killer cells and CD8+ T cells: mechanism of immune evasion. PLoS ONE. 2014;9(9):e108925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Clayton A, Mitchell JP, Court J, Mason MD, Tabi Z. Human tumor‐derived exosomes selectively impair lymphocyte responses to interleukin‐2. Cancer Res. 2007;67(15):7458‐7466. [DOI] [PubMed] [Google Scholar]
- 176. Seitz AK, Thoene S, Bietenbeck A, et al. AR‐V7 in peripheral whole blood of patients with castration‐resistant prostate cancer: association with treatment‐specific outcome under abiraterone and enzalutamide. Eur Urol. 2017;72(5):828. [DOI] [PubMed] [Google Scholar]
- 177. Del Re M, Biasco E, Crucitta S, et al. The detection of androgen receptor splice variant 7 in plasma‐derived exosomal RNA strongly predicts resistance to hormonal therapy in metastatic prostate cancer patients. Eur Urol. 2017;71(5):680‐687. [DOI] [PubMed] [Google Scholar]
- 178. Seifalian A. The application of exosomes as a nanoscale cancer vaccine. Int J Nanomed. 2010;5(1):889‐900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Rountree RB, Mandl SJ, Nachtwey JM, et al. Exosome targeting of tumor antigens expressed by cancer vaccines can improve antigen immunogenicity and therapeutic efficacy. Cancer Res. 2011;71(15):5235‐5244. [DOI] [PubMed] [Google Scholar]
- 180. Corcoran C, Rani S, O’Brien K, et al. Docetaxel‐resistance in prostate cancer: evaluating associated phenotypic changes and potential for resistance transfer via exosomes. PLoS ONE. 2012;7(12):e50999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Lötvall J, Hill AF, Hochberg F, et al. Minimal experimental requirements for definition of extracellular vesicles and their functions: a position statement from the International Society for Extracellular Vesicles. J Extracell Vesicles. 2014;3:26913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Thery C, Witwer KW, Aikawa E, et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J Extracell Vesicles. 2018;7(1):1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Chen G, Huang AC, Zhang W, et al. Exosomal PD‐L1 contributes to immunosuppression and is associated with anti‐PD‐1 response. Nature. 2018;560(7718):382‐386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Poggio M, Hu T, Pai C‐C, et al. Suppression of exosomal PD‐L1 induces systemic anti‐tumor immunity and memory. Cell. 2019;177(2):414‐427.e413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Ha D, Yang N, Nadithe V. Exosomes as therapeutic drug carriers and delivery vehicles across biological membranes: current perspectives and future challenges. Acta pharmaceutica Sinica B. 2016;6(4):287‐296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186. Vader P, Mol EA, Pasterkamp G, Schiffelers RM. Extracellular vesicles for drug delivery. Adv Drug Deliv Rev. 2016;106(Pt A):148‐156. [DOI] [PubMed] [Google Scholar]
- 187. Willms E, Cabañas C, Mäger I, Wood M, Vader P. Extracellular vesicle heterogeneity: subpopulations, isolation techniques, and diverse functions in cancer progression. Front Immunol. 2018;9:738. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Research data are not shared.


