Abstract
Amyotrophic lateral sclerosis (ALS) is a severe neurodegenerative disease, which primarily targets the motor system. The structural integrity of the motor network and the way it is embedded in the overall brain network is essential for motor functioning. We studied the longitudinal effects of ALS on the brain network using diffusion tensor imaging and questioned whether over time an increasing number of connections become involved or whether there is progressive impairment of a limited number of connections. The brain network was reconstructed based on “whole brain” diffusion tensor imaging data. We examined: (1) network integrity in 24 patients with ALS at baseline (T = 1) and at a more advanced stage of the disease (T = 2; interval 5.5 months) compared with a group of healthy controls and (2) progressive brain network impairment comparing patients at two time‐points in a paired‐analysis. These analyses demonstrated an expanding subnetwork of affected brain connections over time with a central role for the primary motor regions (P‐values T = 1 0.003; T = 2 0.001). Loss of structural connectivity mainly propagated to frontal and parietal brain regions at T = 2 compared with T = 1. No progressive impairment of the initially affected (motor) connections could be detected. The main finding of this study is an increasing loss of network structure in patients with ALS. In contrast to the theory of ALS solely affecting a fixed set of primary motor connections, our findings show that the network of impaired connectivity is expanding over time. These results are in support of disease spread along structural brain connections. Hum Brain Mapp 35:1351–1361, 2014. © 2013 Wiley‐Periodicals, Inc.
Keywords: amyotrophic lateral sclerosis, magnetic resonance imaging, diffusion tensor imaging, neuroimaging, neural networks, connectivity
Abbreviations
- ALS
amyotrophic lateral sclerosis
- ALSFRS‐R
revised ALS Functional Rating Scale
- FA
fractional anisotropy
- NBS
Network Based Statistics
INTRODUCTION
Amyotrophic Lateral Sclerosis (ALS) is a severe neurodegenerative disease progressively affecting both upper motor neurons in the motor cortex and lower motor neurons in the brainstem and spinal cord, with a median survival time of as little as 3 years after onset of symptoms [del Aguila et al., 2003; Hardiman et al., 2011]. Progressive motor neuron loss results in declining motor function involving more and more body regions and increasing functional impairment over time. Based on clinical observations, it has been hypothesized that ALS is a focal process spreading contiguously through the nervous system [Ravits and La Spada, 2009; Ravits et al., 2007]. In vivo insight into the pattern of disease spread within the brain—and the motor system in particular—is crucial as it could provide new therapeutic targets and novel disease activity biomarkers to facilitate therapeutic development.
Recent imaging studies in ALS have revealed neurodegenerative effects in specific white matter motor tracts, showing consistent involvement of bilateral corticospinal tracts and corpus callosum [Agosta et al., 2010; Filippini et al., 2010; Grosskreutz et al., 2008; Nair et al., 2010; Verstraete et al., 2010]. However, the degree of corticospinal tract integrity correlates poorly with clinical markers and progression over time has not been consistently demonstrated [Blain et al., 2007; Mitsumoto et al., 2007; Senda et al., 2011; van der Graaff et al., 2011]. These observations cast doubt on whether progressive degeneration of these main motor tracts is the leading cause of deteriorating motor functions in ALS.
The brain should be regarded as an integrative complex system, rather than a set of independently operating regions [Sporns, 2006; Stam and Reijneveld, 2007; van den Heuvel and Hulshoff Pol, 2010]. In this context, the structural integrity of the motor network and the way it is embedded in the overall brain network is essential for normal motor functioning. It is, therefore, highly plausible that dysfunction of the motor network underlies ALS, but the extent and evolution of dysfunction has not yet been elucidated [Rose et al., 2012; Verstraete et al., 2011]. Using diffusion tensor imaging (DTI) in a longitudinal setting, we investigated how disease progression in ALS is reflected in the reconstructed structural brain network and questioned whether, over time, an increasing number of connections become involved or whether the same initially involved (motor) connections become progressively impaired.
MATERIALS AND METHODS
Subjects
Twenty‐four patients with sporadic ALS (22 males; mean age/SD: 57.3/12.0 years) and 19 healthy controls (14 males; mean age/SD: 52.8/10.6 years) participated in this longitudinal study (see Table 1 for full demographics and clinical characteristics) [Verstraete et al., 2011]. Patients, recruited from the outpatient clinic for motor neuron diseases of the University Medical Center Utrecht and Radboud University Medical Center, Nijmegen (The Netherlands), were diagnosed with ALS according to the El Escorial criteria. Subjects did not have a history of brain injury, epilepsy, psychiatric illness, or neurodegenerative diseases other than ALS. Clinical status of the patients was evaluated using the revised ALS Functional Rating Scale (ALSFRS‐R). Rate of disease progression was defined as the average decline in ALSFRS‐R score per month since onset of symptoms. After an average interval of 5.5 months, patients were examined again. Clinical status and follow‐up times are provided in Supporting Information Table S1 online.
Table 1.
Subject demographics and clinical characteristics
| Healthy control subjects (n = 19) | ALS patients (n = 24) | ||
|---|---|---|---|
| Mean ± SD (range) | Mean ± SD (range) | ||
| Age (years) | 52.8 ± 10.6 (33−70) | 56.8 ± 11.9 (38−74) | |
| Male/Female | 14/5 | 22/2 | |
| Site of onset [n (%)] | Bulbar | 2 (8%) | |
| Upper limbs | 15 (63%) | ||
| Lower limbs | 7 (29%) | ||
| Diagnostic category [n (%)] | Possible | 2 (8%) | |
| Probable‐laboratory‐supported | 8 (33%) | ||
| Probable | 12 (50%) | ||
| Definite | 2 (8%) | ||
| Disease duration at baseline scan (months) | 15.8 ± 11.8 (3−59) | ||
| Longitudinal change in ALSFRS‐R | 4.3 ± 4.0 (0−13) | ||
SD, standard deviation.
The Ethical Committee for research in humans of the University Medical Center Utrecht approved study protocols. All subjects provided informed written consent according to the Declaration of Helsinki.
Image Acquisition
During each of the two sessions, participants underwent a 35‐min scan using a 3 Tesla Philips Achieva Clinical scanner at the University Medical Center Utrecht with a SENSE receiver head‐coil. High resolution DTI was performed to reconstruct the white matter tracts of the brain network. Within each scanning session, 2 DTI sets, each consisting of 30 weighted diffusion scans and 5 unweighted B0 scans, were acquired (DTI‐MR using parallel imaging SENSE p‐reduction 3; high angular gradient set of 30 different weighted directions, TR/TE = 7035/68 ms, 2 × 2 × 2 mm, 75 slices, b = 1000 s/mm2, second set with reversed k‐space read‐out). Then, directly after the acquisition of the DTI scans, an anatomical T1 weighted image (3D FFE using parallel imaging; TR/TE 10/4.6 ms; FOV 240 × 240 mm, 200 slices, 0.75 mm isotropic voxel size) was obtained for anatomical reference.
Image Preprocessing
DTI preprocessing
DTI preprocessing included the following steps [van den Heuvel and Sporns, 2011]. First of all, susceptibility distortions, often reported in single‐shot EPI DTI images, were corrected for by computing a field distortion map based on the two B = 0 images, which were acquired with an opposite k‐space direction. This map was then applied to the two sets of 30 weighted images [Andersson et al., 2003], resulting in a single set of 30 corrected weighted directions. Second, images were realigned with the corrected B = 0 image, correcting for small head movements and eddy‐current distortions [Andersson and Skare, 2002]. Third, the diffusion profile within each voxel was fitted a tensor (based on robust estimation of tensors by outlier rejection or RESTORE) [Chang et al., 2005] and the preferred diffusion direction was determined as the principal eigenvector of the eigenvalue decomposition of the fitted tensor. To provide information on the microstructural organization of white matter, for each voxel the fractional anisotropy (FA) was computed, a measure often used as a marker for white matter integrity [Basser and Pierpaoli, 1996; Beaulieu and Allen, 1994]. Fourthly, streamline tractography was applied to reconstruct white matter tracts [Mori and van Zijl, 2002; Mori et al., 1999]. Within each voxel, a single streamline was initiated, which then followed the main diffusion direction from voxel to voxel, reconstructing a white matter fiber. A streamline was terminated when the fiber track reached a voxel with an FA value <0.1 (indicating low levels of preferred diffusion), when the streamline made an angle of more than 45°, or when the trajectory of the traced fiber exceeded the brain mask. To assess the integrity of a streamline, each reconstructed fiber tract was assigned the FA values of the voxels crossed [Mandl et al., 2008; Van den Heuvel et al., 2008, 2009a, 2009b].
T1 preprocessing
T1 images were realigned with the corrected B = 0 images [Andersson et al., 2003] to obtain spatial overlap between the anatomical and DTI images. This registration procedure was performed based on “mutual information” and manually verified for all datasets. T1 images were used to select cortical and subcortical regions, resulting from parcellating the brain into distinct regions, using the well‐validated Freesurfer suite (V4.5, http://surfer.nmr.mgh.harvard.edu/). This analysis included the automatic segmentation of gray and white matter tissue, followed by a parcellation of the segmented gray matter mask into 68 distinct cortical regions (34 within each hemisphere), 14 subcortical regions (bilateral thalamus, hippocampus, amygdala, accumbens, caudate, pallidum, and putamen) and the brainstem.
Reconstruction of Structural Brain Networks
A graph is a mathematical description of a network, consisting of a collection of nodes and a collection of connections. The nodes were selected as the 83 segmented regions; the connections were taken as the white matter pathways that interlink the regions. For each dataset (both patients and healthy controls), and for each time‐point (i.e., longitudinal measurement) an individual brain network was reconstructed [van den Heuvel and Sporns, 2011; Verstraete et al., 2011]. This procedure included the following steps. First of all, from the total collection of reconstructed streamlines, those that touched both region i and region j were selected. Second, from the resulting fiber streamlines, the average FA values were computed as the average of all points and all included streamlines. This value was incorporated into cell c(i,j) in connectivity matrix M. If no tracts were found between regions i and j, the value of zero was assigned to matrix M, reflecting the nonexistence of a direct tract between regions i and j in the brain network. This procedure was repeated for all regions in the total collection of regions (V), resulting in a connected, undirected, weighted graph G.
Statistical Analyses
To examine the status of the brain's network during different stages of disease, Network Based Statistics (NBS) was used [Verstraete et al., 2011; Zalesky et al., 2010]. NBS is based on the idea of Statistical Parameter Mapping, extending the notion that changes in connections that occur together—by forming a connected component—are more likely to indicate true network abnormality than effects that occur in relative isolation (i.e., solitary connections). Two types of analyses were performed: (1) examining the status of the brain network during different stages of disease compared to healthy controls and (2) studying the effect of disease progression on the brain networks of patients (= within subject effect). These two analyses detect different types of longitudinal connectivity changes. We hypothesized four different scenarios: (1) progressive degeneration within the initially impaired structural connections (Fig. 1, scenario A) will be detected by the paired analysis (only positive results with Analysis 2); (2) degenerative changes over time show spatial spread (Fig. 1, scenario B), this will be detected by Analysis 1 as these changes will result in an expanding subnetwork of impaired connections and leave negative results with Analysis 2; (3) both progressive impairment of initially impaired structural connections, as well as expansion of the impaired connectivity (positive results on both Analysis 1 and 2; Fig. 1, scenario A + B); (4) longitudinal degeneration involving brain regions distant to the initially impaired subnetwork or multifocal degeneration (negative results on both Analysis 1 and 2, Fig. 1, scenario C). The NBS analysis is designed to detect changes within a network and therefore both Analysis 1 and 2 will be negative when structural connections distant to the initial degenerative process become impaired over time. Finally, if no longitudinal changes occur, both analyses will turn out negative as well. The two types of analyses included the following steps.
Figure 1.
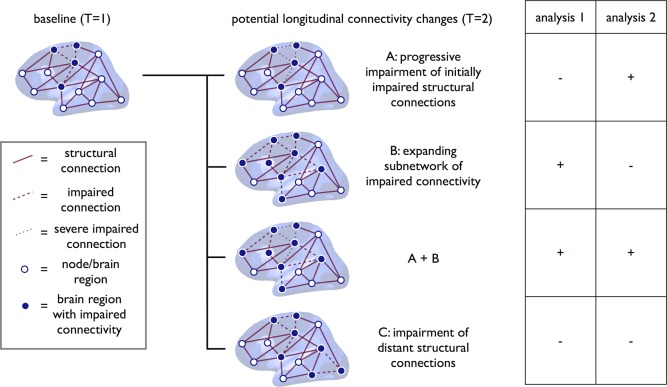
Potential longitudinal connectivity changes in ALS. This figure shows four potential scenarios for longitudinal neurodegeneration in ALS and the hypothesized results of the two types of analyses. A: Progressive impairment within a subnetwork of initially impaired structural connections and regions will be detected by Analysis 2. Analysis 1 will not detect this type of progressive degeneration. B: If the degenerative changes over time show spatial spread within the brain, this will be detected by Analysis 1 as this will result in an expanding subnetwork of impaired connections. (A + B) The third scenario is a combination of both progressive impairment of initially impaired structural connections, as well as expansion of the impaired connectivity (i.e., scenario A and B combined). In this scenario, both analyses show positive results. C: The fourth scenario is longitudinal degeneration involving distant structural connections to the initially impaired subnetwork or multifocal degeneration. As the NBS analysis is designed to detect changes within a network, both Analysis 1 and 2 will show negative results when distant structural connections become impaired over time. [Color figure can be viewed in the online issue, which is available at http://wileyonlinelibrary.com.]
Analysis 1
To assess the longitudinal changes in the brain network, the connectivity profiles of the 24 patients at baseline (T = 1) and at a more advanced stage of the disease (T = 2) were compared with the reconstructed brain networks of healthy controls using NBS (Fig. 2). DTI data of the group of 19 healthy controls was used to obtain a good stable estimate of normal brain connectivity. First of all, for each time‐point, a difference matrix, D1, between controls and the brain networks of patients was created by two‐sample t‐testing across the two groups (controls vs. patients) for all entries (i.e., connections) in M. Second, effects showing a statistical difference larger than a set threshold of alpha < 0.01 were set to 1, and otherwise to zero. Third, from the resulting difference matrix, D1, the size(s) of the largest component(s) of connections that formed an interconnected subcluster of nodes was computed. Fourth, permutation testing, randomly mixing group assignment, was performed to obtain a null‐distribution of component size, independent of group status (10,000 permutations; Fig. 2). The component(s) of reduced connectivity observed between patients and controls was assigned a P‐value (corrected for multiple comparisons) based on the proportion of the null‐distribution that showed a larger component size than the observed subnetwork of impaired connectivity. The NBS procedure is known to show good control over type‐I errors and to correct properly for multiple testing [Zalesky et al., 2010].
Figure 2.
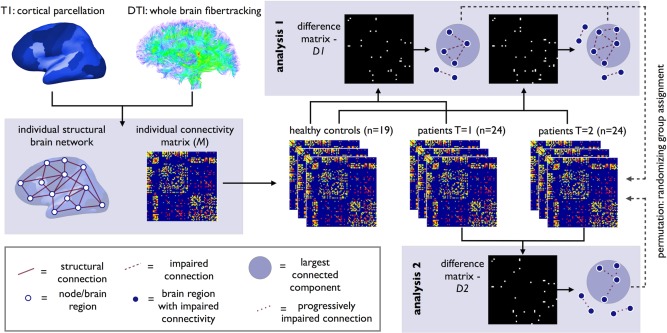
Overview of longitudinal connectome imaging methods. Cortical and subcortical brain regions were selected by automatic parcellation of the cerebrum. Using the DTI data, white matter tracts of the brain were reconstructed. An individual brain network was defined consisting of nodes (i.e., the parcellated brain regions) and white matter connections between the nodes resulting in a (FA‐weighted) connectivity matrix M. Next, using NBS, the connectivity matrices of ALS patients at T = 1 and T = 2 were compared with healthy controls (Analysis 1). Using t‐statistics each connection was tested resulting in a binary difference matrix (D1); connections with a statistical difference larger than a set threshold of alpha < 0.01 were set to 1, and otherwise to zero. The size of the (largest) connected component in the difference matrix was computed, revealing a subnetwork of affected connectivity in patients. Permutation testing was used to define the p‐value of this component based on the computed null‐distribution of (largest) connected component sizes as can occur under the null‐hypothesis (no difference between patients and controls). An identical analysis was performed comparing brain connectivity at T = 1 and T = 2 in patients (Analysis 2). [Color figure can be viewed in the online issue, which is available at http://wileyonlinelibrary.com.]
Analysis 2
The individual connectivity matrices (M) of the patients, one for each time point, were compared with detect possible progressive changes in connectivity strength of connections between regions i and j over time (Fig. 2). First, a progressive difference matrix, D2, was created by performing a paired sample t‐test for each of the cells in M. Differences showing a statistical difference of alpha < 0.01 were marked by 1 in D2 and otherwise zero. Second, from the resulting matrix, D2, the largest connected component was extracted, forming the subnetwork of progressively involved structural brain connections. Third, following the same procedure as in Analysis 1, a null‐distribution of component sizes that could occur by chance, rather than related to time effects, was created using permutation (Fig. 2). For this procedure, each measurement was randomly assigned to one of two pseudo time‐points, effectively randomizing T = 1 and T = 2 time‐points across subjects. A null‐distribution of component size that could occur under the null hypothesis was established by performing 10,000 permutations. Fourth, given the computed null‐distribution, the size of component(s) observed in the original longitudinal difference matrix, D2 (reflecting longitudinal changes in connectivity strength in ALS) were assigned a P‐value representing the proportion of the null‐distribution that showed a larger component size than the size of the observed sub‐network [Zalesky et al., 2010].
Network integrity and clinical characteristics
To study the relation between clinical markers (ALSFRS‐R and progression rate) and network integrity, we correlated the cumulative FA of all the structural connections included in the NBS network with clinical markers using Pearson's correlation testing.
RESULTS
Analysis 1: Structural Brain Networks in Patients at Two Time‐Points Compared with Healthy Controls
Using “whole‐brain” DTI data we reconstructed the structural brain networks of patients (at different time points) and healthy controls. Network impairment at baseline (T = 1) was assessed and compared with the affected subnetwork at a more advanced stage of the disease (T = 2). The methods applied are schematically displayed in Figure 2.
Figure 3 shows the subnetwork of affected connectivity in ALS (as compared with the brain networks of healthy controls) at the two measured time points T = 1 and T = 2. The affected network (NBS‐corrected P = 0.003, 10,000 permutations) at T = 1 consisted of 18 regions and 19 affected connections. The following regions were included: cortical motor (precentral gyrus and paracentral lobule left and right), subcortical (thalamic nucleus right; pallidum left; hippocampus left; caudate right), frontal (caudal middle frontal left and right; superior frontal left), parietal (precuneus left and right; posterior cingulate gyrus right; inferior parietal left; postcentral left), temporal (supramarginal left), and the brainstem (the impaired connections are specified in Supporting Information Table S2).
Figure 3.
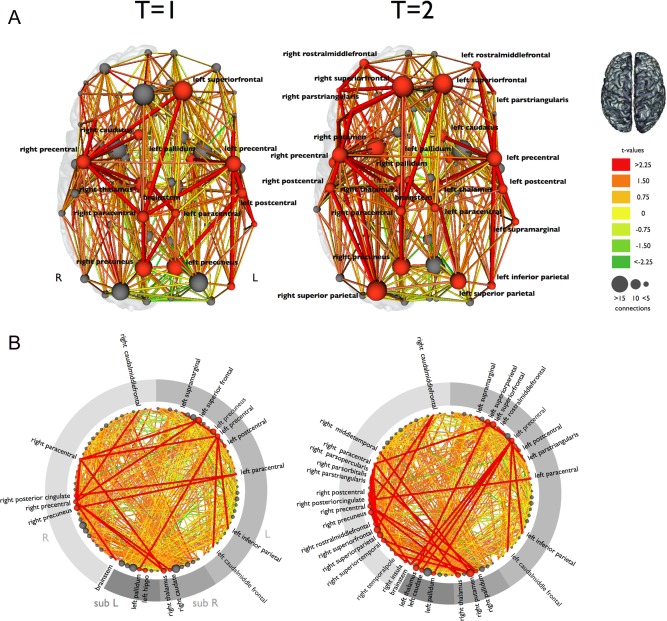
The reconstructed brain network and the affected structural connections in ALS at two time points. A: The affected connections and interconnecting nodes are displayed comparing patients at T = 1 and T = 2 with the group of healthy controls. The size of the nodes is determined by their degree (number of structural connections). Nodes and connections were colored red if they were part of the affected subnetwork. The affected subnetwork at T = 2 included more structural connections, extending mainly to frontal and parietal brain regions. B: This figure shows all regions arranged on a ring. The red colored nodes and connections are part of the affected subnetwork, showing an increasing number of affected connections and regions at T = 2 compared with T = 1. L: left hemisphere; R: right hemisphere; sub L: left subcortical regions; sub R: right subcortical regions. [Color figure can be viewed in the online issue, which is available at http://wileyonlinelibrary.com.]
The affected network at T = 2 (after on average 5.5 months) revealed a more extensive network of 38 affected structural connections and 33 regions (NBS‐corrected P = 0.001, 10,000 permutations). The impaired subnetwork at T = 2 consisted of the following regions: cortical motor (precentral gyrus and paracentral lobule left and right), subcortical (thalamic nuclei and pallidum left and right; putamen right; caudate nucleus left), frontal (superior frontal gyrus, caudal middle frontal, rostral middle frontal and parstriangularis left and right; parsorbitalis right; parsopercularis right), parietal (superior parietal and postcentral left and right; precuneus right; posterior cingulate right; inferior parietal left), temporal (supramarginal left; middle temporal right; superior temporal right; temporal pole right), insula right, and the brainstem (the impaired connections are specified in Supporting Information Table S3).
The cortical regions included in the affected network at T = 1 and T = 2 are displayed in Figure 4. Comparing the regions involved in the network at T = 2 with those involved at T = 1, reveals that it is mainly the connections with frontal and parietal brain regions, which are newly affected as disease progresses, indicating spatial spread of disease along motor‐related structural connections of the brain's network (Fig. 5).
Figure 4.
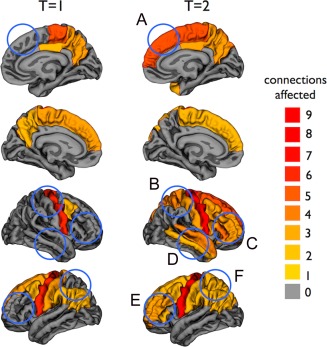
Cortical regions included in the affected sub‐network at T = 1 and T = 2. The regions are colored based on the number of affected connections. The blue circles indicate the newly affected cortical regions at T = 2 compared with T = 1 (A: superior frontal right; B: postcentral and superior parietal right; C: rostral middle frontal, pars orbitalis, pars triangularis, and pars opercularis right; D: temporal pole, superior, and middle temporal right, insula right [not visible]; E: rostral middle frontal and pars triangularis left; and F: superior parietal left). [Color figure can be viewed in the online issue, which is available at http://wileyonlinelibrary.com.]
Figure 5.
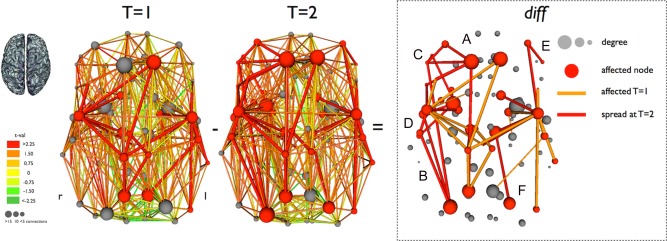
Longitudinal reduction of structural connectivity in ALS. The differences are shown (diff, third panel of figure) between the affected subnetworks at T = 1 and T = 2. Connections affected at T = 1 (and T = 2) are colored orange and newly affected connections at T = 2 are colored red. The newly involved cortical regions are indicated (A: superior frontal right; B: postcentral and superior parietal right; C: rostral middle frontal, pars orbitalis, pars triangularis, and pars opercularis right; D: temporal pole, superior and middle temporal right, insula right; E: rostral middle frontal and pars triangularis left; and F: superior parietal left). This figure marks the spatial spread of disease, resulting in expanding motor network disintegration. [Color figure can be viewed in the online issue, which is available at http://wileyonlinelibrary.com.]
Analysis 2: Longitudinal Structural Brain Network Changes in Patients with ALS
To study the progressiveness of degenerative effects within structural connections, we compared the brain networks of the group of 24 patients at T = 1 and T = 2 using paired‐NBS (Fig. 2). This analysis did not reveal a subnetwork of progressively affected connections and regions, indicating that the initially affected motor network is not progressively affected in ALS (Fig. 1). A post‐hoc analysis comparing individual structural connections at T = 1 and T = 2, rather than focusing on degenerative changes occurring in a network, showed a number of connections with a significantly lower FA at T = 2 compared with T = 1. The majority (67%) of these newly affected connections was linked to the impaired network found at T = 1 (analysis 1). This finding is consistent with the spatial spread of degenerative effects as suggested by the results in analysis 1. No significant correlation was found between the integrity of the NBS network (at both T = 1 and T = 2) and clinical markers.
DISCUSSION
The main finding of this study is an expanding loss of network structure in patients with ALS. Previous studies have suggested reduced motor network integrity in patients with ALS [Rose et al., 2012; Verstraete et al., 2011]. Our findings now show that the network of impaired connectivity is expanding over time, involving more and more connections and regions (as shown by Analysis 1), rather than progressively affecting a fixed set of impaired connections (as shown by Analysis 2; Fig. 1). Therefore, these results suggest spread of disease following the connectivity architecture of the brain network.
The observation of progressive brain network impairment in ALS puts previous findings in a new perspective. Longitudinal DTI studies using a voxel‐wise comparison in a similar number of patients demonstrated widespread FA reduction over time [Senda et al., 2011; van der Graaff et al., 2011] as well as progressive loss of global FA [Sage et al., 2007]. These widespread effects may be part of an expanding network of affected white matter connections, resulting in disintegration of the motor network as is illustrated by scenario B in Figure 1. Previous longitudinal imaging studies in ALS, examining specific white matter tracts, revealed no progressive degeneration of corticospinal tracts, the primary efferent motor fiber pathways from the motor cortex to the spinal cord [Agosta et al., 2009; Blain et al., 2007; Mitsumoto et al., 2007]. This supports our findings that the limited number of initially involved white matter tracts are not progressively affected (Analysis 2). In addition, connectivity measures seem to be more sensitive to the degenerative changes occurring over time in ALS compared with cortical atrophy patterns. Cortical thinning in motor regions has been demonstrated but has not shown to be progressive or spreading over time [Verstraete et al., 2012]. We performed a post‐hoc analysis studying longitudinal cortical thickness changes in this cohort as previously described [Verstraete et al., 2012]. Extending our current study, we compared cortical thickness measures between all cortical regions in patients at baseline (T = 1) with controls and patients at follow‐up (T = 2) with controls. In addition, we performed a paired analysis between baseline measures in patients and follow‐up (T = 1 vs. T = 2). We found significant cortical thinning of the precentral gyrus in patients with ALS as demonstrated previously [Verstraete et al., 2012]. Using an explorative (uncorrected) P‐value of 0.05, we found several more regions showing cortical thinning at baseline in ALS, however, the number of (potentially) affected regions did not increase over time. In line with this finding, the paired analysis revealed no significantly progressive cortical thinning. Summarizing, the degenerative effects observed in the corticospinal tracts and cortical motor regions appear to be an early sign of the degenerative process in ALS. However, our data suggest that clinical deterioration over time might result from increasing disconnection of the motor system as a whole.
Disease spread in ALS has been a topic of interest for some time now. Clinical observations have given rise to the hypothesis that there might be a focal onset with subsequent spread to other body regions [Ravits and La Spada, 2009; Ravits et al., 2007]. In support of this hypothesis, we now provide in vivo, imaging‐based evidence for spatial spread of disease following the architecture of the brain network. A similar type of spread has been proposed for other neurodegenerative diseases involving protein aggregates, including Alzheimer's disease [Eisele et al., 2010; Jucker and Walker, 2011]. Recent imaging studies in subtypes of dementia have shown that neurodegenerative diseases target specific neural networks [Seeley et al., 2009], and brain regions with high functional and structural connectedness to “epicenters” for disease (i.e., primarily affected regions) have a greater disease‐related vulnerability [Raj et al., 2012; Zhou et al., 2012]. It is notable that the brain regions, included in the impaired subnetwork we found, show great overlap with the activation patterns observed with functional MRI during motor tasks [Poujois et al., 2012]. The view that ALS potentially has a focal onset with subsequent spread along brain connections might open the way for development of new treatment strategies.
We were not able to perform a subgroup analysis on bulbar versus spinal site of onset due to the underrepresentation of bulbar onset patients in this longitudinal cohort. In addition, we did not include elaborate neuropsychological testing to assess possible cognitive decline, however, none of the patients in our study actually fulfilled the clinical criteria of frontotemporal dementia [Neary et al., 1998]. In future studies, it would be of great interest to examine a possible link between ALS‐related cognitive impairment and the extent of brain network impairment, as suggested by reports on a direct link between brain connectivity and cognitive performance [Bassett et al., 2010; van den Heuvel et al., 2009a, 2009b]. Also, analysis of specific differences in the extent or spread of degeneration in (presymptomatic) carriers of the intronic repeat expansion in C9orf72, compared with other patients with ALS and their cognitive profile, would be of great value [DeJesus‐Hernandez et al., 2011].
DTI measurement of structural brain network integrity is influenced by multiple factors including crossing fibers, fiber reorganization, increased membrane permeability, destruction of intracellular compartments, and glial alterations [Acosta‐Cabronero et al., 2010]. Specifically, crossing fibers might introduce important bias as univocal information on the diffusion direction can lead to low FA values and premature termination of a fiber streamline. As a result, the streamline count might be reduced and some key fiber pathways may be missed at all. In addition, if the crossing fibers are part of the degenerative process this might cause a paradoxical increase in FA of the tract studied. Such an increase in FA was not observed in our study. Alternative methods have been suggested, including probabilistic fiber tracking [Behrens et al., 2003]. However, probabilistic fiber tracking complicates the measurement of FA values along a tract, which was our main topic of investigation. In addition to false negatives, streamline tractography may sometimes lead to detection of false positive streamlines. To prevent inclusion of false positive tracts in the NBS analysis, connections were included only if they were present in more than half of the individuals in the groups compared. Second, the examined structural brain network comprised 83 segmented brain regions. Future work examining connectome abnormalities in ALS using a higher resolution of the brain's network may result in a more finely grained representation of the motor connections. In addition, it would have been appropriate to include the cerebellum as a node of the structural brain network as this structure is part of the motor network and has been suggested to be involved in ALS [Keil et al., 2012]. Third, it is important to note that only few of the transcallosal motor connections were deemed affected in the NBS‐analysis while the corpus callosum has been consistently found involved in ALS [Filippini et al., 2010]. This apparent discrepancy is likely related to the stringent settings of the connectome imaging protocol, limiting the number of corpus callosum streamlines found across healthy controls and ALS patients, making it hard for white matter changes to survive the set NBS threshold. In addition, crossing fibers—specifically from the corona radiata—might have impeded the streamline tractography for the callosal motor connections. However, specifically examining the corpus callosum in a post hoc analysis did show significant—but not progressive—involvement of the motor related corpus callosum tracts, which is in line with previous studies (P < 0.05 at T = 2) [Filippini et al., 2010; Verstraete et al., 2010]. Fourth, NBS‐analyses, by definition, are not sensitive to (multi) focal changes within the brain network as illustrated by scenario C (Fig. 1). It is important to note that newly degenerative connections at the borders of a network will not result in positive results from Analysis 2 since they do not form a connected component together. Fifth, following the procedures of previous longitudinal studies in ALS [Senda et al., 2011; van der Graaff et al., 2011], we did not perform routine longitudinal measures in healthy controls, as aging effects in this short time period are unlikely. To provide insight into possible scanner effects, a subset (n = 7) was evaluated after an average of 7 months, performing an analysis identical to that used in the group of patients. No significant network changes were found in controls, which is in line with recent studies showing a high reproducibility of connectome imaging [Cammoun et al., 2012]. For the future, it is important to reproduce our findings in a larger cohort of longitudinally assessed patients with ALS, as will be facilitated by currently emerging international collaboration [Turner et al., 2011].
The observations in this study, which examines longitudinal spread of ALS within the nervous system, suggest increasing loss of motor network connectivity, consisting of spatial spread along the architecture of the brain's network. The idea that ALS is a progressive disease of the brain network, potentially with a focal onset in primary motor regions, is important in helping to understand the progressive nature of the disorder and providing new leads for the future development of therapeutic targets.
Supporting information
Supporting Information Table 1.
Supporting Information Tables 2 and 3.
ACKNOWLEDGMENTS
The authors thank all patients and healthy control subjects for their generous contribution by participating in this study.
References
- Acosta‐Cabronero J, Williams GB, Pengas G, Nestor PJ (2010): Absolute diffusivities define the landscape of white matter degeneration in Alzheimer's disease. Brain 133:529–539. [DOI] [PubMed] [Google Scholar]
- Agosta F, Rocca MA, Valsasina P, Sala S, Caputo D, Perini M, Salvi F, Prelle A, Filippi M (2009): A longitudinal diffusion tensor MRI study of the cervical cord and brain in amyotrophic lateral sclerosis patients. J Neurol Neurosurg Psychiatry 80:53–55. [DOI] [PubMed] [Google Scholar]
- Agosta F, Pagani E, Petrolini M, Caputo D, Perini M, Prelle A, Salvi F, Filippi M (2010): Assessment of white matter tract damage in patients with amyotrophic lateral sclerosis: A diffusion tensor MR imaging tractography study. AJNR Am J Neuroradiol 31:1457–1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson JL, Skare S (2002): A model‐based method for retrospective correction of geometric distortions in diffusion‐weighted EPI. NeuroImage 16:177–199. [DOI] [PubMed] [Google Scholar]
- Andersson JL, Skare S, Ashburner J (2003): How to correct susceptibility distortions in spin‐echo echo‐planar images: Application to diffusion tensor imaging. NeuroImage 20:870–888. [DOI] [PubMed] [Google Scholar]
- Basser PJ, Pierpaoli C (1996): Microstructural and physiological features of tissues elucidated by quantitative‐diffusion‐tensor MRI. J Magn Reson 111:209–219. [DOI] [PubMed] [Google Scholar]
- Bassett DS, Greenfield DL, Meyer‐Lindenberg A, Weinberger DR, Moore SW, Bullmore ET (2010): Efficient physical embedding of topologically complex information processing networks in brains and computer circuits. PLoS Comput Biol 6:e1000748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaulieu C, Allen PS (1994): Water diffusion in the giant axon of the squid: Implications for diffusion‐weighted MRI of the nervous system. Magn Reson Med 32:579–583. [DOI] [PubMed] [Google Scholar]
- Behrens TE, Woolrich MW, Jenkinson M, Johansen‐Berg H, Nunes RG, Clare S, Matthews PM, Brady JM, Smith SM (2003): Characterization and propagation of uncertainty in diffusion‐weighted MR imaging. Magn Reson Med 50:1077–1088. [DOI] [PubMed] [Google Scholar]
- Blain CR, Williams VC, Johnston C, Stanton BR, Ganesalingam J, Jarosz JM, Jones DK, Barker GJ, Williams SC, Leigh NP, Simmons A (2007): A longitudinal study of diffusion tensor MRI in ALS. Amyotroph Lateral Scler 8:348–355. [DOI] [PubMed] [Google Scholar]
- Cammoun L, Gigandet X, Meskaldji D, Thiran JP, Sporns O, Do KQ, Maeder P, Meuli R, Hagmann P (2012): Mapping the human connectome at multiple scales with diffusion spectrum MRI. J Neurosci Methods 203:386–397. [DOI] [PubMed] [Google Scholar]
- Chang LC, Jones DK, Pierpaoli C (2005): RESTORE: Robust estimation of tensors by outlier rejection. Magn Reson Med 53:1088–1095. [DOI] [PubMed] [Google Scholar]
- DeJesus‐Hernandez M, Mackenzie IR, Boeve BF, Boxer AL, Baker M, Rutherford NJ, Nicholson AM, Finch NA, Flynn H, Adamson J, Kouri N, Wojtas A, Sengdy P, Hsiung GY, Karydas A, Seeley WW, Josephs KA, Coppola G, Geschwind DH, Wszolek ZK, Feldman H, Knopman DS, Petersen RC, Miller BL, Dickson DW, Boylan KB, Graff‐Radford NR, Rademakers R (2011): Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p‐linked FTD and ALS. Neuron 72:245–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Aguila MA, Longstreth WT Jr., McGuire V, Koepsell TD, van Belle G (2003): Prognosis in amyotrophic lateral sclerosis: A population‐based study. Neurology 60:813–819. [DOI] [PubMed] [Google Scholar]
- Eisele YS, Obermuller U, Heilbronner G, Baumann F, Kaeser SA, Wolburg H, Walker LC, Staufenbiel M, Heikenwalder M, Jucker M (2010): Peripherally applied Abeta‐containing inoculates induce cerebral beta‐amyloidosis. Science 330:980–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippini N, Douaud G, Mackay CE, Knight S, Talbot K, Turner MR (2010): Corpus callosum involvement is a consistent feature of amyotrophic lateral sclerosis. Neurology 75:1645–1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosskreutz J, Peschel T, Unrath A, Dengler R, Ludolph AC, Kassubek J (2008): Whole brain‐based computerized neuroimaging in ALS and other motor neuron disorders. Amyotroph Lateral Scler 9:238–248. [DOI] [PubMed] [Google Scholar]
- Hardiman O, van den Berg LH, Kiernan MC (2011): Clinical diagnosis and management of amyotrophic lateral sclerosis. Nat Rev Neurol 7:639–649. [DOI] [PubMed] [Google Scholar]
- Jucker M, Walker LC (2011): Pathogenic protein seeding in alzheimer disease and other neurodegenerative disorders. Ann Neurol 70:532–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keil C, Prell T, Peschel T, Hartung V, Dengler R, Grosskreutz J (2012): Longitudinal diffusion tensor imaging in amyotrophic lateral sclerosis. BMC Neurosci 13:141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandl RC, Schnack HG, Zwiers MP, van der Schaaf A, Kahn RS, Hulshoff Pol HE (2008): Functional diffusion tensor imaging: measuring task‐related fractional anisotropy changes in the human brain along white matter tracts. PLoS ONE 3:e3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsumoto H, Ulug AM, Pullman SL, Gooch CL, Chan S, Tang MX, Mao X, Hays AP, Floyd AG, Battista V, Montes J, Hayes S, Dashnaw S, Kaufmann P, Gordon PH, Hirsch J, Levin B, Rowland LP, Shungu DC (2007): Quantitative objective markers for upper and lower motor neuron dysfunction in ALS. Neurology 68:1402–1410. [DOI] [PubMed] [Google Scholar]
- Mori S, van Zijl PC (2002): Fiber tracking: principles and strategies: A technical review. NMR Biomed 15:468–480. [DOI] [PubMed] [Google Scholar]
- Mori S, Crain BJ, Chacko VP, van Zijl PC (1999): Three‐dimensional tracking of axonal projections in the brain by magnetic resonance imaging. Ann Neurol 45:265–269. [DOI] [PubMed] [Google Scholar]
- Nair G, Carew JD, Usher S, Lu D, Hu XP, Benatar M (2010): Diffusion tensor imaging reveals regional differences in the cervical spinal cord in amyotrophic lateral sclerosis. NeuroImage 53:576–583. [DOI] [PubMed] [Google Scholar]
- Neary D, Snowden JS, Gustafson L, Passant U, Stuss D, Black S, Freedman M, Kertesz A, Robert PH, Albert M, Boone K, Miller BL, Cummings J, Benson DF (1998): Frontotemporal lobar degeneration: a consensus on clinical diagnostic criteria. Neurology 51:1546–1554. [DOI] [PubMed] [Google Scholar]
- Poujois A, Schneider FC, Faillenot I, Camdessanche JP, Vandenberghe N, Thomas‐Anterion C, Antoine JC (2012): Brain plasticity in the motor network is correlated with disease progression in amyotrophic lateral sclerosis. Hum Brain Mapp (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj A, Kuceyeski A, Weiner M (2012): A network diffusion model of disease progression in dementia. Neuron 73:1204–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravits J, Paul P, Jorg C (2007): Focality of upper and lower motor neuron degeneration at the clinical onset of ALS. Neurology 68:1571–1575. [DOI] [PubMed] [Google Scholar]
- Ravits JM, La Spada AR (2009): ALS motor phenotype heterogeneity, focality, and spread: Deconstructing motor neuron degeneration. Neurology 73:805–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose S, Pannek K, Bell C, Baumann F, Hutchinson N, Coulthard A, McCombe P, Henderson R (2012): Direct evidence of intra‐ and interhemispheric corticomotor network degeneration in amyotrophic lateral sclerosis: An automated MRI structural connectivity study. NeuroImage 59:2661–2669. [DOI] [PubMed] [Google Scholar]
- Sage CA, Peeters RR, Gorner A, Robberecht W, Sunaert S (2007): Quantitative diffusion tensor imaging in amyotrophic lateral sclerosis. NeuroImage 34:486–499. [DOI] [PubMed] [Google Scholar]
- Seeley WW, Crawford RK, Zhou J, Miller BL, Greicius MD (2009): Neurodegenerative diseases target large‐scale human brain networks. Neuron 62:42–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senda J, Kato S, Kaga T, Ito M, Atsuta N, Nakamura T, Watanabe H, Tanaka F, Naganawa S, Sobue G (2011): Progressive and widespread brain damage in ALS: MRI voxel‐based morphometry and diffusion tensor imaging study. Amyotroph Lateral Scler 12:59–69. [DOI] [PubMed] [Google Scholar]
- Sporns O (2006): Small‐world connectivity, motif composition, and complexity of fractal neuronal connections. Bio Systems 85:55–64. [DOI] [PubMed] [Google Scholar]
- Stam CJ, Reijneveld JC (2007): Graph theoretical analysis of complex networks in the brain. Nonlinear Biomed Phys 1:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner MR, Grosskreutz J, Kassubek J, Abrahams S, Agosta F, Benatar M, Filippi M, Goldstein LH, van den Heuvel M, Kalra S, Lule D, Mohammadi B, for the first Neuroimaging Symosium in ALS (NISALS) (2011): Towards a neuroimaging biomarker for amyotrophic lateral sclerosis. Lancet Neurol 10:400–403. [DOI] [PubMed] [Google Scholar]
- van den Heuvel MP, Hulshoff Pol HE (2010): Exploring the brain network: A review on resting‐state fMRI functional connectivity. Eur Neuropsychopharmacol 20:519–534. [DOI] [PubMed] [Google Scholar]
- van den Heuvel MP, Sporns O (2011): Rich‐club organization of the human connectome. J Neurosci 31:15775–15786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Heuvel MP, Mandl RC, Luigjes J, Hulshoff Pol HE (2008): Microstructural organization of the cingulum tract and the level of default mode functional connectivity. J Neurosci 28:10844–10851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Heuvel MP, Mandl RC, Kahn RS, Hulshoff Pol HE (2009a): Functionally linked resting‐state networks reflect the underlying structural connectivity architecture of the human brain. Hum Brain Mapp 30:3127–3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Heuvel MP, Stam CJ, Kahn RS, Hulshoff Pol HE (2009b): Efficiency of functional brain networks and intellectual performance. J Neurosci 29:7619–7624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Graaff MM, Sage CA, Caan MW, Akkerman EM, Lavini C, Majoie CB, Nederveen AJ, Zwinderman AH, Vos F, Brugman F, van den Berg LH, de Rijk MC, van Doorn PA, Van Hecke W, Peeters RR, Robberecht W, Sunaert S, de Visser M (2011): Upper and extra‐motoneuron involvement in early motoneuron disease: A diffusion tensor imaging study. Brain 134:1211–1228. [DOI] [PubMed] [Google Scholar]
- Verstraete E, van den Heuvel MP, Veldink JH, Blanken N, Mandl RC, Hulshoff Pol HE, van den Berg LH (2010): Motor network degeneration in amyotrophic lateral sclerosis: a structural and functional connectivity study. PLoS One 5:e13664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verstraete E, Veldink JH, Mandl RC, van den Berg LH, van den Heuvel MP (2011): Impaired structural motor connectome in amyotrophic lateral sclerosis. PLoS One 6:e24239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verstraete E, Veldink JH, Hendrikse J, Schelhaas HJ, van den Heuvel MP, van den Berg LH (2012): Structural MRI reveals cortical thinning in amyotrophic lateral sclerosis. J Neurol Neurosurg Psychiatry 83:383–388. [DOI] [PubMed] [Google Scholar]
- Zalesky A, Fornito A, Bullmore ET (2010): Network‐based statistic: Identifying differences in brain networks. NeuroImage 53:1197–1207. [DOI] [PubMed] [Google Scholar]
- Zhou J, Gennatas ED, Kramer JH, Miller BL, Seeley WW (2012): Predicting regional neurodegeneration from the healthy brain functional connectome. Neuron 73:1216–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information Table 1.
Supporting Information Tables 2 and 3.


