Abstract
Previous studies have shown that hippocampal volume is an early marker for dementia. We investigated whether hippocampal shape characteristics extracted from MRI scans are predictive for the development of dementia during follow up in subjects who were nondemented at baseline. Furthermore, we assessed whether hippocampal shape provides additional predictive value independent of hippocampal volume. Five hundred eleven brain MRI scans from elderly nondemented participants of a prospective population‐based imaging study were used. During the 10‐year follow‐up period, 52 of these subjects developed dementia. For training and evaluation independent of age and gender, a subset of 50 cases and 150 matched controls was selected. The hippocampus was segmented using an automated method. From the segmentation, the volume was determined and a statistical shape model was constructed. We trained a classifier to distinguish between subjects who developed dementia and subjects who stayed cognitively healthy. For all subjects the a posteriori probability to develop dementia was estimated using the classifier in a cross‐validation experiment. The area under the ROC curve for volume, shape, and the combination of both were, respectively, 0.724, 0.743, and 0.766. A logistic regression model showed that adding shape to a model using volume corrected for age and gender increased the global model‐fit significantly (P = 0.0063). We conclude that hippocampal shape derived from MRI scans is predictive for dementia before clinical symptoms arise, independent of age and gender. Furthermore, the results suggest that hippocampal shape provides additional predictive value over hippocampal volume and that combining shape and volume leads to better prediction. Hum Brain Mapp 35:2359–2371, 2014. © 2013 Wiley Periodicals, Inc.
Keywords: early diagnosis, hippocampus, magnetic resonance imaging, cohort studies, prospective studies, mild cognitive impairment, Alzheimer disease, classification
INTRODUCTION
There are 5.4 million individuals suffering from Alzheimer's disease (AD) in the USA [Thies and Bleiler, 2011]. Furthermore, it is estimated that on average patients suffering from dementia use three times more medical care compared to people not suffering from dementia in the same age range. Although there are currently no cures or drugs to prevent dementia, recent studies show the promise of better drugs to slow or halt the progression of dementia [Neugroschl and Sano, 2010; Scarpini et al., 2003]. Still, all damage suffered by the brain as a result of dementia is irreversible. This makes early detection—preferably before clinical symptoms appear—of great importance.
Experienced radiologists can recognize the typical brain atrophy patterns associated with dementia and can support the traditional diagnosis based on cognitive tests [Scheltens et al., 1992]. In recent years, a large number of articles have explored the possibilities of computer‐aided diagnosis and prediction of dementia in MRI.
Chan et al. [2001] showed that atrophy in demented subjects is most pronounced in the hippocampus, amygdala, and entorhinal cortex. Also, it has been shown that the hippocampal subregions are not equally affected by dementia and that localized atrophy within the hippocampus can be linked to dementia [Apostolova et al., 2006; Csernansky et al., 2005; Scher et al., 2007]. Hippocampal shape can be used as a measure for very localized atrophy. Hippocampal shape in relation to dementia has been studied and shown to contain information to distinguish demented subjects from healthy controls [Ferrarini et al., 2009; Gerardin et al., 2009].
It is well established that gray matter atrophy related to dementia is visible on MRI, even before clinical symptoms become apparent [den Heijer et al., 2006; Jack et al., 1999; Philip Scheltens et al., 2002]. Additionally, histology studies show that certain hippocampal subfields are affected stronger and earlier by atrophy [West et al., 2004], suggesting that local analysis is best suited to detect dementia in an early stage. Considering that the hippocampus is one of the brain structures affected earliest and strongest by dementia, in this study we used hippocampal shape to investigate localized hippocampal atrophy.
Many studies investigating dementia using MRI, have focused on distinguishing demented subjects from controls [DeCarli et al., 1995; Li et al., 2007]. Other studies compared MR images of cognitively normal subjects to subjects with mild cognitive impairment (MCI), which is considered a precursor of dementia. However, not all MCI subjects develop dementia; some remain MCI for a long time and some even revert to cognitively normal.
To be able to create a model for dementia prediction, longitudinal clinical data is required. This allows researchers to check the cognitive function of subjects over time. When this data is available, new subject groups can be identified: MCI converters (MCI‐c), subjects with MCI who develop dementia during a follow‐up period, and MCI nonconverters (MCI‐nc). Some studies try to distinguish between MCI‐c and MCI‐nc [Apostolova et al., 2006; Ferrarini et al., 2009; Jack et al., 1999]. Other studies try to detect very early dementia by comparing MCI‐c subjects to controls [Davatzikos et al., 2008]. In contrast to these studies, the subjects in our population were all nondemented at baseline. Furthermore, they form a cross section of middle aged and elderly people from the general population.
This article aims to answer the following questions: Is hippocampal shape extracted from MRI scans predictive for the development of dementia before clinical symptoms? If so, does hippocampal shape provide extra predictive value over hippocampal volume?
A preliminary version of this study has already been made available in a workshop article [Achterberg et al., 2010]. In the current study, we included 511 instead of 94 subjects and evaluated the models more extensively.
MATERIALS AND METHODS
MRI scans of the brain were acquired from 511 nondemented, elderly persons (Data section). During the 10‐year clinical follow‐up period, 52 subjects (10%) developed dementia (any type). For these subjects, MRI scans taken up to 10 years before the clinical diagnosis were available, allowing the evaluation of the predictive value of imaging biomarkers.
From the MRI scans, the hippocampus was segmented using an automated method (Hippocampus Segmentation section). Subsequently, a statistical shape model was constructed using the segmented data (Shape Representation section). Features from this model were used to train a classifier that distinguishes subjects who will develop dementia from subjects who will stay cognitively healthy (Classification section). The performance of this classifier was evaluated in a cross‐validation manner (Evaluation section). A schematic overview of the methods is shown in Figure 1. The predictive value of hippocampal volume, shape, and the combination of both shape and volume were compared to each other.
Figure 1.
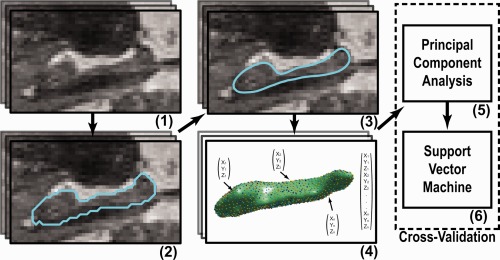
Overview of methods used: (1) MRI scans of the brain were acquired. (2) In each scan, the left and right hippocampus was segmented. (3) The segmentations were postprocessed. (4) Points were distributed over each surface, such that points on a different scans correspond with each other, and were concatenated to create one feature vector per scan. (5) The dimensionality of the feature vectors was reduced using principal component analysis. (6) A Support Vector Machine classifier was used to predict dementia development for each scan. Step (5) and (6) were performed in a cross‐validation manner (for a colored delineation in the figure, refer to the web version of this article.) [Color figure can be viewed in the online issue, which is available at http://wileyonlinelibrary.com.]
Data
The imaging data used in this study was a subset taken from the Rotterdam Scan Study: a prospective, population‐based MRI study on age‐related neurological diseases [den Heijer et al., 2003; Ikram, 2011]. For 511 nondemented, elderly subjects, MRI scans and the age, gender, dementia diagnosis, and time of follow‐up were available.
All subjects were scanned in 1995–1996 on a Siemens 1.5T scanner. The sequence used was a custom designed, inversion recovery, three‐dimensional (3D) half‐Fourier acquisition single‐shot turbo spin echo sequence. This sequence had the following characteristics: inversion time 4,400 ms, repetition time 2,800 ms, effective echo time 29 ms, matrix size 192 × 256, flip angle 180°, slice thickness 1.25 mm, acquired in sagittal direction. The images were reconstructed to a 128 × 256 × 256 matrix with a voxel dimension of 1.25 × 1.0 × 1.0 mm.
Study participants were followed during a 10‐year period. During this period, they were invited for four cognitive follow‐up tests, and the general practitioners records were tracked for diagnosis of dementia. Dementia screening followed a strict two‐step protocol [den Heijer et al., 2006]; initially, participants were cognitively screened with the Mini Mental State Examination (MMSE) and the Geriatric Mental Schedule. If the results of this initial screening indicated possible dementia, a more thorough cognitive testing was performed for verification. During the study period, 52 persons were diagnosed with dementia. The median interval between MRI acquisition and dementia diagnosis was 4.0 years with an interquartile range of 4.8 years.
The entire dataset, hereafter referred to as the cohort set, contained 52 prodromal dementia cases and 459 persons who did not develop dementia. To train and test a model independent of age and gender, an age‐ and gender‐matched subset of 50 prodromal dementia subjects and 150 controls was identified, hereafter referred to as the matched set. Characteristics of the cohort set and matched set can be found in Table 1. None of the subjects were demented at the time the MRI scan was taken.
Table 1.
Characteristics of the subjects in cohort set and matched set
| Cohort set | Matched set | |||
|---|---|---|---|---|
| Prodromal dementia (N = 52) | Controls (N = 459) | Prodromal dementia (N = 50) | Controls (N = 150) | |
| Women (%) | 61.5 | 48.6 | 60.0 | 60.0 |
| Age (years) | 79.02 (6.44 std) | 72.87 (7.81 std) | 78.75 (6.41 std) | 78.72 (6.43 std) |
| [64.37–88.73] | [58.96–89.83] | [64.37–88.73] | [64.37–89.83] | |
| Memory complainers (%) | 57.7 | 26.8 | 58.0 | 27.3 |
| MMSE | 27 (3 iqr) | 28 (2 iqr) | 27 (3 iqr) | 28 (2 iqr) |
| [20–30] | [14–30] | [20–30] | [21–30] | |
| Time to diagnosis | 4.0 (4.8 iqr) | n/a | 4.0 (4.9 iqr) | n/a |
| [0.73–10.28] | n/a | [0.73–10.28] | n/a | |
Values given as mean (standard deviation) or median (interquartile range), and range given as [min–max].
Because memory impairment is the first detectable neuropsychological sign of incipient dementia, we questioned persons on subjective memory complaints. This was done by a single question: “Do you have complaints about your memory performance?” Furthermore, objective memory performance was assessed using a 15‐word verbal learning task [den Heijer et al., 2006] resulting in a memory score.
To increase the sample size in the matched set, we selected three unique controls per case; this was possible for 50 cases. The matching was performed using the following criteria: the gender had to be the same, the follow‐up time of the controls should be at least as long as the time to diagnosis of the corresponding case, and the age could not differ more than 1.5 years. To avoid significant age differences, the mean age of the controls was kept as close as possible to the age of the case. We verified that the age matching resulted in no significant difference between groups with a paired t‐test.
Hippocampus Segmentation
Hippocampi were automatically segmented using a segmentation method based on multiatlas registration, a statistical intensity model, and a regularizer to promote smooth segmentations [van der Lijn et al., 2008]. These components were combined in an energy model, which is globally optimized using graph cuts. As training data, we used manually delineated images from 20 participants from the same population. Leave‐one‐out experiments on the training images showed mean Dice similarity indices of 0.85 ± 0.04 and 0.86 ± 0.02 for the left and right side. The final segmentation results of the 511 images used in this study were inspected by a trained observer (TdH) and manually corrected in case of large errors; two cases and 69 controls were manually corrected.
Because the creation of the shape model requires one single‐body object, we extracted the largest single body from the segmentation and applied a hole‐fill. This ensures that the object can be described by one single surface. Furthermore, an antialiasing step was used to smooth the binary segmentation [Whitaker, 2000].
From the segmentation, the hippocampal volume was calculated, which was subsequently corrected for intracranial volume. The intracranial volume was calculated by registration of a single brain mask to the target image and calculating the volume inside the brain mask [Ikram et al., 2008].
Shape Representation
The hippocampal shapes were described by corresponding points on the surfaces using the entropy‐based particle system as presented before [Cates et al., 2006, 2007] . This method aims at finding a uniform sampling of the shapes while minimizing the information content of the resulting shape model, leading to a compact model with optimal point correspondences. By describing both these criteria as entropies, they can be combined in a single model in a natural way.
When describing shapes by a set of N points on their surface, shapes can be seen as points in a 3N‐dimensional space and a collection of shapes forms a distribution in this space. The variance in this distribution can be caused by real shape differences or by errors in the point correspondence between the shapes. By moving the points over the surface for the individual shapes, the point correspondence error can be reduced. However, when the sampling is approximately uniform, the real shape differences will not change. While minimizing the variance in the distribution of shapes will thus change the description of the shape, in our case the point sampling, in such a way the correspondence is optimal without losing the real variation between shapes. This optimization is performed using a gradient descent algorithm. During gradient descent optimization, the points are constrained to lie on the surfaces of original segmentations.
All hippocampus segmentations were isotropically scaled to have equal volumes before the creation of the shape model, to exclude any volume information from the model. We created a shape model for both the left and the right hippocampus separately. The number of points (N) to represent each shape was set to 1,024. The shapes were aligned rigidly using Procrustes analysis [Goodall, 1991] at regular intervals of 25 iterations. To determine the number of iterations required for convergence, one optimization was run for 1,600 iterations, saving intermediate results every 10 iterations. For every intermediate output, the point displacements in the last 10 iterations were calculated. The optimization converged after 150–200 iterations. In all our following experiments, we ran the optimization for 200 iterations.
Classification
We trained statistical pattern classifiers to discriminate between subjects who developed dementia and those who remained cognitively healthy during the follow up period. This resulted in an estimated class for each subject, allowing us to evaluate how well we can predict dementia development in our dataset.
For classification three different feature sets were used: volume (normalized for intracranial volume), shape and a combination of both (hereafter referenced to as shape+volume). Volume and shape were combined by scaling the volume and shape feature vectors to have equal total variance and then concatenating them. All feature sets were created by concatenating the feature vectors derived from left and right hippocampus.
The dense sampling of points on a shape leads to a high dimensional feature space: two shapes with 1,024 points in a 3D space results in a 6,144 dimensional feature space. To reduce the dimensionality of the feature space a principal component analysis (PCA) retaining 99% of the variance was applied. After PCA, the number of dimensions was reduced to around 175.
A Support Vector Machine (SVM) classifier was used in all experiments. For completeness, we tested other classifiers, but none outperformed the SVM. For the shape and shape+volume features, the SVM classifier used a radial basis kernel. For the volume features, the SVM classifier used a linear kernel; the dimensionality was only two (right and left hippocampal volume) and a radial basis kernel did not improve classification performance.
The slack parameter (controlling the trade‐off between a large margin and small error on the training data) and the scale parameter of the radial basis function were estimated automatically by a grid search using leave‐group‐out cross‐validation on the training data. Each fold of the cross‐validation contained one case and its corresponding controls, thus preserving the age‐ and gender‐matching. An SVM results in a signed distance to the decision boundary for each subject. To convert this distance to the posterior probability that a subject belongs to a class, we use the inverse logit function
with d the distance to the decision boundary. We used these posterior probabilities both to compute receiver operating characteristic (ROC) curves and for regression analysis. All classification tests were performed using the PRTools [Duin et al., 2005] Matlab toolbox and libsvm [Chang and Lin, 2001].
Evaluation
All classification tests were performed in a leave‐group‐out cross‐validation loop. We trained (including the dimension reduction by PCA and parameter estimation of the SVM) on the matched dataset, except for one case and its matching controls, and estimated classification rate on the left out subjects. This was repeated for every case in the set. This keeps the age‐and gender‐matching intact, but still allows for as many cross‐validation folds as possible.
The results were stratified for time to diagnosis; using only results of cases (and the corresponding controls) who developed dementia within a certain time interval. The following time intervals were used: less than 3 years (N = 72), 3–6 years (N = 60), and more than 6 years (N = 68) until diagnosis.
To evaluate the predictive value of hippocampal volume and shape in a population setting, we added the remaining 311 subjects who were not in the matched set. The class label for these subjects was estimated using a classifier trained on the entire matched set. These results combined with the cross‐validation results on the matched set, constituted a predicted class label and posterior probability for all 511 subjects in the cohort. Even though the vast majority of the additional 311 subjects were controls, we add these subjects so we can evaluate a cross section of a normal, elderly population.
For all classification experiments, ROC curves were computed. We used the area under the ROC curve (AUC) as a measure of predictive value. Furthermore, we calculated the sensitivity, specificity, and risk ratio for specific points on the ROC curve. These points were chosen so that they had either a sensitivity or specificity of 0.8.
In practice, any imaging biomarker for dementia will be used in combination with other variables. To investigate the predictive value of the posterior probabilities in such a situation, we used SAS 9.2 to fit a logistic regression model with the posterior probabilities of the cohort set as an independent variable and the future development of dementia as the dependent variable. We considered three models: the classical model with only volume posterior as an independent variable, the classical model extended with shape information, and finally, the new model using the shape+volume posterior. All models were corrected for age and gender by including these as covariates in the model. These regressions provide insight in the relation between posterior probabilities and actual development of dementia. In addition, the significance level can be computed for every term in every model, indicating the value of a posterior as an imaging biomarker. Finally, we investigated the overall fit of the model by comparing the log‐likelihood of the models; a higher log‐likelihood means a better model fit. A likelihood‐ratio test was used to estimate the significance of model fit improvements. The likelihood‐ratio test is based on the fact that the log‐likelihood ratio of two comparable models follows a χ 2 1 distribution.
Our data were taken from a population‐based study and includes subjects who have varying cognitive abilities; there were no demented subjects present at inclusion, but some subjects might have memory complaints or lesser cognitive impairment. To investigate the predictive value of hippocampal shape and volume before any symptoms arise, we stratified the data into groups that exclude different forms of decreased cognitive function. We created four subgroups by excluding groups with various levels of cognitive impairment: (1) subjective memory complaints, (2) a memory score lower than the average memory score minus one standard deviation, (3) a memory score lower than the average memory score minus one and a half standard deviations, and (4) considered having MCI (see Fig. 4). We defined subjects to belong to the MCI group if they have both a memory complaint and a memory score lower than the average minus one and a half standard deviation.
Figure 4.
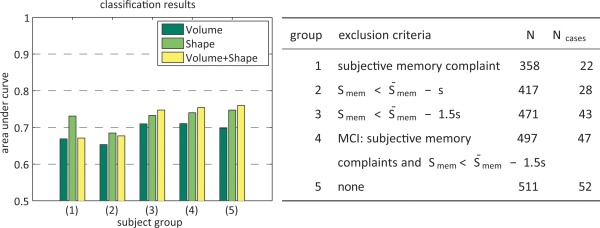
Left: the AUC results on subsets of the cohort. Right: information about subsets used. The subsets were created by excluding: (1) subjects with memory complaints, (2) subjects with a Smem (memory score) lower than the population mean minus a standard deviation, (3) subjects with a Smem 1.5 standard deviations lower than the population mean, (4) subjects considered MCI (memory complaints and Smem 1.5 standard deviations lower than the population mean), and finally (5) no one, the entire cohort without exclusions. [Color figure can be viewed in the online issue, which is available at http://wileyonlinelibrary.com.]
The experiments described above do not make a distinction between dementia subtypes. Our database does not have a complete subtype differential diagnosis, but we know which subjects were clinically diagnosed with AD. To see if predictive value is different for the more homogenous group of patients with AD, we repeated the classifier training and testing for a subset of the data, including only the subjects with AD.
To investigate left–right hippocampal asymmetry, we performed the classification using only one of the two hippocampi. We did this for the left and the right hippocampus and with each feature set (volume, shape and the combination shape+volume).
Finally, we visualized the areas (see Fig. 7) that contribute most to the classifier by calculating the discriminative direction of the classifier. We used the method introduced by Golland [2002] to locally approximate the discriminative direction at a representative point. As a representative point, we selected the point where the line between the class means intersects the decision boundary.
Figure 7.
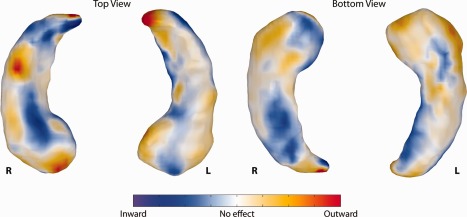
The discriminative direction of the classifier. The colors represent coefficients of the classifier localized on the hippocampal surface. The posterior probability of developing dementia increases if the points move in the direction indicated by the colors: blue points further inward and red/yellow points further outward indicate a higher chance of developing dementia. [Color figure can be viewed in the online issue, which is available at http://wileyonlinelibrary.com.]
We standardized the elements of the discriminative direction by multiplying the coefficients with the standard deviation of the feature. These standardized coefficients indicate how much each feature contributes to the final classification results. The strength and sign of the contribution for each surface‐point on the classifier is color‐coded (see Fig. 7). The color contains the contribution of the point when moving in the direction of the surface normal.
RESULTS
The ROC curve for both the matched set and the cohort set using volume, shape, and the shape+volume features is shown in Figure 2. Shape performs best performance when the specificity is high, whereas volume performs best when the sensitivity is around 80%. Overall, the combination shape+volume outperforms both volume and shape individually. On the matched set, the AUC for volume, shape, and shape+volume posteriors was, respectively, 0.734, 0.715, and 0.769. On the cohort set, the AUC was, respectively, 0.724, 0.743, and 0.766. Figure 3 shows the AUC for the matched set stratified by time to diagnosis. It can be seen that the predictive value of the volume features is very good for a short time to diagnosis, whereas more than 3 years before diagnosis the predictive value of volume decreases. For shape and especially the shape+volume features, the predictive value does not decrease when the time to diagnosis increases.
Figure 2.
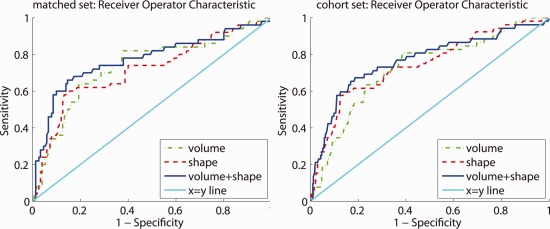
The receiver operating characteristic (ROC) curve. Left: the matched set. right: the cohort set. [Color figure can be viewed in the online issue, which is available at http://wileyonlinelibrary.com.]
Figure 3.
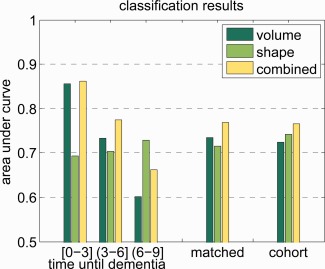
The area under the ROC curve. The results on the matched set stratified by time to diagnosis in years, and the results on the complete matched and cohort sets. Time until dementia diagnosis is a continuous variable, and the interval is defined in interval notation. In interval notation, a bracket means inclusive and a parentheses means exclusive. [Color figure can be viewed in the online issue, which is available at http://wileyonlinelibrary.com.]
The sensitivity and specificity for the specified points on the ROC curve are presented in Table 2. In most cases, volume has a better specificity, sensitivity, and risk ratio than shape at the two selected points on the ROC curve. Combining volume and shape improves the sensitivity when the specificity is fixed at 80%, but only has values in between shape and volume when fixing the sensitivity at 80%. Figure 2 however shows that although at 80% sensitivity volume shows a high specificity, overall shape+volume will still outperform volume only.
Table 2.
Senstivity, specificity, and risk ratio for fixed locations on the ROC curve
| Feature set | Cohort set | Matched set | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Volume | Shape | Volume + shape | Volume | Shape | Volume + shape | |||||||
| Sensitivity | 53.8 | 80.0 | 61.5 | 80.0 | 67.3 | 80.0 | 64.0 | 80.0 | 62.0 | 80.0 | 68.0 | 80.0 |
| Specificity | 80.0 | 61.4 | 80.0 | 45.8 | 80.0 | 56.0 | 80.0 | 62.7 | 80.0 | 38.7 | 80.0 | 53.3 |
| Risk ratio | 3.92 | 5.60 | 5.05 | 3.17 | 6.29 | 4.60 | 4.05 | 4.33 | 3.72 | 2.06 | 4.52 | 3.27 |
The points on the ROC used were those that had either a sensitivity or specificity of 0.8.
For the logistic regression, we present three models in Table 3. In the first model, we show the results of a regression of volume, corrected for age and gender. In this model, volume is highly significant (P < 0.0001). In the second model, shape is added as an extra term to the regression. In the new model, volume posteriors (P = 0.0012) and shape posteriors (P = 0.0071) both are significant. In the last model, we show a model with the combined shape+volume feature posterior, corrected for age and gender. The shape+volume posterior is highly significant (P < 0.0001).
Table 3.
Logistic regression models fitted on the cohort set
| Parameter | Estimate | Wald 95% confidence limits | P value | ||
|---|---|---|---|---|---|
| Model 1: volume posterior (Log Likelihood: −139.0860) | |||||
| Volume | 0.0575 | (0.0342−0.0809) | <0.0001 | ||
| Model 2: volume and shape posterior (Log Likelihood: −135.3668) | |||||
| Volume | 0.0423 | (0.0167−0.0678) | 0.0012 | ||
| Shape | 0.0283 | (0.0077−0.0489) | 0.0071 | ||
| Model 3: shape + volume posterior (Log Likelihood: −132.0318) | |||||
| Shape + volume | 0.0478 | (0.0322−0.0633) | <0.0001 | ||
All models are corrected for age and gender; in all cases, age and gender were significant (P < 0.05).
For the model with the volume, the log likelihood was −139.1; for the model with volume and shape, it was −135.4; and for the model with shape+volume features, it was −132.0. A likelihood‐ratio test revealed that the model improvement when adding shape to the first model is significant (P = 0.0063). When replacing the separate shape and volume posteriors with the shape+volume posterior, the model again improves significantly (P = 0.0098).
In Figure 4, it can be seen that excluding the subjects considered to have MCI or with a memory score of lower than the mean minus 1.5 standard deviations has little effect on the results; for shape+volume features AUC decreases by 0.007 and 0.006, respectively. When excluding subjects with subjective memory complaints or a memory score lower than the mean minus 1.0 standard deviation, the AUC decreases with 0.057 and 0.072, respectively, for shape+volume features. However, even with a loss of 0.072, there is still significant predictive value with an AUC of 0.69.
In Figure 5, we show a figure similar to Figure 3, but using only the cases that were diagnosed with dementia of Alzheimer's type. Cases with other types of dementia, and their controls, were excluded from the dataset. This resulted in a dataset with 41 cases in the cohort set and 39 cases in the matched set. On that matched set, the AUC was 0.690, 0.662, and 0.722 for volume, shape, and shape+volume, respectively. This is lower in than in the original dataset.
Figure 5.
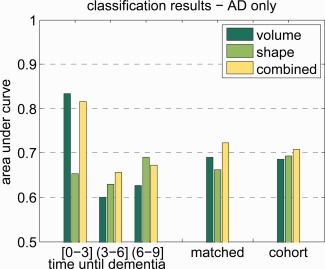
The area under the ROC curve. The dataset only contained cases that have AD, other types of dementia were excluded. The results on the matched set stratified by time to diagnosis in years, and the results on the complete matched and cohort sets. Time until dementia diagnosis is a continuous variable and the interval is defined in interval notation. In interval notation, a bracket means inclusive and a parentheses means exclusive. [Color figure can be viewed in the online issue, which is available at http://wileyonlinelibrary.com.]
Figure 6 shows the classification results when only using the left or right hippocampus, as well as the combination of both.
Figure 6.
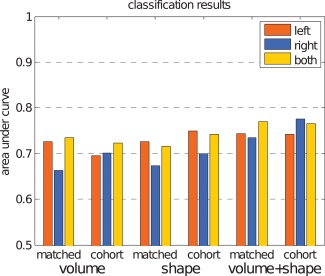
The area under the ROC curve for left and right hippocampus separate and combined. [Color figure can be viewed in the online issue, which is available at http://wileyonlinelibrary.com.]
Finally, the discriminative direction of the classifier is presented in Figure 7. For the left hippocampus, the most influential points appear to be in the CA1 and subiculum subfields. Also, the tip of the hippocampal tail contains points with very high coefficients. For the right hippocampus, the pattern is different; there are influential points located on the inferior side of the subiculum and CA2 subfield. Additionally, there is a group of points on the head of the right hippocampus, which appears to be on the interface of the CA1 and subiculum. Lastly, the right hippocampus also shows very high coefficients at the tip of the tail.
DISCUSSION
Hippocampal shape extracted from MRI scans is predictive for dementia before clinical symptoms arise, independent of age, gender, and hippocampal volume: we clearly show that shape contains predictive information with an AUC for shape of 0.715 on the matched set and 0.743 on the cohort set.
Stratification of the results on the time to dementia diagnosis shows that the predictive value of hippocampal volume is largest for the subjects who developed dementia soon after scan time, while with an increasing time between scan and diagnosis, the predictive value decreases. In contrast, for shape, there is no clear relation between the predictive value and time to diagnosis; this might indicate that shape features are less dependent on the time to diagnosis. Therefore, shape seems to provide valuable information for early detection. This is in line with the findings on histology [West et al., 2004]; the fact that the early neuronal loss caused by AD is localized in the CA1 and subiculum subfields of the hippocampus, indicates that hippocampal shape might be more suited to detect dementia in a prodromal phase than hippocampal volume. In Figure 7, it appears indeed that the CA1 and subiculum subfields play a role in the early prediction of dementia.
It is not only important to identify subjects who will develop dementia early, but also before any symptoms arise; subjects might have memory complaints or MCIs years before dementia diagnosis, and this damage is irreversible. Excluding subjects with MCI or a very low memory score (more than 1.5 standard deviations lower than the mean) does not change the results considerably, indicating that hippocampal shape is also predictive for the development of dementia in subjects who did not yet have MCI. When we exclude subjects with subjective memory complaints or with a memory score lower than the mean minus a standard deviation, the predictive value is not as high as in the complete dataset, but we can still predict dementia with an AUC of over 0.67. This shows that hippocampal volume and shape are predictive for dementia development in cognitively normal subjects.
Hippocampal shape provides additional predictive value over hippocampal volume: Figure 2 shows that classification based on shape has similar predictive value as volume and that combining shape and volume increases predictive value. When looking at the AUC's this is even more clear: shape has an AUC of 0.715 on the matched set and 0.743 on the cohort set, and volume has 0.734 on the matched set and 0.724 on the cohort set. This suggests that on the matched set volume has more predictive value than shape, while on the cohort set, shape has more predictive value than volume. However, when shape and volume features are combined in a single classifier, in both datasets the AUC increases. These observations are reinforced when taking into account the ROC curves and the sensitivity, specificity, and risk ratio for predefined points on the ROC curve.
If hippocampal volume and shape would be used in the prediction of development of dementia, it would be combined with other predictors (e.g., genetic biomarkers) which might increase the predictive value of a model. We created simple models, corrected for age and gender, to estimate the added value of using both volume and shape over just volume (see Table 3). In the first model, the volume posterior probability was a highly significant term. When we added the shape posterior probability to this model, the model improved significantly and both volume and shape were significant. In the third model, the separate volume and shape posterior probabilities where substituted by the shape+volume posterior probability, which resulted in a model with a better global fit.
When considering only cases who develop AD instead of cases who develop any type of dementia, the trends in the results appear to remain the same, but the absolute performance seems to be worse. This may be explained by the smaller number of shapes available for training in these experiments. For a reliable subgroup analysis, more data will be required.
The regression analysis indicates that shape is more predictive than volume, just as the classification results show in the cohort set. However, in all cases the shape+volume features resulted in a better predictive value than volume alone; in the regression using both volume and shape posterior probabilities even increased the model fit significantly.
Relation to Literature
In this article, we predicted dementia development with pattern recognition methods using hippocampal shape and volume, in a normal, elderly population. Dementia classification based on hippocampal shape has been investigated in various diagnostic studies before. Li et al. [2007] have performed classification to distinguish AD subjects from controls with up to 94.9% accuracy. Gerardin et al. [2009] reported accuracies of up to 83%, sensitivity 83%, and specificity 84% for MCI versus control classification. Ferrarini et al. [2009] reported accuracies of up to 90%, sensitivity 88%, specificity 92% for AD versus control classification and accuracies of up to 80%, sensitivity 80%, specificity 80% on MCI‐converter (MCI‐c) versus MCI‐non converter (MCI‐nc). Leung et al. [2010b] reported an AUC of up 0.67 on MCI‐c versus MCI‐nc subjects.
These studies use similar methods to our study, but given their case–control design were diagnostic of nature. Therefore, our work cannot be compared to the mentioned studies. Diagnostic accuracy in extreme groups (dementia or MCI patients vs. healthy controls) is very different from prediction of disease development in an asymptomatic population, as differences between subjects in the latter group are much more subtle. In our study, all persons were nondemented at scan time and developed dementia only later. Therefore, our results support the use of shape as a predictive marker.
The most notable other imaging methods used for extracting features for dementia classification are based on voxel‐based morphometry (VBM) [e.g., Fan et al., 2007; Klöppel et al., 2008] and cortical thickness [e.g., Desikan et al., 2009; Querbes et al., 2009]. Cuingnet et al. [2010] compared these methods of dementia classification, using a large dataset from the AD Neuroimaging Initiative database. They compared three groups of methods: VBM, cortical thickness measurements and hippocampus volume/shape based methods. They found that for AD versus control classification the whole brain methods outperformed the hippocampus‐based methods. However, for MCI‐c versus control classification the hippocampal methods were competitive with the whole‐brain methods. This result confirms that the hippocampus is one of the regions in the brain where atrophy is noticeable first in subjects with dementia.
We are not aware of any work using pattern recognition techniques to evaluate predictive value of hippocampal shape on similar data used in our study. There are studies which use statistical methods (e.g., regression or analysis of variance) to evaluate predictive value. Csernansky et al. [2005] and Apostolova et al. [2010] studied hippocampal shape using comparable subject groups. Their studies were more descriptive of nature making it impossible to quantitatively compare their results to our study. We can, however, qualitatively compare the discriminative direction obtained in our study to the maps obtained by Csernansky et al. [2005, Fig. 3] and Apostolova et al. [2010, Fig. 1]. For the left hippocampus, the discriminative direction maps presented in Figure 7 appears to match the atrophy and significance maps presented by Csernansky and Apostolova respectively: most influential points are found in the CA1 and Subiculum subfields. Csernansky also provides the direction of change, which corresponds with our results. For the right hippocampus, the similarity between the studies is lower: there are areas which contribute to our classification in the CA2 subfield that Csernansky or Apostolova do not find. This may be partly due to the fact that the discriminative direction in our work is based on the classifier that uses all points jointly, rather than the group differences per point as used by Csernasky and Apostolova. Also, Figure 6 shows asymmetry in the classification performance of the left and right hippocampus, indicating that the right hippocampus might not contribute much discriminative information to the classifier.
Many studies have shown asymmetry in hippocampal volume [Karas et al., 2004; Morra et al., 2009b; Scher et al., 2011], atrophy rates [Morra et al., 2009a; Zhou et al., 2009], or report differences in the diagnostic value of the left and right hippocampus [Csernansky et al., 2005; Tepest et al., 2008]. However, the asymmetry and the direction of asymmetry are not consistent across studies. It has been suggested that the asymmetry depends on the stage of dementia; the left hippocampus is affected first by dementia related atrophy and the right hippocampus follows with a time lag [Morra et al., 2009b; Thompson et al., 2003, 2004; Zhou et al., 2009]. In our data, the left hippocampus was found to be more predictive for dementia, which fits the suggested pattern for asymmetry; in our subjects, the disease is in a very early stage, and it is possible that the left hippocampus is already affected, while the right hippocampus is still unaffected. The combination of the left and the right hippocampal features in one classifier generally improves the classification result, indicating that asymmetry might be relevant for the prediction of dementia.
The quality of the segmentation method obviously has an important influence on the accuracy of the predictions. The automatic method used in this work has been shown to produce accurate segmentations; in a leave‐one‐out experiment on a manually labeled subset of the dataset used in our experiments a mean SI of 0.86 was obtained [van der Lijn et al., 2008]. The technique did occasionally label parts of the parahippocampal gyrus or the entorhinal cortex as foreground. However, these errors were manually corrected so we expect their influence on the classification results to be negligible. Furthermore, the leave one out experiments showed that as an atlas based method, the segmentation method has a tendency to underestimate the volume of large hippocampi and to overestimate the volume of small hippocampi. This bias toward the population mean is also likely to affect the shape of the segmentations, and could therefore negatively influence the classification accuracy. This effect may be reduced by selecting a more representative subset of atlases from a larger library. Segmentation methods based on this strategy tend to yield a higher overall accuracy than using all atlases [Aljabar et al., 2009; Barnes et al., 2008; Collins and Pruessner, 2010; Leung et al., 2010a]. Unfortunately, we could not experimentally verify this, since we did not have access to a larger template library.
In our work, we represented hippocampal shape by points on the surfaces of the hippocampus and optimized point correspondence using an entropy‐based particle method [Cates et al., 2007]. Ferrarini et al. [2009] and Leung et al. [2010b] also use point clouds to represent hippocampal shape but optimize correspondence by using, respectively, adaptive mesh optimization [Ferrarini et al., 2007] and minimal description length [Davies et al., 2002].
Besides other correspondence optimization methods, also different shape descriptions have been used. Brechbühler et al. [1995] described shapes using spherical harmonics, which was the representation used in Gerardin et al. [2009] to describe hippocampi. Both Morra et al. [2009a] and Qiu et al. [2009] use a map‐based representation for hippocampal shape, creating a deformation map to a template surface for each subject. Morra et al. [2009a] use a medial axis method for creating correspondence, where Qiu et al. [2009] uses a large diffeomorphic deformation metric matching method (LDDMM). In Qiu et al. [2008], shape differences derived by LDDMM are described directly by the deformation field, via the Jacobian of the transformation.
We decided to use the entropy‐based particles method for shape correspondence modeling in our study, as it allows explicit optimization of correspondence. Moreover, an implementation of this method is publicly available and has been successfully used to study the shape of the hippocampus in patients with schizophrenia [Cates et al., 2007].
CONCLUSIONS
In this article, we have shown that hippocampal shape extracted from MRI scans is predictive for dementia before clinical symptoms arise, independent of age and gender. Furthermore, hippocampal shape provides additional predictive value over hippocampal volume.
Shape seems to have more predictive value for subjects who develop dementia more than 6 years after scan time or exhibit less symptoms, while volume is more predictive for subjects who develop dementia shortly after scan time or exhibit more symptoms. Therefore, shape can be of great value for prodromal prediction of dementia onset.
Future treatment trials in AD will include persons with the most early and maybe even presymptomatic stage of the disease. It will therefore be increasingly important to detect persons in these stages. We show in this article that hippocampal shape may be used to partially identify these stages. Combination of hippocampal shape with other early biomarkers such as PET imaging and cerebrospinal fluid markers will be ultimately necessary for most optimal prodromal diagnosis.
Biography
Wiro Niessen is co‐founder, share holder and part‐time employed as scientific director of Quantib BV.
Wiro Niessen is co‐founder, share holder and part‐time employed as scientific director of Quantib BV.
Footnotes
We used the software provided by the authors. ShapeWorks: An open‐source tool for constructing compact statistical point‐based models of ensembles of similar shapes. Scientific Computing and Imaging Institute (SCI). Can be found at http://www.sci.utah.edu/software.html
REFERENCES
- Achterberg HC, van der Lijn F, den Heijer T, van der Lugt A, Breteler MMB, Niessen WJ, de Bruijne M (2010): Prediction of dementia by hippocampal shape analysis In: Wang F, Yan P, Suzuki K, Shen D, editors. Machine Learning in Medical Imaging. Vol. 6357 of Lecture Notes in Computer Science. Berlin/Heidelberg: Springer; pp 42–49. [Google Scholar]
- Aljabar P, Heckemann Ra, Hammers A, Hajnal JV, Rueckert D (2009): Multi‐atlas based segmentation of brain images: atlas selection and its effect on accuracy. NeuroImage 46:726–38. [DOI] [PubMed] [Google Scholar]
- Alzheimer's Association , Thies W, Bleiler L (2011): 2011 Alzheimer's disease facts and figures. Alzheimer's Dementia 7:208–244. [DOI] [PubMed] [Google Scholar]
- Apostolova LG, Dutton RA, Dinov ID, Hayashi KM, Toga AW, Cummings JL, Thompson PM (2006): Conversion of mild cognitive impairment to Alzheimer disease predicted by hippocampal atrophy maps. Arch Neurol 63:693–699. [DOI] [PubMed] [Google Scholar]
- Apostolova LG, Mosconi L, Thompson PM, Green AE, Hwang KS, Ramirez A, Mistur R, Tsui WH, de Leon MJ (2010): Subregional hippocampal atrophy predicts Alzheimer's dementia in the cognitively normal. Neurobiol Aging 31:1077–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes J, Foster J, Boyes RG, Pepple T, Moore EK, Schott JM, Frost C, Scahill RI, Fox NC (2008): A comparison of methods for the automated calculation of volumes and atrophy rates in the hippocampus. NeuroImage 40:1655–1671. [DOI] [PubMed] [Google Scholar]
- Brechbühler C, Gerig G, Kübler O (1995): Parametrization of closed surfaces for 3‐D shape description. CVIU 61:154–170. [Google Scholar]
- Cates J, Fletcher PT, Whitaker R (2006): Entropy‐Based Particle Systems for Shape Correspondence. Mathematical Foundations of Computational Anatomy Workshop, MICCAI 2006. pp 90–99. Available at: http://www-sop.inria.fr/asclepios/events/MFCA06/.
- Cates JE, Fletcher PT, Styner MA, Shenton ME, Whitaker RT (2007): Shape modeling and analysis with entropy‐based particle systems. In: Karssemeijer N, Lelieveldt BPF, editors. IPMI, Springer; pp 333–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan D, Fox NC, Scahill RI, Crum WR, Whitwell JL, Leschziner G, Rossor AM, Stevens JM, Cipolotti L, Rossor MN (2001): Patterns of temporal lobe atrophy in semantic dementia and Alzheimer's disease. Ann Neurol 49:433–442. [PubMed] [Google Scholar]
- Chang CC, Lin CJ (2001): LIBSVM: a library for support vector machines. Software available at http://www.csie.ntu.edu.tw/~cjlin/libsvm/. doi: 10.1145/1961189.1961199.
- Collins DL, Pruessner JC (2010): Towards accurate, automatic segmentation of the hippocampus and amygdala from MRI by augmenting ANIMAL with a template library and label fusion. NeuroImage 52:1355–66. [DOI] [PubMed] [Google Scholar]
- Csernansky JG, Wang L, Swank J, Miller JP, Gado M, McKeel D, Miller MI, Morris JC (2005): Preclinical detection of Alzheimer's disease: hippocampal shape and volume predict dementia onset in the elderly. Neuroimage 25:783–792. [DOI] [PubMed] [Google Scholar]
- Cuingnet R, Gerardin E, Tessieras J, Auzias G, Lehéricy S, Habert MO, Chupin M, Benali H, Colliot O, The Alzheimer's Disease Neuroimaging Initiative (2010): Automatic classification of patients with alzheimer's disease from structural MRI: A comparison of ten methods using the ADNI database. Neuroimage 56:766–781. [DOI] [PubMed] [Google Scholar]
- Davatzikos C, Fan Y, Wu X, Shen D, Resnick SM (2008): Detection of prodromal Alzheimer's disease via pattern classification of magnetic resonance imaging. Neurobiol Aging 29:514–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies RH, Twining CJ, Cootes TF, Waterton JC, Taylor CJ (2002): A minimum description length approach to statistical shape modeling. IEEE Trans Med Imaging 21:525–537. [DOI] [PubMed] [Google Scholar]
- DeCarli C, Murphy DG, McIntosh AR, Teichberg D, Schapiro MB, Horwitz B (1995): Discriminant analysis of MRI measures as a method to determine the presence of dementia of the Alzheimer type. Psychiatry Res 57:119–130. [DOI] [PubMed] [Google Scholar]
- Desikan RS, Cabral HJ, Hess CP, Dillon WP, Glastonbury CM, Weiner MW, Schmansky NJ, Greve DN, Salat DH, Buckner RL, Fischl B, Initiative ADN (2009): Automated MRI measures identify individuals with mild cognitive impairment and Alzheimer's disease. Brain 132:2048–2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duin R, Juszczak P, de Ridder D, Paclík P, Pekalska E, Tax DMJ (2005): PR‐Tools. http://prtools.org.
- Fan Y, Shen D, Gur RC, Gur RE, Davatzikos C (2007): Compare: classification of morphological patterns using adaptive regional elements. IEEE Trans Med Imaging 26:93–105. [DOI] [PubMed] [Google Scholar]
- Ferrarini L, Frisoni GB, Pievani M, Reiber JHC, Ganzola R, Milles J (2009): Morphological hippocampal markers for automated detection of Alzheimer's disease and mild cognitive impairment converters in magnetic resonance images. J Alzheimers Dis 17:643–659. [DOI] [PubMed] [Google Scholar]
- Ferrarini L, Olofsen H, Palm WM, van Buchem MA, Reiber JHC, Admiraal‐Behloul F (2007): GAMEs: growing and adaptive meshes for fully automatic shape modeling and analysis. Med Image Anal 11:302–314. [DOI] [PubMed] [Google Scholar]
- Gerardin E, Chételat G, Chupin M, Cuingnet R, Desgranges B, Kim HS, Niethammer M, Dubois B, Lehéricy S, Garnero L, Eustache F, Colliot O, Alzheimer's Disease Neuroimaging Initiative (2009): Multidimensional classification of hippocampal shape features discriminates Alzheimer's disease and mild cognitive impairment from normal aging. Neuroimage 47:1476–1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golland P (2002): Discriminative direction for kernel classifiers. Adv Neural Inf Process Syst 14:745–752. [Google Scholar]
- Goodall C (1991): Procrustes methods in the statistical analysis of shape. J R Stat Soc Ser B Methodol 53:285–339. [Google Scholar]
- den Heijer T, Geerlings MI, Hoebeek FE, Hofman A, Koudstaal PJ, Breteler MMB (2006): Use of hippocampal and amygdalar volumes on magnetic resonance imaging to predict dementia in cognitively intact elderly people. Arch Gen Psychiatry 63:57–62. [DOI] [PubMed] [Google Scholar]
- den Heijer T, Vermeer S, Clarke R, Oudkerk M, Koudstaal P, Hofman A, Breteler M (2003): Homocysteine and brain atrophy on MRI of non‐demented elderly. Brain 126:170–175. [DOI] [PubMed] [Google Scholar]
- Ikram MA, van der Lugt A, Niessen WJ, Krestin GP, Koudstaal PJ, Hofman A, Breteler MMB, Vernooij MW (2011): The Rotterdam Scan Study: design and update up to 2012. Eur J Epidemiol 26:811–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikram MA, Vrooman HA, Vernooij MW, van der Lijn F, Hofman A, van der Lugt A, Niessen WJ, Breteler MMB (2008): Brain tissue volumes in the general elderly population The Rotterdam Scan Study. Neurobiol Aging 29:882–890. [DOI] [PubMed] [Google Scholar]
- Jack CR, Petersen RC, Xu YC, O'Brien PC, Smith GE, Ivnik RJ, Boeve BF, Waring SC, Tangalos EG, Kokmen E (1999): Prediction of AD with MRI‐based hippocampal volume in mild cognitive impairment. Neurology 52:1397–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karas GB, Scheltens P, Rombouts SaRB, Visser PJ, van Schijndel Ra, Fox NC, Barkhof F (2004): Global and local gray matter loss in mild cognitive impairment and Alzheimer's disease. NeuroImage 23:708–16. [DOI] [PubMed] [Google Scholar]
- Klöppel S, Stonnington CM, Chu C, Draganski B, Scahill RI, Rohrer JD, Fox NC, Jack CR, Ashburner J, Frackowiak RSJ (2008): Automatic classification of MR scans in Alzheimer's disease. Brain 131:681–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung KK, Barnes J, Ridgway GR, Bartlett JW, Clarkson MJ, Macdonald K, Schuff N, Fox NC, Ourselin S (2010a): Automated cross‐sectional and longitudinal hippocampal volume measurement in mild cognitive impairment and Alzheimer's disease. NeuroImage 51:1345–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung KK, Shen KK, Barnes J, Ridgway GR, Clarkson MJ, Fripp J, Salvado O, Meriaudeau F, Fox NC, Bourgeat P, Ourselin S (2010b): Increasing power to predict mild cognitive impairment conversion to Alzheimer's disease using hippocampal atrophy rate and statistical shape models. Med Image Comput Comput Assist Interv 13:125–132. [DOI] [PubMed] [Google Scholar]
- Li S, Shi F, Pu F, Li X, Jiang T, Xie S, Wang Y (2007): Hippocampal shape analysis of Alzheimer disease based on machine learning methods. AJNR Am J Neuroradiol 28:1339–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Lijn F, den Heijer T, Breteler MMB, Niessen WJ (2008): Hippocampus segmentation in MR images using atlas registration, voxel classification, and graph cuts. Neuroimage 43:708–720. [DOI] [PubMed] [Google Scholar]
- Morra JH, Tu Z, Apostolova LG, Green AE, Avedissian C, Madsen SK, Parikshak N, Hua X, Toga AW, Jack CR, Schuff N, Weiner MW, Thompson PM, Alzheimer's Disease Neuroimaging Initiative (2009a): Automated 3D mapping of hippocampal atrophy and its clinical correlates in 400 subjects with Alzheimer's disease, mild cognitive impairment, and elderly controls. Hum Brain Mapp 30:2766–2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morra JH, Tu Z, Apostolova LG, Green AE, Avedissian C, Madsen SK, Parikshak N, Toga AW, Jack CR, Schuff N, Weiner MW, Thompson PM, Alzheimer's Disease Neuroimaging Initiative (2009b): Automated mapping of hippocampal atrophy in 1‐year repeat mri data from 490 subjects with Alzheimer's disease, mild cognitive impairment, and elderly controls. Neuroimage 45:S3–S15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neugroschl J, Sano M (2010): Current treatment and recent clinical research in Alzheimer's disease. Mt Sinai J Med 77:3–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu A, Fennema‐Notestine C, Dale AM, Miller MI, Alzheimer's Disease Neuroimaging Initiative (2009): Regional shape abnormalities in mild cognitive impairment and Alzheimer's disease. Neuroimage 45:656–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu A, Younes L, Miller MI, Csernansky JG (2008): Parallel transport in diffeomorphisms distinguishes the time‐dependent pattern of hippocampal surface deformation due to healthy aging and the dementia of the Alzheimer's type. Neuroimage 40:68–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Querbes O, Aubry F, Pariente J, Lotterie JA, Démonet JF, Duret V, Puel M, Berry I, Fort JC, Celsis P, Alzheimer's Disease Neuroimaging Initiative (2009): Early diagnosis of Alzheimer's disease using cortical thickness: impact of cognitive reserve. Brain 132:2036–2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarpini E, Scheltens P, Feldman H (2003): Treatment of Alzheimer's disease: current status and new perspectives. Lancet Neurol 2:539–547. [DOI] [PubMed] [Google Scholar]
- Scheltens P, Fox N, Barkhof F, Carli CD (2002): Structural magnetic resonance imaging in the practical assessment of dementia: beyond exclusion. Lancet Neurol 1:13–21. [DOI] [PubMed] [Google Scholar]
- Scheltens P, Leys D, Barkhof F, Huglo D, Weinstein HC, Vermersch P, Kuiper M, Steinling M, Wolters EC, Valk J (1992): Atrophy of medial temporal lobes on MRI in “probable” Alzheimer's disease and normal ageing: diagnostic value and neuropsychological correlates. J Neurol Neurosurg Psychiatry 55:967–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scher AI, Xu Y, Korf ESC, Hartley SW, Witter MP, Scheltens P, White LR, Thompson PM, Toga AW, Valentino DJ, Launer LJ (2011): Hippocampal morphometry in population‐based incident Alzheimer's disease and vascular dementia: the HAAS. J Neurol Neurosurg Psychiatry 82:373–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scher AI, Xu Y, Korf ESC, White LR, Scheltens P, Toga AW, Thompson PM, Hartley SW, Witter MP, Valentino DJ, Launer LJ (2007): Hippocampal shape analysis in Alzheimer's disease: a population‐based study. Neuroimage 36:8–18. [DOI] [PubMed] [Google Scholar]
- Tepest R, Wang L, Csernansky JG, Neubert P, Heun R, Scheef L, Jessen F (2008): Hippocampal surface analysis in subjective memory impairment, mild cognitive impairment and Alzheimer's dementia. Dement Geriatr Cogn Disord 26:323–329. [DOI] [PubMed] [Google Scholar]
- Thompson PM, Hayashi KM, de Zubicaray GI, Janke AL, Rose SE, Semple J, Herman D, Hong MS, Dittmer SS, Doddrell DM, Toga AW (2003): Dynamics of gray matter loss in Alzheimer's disease. J Neurosci 23:994–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson PM, Hayashi KM, de Zubicaray GI, Janke AL, Rose SE, Semple J, Hong MS, Herman D, Gravano D, Doddrell DM, Toga AW (2004): Mapping hippocampal and ventricular change in Alzheimer disease. Neuroimage 22:1754‐1766. [DOI] [PubMed] [Google Scholar]
- West MJ, Kawas CH, Stewart WF, Rudow GL, Troncoso JC (2004): Hippocampal neurons in pre‐clinical alzheimer's disease. Neurobiol Aging 25:1205–1212. [DOI] [PubMed] [Google Scholar]
- Whitaker RT (2000): Reducing aliasing artifacts in iso‐surfaces of binary volumes. IEEE Symp Vol Visual Graph 23‐32. [Google Scholar]
- Zhou L, Lieby P, Barnes N, Réglade‐Meslin C, Walker J, Cherbuin N, Hartley R (2009): Hippocampal shape analysis for Alzheimer's disease using an efficient hypothesis test and regularized discriminative deformation. Hippocampus 19:533–40. [DOI] [PubMed] [Google Scholar]


