Abstract
The progression to AIDS is influenced by changes in the biology of heterogeneous monocyte subsets. Classical (CD14++CD16–), intermediate (CD14++CD16+), and nonclassical (CD14+CD16++) monocytes may represent progressive stages of monocyte maturation or disparate myeloid lineages with different turnover rates and function. To investigate the relationship between monocyte subsets and the response to SIV infection, we performed microarray analysis of monocyte subsets in rhesus macaques at three time points: prior to SIV infection, 26 days postinfection, and necropsy with AIDS. Genes with a 2‐fold change between monocyte subsets (2023 genes) or infection time points (424 genes) were selected. We identify 172 genes differentially expressed among monocyte subsets in both uninfected and SIV‐infected animals. Classical monocytes express genes associated with inflammatory responses and cell proliferation. Nonclassical monocytes express genes associated with activation, immune effector functions, and cell cycle inhibition. The classical and intermediate subsets are most similar at all time points, and transcriptional similarity between intermediate and nonclassical monocytes increases with AIDS. Cytosolic sensors of nucleic acids, restriction factors, and IFN‐stimulated genes are induced in all three subsets with AIDS. We conclude that SIV infection alters the transcriptional relationship between monocyte subsets and that the innate immune response to SIV infection is conserved across monocyte subsets.
Keywords: AIDS, CD14, CD16, HIV, immune activation, pathogenesis
Abbreviations
- AFC
absolute fold change
- CX3CR
chemokine (C‐X3‐C motif) receptor
- DC
dendritic cell
- dpi
days postinfection
- GO
gene ontology
- HIVE
HIV encephalitis
- ISG
IFN‐stimulated gene
- IACUC
Institutional Animal Care and Use Committee
- MFI
mean fluorescence intensity
- qRT‐PCR
quantitative real‐time PCR
- SIVE
SIV encephalitis
1. INTRODUCTION
In HIV and SIV infection, the progression to AIDS is associated with changes in monocyte biology including increased egress from the bone marrow, accelerated turnover, and increased activation.1, 2, 3, 4, 5, 6, 7 Importantly, monocytes are phenotypically and functionally heterogeneous, and subpopulations differ with regard to immune function, activation status, migratory properties, and susceptibility to HIV or SIV infection.4, 8, 9, 10, 11 Due to functional differences between monocyte subsets, perturbations to normal monocyte homeostasis and the relative proportion of each subset may influence disease progression. In humans and nonhuman primates, three populations of monocyte exist, defined by expression of CD14 (LPS coreceptor) and CD16 (FcγRIII): classical (CD14++CD16–), intermediate (CD14++CD16+), and nonclassical (CD14+CD16++) monocytes.4, 12, 13 The developmental relationship between monocyte subsets and the specific role of each in HIV and SIV pathogenesis are not known.
Current understanding of the biology of monocyte subpopulations comes from studies in humans and macaques and homologous monocyte populations in mice.14, 15 In humans, circulating monocytes are composed of approximately 80% classical monocytes, 5% intermediate monocytes, and 10% nonclassical monocytes.4, 13, 16 Classical monocytes exhibit high phagocytic ability and produce reactive oxygen species and IL‐10 in response to LPS.9, 16, 17 Additionally, classical monocytes express CCR2 and CD62L, migrate in response to inflammation, and may differentiate into classically activated (M1 polarized) macrophages or dendritic cells (DCs) in inflamed tissues.13, 14, 15, 18, 19 Intermediate monocytes are capable of antigen presentation and T‐cell activation, and may be a transitional population between classical and nonclassical monocytes.4, 13 In response to LPS, intermediate monocytes produce TNF‐α and IL‐1β.16 Nonclassical monocytes migrate along the luminal face of the vasculature, express chemokine (C‐X3‐C motif) receptor (CX3CR1; fractalkine receptor), and may extravasate into tissue to replenish resident macrophage populations.4, 12, 13, 16, 18 In response to TLR7/TLR8 agonists (viruses, nucleic acids), nonclassical monocytes produce proinflammatory cytokines TNF‐α, IL‐1β, and CCL3/MIP‐1α.16 Thus, monocyte subsets differ in abundance, phenotype, migratory ability, and response to immune stimuli.
The progression to AIDS and the incidence of CNS disease are directly influenced by monocyte dysregulation in HIV and SIV infection.6, 20, 21, 22 Previous gene array studies regarding gene regulation in ex vivo monocytes in the context of HIV infection have identified a number of affected pathways including macrophage activation, chemotaxis, cell cycle, apoptosis, lipid metabolism, proteasome function, and transcriptional regulation.2, 23, 24, 25, 26, 27 With HIV or SIV infection, increased numbers of circulating monocytes and increased monocyte activation correlate with the development of viral encephalitis (HIV encephalitis [HIVE] and SIV encephalitis [SIVE]) and dementia.3, 28, 29, 30, 31 The gene expression profile of CD14+ monocytes from HIV‐1+ patients is similar to the gene expression profiles of uninfected monocytes stimulated with type‐I IFN and uninfected monocytes stimulated with Staphylococcus aureus, a TLR2 agonist.27, 32 Additionally, CD14+ monocytes from HIV‐1+ individuals with high viral loads were shown to upregulate macrophage‐associated genes involved in chemotaxis and inflammatory response, underscoring the role of activated monocytes as precursors to tissue macrophages in HIVE.2
In vivo, the frequency of productively infected monocytes is low (<0.1%); however, the mean half‐life of HIV‐1 DNA in monocytes was found to be greater than 40 months in spite of effective antiretroviral therapy.10, 33, 34, 35, 36 Given the low frequency of infection, changes in monocyte biology are most likely the result of systemic immune activation, altered cytokine signaling, and other indirect mechanisms resulting from chronic HIV or SIV infection. CD16+ monocytes are more permissive to HIV and SIV infection compared to CD16– monocytes, and are preferentially infected in vivo.6, 10 Historically in HIV research, many studies have focused on the role of activated CD14+CD16+ monocytes (intermediate monocytes), which expand with inflammation and are precursors of tissue macrophages.3, 5, 7, 28, 30, 31, 37, 38 In HIVE, increased activation and expansion of the intermediate subset is thought to be associated with traffic of virus across the blood‐brain barrier and accumulation of inflammatory macrophages in the CNS.6, 37, 39 Specific roles for classical and nonclassical monocytes in HIV and SIV pathogenesis are not well characterized. With a new view of monocyte heterogeneity, it is necessary to define the shared and unique biology of the three monocyte subsets and the role each plays in HIV pathogenesis.
We performed transcriptional analysis of the three monocyte subpopulations in rhesus macaques (n = 4) at three time points: (1) prior to SIV infection, (2) at 26 days postinfection (dpi), and (3) at necropsy, terminally with AIDS. We found that the majority of markers that differentiate monocyte subsets in uninfected animals are not differentially expressed between monocyte subsets in SIV‐infected animals. Transcripts that identify classical and nonclassical monocytes in humans and mice were conserved in rhesus macaques. We found that classical monocytes expressed transcripts associated with proliferation, wound repair, and response to immune stimuli, while nonclassical monocytes expressed transcripts associated with macrophage phenotype, withdrawal from cell cycle, and immune effector functions. Intermediate monocytes were more similar to classical monocytes than nonclassical monocytes, but showed increased transcriptional similarity to nonclassical monocytes terminally with AIDS. Changes to monocyte gene expression occurred within weeks of primary infection and were sustained throughout the course of infection. Transcripts associated with innate immune response, particularly IFN‐stimulated genes (ISGs), were induced in all monocyte subsets in SIV‐infected animals. These studies indicate that the set of expressed genes associated with each monocyte subset is altered with SIV infection, but each subset retains its putative functional characteristics. Additionally, the innate immune response to SIV infection, including induction of ISGs, is conserved across monocyte subsets.
2. MATERIALS AND METHODS
2.1. Ethics statement
All animals were handled in accordance with good animal practice as defined by the Tulane National Primate Research Center's Institutional Animal Care and Use Committee (IACUC). All animal work was approved by this committee and the IACUC of Boston College.
2.2. Animals and treatments
Four adult male rhesus macaques (animal ID: CM07, DB79, FD05, FB92) were infected i.v. with 1 ng of SIVp27 of SIVmac251, a generous gift from by Ronald Desrosiers. CD8+ lymphocytes were depleted in order to achieve rapid progression to AIDS with a >75% incidence of SIVE.3, 7, 40, 41, 42 The chimeric anti‐CD8 antibody cM‐T807 (NIH Nonhuman Primate Reagent Resource) was administered s.c. (10 mg/kg) at 6 dpi and i.v. (5 mg/kg) on 8 dpi and 12 dpi as previously described.3, 30 Complete blood counts and flow cytometry to monitor leukocyte populations were performed prior to infection and weekly throughout the course of the disease.
SIV‐infected animals were sacrificed with any of the following criteria, indicative of AIDS when present: >15% decrease in body weight in 2 weeks or >30% decrease in body weight in 2 months; documented opportunistic infection; persistent anorexia >3 days without explicable cause; severe, intractable diarrhea; progressive neurological symptoms; significant cardiac or pulmonary symptoms. Prior to sacrifice, animals were anesthetized with ketamine‐HCl and euthanized by an intravenous pentobarbital overdose and exsanguinated. A postmortem diagnosis of AIDS was confirmed by the presence of AIDS defining lesions: Pneumocystis pneumonia, Mycobacterium avium infection, and intestinal adenovirus infection. SIVE was defined by the presence of multinucleated giant cells, accumulation of macrophages in the CNS, and productive viral infection.3, 43, 44, 45
2.3. Viral load determination
SIV RNA in the plasma was quantitated by real‐time PCR as previously described.46 SIV virions were pelleted from 0.5 ml EDTA plasma by centrifugation at 20,000 g for 1 h. The fluorescently labeled, real‐time PCR probe employed contained a nonfluorescent quencher, BHQ‐1, at its 3′ end. The threshold of sensitivity was 100 copy equivalents/ml, with an average interassay coefficient of variation of less than 25%.
2.4. Isolation of monocyte subpopulations for microarray analysis
Prior to infection, at 26 dpi, and at necropsy, mononuclear cells were isolated from 10 ml EDTA‐treated whole blood (n = 4 animals: CM07, DB79, FD05, FB92) by density gradient centrifugation over Ficoll‐Hypaque (GE Healthcare, Chicago, IL). Buffy coats were collected and washed in calcium‐ and magnesium‐free PBS and stained for 15 min with anti‐CD14‐PacificBlue, anti‐CD16‐PE, anti‐HLA‐DR‐PerCP‐Cy‐5.5, anti‐CD3‐FITC, and anti‐CD20‐APC (BD Biosciences) in PBS containing 2% FBS. Monocytes were isolated by forward and side scatter characteristics and CD14 antigen expression. HLA‐DR+CD3‐CD20– cells were selected to exclude B‐ and T‐cells (Figure 2A). Nonoverlapping populations of classical, intermediate, and nonclassical monocytes were isolated based on expression of CD14 and CD16 on a BD FACS ARIA (BD Biosciences, San Jose, CA). FACS‐isolated monocytes were washed in PBS to remove residual FACS buffer and vacuum pelleted to preserve RNA integrity.
Figure 2.
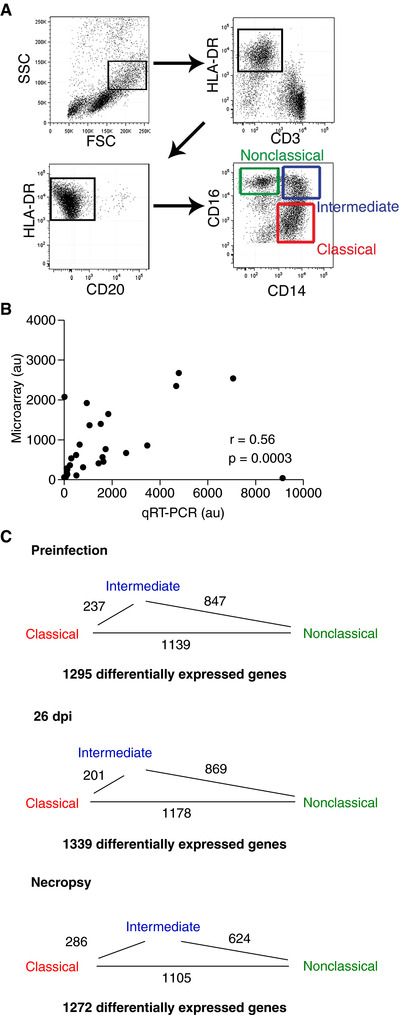
Gating strategy, microarray validation, and identification of differentially expressed genes. (A) Classical, intermediate, and nonclassical monocytes were isolated from total PBMCs from each animal at each infection time point. Monocytes were selected based on high forward scatter and intermediate side scatter. HLA‐DR+ cells were positively selected and CD3+ T‐cells and CD20+ B‐cells were excluded. Nonoverlapping populations of classical (CD14++CD16–, red gate), intermediate (CD14++CD16+, blue gate), and nonclassical (CD14+CD16++, green gate) monocytes were sorted based on expression of CD14 and CD16. (B) Paired XY values for gene expression values determined by microarray and qRT‐PCR. For qRT‐PCR analysis, cDNA was made from the same RNA used in the microarray analysis. Nine samples had sufficient RNA remaining to validate four targets each. GAPDH was used as a housekeeping gene. Gene expression values determined by qRT‐PCR significantly correlated with the gene expression values determined by microarray (Spearman r = 0.56, P = 0.0003). (C) The number of gene expression comparisons with ≥2.0 AFC between monocyte subsets prior to infection, at 26 dpi, and at necropsy. The length of the line between monocyte subsets is proportional to the number of differentially expressed genes. Intermediate monocytes are most similar to classical monocytes, but increase in similarity to nonclassical monocytes terminally with AIDS
2.5. Microarray processing
RNA extraction, labeling, and hybridization were performed by the Beth Israel Deaconess Medical Center Genomics Core Laboratory as previously described.47, 48 Briefly, total RNA was extracted with TRIzol (Life Technologies, Carlsbad, CA) according to the manufacturer's instructions. RNA quality at all steps was assayed on an Agilent 2100 Bioanlayzer (Agilent Technologies). RNA (>500 pg) was amplified using the Ovation pico WTA system (NuGEN Technologies Inc., San Carlos, CA). cDNA (20 μg) was fragmented as described and hybridized with a preequilibrated Affymetrix GeneChip Rhesus Macaque Genome Array (Affymetrix, Santa Clara, CA) at 45°C for 16 h. After the hybridization cocktails were removed, the chips were washed, stained, and scanned according to the manufacturer's protocols as previously described.47 The microarray data files were deposited in the Gene Expression Omnibus, accession GSE98717 (https://www.ncbi.nlm.nih.gov/geo).
2.6. Microarray data analysis
The gene expression data generated (>47,000 rhesus transcripts interrogated per array) were analyzed with Partek Express Genomics Suite software (Partek Inc., Chesterfield, MO). The data were normalized by using a robust multichip average algorithm.49, 50 Quality control measures included assessment of labeling controls, hybridization controls, 3′/5′ ratio, mean raw probe intensity, mean absolute deviation across chips, and mean relative log expression. Due to the low abundance of the nonclassical monocyte subset, insufficient RNA was isolated from this population at preinfection from animals FD05 and FB92, and at necropsy from animal CM07. These samples did not meet quality control parameters and were excluded from further analysis.
2.7. Statistical analysis
Partek Express was used to identify genes that were that were significantly (P < 0.05), differentially expressed across monocyte subset or SIV infection time point with a 5% false discovery rate. ANOVA was performed followed by pairwise comparisons using the Benjamini‐Hochberg step‐up correction for multiple tests. A report of differentially expressed probes with fold‐change and P‐values was generated from this analysis. This list was used to select probes with a fold change ≥|2| in 1 monocyte subset or at one SIV infection time point. Affymetrix probe IDs were annotated to the human official gene symbol based on information provided by the manufacturer and supplementary annotation data from the Norgren lab, which collaborated in the development of the Affymetrix rhesus macaque genome array (http://www.unmc.edu/rhesusgenechip/#RhesusGeneChip). Heatmaps were generated using dChip software.51 Gene lists were subjected to functional annotation clustering with DAVID bioinformatics tools to identify enrichment clusters associated with specific biological processes according to gene ontology (GO) annotations.52, 53 Statistical significance of the expansion of monocyte subsets (Figure 1) was determined by Mann‐Whitney U. Statistical significance of the correlations between probe intensity and quantitative real‐time PCR (qRT‐PCR) (Figure 2) and mean fluorescence intensity (MFI) by FACS (Table 1) was determined by t test using Spearman's R.
Figure 1.
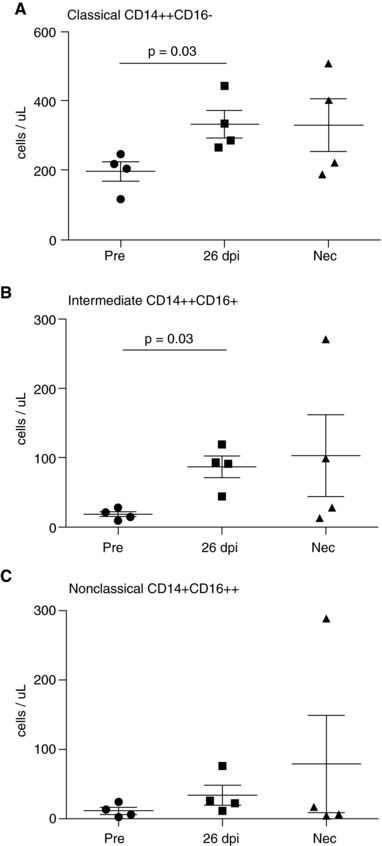
Monocyte populations expand with SIV infection. The absolute number of classical, intermediate, and nonclassical monocytes (mean ± sem, n = 4 animals) was determined prior to infection. Gene expression analysis was performed on these cell populations at the same time points. These monocytes were isolated for gene expression analysis prior to infection, at 26 dpi, and at necropsy with AIDS. (A) Prior to infection classical monocytes were 196 ± 56 cells/μl. With SIV, the number of classical monocytes increased to 332 ± 40 cells/μl at 26 dpi (P = 0.03 26 dpi vs. preinfection) and 329 ± 75 cells/μl terminally with AIDS. (B) Prior to infection intermediate monocytes were 18 ± 4 cells/ μl. With SIV, the number of intermediate monocytes increased to 87 ± 16 cells/μl at 26 dpi (P = 0.03 26 dpi vs. preinfection) and 103 ± 60 cells/μl terminally with AIDS. (C) Prior to infection, nonclassical monocytes were 11 ± 5 cells/μl. With SIV, the number of nonclassical monocytes increased to 34 ± 15 cells/μl at 26 dpi and 79 ± 70 cells/μl terminally with AIDS. Significance of pairwise comparisons was determined by Mann‐Whitney U
Table 1.
Microarray signal intensity correlates with mean fluorescence intensity (MFI) by FACS
| Gene/protein | Spearman's R a | P‐valueb |
|---|---|---|
| CD14 | 0.75 | <0.0001 |
| FCGR3A/CD16c | 0.80 | <0.0001 |
| FCGR3B/CD16c | 0.48 | <0.0001 |
| CD68 | 0.68 | <0.0001 |
| CD163 | 0.67 | 0.003 |
| S100A8 | 0.46 | 0.04 |
| HLA‐DR | 0.64 | 0.001 |
| CD44/CD44v6c | 0.48 | 0.03 |
| CX3CR1 | 0.48 | <0.0001 |
| CCR2 | 0.48 | NS |
The normalized log2 microarray signal intensity was plotted versus the median fluorescence intensity (MFI) as determined by FACS for each target. Correlation was determined using Spearman's R.
Significance was determined by t test using Spearman's R.
For these gene/protein target pairs, the microarray probe or antibody recognizes a specific isoform of the gene or protein, as indicated.
NS, not significant.
2.8. qRT‐PCR for microarray validation
RNA from each sample was extracted with TRIzol as indicated above. First‐strand cDNA was generated from >40 ng total RNA with Superscript III first strand synthesis kit (Invitrogen, Carlsbad, CA). qRT‐PCR was performed on 3 ng of cDNA with the Platinum qPCR SuperMix‐UDG kit (Invitrogen) using a Step One Plus system (Life Technologies). Samples were analyzed in duplicate and normalized to the GAPDH housekeeping gene. Relative gene expression was determined using the ΔΔCT method and converted to arbitrary units relative to a reference RNA sample from pooled CD14+ monocytes. qPCR expression data were compared to normalized log2 expression data from the microarray also normalized to GAPDH. Oligonucleotides (IDT Inc., Coralville, IN) used are listed in Supplemental Table 2.
3. RESULTS
3.1. Monocyte populations expand with SIV infection
The absolute number and relative percentage of each monocyte subset were determined for each animal (n = 4): (1) prior to infection, (2) at 26 dpi when increased monocyte egress from the bone marrow is consistently observed, and (3) at necropsy, terminally with AIDS (Supplemental Table 1). At the same time points, monocytes were isolated for gene expression analysis. We chose to include 26 dpi because of our previous BrdU labeling studies that indicate this is a pathogenically important time point in early SIV infection when monocyte expansion from bone marrow increases.3, 7 Animals that have greater than 5% BrdU positive monocytes in blood 24 h after BrdU pulse consistently develop rapid AIDS with SIVE and cardiovascular lesions.3, 7 Prior to infection, classical monocytes accounted for 196 ± 56 cells/μl (87.7 ± 4.3% of total monocytes), intermediate monocytes were 18 ± 4 cells/μl (7.8 ± 1.2% of total monocytes), and nonclassical monocytes were 11 ± 5 cells/μl (4.4 ± 1.5% of total monocytes) (Figure 1). With SIV infection, the absolute number of each of the three monocyte subpopulations increased as well as the percentage of both intermediate and nonclassical monocytes. At 26 dpi, classical, intermediate, and nonclassical monocytes expanded to 332 ± 40 cells/μl (73.9 ± 3.4%, P = 0.03 26 dpi vs. preinfection), 87 ± 16 cells/μl (19.3 ± 3.1%, P = 0.03 26 dpi vs. preinfection), and 34 ± 15 cells/μl (6.8 ± 2%), respectively (Figure 1). An increased absolute number of monocytes were also observed at necropsy terminally with AIDS: classical monocytes 329 ± 75 cells/μl (74.1 ± 10.7%), 103 ± 60 cells/μl (16.9 ± 6.1%), and 79 ± 70 cells/μl (9.0 ± 6.1%) (Figure 1). These data indicate that the absolute number of total monocytes increases with SIV infection and that intermediate and nonclassical monocytes account for a greater percentage of total monocytes in SIV‐infected animals compared to uninfected animals.
3.2. Isolation of monocyte populations and microarray analysis
At each collection time point, PBMCs were isolated from EDTA anticoagulated whole blood by FACS (Supplemental Table 1). Monocytes were selected based on high forward scatter and low side scatter, HLA‐DR+ cells were selected, and CD3 and CD20 were used to exclude T‐ and B‐cells, respectively (Figure 2A). Classical, intermediate, and nonclassical monocyte populations were sorted based on expression of CD14 and CD16 from each animal at each time point (Supplemental Table 1, Figure 2A). cDNA amplified from each sample (n = 33 samples, Supplemental Table 1) was hybridized to Affymetrix rhesus macaque genome arrays (GEO accession # GSE09717 on NCBI). Each array interrogates approximately 47,000 transcripts per sample using probes designed from the Baylor School of Medicine rhesus macaque whole‐genome shotgun assembly and from the Affymetrix Human Genome U133 Plus 2.0 Array. Partek Express Genomic Suite software was used to normalize the raw microarray signal intensities and analyze the gene expression data.
The total variation in gene expression across all samples can be measured by the effect size (F ratio) of the three variables that define each sample: animal (F = 3.01), monocyte subset (F = 2.98), and infection status (F = 1.83). The differences in effect size indicate that, across all samples, inherent differences between individual animals and differences between monocyte subsets account for a greater proportion of the total variation in gene expression compared to differences in gene expression due to SIV infection.
3.3. Identification of transcripts that differentiate monocyte subsets or infection time points
To identify differentially expressed transcripts, gene expression data from each of the four animals were grouped and compared based on the variables of interest: monocyte subset and infection time point. A total of 3418 probe sets were identified as significantly, differentially expressed between monocyte subsets and 665 were identified as significantly, differentially expressed between infection time points. This list was then filtered based on an absolute fold change (AFC) ≥2.0 between monocyte subsets or infection time points and a P‐value < 0.05 by ANOVA followed by pairwise comparisons with Benjamini‐Hochberg correction. Probe sets were annotated with the corresponding human official gene symbol and redundant probe sets representing the same gene and probe sets representing hypothetical genes were removed. In agreement with the observed effect sizes, we found 2023 genes that were significantly, differentially expressed between monocyte subsets in at least one time point and 424 genes that were significantly differentially expressed between infection time points in at least 1 monocyte subset.
3.4. Validation of microarray data by qRT‐PCR and FACS
To validate the microarray expression data, we generated cDNA from available RNA and performed qRT‐PCR. Gene expression as determined by qRT‐PCR was significantly correlated with gene expression as determined by microarray (r = 0.56, P = 0.0003) (Figure 2B). Additionally, we compared gene expression as determined by microarray to protein expression as determined by flow cytometry for a panel of monocyte‐related proteins (CD14, CD16, CD68, CD163, S100A8, HLA‐DR, CD44v6, CX3CR1, CCR2). Protein expression (MFI by FACS) was significantly correlated with gene expression determined by microarray for 8 of the 9 target genes investigated (Table 1).
3.5. Intermediate monocytes resemble classical monocytes and become more transcriptionally similar to nonclassical monocytes in terminal AIDS
To investigate the relationship between 3 monocytes subsets in uninfected and SIV‐infected animals, we compared the number of genes differentially expressed in one or more comparisons between monocyte subsets (classical vs. intermediate, classical vs. nonclassical, or intermediate vs. nonclassical), at each infection time point (Figure 2C). Of the 2023 genes identified as differentially expressed between monocyte subsets, 1295 genes were differentially expressed prior to infection, 1339 genes were differentially expressed at 26 dpi, and 1272 genes were differentially expressed at necropsy (Figure 2C). Intermediate monocytes expressed 69% (890 genes, preinfection), 68% (914 genes, 26 dpi), and 75% (960 genes, necropsy) of differentially expressed genes at levels between classical and nonclassical monocytes. Based on the number of differentially expressed genes, intermediate monocytes were more similar to classical monocytes than nonclassical monocytes, and classical monocytes and nonclassical monocytes were most dissimilar at all time points (Figure 2C). We observed that terminally with AIDS, the transcriptomes of intermediate and nonclassical monocytes increase in similarity. Intermediate and nonclassical monocytes were differentiated by 847 genes preinfection, 869 genes at 26 dpi, and 624 genes at necropsy (Figure 2C). Conversely, the number of genes differentiating intermediate and classical monocytes increased from 237 genes in uninfected animals and 201 genes at 26 dpi to 286 genes at necropsy (Figure 2C). The number of genes differentially expressed between classical and nonclassical monocytes was similar at all three time points: 1139 genes preinfection, 1178 genes at 26 dpi, and 1105 genes at necropsy (Figure 2C). Increased similarity between the intermediate and nonclassical monocyte subsets with SIV infection may reflect increased activation or maturation of the intermediate subset in chronic SIV infection and suggests that the set of genes that differentiate monocyte subsets may exhibit plasticity in response to SIV infection and inflammation. Despite changes in gene expression with SIV infection, intermediate monocytes were more similar to classical monocytes than nonclassical monocytes at all time points investigated.
3.6. Gene signature of monocyte subsets in uninfected animals
In order to characterize genes specific to each monocyte subset in uninfected animals, we selected genes that were differentially expressed in each subset compared to the other two subsets (Figure 3, Supplemental Tables 3–7). Prior to infection, we identified that 99 genes expressed ≥2.0‐fold by classical monocytes relative to intermediate monocytes (range: 2.0‐ to 5.3‐fold, median: 2.6) and nonclassical monocytes (range: 2.0‐ to 80‐fold, median: 4.2) (Figure 3A, Supplemental Table 3). Only 17 genes were expressed ≥2.0‐fold by intermediate monocytes relative to classical (range: 2.1‐ to 3.8‐fold, median: 2.5) and nonclassical monocytes (range: 2.2‐ to 20‐fold, median: 3.4) (Figure 3A, Supplemental Table 4). Nonclassical monocytes expressed 242 genes ≥2.0‐fold relative to classical (range: 2.0‐ to 82‐fold; median: 4.0) and intermediate monocytes (range: 2.0‐ to 27‐fold; median: 3.1) (Figure 3A, Supplemental Table 5). The low number of genes expressed ≥2.0‐fold uniquely by CD14++CD16+ intermediate monocytes is the result of an overlap in shared gene expression with both classical and nonclassical monocytes. There were 426 genes expressed ≥2.0‐fold in both classical (range: 2.0‐ to 148.2‐fold; median: 3.6) and intermediate monocytes (range: 2.0‐ to 86.7‐fold; median: 3.2) compared to nonclassical monocytes (Figure 3A, Supplemental Table 6). Relative to classical monocytes, 29 genes were expressed ≥ 2.0‐fold in the intermediate (range: 2.0‐ to 22.6‐fold; median: 2.9) and nonclassical monocytes (range: 2.1‐ to 46.5‐fold; median: 4.4) (Figure 3A, Supplemental Table 7). Because intermediate monocytes shared a greater number of expressed genes with classical monocytes than nonclassical monocytes, and because nonclassical monocytes had the greatest number of uniquely expressed genes, we conclude that classical and intermediate monocytes are more similar to one another than to the nonclassical monocytes, which are more transcriptionally distinct.
Figure 3.
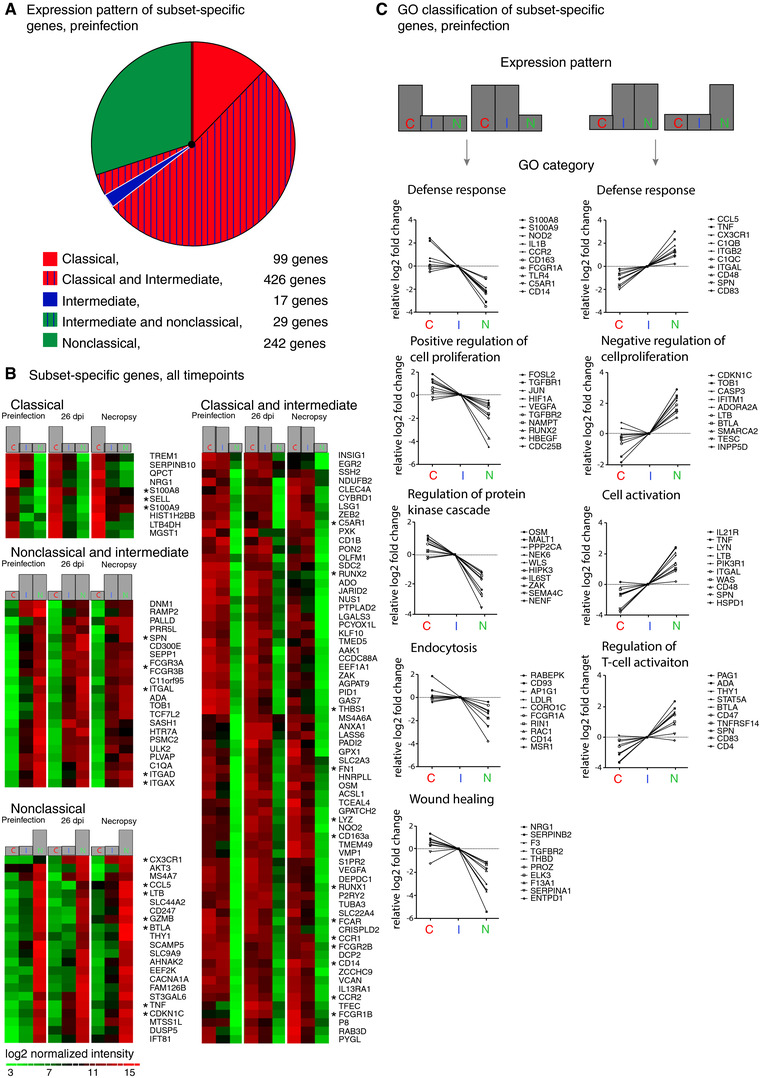
Identification of subset‐specific genes reveals the putative functional character of monocyte subsets. (A) Eight hundred thirteen genes found to differentiate monocyte subsets in uninfected animals were grouped by expression pattern: classical (red), intermediate (blue), and nonclassical (green) monocytes. Striping indicates expression by two subsets. The size of each segment is proportional to the number of genes specific to the indicated subset(s). Nonclassical monocytes had more unique genes (242 genes) than classical monocytes (99 genes) and intermediate monocytes (17 genes); 426 genes were expressed by both classical and intermediate monocytes, and 29 genes were expressed by both nonclassical and intermediate monocytes. Classical and intermediate monocytes express a large number of the same phenotypic markers, while nonclassical monocytes are more dissimilar. (B) Heat maps of the 172 genes that differentiate the three monocyte subsets both preinfection and in SIV‐infected animals. Rows are the average normalized intensity for the gene indicated. Green indicates lower expression, and red indicates higher expression. Columns are the different monocyte subsets for each of the three infection time points. At all time points, 10 genes were expressed ≥2.0‐fold more by classical monocytes, 22 genes were expressed ≥2.0‐fold more by nonclassical monocytes, and two genes were expressed ≥2.0‐fold more by intermediate monocytes (data not shown) than by the other subsets. Additionally, 22 genes were expressed ≥2.0‐fold more by both intermediate and nonclassical monocytes compared to classical monocytes, and 116 genes were expressed ≥2.0‐fold more by both classical and intermediate monocytes compared to nonclassical monocytes; for simplicity only the top 70 genes are shown. Asterisks indicate genes of interest regarding macrophage immune function and phenotype. (C) Representative GO terms for biological processes associated with genes expressed ≥2.0‐fold by classical monocytes or both classical and intermediate monocytes (left panels) and nonclassical or both intermediate and nonclassical monocytes (right panels) prior to infection are shown. Gene expression values are the log2 transformed fold change in classical, intermediate or nonclassical monocytes normalized to expression in intermediate monocytes. Representative genes are shown for each cluster. Complete data for functional enrichment categories at each infection time point are presented in Table 2
With regard to maturation, nonclassical monocytes expressed transcripts associated with macrophage or DC phenotypes (e.g., CD16, CD83, SPN, ITGAL, and BTLA), and classical and intermediate monocytes expressed transcripts associated with myeloid and granulocyte phenotype (e.g., CD14, TREM1, CD93, CD114, CD116). This may indicate that classical monocytes have a more “immature” granulocyte/monocyte phenotype while nonclassical monocytes have a more “mature” macrophage‐like phenotype. Additionally, expression of genes associated with proliferation (JUN, FOSL2, RUNX1, RUNX2) in classical monocytes and genes associated with inhibition of cell cycle (CDKN1C) in nonclassical monocytes may indicate that nonclasscial monocytes are more differentiated than classical monocytes.
3.7. Invariant gene signature of monocyte subsets with SIV infection
Of the 813 genes expressed in a subset‐specific manner preinfection, only 172 genes were also subset‐specific at 26 dpi and terminally with AIDS (Figure 3B). Ten genes were consistently expressed ≥2.0‐fold by classical monocytes than by other subsets, including S100A8, S100A9, and SELL (L‐selectin, CD62L) (Figure 3B). These genes are expressed by classical monocytes in humans and mice (Figure 3B). Only the complement receptor C3AR1 (C3a receptor 1) and MERTK tyrosine kinase were consistently expressed ≥2.0‐fold by intermediate monocytes compared to other subsets at all time points (data not shown). Nonclassical monocytes had consistently higher expression of 22 genes involved with inflammatory response (CX3CR1, CCL5, TNF, BTLA, GZMB) and inhibition of cell cycle (CDKN1C) than did the other two monocyte subsets (Figure 3B). Both intermediate and nonclassical monocytes were consistently differentiated by expression of 22 genes relative to classical monocytes including CD16 and integrins ITGAL, ITGAD, and ITGAX (Figure 2B). We identified 116 genes expressed ≥2.0‐fold by classical and intermediate monocytes relative to nonclassical monocytes, including many immune receptors (CD14, CD163, FCGR1B, FCGR2B, FCAR, CCR1, CCR2, C5AR1). These gene sets suggest candidate markers that may be useful for differentiating the three monocyte subsets. In particular, we have identified C3AR1 and MERTK as potential surface markers of intermediate monocytes. These data also indicate that SELL/CD62L is expressed by classical monocytes and CX3CR1 is expressed by nonclassical monocytes in rhesus macaques, as has previously been demonstrated in humans and mice.
3.8. In silico analysis of gene‐associated functions suggests classical monocytes are immune sensors and nonclassical monocytes are immune effectors
In order to investigate whether the genes that were differentially expressed between the monocyte subsets were associated with specific biological processes, we performed an in silico analysis using DAVID bioinformatics tools.52, 53 Human official gene symbols were used in order to take advantage of the relative abundance of gene annotation data for humans relative to macaques. With DAVID tools, gene lists were annotated with the GO terms for biological process—a controlled vocabulary for describing the biological function of gene products—and statistically overrepresented GO terms were identified. The analysis was performed for each of the infection time points (Table 2). Interestingly, we found that the GO terms that were enriched for each subset were generally the same in uninfected and SIV‐infected animals, suggesting that the functional role of each subset is maintained with SIV infection and inflammation (Table 2).
Table 2.
GO terms for biological process enriched in the set of genes differentially expressed across monocyte subsets
| Preinfection | 26 dpi | Necropsy | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| GO term | Subset | # | % | P | # | % | P | # | % | P |
| Defense response | C | 38 | 7.6 | *** | 36 | 5.8 | ** | 41 | 7.7 | **** |
| GO:0006952 | I | 40 | 8.8 | **** | 40 | 6.4 | *** | 36 | 8.1 | **** |
| N | 25 | 9.5 | **** | 22 | 13.4 | **** | 9 | 8.0 | * | |
| Inflammatory response | C | 24 | 4.8 | *** | 27 | 4.3 | *** | 28 | 5.2 | **** |
| GO:0006954 | I | 24 | 5.3 | **** | 29 | 4.6 | **** | 27 | 6.1 | **** |
| N | 13 | 4.9 | ** | 13 | 7.9 | *** | 7 | 6.3 | * | |
| Regulation of T cell activation | C | NS | NS | NS | NS | NS | NS | NS | NS | NS |
| GO:0050863 | I | 9 | 2.0 | * | 10 | 1.6 | * | 8 | 1.8 | * |
| N | 13 | 4.9 | **** | 9 | 5.5 | **** | 7 | 6.3 | **** | |
| Cell activation | C | NS | NS | NS | NS | NS | NS | NS | NS | NS |
| GO:0001775 | I | NS | NS | NS | NS | NS | NS | NS | NS | NS |
| N | 17 | 6.4 | **** | 8 | 4.9 | * | 11 | 9.8 | **** | |
| Positive regulation of cell proliferation | C | 27 | 5.4 | *** | 26 | 4.2 | ** | 26 | 4.9 | ** |
| GO:0008284 | I | 21 | 4.6 | * | 28 | 4.5 | ** | 21 | 4.7 | ** |
| N | NS | NS | NS | NS | NS | NS | NS | NS | NS | |
| Negative regulation of cell proliferation | C | NS | NS | NS | NS | NS | NS | NS | NS | NS |
| GO:0008285 | I | NS | NS | NS | NS | NS | NS | NS | NS | NS |
| N | 15 | 5.7 | ** | 12 | 7.3 | ** | 10 | 8.9 | *** | |
| Positive regulation of apoptosis | C | 24 | 4.8 | ** | NS | NS | NS | NS | NS | NS |
| GO:0043065 | I | NS | NS | NS | 26 | 4.1 | * | NS | NS | NS |
| N | 15 | 5.7 | ** | NS | NS | NS | 8 | 7.1 | * | |
| Negative regulation of apoptosis | C | 25 | 5.0 | *** | 22 | 3.5 | * | 22 | 4.1 | ** |
| GO:0043066 | I | 20 | 4.4 | ** | 21 | 3.3 | * | 18 | 4.0 | * |
| N | 12 | 4.5 | * | 9 | 5.5 | * | 7 | 6.3 | * | |
| Positive regulation of cell differentiation | C | 17 | 3.4 | ** | 16 | 2.6 | * | 17 | 3.2 | ** |
| GO:0045597 | I | 15 | 3.3 | ** | 16 | 2.5 | * | 13 | 2.9 | * |
| N | 10 | 3.8 | * | NS | NS | NS | 5 | 4.5 | * | |
| Wound healing | C | 21 | 4.2 | **** | 15 | 2.4 | ** | 17 | 3.2 | *** |
| GO:0042060 | I | 18 | 4.0 | **** | 13 | 2.1 | * | 11 | 2.5 | * |
| N | NS | NS | NS | NS | NS | NS | NS | NS | NS | |
| Angiogenesis | C | 11 | 2.2 | * | 13 | 2.1 | ** | 12 | 2.2 | ** |
| GO:0001525 | I | NS | NS | NS | 12 | 1.9 | * | 11 | 2.5 | ** |
| N | NS | NS | NS | NS | NS | NS | NS | NS | NS | |
| Endocytosis | C | 16 | 3.2 | ** | 20 | 3.2 | *** | 18 | 3.4 | *** |
| GO:0006897 | I | 16 | 3.5 | ** | 21 | 3.3 | *** | 16 | 3.6 | *** |
| N | NS | NS | NS | NS | NS | NS | NS | NS | NS | |
| Membrane organization | C | 29 | 5.8 | **** | 31 | 5.0 | **** | 27 | 5.0 | *** |
| GO:0016044 | I | 27 | 6.0 | **** | 32 | 5.1 | **** | 24 | 5.4 | *** |
| N | NS | NS | NS | NS | NS | NS | NS | NS | NS | |
| Vesicle‐mediated transport | C | 35 | 7.0 | *** | 39 | 6.3 | *** | 33 | 6.2 | ** |
| GO:0016192 | I | 30 | 6.6 | ** | 40 | 6.4 | *** | 29 | 6.5 | ** |
| N | NS | NS | NS | NS | NS | NS | NS | NS | NS | |
| Phagocytosis | C | NS | NS | NS | 7 | 1.1 | ** | 6 | 1.1 | * |
| GO:0006909 | I | 6 | 1.3 | ** | 7 | 1.1 | ** | 6 | 1.3 | ** |
| N | NS | NS | NS | NS | NS | NS | NS | NS | NS | |
| Regulation of protein kinase cascade | C | 23 | 4.6 | **** | 18 | 2.9 | ** | 19 | 3.6 | ** |
| GO:0010627 | I | 18 | 4.0 | *** | 18 | 2.9 | * | 13 | 2.9 | * |
| N | NS | NS | NS | NS | NS | NS | NS | NS | NS | |
| Regulation of MAP kinase activity | C | 12 | 2.4 | ** | 12 | 1.9 | ** | NS | NS | NS |
| GO:0043405 | I | 9 | 2.0 | * | 13 | 2.1 | ** | 16 | 3.6 | * |
| N | NS | NS | NS | NS | NS | NS | NS | NS | NS | |
| Regulation of I‐kappaB kinase/ NFkB cascade | C | 10 | 2.0 | ** | NS | NS | NS | 10 | 1.9 | ** |
| I | 9 | 2.0 | ** | NS | NS | NS | NS | NS | NS | |
| GO:0043122 | N | NS | NS | NS | NS | NS | NS | NS | NS | NS |
| Positive regulation of cytokine production | C | 12 | 2.4 | **** | 10 | 1.6 | ** | NS | NS | NS |
| GO:0001819 | I | 12 | 2.6 | **** | 11 | 1.7 | ** | NS | NS | NS |
| N | NS | NS | NS | NS | NS | NS | NS | NS | NS | |
| Cell adhesion | C | 31 | 6.2 | * | NS | NS | NS | NS | NS | NS |
| GO:0007155 | I | 29 | 6.4 | * | NS | NS | NS | NS | NS | NS |
| N | 20 | 7.6 | * | NS | NS | NS | 10 | 8.9 | * | |
| Chemotaxis | C | 12 | 2.4 | ** | 12 | 1.9 | * | 12 | 2.2 | * |
| GO:0006935 | I | 12 | 2.6 | ** | 13 | 2.1 | * | NS | NS | NS |
| N | NS | NS | NS | 7 | 4.3 | ** | NS | NS | NS | |
| Cell migration | C | 19 | 3.8 | ** | 18 | 2.9 | * | 18 | 3.4 | ** |
| GO:0016477 | I | 14 | 3.1 | * | NS | NS | NS | 15 | 3.4 | * |
| N | NS | NS | NS | NS | NS | NS | NS | NS | NS | |
| Actin cytoskeleton organization | C | 13 | 2.6 | * | 15 | 2.4 | * | 15 | 2.8 | * |
| GO:0030036 | I | 13 | 2.9 | * | 12 | 1.9 | * | 14 | 3.1 | ** |
| N | 10 | 3.8 | ** | NS | NS | NS | NS | NS | NS | |
| Posttranscriptional regulation of gene expression | C | NS | NS | NS | 18 | 2.9 | ** | 15 | 2.8 | ** |
| GO:0010608 | I | NS | NS | NS | 17 | 2.7 | ** | 12 | 2.7 | * |
| N | NS | NS | NS | NS | NS | NS | NS | NS | NS | |
C, classical; I, intermediate; N, nonclassical; #, number of queried genes associated with the GO term; %, (# GO associated genes/total genes queried) × 100; NS, not significant; P‐value for enrichment of the GO term: * P ≤ 0.05, ** P < 0.01 *** P < 0.001, P < 0.0001.
In uninfected and SIV‐infected animals, the genes specifically expressed by classical monocytes or both classical and intermediate monocytes were enriched for biological processes including defense response, positive regulation of cell proliferation, regulation of protein kinase cascade, endocytosis, and wound healing (Figure 3C and Table 2). Defense response genes expressed in classical monocytes include inflammatory mediators (e.g., IL‐8, S100A8, and S100A9). Interestingly, S100A9 and the S100A8/S100A9 heterodimer comprise the epitope recognized by the antibody MAC387, which defines a subset of macrophages that is recruited in response to inflammation.41, 54 Both classical and intermediate monocytes express receptors associated with detection of immune ligands including TLR4 and CD14 (LPS receptor), FCGR1A (high affinity FcγRIa), CCR1, CCR2, and IL‐6R. Genes associated with regulation of protein kinase cascade include genes involved in MAPK and NF‐κB signaling, which are important pathways in mitogenic signaling, cell survival, and cytokine secretion. Enrichment of MAPK‐associated genes and transcription factors involved in proliferation may indicate that the classical subset may be or has recently been progressing through the cell cycle. Thus, biological processes associated with genes enriched in classical monocytes include detection of innate immune stimuli and remediation of injured tissue.
In uninfected and SIV‐infected animals, genes specifically expressed by nonclassical monocytes or by intermediate and nonclassical monocytes were enriched for biological processes including defense response, negative regulation of cell proliferation, regulation of T‐cell activation, and cellular activation (Figure 3C and Table 2). Defense response genes in nonclassical monocytes are associated with a range of effector functions including initiation of complement cascades (C1QA, C1QB, C1QC), cell‐mediated cytotoxicity (GZMA, GZMB, GZMH, LTB, GNLY), and T‐cell activation (CD4, CD47, CD83, SPN). Nonclassical monocytes may be able to contribute to proinflammatory responses and recruit monocytes and T cells to sites of inflammation through production of TNF‐α and CCL5. Additionally, nonclassical monocytes express the fractalkine receptor (CX3CR1) and integrins (ITGAD, ITGAL, ITGAX, ITGB2). Nonclasscial monocytes also express transcripts associated with negative regulation of cell cycle; maturation of monocytes into macrophages is closely associated with exit from the cell cycle.55 Biological processes associated with genes enriched in nonclassical monocytes are consistent with this subset having exited the cell cycle, mediating immune effector functions, producing chemoattractant ligands, and being able to extravasate vessels to become tissue macrophages.
3.9. Changes in monocyte gene expression occur early in SIV infection
To investigate subset‐specific changes in gene expression with SIV infection, we identified genes that were differentially expressed (≥2.0 AFC) between infection time points for each monocyte subset (Figure 4A). Between preinfection and necropsy, more genes were differentially expressed in the nonclassical subset (265 genes) than in the classical (227 genes) or intermediate subsets (202 genes) (Figure 4A). In all subsets, the majority of changes in gene expression had occurred by 26 dpi, indicating that changes in gene expression due to SIV infection are established early (Figure 4A). The majority of genes differentially expressed at 26 dpi were upregulated relative to preinfection in the classical and intermediate subsets. In the nonclassical subset, upregulated and downregulated genes were present in approximately equal numbers (Figure 4A). Between 26 dpi and necropsy, the majority of differentially expressed genes were downregulated relative to 26 dpi (Figure 4A). Many of these downregulated genes were transcription factors. These data suggest that changes to monocyte gene expression occur early in SIV infection. Additionally, we observed that the pattern of changes in gene expression with SIV infection is more similar between classical and intermediate monocytes compared to nonclassical monocytes.
Figure 4.
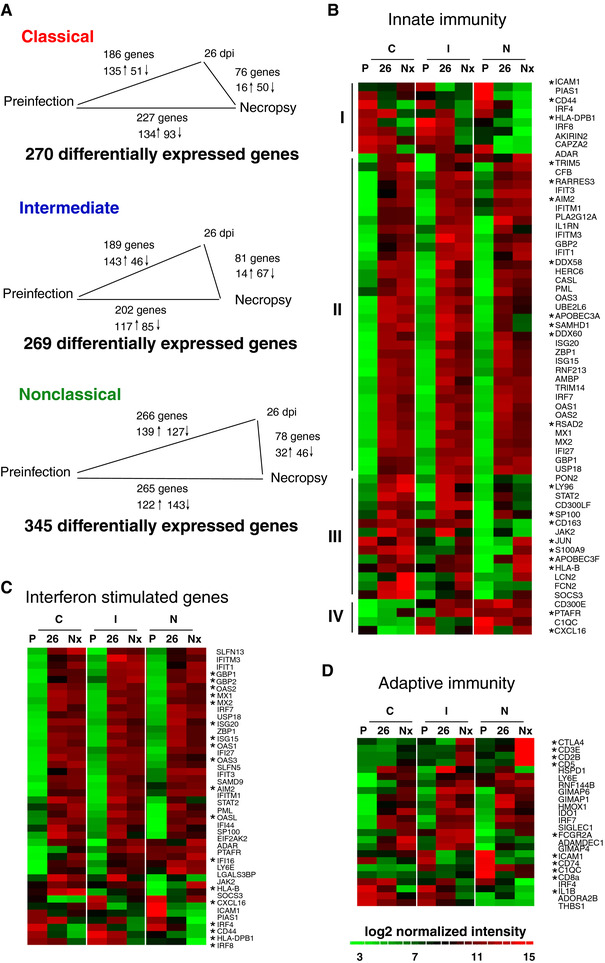
Changes in gene expression with SIV infection primarily occur prior to 26 dpi and include induction of interferon stimulated and innate immune genes. (A) The number of comparisons with ≥2.0 AFC between infection time points in each monocyte subset. The length of the line between infection time points is proportional to the number of differentially expressed genes. The majority of changes in gene expression occur between preinfection and 26 dpi in all subsets. (B–D) Heat maps of immune‐associated genes that are differentially expressed with SIV infection. Rows are the average normalized intensity for the gene indicated. Green indicates lower expression, and red indicates higher expression. Columns are the different infection time points (P: preinfection, 26: 26 dpi, Nx: Necropsy) for each of the three monocyte subsets (C: classical, I: intermediate, N: nonclassical). Genes were selected based on GO annotation for innate immune response (B), response to interferon (C), and adaptive immune response (D). Asterisks indicate genes of interest regarding macrophage immune function and phenotype. (B) Sixty‐two genes associated with innate immune response are differentially expressed with SIV infection. Hierarchical clustering reveals four distinct patterns of gene expression (I–IV). The majority of innate immune genes are induced in all three subsets at 26 dpi and necropsy. (C) Forty‐two interferon stimulated genes are differentially expressed with SIV infection. The majority of ISG are upregulated in all three monocyte subsets with SIV infection. (D) Twenty‐four genes associated with adaptive immune response are differentially expressed with SIV infection. The majority of these are upregulated in all three monocyte subsets or both intermediate and nonclassical monocytes
3.10. Characterization of the monocyte immune response to SIV infection
We next sought to characterize the immune response of each monocyte subset to SIV infection. Genes with GO annotation for innate immune response (62 genes, GO: 0045087), ISGs (44 genes, GO: 0034340, 0034341, or previous description in the literature), and adaptive immune response (24 genes, GO:0002250) were selected from the set of genes modulated with SIV infection (Figure 4B–D). Almost all immune associated genes were upregulated relative to the preinfection time point. Additionally, most genes modulated with SIV infection had a similar pattern of expression in classical, intermediate, and nonclassical monocytes (Figure 4B–D).
Hierarchical clustering was used to identify patterns of expression of innate immune response genes across monocyte subsets (Figure 4B). The largest cluster (Group II) was composed of genes that were upregulated in all monocyte subsets with SIV infection. ISGs as well as cytosolic sensors of nucleic acids (DDX58, DDX60, RARRES3, AIM2), restriction factors (APOBEC3A, TRIM5, SAMHD1), and ubiquitin‐associated genes (HERC6, TRIM14, UBE2L6) are representative of this group. The second largest cluster (Group III) consisted of genes that were upregulated primarily in classical and intermediate monocytes. These include genes associated with defense response to bacteria or viruses (CD163, APOBEC3F, S100A9, HLA‐B, LY96, SP100). Two minor clusters consisting of genes downregulated in all monocyte subsets (Group I), and genes upregulated primarily in intermediate and nonclassical monocytes (Group IV) contained transcription factors (PIAS1, IRF4, IRF8, AKIRIN2) and genes associated with chemotaxis (PTAFR, CXCL16), respectively. Upregulation of defense response genes, restriction factors, and cytosolic sensors of nucleic acids may suggest a mechanism of limiting SIV infection in monocytes. Immune‐associated genes were upregulated at 26 dpi and terminally with AIDS, which indicates these changes in gene expression in monocytes occur early and are sustained throughout infection. Additionally, the pattern of gene expression was similar across the three subsets, which suggests that the innate response to virus is conserved in spite of subset heterogeneity.
Induction of ISGs is part of the innate immune response to HIV and SIV infection and has been implicated in viral pathogenesis. We identified 44 ISGs that changed expression in response to SIV infection (Figure 4C). Most ISGs (38/44 genes) were upregulated in all three subsets and were associated with an antiviral function (OAS1‐3, GBP1‐2, MX1‐2, ISG15, ISG20). The ISGs IRF4 and PIAS1, which are downregulated with infection, are negative regulators of immune activation.56, 57 ISGs were also associated with myeloid cell differentiation (IRF4, IRF8, IFI16), JAK‐STAT signaling (JAK2, PIAS1, STAT2), and response to cytokine stimulus (SP100, CXCL16, PML, SOCS3). Sustained induction of IFN signaling in monocytes may be a mechanism of monocyte dysregulation and chronic immune activation, which contribute to SIV pathogenesis.
We identified 24 adaptive immune response genes that changed expression with SIV infection (Figure 4D), of which 16 were upregulated in all subsets or intermediate and nonclassical subsets. These genes were associated with Th1 polarization (GIMAP1, GIMAP4) and T‐cell binding or regulation (IDO1, SIGLEC1, ADAMDEC1). Additional genes associated with modulation of T‐cell function were upregulated in intermediate and nonclassical monocytes terminally with AIDS (CD3E, CD28, CD5, CTLA4). Downregulated genes include the MHC class II invariant chain (CD74) and cytokine‐associated genes (IRF4, IL‐1β, ADORA2B). Induction of genes associated with a Th1 type adaptive response and costimulatory molecules, particularly in intermediate and nonclassical monocytes, implies that SIV infection affects the ability of monocytes to regulate T‐cell function.
4. DISCUSSION
In this study, we identify changes in the gene expression profile of classical, intermediate, and nonclassical monocytes with SIV infection. We found that only 172 of 813 genes differentially expressed between monocyte subsets prior to infection remained differentially expressed in SIV‐infected animals. In this study, we show that in uninfected and SIV‐infected macaques classical monocytes express CD62L/SELL, S100A8, and S100A9, while nonclassical monocytes express CX3CR1. These markers are also differentially expressed between classical and nonclassical monocytes in humans and mice.13, 18 The chemokine and Fc receptors FcγRIa, CCR1, and CCR2, which are expressed on classical monocytes and not intermediate monocytes in humans and mice, were expressed on both classical and intermediate monocytes in rhesus macaques.13, 15 Few markers uniquely differentiate the intermediate subset as they share markers with classical and nonclassical monocytes. We identified two genes in macaques specific to intermediate monocytes whose proteins are known to be expressed on the cell surface: MERTK and C3AR1. In humans, the receptor tyrosine kinase MERTK is a marker of intermediate monocytes, and C3AR1 was reported to be more highly expressed in CD16+ monocytes.11, 13 MERTK is involved in leukocyte migration, clearance of apoptotic cells, inhibition of lymphocyte activation, and has been linked to susceptibility to multiple sclerosis.58 The subset‐specific genes identified herein may provide useful markers of the individual monocyte subsets in uninfected and SIV‐infected animals.
Genes expressed by classical monocytes were associated with defense response, proliferation, and wound healing. Expression of CCR1, CCR2, and CD62L suggests that classical monocytes can be mobilized in response to CCR1 and CCR2 ligands (CCL2/MCP‐1, CCL7/MCP‐3, CCL5/RANTES), home to lymphoid organs or sites of inflammation (via CD62L/SELL), and mediate inflammation or wound healing. Classical monocytes also express transcripts for S100A8 and S100A9 whose proteins comprise the antigen (calprotectin, S100A8/S100A9) recognized by the antibody MAC387.54, 59 MAC387 has been used to identify a population of inflammatory macrophages that is recruited to the CNS in response to acute inflammation.41, 54, 59 Notably, MAC387+ macrophages present in SIVE lesions were found to have recently emigrated from the bone marrow, as indicated by BrdU labeling of proliferating monocyte precursors.3, 41 Expression of genes associated with cell proliferation in classical monocytes also may suggest their recent proliferation in the bone marrow. Thus, the classical monocyte pool may represent precursors of inflammatory MAC387+ macrophages found in tissues. Though we did not perform functional studies in macaques, the biological functions suggested by genes enriched in the classical monocyte subset appear to recapitulate the known biology of mouse classical (Ly6C+) monocytes, which are mobilized from the bone marrow in a CCR2‐dependent manner, are recruited to sites of inflammation, and differentiate into classically activated (M1 polarized) macrophages in inflamed tissue.19
Nonclassical monocytes express transcripts associated with defense response, cell cycle inhibition, cellular activation, and macrophage phenotype. The withdrawal from the cell cycle and similarities to tissue macrophages may suggest that nonclassical monocytes are at a later stage of maturation than classical monocytes.55 Genes expressed by nonclassical monocytes, or both intermediate and nonclassical monocytes, suggest the ability to mediate a range of immune effector functions including cell‐mediated cytotoxicity and T‐cell activation. Notably, intermediate and nonclassical monocytes were more transcriptionally similar terminally with AIDS compared to early infection or preinfection time points. This may indicate increased activation or maturation of the intermediate subset with fulminant disease and reflect a phenotypic plasticity in response to the pathogenic microenvironment. Expression of CX3CR1 on nonclassical monocytes suggests that they are able to migrate in response to fractalkine, which is upregulated in the CNS with AIDS and HIVE.60 Additionally, expression of integrins on nonclassical monocytes likely indicates the ability to bind activated endothelial cells and migrate into tissues. Recruited nonclassical monocytes could contribute to neurodegeneration by elaborating proinflammatory mediators and recruiting additional leukocytes through production of TNFα and CCL5. The expansion of both nonclassical and intermediate monocytes may negatively contribute to AIDS progression and SIVE through increased traffic of activated and infected monocytes into the CNS and elaboration of chemoattractant and proinflammatory cytokines.
We found that intermediate monocytes and classical monocytes share more expressed genes in macaques, whereas intermediate monocytes and nonclassical monocytes share more expressed genes in humans.9 This suggests that the biology of monocyte subsets may vary between humans and nonhuman primates. We found high interanimal variability in gene expression, which indicates that the genes identified as differentially expressed in a given study will be greatly affected by the subject pool. Additionally, the genes identified in these studies may be affected by the depletion of CD8+ lymphocytes. Studies of SIV infection without CD8+ depletion and in HIV‐infected humans are necessary to validate the findings herein. Studies that evaluate the effect of antiretroviral therapy on gene expression in monocyte subsets and associated changes in disease progression are also needed.
Comparing early and end‐stage disease, we observed that the majority of changes in gene expression were present by 26 dpi and were then sustained throughout infection. We have previously shown that increased monocyte egress from the bone marrow is detectable within weeks of primary infection.3 The magnitude of the expansion of blood monocytes is predictive of how rapidly the animals progress to AIDS and the severity of macrophage‐mediated, AIDS‐related pathogenesis.3 We can consistently detect or predict by day 26 postinfection which animals will develop rapid AIDS and CNS and cardiac pathology.3, 61 This suggests that monocyte dysregulation with SIV infection is present early after infection and prior to AIDS onset. Although intermediate monocytes have been the primary focus of previous research, we observed that the nonclassical subset had the greatest number of genes that were differentially expressed with SIV infection. Moreover, expansion of these cells correlates with AIDS‐related pathologies including CNS and cardiac disease.3, 28, 61, 62, 63 This suggests that nonclassical monocytes merit additional focus going forward. Because nonclassical monocytes are innate effector cells, likely precursors to tissue macrophages, and regulators of adaptive immunity, changes to nonclassical monocyte biology may have varied effects. It is likely that these cells expand in response to macrophage turnover in the lymph nodes with AIDS and as a general innate immune response to tissue damage.3, 7 Further studies to elucidate the mechanisms and consequences of monocyte dysregulation in early SIV infection are needed.
We found that ISGs were upregulated in all three monocyte subsets by 26 dpi and terminally with AIDS indicating a sustained IFN response that likely indicates ongoing immune activation that may contribute to exhaustion of the immune system with chronic SIV infection. IFNα, which is produced primarily by plasmacytoid DCs in response to TLR signaling, has been implicated as a major mediator of chronic immune activation in HIV and SIV infection.25, 27, 64, 65, 66 Although the direct effect of HIV or SIV on monocytes is likely limited by the low frequency of infected monocytes, the effects of IFN‐α signaling on monocytes may be more widespread. Gene array studies of CD14+ monocytes from HIV‐infected patients and whole blood from SIV‐infected rhesus macaques have demonstrated a robust and sustained induction of ISG.26, 27, 64, 65 In humans, greater monocyte ISG induction is associated with higher viral load and increased neuronal damage, suggesting a link between type‐I IFN responses and HIVE.26, 27 Importantly, comparisons between pathogenic infection of humans and rhesus macaques, and nonpathogenic infection of sooty mangabeys or African green monkeys have demonstrated that increased IFN‐α production and failure to downregulate ISG induction after acute viremia are associated with pathogenic infection.64, 65, 66 We also observed upregulation of genes associated with detection of cytosolic nucleic acids and host restriction factors (APOBEC3A, TRIM5, SAMHD1), which may contribute to limited infection of monocytes in vivo. Despite heterogeneity of monocyte subsets, the pattern of induction of ISG and genes associated with innate immunity was similar between the three monocyte subsets, indicating a conserved immune response to SIV infection.
Supporting information
SuppTable 1
Tables 27
AUTHORSHIP
Conceived and designed the experiments: K.W. Performed the experiments: P.A., B.N., and J.W. Analyzed the data: B.N., T.B., and K.W. Critique of paper and intellectual input: T.B. and J.S. Wrote the paper: B.N. and K.W.
ACKNOWLEDGMENTS
This work was supported by grants NS040237 (K.C.W.), NS063897 (K.C.W.), NS082116 (T.H.B.), and OD011104 (base grant to TNPRC). The in vivo CD8 T lymphocyte depletion antibodies used in these studies were provided by the NIH Nonhuman Primate Reagent Resource (R24 OD010976, and NIAID contract HHSN 272201300031C). We thank the veterinary staff at the TNPRC for animal care, Cecily Conerly Midkiff and staff for assisting with tissue collection. We thank Michael Piatak and Jeffrey Lifson for plasma viral load determination and Ronald Desrosiers for providing the SIVmac251.
DISCLOSURES
The authors declare no conflicts of interest.
Nowlin BT, Wang J, Schafer J, Autissier P, Burdo TH, Williams KC. Monocyte subsets exhibit transcriptional plasticity and a shared response to interferon in SIV‐infected rhesus macaques. J Leukoc Biol. 2018;103:141–155. 10.1002/JLB.4A0217-047R
REFERENCES
- 1. Thieblemont N, Weiss L, Sadeghi HM, Estcourt C, Haeffner‐Cavaillon N. CD14lowCD16high: a cytokine‐producing monocyte subset which expands during human immunodeficiency virus infection. Eur J Immunol. 1995;25:3418–3424. [DOI] [PubMed] [Google Scholar]
- 2. Pulliam L, Sun B, Rempel H. Invasive chronic inflammatory monocyte phenotype in subjects with high HIV‐1 viral load. J Neuroimmunol. 2004;157:93–98. [DOI] [PubMed] [Google Scholar]
- 3. Burdo TH, Soulas C, Orzechowski K, et al. Increased monocyte turnover from bone marrow correlates with severity of SIV encephalitis and CD163 levels in plasma. PLoS Pathog. 2010;6:e1000842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kim WK, Sun Y, Do H, et al. Monocyte heterogeneity underlying phenotypic changes in monocytes according to SIV disease stage. J Leukoc Biol. 2010;87:557–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ansari AW, Meyer‐Olson D, Schmidt RE. Selective expansion of pro‐inflammatory chemokine CCL2‐loaded CD14+CD16+ monocytes subset in HIV‐infected therapy naive individuals. J Clin Immunol. 2013;33:302–306. [DOI] [PubMed] [Google Scholar]
- 6. Kim WK, Corey S, Alvarez X, Williams K. Monocyte/macrophage traffic in HIV and SIV encephalitis. J Leukoc Biol. 2003;74:650–656. [DOI] [PubMed] [Google Scholar]
- 7. Hasegawa A, Liu H, Ling B, et al. The level of monocyte turnover predicts disease progression in the macaque model of AIDS. Blood. 2009;114:2917–2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ziegler‐Heitbrock HW, Fingerle G, Strobel M, et al. The novel subset of CD14+/CD16+ blood monocytes exhibits features of tissue macrophages. Eur J Immunol. 1993;23:2053–2058. [DOI] [PubMed] [Google Scholar]
- 9. Wong KL, Yeap WH, Tai JJ, Ong SM, Dang TM, Wong SC. The three human monocyte subsets: implications for health and disease. Immunol Res. 2012;53:41–57. [DOI] [PubMed] [Google Scholar]
- 10. Ellery PJ, Tippett E, Chiu YL, et al. The CD16+ monocyte subset is more permissive to infection and preferentially harbors HIV‐1 in vivo. J Immunol. 2007;178:6581–6589. [DOI] [PubMed] [Google Scholar]
- 11. Ancuta P, Liu KY, Misra V, et al. Transcriptional profiling reveals developmental relationship and distinct biological functions of CD16+ and CD16‐ monocyte subsets. BMC Genomics. 2009;10:403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ziegler‐Heitbrock L, Ancuta P, Crowe S, et al. Nomenclature of monocytes and dendritic cells in blood. Blood. 2010;116:e74–e80. [DOI] [PubMed] [Google Scholar]
- 13. Wong KL, Tai JJ, Wong WC, et al. Gene expression profiling reveals the defining features of the classical, intermediate, and nonclassical human monocyte subsets. Blood. 2011;118, e16–e31. [DOI] [PubMed] [Google Scholar]
- 14. Sunderkotter C, Nikolic T, Dillon MJ, et al. Subpopulations of mouse blood monocytes differ in maturation stage and inflammatory response. J Immunol. 2004;172:4410–4417. [DOI] [PubMed] [Google Scholar]
- 15. Ingersoll MA, Spanbroek R, Lottaz C, et al. Comparison of gene expression profiles between human and mouse monocyte subsets. Blood. 2010;115:e10–e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cros J, Cagnard N, Woollard K, et al. Human CD14dim monocytes patrol and sense nucleic acids and viruses via TLR7 and TLR8 receptors. Immunity. 2010;33:375–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Passlick B, Flieger D, Ziegler‐Heitbrock HW. Identification and characterization of a novel monocyte subpopulation in human peripheral blood. Blood. 1989;74:2527–2534. [PubMed] [Google Scholar]
- 18. Geissmann F, Jung S, Littman DR. Blood monocytes consist of two principal subsets with distinct migratory properties. Immunity. 2003;19:71–82. [DOI] [PubMed] [Google Scholar]
- 19. Robbins CS, Swirski FK. The multiple roles of monocyte subsets in steady state and inflammation. Cell Mol Life Sci. 2010;67:2685–2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Williams KC, Hickey WF. Central nervous system damage, monocytes and macrophages, and neurological disorders in AIDS. Annu Rev Neurosci. 2002;25:537–562. [DOI] [PubMed] [Google Scholar]
- 21. Crowe S, Zhu T, Muller WA. The contribution of monocyte infection and trafficking to viral persistence, and maintenance of the viral reservoir in HIV infection. J Leukoc Biol. 2003;74:635–641. [DOI] [PubMed] [Google Scholar]
- 22. Williams K, Burdo TH. Monocyte mobilization, activation markers, and unique macrophage populations in the brain: observations from SIV infected monkeys are informative with regard to pathogenic mechanisms of HIV infection in humans. J Neuroimmune Pharmacol. 2012;7:363–371. [DOI] [PubMed] [Google Scholar]
- 23. Giri MS, Nebozyhn M, Raymond A, et al. Circulating monocytes in HIV‐1‐infected viremic subjects exhibit an antiapoptosis gene signature and virus‐ and host‐mediated apoptosis resistance. J Immunol. 2009;182:4459–4470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Van den Bergh R, Florence E, Vlieghe E, et al. Transcriptome analysis of monocyte‐HIV interactions. Retrovirology. 2010;7:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Woelk CH, Ottones F, Plotkin CR, et al. Interferon gene expression following HIV type 1 infection of monocyte‐derived macrophages. AIDS Res Hum Retroviruses. 2004;20:1210–1222. [DOI] [PubMed] [Google Scholar]
- 26. Pulliam L, Rempel H, Sun B, Abadjian L, Calosing C, Meyerhoff DJ. A peripheral monocyte interferon phenotype in HIV infection correlates with a decrease in magnetic resonance spectroscopy metabolite concentrations. AIDS. 2011;25:1721–1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rempel H, Sun B, Calosing C, Pillai SK, Pulliam L. Interferon‐alpha drives monocyte gene expression in chronic unsuppressed HIV‐1 infection. AIDS. 2010;24:1415–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pulliam L, Gascon R, Stubblebine M, McGuire D, McGrath MS. Unique monocyte subset in patients with AIDS dementia. Lancet. 1997;349:692–695. [DOI] [PubMed] [Google Scholar]
- 29. Sanchez‐Ramon S, Bellon JM, Resino S, et al. Low blood CD8+ T‐lymphocytes and high circulating monocytes are predictors of HIV‐1‐associated progressive encephalopathy in children. Pediatrics. 2003;111:E168–E175. [DOI] [PubMed] [Google Scholar]
- 30. Williams K, Westmoreland S, Greco J, et al. Magnetic resonance spectroscopy reveals that activated monocytes contribute to neuronal injury in SIV neuroAIDS. J Clin Invest. 2005;115:2534–2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Martin GE, Gouillou M, Hearps AC, et al. Age‐associated changes in monocyte and innate immune activation markers occur more rapidly in HIV infected women. PLoS One. 2013;8:e55279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gekonge B, Giri MS, Kossenkov AV, et al. Constitutive gene expression in monocytes from chronic HIV‐1 infection overlaps with acute Toll‐like receptor induced monocyte activation profiles. PLoS One. 2012;7:e41153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. McElrath MJ, Pruett JE, Cohn ZA. Mononuclear phagocytes of blood and bone marrow: comparative roles as viral reservoirs in human immunodeficiency virus type 1 infections. Proc Natl Acad Sci USA. 1989;86:675–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sonza S, Mutimer HP, Oelrichs R, et al. Monocytes harbour replication‐competent, non‐latent HIV‐1 in patients on highly active antiretroviral therapy. AIDS. 2001;15:17–22. [DOI] [PubMed] [Google Scholar]
- 35. Bergamaschi A and Pancino G. Host hindrance to HIV‐1 replication in monocytes and macrophages. Retrovirology. 2010;7:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhu T, Muthui D, Holte S, et al. Evidence for human immunodeficiency virus type 1 replication in vivo in CD14(+) monocytes and its potential role as a source of virus in patients on highly active antiretroviral therapy. J Virol. 2002;76:707–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kim WK, Alvarez X, Fisher J, et al. CD163 identifies perivascular macrophages in normal and viral encephalitic brains and potential precursors to perivascular macrophages in blood. Am J Pathol. 2006;168:822–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Han J, Wang B, Han N, et al. CD14(high)CD16(+) rather than CD14(low)CD16(+) monocytes correlate with disease progression in chronic HIV‐infected patients. J Acquir Immune Defic Syndr. 2009;52:553–559. [DOI] [PubMed] [Google Scholar]
- 39. Fischer‐Smith T, Bell C, Croul S, Lewis M, Rappaport J Monocyte/macrophage trafficking in acquired immunodeficiency syndrome encephalitis: lessons from human and nonhuman primate studies. J Neurovirol. 2008;14:318–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Campbell JH, Burdo TH, Autissier P, et al. Minocycline inhibition of monocyte activation correlates with neuronal protection in SIV neuroAIDS. PLoS One. 2011;6:e18688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Soulas C, Conerly C, Kim WK, et al. Recently infiltrating MAC387(+) monocytes/macrophages a third macrophage population involved in SIV and HIV encephalitic lesion formation. Am J Pathol. 2011;178:2121–2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Williams K, Burdo TH. Monocyte mobilization, activation markers, and unique macrophage populations in the brain: observations from SIV infected monkeys are informative with regard to pathogenic mechanisms of HIV infection in humans. J Neuroimmune Pharmacol. 2011;7:363–371. [DOI] [PubMed] [Google Scholar]
- 43. Westmoreland SV, Halpern E, Lackner AA. Simian immunodeficiency virus encephalitis in rhesus macaques is associated with rapid disease progression. J Neurovirol. 1998;4:260–268. [DOI] [PubMed] [Google Scholar]
- 44. Sasseville VG, Lackner AA. Neuropathogenesis of simian immunodeficiency virus infection in macaque monkeys. J Neurovirol. 1997;3:1–9. [DOI] [PubMed] [Google Scholar]
- 45. Bell JE The neuropathology of adult HIV infection. Rev Neurol (Paris). 1998;154:816–829. [PubMed] [Google Scholar]
- 46. Lifson JD, Rossio JL, Piatak M, Jr. , et al. Role of CD8(+) lymphocytes in control of simian immunodeficiency virus infection and resistance to rechallenge after transient early antiretroviral treatment. J Virol. 2001;75:10187–10199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Mitsiades N, Mitsiades CS, Poulaki, V , et al. Molecular sequelae of proteasome inhibition in human multiple myeloma cells. Proc Natl Acad Sci USA. 2002;99:14374–14379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Jones J, Otu H, Spentzos D, et al. Gene signatures of progression and metastasis in renal cell cancer. Clin Cancer Res. 2005;11:5730–5739. [DOI] [PubMed] [Google Scholar]
- 49. Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. [DOI] [PubMed] [Google Scholar]
- 50. Irizarry RA, Hobbs B, Collin F, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. [DOI] [PubMed] [Google Scholar]
- 51. Li C, Wong WH. Model‐based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci USA. 2001;98:31–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. [DOI] [PubMed] [Google Scholar]
- 53. Huang da W, Sherman BT, Zheng X, et al. Extracting biological meaning from large gene lists with DAVID. Curr Protoc Bioinformatics. 2009;Chapter 13, Unit.;13.11. [DOI] [PubMed] [Google Scholar]
- 54. Hessian PA, Fisher L. The heterodimeric complex of MRP‐8 (S100A8) and MRP‐14 (S100A9). Antibody recognition, epitope definition and the implications for structure. Eur J Biochem. 2001;268:353–363. [DOI] [PubMed] [Google Scholar]
- 55. Auffray C, Sieweke MH, Geissmann F. Blood monocytes: development, heterogeneity, and relationship with dendritic cells. Annu Rev Immunol. 2009;27:669–692. [DOI] [PubMed] [Google Scholar]
- 56. Negishi H, Ohba Y, Yanai H, et al. Negative regulation of Toll‐like‐receptor signaling by IRF‐4. Proc Natl Acad Sci USA. 2005;102:15989–15994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Liu B, Yang Y, Chernishof V, et al. Proinflammatory stimuli induce IKK‐alpha‐mediated phosphorylation of PIAS1 to restrict inflammation and immunity. Cell. 2007;129:903–914. [DOI] [PubMed] [Google Scholar]
- 58. Ma GZ, Stankovich J, Kilpatrick TJ, Binder MD, Field J. Polymorphisms in the receptor tyrosine kinase MERTK gene are associated with multiple sclerosis susceptibility. PLoS One. 2011;6:e16964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Goebeler M, Roth J, Teigelkamp S, Sorg C. The monoclonal antibody MAC387 detects an epitope on the calcium‐binding protein MRP14. J Leukoc Biol. 1994;55:259–261. [DOI] [PubMed] [Google Scholar]
- 60. Tong N, Perry SW, Zhang Q, et al. Neuronal fractalkine expression in HIV‐1 encephalitis: roles for macrophage recruitment and neuroprotection in the central nervous system. J Immunol. 2000;164:1333–1339. [DOI] [PubMed] [Google Scholar]
- 61. Walker J, Sulciner M, Nowicki K, Miller A, Burdo T, Williams K. Elevated numbers of CD163+ macrophages in hearts of simian immunodeficiency virus‐infected monkeys correlate with cardiac pathology and fibrosis. AIDS Res Hum Retroviruses. 2014;30:685–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Burdo TH, Lo J, Abbara S, et al. Soluble CD163, a novel marker of activated macrophages, is elevated and associated with noncalcified coronary plaque in HIV‐infected patients. J Infect Dis. 2011;204:1227–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Subramanian S, Tawakol A, Burdo TH, et al. Arterial inflammation in patients with HIV. JAMA. 2012;308:379–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Bosinger SE, Li Q, Gordon SN, et al. Global genomic analysis reveals rapid control of a robust innate response in SIV‐infected sooty mangabeys. J Clin Invest. 2009;119:3556–3572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Jacquelin B, Mayau V, Targat B, et al. Nonpathogenic SIV infection of African green monkeys induces a strong but rapidly controlled type I IFN response. J Clin Invest. 2009;119:3544–3555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Mandl JN, Barry AP, Vanderford TH, et al. Divergent TLR7 and TLR9 signaling and type I interferon production distinguish pathogenic and nonpathogenic AIDS virus infections. Nat Med. 2008;14:1077–1087. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SuppTable 1
Tables 27


