Abstract
The fusion of intracellular vesicles with target membranes is mediated by two classes of conserved molecules – soluble N-ethylmaleimide-sensitive factor attachment protein receptors (SNAP receptors or SNAREs) and Sec1/Munc18 (SM) proteins. A conserved function of SM proteins is to recognize their cognate trans-SNARE complexes and accelerate fusion kinetics. Here, we describe a physiologically relevant reconstitution system in which macromolecular crowding agents are included to recapitulate the crowded intracellular environment. Through this system, we elucidate the molecular mechanisms by which SNAREs and SM proteins drive vesicle fusion.
Keywords: SNARE, SM protein, vesicle fusion, membrane fusion, reconstitution, lipid mixing, content mixing, macromolecular crowding
1. INTRODUCTION
The machinery mediating intracellular vesicle fusion consists of two classes of conserved molecules –SNAREs and SM proteins (Fig. 1) (1). The fusion reaction is initiated when the vesicular (v-) SNARE pairs with the target membrane (t-) SNAREs to form a trans-SNARE complex (also known as SNAREpin) (2–5). The trans-SNARE complex zippers progressively toward the membranes (6–10), bringing the lipid bilayers into close apposition to fuse (11,12). SM proteins are soluble factors of 60–70 kDa that control vesicle fusion by binding to their cognate SNAREs (13–17). SM proteins exhibit similar loss-of-function phenotypes as SNAREs (i.e., abrogation of fusion) and are essential for every vesicle fusion event in the cell (18,19).
Figure 1. Model showing the vesicle fusion machinery comprised of SNAREs and SM protein.
The image was modeled from the atomic structures of the SNARE core bundle (24,42), and unpaired VAMP2 (43). Yellow: SM protein; Green: t-SNARE heavy chain (syntaxin-1 in synaptic exocytosis); Blue: t-SNARE light chains (SNAP-25 in synaptic exocytosis); Pink: v-SNARE. Structures were edited in PyMol. For clarity, regulatory sequences of SNAREs such as the Habc domain are not shown.
The best studied vesicle fusion pathway is synaptic exocytosis (neurotransmitter release), which requires syntaxin-1 and SNAP-25 as the t-SNAREs (also known as Q-SNAREs), VAMP2/synaptobrevin as the v-SNARE (also known as R-SNARE), and Munc18-1/STXBP1/nSec1 as the cognate SM protein (2,20–26). The first molecular target of Munc18-1 identified was the “closed” syntaxin-1 monomer (27). Disruption of this closed syntaxin binding mode, however, had little effect on Munc18-1 function in neurons (28,29). Moreover, binding to a syntaxin monomer is not a conserved feature of SM family proteins (1). Thus, regulation of the syntaxin monomer cannot explain the crucial role of SM proteins in vesicle fusion.
Using reconstituted liposome fusion assays, we discovered a stimulatory function of Munc18-1 in SNARE-dependent membrane fusion reactions (30). Recognizing both the v- and t-SNAREs, Munc18-1 promotes trans-SNARE zippering and strongly accelerates the fusion kinetics (28,30–33). Our reconstitution analysis of Munc18c/Munc18–3, an unrelated SM protein involved in insulin-stimulated GLUT4 exocytosis (34), suggests that the trans-SNARE-regulating function of Munc18-1 is conserved among SM family proteins (33,35). The biological relevance of this trans-SNARE-regulating function of SM proteins is supported by multiple lines of evidence. First, efficient liposome fusion is observed only when SM proteins are included (30,33,35), correlating with the essential roles of SM proteins in vesicle transport. Second, SM proteins selectively activate the reconstituted fusion reactions driven by their cognate SNARE isoforms (28,30,32,35), in agreement with the compartmental specificity of SM proteins in vivo (1). Third, targeted mutations that impair the trans-SNARE-regulating functions of SM proteins compromise vesicle fusion in intact cells (28–30,33,36,37). Finally, recent crystal structures revealed that SM proteins recognize both v- and t-SNAREs (38), supporting the notion that SM proteins regulate trans-SNARE zippering. Altogether, these findings established that the trans-SNARE-regulating function of SM proteins uncovered in our reconstitution studies provides a molecular explanation for the conserved role of SM proteins in intracellular vesicle fusion.
We introduced macromolecular crowding agents into the reconstituted fusion reactions to recapitulate the crowded cellular environment. This functional reconstitution system described here more closely recapitulates the physiological environment and enabled us to establish fundamental features of SM proteins in SNARE-dependent intracellular vesicle fusion. We anticipate that this reconstitution approach will be instrumental in delineating the molecular basis of additional regulatory factors in vesicle fusion such as the SNARE-binding regulators involved in insulin-stimulated GLUT4 exocytosis (34,39,40).
2. MATERIALS
2.1. Plasmids
Synaptic t-SNARE complex: genes encoding untagged rat syntaxin-1 and N-terminally His6-tagged mouse SNAP-25 were subcloned into a pET-28a-based bicistronic expression vector (3,41).
GLUT4 exocytic t-SNARE complex: genes encoding untagged rat syntaxin-4 and N-terminally His6-tagged mouse SNAP-23 were subcloned into pET-28a and pET-15b vectors, respectively (30,35).
VAMP2: the mouse Vamp2 gene was subcloned into a pET-28a-His6-SUMO vector (30).
Munc18-1: the rat Munc18-1/Stxbp1 gene was subcloned into the pET-28a-His6-SUMO vector (28,30).
Munc18c: the mouse Munc18c/Stxbp3 gene was subcloned into the baculovirus transfer vector pFastBac HT A (35). The resulted plasmid encodes an N-terminally His6-tagged Munc18c protein with a Tobacco Etch Virus (TEV) cleavage site between the His6 tag and Munc18c.
2.2. Buffers
E. coli lysis buffer: 25 mM HEPES (pH 7.4), 400 mM KCl, 10% glycerol (v/v), 20 mM imidazole, 2 mM β-mercaptoethanol (freshly added), and protease inhibitor cocktail (Roche, one tablet per 100 mL buffer, freshly added). In a glass beaker of ~700 mL water, dissolve 5.96 g HEPES, 29.82 g KCl, and 1.36 g imidazole. Add 100 mL glycerol and adjust the pH to 7.4 using HCl. Add water to adjust the volume to 1 L.
Insect cell lysis buffer: 25 mM HEPES (pH 7.4), 400 mM KCl, 10% glycerol, 20 mM imidazole, 1% Triton, 2 mM β-mercaptoethanol, and protease inhibitor cocktail. Dissolve 5.96 g HEPES, 29.82 g KCl, and 1.36 g imidazole in ~700 mL water. Add 100 mL glycerol and adjust the pH to 7.4 using HCl. Add 10 mL Triton and adjust the volume to 1L with water.
SNARE storage buffer: 25 mM HEPES (pH 7.4), 400 mM KCl, 1% n-octyl-β-D-glucoside (OG), 10% glycerol, and 0.5 mM Tris(2-carboxyethyl)phosphine (TCEP). Dissolve 5.96 g HEPES, 29.82 g KCl and 10 g OG in ~700 mL water. Add 100 mL glycerol and adjust the pH to 7.4 using HCl. Adjust the final volume to 1 L using water.
SM protein storage buffer: 25 mM HEPES (pH 7.4), 150 mM KCl, 10% glycerol, and 0.5 mM TCEP. Dissolve 5.96 g HEPES and 11.18 g KCl in ~700 mL water. Add 100 mL glycerol and adjust the pH to 7.4 using HCl. Add water to achieve a final volume of 1 L.
Reconstitution buffer: 25 mM HEPES (pH 7.4), 100 mM KCl, 10% Glycerol, and 0.5 mM TCEP. Dissolve 5.96 g HEPES and 7.45 g KCl in ~700 mL water. Add 100 mL glycerol and adjust the pH to 7.4 using HCl. Add water to achieve a final volume of 1 L.
Nycodenz solution: 20%, 30% and 80% Nycodenz (w/v) in reconstitution buffer. Dissolve 0.596 g HEPES, 0.745 g KCl, and 20 g (20%) or 30g (30%) or 80 g (80%) Nycodenz in ~70 mL water. Add 10 mL glycerol, and adjust pH of the solution to 7.4 using HCl. Add water to achieve a final volume of 100 mL.
Macromolecular crowding agent: Ficoll 70 was dissolved in the reconstitution buffer at the concentration of 200 mg/mL. Dissolve 0.596 g HEPES, 0.745 g KCl, and 20 g Ficoll 70 in ~70 mL water. Add 10 mL glycerol and adjust the pH to 7.4 using HCl. Add water to achieve a final volume of 100 mL.
2.3. Lipids
Unlabeled lipid mixture (15 mM): 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (POPE), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoserine (POPS) and cholesterol were dissolved in chloroform and mixed at a molar ratio of 60:20:10:10.
Labeled lipid mixture (3 mM): POPC, POPE, POPS, cholesterol, (N-(7-nitro-2,1,3-benzoxadiazole-4-yl)-1,2-dipalmitoyl phosphatidylethanolamine (NBD-DPPE), and N-(Lissamine rhodamine B sulfonyl)-1,2-dipalmitoyl phosphatidylethanolamine (rhodamine-DPPE) were dissolved in chloroform and mixed at a molar ratio of 60:17:10:10:1.5:1.5.
3. METHODS
3.1. Recombinant protein expression and purification
3.1.1. Synaptic t-SNAREs
Transform the expression plasmid into BL21-Gold (DE3) competent cells and plate the cells on a Luria-Bertani (LB) agar plate with 100 μg/mL kanamycin.
Pick a single colony and inoculate it into 100 mL LB preculture. Grow the preculture overnight at 37 °C in a shaker set at 220 rpm.
The next morning, add 50 mL of the preculture into 2 L LB culture with 100 μg/mL kanamycin. Grow the culture at 37 °C in a shaker set at 220 rpm.
When the O.D. 600 of the culture reaches 0.6, add isopropyl β-D-1-thiogalactopyranoside (IPTG) to the final concentration of 1 mM.
After another four hours at 37 °C, centrifuge the culture at 4,200 rpm for 20 minutes.
Resuspend the bacteria pellet in 40 mL E. coli lysis buffer.
Add 10 mL of 20% Triton (v/v in water) to the lysate and mix well.
Lyse the cells using a homogenizer.
Centrifuge the lysate for 30 minutes at 4 °C at 180,000 g in an ultracentrifuge (tube dimension: 38×102 mm).
Collect the supernatant and purify the protein using nickel affinity chromatography.
Aliquot the purified proteins in the SNARE storage buffer and store at −70 °C.
3.1.2. GLUT4 exocytic t-SNAREs
Transform the two expression plasmids into BL21-Gold (DE3) competent cells and plate the cells on a LB agar plate with 100 μg/mL kanamycin and 100 μg/mL ampicillin.
Pick a single colony and inoculate it into 100 mL LB preculture. Grow the preculture overnight at 37 °C in a shaker set at 220 rpm.
The next morning, add 50 mL of the preculture into 2 L LB culture with 100 μg/mL kanamycin and 100 μg/mL ampicillin. Grow the culture at 37 °C in a shaker set at 220 rpm.
When the O.D. 600 of the culture reaches 0.6, add IPTG to the final concentration of 0.2 mM.
After overnight induction at 16 °C, centrifuge the culture at 4,200 rpm for 20 minutes.
Purify the proteins as described in 3.1.1.
3.1.3. VAMP2
Express His6-SUMO-VAMP2 in E. coli in a similar way as synaptic t-SNAREs.
Instead of eluting the protein, add 80 μg His6-tagged SUMO protease to the VAMP2-bound nickel beads in the SNARE storage buffer.
Rotate overnight at 4 °C and collect the cleaved VAMP2 proteins (with no extra residues). His6-SUMO moiety and His6-tagged SUMO protease are retained on the nickel beads.
3.1.4. Munc18-1 protein
Transform the pET-28a-His6-SUMO-Munc18-1 plasmid into BL21-Gold (DE3) competent cells and plate the cells on a LB agar plate with 100 μg/mL kanamycin.
Pick a single colony and inoculate it into 100 mL LB preculture. Grow the preculture overnight at 37 °C in a shaker set at 220 rpm.
The next morning, add 50 mL of the preculture into 2 L LB culture with 100 μg/mL kanamycin. Grow the culture at 37 °C in a shaker set at 220 rpm.
When the O.D. 600 of the culture reaches 0.6, add IPTG to the final concentration of 1 mM.
After another two hours at 37 °C, centrifuge the culture at4200 rpm.
Purify Munc18-1 as described in 3.1.1 except that no detergent is added.
Add 50 μg SUMO protease to the eluted proteins and dialyze (molecular weight cutoff: 6–8 kDa) the sample overnight at 4 °C against 2 L SM protein storage buffer.
Remove the His6–SUMO moiety from Munc18-1 proteins using gel filtration chromatography.
3.1.5. Munc18c expression and purification from Sf9 insect cells
Transform the pFastBac-Munc18c plasmid into DH10Bac competent cells and plate the cells on a LB plate with 50 μg/mL kanamycin, 7 μg/mL gentamicin, 10 μg/mL tetracycline, 100 μg/mL Bluo-Gal, and 40 μg/mL IPTG.
After 48 hours, pick one white colony and inoculate into 100 mL LB culture supplemented with 50 μg/mL kanamycin, 7 μg/mL gentamicin, and 10 μg/mL tetracycline.
Isolate Bacmid DNA using a Midiprep kit.
Mix 0.5 μg Bacmid DNA and 8 μL Cellfectin II transfection reagent in 200 μL Sf-900 III media without antibiotics and incubate at room temperature for 20 minutes.
Add the mixture to 0.9 million Sf9 cells grown in 0.8 mL Sf-900 III media without antibiotics.
Grow the cells at 27 °C for five hours without shaking.
Change to fresh media and continue to grow the cells for 96 hours.
Collect the supernatant and centrifuge at 1,000 rpm for five minutes.
Use the virus-containing supernatant to infect Sf9 cells to amplify the viruses.
After two rounds of amplification, collect the virus-containing supernatant and use it to infect 6 × 108 Sf9 cells in 300 mL Sf-900 III media supplemented with 100 μg/mL penicillin and 100 μg/mL streptomycin.
Grow the cells for 48 hours in a shaker set at 130 rpm.
Centrifuge the cell culture at 1,000 rpm.
Add 50 mL insect cell lysis buffer to the pellet and rotate the sample at 4 °C for one hour.
Centrifuge the lysate at 40,000 g for 30 minutes in an ultracentrifuge at 4 °C (tube dimension: 38 × 102 mm).
Purify Munc18c protein using nickel affinity chromatography.
Add 50 μg TEV protease to the eluted protein and dialyze (molecular weight cutoff: 6–8 kDa) the sample overnight at 4 °C against 2 L SM protein storage buffer.
3.2. Reconstitution of proteoliposomes for lipid mixing assays
Add 100 μL unlabeled lipid mixtures to 12 × 75 mm glass tubes and 100 μL labeled lipid mixtures to 10 × 75 mm glass tubes.
Dry the lipids using Argon air flow for 15 minutes.
Further dry the samples using vacuum pump for one hour.
Add 500 μL t-SNARE proteins (125 μg) to the dried unlabeled lipid film and 100 μL v-SNARE protein (37.5 μg) to the dried labeled lipid film.
Dissolve the lipid films by mixing the samples for 15 minutes at room temperature on a vortex mixer.
Add 1 mL reconstitution buffer to the t-SNARE sample and 200 μL reconstitution buffer to the v-SNARE sample while vigorously shaking on a vortex mixer. Liposomes will form when OG concentration drops below its micellar critical concentration.
Dialyze the liposome samples overnight at 4 °C in dialysis tubes (molecular weight cutoff: 12–14 kDa) against 2 L reconstitution buffer.
Add equal volumes of 80% Nycodenz solution to the liposome samples and mix well using a rotating mixer.
Load 3 mL t-SNARE liposomes into a 11 × 60 mm ultracentrifuge tube. Overlay the sample with 750 μL 30% Nycodenz, followed by 250 μL reconstitution buffer without glycerol.
Load 300 μL v-SNARE liposomes into a 5 × 41 mm ultracentrifuge tube. Overlay the sample with 300 μL 30% Nycodenz, followed by 50 μL reconstitution buffer without glycerol.
Centrifuge the t-SNARE liposome samples for three hours and 40 minutes at 400,000 g in an ultracentrifuge. Collect 400 μL t-SNARE liposomes from the 0/30% Nycodenz interface.
Centrifuge the v-SNARE liposome samples for four hours at 320,000 g in an ultracentrifuge. Collect 75 μL v-SNARE liposome samples from the 0/30% Nycodenz interface.
Aliquot the liposomes and store them at −70 °C.
3.3. Reconstitution of proteoliposomes for content mixing assays
Prepare unlabeled t-SNARE liposomes as described in 3.2.
To prepare labeled v-SNARE liposomes for content mixing assays, add 20 μL unlabeled lipid mixture to 10 × 75 mm glass tubes
Dry the lipids using Argon air flow for 15 minutes.
Further dry the samples using vacuum pump for one hour.
Add 100 μL v-SNARE proteins (37.5 μg) to the dried labeled lipid film.
Dissolve the lipid films by mixing the samples for 15 minutes on a vortex mixer at room temperature.
Add 200 μL reconstitution buffer containing 75 mM sulforhodamine B to the v-SNARE sample while vigorously shaking on a vortex mixer.
Dialyze the liposome samples overnight at 4 °C in dialysis tubes (molecular weight cutoff: 12–14 kDa) against 2 L reconstitution buffer.
Add equal volumes of 80% Nycodenz solution to the liposome samples and mix well using a rotating mixer.
Load 300 μL v-SNARE liposomes into a 5 × 41 mm ultracentrifuge tube. Overlay the sample with 200 μL 30% Nycodenz and 100 μL 20% Nycodenz, followed by 50 μL reconstitution buffer without glycerol.
Centrifuge the v-SNARE liposome samples for four hours at 320,000 g in an ultracentrifuge. Collect 75 μL v-SNARE liposome samples from the 0/20% Nycodenz interface.
Use the sulforhodamine B-labeled v-SNARE liposomes immediately after preparation. Do not freeze.
3.4. Liposome lipid mixing assay
Mix 45 μL unlabeled t-SNARE liposomes and 5 μL NBD/rhodamine labeled v-SNARE liposomes with and without the cognate SM protein (5 μM).
Add equal volumes of 200 mg/mL Ficoll 70 such that the final concentration of Ficoll 70 is 100 mg/mL. In the control experiments, add 20 μM VAMP2 cytoplasmic domain (CD, a.a. 1–92) to block SNARE zippering. Keep the ingredients on ice prior to mixing.
Immediately load the mixed samples into a 96-well microplate (such as BioTek Synergy 2) preheated to 37 °C.
Measure the NBD fluorescence (excitation: 460 nm; emission: 538 nm) of the samples every two minutes. Allow the fusion reactions to proceed for 60 to 120 minutes.
Add 10 μL 10% CHAPS to each sample. Continue to measure NBD fluorescence every two minutes for another 20 minutes.
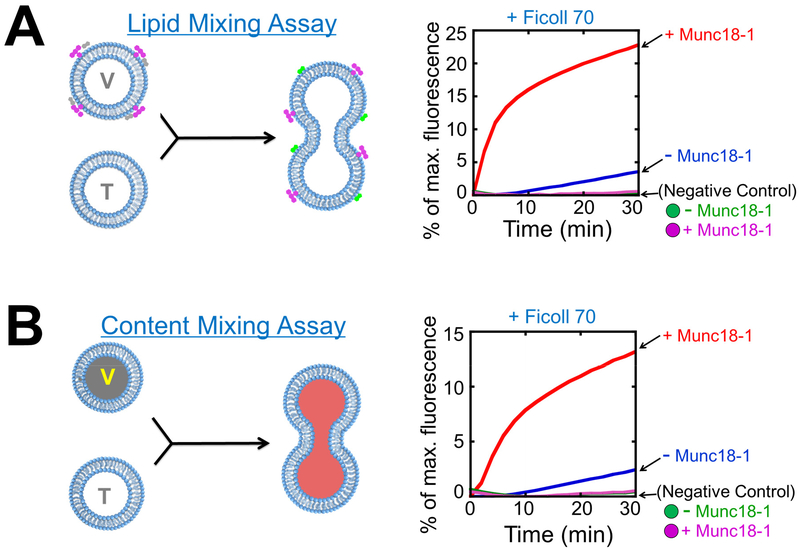
Normalize the data to maximum fluorescence after CHAPS treatment (Fig. 2A).
Figure 2. Reconstitution of SNAREs and SM proteins in liposome fusion assays.
A) Left: diagram showing the Förster/fluorescence resonance energy transfer (FRET)-based assay that measures the lipid mixing of liposome fusion reaction. When unlabeled t-SNARE liposomes fuse with NBD/rhodamine-labeled v-SNARE liposomes, NBD fluorescence increases due to diminished NBD-rhodamine FRET. For clarity, proteins and crowding agents are not shown. Right: liposome lipid mixing reactions. The t-SNARE liposomes containing syntaxin-1 and SNAP-25 were directed to fuse with VAMP2 liposomes with or without 5 μM Munc18-1. The reactions were carried out in the presence of 100 mg/mL Ficoll 70. Ingredients of the samples were mixed and immediately loaded into a pre-warmed microplate to initiate fusion. In negative controls, VAMP2 CD was added to the reactions to the final concentration of 20 μM. B) Left: diagram showing the assay that measures the content mixing of liposome fusion reaction. In these assays, unlabeled t-SNARE liposomes are mixed with sulforhodamine B-loaded v-SNARE liposomes in which the sulforhodamine B fluorescence is inhibited by self-quenching. The fusion of the liposomes leads to the mixing of their contents and the dequenching of sulforhodamine B fluorescence. Right: liposome content mixing reactions. The fusion reactions were performed as in A. Adapted with permission from reference (33). Copyright 2015 American Chemical Society.
3.5. Liposome content mixing assay
Mix 45 μL unlabeled t-SNARE liposomes and 5 μL sulforhodamine B-loaded v-SNARE liposomes in the presence or absence of 5 μM SM protein.
Add equal volumes of 200 mg/ml Ficoll 70 such that the final concentration is 100 mg/mL.
Immediately load the mixed samples into a 96-well microplate (preheated to 37 °C) to initiate fusion.
Measure sulforhodamine B fluorescence (excitation: 565; emission: 585 nm).
At the end of the reaction (60–120 minutes), add 10 μL 10% CHAPS to each sample. Allow the reaction to continue for another 20 minutes.
Normalize the data to the maximum fluorescence after CHAPS treatment (Fig. 2B).
3.6. Trans-SNARE assembly assay
Mix 45 μL t-SNARE liposomes and 5 μL v-SNARE liposomes in the presence or absence of 5 μM SM protein.
Add equal volumes of 200 mg/mL Ficoll 70 such that the final concentration is 100 mg/mL.
After incubation at 4 °C for various time periods, add ten-fold excess amount of GST-tagged VAMP2 CD and mix the sample on a rotating mixer for 20 minutes.
Add CHAPS to the final concentration of 1% and mix the sample on a rotating mixer for 20 minutes.
Add 10 μL nickel beads to each sample and mix them on a rotating mixer for 30 minutes.
Wash the nickel beads three times using the reconstitution buffer supplemented with 1% CHAPS.
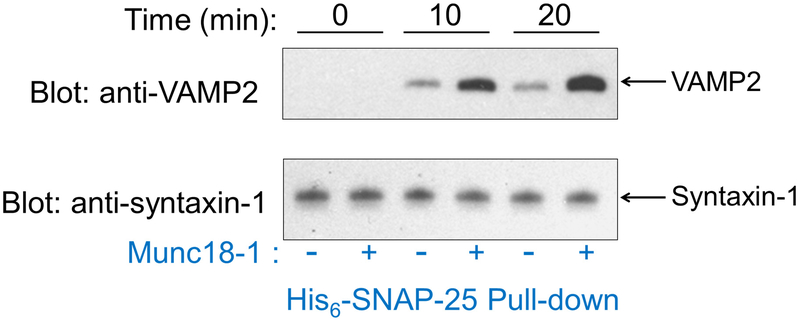
Measure full-length (FL) VAMP2 levels in the precipitates by immunoblotting using monoclonal anti-VAMP2 antibodies, which reflect the amounts of trans-SNARE complexes formed between liposomes (Fig. 3).
Figure 3. Trans-SNARE assembly assay.
Reconstituted t- and v-SNARE liposomes were incubated at 4 °C for the indicated time periods in the presence or absence of 5 μM Munc18-1 before 10-fold excess amount of inhibitory VAMP2 CD was added to block unpaired t-SNAREs. The liposomes were subsequently solubilized and the t-SNAREs were precipitated using nickel sepharose beads. Presence of FL VAMP2 in the precipitates was probed by immunoblotting, which was used as an indicator for trans-SNARE assembly between liposomes. The reactions were performed in the presence of 100 mg/mL Ficoll 70. Adapted with permission from reference (33). Copyright 2015 American Chemical Society.
Notes
β-mercaptoethanol, TCEP and protease inhibitor cocktail were freshly added into the buffer.
To best preserve SM protein activities, immediately freeze purified SM proteins by dipping them into liquid nitrogen and store the proteins at −70 °C. The activities of SM proteins tend to decrease even stored at −70 °C.
SM proteins promote fusion with such potency that it is critical to start all fusion reactions immediately after mixing (less than one minute). Otherwise, SM protein-containing liposomes would fuse during the preparation period, yielding inaccurate initial fluorescence readings.
ACKNOWLEDGEMENTS
This work was supported by National Institutes of Health grants GM102217 and DK095367 (JS).
REFERENCES
- 1.Sudhof TC, and Rothman JE (2009) Membrane fusion: grappling with SNARE and SM proteins. Science (New York, N.Y.) 323, 474–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sollner T, Whiteheart SW, Brunner M, Erdjument-Bromage H, Geromanos S, Tempst P, and Rothman JE (1993) SNAP receptors implicated in vesicle targeting and fusion. Nature 362, 318–324 [DOI] [PubMed] [Google Scholar]
- 3.Weber T, Zemelman BV, McNew JA, Westermann B, Gmachl M, Parlati F, Sollner TH, and Rothman JE (1998) SNAREpins: minimal machinery for membrane fusion. Cell 92, 759–772 [DOI] [PubMed] [Google Scholar]
- 4.Wickner W, and Schekman R (2008) Membrane fusion. Nat Struct Mol Biol 15, 658–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jahn R, and Scheller RH (2006) SNAREs--engines for membrane fusion. Nature reviews. Molecular cell biology 7, 631–643 [DOI] [PubMed] [Google Scholar]
- 6.Gao Y, Zorman S, Gundersen G, Xi Z, Ma L, Sirinakis G, Rothman JE, and Zhang Y (2012) Single reconstituted neuronal SNARE complexes zipper in three distinct stages. Science (New York, N.Y.) 337, 1340–1343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Melia TJ, Weber T, McNew JA, Fisher LE, Johnston RJ, Parlati F, Mahal LK, Sollner TH, and Rothman JE (2002) Regulation of membrane fusion by the membrane-proximal coil of the t-SNARE during zippering of SNAREpins. The Journal of cell biology 158, 929–940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu T, Rammner B, Margittai M, Artalejo AR, Neher E, and Jahn R (1999) Inhibition of SNARE complex assembly differentially affects kinetic components of exocytosis. Cell 99, 713–722 [DOI] [PubMed] [Google Scholar]
- 9.Pobbati AV, Stein A, and Fasshauer D (2006) N- to C-terminal SNARE complex assembly promotes rapid membrane fusion. Science (New York, N.Y.) 313, 673–676 [DOI] [PubMed] [Google Scholar]
- 10.Li F, Pincet F, Perez E, Eng WS, Melia TJ, Rothman JE, and Tareste D (2007) Energetics and dynamics of SNAREpin folding across lipid bilayers. Nat Struct Mol Biol 14, 890–896 [DOI] [PubMed] [Google Scholar]
- 11.Zhou P, Bacaj T, Yang X, Pang ZP, and Sudhof TC (2013) Lipid-Anchored SNAREs Lacking Transmembrane Regions Fully Support Membrane Fusion during Neurotransmitter Release. Neuron 80, 470–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu H, Zick M, Wickner WT, and Jun Y (2011) A lipid-anchored SNARE supports membrane fusion. Proc Natl Acad Sci U S A 108, 17325–17330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hata Y, Slaughter CA, and Sudhof TC (1993) Synaptic vesicle fusion complex contains unc-18 homologue bound to syntaxin. Nature 366, 347–351 [DOI] [PubMed] [Google Scholar]
- 14.Dulubova I, Khvotchev M, Liu S, Huryeva I, Sudhof TC, and Rizo J (2007) Munc18-1 binds directly to the neuronal SNARE complex. Proc Natl Acad Sci U S A 104, 2697–2702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Novick P, and Schekman R (1979) Secretion and cell-surface growth are blocked in a temperature-sensitive mutant of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A 76, 1858–1862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pevsner J, Hsu SC, and Scheller RH (1994) n-Sec1: a neural-specific syntaxin-binding protein. Proc Natl Acad Sci U S A 91, 1445–1449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garcia EP, Gatti E, Butler M, Burton J, and De Camilli P (1994) A rat brain Sec1 homologue related to Rop and UNC18 interacts with syntaxin. Proc Natl Acad Sci U S A 91, 2003–2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carr CM, and Rizo J (2010) At the junction of SNARE and SM protein function. Current opinion in cell biology 22, 488–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Burgoyne RD, Barclay JW, Ciufo LF, Graham ME, Handley MT, and Morgan A (2009) The functions of Munc18-1 in regulated exocytosis. Ann N Y Acad Sci 1152, 76–86 [DOI] [PubMed] [Google Scholar]
- 20.Bennett MK, Calakos N, and Scheller RH (1992) Syntaxin: a synaptic protein implicated in docking of synaptic vesicles at presynaptic active zones. Science (New York, N.Y.) 257, 255–259 [DOI] [PubMed] [Google Scholar]
- 21.Elferink LA, Trimble WS, and Scheller RH (1989) Two vesicle-associated membrane protein genes are differentially expressed in the rat central nervous system. J Biol Chem 264, 11061–11064 [PubMed] [Google Scholar]
- 22.Oyler GA, Higgins GA, Hart RA, Battenberg E, Billingsley M, Bloom FE, and Wilson MC (1989) The identification of a novel synaptosomal-associated protein, SNAP-25, differentially expressed by neuronal subpopulations. The Journal of cell biology 109, 3039–3052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sudhof TC, Baumert M, Perin MS, and Jahn R (1989) A synaptic vesicle membrane protein is conserved from mammals to Drosophila. Neuron 2, 1475–1481 [DOI] [PubMed] [Google Scholar]
- 24.Sutton RB, Fasshauer D, Jahn R, and Brunger AT (1998) Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 A resolution. Nature 395, 347–353 [DOI] [PubMed] [Google Scholar]
- 25.Weimer RM, Richmond JE, Davis WS, Hadwiger G, Nonet ML, and Jorgensen EM (2003) Defects in synaptic vesicle docking in unc-18 mutants. Nat Neurosci 6, 1023–1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Verhage M, Maia AS, Plomp JJ, Brussaard AB, Heeroma JH, Vermeer H, Toonen RF, Hammer RE, van den Berg TK, Missler M, Geuze HJ, and Sudhof TC (2000) Synaptic assembly of the brain in the absence of neurotransmitter secretion. Science (New York, N.Y.) 287, 864–869 [DOI] [PubMed] [Google Scholar]
- 27.Misura KM, Scheller RH, and Weis WI (2000) Three-dimensional structure of the neuronal-Sec1-syntaxin 1a complex. Nature 404, 355–362 [DOI] [PubMed] [Google Scholar]
- 28.Rathore SS, Bend EG, Yu H, Hammarlund M, Jorgensen EM, and Shen J (2010) Syntaxin N-terminal peptide motif is an initiation factor for the assembly of the SNARE-Sec1/Munc18 membrane fusion complex. Proc Natl Acad Sci U S A 107, 22399–22406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou P, Pang ZP, Yang X, Zhang Y, Rosenmund C, Bacaj T, and Sudhof TC (2012) Syntaxin-1 N-peptide and H(abc)-domain perform distinct essential functions in synaptic vesicle fusion. EMBO J [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shen J, Tareste DC, Paumet F, Rothman JE, and Melia TJ (2007) Selective Activation of Cognate SNAREpins by Sec1/Munc18 Proteins. Cell 128, 183–195 [DOI] [PubMed] [Google Scholar]
- 31.Rathore SS, Ghosh N, Ouyang Y, and Shen J (2011) Topological arrangement of the intracellular membrane fusion machinery. Molecular biology of the cell 22, 2612–2619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shen J, Rathore S, Khandan L, and Rothman JE (2010) SNARE bundle and syntaxin N-peptide constitute a minimal complement for Munc18-1 activation of membrane fusion. J Cell Biology 190, 55–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu H, Rathore SS, Shen C, Liu Y, Ouyang Y, Stowell MH, and Shen J (2015) Reconstituting Intracellular Vesicle Fusion Reactions: The Essential Role of Macromolecular Crowding. J Am Chem Soc 137, 12873–12883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bryant NJ, Govers R, and James DE (2002) Regulated transport of the glucose transporter GLUT4. Nature reviews. Molecular cell biology 3, 267–277 [DOI] [PubMed] [Google Scholar]
- 35.Yu H, Rathore SS, Lopez JA, Davis EM, James DE, Martin JL, and Shen J (2013) Comparative studies of Munc18c and Munc18-1 reveal conserved and divergent mechanisms of Sec1/Munc18 proteins. Proc Natl Acad Sci U S A 110, E3271–3280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shen C, Rathore SS, Yu H, Gulbranson DR, Hua R, Zhang C, Schoppa NE, and Shen J (2015) The trans-SNARE-regulating function of Munc18-1 is essential to synaptic exocytosis. Nat Commun 6, 8852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Deak F, Xu Y, Chang WP, Dulubova I, Khvotchev M, Liu X, Sudhof TC, and Rizo J (2009) Munc18-1 binding to the neuronal SNARE complex controls synaptic vesicle priming. The Journal of cell biology 184, 751–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baker RW, Jeffrey PD, Zick M, Phillips BP, Wickner WT, and Hughson FM (2015) A direct role for the Sec1/Munc18-family protein Vps33 as a template for SNARE assembly. Science (New York, N.Y.) 349, 1111–1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gulbranson DR, Davis EM, Demmitt BA, Ouyang Y, Ye Y, Yu H, and Shen J (2017) RABIF/MSS4 is a Rab-stabilizing holdase chaperone required for GLUT4 exocytosis. Proc Natl Acad Sci U S A 114(39), E8224–E8233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Leto D, and Saltiel AR (2012) Regulation of glucose transport by insulin: traffic control of GLUT4. Nature reviews. Molecular cell biology 13, 383–396 [DOI] [PubMed] [Google Scholar]
- 41.Weber T, Parlati F, McNew JA, Johnston RJ, Westermann B, Sollner TH, and Rothman JE (2000) SNAREpins are functionally resistant to disruption by NSF and alphaSNAP. The Journal of cell biology 149, 1063–1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stein A, Weber G, Wahl MC, and Jahn R (2009) Helical extension of the neuronal SNARE complex into the membrane. Nature 460, 525–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ellena JF, Liang B, Wiktor M, Stein A, Cafiso DS, Jahn R, and Tamm LK (2009) Dynamic structure of lipid-bound synaptobrevin suggests a nucleation-propagation mechanism for trans-SNARE complex formation. Proc Natl Acad Sci U S A 106, 20306–20311 [DOI] [PMC free article] [PubMed] [Google Scholar]